Abstract
Determining the structural elements that define substrates and inhibitors at the monoamine transporters is critical to elucidating the mechanisms underlying these disparate functions. In this study, we addressed this question directly by generating a series of N-substituted 3,4-methylenedioxyamphetamine analogs that differ only in the number of methyl substituents on the terminal amine group. Starting with 3,4-methylenedioxy-N-methylamphetamine, 3,4-methylenedioxy-N,N-dimethylamphetamine (MDDMA) and 3,4-methylenedioxy-N,N,N-trimethylamphetamine (MDTMA) were prepared. We evaluated the functional activities of the compounds at all three monoamine transporters in native brain tissue and cells expressing the transporters. In addition, we used ligand docking to generate models of the respective protein-ligand complexes, which allowed us to relate the experimental findings to available structural information. Our results suggest that the 3,4-methylenedioxyamphetamine analogs bind at the monoamine transporter orthosteric binding site by adopting one of two mutually exclusive binding modes. 3,4-methylenedioxyamphetamine and 3,4-methylenedioxy-N-methylamphetamine adopt a high-affinity binding mode consistent with a transportable substrate, whereas MDDMA and MDTMA adopt a low-affinity binding mode consistent with an inhibitor, in which the ligand orientation is inverted. Importantly, MDDMA can alternate between both binding modes, whereas MDTMA exclusively binds to the low-affinity mode. Our experimental results are consistent with the idea that the initial orientation of bound ligands is critical for subsequent interactions that lead to transporter conformational changes and substrate translocation.
Introduction
The principal role of neurotransmitter transporters is to retrieve previously released neurotransmitter molecules from the extracellular space (Kristensen et al., 2011). This is an economical way of regulating synaptic transmission and neurotransmitter homeostasis because it inactivates extracellular monoamine signaling and replenishes intracellular stores of the vesicular transmitter. The physiologic significance of transporter-mediated uptake is clinically relevant since ligands that interact with monoamine transporters [serotonin transporter (SERT), dopamine transporter (DAT), and norepinephrine transporter (NET)] are used as medications to treat a number of psychiatric disorders that involve an underlying imbalance of monoamine signaling and homeostasis (Iversen, 2000; Rothman and Baumann, 2003). On the other hand, some transporter ligands are used for nonmedical purposes and can be addictive, therefore blurring the borders between therapeutically and illicitly used drugs (Kristensen et al., 2011).
The structurally diverse array of monoamine transporter ligands can be classified with respect to their mode of action at the transporter. Inhibitors (e.g., cocaine) elevate extracellular monoamine concentrations by blocking reuptake of endogenous substrate from the extracellular space. Most of these inhibitors are competitive, and their binding sites overlap with the substrate-binding sites (Beuming et al., 2008; Penmatsa et al., 2013, 2015; Wang et al., 2015). Substrates (e.g., D-amphetamine), in contrast, increase extracellular monoamine concentrations via two mechanisms: 1) they compete with the endogenous substrate for reuptake from the extracellular space; and 2) they induce release of endogenous substrate molecules from intracellular stores by reversing the normal direction of transporter flux (Sitte and Freissmuth, 2015). The mechanistic difference between competitive inhibitors and substrates raises questions about the structural determinants that distinguish both compound groups. It also suggests the possibility that some compounds might have mixed substrate and inhibitory properties at different transporters (Blough et al., 2014; Reith et al., 2015; Saha et al., 2015).
Recently, crystal structures of the Drosophila melanogaster DAT (dDAT) bound to different substrates and inhibitors have been reported (Penmatsa et al., 2013, 2015; Wang et al., 2015). These studies have advanced our understanding of how the monoamine transporters coordinate ligand binding. Yet the structural features that separate substrates from inhibitors remain ill-defined, in part because conclusions are difficult to draw when comparing the actions of substrate drugs based on dissimilar chemical scaffolds.
Here, we tackled this issue by synthesizing and investigating a series of 3,4-methylenedioxy ring–substituted amphetamine compounds that share the same structural scaffold and differ only in the number of methyl substituents at the terminal amine. Specifically, 3,4-methylenedioxyamphetamine (MDA; “Sally”) and 3,4-methylenedioxy-N-methylamphetamine (MDMA; “Ecstasy” or “Molly”) are synthetic drugs that are consumed for recreational purposes because they exert psychostimulatory and entactogenic effects (Steinkellner et al., 2011). It is well established that MDA and MDMA are transporter substrates that induce transporter-mediated release of serotonin (5-HT), dopamine, and norepinephrine from neurons (Baumann et al., 2007; Rickli et al., 2015b). Insertion of additional N-methyl groups on MDMA produces enlarged ligands, with altered hydrophobicity and charge density. Importantly, these structural modifications convert the parent compound from a substrate to an inhibitor. Thorough investigation of the pharmacological properties of these amphetamine analogs allowed us to systematically define structural features that distinguish substrates from inhibitors.
Materials and Methods
Drugs and Reagents
For uptake and release assays in synaptosomes, [3H]dopamine, [3H]norepinephrine, [3H]5-HT, and [3H]1-methyl-4-phenylpyridinium ([3H]MPP+) were purchased from Dupont New England Nuclear (Boston, MA). S-(+)-3,4-methylenedioxyamphetamine (MDA) and S-(+)-3,4-methylenedioxy-N-methylamphetamine (MDMA) were provided by RTI International (Research Triangle Park, NC) through the National Institute on Drug Abuse Drug Supply Program.
S-(+)-3,4-methylenedioxy-N,N-dimethylamphetamine (MDDMA) and S-(−)-3,4-methylenedioxy-N,N,N-trimethylamphetamine (MDTMA) were synthesized, purified, and characterized according to the methods described in the Supplemental Data. All other reagents, buffer salts, and chemicals were obtained from Sigma Chemical (St. Louis, MO) unless otherwise noted. Reagents used in the experiments for uptake and efflux assays in cells were purchased and used according to previous work (Hofmaier et al., 2014). Plasmids encoding human SERT were a generous gift of Dr. Randy D. Blakely.
Animals and Housing
Male Sprague-Dawley rats (Charles River, Wilmington, MA) weighing 250–350 g were housed in standard conditions (lights on from 7:00 a.m. to 7:00 p.m.), with food and water freely available. Rats were maintained in facilities fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care, and experiments were performed in accordance with the Institutional Care and Use Committee of the National Institute on Drug Abuse Intramural Research Program (IRP).
Uptake and Release Assay in Rat Brain Synaptosomes
Uptake and release assays were carried out in rat brain synaptosomes as previously described (Baumann et al., 2013). Synaptosomes were prepared from rat striatum for DAT assays, whereas synaptosomes were prepared from whole brain minus striatum and cerebellum for NET and SERT assays.
For uptake inhibition assays, 5 nM [3H]dopamine, [3H]norepinephrine, and [3H]5-HT were used to assess transport activity at DAT, NET, and SERT, respectively. Where necessary, the selectivity of uptake assays was optimized for a single transporter of interest by including unlabeled blockers to prevent uptake of a [3H]transmitter by competing transporters. Specifically, NET assays were carried out in the presence of 50 nM GBR12935 to prevent uptake of [3H]norepinephrine by DAT, whereas SERT assays were carried out in the presence of 50 nM GBR12935 and 100 nM nomifensine to prevent uptake of [3H]5-HT by DAT and NET, respectively. No unlabeled blockers are required for DAT uptake assays because no measurable uptake of [3H]dopamine by NET and SERT occurs in caudate tissue. Uptake inhibition assays were initiated by adding 100 µl of tissue suspension to 900 µl of Krebs-phosphate buffer containing the test drug and [3H]transmitter. Uptake inhibition assays were terminated by rapid vacuum filtration through Whatman GF/B filters (Sigma-Aldrich Handels GmbH, Vienna, Austria), and the retained radioactivity was quantified by liquid scintillation counting.
For release assays, 9 nM [3H]MPP+ was used as the radiolabeled substrate for DAT and NET, whereas 5 nM [3H]5-HT was used as the radiolabeled substrate for SERT. All buffers used in the release assays contained 1 µM reserpine to block vesicular uptake of substrates.
The selectivity of release assays was optimized for a single transporter by including unlabeled blockers to prevent the uptake of [3H]MPP+ or [3H]5-HT by competing transporters. Specifically, DAT release assays were carried out in the presence of 100 nM desipramine and 100 nM citalopram to block NET and SERT. NET release assays were carried out in the presence of 100 nM citalopram and 50 nM GBR12935 to block SERT and DAT. SERT release assays were carried out in the presence of 50 nM GBR12935 and 100 nM nomifensine to block DAT and NET. Synaptosomes were preloaded with radiolabeled substrate in Krebs-phosphate buffer for 1 hour (steady state). Release assays were initiated by adding 850 µl of preloaded synaptosomes to 150 µl of the test drug. Release was terminated by vacuum filtration and the retained radioactivity was quantified as described for uptake inhibition.
