Abstract
Calcium-dependent protein kinases (CDPKs) are plant-specific calcium sensors that play important roles in various aspects of plant physiology. Here, we investigated phylogenic relationships, chromosomal locations, gene structures, and tissue-specific, herbivory- and drought-induced expression profiles of soybean (Glycine max) GmCDPKs. Fifty GmCDPK genes were identified, which phylogenetically grouped into 4 distinct clusters and distributed across 13 sub-clusters. Individual classes of GmCDPKs harbor highly conserved mRNA splicing sites, and their exon numbers and lengths were consistent with the phylogenetic relationships, suggesting that at least 13 ancestral CDPK genes had emerged before the split of monocots and eudicots. Gene expression analysis indicated that several GmCDPKs were tissue-specific expressed. GmCDPKs’ transcript levels changed after wounding, exhibited specific expression patterns after simulated Spodoptera exigua feeding or soybean aphid (Aphis glycines) herbivory, and were largely independent of the phytohormones jasmonic acid and salicylic acid. The most pronounced transcriptional responses were detected after drought and abscisic acid treatments with more than half of all GmCDPKs being upregulated, suggesting their important roles during abiotic stress responses in soybean. Our data provide an important foundation for further functional dissection of GmCDPKs, especially in the context of soybean-insect interactions and drought stress adaptation.
Ca2+ is a ubiquitous second messenger that is critical for signal transduction in eukaryotes by activating various cellular processes in response to developmental and stress stimuli1,2,3. In plants, transient Ca2+ changes in the cytoplasm are sensed and decoded by an array of calcium sensors, such as calmodulins, calmodulin-binding proteins, calcineurin B-like proteins, and calcium-dependent protein kinases (CDPKs or CPKs), and these Ca2+ sensors further interact with their downstream target proteins and in turn trigger Ca2+ signature-specific responses.
CDPKs have been identified throughout the plant kingdom and in some protozoans, but not in animals or fungi. A typical CDPK consists of five domains: a variable N-terminal domain, a protein kinase domain, an autoinhibitory junction domain, a calmodulin-like domain, and a C-terminal domain. The N-terminal domain shows the highest sequence divergence among CDPKs and often contains myristoylation or palmitoylation sites that are believed to be associated with subcellular targeting4. The protein kinase domain is the catalytic domain with an ATP binding site, and is adjacent to the autoinhibitory junction domain5 and the consecutive calmodulin-like domain that contains EF hands for calcium binding4. The C-terminal domain is also variable and differs in lengths and amino acid compositions amongst CDPKs. It has been suggested that the N- and C-terminal variable domains determine the specific function of individual CDPK6. Binding of Ca2+ to the calmodulin-like domain results in conformational changes that in turn displace the autoinhibitory domain from the kinase domain, and thereby activate CDPKs.
Recent findings have pointed to the roles of CDPKs in wounding- and herbivory-induced plant responses. The expression and enzymatic activity of maize (Zea mays) ZmCDPK11 are up-regulated by wounding, linolenic acid, and methyl jasmonate (MeJA), and the wound-induced activation of ZmCPK11 is dependent on phosphatidic acid produced in the phospholipase D (PLD) pathway7,8. Arabidopsis AtCPK3 and AtCPK13 phosphorylate the transcription factor (TF) HsfB2a, and HsfB2a in turn regulates the transcript accumulation of several stress-related genes after Spodoptera littoralis feeding9. Tomato (Solanum lycopersicum) LeCPK1 inhibits the plasma membrane H+-ATPase to induce extracellular alkalization as a part of the wound and pathogen defense signaling cascades10, and LeCDPK2 phosphorylates LeACS2 in response to wounding, stabilizing this rate-limiting enzyme for ethylene (ET) biosynthesis by direct phosphorylation11. In addition, Zhang et al. showed that in grape wine, several CDPK genes’ expression was increased by ET-treatment12. N. attenuata CDPKs, NaCDPK4 and NaCDPK5, play redundant and negative roles in mediating wounding- and herbivory-elicited JA accumulations, and silencing NaCDPK4 and NaCDPK5 greatly elevates plant resistance to the specialist insect Manduca sexta13,14.
Drought stress poses another major threat to plants and recent research has shown that CDPKs play important roles in regulating plant adaptation to drought. The transcript levels of ZmCPK12 are highly induced by drought in maize seedlings, and overexpression of ZmCPK12 in Arabidopsis improves plant survival under drought conditions15. Similarly, Populus euphratica PeCPK10 confers drought tolerance and enhances the expression of several ABA-responsive genes when being overexpressed in Arabidopsis16. OsCPK9 and OsCDPK7 positively regulate drought stress tolerance in rice, most likely by affecting the transcript levels of ABA- and stress-responsive genes17,18. Importantly, guard cell-expressed Arabidopsis CPK21 was identified as a major interacting partner of the guard cell anion channel SLAC1 and thus regulates stomatal ABA signal transduction19.
CDPKs constitute a large multigene family in higher plants with 34, 35, 31, 30, and 26 CDPK genes found in Arabidopsis, maize, rice (Oryza sativa), poplar (Populus trichocarpa), and wheat (Triticum aestivum), respectively4,20,21,22,23. Soybean (Glycine max) is one of the most important crop plants worldwide, providing oil and protein to human and livestock. Valmonte et al. identified 47 CDPK genes from soybean and revealed extensive sequence conservation amongst them24. Soybean CDPKα and CDPKγ phosphorylate a serine acetyltranferase involved in cysteine biosynthesis after oxidative stress25. In soybean nodules, certain CDPKs phosphorylate the membrane protein nodulin-26 affecting its voltage-sensitive channel activity26, and nodulin-100, a sucrose synthase that is essential for the cleavage of sucrose translocated from the shoots to the roots in support of nodule C/N-metabolism27,28. However, still little is known about the roles of GmCDPKs in soybean development, growth, and in adaptation to biotic and abiotic stresses.
In this study, we carried out a genome-wide analysis and identified 50 GmCDPKs. Phylogenetic analysis was performed and GmCDPKs gene structures were compared to reveal their evolutionary relationships. Furthermore, we examined the expression of all GmCDPKs after wounding, simulated Spodoptera exigua herbivory, Aphis glycines feeding, and treatments of JA, ET, and SA, phytohormones important for plant defense against insects, as well as in response to drought and ABA. We found that the transcript levels of many GmCDPKs were not dependent on JA or SA signaling but were specifically altered in response to herbivory, suggesting that GmCDPKs may play important roles in soybean defense against insects. Moreover, almost 80% of all GmCDPKs were induced after drought or ABA treatments, highlighting their important role in regulating drought stress responses.
Results
Identification and characteristics of GmCDPK genes
To find all CDPKs in soybean, a domain search against all predicted proteins in the soybean genome (http://www.phytozome.net/soybean) was performed. A total of 68 proteins containing a protein kinase domain and at least one EF hand domain were identified, amongst which 5 showed high similarities to CRKs, CaMKs, and CCaMKs and were thus eliminated, and the remaining proteins were designated as GmCDPKs. Further database search using these GmCDPKs as queries against the draft genome and predicted mRNAs resulted in no further hits. In total, 63 proteins, including the alternatively spliced forms, from 50 gene loci were identified as GmCDPKs, and their coding genes were designated as GmCDPK1 to GmCDPK50, according to their locations on soybean chromosomes (Supplementary Table S1). The alternatively spliced regions in GmCDPK1, GmCDPK9, GmCDPK35 and GmCDPK46 contain the EF hand domains, resulting in protein isoforms with less than 4 EF hands (Supplementary Table S1). GmCDPK50 lacked ~600 bp at the 5′ end, which normally accounts for the variable N-terminal domain and a part of the protein kinase domain. We inspected the 5′ intergenic region (about 4 kb) and did not find any fragment encoding the conserved protein kinase domain. These findings strongly indicate that GmCDPK50 is a pseudogene.
