Abstract
Mycobacterium tuberculosis is known to be associated with several autoimmune diseases such as systemic lupus erythematous, rheumatoid arthritis and multiple sclerosis. This is attributed to sequence similarity between virulent factors and human proteins. Therefore, it is of interest to identify such regions in the virulent factors to assess potential autoimmune related information. M. tb specific virulent factors were downloaded from the VFDB database and its human homologs were identified using the sequence comparison search tool BLASTP. Both virulent proteins and their corresponding human homologs were further scanned for epitopes (B cell and HLA class I and II allele specific) using prediction programs (BCPRED and NETMHC). Data shows the presence of matching 22 B-cell, 79 HLA class II and 16 HLA class I specific predicted epitopes in these virulent factors having human homologs. A known peptide (HAFYLQYKNVKVDFA) associated with autoimmune atopic dermatitis is shown in the superoxide dismutase homolog structures of the bacterium (PDB ID: 1IDS) and human (PDB ID: 2QKC). This data provides insight into the understanding of infection-associated auto-immunity
Background
Pathogenic intracellular organisms have strategies of evading or suppressing the host׳s immune response. Strategies against acquired immunity include antigenic variation, immune suppression and molecular mimicry. Molecular mimicry is well documented in viruses such as HIV, monkey pox and cow pox and its primary function is camouflage [1, 2]. Molecular mimicry can be defined as sequence or structural similarity between host and pathogen peptides resulting in immune evasion or cross reactivity leading to autoimmune response. Pathogens may also mimic host molecules to manipulate factors in signal transduction pathways via their receptors [3–6]. Previous studies have shown that cross reactive antibodies are produced in response to bacterial infections causing tissue damage [7–9]. Tuberculosis (TB) has been associated with several autoimmune diseases such as systemic lupus erythematous, rheumatoid arthritis and multiple sclerosis [10–18]. M. tb induced T cell reactivity with foreign and selfantigens lead to autoimmune responses [11, 14, 15, 17]. Thus, detecting epitopes involved in cross reactivity could help in comprehending TB immuno-pathogenesis. The present study identified epitopes with sequence and structural similarities between M. tuberculosis virulent factors and host homologs for B-cells and T-cells (class I and II HLA alleles) specificity [19, 20]
Methodology
Virulent Factors Database (VFDB):
M.tb specific virulent factors (number) were downloaded in FASTA format from VFDB (a database of virulent factors) [21]. Basic Local Alignment Search Tool (BLAST) - 2.2.28 The Basic Local Alignment Search Tool (BLAST) is used to find regions of local similarity between M.tb virulent factor and human proteome [22].
Phobius 1.0.1:
Phobius was used to identify and exclude the signal region of the homologous proteins. [23].
Conserved Domain Database (CDD):
This database was used to identify conserved domains in homologous proteins of M.tb virulent factors and human.
B-cell epitope prediction server (BCPREDS):
Prediction of B cell epitopes (Table 1) for M.tb specific virulent factors and its corresponding human homologs using B cell epitope prediction server (BCPREDS) [24].
T-cell epitope prediction:
Prediction of HLA class-I (Table 2) Table 1and class-II (Table 1) T cell epitopes was completed for virulent factors and its corresponding human homologs using NetMHC 2.2 and 3.0 [25].
Visual Molecular Dynamics 1.9.1(VMD):
This program was used to visualize the 3D structures of M. tb and human superoxide dismutase [26]
Workflow:
M. tb virulent factors are obtained from VFDB and human proteins from Ensembl. Virulent factors are BLAST searched against human proteome using BLAST (version 2.2.28). Then homologs are extracted at E-value ≤ 0.01. The remaining M. tb proteins were run through Phobius to remove predicted Nterminal signal peptides from the protein sequence. Then sequences are run through CDD for getting domain coordinates. Further the collected sequences are run through BCPRED server for B cell epitopes of 20 amino acids length and the classifier specificity was 75% and overlap filter was used for analysis. Based on prior BLAST results, regions of amino acids (small peptides) that were similar between the human and M. tuberculosis proteins were selected for further analysis. BCPRED score of greater than 0.9 is considered for blast matched peptides in both pathogen and host homologs.