Uptake and Release Assay in Human Embryonic Kidney 293 Cells
The uptake and release assays in human embryonic kidney (HEK) 293 cells were carried out as previously described (Hofmaier et al., 2014). For uptake assays, cells were washed twice with Krebs HEPES buffer (KHB) (composition in mM: 10 HEPES, 130 NaCl, 1.3 KH2PO4, 1.5 CaCl2, and 0.5 MgSO4, 10 glucose, pH 7.4 adjusted with NaOH). Test drugs were added to cells for 5 minutes, allowing equilibration with transporters. Subsequently, 0.1 µM of either [3H]5-HT or [3H]dopamine were added, and the reaction was stopped after allowing uptake for 1 minute. The uptake was terminated by washing with 500 µl of ice-cold KHB, cells were lysed with 500 µl of 1% sodium dodecyl sulfate, and tritium was counted on a Packard 2300TR TriCarb Liquid Scintillation Analyzer (PerkinElmer, Brunn, Austria). For release studies, HEK293 cells expressing human SERT (hSERT) or human DAT (hDAT) were grown overnight on round glass coverslips (5-mm diameter, 40,000 cells per coverslip) placed in a 96-well plate and preloaded with 0.4 µM [3H]5-HT or 0.03 µM [3H]MPP+ for 20 minutes at 37°C in a final volume of 0.1 ml/well. Coverslips were transferred to small chambers (0.2 ml) and superfused with KHB (25°C, 0.7 ml/min). The 40-minute baseline for efflux of radioactivity was followed by the addition of test drugs and collection of fractions every 2 minutes. The experiment was terminated by lysis of the cells with 1% sodium dodecyl sulfate and counted.
Measurements of Conformational Change of Fluorescently Tagged SERT Expressed in HEK293 Cells
Fluorescence resonance energy transfer microscopy (FRET) was measured with a Carl Zeiss Axiovert 200 epifluorescence microscope (Carl Zeiss, Vienna, Austria). We used HEK293 cells transiently transfected with plasmid cDNA (1.7 μg) by means of the calcium phosphate coprecipitation method as described previously (Fenollar-Ferrer et al., 2014). Cells were transfected directly in ibidi µ-Slide chambered coverslips with eight wells (Martinsried, Germany). Directly before each FRET microscopy experiment, every well was washed with 300 µl of KHB and incubated in 150 µl of KHB. The “three-filter method” was performed as previously described. Images were acquired using a 63× oil immersion objective under continuous usage of a gray filter (20% density). Ludl filter wheels (Ludl Electronic Products, Hawthorne, NY) allowed for a rapid excitation and emission filter exchange (system assembled by Visitron Systems GmbH, Puchheim, Germany). The Ludl filter wheels were configured as follows: cyan fluorescent protein (CFP) (IDonor; excitation: 436 nm; emission: 480 nm; and dichroic mirror: 455 nm), yellow fluorescent protein (YFP) (IAcceptor; excitation: 500 nm; emission: 535 nm; and dichroic mirror: 515 nm), and FRET (IFRET; excitation: 436 nm; emission: 535 nm; and dichroic mirror: 455 nm). Images were acquired with a charge-coupled device camera (Coolsnap fx; Roper Scientific, Tucson, AZ) using the MetaMorph of MetaSeries software package (release 4.6; Universal Imaging Corp., Downingtown, PA). Pixel shift was corrected whenever necessary by using the following combination of ImageJ-plugins: TurboReg and StackReg (Thevenaz et al., 1998). Background fluorescence was subtracted from all images. We analyzed the images pixel by pixel using ImageJ (version 1.43b; Wayne Rassband, National Institutes of Health, Bethesda, MD) and the ImageJ plugin PixFRET (pixel by pixel analysis of FRET with ImageJ, version 1.6.0_10) (Feige et al., 2005) spectral bleed-through parameters were determined for the donor bleed through and acceptor bleed through. Next, normalized FRET was computed at the plasma membrane (predefined as the region of interest) using the computed FRET efficiency image. The regions of interest were selected in the CFP (donor) or YFP (acceptor) image (to avoid bleaching-associated bias) and transmitted to the FRET image by the ImageJ Multi Measure Tool. All experiments were conducted for several individual transfections; three to five wide-field images were captured during each experiment and one to seven transfected cells per image were included in the study to reach an n number of 18–64.
Measurements of Conformational Change of SERT Expressed in HeLa Cells
HeLa cells were transfected with SERT C109A-S404C, treated with 2-aminoethyl methanethiosulfonate hydrobromide (MTSEA), and assayed as described previously (Jacobs et al., 2007). Cys109 is the primary reactive cysteine on the extracellular surface of SERT. Replacing it with alanine renders SERT relatively resistant to MTSEA treatment (Chen et al., 1997). In S404C, the inserted cysteine reacts more rapidly when SERT is in an outward-open conformation and more slowly in an inward-open conformation. Briefly, cells expressing this SERT mutant were exposed to a range of MTSEA concentrations in the presence or absence of 20 µM methylenedioxyamphetamine derivative for 15 minutes, washed to remove excess unreacted MTSEA and drug, and then assayed for [3H]5-HT influx in a 5-minute incubation.
Electrophysiological Measurements: Whole-Cell Patch Clamp
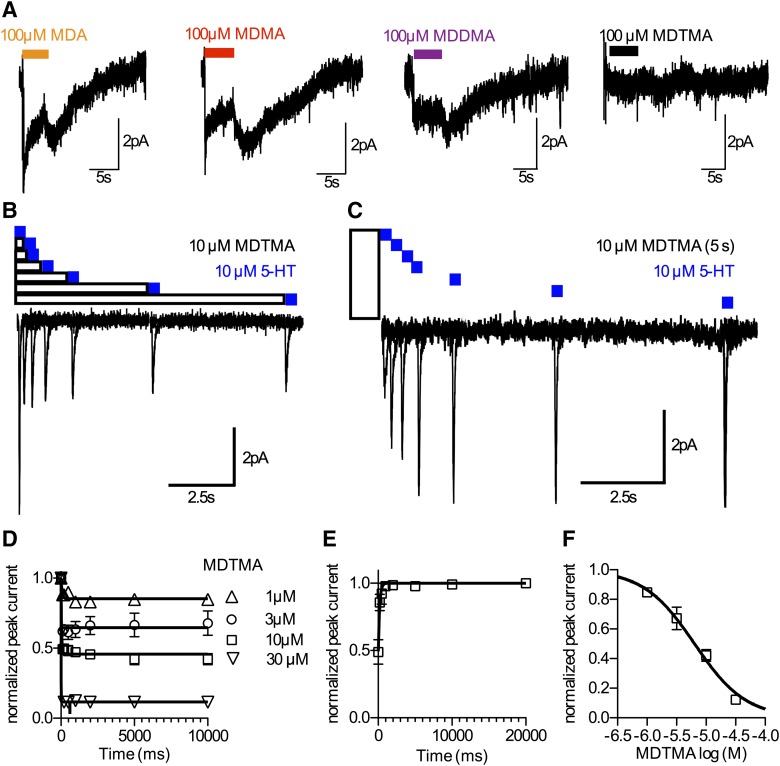
For patch clamp recordings, HEK293 cells stably expressing hSERT (Hilber et al., 2005) were seeded at low density for 24 hours before measuring currents. To measure substrate-induced hSERT currents, cells were voltage clamped using the whole-cell patch-clamp technique. For measurements of the substrate-induced steady-state current, glass pipettes were filled with a solution consisting of 133 mM K-gluconate, 5.9 mM NaCl, 1 mM CaCl2, 0.7 mM MgCl2, 10 mM EGTA, and 10 mM HEPES adjusted to pH 7.2 with KOH. For measurements that required isolation of the substrate-induced peak currents, the pipette solution consisted of 152 mM NaCl, 1 mM CaCl2, 0.7 mM MgCl2, 10 mM EGTA, and 10 mM HEPES (pH 7.2 with NaOH). Although substrate-induced steady-state currents were measured at −60 mV, the membrane potential was clamped to 0 mV for the measurement of MDTMA association and dissociation (Hasenhuetl et al., 2015). The cells were continuously superfused with an external solution of 140 mM NaCl, 3 mM KCl, 2.5 mM CaCl2, 2 mM MgCl2, 20 mM glucose, and 10 mM HEPES adjusted to pH 7.4 with NaOH. Currents were recorded at room temperature (20–24°C) using an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA) and pClamp 10.2 software (Molecular Devices, Sunnyvale, CA). MDA, MDMA, MDDMA, and MDTMA were applied for 5 seconds once every 60 seconds, respectively.
Current traces were filtered at 1 kHz and digitized at 10 kHz using Digidata 1320A (MDS Analytical Technologies). The liquid junction potential was taken into account, and the measurements were accordingly compensated. Drugs were applied using a DAD-12 (Adams & List, Westbury, NY), which permits complete solution exchange around the cells within 100 milliseconds (Boehm, 1999). Current amplitudes in response to substrate application were quantified using Clampfit 10.2 software (Molecular Devices, Sunnyvale, CA). Passive holding currents were subtracted, and the traces were filtered using a 100-Hz digital Gaussian low-pass filter (Molecular Devices, Sunnyvale, CA).