Myristoylation and palmitoylation at the N-terminal regions of CDPKs are correlated with their subcellular localization and substrate specificity29,30,31. Forty-nine of the 63 GmCDPK proteins possess myristoylation sites that are, with the exception for GmCDPK2, GmCDPK38, and GmCDPK41, located in the first 32 amino acids. Seven of the 63 GmCDPK proteins have no the palmitoylation sites. Most palmitoylation sites are located within the first 7 amino acids, albeit those of 7 GmCDPKs were predicted to be located close to the 50th amino acid of the N-terminus (Supplementary Table S1). We further predicted the organelle-specific localization using TargetP and NucPred32, and found 10 GmCDPKs to be localized in chloroplasts, and that 2 GmCDPKs, GmCDPK29 and GmCDPK49, contain mitochondrion-specific targeting sequences (Supplementary Table S1). GmCDPK37.2 and GmCDPK41 were predicted to be located in the nucleus.
Phylogenetic relationships, gene structures, and potential functional divergence of GmCDPKs
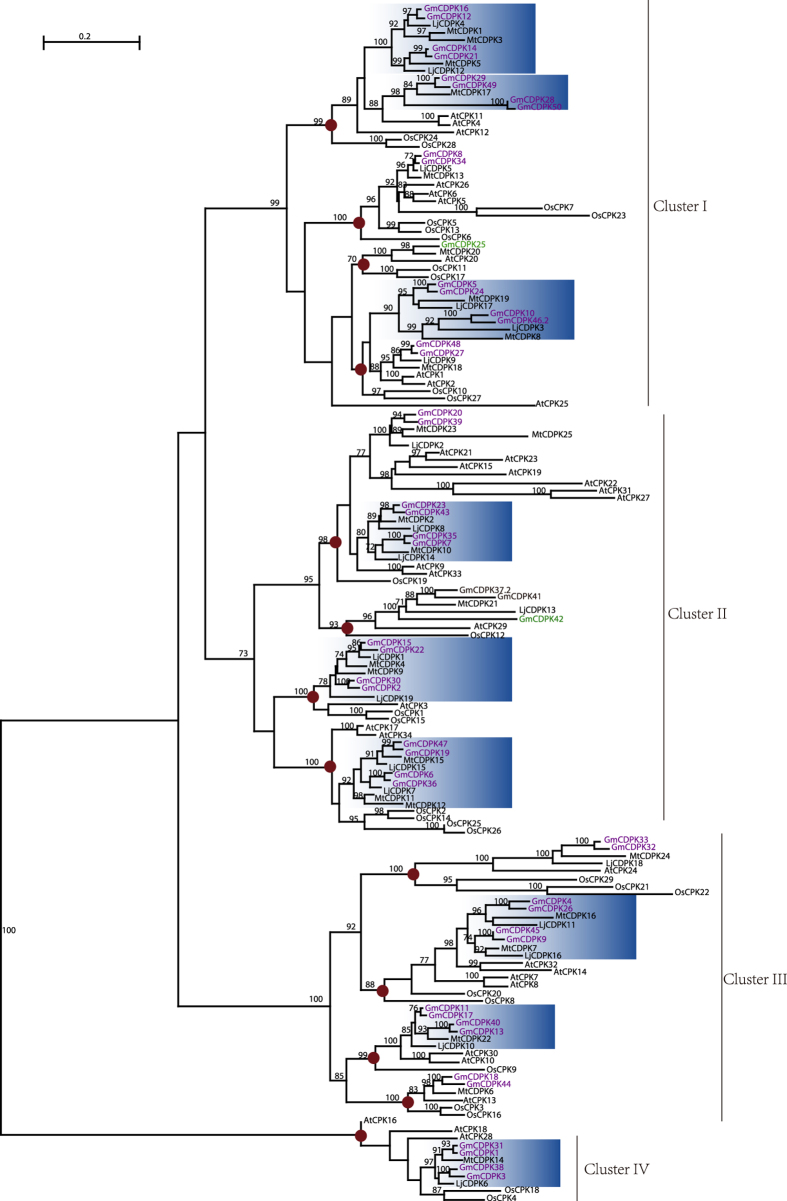
To gain insights into the evolution of GmCDPKs, we also identified the CDPK genes from two other legume species, Lotus japonicas and Medicago truncatula, using the same procedure and found 19 and 25 CDPK genes, respectively (Supplementary Table S2). A maximum likelihood tree including all CDPKs from these three legumes, Arabidopsis, and rice was constructed (Fig. 1). The phylogenetic analysis indicated that, consistent with the previous reports20,24,33, all CDPKs from these 5 species grouped into 4 major clusters (Fig. 1, Clusters I–IV). Detailed inspection revealed that the CDPKs are distributed among 13 sub-clusters (Fig. 1, indicated by the brown dots), each containing CDPKs from eudicots and monocots. Therefore, at least 13 ancestral CDPK genes existed prior to the split of eudicots and monocots. As for the 3 legume species, 9 out of the 13 sub-clusters (Fig. 1, blue background) contain at least 2 paralogous CDPKs of L. japonicas and M. truncatula and 4 paralogs of soybean. The paralogs in these 9 sub-clusters probably resulted from the genome duplication event that occurred in the common ancestor of legumes and the consequent genome duplication that occurred uniquely in soybean34. Notably, GmCDPK25 clustered as a singleton in Cluster I and GmCDPK37/41/42 grouped as a triad in Cluster II (Fig. 1 and Supplementary Fig. S1), suggesting that gene loss may have had occurred after the genome duplication events.
Figure 1. Phylogenetic relationship of soybean, Arabidopsis, Lotus japonicas, Medicago truncatula, and rice CDPKs.
A phylogenetic tree was constructed for 50 soybean (Glycine max), 19 Lotus japonicas, 25 Medicago truncatula, 29 rice (Oryza sativa), and 34 Arabidopsis thaliana CDPKs. The 4 phylogenetic clusters designated as I–IV are marked with vertical bars. Thirteen sub-clusters containing both eudicots and monocots are highlighted with brown dots. The legume CDPKs that probably resulted from the genome duplication event (GDE) occurred in the legume common ancestor are shaded in blue. The genes retained after the soybean GDE are in purple letters, except that GmCDPK25 and GmCDPK42 without paralogs are in green and the paralog pair, GmCDPK37 and GmCDPK41 (88% sequence identity) that is not from the soybean GDE, is in black. Numbers above branches show bootstrap support values from maximum likelihood analyses.
To correlate the phylogenetic relationships with the patterns of GmCDPK gene structures, we analyzed the intron-exon characteristics of GmCDPKs. The exon numbers and lengths within all 13 sub-clusters are consistent with their phylogenetic relationships, although the intron lengths are variable (Supplementary Fig. S1). The median length of all introns is 265 bp, the minimum is 31 bp in GmCDPK41, and the maximum is 4168 bp in GmCDPK6. Most GmCDPKs in Clusters I-III have 6 to 9 exons, while GmCDPKs in Cluster IV have 12 or 14 exons. The complex gene structures of Cluster IV GmCDPKs suggest a different evolutionary history and probably functional divergence from the other GmCDPKs.