NetMHC (version 2.2) was used for HLA class II and NetMHC (version 3.0) for HLA class I binding peptide prediction. Peptides were selected based on IC50 values <50 nM as high affinity, <500 nM as intermediate affinity and <5000 nM as low affinity [27]. The matched peptides in both pathogen and host with a binding score less than IC50 ≤ 50 are considered as strong binders. 3D structures of protein sequences matched to host are viewed and aligned structurally to find out whether these peptides are on the surface of the protein. The similarity between the predicted epitopes of virulent factors was found by multiple structural alignments using the STAMP algorithm in VMD. The detection of epitopes is shown in Figure 1. All calculations were performed using the local Linux server.
Figure 1.
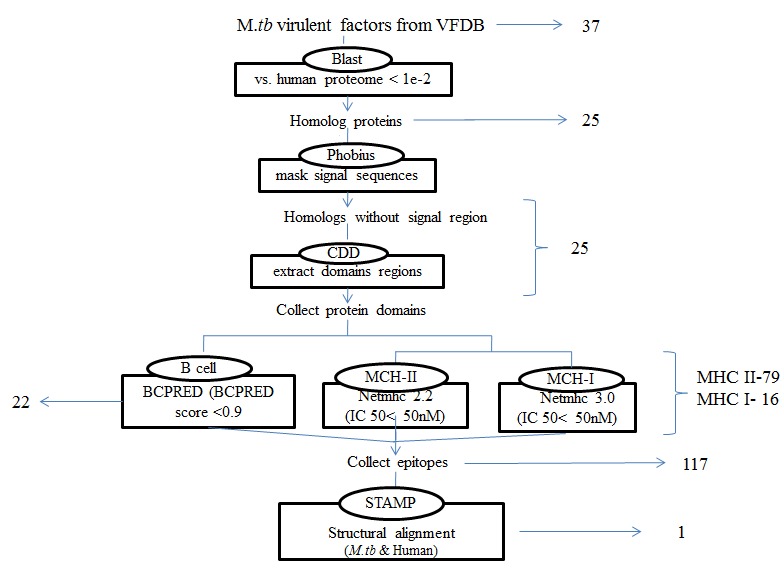
A workflow showing steps involved in the identification of epitopes in the virulent factors of M. tb having human homologs. Epitopes were predicted in both virulent factors and their corresponding homologs. Both virulent proteins and their corresponding human homologs were further scanned for epitopes (B cell (BCPRED with score > 0.9) and HLA class I and II alleles (NETMHC with binding score < 50nM) prediction.
Results & Discussion
The analysis of data obtained from the search between M. tuberculosis virulence factors and the human proteome revealed considerable similarities in sequences. A total of 25 best-hit homologous proteins with E-value cut off 1-E02 and similar regions of 9 or more amino acids were identified. The classification of the homologous virulent factor proteins are 21 metabolic proteins 3 membrane associated protein and a protein kinase (Table 3). Binding affinities of M.tb virulent factors vs. B cell epitopes and HLA class II and I alleles were measured by BCPRED and NetMHC. A peptide was considered having significant affinity to virulent factors if it had a BCPRED score ≥ 0.9 for B cell epitope (Table 4) and IC50 value ≤ 50 for HLA class II and I epitopes. The analysis of binding affinities of HLA class II peptides is 83% as compared to HLA class I (17%). Of 79 HLA class II host-pathogen epitopes highest affinity was to HLA-DRB10101 (57% followed byHLADRB10701 (14%), HLA-DRB10401 (11%), HLA-DRB10301 (6%), HLA-DRB11101 (5%), HLA-DRB10302 (2.5%) and HLADRB11501 (2.5%). The analysis of HLA class I peptides indicated a maximum affinity of peptides binding to allele A*0201 (44%) followed by B*0702 (31%), 2 for A*1101 (13%) and 6% each for A*0101 & A*2402 (Figure 2, Table 1 & Table 4). HLA class II has significant number of high affinity binding peptides which could be involved in dys-regulation of T cell function and or autoimmunity [28]. The virulent factors binding to host tissue antigens could influence signaling and immune evasion [29].