Pharmacoinformatics
Model Generation.
The protein models were created following the procedure published earlier (Stockner et al., 2013, 2014). The models were based on the outward-facing structures of the leucine transporter, LeuTAa, crystallized from Aquifex aeolicus (Protein Data Bank identity 3F3A) (Singh et al., 2007) because the models created from the occluded conformation resulted in a potentially too narrow S1-binding site. In brief, homology models were constructed using MODELLER (Sali and Blundell, 1993). The 10 best scoring models were inserted into a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine membrane, equilibrated, and simulated using Gromacs (Hess et al., 2008) and applying the Optimized Potentials for Liquid Simulations (OPLS) force field (Kaminski et al., 2001). For each transporter, we selected the equilibrated structures of the three models that behaved best in 50-ns-long MD simulations.
Ligand Docking.
The ligands were built as the (S)-enantiomer (Seddik et al., 2013), and protonation states for pH 7 were calculated with Protonate3D using Molecular Operating Environment (MOE) 2012 (Chemical Computing Group, Montreal, QC, Canada). All four ligands carry an overall charge of +1. We estimated the total van der Waals surface area for atoms with a partial change above 0.2 using the molecular descriptor “PEOE_VSA_PPOS” of MOE that applies the partial equalization of the orbital electronegativities method (Gasteiger and Marsili, 1980) and uses the connection table approximation.
Using the docking software GOLD (Jones et al., 1997), 500 poses of each ligand were generated with the ChemPLP scoring function. ChemPLP employs a piecewise linear potential optimized for pose prediction and virtual screening (Korb et al., 2009). Side chain dihedrals in the substrate-binding site were allowed to rotate according to a rotamer database. In SERT, residues Tyr95, Trp103, Arg104, Ile172, Tyr172, Tyr176, Phe335, Phe341, Thr439, and Thr497 were selected. In DAT, the corresponding residues were Phe76, Trp84, Arg85, Val152, Phe155, Tyr156, Phe320, and Phe326. No restraints were imposed. The electrostatic surface maps of the ligands were calculated with MOE using the Poisson-Boltzmann solver. The color scale (red-white-blue) was set to range from −40 to 40 kcal mol−1e−1. These distances were calculated with python, and heat maps were created using Matplotlib (Hunter, 2007) and Plotly (for the code, data, and interactive plot, see https://plot.ly/∼eymayr/143/sert/) packages.
Statistical Analysis
Statistical analyses were carried out using GraphPad Prism (version 5.0; GraphPad Scientific, San Diego, CA). IC50 values for the inhibition of uptake and EC50 values for stimulation of release were calculated based on a nonlinear regression analysis.
Results
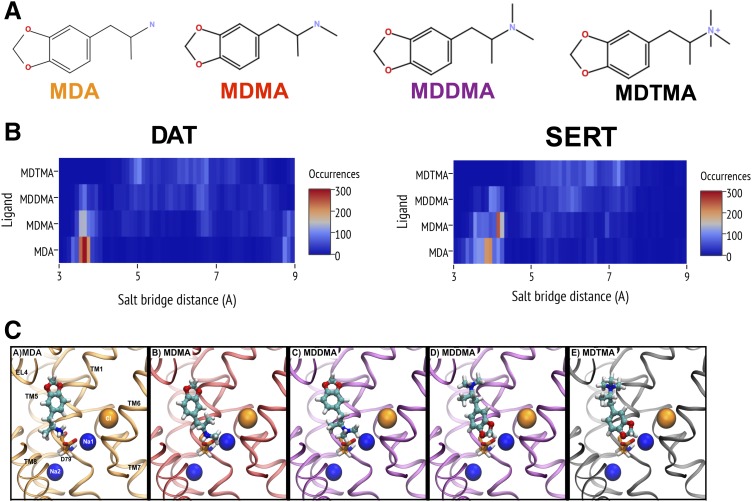
We generated a series of compounds that share the same structural scaffold but differ by the degree of methylation at the terminal amine group (Supplemental Fig. 1). The chemical structures of all compounds are shown in Fig. 1A. Starting from MDA, methyl groups were successively added to the primary amine to generate MDMA, MDDMA, and MDTMA. Addition of the first methyl group increased the solvent accessible surface area from 2.79 (MDA) to 3.17 nm2 (MDMA). The primary, secondary, and tertiary amino groups of MDA, MDMA, and MDDMA, respectively, are protonatable, and these compounds exist in positively charged and neutral forms. The pKa values for MDA, MDMA, and MDDMA are predicted to be 10.0, 10.1, and 9.4, respectively. Hence, the prevalent species at a physiologic pH is protonated and charged. The quaternary amine of MDTMA carries a permanent positive charge. All compounds used in either pharmacoinformatic or biochemical experiments consisted of the S(+)-enantiomer.
Fig. 1.
(A) Chemical structures of MDA, MDMA, MDDMA, and MDTMA. (B) Heat map of the distance distributions between the nitrogen atom of the ligand and Cδ of the aspartate residue in the respective binding site of DAT-D79 (left panel) and SERT-D98 (right panel). The distances were derived from 500 docking poses for each ligand grouped into 0.01-nm-wide bins. We observed the high population of poses with short nitrogen-Asp distances for MDA, decreasing with an increasing degree of methylation. Wide distributions and generally longer distances were found in the case of MDDMA and MDTMA. (C) Representative poses are shown for MDA, MDMA, MDDMA, and MDTMA docked into DAT. Two poses are depicted for MDDMA, as we found that docking poses of MDDMA had MDA-like and MDTMA-like characteristics.
Ligand Docking Poses Point to Different Binding Modes of the Series.
We previously described procedures to generate free outward-facing homology models for DAT inserted into a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine membrane, which were optimized and tested for stability in 200 ns molecular dynamics simulations (Stockner et al., 2013, 2014). At the end of the simulations, the three best-scored models were selected for docking. The homology models for SERT were produced based on the DAT template. Because SERT and DAT are similar in sequence, with 52% identity in the transmembrane domain, we assume that their structures are very similar. MDA and its analogs were docked into the central binding site S1, creating 500 poses for each of the three SERT and DAT models (see Supplemental Data for poses). All poses were retained for further analyses.
It is well established that the charged amino groups of DAT and SERT ligands interact with a conserved aspartate in the binding pocket that may also coordinate Na+ (Asp79 in DAT and Asp98 in SERT; these residues will be subsequently referred to as “aspartate” for the sake of simplicity) (Barker et al., 1999; Celik et al., 2008). In contrast, the corresponding position in amino acid transporters, such as the leucine transporter of Aquifex aeolicus, LeuTAa, and a transporter for GABA, GAT1, is a glycine residue, and Na+ is coordinated by the carboxylate group of the substrate (Yamashita et al., 2005). Likewise, the nitrogen in the dopamine molecule is positioned directly adjacent to the aspartate in the dDAT crystal structure (Wang et al., 2015). Given the importance of this interaction, we determined the distance between the nitrogen and the aspartate side chain in the complete set of docked poses. The results are visualized in a heat map representation (Fig. 1B) using a bin width of 0.01 nm.
A distance of ∼3.5–4.0 Å indicates direct contact. The heat map revealed that the two smaller compounds, MDA and MDMA, showed one peak in the distance distribution derived from all poses at ∼3.5–4.5 Å. It is noteworthy that poses displaying longer distances occurred sparsely. For MDDMA and MDTMA, we found a wider distance distribution. In the case of MDDMA, we identified a small population of poses in which the nitrogen was in direct contact with the aspartate. In contrast to the other three compounds, the docking poses of trimethylated MDTMA lacked poses, where the trimethylated nitrogen was in contact with the aspartate. Here, the poses showed a distribution of several conformations and this behavior was similar for DAT and SERT. Recently, dDAT has been cocrystallized in complex with several substrates (Wang et al., 2015). When comparing the present docking results with these structures, it is evident that the charged nitrogen of the docked methylenedioxyamphetamines interacts with the aspartate similarly to the substrates that were cocrystallized with dDAT. However, the ring structure of the methylenedioxyamphetamines showed the same orientation as the cocrystallized amphetamines only in a subset of all docked poses. Several factors could contribute to the observed differences. For example, Asp121 of dDAT, which interacts with the cocrystallized amphetamines, is a glycine in hDAT and an alanine in hSERT. Hence, this interaction is likely lost in the human transporters. In addition, the methylenedioxyamphetamines are larger and more extended than the amphetamines. Finally, the β-factors of the cocrystallized amphetamine ligands in the dDAT structures are higher than for the surrounding residues in the S1 site and their electron densities are not well resolved. They might therefore adopt more than one conformation.