Chromosomal distributions and development-related expression profiles of GmCDPKs
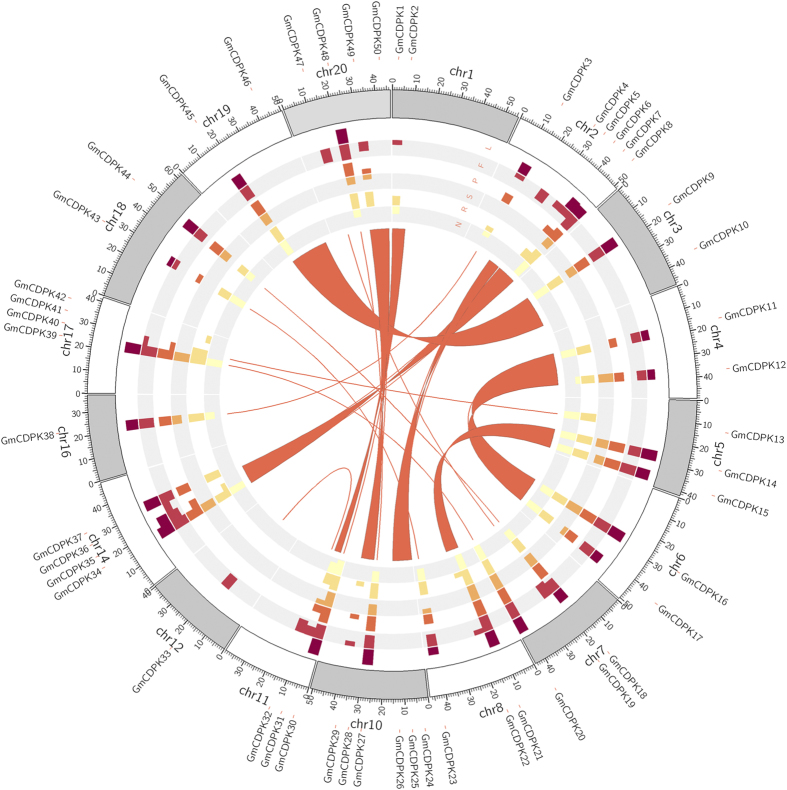
At least two genome duplication events occurred in the evolutionary history of soybean, which were followed by numerous chromosome rearrangements, gene structure diversification, and multitudinous gene loss, and these events led to rather complex gene families. Among the 20 chromosomes, chromosomes 9, 13, and 15 carry no GmCDPKs, chromosomes 12 and 16 each contain only 1 GmCDPK gene, chromosomes 2 and 10 each contain 6 GmCDPK genes, and all other chromosomes have 2–4 GmCDPKs (Fig. 2). All GmCDPKs, except GmCDPK26, are located within the chromosome ends, in agreement with the data from soybean genome sequencing: ~78% of all soybean genes were predicted to be located in these regions34. At least 7 segment duplications contributed to the expansion of the GmCDPK gene family (Fig. 2 and Supplementary Fig. S1). For example, GmCDPK1 is adjacent to GmCDPK2 on Chromosome 1 and GmCDPK31 is close to GmCDPK30 on Chromosome 11; GmCDPK1 and GmCDPK31 are paralogs, as are GmCDPK2 and GmCDPK30 (Fig. 2). Inspection of the genes between the individual CDPKs showed high similarity between the two chromosomal regions, suggesting that they originated from segment duplication. Another case connects Chromosome 10 and Chromosome 20: It has been reported that the long arms of Chromosome 10 and Chromosome 20 very likely derived from segment duplication34; supporting this scenario, GmCDPK28 on Chromosome 10 and GmCDPK49 on Chromosome 20 showed high sequence identity, and so did their respective adjacent genes GmCDPK29 and GmCDPK50 (Fig. 2 and Supplementary Fig. S2). The physical distance between GmCDPK41 and GmCDPK42 on Chromosome 17 is ~ 3 kb, and their sequence identity is 68% which suggests that these two genes may have originated from a tandem duplication event.
Figure 2. GmCDPKs’ Chromosome distributions, development-related expression profiles, and paralogous relationships resulted from the soybean genome duplication event.
The distribution of GmCDPK genes on 17 soybean chromosomes are given in the outer circle (the 3 chromosomes containing no GmCDPKs are not shown), where the numbers represent the chromosome length in Mb. GmCDPKs’ expression levels in 6 different tissues and developmental stages are displayed as filled blocks in yellow to dark red, with the block height proportional to the expression level (LOG2 transformed) (L, young leaves (0.4 Leaflets unfurled); F, flowers; P, one cm pods; S, seeds 14 days after flowering; R, roots; N, nodules). GmCDPK pairs with sequence identities more than 94%, which had most likely derived from the recent soybean genome duplication (SGD), are connected by orange arcs in the inner circle. Their adjacent chromosomal regions that had derived from SGD are indicated by arcs filled with orange. For the convenience of display, the localization of GmCDPK genes on the chromosomes is disproportional to their real positions.
A gene’s function is often correlated with its transcriptional profile. To gain insights into the potential functions of CDPK genes in soybean development and growth, RNA-Seq data from expression profiles of 6 different soybean tissues and developmental stages obtained from SoyBase35 were analyzed and aligned with the phylogeny of the 50 GmCDPKs (Fig. 2). To determine potential functional divergence, we divided the paralogous genes into two types - those derived from the early and those from the later genome duplication event respectively, according to their clustering in the phylogenetic tree and their sequence identities. Most paralogous genes from the later genome duplication event, which clustered as sister branches in the phylogenetic tree (Fig. 1) and shared sequence identities no less than 94% (Supplementary Fig. S1), presented largely similar expression patterns. Notably, GmCDPK47 and GmCDPK19, GmCDPK6 and GmCDPK36, and GmCDPK32 and GmCDPK33 were almost exclusively expressed in flowers, and GmCDPK13 and GmCDPK40 were highly expressed in roots, suggesting that they may be involved in these organs’ development and growth (Fig. 2). However, some paralogous GmCDPKs exhibited rather different patterns of expression. GmCDPK22 and GmCDPK2 had very low expression, while their paralogs (GmCDPK15 and GmCDPK30, respectively) appeared to have relatively high expression throughout the whole sample set; GmCDPK28 and GmCDPK50 showed very low expression across all 6 samplings, whereas their paralogous counterparts from the same tetrad (77% sequence identity), GmCDPK29 and GmCDPK49, were transcribed in flowers, seeds, and roots (Fig. 2). In contrast, the majority of paralogous genes from the early genome duplication event clustered as tetrads with sequence identities lower than 94% and showed distinct transcriptional profiles (Supplementary Fig. S2).
GmCDPKs transcriptional response to wounding and herbivore feeding
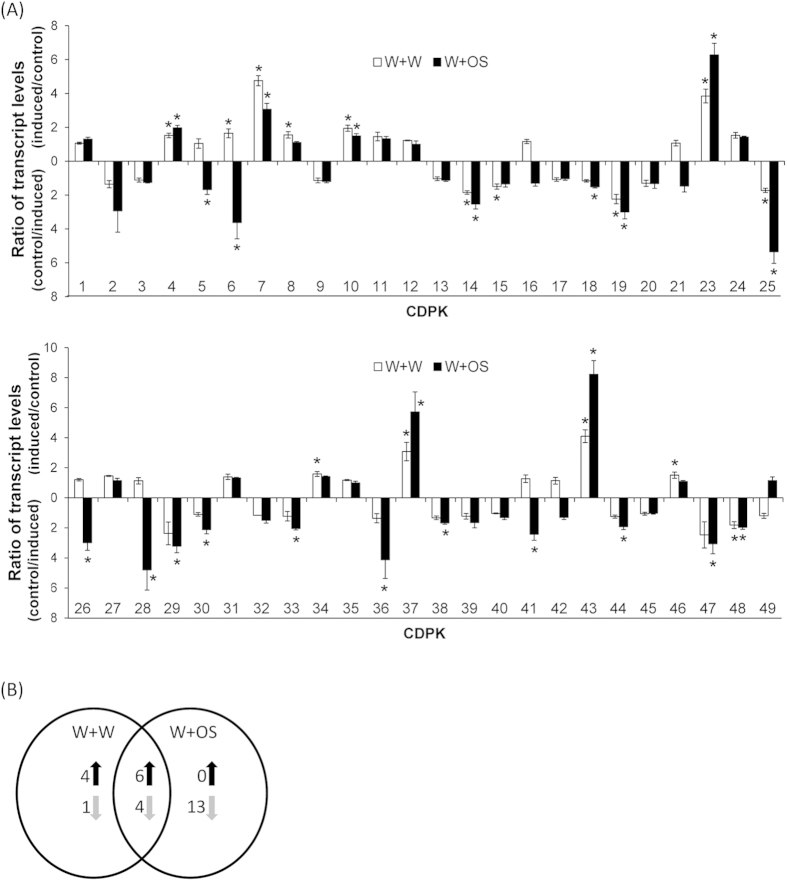
Several CDPKs have been demonstrated to be involved in plant-herbivore interactions9,13. S. exigua is a generalist insect herbivore that causes damages to soybean. To gain insights into the roles of GmCDPKs in soybean response to S. exigua herbivory, leaves were wounded with a pattern wheel, and the generated puncture wounds were immediately treated with water (W+W) or S. exigua oral secretions (W+OS) to cause mechanical damages or to simulate herbivore feeding (the feeding behavior of S. exigua cannot be synchronized, thus simulated herbivory was used). Given that the transcript levels of CDPKs usually reach their highest 1.5 h after these treatments13,36 (and see below), samples were harvested 1.5 h after treatments and the expression levels of GmCDPKs were analyzed by q-PCR (quantitative real-time PCR). Mechanical wounding significantly increased the transcript abundances of 10 out of 48 GmCDPKs (the truncated and the putatively pseudogenized GmCDPK22 and GmCDPK50 were excluded from further analyses), whereas 5 GmCDPKs showed decreased transcript levels (Fig. 3A). In contrast, W+OS treatment only increased the expression of 6 GmCDPKs, whereas 17 GmCDPKs’ transcript levels were suppressed (Fig. 3A). Notably, GmCDPKs whose levels were upregulated by W+OS treatment also showed elevated levels after W+W, and 4 out of 5 GmCDPKs with decreased expression levels after wounding were also suppressed following W+OS treatment (Fig. 3B). Compared with their levels after W+W treatment, greater (about 2-fold) transcription levels of GmCDPK23 and GmCDPK43 were detected after W+OS treatment; in contrast, 10 GmCDPKs (GmCDPK6, 7, 25, 26, 28, 30, 36, 41, 44 and 47) had significantly (t-test, p < 0.05) lower expression levels (about 6-fold for GmCDPK6 and GmCDPK28) after induction with W+OS compared to W+W treatments. Notably, GmCDPK23 and GmCDPK43 are paralogs and share high sequence identity (97% on the protein level). Similarly, GmCDPK6 and GmCDPK36 are paralogous genes (98% identical) and both genes were suppressed by W+OS treatment. In contrast, GmCDPK4 and GmCDPK26, as well as GmCDPK37 and GmCDPK41, may have also derived from gene duplications (95 and 88% sequence identity, respectively) but these pairs responded in opposite ways to W+OS treatment (Fig. 3A).