Figure 2.
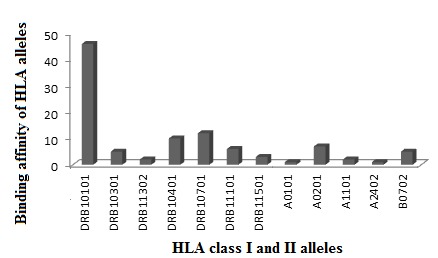
Frequency of epitopes with strong binding affinity for HLA class I and II T cell alleles that shared similarity with M.tb virulent factors. Legend:HLA class II host-pathogen epitopes binding to HLA-DRB10101 (57%) show highest affinity over HLA class Iepitopes indicating their significant role in pathogenesis of TB.
The myco-bacterial virulent proteins of this study were classified into categories such as structural, metabolic, catalytic, kinases and transport proteins (Table 4, 1 & 2). Majority of the virulent factor epitopes having binding affinity for B and HLA class I and II alleles were involved in (i) lipid, protein and nucleotide metabolism/degradation pathways (ii) free radical mediated damage pathway (iii) ion transport (iv) degradation of proteins glycosylation/phosphorylation pathways. These similarities could impact metabolic rate of reactions, interfere in homeostasis of cell and could trigger cell damage by free radical mediated reactions [29]. Peptides, which have binding capacity to more than one allele of HLA class-I and-II, are called promiscuous peptides. Promiscuous peptides for HLA class II were 24% (19/79) and none for HLA class I molecules. Interestingly, the presence of promiscuous peptides for HLA class II suggest that these peptides could have role in presentation of antigens for immune recognition and amplification of response against M.tb (Table 3 and 4) [30]. Genetic studies on HLA class I and II alleles are associated with susceptibility to disease and the present study indicates their similarities/binding to M.tb virulent factors.
This study identified a host peptide human manganese superoxide dismutase (MnSOD) sharing structural similarity with M. tb Superoxide dismutase (M. tb SOD) virulent factor. This epitope was previously implicated in diseases such as atopic dermatitis (AD), autoimmune hepatitis (AIH), Epstein- Barr virus (EBV) infection and fumigatus-allergy [31–34]. A peptide from M.tb Superoxide dismutase (M.tb SOD) HAFYLQYKNVKVDFA, bound to allele HLA-DRB1*15:01 allele with high affinity is identified (Table 3). HLADRB1* 15:01 is known to be responsible for susceptibility to tuberculosis. Further there was a high structural similarity of M.tb SOD and human MnSOD at both primary and tertiary structure level Figure 3 [35]. Clinical studies identified MnSOD cross-reactive autoimmune antibodies in patients with atopic dermatitis (AD) and has been implicated in disease pathogenesis [31]. This epitope is conserved and well investigated in Aspergillus fumigatus Mn SOD (1KKC) in relation to various autoimmune conditions [31–34]. Identifying the key homologous peptides of host pathogen similarity could help us design highly selective peptide blockers, which would be a valuable addition to complement the understanding of autoimmune diseases.
Figure 3.
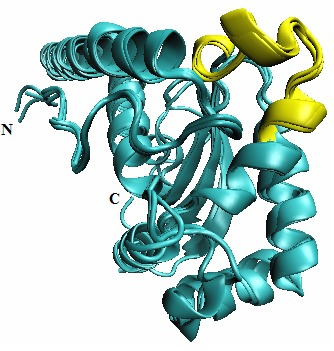
Multiple structural alignments of M.tbsuperoxide dismutase (1IDS), Aspergillus fumigatus Mn superoxide dismutase (1KKC) and human Mn superoxide dismutase (2QKC) by STAMP in VMD-multiseq window. Legend:visualization panel of VMD shows structurally conserved epitope ‘HAFYLQYKNVKVDFA’ in yellow among M.tbsuperoxide dismutase (1IDS), Aspergillus fumigatus Mn superoxide dismutase (1KKC) and human Mn superoxide dismutase (2QKC)
PDB crystal structures of superoxide dismutase M.tb, human and Aspergillus fumigatus were available. Superposition revealed a high measure of structural conservation and similarity with low RMSD having Qres value of 0.9 and showing high measure of the similarity of the ‘C-C alpha’ distances between residues of aligned proteins (Figure 3) [36]. These structurally similar regions of these three epitopes (which is known to cause atopic dermatitis) could be significant in tuberculosis in causing immune inflammatory processes characteristic of TB (Figure 3). It can be noted that many other mycobacterial antigens have been associated with autoimmune diseases [37– 39]. There is no clear evidence that M.tb virulent factors are involved and further clinical investigations on epitope specificities involved in autoimmunity are warranted.