The main results obtained from our docking runs indicated that MDA and MDMA adopt a single conformation, with an orientation expected for a substrate. Representative docking poses are displayed in Fig. 1C. Notably, MDTMA did not show this particular conformation. The docking poses revealed the positively charged nitrogen to be distant from the aspartate. In fact, most poses are in an inverted orientation compared with the substrates. This places the MDTMA tail next to the aspartate (Fig. 1C). The dimethylated MDDMA showed an intermediate behavior where we found a subset of the poses (30% in SERT; 17% in DAT), with a direct nitrogen-aspartate interaction.
A representative pose of both binding modes of MDDMA is shown in Fig. 1C to exemplify this hybrid behavior. This observation was unexpected because the similarity of the compound series had suggested that the binding pose would be highly similar. Based on the change in the binding mode, we hypothesized that substrates assume an orientation where the (methylated) nitrogen contacts the aspartate directly, whereas the analogs in an alternative orientation would act as inhibitors. The change in orientation also implies that the binding affinity changes. These predictions were subsequently tested.
Each Member of the Series Can Bind to DAT, SERT, and NET.
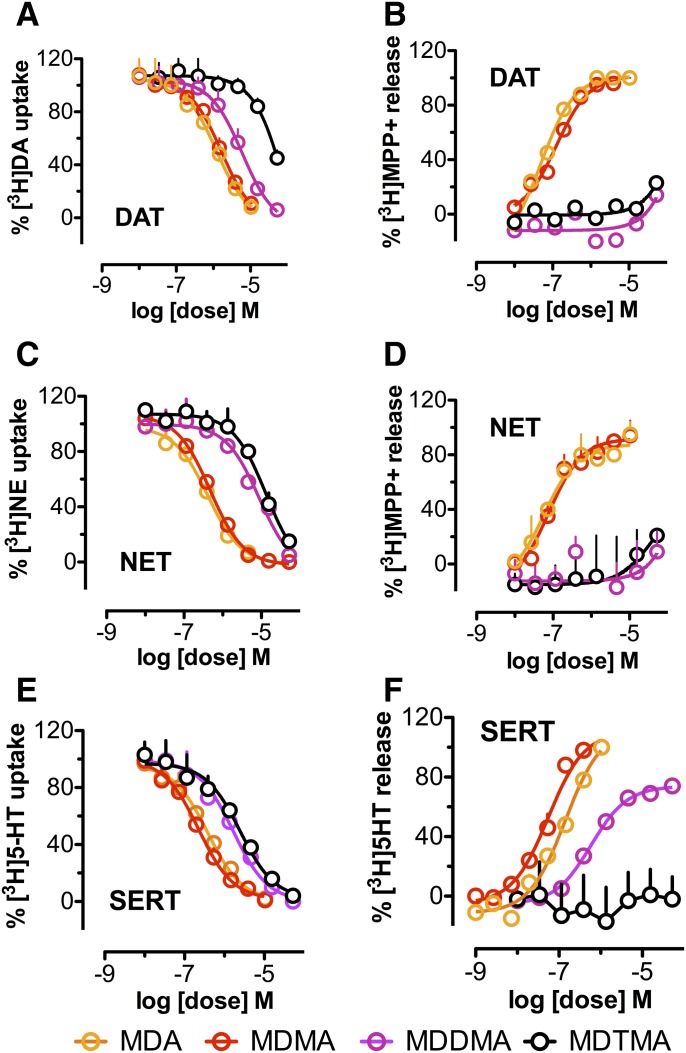
As shown in Fig. 2, A–C, we tested the ability of each member of the series to inhibit [3H]substrate uptake into synaptosomes prepared from the rat brain under assay conditions optimized for DAT, NET, or SERT. Uptake was inhibited in a concentration-dependent manner, suggesting that all tested compounds are able to interact with all three transporters. We found that MDA and MDMA inhibited uptake at DAT, NET, and SERT in a nonselective manner with similar potencies, whereas MDDMA and MDTMA blocked uptake at the monoamine transporters much less potently. Nevertheless, all compounds were able to fully block uptake.
Fig. 2.
Uptake inhibition by MDA, MDMA, MDDMA, and MDTMA of (A) DAT, (C) NET, and (E) SERT measured in synaptosomes (see Materials and Methods). Substrate release by MDA, MDMA, MDDMA, and MDTMA via (B) DAT, (D) NET, and (F) SERT measured in the same preparation. The respective IC50 for uptake inhibition and the EC50 values for substrate release are displayed in Table 1.
TABLE 1.
Uptake inhibition and release in rat brain synaptosomes
| MDA | MDMA | MDDMA | MDTMA | |
|---|---|---|---|---|
| Uptake | ||||
| DAT (IC50 in nM) mean ± S.D. | 1177 ± 252 | 1654 ± 274 | 6010 ± 1028 | 33,300 ± 10,370 |
| NET (IC50 in nM) mean ± S.D. | 411 ± 49 | 475 ± 33 | 8814 ± 1742 | 13,500 ± 2495 |
| SERT (IC50 in nM) mean ± S.D. | 384 ± 44 | 227 ± 30 | 1661 ± 289 | 2476 ± 491 |
| Release | ||||
| DAT (EC50 in nM) mean ± S.D. | 50.1 ± 6.5 | 125.6 ± 13.7 | Inactive | Inactive |
| NET (EC50 in nM) mean ± S.D. | 59.1 ± 14.2 | 78.0 ± 15.8 | Inactive | Inactive |
| SERT (EC50 in nM) mean ± S.D. | 141.2 ± 17.6 | 55.2 ± 6.7 | 588.8 ± 68.0 | Inactive |
These observations prompted us to test whether the interaction of MDDMA and MDTMA with the transporter were qualitatively different from MDA and MDMA. Specifically, we examined whether the additional methyl groups had hampered these compounds’ ability to act as a substrate, as has been shown previously for other monoamine transporter substrates (Simmler et al., 2013; Rickli et al., 2015a).
Only MDA and MDMA Are True Substrates of Monoamine Transporters.
Direct and competitive inhibition of [3H]substrate uptake by a ligand without concomitant alteration of transporter surface expression is a formal proof of binding. However, the observed inhibition in the radioligand uptake assay does not provide insights into the nature of the interaction, i.e., it does not address the question whether a compound acts as substrate or inhibitor. Inhibitors block transport, whereas substrates can induce nonexocytotic substrate release. Hence, it is possible to infer whether a given compound acts as a substrate or inhibitor by testing the capacity of a ligand to induce substrate release.
In agreement with previous findings (Baumann et al., 2007; Rickli et al., 2015a), the present data show that MDA and MDMA induce [3H]substrate release via DAT, NET, and SERT in a nonselective manner (Fig. 2, D–F), suggesting that these compounds are “full” substrates of all three transporters. Conversely, MDDMA and MDTMA failed to induce [3H]MPP+ release via DAT and NET, indicating that these compounds act as transport inhibitors at DAT, NET, and SERT. Moreover, in the case of SERT, MDDMA induced substrate release, whereas MDTMA did not, indicating that interaction at the three monoamine transporters was not identical. It is worth noting that even at saturating concentrations, MDDMA released less substrate than MDA and MDMA. We have previously identified a number of compounds that also show this property of “partial release”. Analogous to partial agonism at neurotransmitter receptors, these ligands were classified as “partial releasers” (Rothman et al., 2012). Although partial release by a compound is frequently observed in synaptosomal preparations, this phenomenon is not mechanistically understood and yet constitutes a subject for future investigation.
To rule out that the synaptosomal tissue preparation influenced the observed substrate/inhibitor behavior, we tested MDMA and MDTMA in intact hippocampal slices and recorded substrate behavior for MDMA and inhibitor behavior for MDTMA (Supplementary Fig. 2).
Inhibition of Substrate Uptake and Induction of Substrate Release Measured in HEK293 Cells Stably Expressing SERT.