Figure 3. Wounding and simulated herbivory change the transcript levels of various GmCDPKs.
Plants were wounded with a pattern wheel, and 20 μL of water (W+W) or S. exigua oral secretions (W+OS) were immediately applied to the wounds; untreated plants served as controls. Individual leaves from 5 replicate plants were harvested 1.5 h after the treatments, and GmCDPKs’ expression levels were analyzed by q-PCR. (A) Ratios (mean ± SE) between the GmCDPK transcript levels (Log2 transformed) in W+W- or W+OS-treated and untreated controls. Asterisks indicate significant changes (t-test, p < 0.05) of more than 1.5-fold up- or down-regulated genes. (B) Venn diagram depicting the number of genes having similar or differential (t-test, p < 0.05) expression in response to W+W and W+OS treatments.
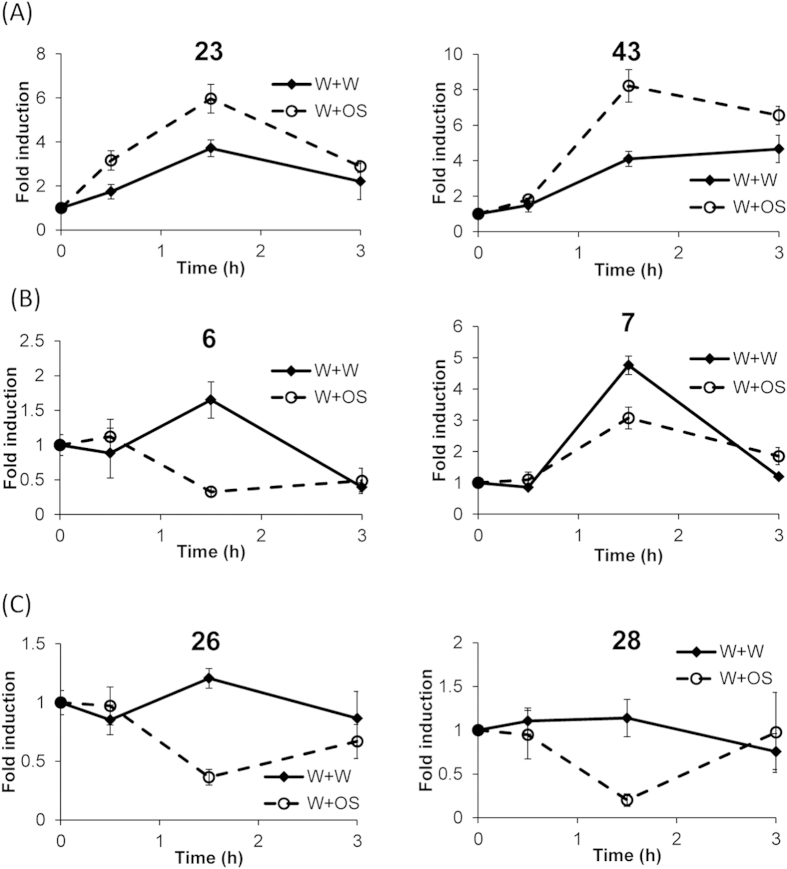
W+W and W+OS treatment induced distinct expression patterns of GmCDPK6, 7, 23, 43, 26 and GmCDPK28 (Fig. 3A). To further investigate the transcriptional responses of GmCDPKs to simulated S. exigua herbivory, these genes were selected and analyzed in detail in samples harvested 0.5, 1.5, and 3 h after elicitations. All these GmCDPKs showed the highest differences compared to untreated controls 1.5 h after the elicitations and the transcript levels mostly returned to the uninduced state 3 h after the initial treatments. GmCDPK23 and GmCDPK43 were up-regulated after both W+W and W+OS treatment, and were higher expressed after W+OS than after W+W treatment (Fig. 4A). W+W treatment induced the expression levels of GmCDPK6 and GmCDPK7, but adding OS to wounds suppressed the wounding-induced responses (Fig. 4B). While W+W barely changed the levels of GmCDPK26 and GmCDPK28, W+OS repressed their transcript levels (Fig. 4C).
Figure 4. Transient changes of GmCDPK transcript levels following wounding and simulated herbivory.
Plants were wounded with a pattern wheel, and 20 μL of water (W+W) or S. exigua oral secretions (W+OS) were immediately applied to the wounds; untreated plants served as controls. Individual leaves from 5 replicate plants were harvested at the indicated times. Mean (±SE) transcript levels (LOG2 transformed) of GmCDPK23 and GmCDPK43 (A), GmCDPK6 and GmCDPK7 (B), and GmCDPK26 and GmCDPK28 (C) were normalized to their levels in untreated controls.
Next, we investigated whether not only chewing herbivores but also aphids, as piercing-sucking feeders, could affect GmCDPKs. Leaves were exposed to 10 nymphs and 10 adult soybean aphids, Aphis glycines, and after feeding for 8 h, samples were collected and GmCDPK transcript levels were analyzed by q-PCR. Ten GmCDPKs (GmCDPK9, 10, 12, 19, 29, 41, and the paralogs GmCDPK5 and GmCDPK24, as well as GmCDPK6 and GmCDPK36) exhibited significantly increased transcript levels (Fig. 5). In contrast to W+W and W+OS treatments, no GmCDPKs were negatively regulated. Notably, the expression levels of GmCDPK6 and GmCDPK36 were suppressed by W+OS treatment but increased after A. glycines herbivory, and this pattern was also found for GmCDPK5, GmCDPK29 and GmCDPK41, suggesting that these GmCDPKs may function very differently in soybean defense against S. exigua and A. glycines.
Figure 5. Aphid (Aphis glycines) herbivory-induced changes in GmCDPKs’ transcript abundances.