Although, computational tools have been used in the past to examine molecular mimics in other diseases [40]; the understanding of these epitopes need to be further probed. Utilizing these methods, we have identified potential autoreactive B cell, HLA class II and class I epitopes that may elicit autoimmune response during M. tuberculosis infection. The findings of this study are as follows: (i) there were 95 auto reactive B cell, HLA class II and class I epitopes that are similar to peptides of myco-bacterial virulent factors; (ii) 22% of similarities were promiscuous that are binding to HLA class II cell epitopes (iii) high Qres score of 0.9 suggesting structural similarity between M.tb SOD and human Mn SOD and the epitope has an established evidence of autoimmunity. The similarities were observed across the spectrum of metabolic activities of host cell suggesting M.tb could use multiple split approach in causing tuberculosis.
Conclusions
We report regions in the M.tb virulent factors having human homologs sharing predicted B-cell and T-cell epitopes. Data shows the presence of 22 B-cell, 79 HLA class II and 16 HLA class I specific predicted peptides in these virulent factors having human homologs. A known peptide (HAFYLQYKNVK VDFA) associated with autoimmune atopic dermatitis is shown in the superoxide dismutase homolog structures of the bacterium (PDB ID: 1IDS) and human (PDB ID: 2QKC). This data provides insights in understanding infection-associated auto-immunity.
Supplementary material
Acknowledgments
The authors gratefully thank CODEWEL Nireekshana-ACET for support.
Footnotes
Citation:Gutlapalli et al, Bioinformation 11(12): 517-524 (2015)
References
- 1.Lambris JD, et al. Nat Rev Microbiol. 2008;6:132. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srinivasappa J, et al. J Virol. 1986b;57:397. doi: 10.1128/jvi.57.1.397-401.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hide G, et al. Mol Biochem Parasitol. 1989;36:51. doi: 10.1016/0166-6851(89)90199-0. [DOI] [PubMed] [Google Scholar]
- 4.Ghansah TJ, et al. J Eukaryot Microbiol. 2002;49:383. doi: 10.1111/j.1550-7408.2002.tb00216.x. [DOI] [PubMed] [Google Scholar]
- 5.Spiliotis M, et al. Gene. 2003;323:57. doi: 10.1016/j.gene.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Vicogne J, et al. J Biol Chem. 2004;279:37407. doi: 10.1074/jbc.M313738200. [DOI] [PubMed] [Google Scholar]
- 7.Sfriso P, et al. J Leukoc Biol. 2010;87:385. doi: 10.1189/jlb.0709517. [DOI] [PubMed] [Google Scholar]
- 8.Shahrizaila N, Yuki N. J Biomed Biotechnol. 2011;2011:829129. doi: 10.1155/2011/829129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vardhini D, et al. Infect Genet Evol. 2004;4:21. doi: 10.1016/j.meegid.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Birnbaum G, Kotilinek L. Ann N Y Acad Sci. 1997;835:157. doi: 10.1111/j.1749-6632.1997.tb48627.x. [DOI] [PubMed] [Google Scholar]
- 11.Birnbaum G, et al. Ann Neurol. 1993;34:18. doi: 10.1002/ana.410340106. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh K, et al. Rheumatol Int. 2009;29:1047. doi: 10.1007/s00296-009-0903-x. [DOI] [PubMed] [Google Scholar]