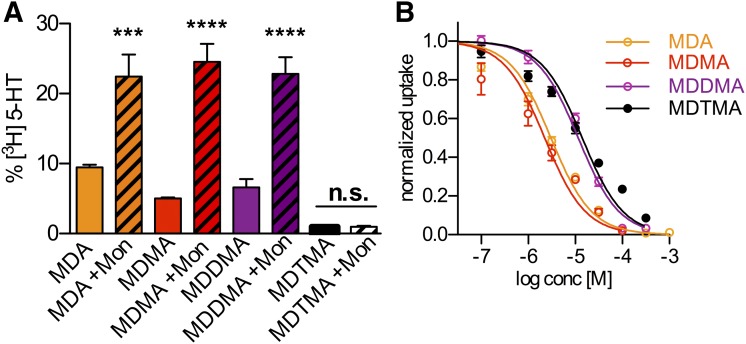
MDA, MDMA, and MDDMA induced substrate release from both synaptosomal and hippocampal slice preparations containing SERT, whereas MDTMA did not. However, both preparations also comprise components of the exocytotic machinery. Hence, we could not rule out the contribution of vesicle fusion to our results. HEK293 cells stably transfected with SERT are devoid of substrate-filled vesicles. Accordingly, in this system, SERT is expected to be the sole carrier of the released substrate. Thus, we aimed at reproducing our results in this overexpression system based on the following premise: substrate-induced, carrier-mediated release depends on the intracellular Na+ concentration (Scholze et al., 2000); therefore, increasing the intracellular Na+ concentration by the addition of monensin (10 µM) will further discriminate substrates from inhibitors (Baumann et al., 2013). As shown in Fig. 3A, we reproduced the results obtained from the synaptosomal preparation in HEK293 cells and thereby confirmed that MDA, MDMA, and MDDMA induced substrate efflux directly via SERT. Furthermore, the elevation of the intracellular sodium concentration facilitates basal efflux (Scholze et al., 2000) and strongly enhances only efflux elicited by MDA, MDMA, and MDDMA, but fails to change the effects seen with MDTMA (Fig. 3A). This is in support of the conclusion that MDA, MDMA, and MDDMA are substrates of SERT. Importantly, MDTMA also failed to induce substrate release in HEK293 cells. This is consistent with the idea that the introduction of the third methyl group at the amine had transformed this compound into an inhibitor. In uptake inhibition experiments, we confirmed that MDTMA is able to bind to SERT with the same affinity as MDDMA, assuring that impaired binding was not the cause of absent substrate release in HEK293 cells (Fig. 3B). Note that the relatively low potency of MDDMA and MDTMA at uptake inhibition in HEK293 stably expressing SERT appears remarkably similar to the results obtained from SERT uptake in rat brain synaptosomes.
Fig. 3.
(A) Nonexocytotic [H3]5-HT release from HEK293 cells stably transfected with SERT induced by MDA (10 µM), MDMA (3 µM), MDDMA (30 µM), and MDTMA (30 µM), with and without monensin (Mon). The respective values are MDA (9.46 ± 0.99%; n = 6), MDA + Mon (22.44 ± 7.71%; n = 6), MDMA (5.02 ± 0.33%; n = 4), MDMA + Mon (24.52 ± 5.81%; n = 5), MDDMA (6.63 ± 4.14%; n = 12), MDDMA + Mon (22.81 ± 8.00%; n = 11), MDTMA (0.98 ± 0.45%; n = 9), and MDTMA + Mon (0.98 ± 0.44%; n = 9). ***P < 0.001 and ****P < 0.0001, one-way analysis of variance with Bonferroni’s multiple comparison test. (B) Inhibition of [H3]5-HT uptake by MDA, MDMA, MDDMA, and MDTMA as a function of concentration measured in HEK293 cells stably transfected with SERT. The IC50 values for MDA, MDMA, MDDMA, and MDTMA are 3.04 µM (2.67–3.45 µM), 2.24 µM (1.71–2.93 µM), 11.56 µM (10.02–13.17 µM), and 14.06 µM (11.95–16.53 µM), respectively.
Conformational Equilibrium of SERT Is Shifted toward the Inward-Facing Conformation by MDA, MDMA, and MDDMA but Not by MTDMA.
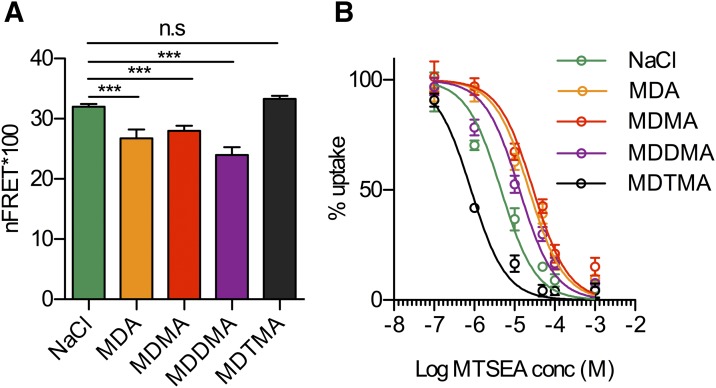
We have recently shown that a double-tagged version of SERT, in which a fluorescence donor (CFP) and acceptor (YFP) are attached to the N and C terminus, respectively, allows for measuring intramolecular distances within SERT by FRET (Just et al., 2004). We demonstrated that substrate application to this construct (which we refer to as C-SERT-Y in this work) decreased FRET efficiency due to a greater separation between the N and C termini in the inward-open state (Schicker et al., 2012). This observation is consistent with earlier reports (Just et al., 2004; Yamashita et al., 2005; Zhang and Rudnick, 2006; Forrest et al., 2008) and a computational study that used a homology model of SERT (Fenollar-Ferrer et al., 2014). The latter predicted the separation of the C and N termini upon opening of the cytoplasmic pathway. In accordance with this notion, manipulations that are known to stabilize the outward-facing conformation and close the cytoplasmic pathway, such as high extracellular Na+ concentration or the presence of a competitive inhibitor, resulted in higher FRET values (Schicker et al., 2012). Here, we employed C-SERT-Y to probe how the members of the series affect the conformational equilibrium. We observed decreased FRET efficiency in the presence of MDA (26.7 ± 1.5%; n = 18), MDMA (28.2 ± 0.9%; n = 37), or MDDMA (24.0 ± 1.3%; n = 21) compared with control buffer conditions (32.0 ± 0.4%; n = 63). This finding indicates that the equilibrium was shifted toward the inward-facing conformation and is expected for substrates (Fig. 4A). In contrast, in the presence of MDTMA, the FRET values (33.3 ± 0.5%; n = 52) were consistently higher and did not significantly differ from control, suggesting that this compound is not a substrate.
Fig. 4.
(A) Calculated normalized FRET (nFRET) values determined from HEK293 cells stably expressing C-SERT-Y. Addition of 100 µM MDA, MDMA, and MDDMA to a Na+ (100 mM) containing bath solution significantly reduced nFRET. The latter effect is rescued by 100 µM MDTMA (Na+, 32.0 ± 1.5; MDA, 26.7 ± 1.5%; MDMA, 28.2 ± 0.9%; MDDMA, 24.0 ± 1.3%; and MDTMA, 33.3 ± 0.5%). ***P < 0.001, n = 18–64, one-way analysis of variance with Tukey’s multiple comparison test. (B) HeLa cells transfected with SERT S404C in the C109A background were treated for 15 minutes in 100 mM Na+ with the indicated concentrations of MTSEA either alone or with 20 µM MDA, MDMA, MDDMA, or MDTMA. From the IC50 values for MTSEA, rate constants for the reaction of MTSEA with Cys404 were calculated. Compared with the control (NaCl) rate of 218 ± 13 M–1seconds−1, MDA (43.6 ± 1.0 M–1s−1), MDMA (31.2 ± 1.3 M–1s−1), and MDDMA (78.4 ± 2.8 M–1s−1) all decreased the reaction rate, indicating an increased fraction of SERT in an inward-facing form, in which Cys404 is less accessible. Addition of MDTMA increased the rate to 1070 ± 23 M–1s−1, which is consistent with a greater fraction in the outward-open form. All of the changes in reaction rate were significantly different from NaCl alone at the P < 0.006 level in a paired t test.
Another method to investigate the conformational equilibrium entails measurements of the accessibility of a cysteine placed in one of the SERT permeation pathways, as measured by its rate of reaction with an aqueous sulfhydryl reagent. We replaced the only reactive endogenous cysteine accessible from the extracellular side (Cys109) with alanine (Chen et al., 1997). In this background, we replaced Ser404 in the SERT extracellular pathway (Yamashita et al., 2005) with cysteine (Mitchell et al., 2004; Jacobs et al., 2007). The reactivity of Cys404 with MTSEA decreased when SERT was stabilized in an inward-open conformation by ibogaine or the substrate (Jacobs et al., 2007; Henry et al., 2011). In structures of LeuTAa, the corresponding residue (Asp321) is more accessible in outward-open or occluded conformations than in the inward-open state (Krishnamurthy and Gouaux, 2012).
Figure 4B shows the results from HeLa cells expressing SERT Cys109Ala-Ser404Cys. Compared with the control (100 mM NaCl), the presence of MDA, MDMA, and MDDMA decreased SERT Ser404Cys accessibility, requiring higher concentrations of MTSEA for modification of Cys404, as shown here by its inhibition of transport. MDTMA, however, increased the sensitivity of Cys-404 to MTSEA, as shown by the left-shifted profile in Fig. 4B, indicating its greater accessibility, which is characteristic of outward-open conformations. These results agree well with those of the FRET experiments, in which only MDTMA acts as a SERT inhibitor, whereas the other tested compounds are substrates. It is worth pointing out that both FRET and the accessibility assay were conducted in intact cells with physiologic ion gradients in place.
Electrophysiological Interrogation of the Series Confirms SERT Substrates and Inhibitors.