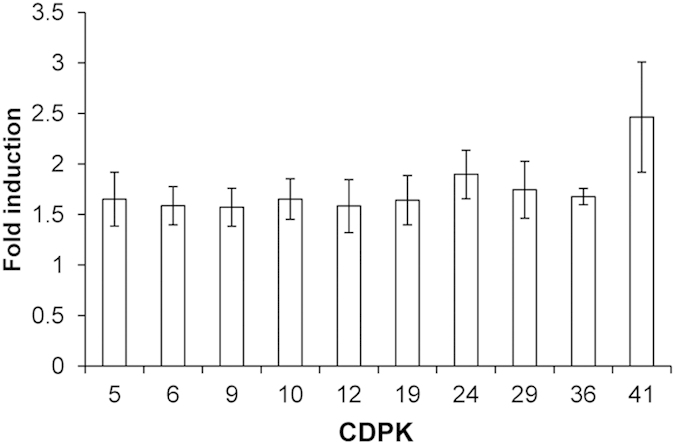
10 nymphs and 10 adult aphids were placed together on leaves of the second trifoliate and leaves were immediately enclosed between two 50 mL food-quality plastic containers secured with miniature claw-style hair clips. Uninfested enclosed leaves served as controls. GmCDPK expression was measured with q-PCR in 6 replicated samples collected 8 h after the start of the treatment. Values represent significantly (t-test, p < 0.05) and more than 1.5-fold induced GmCDPK transcripts normalized to their respective expressions in control samples (mean ± SE); GmCDPKs whose expression levels did not change are not shown.
JA-, SA-, and ethylene-induced transcriptional changes
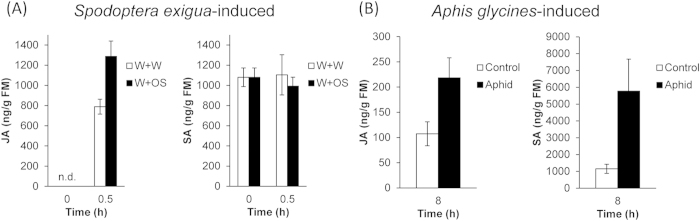
Plant hormones JA, SA, and ET play central roles in plant-herbivore interactions by regulating a large number of genes related to plant defense responses. We determined hormonal changes after W+W and W+OS treatments and after aphid herbivory. By 30 min, W+W- and W+OS-treated leaves accumulated 790 and 1,288 ng/g of JA, respectively, indicating that soybean recognized certain elicitors in S. exigua OS and accumulated higher levels of JA to counteract S. exigua attack (Fig. 6A). In contrast to W+W and W+OS treatments, 8 h aphid feeding highly increased SA levels from 1,150 ng/g to 5,770 ng/g, but the JA contents were only slightly elevated (Fig. 6B).
Figure 6. Herbivory-induced changes in JA and SA levels.
(A) Hormones induced by wounding and simulated S. exigua herbivory. Plants were wounded with a pattern wheel, 20 μL of water (W+W) or S. exigua oral secretions (W+OS) were immediately applied to the wounds, and the JA and SA contents were measured in samples from 5 replicated plants collected 0.5 h after treatments. Non-treated plants served as controls. (B) A. glycines-induced hormone levels. 10 nymphs and 10 adult aphids were placed together on leaves of the second trifoliate and leaves were immediately enclosed between two 50 mL food-quality plastic containers secured with miniature claw-style hair clips. Uninfested enclosed leaves served as controls. Samples were harvested after 8 h, and their JA and SA contents (mean ± SE; N = 6) were analyzed on a high-performance liquid chromatograph-tandem mass spectrometer (HPLC-MS/MS).
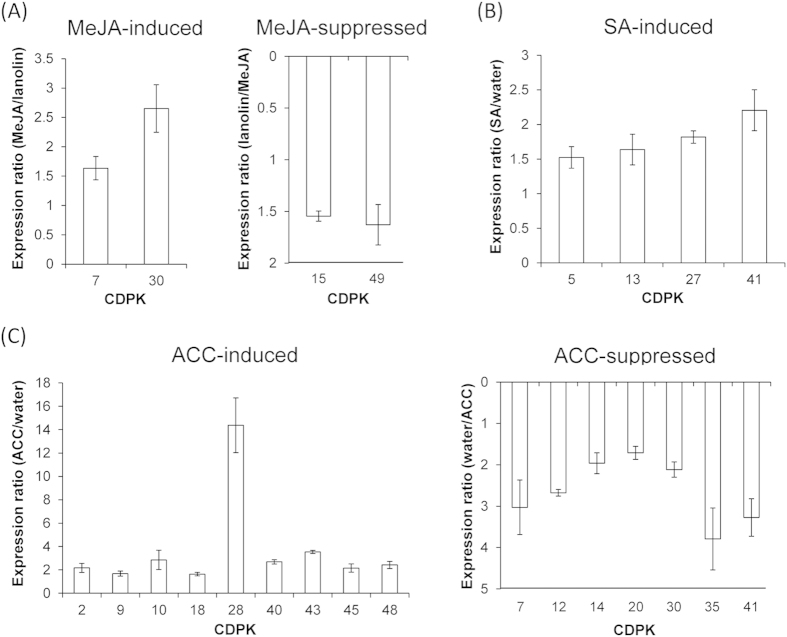
To study whether herbivory-induced JA, SA, and ET play a role in modulating the transcript levels of GmCDPKs, we analyzed GmCDPK transcript accumulations in plants that had been treated with methyl jasmonate (MeJA) that readily diffuses into plants to release JA and SA or 1-aminocyclopropane-1-carboxylic acid (ACC; the precursor of ET), which were sprayed onto the leaves. Only a few GmCDPKs showed significantly altered transcript levels after JA and SA treatments: In contrast to W+W and W+OS treatment, supplementation of MeJA only elevated the levels of GmCDPK7 and GmCDPK30, while two other genes, GmCDPK15 and GmCDPK49, showed reduced expression levels (Fig. 7A). GmCDPK5, 13, 27, and GmCDPK41 were slightly elicited by SA, but SA did not down-regulate any GmCDPKs (Fig. 7B). ET treatment induced more extensive transcriptional changes, with 9 up- and 7 down-regulated GmCDPKs. Amongst them, the paralogous GmCDPK9 and GmCDPK45 were upregulated and the pair GmCDPK7/GmCDPK35 was down-regulated after ET treatment. Interestingly, GmCDPK 7 and 30, which were down-regulated by W+OS but induced by MeJA, were suppressed by ACC treatment, indicating that OS-induced ET may dominate the transcriptional responses for these two GmCDPKs. In contrast, GmCDPK28 that showed the strongest induction by ET (14-fold; Fig. 7C) was 5-fold down-regulated after W+OS treatments and neither responded to JA nor SA, implying additional regulators of GmCDPK28 expression following S. exigua herbivory. Overall, 38 out of 48 GmCDPKs showed transcriptional responses to herbivory or herbivore-related hormone treatments, underlining the important roles of GmCDPKs in plants responses to insect feeding (Supplementary Fig. S3).
Figure 7. MeJA-, SA- and ET-induced changes in GmCDPK transcription.
CDPK expression was measured in leaves from five replicate plants at 8 h post-treatment. Leaves were treated with 150 μg MeJA in lanolin paste, with lanolin as control (A) or were sprayed with either 1 mM SA (B) or 1 mM ACC (C) with water as control. Values represent mean ( ± SE; N = 5) expression ratios of significantly (t-test, p < 0.05) and more than 1.5-fold up- or down-regulated GmCDPK transcript levels normalized to the GmCDPK levels in the respective controls; GmCDPKs whose expression levels did not change are not shown.
GmCDPK expression profiling after drought and ABA treatment
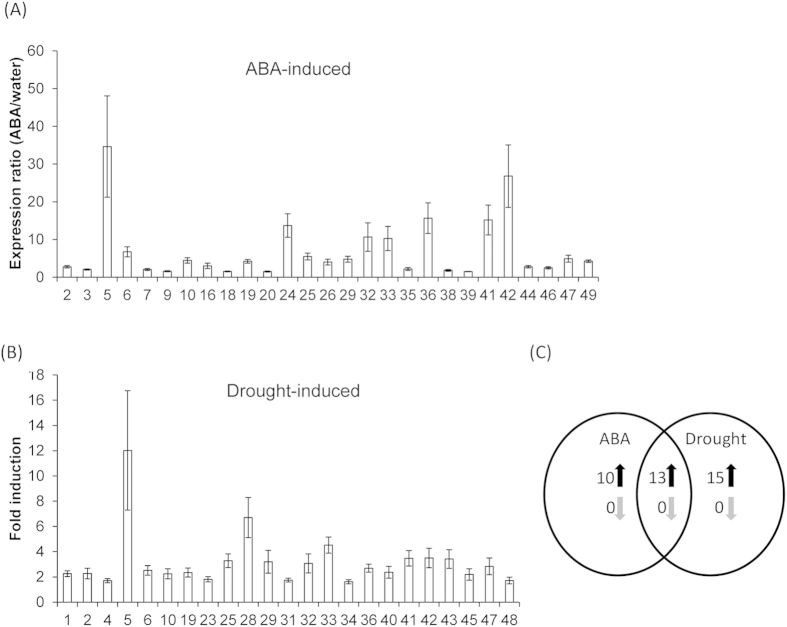
To gain insight into the functions of GmCDPKs in abiotic stress responses, we investigated the patterns of GmCDPKs’ expression after ABA and drought treatments (Fig. 8). With 23 and 27 up-regulated GmCDPKs respectively, about half of the GmCDPKs responded to the individual treatments and only 10 were not regulated by at least one of them. As expected, the transcriptional responses partially overlapped (Fig. 8C) and many paralogous GmCDPKs, such as GmCDPK19 and GmCDPK47 and GmCDPK6 and GmCDPK36 showed consistent expression patterns. These findings confirm the importance of CDPKs in drought stress and suggest that the transcriptional regulation of GmCDPKs in response to drought is of ancient nature and existed long before the recent soybean genome duplication event.