- 13.Mor F, Cohen IR. J Clin Invest. 1992;90:2447. doi: 10.1172/JCI116136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Res PC, et al. Lancet. 1988;22:478. doi: 10.1016/s0140-6736(88)90123-7. [DOI] [PubMed] [Google Scholar]
- 15.Salvetti M, et al. J Autoimmun. 1992;5:691. doi: 10.1016/0896-8411(92)90186-t. [DOI] [PubMed] [Google Scholar]
- 16.Salvetti M, et al. J Neuroimmunol. 1996;65:143. doi: 10.1016/0165-5728(96)00013-6. [DOI] [PubMed] [Google Scholar]
- 17.van Eden W, et al. Proc Natl Acad Sci U S A. 1985;82:5117. doi: 10.1073/pnas.82.15.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Eden W, et al. Nature. 1988;331:171. doi: 10.1038/331171a0. [DOI] [PubMed] [Google Scholar]
- 19.Agrewala JN, Wilkinson RJ. Eur J Immunol. 1999;29:1753. doi: 10.1002/(SICI)1521-4141(199906)29:06<1753::AID-IMMU1753>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 20.Gowthaman U, Agrewala JN. J Proteome Res. 2008;7:154. doi: 10.1021/pr070527b. [DOI] [PubMed] [Google Scholar]
- 21.Chen L, et al. Nucleic Acids Res. 2012;40:D641. doi: 10.1093/nar/gkr989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boratynet GM, et al. Nucleic Acids Res. 2013;41:W29. [Google Scholar]
- 23.Käll L, et al. J Mol Biol. 2004;338:1027. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 24.El-Manzalawyet Y, et al. Comput Syst Bioinformatics Conf. 2008;7:121. [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen M, et al. Bioinformatics. 2004;20:1388. doi: 10.1093/bioinformatics/bth100. [DOI] [PubMed] [Google Scholar]
- 26.Humphrey W, et al. J Mol Graph. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 27.Lundegaard C, et al. Bioinformatics. 2008;24:1397. doi: 10.1093/bioinformatics/btn128. [DOI] [PubMed] [Google Scholar]
- 28.Sundberg EJ, et al. Semin. mmunol. 2007;19:262. doi: 10.1016/j.smim.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmid-Hempel P, et al. Philos Trans R Soc Lond B Biol Sci. 2009;364:85. doi: 10.1098/rstb.2008.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mustafaet AS, et al. PLoS One. 2014;9:e103679. [Google Scholar]
- 31.Schmid-Grendelmeier P, et al. J Allergy Clin Immunol. 2005;15:1068. doi: 10.1016/j.jaci.2005.01.065. [DOI] [PubMed] [Google Scholar]
- 32.Crameri R, et al. J Exp Med. 1996;184:265. doi: 10.1084/jem.184.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyata M, et al. Clin Rheumatol. 1995;14:673. doi: 10.1007/BF02207935. [DOI] [PubMed] [Google Scholar]
- 34.Dalpke AH, et al. J Med Virol. 2003;71:408. doi: 10.1002/jmv.10501. [DOI] [PubMed] [Google Scholar]
- 35.Flückiger S, et al. J Immunol. 2002;168:1267. doi: 10.4049/jimmunol.168.3.1267. [DOI] [PubMed] [Google Scholar]
- 36.Eargle J, et al. Bioinformatics. 2006;22:504. doi: 10.1093/bioinformatics/bti825. [DOI] [PubMed] [Google Scholar]
- 37.Wucherpfennig KW, et al. J Clin Invest. 2001;108:1097. doi: 10.1172/JCI14235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Esaguy N, et al. Infect Immun. 1991;59:1117. doi: 10.1128/iai.59.3.1117-1125.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Eden W, et al. Proc Natl Acad Sci U S A. 1985;82:5117. doi: 10.1073/pnas.82.15.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kovvali G, et al. FEBS Lett. 2005;579:2261. doi: 10.1016/j.febslet.2005.02.073. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