Administration of a substrate (e.g., 5-HT and MDMA) to a cell expressing SERT gives rise to an inwardly directed current (Mager et al., 1994). Conversely, the application of an inhibitor fails to induce such a current. Accordingly, electrophysiological measurements allow for deciding whether a compound is a substrate or an inhibitor. Figure 5A shows example traces of currents triggered by 100 µM MDA, MDMA, and MDDMA, respectively. The recorded currents resemble substrate-induced currents that were reported previously (Sandtner et al., 2014), supporting the notion that these compounds act as substrates on SERT. However, when 100 µM MDTMA was applied, no current could be observed, confirming that MDTMA is not a substrate for SERT.
Fig. 5.
(A) Electrophysiological properties of the series. MDA, MDMA, and MDDMA induce currents in SERT, whereas MDTMA does not. This finding is in accordance with the properties of MDTMA as the inhibitor. (B) Protocol to assess kinetics of MDTMA association. (C) Protocol to measure koff of MDTMA. (D) Time course of MDTMA association to SERT. (E) Direct measurement of koff of MDTMA, 5.6 seconds−1 (95% confidence interval: 4.15–7.02 s−1). (F) The kinetics of association cannot be resolved by the approach shown in (D and E); however, the steady-state values of this experiment were plotted as a titration curve to yield the KD of MDTMA on SERT. The calculated KD value is 6.35 µM (95% confidence interval: 5.70–7.12 µM). Data are the mean ± S.D. from four to eight independent measurements.
Inhibitor binding to SERT is electrically silent. However, we have recently developed an approach to assess inhibitor binding to the outward facing conformation of SERT and DAT. We measured charge movements induced by their cognate substrates (Hasenhuetl et al., 2015). Occupancy of the orthosteric binding site by an inhibitor prevents substrate binding and the concomitant generation of the substrate-induced peak current (Fig. 5B). The resulting reduction in the peak current amplitude can be used to estimate the occupancy of the transporter by the inhibitor and thus provides a means to study inhibitor binding. Rapid application of 10 µM 5-HT elicits a peak current. Because 10 µM 5-HT is a saturating concentration, the respective peak current amplitude is a measure of the maximal number of binding sites available on the measured cell. Subsequently, we applied MDTMA for 0.2, 0.5, 1, 2, 5, and 10 seconds, each prior to application of 10 µM 5-HT. The reduction of the peak current as a function of the exposure time and MDTMA concentration is shown in Fig. 5D. Association of MDTMA to SERT occurred at a very fast rate. As can be seen in Fig. 5D, at the first measuring point (0.2 seconds), inhibition had already reached steady state. Thus, in the case of MDTMA, the temporal resolution provided by our assay is not sufficient to reliably resolve the rate of association (kapp). Nevertheless, the steady-state values of peak current inhibition in Fig. 5D can be used to plot a binding curve (Fig. 5F). Importantly, the calculated KD [6.3 µM (95% confidence interval: 5.7–7.1 µM)] is in excellent agreement with the IC50 values reported above.
As described previously by Hasenhuetl et al. (2015), recording the time-course of recovery from the peak current inhibition can be used to measure inhibitor dissociation. First, we applied 10 µM MDTMA for 5 seconds to permit equilibration in the binding site. Then, MDTMA dissociation was assessed by washing the cell for 0.2, 0.5, 1, 2, 5, 10, and 20 seconds, each prior to application of 10 µM 5-HT to probe the fraction of transporters available for 5-HT binding (Fig. 5C). As shown in Fig. 5E, MDTMA exhibits a rapid dissociation rate constant [koff: 5.58 seconds−1 (95% confidence interval: 4.15–7.02 µM)]
Given that KD can be calculated as koff divided by kon, one can derive kon by knowing KD and koff (i.e., kon = koff/KD). We calculated a kon of approximately 9*105 M−1 seconds−1. This rate constant is comparable to association rate constants of other SERT inhibitors (Sucic et al., 2010; Hasenhuetl et al., 2015).
Discussion
The present study was designed to identify structural determinants that separate substrates from inhibitors of monoamine transporters. To ensure comparability of results and simplify the interpretation of possible outcomes, we generated a series of compounds that shared the same structural scaffold but differed solely by the number of methyl substituents on the terminal amine (see Fig. 1). Initially, we docked the compounds into homology models of DAT and SERT to relate our experimental findings to available structural information. We tested if the resulting models of the respective transporter-ligand complexes could plausibly predict the experimental findings. These predictions were then scrutinized intensively by employing various functional assays that allow for an assessment of ligand-transporter interactions. These include uptake and release assays, cysteine accessibility studies, FRET-based assays, and electrophysiological recordings.
MDA and MDMA show similar binding affinities across monoamine transporters, which were considerably higher as compared with the more sterically bulky compounds MDDMA and MDTMA. The simplest explanation for why MDDMA and MDTMA have lower affinity is a change in their binding mode. This was indeed predicted by our docking results. Successive methylation of the terminal amine gradually changes the following physicochemical properties: 1) it enlarges the volume surrounding the nitrogen; 2) it increases the hydrophobic surface area and; and 3) it lowers the surface charge density around the nitrogen. Each of these changes or a combination thereof may therefore account for our experimental findings. We found that our trimethylated compound, MDTMA, is an inhibitor of SERT and DAT. However, the increase in steric bulk due to the number of methyl groups attached to the amine nitrogen alone does not sufficiently explain our results as other compounds exist that have sterically bulky N-substituents and still behave like substrates (Rothman et al., 2012). Our analysis suggests that a change in the binding mode is mainly caused by a decrease in surface charge density. The surface of the S1 binding site in SERT is largely hydrophobic, but contains a charged aspartate and a sodium ion, with which ligands are known to interact via electrostatic forces. While the surface charge density at the nitrogen is high for MDA and MDMA, it is considerably lower in the case of MDDMA and MDTMA.
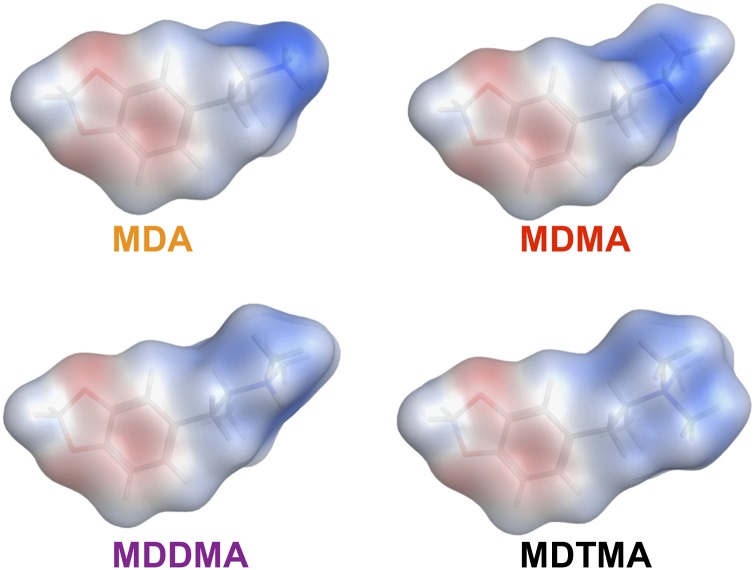
Hence, we surmised that electrostatic interactions might play an important role in ligand binding. We determined the surface charge density for the members of the compound series using the Poisson-Boltzmann solver of MOE and mapped the charge density onto the solvent accessible surface area (Fig. 6). The positive potential of the nitrogen becomes successively shielded with the increasing degree of methylation. This is shown by the fading blue color at the position of the nitrogen (Fig. 6). We also quantified the total van der Waals surface area of ligand atoms with a partial charge exceeding 0.2. MDA shows the largest surface area (0.25 nm2) of atoms with a high partial charge, followed by MDMA (0.17 nm2), MDDMA (0.8 nm2), and MDTMA (0.0 nm2). Lowering the surface charge density weakens the electrostatic interaction to the extent that the binding poses are no longer dominated by the electrostatic interaction. It is worth mentioning that the interaction between the amino moiety of the substrate and the aspartate in the binding site was shown to be critical for transport (Barker et al., 1999; Celik et al., 2008). The crystal structures of dDAT show that the charged nitrogen of transporter substrates interacts specifically with the aspartate within the binding site, whereas the hydrophobic core of the compounds binds the hydrophobic subpocket B within the S1 binding site (Wang et al., 2015). Crystal structures of dDAT cocrystallized with cocaine and the related compounds (−)-2b-carbomethoxy-3b-(4-fluorophenyl)tropane and 2b-carbomethoxy-3b-(4-iodophenyl)tropane show that these inhibitors do not allow for occlusion and subsequent transport after binding, while the overall pattern of interaction with the transporter is maintained.
Fig. 6.
Electrostatic potential mapped on the methylenedioxyamphetamine surfaces as calculated by MOE using the Poisson-Boltzmann solver. The color shows the potential ranging from −40 (red) to +40 (blue) kcal mol–1e−1.