Figure 8. Drought- and ABA-induced changes in GmCDPK transcript levels.
(A) Leaves were sprayed with 1 mM ABA or water as control and the transcript levels of GmCDPKs were measured in leaves from five replicate plants at 8 h post-treatment. (B) For drought treatments, watering was stopped for 5 days and samples were harvested when leaves begun to lose turgor and wilted. Values represent mean ( ± SE; N = 5) expression ratios of significantly (t-test, p < 0.05) and more than 1.5-fold up- or down-regulated GmCDPK transcript levels normalized to the GmCDPK levels in the respective controls; GmCDPKs whose expression levels did not change are not shown. (C) Venn diagram illustration of overlapping and specific transcriptional responses after ABA and drought treatments. Numbers show significantly (t-test, p < 0.05) and more than 1.5-fold up- or down-regulated genes in the respective group.
Promoter cis-elements analysis of GmCDPKs
Promoter cis-elements play critical roles in the initiation of gene expression. Thus, bioinformatic analysis was done to identify possible cis-elements in the promoter sequences of GmCDPKs (Supplementary Dataset 1). Consistent with the proposed roles of CDPKs to be involved in multiple signaling pathways, most GmCDPKs carry several cis-elements related to stress responses; furthermore, most paralogs only showed moderate consistency in distributions of the cis-elements, indicating that after genome duplication events the promoters of the paralogous GmCDPKs have diverged (Supplementary Dataset 1). Notably, among the 48 GmCDPK promoters, 38 possess the HSE element (heat stress responsiveness), and 38 contain the TC-rich repeats (defense and stress responsiveness). Most of the ABA- or drought-inducible GmCDPK genes were found to contain ABRE (abscisic acid responsiveness) and/or MBS (MYB binding site involved in drought-inducibility) elements in their promoters. However, even SA treatment induced only 4 GmCDPKs, 37 GmCDPK promoters carry the TCA-elements (SA-responsive), and even 15 genes responded to wounding, only the GmCDPK43 promoter consists of a WUN-motif (wound-responsive). cis-Element analysis also did not well predict the responsiveness of GmCDPKs to ET. More research is needed for a better understanding of transcriptional regulation in soybean plants, including TFs and their specific cis-elements.
Discussion
A genome-wide database search, based on conserved domains and sequence similarities to known CDPKs, revealed 50 GmCDPK genes in the soybean genome. Thus we identified 3 additional GmCDPK genes, GmCDPK2, GmCDPK24, and GmCDPK50, compared with the GmCDPKs report by Valmonte et al.24. Similar to those in wheat21, all GmCDPK genes encoded at least one form of CDPK protein with 4 EF hands; in contrast, Arabidopsis4, rice20, and maize22 have CDPKs with less than 4 EF hands.
The pattern of GmCDPK chromosomal distribution is caused by two recent whole-genome duplication events, the early-legume duplication and the soybean-lineage-specific duplication, and subsequent chromosomal rearrangements and segment losses37. Most closely related homologs are not physically adjacent to each other or are even located at different chromosomes, indicating that segment duplication or genome rearrangement occurred during soybean evolution. We found evidence for two tandem duplication events: 1) GmCDPK28 and GmCDPK29 on Chromosome 10 are tandem duplicates and so are their respective paralogs GmCDPK49 and GmCDPK50 on Chromosome 20; 2) another tandem duplication event and consecutive segment duplication originated the homologs GmCDPK41 and GmCDPK42 on Chromosome 17 and GmCDPK37 on Chromosome 14 but the paralog of GmCDPK42 was lost over time.
Wounding regulates CDPK expression in several plant species38,39. However, changes in CDPK expression levels after herbivore challenges were only reported in the wild tobacco N. attenuata, in which 4 NaCDPKs were induced by wounding or adding M. sexta OS to wounds13,36. Plants recognize insect-specific factors to induce specific defense responses. S. exigua OS contain various fatty acid-amino acid conjugates (FACs)40, which are the best studied group of elicitors in lepidopteran insect OS41. Adding different synthetic FACs to wounded soybean leaves highly elicits ET and JA production42. Thus, it is likely that the FACs in the S. exigua OS are among the elicitors that induced OS-specific transcriptional changes of GmCDPKs, although other elicitors, such as proteins, cannot be ruled out. Feeding from a single-cell type, the phloem sieve element, aphids induce defenses in host plants which are distinct from those induced by chewing insects43. Consistently, the GmCDPK expression profiles induced by simulated S. exigua feeding and aphid treatment overlapped very little, and some GmCDPKs even exhibited opposite patterns: GmCDPK5, 6, 36, and GmCDPK41 were suppressed by simulated S. exigua herbivory but induced after A. glycines feeding (Figs. 3 and 5).
Some CDPKs are known to function up- or downstream of hormone signaling to regulate plant wound- and herbivore-elicited defense responses. S. exigua feeding induced high levels of JA in soybean leaves (Fig. 6A); however, our analysis indicated that the transcriptional changes of almost all GmCDPKs were independent of JA signaling. GmCDPK7 was wound- and also MeJA-induced but this response was likely not from JA alone, because after W+OS treatment, which elicited higher JA levels than did W+W, GmCDPK7 transcript levels were down-regulated. Similarly, GmCDPK30 was suppressed after W+OS treatment but induced by MeJA. Moreover, compared with simulated S. exigua herbivory, MeJA treatment only weakly changed GmCDPKs’ expression levels, suggesting that transcriptional regulation of most GmCDPKs is downstream of perception of wounding or S. exigua herbivory (probably FACs) but not downstream of the JA signaling. Besides JA, ET is also one of the main signaling molecules mediating plant defense against herbivores44. ET treatment induced extensive transcriptional changes and certain W+OS-elicited GmCDPK expression patterns, such as the up-regulation of GmCDPK23 or the suppression of GmCDPKs 5, 30 and 41 might be dependent on W+OS-elicited ET. Interestingly, MeJA-induced GmCDPK7 and GmCDPK30 were both suppressed by ET (Fig. 7C), indicating a potential antagonism of JA and ET in regulating these transcripts. In contrast, the responses of GmCDPKs to aphid feeding partly overlapped with SA-induced GmCDPK expression. As aphid herbivory elicited high levels of SA (Fig. 6B), we suspect that the SA signaling pathway is involved in aphid-induced transcriptional regulation of GmCDPKs.
In addition, CDPKs have been shown to play important roles in plants responses to abiotic stresses, especially ABA and drought45,46,47,48. Our analyses revealed that almost 80% of all GmCDPKs responded to either ABA, drought treatment, or both, supporting the notion that CDPKs are central regulators of plant drought stress responses. ABA- and drought-induced transcriptional patterns of GmCDPK greatly overlap, nevertheless, about half of the drought-induced transcripts were not regulated by ABA, indicating that other regulatory factors are also upstream of the transcriptional regulation of GmCDPKs. As expected, drought- and herbivore-induced expression patterns did not show much overlap.