We deduce from the substantial decrease in inhibitory potency and the results from docking that the mode of inhibition by MDDMA and MDTMA must derive from a change in the mode of binding. Shielding of the positive charge on the nitrogen as the number of bound substituents increases successively decreases its surface charge density such that interaction with the charged aspartate is no longer favorable over alternative competing interactions, resulting in a change in the mode of binding. This being the case, the change in binding mode may not only account for the observed loss in affinity for MDDMA and MDTMA. It may also explain why the larger compounds of the series behave like inhibitors despite sharing the same structural core. Thus, overall, the prediction from docking that the tertiary and quaternary amine compounds prefer a different binding mode agrees well with our findings. The example of MDDMA supports our conclusion. MDDMA assumes both substrate-like and inverted binding modes when docked into SERT, and our results unequivocally show that it acts as a substrate of this transporter. Interestingly, saturating concentrations of MDDMA released less 5-HT than the other compounds in our release assays. Considering the fact that for MDDMA, some poses were identified that docked into SERT in the transport competent binding orientation, we speculate that some SERT molecules are inhibited by MDDMA binding in a reversed orientation, while others are stimulated to release because MDDMA binds to them in a substrate-like orientation. A mechanism for partial release can be envisioned, in which a single compound acts like a mixture of two compounds, with one being a substrate and the other an inhibitor. The insurmountable inhibitory action could then explain why it is not possible to obtain full release by raising the concentration of the compound. Our data suggest the concept of partial release, which is illustrated herein by the activity of MDDMA at SERT. Specifically, the apparent partial releasing activity of this compound at SERT could be related to MDDMA adopting both high-affinity substrate-like and low-affinity inhibitory binding modes at all concentrations. Thus, MDDMA can never be as effective as MDA or MDMA in the SERT release assay because a fraction of SERT molecules will always be inhibited by MDDMA, even at saturating concentrations. Further experiments are required to test this intriguing hypothesis.
MDTMA is a quaternary amine; hence, the charge at the nitrogen cannot be neutralized. However, in the case of the other compounds, deprotonation augments the neutral form. Although our experiments were conducted at pH 7.4 and the pKa values of MDA, MDMA, and MDDMA are in the range of 10, several examples were described where local conditions at the binding site shifted the pKa by up to three orders of magnitude. Therefore, it is conceivable that the reason why MDTMA acts as an inhibitor is because it is permanently charged. However, two lines of evidence refute this possibility: 1) MPP+ is also permanently charged; nonetheless, it is a substrate of SERT, DAT, and NET (Scholze et al., 2001); and 2) MDMA and MDDMA have very similar pKa values, yet they differ considerably in how they interact with the three closely related monoamine transporters. Our experimental results show that the major changes in property occur with the addition of the second methyl group to the amine. This is when the IC50 values are consistently increased for all monoamine transporters and this is also when the compounds are transformed into inhibitors, with the formerly mentioned exception of MDDMA and SERT. If deprotonation was the cause of all this, the transition point should be reached when MDDMA is converted into MDTMA. This, however, was not observed in our experiments.
We have recently shown that the prototypical inhibitors of SERT and DAT, such as cocaine, bind to monoamine transporters at a rate that is below the diffusion-controlled limit (Hasenhuetl et al., 2015). An assessment of the binding kinetics of substrates is currently not possible because, unlike inhibitors that bind and subsequently dissociate, a substrate that is bound can either dissociate or induce a conformational rearrangement of the transporter. It is, therefore, difficult to design a protocol that can separate these two processes. MDTMA is an inhibitor of SERT and DAT, but structurally, it is closely related to the substrate MDMA. We therefore measured the binding kinetics of MDTMA to explore whether this compound displays distinctly different binding kinetics. Although we found a considerably faster dissociation rate when compared with the classic monoamine transport inhibitor cocaine, the association rate constant was very similar (Hasenhuetl et al., 2015). This could either mean that substrates also bind to the transporters at a slow rate or it could mean that the rate of substrate association is controlled by factors that are largely independent of the ligand size. Further studies are required to address this question.
Supplementary Material
Abbreviations
- CFP
cyan fluorescent protein
- DAT
dopamine transporter
- dDAT
Drosophila melanogaster dopamine transporter
- 5-HT
serotonin
- KHB
Krebs HEPES buffer
- FRET
fluorescence resonance energy transfer
- HEK
human embryonic kidney
- MDA
3,4-methylenedioxyamphetamine
- MDDMA
3,4-methylenedioxy-N,N-dimethylamphetamine
- MDMA
3,4-methylenedioxy-N-methylamphetamine
- MDTMA
3,4-methylenedioxy-N,N,N-trimethylamphetamine
- MOE
Molecular Operating Environment
- MPP+
1-methyl-4-phenylpyridinium
- MTSEA
2-aminoethyl methanethiosulfonate hydrobromide
- NET
norepinephrine transporter
- SERT
serotonin transporter
- YFP
yellow fluorescent protein
Authorship Contributions
Participated in research design: Sandtner, Stockner, Hasenhuetl, Steinkellner, Rudnick, Baumann, Ecker, Sitte
Conducted experiments: Sandtner, Stockner, Hasenhuetl, Partilla, Seddik, Zhang, Cao, Holy, Steinkellner, Sitte.
Contributed new reagents or analytic tools: Newman, Cao.
Performed data analysis: Sandtner, Stockner, Hasenhuetl, Partilla, Seddik, Cao, Holy, Steinkellner, Baumann, Sitte.
Wrote or contributed to the writing of the manuscript: Sandtner, Stockner, Hasenhuetl, Partilla, Seddik, Cao, Steinkellner, Rudnick, Baumann, Newman, Sitte.
Footnotes
This work was supported by the Austrian Science Fund/FWF (Fonds zur Förderung der Wissenschaft und Forschung) [Grant W1232] to G.F.E. and H.H.S., Austrian Science Fund/FWF (Fonds zur Förderung der Wissenschaft und Forschung) [Grant F35] to G.F.E., H.H.S., and T.Sto., and a MDPhD fellowship by the Medical University of Vienna to P.S.H. M.B.H. and A.H.N. receive support by the National Institute on Drug Abuse Intramural Research Program [Grant DA000389].
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Barker EL, Moore KR, Rakhshan F, Blakely RD. (1999) Transmembrane domain I contributes to the permeation pathway for serotonin and ions in the serotonin transporter. J Neurosci 19:4705–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann MH, Partilla JS, Lehner KR, Thorndike EB, Hoffman AF, Holy M, Rothman RB, Goldberg SR, Lupica CR, Sitte HH, et al. (2013) Powerful cocaine-like actions of 3,4-methylenedioxypyrovalerone (MDPV), a principal constituent of psychoactive ‘bath salts’ products. Neuropsychopharmacology 38:552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann MH, Wang X, Rothman RB. (2007) 3,4-Methylenedioxymethamphetamine (MDMA) neurotoxicity in rats: a reappraisal of past and present findings. Psychopharmacology (Berl) 189:407–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuming T, Kniazeff J, Bergmann ML, Shi L, Gracia L, Raniszewska K, Newman AH, Javitch JA, Weinstein H, Gether U, et al. (2008) The binding sites for cocaine and dopamine in the dopamine transporter overlap. Nat Neurosci 11:780–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blough BE, Landavazo A, Partilla JS, Baumann MH, Decker AM, Page KM, Rothman RB. (2014) Hybrid dopamine uptake blocker-serotonin releaser ligands: a new twist on transporter-focused therapeutics. ACS Med Chem Lett 5:623–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm S. (1999) ATP stimulates sympathetic transmitter release via presynaptic P2X purinoceptors. J Neurosci 19:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik L, Sinning S, Severinsen K, Hansen CG, Møller MS, Bols M, Wiborg O, Schiøtt B. (2008) Binding of serotonin to the human serotonin transporter. Molecular modeling and experimental validation. J Am Chem Soc 130:3853–3865. [DOI] [PubMed] [Google Scholar]
- Chen JG, Liu-Chen S, Rudnick G. (1997) External cysteine residues in the serotonin transporter. Biochemistry 36:1479–1486. [DOI] [PubMed] [Google Scholar]
- Feige JN, Sage D, Wahli W, Desvergne B, Gelman L. (2005) PixFRET, an ImageJ plug-in for FRET calculation that can accommodate variations in spectral bleed-throughs. Microsc Res Tech 68:51–58. [DOI] [PubMed] [Google Scholar]
- Fenollar-Ferrer C, Stockner T, Schwarz TC, Pal A, Gotovina J, Hofmaier T, Jayaraman K, Adhikary S, Kudlacek O, Mehdipour AR, et al. (2014) Structure and regulatory interactions of the cytoplasmic terminal domains of serotonin transporter. Biochemistry 53:5444–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest LR, Zhang YW, Jacobs MT, Gesmonde J, Xie L, Honig BH, Rudnick G. (2008) Mechanism for alternating access in neurotransmitter transporters. Proc Natl Acad Sci USA 105:10338–10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger J, Marsili M. (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36:3219–3228. [Google Scholar]
- Hasenhuetl PS, Schicker K, Koenig X, Li Y, Sarker S, Stockner T, Sucic S, Sitte HH, Freissmuth M, Sandtner W. (2015) Ligand selectivity among the dopamine and serotonin transporters specified by the forward binding reaction. Mol Pharmacol 88:12–18. [DOI] [PubMed] [Google Scholar]
- Henry LK, Iwamoto H, Field JR, Kaufmann K, Dawson ES, Jacobs MT, Adams C, Felts B, Zdravkovic I, Armstrong V, et al. (2011) A conserved asparagine residue in transmembrane segment 1 (TM1) of serotonin transporter dictates chloride-coupled neurotransmitter transport. J Biol Chem 286:30823–30836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447. [DOI] [PubMed] [Google Scholar]
- Hilber B, Scholze P, Dorostkar MM, Sandtner W, Holy M, Boehm S, Singer EA, Sitte HH. (2005) Serotonin-transporter mediated efflux: a pharmacological analysis of amphetamines and non-amphetamines. Neuropharmacology 49:811–819. [DOI] [PubMed] [Google Scholar]
- Hofmaier T, Luf A, Seddik A, Stockner T, Holy M, Freissmuth M, Ecker GF, Schmid R, Sitte HH, Kudlacek O. (2014) Aminorex, a metabolite of the cocaine adulterant levamisole, exerts amphetamine like actions at monoamine transporters. Neurochem Int 73:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD. (2007) Matplotlib: a 2D graphics environment. Comput Sci Eng 9:90–95. [Google Scholar]
- Iversen L. (2000) Neurotransmitter transporters: fruitful targets for CNS drug discovery. Mol Psychiatry 5:357–362. [DOI] [PubMed] [Google Scholar]
- Jacobs MT, Zhang YW, Campbell SD, Rudnick G. (2007) Ibogaine, a noncompetitive inhibitor of serotonin transport, acts by stabilizing the cytoplasm-facing state of the transporter. J Biol Chem 282:29441–29447. [DOI] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC, Leach AR, Taylor R. (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267:727–748. [DOI] [PubMed] [Google Scholar]
- Just H, Sitte HH, Schmid JA, Freissmuth M, Kudlacek O. (2004) Identification of an additional interaction domain in transmembrane domains 11 and 12 that supports oligomer formation in the human serotonin transporter. J Biol Chem 279:6650–6657. [DOI] [PubMed] [Google Scholar]
- Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. (2001) Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B 105:6474–6487. [Google Scholar]
- Korb O, Stützle T, Exner TE. (2009) Empirical scoring functions for advanced protein-ligand docking with PLANTS. J Chem Inf Model 49:84–96. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy H, Gouaux E. (2012) X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature 481:469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen AS, Andersen J, Jørgensen TN, Sørensen L, Eriksen J, Loland CJ, Strømgaard K, Gether U. (2011) SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacol Rev 63:585–640. [DOI] [PubMed] [Google Scholar]
- Mager S, Min C, Henry DJ, Chavkin C, Hoffman BJ, Davidson N, Lester HA. (1994) Conducting states of a mammalian serotonin transporter. Neuron 12:845–859. [DOI] [PubMed] [Google Scholar]
- Mitchell SM, Lee E, Garcia ML, Stephan MM. (2004) Structure and function of extracellular loop 4 of the serotonin transporter as revealed by cysteine-scanning mutagenesis. J Biol Chem 279:24089–24099. [DOI] [PubMed] [Google Scholar]
- Penmatsa A, Wang KH, Gouaux E. (2013) X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature 503:85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmatsa A, Wang KH, Gouaux E. (2015) X-ray structures of Drosophila dopamine transporter in complex with nisoxetine and reboxetine. Nat Struct Mol Biol 22:506–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith ME, Blough BE, Hong WC, Jones KT, Schmitt KC, Baumann MH, Partilla JS, Rothman RB, Katz JL. (2015) Behavioral, biological, and chemical perspectives on atypical agents targeting the dopamine transporter. Drug Alcohol Depend 147:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickli A, Hoener MC, Liechti ME. (2015a) Monoamine transporter and receptor interaction profiles of novel psychoactive substances: para-halogenated amphetamines and pyrovalerone cathinones. Eur Neuropsychopharmacol 25:365–376. [DOI] [PubMed] [Google Scholar]
- Rickli A, Kopf S, Hoener MC, Liechti ME. (2015b) Pharmacological profile of novel psychoactive benzofurans. Br J Pharmacol 172:3412–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman RB, Baumann MH. (2003) Monoamine transporters and psychostimulant drugs. Eur J Pharmacol 479:23–40. [DOI] [PubMed] [Google Scholar]
- Rothman RB, Partilla JS, Baumann MH, Lightfoot-Siordia C, Blough BE. (2012) Studies of the biogenic amine transporters. 14. Identification of low-efficacy “partial” substrates for the biogenic amine transporters. J Pharmacol Exp Ther 341:251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha K, Partilla JS, Lehner KR, Seddik A, Stockner T, Holy M, Sandtner W, Ecker GF, Sitte HH, Baumann MH. (2015) ‘Second-generation’ mephedrone analogs, 4-MEC and 4-MePPP, differentially affect monoamine transporter function. Neuropsychopharmacology 40:1321–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. [DOI] [PubMed] [Google Scholar]
- Sandtner W, Schmid D, Schicker K, Gerstbrein K, Koenig X, Mayer FP, Boehm S, Freissmuth M, Sitte HH. (2014) A quantitative model of amphetamine action on the 5-HT transporter. Br J Pharmacol 171:1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schicker K, Uzelac Z, Gesmonde J, Bulling S, Stockner T, Freissmuth M, Boehm S, Rudnick G, Sitte HH, Sandtner W. (2012) Unifying concept of serotonin transporter-associated currents. J Biol Chem 287:438–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholze P, Sitte HH, Singer EA. (2001) Substantial loss of substrate by diffusion during uptake in HEK-293 cells expressing neurotransmitter transporters. Neurosci Lett 309:173–176. [DOI] [PubMed] [Google Scholar]
- Scholze P, Zwach J, Kattinger A, Pifl C, Singer EA, Sitte HH. (2000) Transporter-mediated release: a superfusion study on human embryonic kidney cells stably expressing the human serotonin transporter. J Pharmacol Exp Ther 293:870–878. [PubMed] [Google Scholar]
- Seddik A, Holy M, Weissensteiner R, Zdrazil B, Sitte HH, Ecker GF. (2013) Probing the selectivity of monoamine transporter substrates by means of molecular modeling. Mol Inform 32:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmler LD, Buser TA, Donzelli M, Schramm Y, Dieu LH, Huwyler J, Chaboz S, Hoener MC, Liechti ME. (2013) Pharmacological characterization of designer cathinones in vitro. Br J Pharmacol 168:458–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Yamashita A, Gouaux E. (2007) Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature 448:952–956. [DOI] [PubMed] [Google Scholar]
- Sitte HH, Freissmuth M. (2015) Amphetamines, new psychoactive drugs and the monoamine transporter cycle. Trends Pharmacol Sci 36:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkellner T, Freissmuth M, Sitte HH, Montgomery T. (2011) The ugly side of amphetamines: short- and long-term toxicity of 3,4-methylenedioxymethamphetamine (MDMA, ‘Ecstasy’), methamphetamine and D-amphetamine. Biol Chem 392:103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockner T, Jurik A, Freissmuth M, Ecker GF, Sitte HH. (2014) Development of refined homology models: adding the missing information to the medically relevant neurotransmitter transporters, in Membrane Transport Mechanism (Ziegler C, Krämer R, eds) pp 99–120, Springer, Berlin. [Google Scholar]
- Stockner T, Montgomery TR, Kudlacek O, Weissensteiner R, Ecker GF, Freissmuth M, Sitte HH. (2013) Mutational analysis of the high-affinity zinc binding site validates a refined human dopamine transporter homology model. PLOS Comput Biol 9:e1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucic S, Dallinger S, Zdrazil B, Weissensteiner R, Jørgensen TN, Holy M, Kudlacek O, Seidel S, Cha JH, Gether U, et al. (2010) The N terminus of monoamine transporters is a lever required for the action of amphetamines. J Biol Chem 285:10924–10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenaz P, Ruttimann UE, Unser M. (1998) A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7:27–41. [DOI] [PubMed] [Google Scholar]
- Wang KH, Penmatsa A, Gouaux E. (2015) Neurotransmitter and psychostimulant recognition by the dopamine transporter. Nature 521:322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. (2005) Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature 437:215–223. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Rudnick G. (2006) The cytoplasmic substrate permeation pathway of serotonin transporter. J Biol Chem 281:36213–36220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.