Duplicated genes might undergo different evolutionary processes including nonfunctionalization, neofunctionalization, and subfunctionalization, and these are usually associated with divergence of gene expression profiles of the paralogous genes49. Among the 24 paralogous pairs, 3 pairs GmCDPK10/46, GmCDPK8/34 and GmCDPK23/43 were induced by wounding (Fig. 3A); GmCDPK23/43 was also induced by W+OS. Three paralogous pairs, GmCDPK19/47, GmCDPK18/44, and GmCDPK6/36 were suppressed after W+OS (Fig. 3A); 2 paralogous pairs GmCDPK5/24 and GmCDPK6/36 were induced by aphid feeding (Fig. 5); ET increased the expression of the paralogous GmCDPK9/45 but suppressed GmCDPK7/35 (Fig. 7C). ABA induced 5 paralogous gene pairs, 3 of which were also induced by drought, and drought stress increased the expression of 10 pairs in total (Fig. 8). In contrast, the expression patterns of two paralogous pairs, GmCDPK4/26 and GmCDPK37/41, differed dramatically. GmCDPK4 was induced after W+W, W+OS and ABA treatments but the paralog GmCDPK26 was suppressed following W+OS treatment (Fig. 3A) and induced by drought (Fig. 8). Similarly, W+W and W+OS elicited the expression of GmCDPK37, but GmCDPK41 was suppressed by W+OS (Fig. 3A); GmCDPK37 tended to be suppressed by SA, while GmCDPK41 was induced by SA (Fig. 7B). These differences suggest possible neofunctionalization of GmCDPKs following gene duplication. Several other duplicated gene pairs (such as GmCDPK14/21, GmCDPK27/48 and GmCDPK2/30) demonstrated divergent expression patterns, with one gene responding to the treatment and the other remaining at control levels, indicating that they might have had undergone subfunctionalization. Consistent with the deviating expression patterns of these GmCDPK paralogs, in silico promoter analysis indicated that the distributions of cis-elements are different between many paralogs’ promoters, probably due to the divergence of the promoters after genome duplication events (Supplementary Dataset 1).
Recent advances have revealed CDPKs as central regulators of Ca2+ -mediated immune and stress responses that are crucial for plant survival50,51. Various CDPKs mediate transient and sustained transcriptional reprogramming and play key roles in the regulation of JA, SA, ET and ABA signaling, facilitating plant responses to wounding, herbivory and drought. Our data shed light on the potential roles of GmCDPKs in regulating soybean responses to biotic and abiotic stresses and provide evidence that many GmCDPKs may have functionally diverged since the two soybean genome duplications. Future genetic and biochemical studies are needed to unravel GmCDPKs’ functions in plant adaptation to environmental factors and the underlying mechanisms.
Methods
Identification and structure analyses of CDPKs in soybean and two other legume species
The predicted protein sequences of soybean Glycine max were downloaded from Phytozome 9.0 database (http://www.phytozome.net/soybean)52, and domains and functional sites in each protein were analyzed with ps_scan.pl53. All candidates with protein kinase domains (PS50011) and EF-hand calcium-binding domains (PS50222) were extracted and used to search against the GenBank non-redundant (nr) protein database. The sequences with CDPK-related protein kinases (CRK), calcium/calmodulin-dependent protein kinases (CaMK), or calcium and calcium/calmodulin-dependent protein kinases (CCaMK) as the top hits were filtered out, and the remaining proteins were considered as GmCDPKs. These GmCDPKs were further used as queries to search the draft genome and predicted cDNAs of soybean from Phytozome 9.0 database to obtain potentially overlooked GmCDPKs. The molecular masses were calculated using the Compute pI/Mw tool of ExPaSy (http://web.expasy.org/compute_pi/). The schematic GmCDPK gene structure diagrams were drawn on the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/)54. Myristoylation and palmitoylation sites were predicted using the Plant-Specific Myristoylation Predictor (http://plantsp.genomics.purdue.edu/myrist.html) and the CSS-Palm 3.0 software55, respectively. The subcellular localization in chloroplasts and mitochondria was predicted using TargetP (http://www.cbs.dtu.dk/services/TargetP) with the cutoff for specificity set to 90%. Nuclear localization was predicted by NucPred (https://www.sbc.su.se/~maccallr/nucpred). The annotated proteins of two other legume species, Medicago truncatula and Lotus japonicas, were retrieved from Phytozome 10 database (http://phytozome.jgi.doe.gov) and Kazusa DNA Research Institute (ftp://ftp.kazusa.or.jp/pub/lotus/), respectively. The same procedure was used to identify the CDPKs in these two legumes.
Phylogenetic analysis of GmCDPK proteins
The putative protein sequences were aligned using the ClustalX2 software56 followed by visual inspection. Gaps and ambiguously aligned sites were removed manually. The optimal model with rate heterogeneity was determined by ModelGenerator57. Phylogenetic analyses were carried out with a maximum likelihood method using PHYML 3.0. Bootstrap analysis used 100 pseudo-replicates.
Expression profile based on the estimation of expression levels from RNA-Seq data
The estimated expression levels, RPKM (reads per kilobase per million reads) values, for each GmCDPKs from 6 different tissues and developmental stages were obtained from SoyBase (http://soybase.org/soyseq), reanalyzed, log-transformed and visualized using Circos. Samples include young leaves (0.4 Leaflets unfurled), flowers, one cm pod, seeds 14 days after flowering (DAF), roots, nodules, which are described in detail at SoyBase (http://soybase.org/soyseq/tables_lists/index.php).
Prediction of the GmCDPK promoter cis-elements
The 2 kb regions upstream of the putative translational initiation sites in all the 48 GmCDPK genes were retrieved from the soybean genome assembly in the Phytozome 9.0 database (Supplemental Dataset 1) and were analyzed to predict the cis-regulatory elements using the PlantCARE58 (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The motifs putatively involved in transcriptional initiation, transcriptional enhancement, and hormone and/or stress responsiveness were summarized.
Plant growth, insect rearing, and sample treatments
Seeds of the soybean genotype Huachun No. 6 were grown in the greenhouse at 24 to 28 °C under 16 h of light supplied by sodium lights. All treatments were performed on leaves of the second trifoliate. Soybean aphids, Aphis glycines, were from a population maintained on soybean in the greenhouse. For treatments, 10 nymphs and 10 adults were placed at the abaxial site of a second trifoliate leaf and confined in clip cages to avoid moving to other uninduced leaves; empty clip cages were used as control treatment. For the collection of S. exigua oral secretions, larvae were reared on soybean until the third to fifth instars. OS were collected on ice using a pipette. For W+W treatments, leaves were wounded with a pattern wheel, and 20 μL of water was rubbed onto each leaf; for W+OS treatments, 20 μL of S. exigua OS was rubbed into wounds. MeJA was dissolved in heat-liquefied lanolin at a concentration of 7.5 mg/mL; 20 μL of the resulting lanolin paste was applied to leaves, and pure lanolin was applied as a control. For SA, ET and ABA treatments, 1 mM SA, ACC, or ABA were sprayed on the top and the bottom of each leaf, water was sprayed as control. After specific times, leaves were excised, immediately frozen in liquid nitrogen, and stored at −80 °C until use. For drought treatment, watering was stopped for 5 days and samples were harvested when leaves begun to loose turgor and wilted.
RNA extraction and q-PCR
Total RNA was extracted from leaves using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. A total of 0.5 μg of total RNA per sample was reverse transcribed using oligo(dT) and Superscript II reverse transcriptase (Invitrogen). q-PCR was performed on a CFX ConnectTM real-time system (BIO-RAD) using iTaqTM Universal SYBR Geen Supermix kits (BIO-RAD). For each analysis, a linear standard curve, threshold cycle number versus log (designated transcript level), was constructed using a series dilution of a specific cDNA standard; the levels of the transcript in all unknown samples were determined according to the standard curve. The soybean actin gene, which is a housekeeping gene, was used as an internal standard for normalizing cDNA concentration variations. Relative transcript levels of genes were obtained by dividing the extrapolated transcript levels of the target genes by the levels of actin from the same sample. Sequences of primers used for q-PCR are listed in Supplementary Table S3.
Analysis of JA and SA concentrations
One milliliter of ethyl acetate spiked with 200 ng of D2-JA and 40 ng of D4-SA, the internal standards for JA and SA, respectively, was added to each crushed leaf sample (approximately 150 mg). Samples were then vortexed for 10 min. After centrifugation at 13,000 g for 10 min at 4 °C, the supernatants were transferred to fresh tubes and evaporated to dryness in a vacuum concentrator (Eppendorf). Each residue was resuspended in 0.5 mL of 70% methanol (v/v), vortexed for 10 min, and then centrifuged at 13,000 g for 15 min at 4 °C to remove particles. The supernatants were analyzed on an HPLC-MS/MS (LCMS-8040 system, Shimadzu).
Statistical analysis
Data were analyzed by unpaired t-test using StatView, version 5.0 (SAS Institute).
Additional Information
How to cite this article: Hettenhausen, C. et al. Genome-wide identification of calcium-dependent protein kinases in soybean and analyses of their transcriptional responses to insect herbivory and drought stress. Sci. Rep. 6, 18973; doi: 10.1038/srep18973 (2016).
Supplementary Material
Acknowledgments
This work was supported the Strategic Priority Research Program of the Chinese Academy of Sciences (CAS) (No. XDB11050200 to JW) and a grant from the Yunnan Recruitment Program of Experts in Sciences (2012HA016, to JW), grants from the Natural Science Foundation of China (No. 31301037 and 31470012 to GS), a grant from the Natural Science Foundation of Yunnan Province (No. 2013FB068 to GS), the Young Academic and Technical Leader Raising Foundation of Yunnan Province (No. 2014HB046 to GS), and the Western Light Talent Culture Project of the CAS (to GS). CH was supported by a “Young International Scientists Program” from the CAS. We also thank the HPC Center and the Biotechnology Experimental Center at the Kunming Institute of Botany, CAS, for supporting computational work and plant cultivation.
Footnotes
Author Contributions C.H., G.S. and J.W. designed this study. G.S., Y.H., T.S., H.Z. performed the bioinformatic work and C.H. and J.Q. did the experimental work and analyzed the data. C.H., G.S. and J.W. wrote the manuscript text. All authors reviewed and approved the manuscript.
References
- Hetherington A. M. & Brownlee C. The generation of Ca2+ signals in plants. Annu Rev Plant Biol 55, 401–427 (2004). [DOI] [PubMed] [Google Scholar]
- Lecourieux D., Ranjeva R. & Pugin A. Calcium in plant defence-signalling pathways. New Phytol 171, 249–269 (2006). [DOI] [PubMed] [Google Scholar]
- Sanders D., Pelloux J., Brownlee C. & Harper J. F. Calcium at the crossroads of signaling. Plant Cell 14, S401–S417 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. H., Willmann M. R., Chen H. C. & Sheen J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129, 469–485 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon A. C., Yoo B. C. & Mccaffery C. Pseudosubstrate inhibition of CDPK, a protein-kinase with a calmodulin-like domain. Biochemistry-US 33, 7278–7287 (1994). [DOI] [PubMed] [Google Scholar]
- Harper J. F., Huang J. F. & Lloyd S. J. Genetic identification of an autoinhibitor in CDPK, a protein-kinase with a calmodulin-like domain. Biochemistry-US 33, 7267–7277 (1994). [DOI] [PubMed] [Google Scholar]
- Szczegielniak J. et al. Maize calcium-dependent protein kinase (ZmCPK11): local and systemic response to wounding, regulation by touch and components of jasmonate signaling. Physiol Plantarum 146, 1–14 (2012). [DOI] [PubMed] [Google Scholar]
- Klimecka M. et al. Regulation of wound-responsive Calcium-Dependent Protein Kinase from maize (ZmCPK11) by phosphatidic acid. Acta Biochim Pol 58, 589–595 (2011). [PubMed] [Google Scholar]
- Kanchiswamy C. N. et al. Regulation of Arabidopsis defense responses against Spodoptera littoralis by CPK-mediated calcium signaling. BMC Plant Biol 10, 97 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A. & Oecking C. Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell 11, 263–272 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyoshihara Y., Iwata M., Fukaya T., Tatsuki M. & Mori H. Turnover of LeACS2, a wound-inducible 1-aminocyclopropane-1-carboxylic acid synthase in tomato, is regulated by phosphorylation/dephosphorylation. Plant J 64, 140–150 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang K. et al. Genome-wide identification and expression analysis of the CDPK gene family in grape, Vitis spp. BMC Plant Biol 15, 10.1186/s12870-015-0552-z (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D. H., Hettenhausen C., Baldwin I. T. & Wu J. Silencing Nicotiana attenuata calcium-dependent protein kinases, CDPK4 and CDPK5, strongly up-regulates wound- and herbivory-induced jasmonic acid accumulations. Plant Physiol 159, 1591–1607 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettenhausen C., Yang D. H., Baldwin I. T. & Wu J. Calcium-dependent protein kinases, CDPK4 and CDPK5, affect early steps of jasmonic acid biosynthesis in Nicotiana attenuata. Plant Signal Behav 8, e22784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. T. & Song W. Calcium-dependent protein kinase gene ZmCPK12 from maize confers tolerance to drought and salt stresses in transgenic plants. Acta Physiol Plant 35, 1659–1666 (2013). [Google Scholar]
- Chen J. H., Xue B., Xia X. L. & Yin W. L. A novel calcium-dependent protein kinase gene from Populus euphratica, confers both drought and cold stress tolerance. Biochem Bioph Res Co 441, 630–636 (2013). [DOI] [PubMed] [Google Scholar]
- Wei S. Y. et al. A rice calcium-dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility. BMC Plant Biol, 14, 133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Hata S., Kyozuka J., Shimamoto K. & Izui K. Over-expression of a single C Ca2+ -dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23, 319–327 (2000). [DOI] [PubMed] [Google Scholar]
- Geiger D. et al. Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. P Natl Acad Sci USA 107, 8023–8028 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T., Tanaka N., Yang G., Hayashi N. & Komatsu S. Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol 46, 356–366 (2005). [DOI] [PubMed] [Google Scholar]
- Li A. L. et al. Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol Biol 66, 429–443 (2008). [DOI] [PubMed] [Google Scholar]
- Kong X. et al. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC genomics 14, 433 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo R. et al. Genome-wide identification, classification, and expression analysis of CDPK and its closely related gene families in poplar (Populus trichocarpa). Mol Biol Rep 40, 2645–2662 (2013). [DOI] [PubMed] [Google Scholar]
- Valmonte G. R., Arthur K., Higgins C. M. & Macdiarmid R. M. Calcium-dependent protein kinases in plants: Evolution, expression and function. Plant Cell Physiol 55, 551–569 (2014). [DOI] [PubMed] [Google Scholar]
- Liu F. L., Yoo B. C., Lee J. Y., Pan W. & Harmon A. C. Calcium-regulated phosphorylation of soybean serine acetyltransferase in response to oxidative stress. J Biol Chem 281, 27405–27415 (2006). [DOI] [PubMed] [Google Scholar]
- Lee J. W. et al. Phosphorylation of nodulin-26 on serine-262 affects its voltage-sensitive channel activity in planar lipid bilayers. J Biol Chem 270, 27051–27057 (1995). [DOI] [PubMed] [Google Scholar]
- Zhang X. Q. & Chollet R. Seryl-phosphorylation of soybean nodule sucrose synthase (nodulin-100) by a Ca2+ -dependent protein kinase. Febs Lett 410, 126–130 (1997). [DOI] [PubMed] [Google Scholar]
- Zhang X. Q. et al. Soybean nodule sucrose synthase (Nodulin-100): Further analysis of its phosphorylation using recombinant and authentic root-nodule enzymes. Arch Biochem Biophys 371, 70–82 (1999). [DOI] [PubMed] [Google Scholar]
- Asai S. et al. The variable domain of a plant calcium-dependent protein kinase (CDPK) confers subcellular localization and substrate recognition for NADPH oxidase. J Biol Chem 288, 14332–14340 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. X. & Hrabak E. M. The myristoylated amino-terminus of an Arabidopsis calcium-dependent protein kinase mediates plasma membrane localization. Plant Mol Biol 82, 267–278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. X. & Hrabak E. M. An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiol 128, 1008–1021 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameier M., Krings A. & MacCallum R. M. NucPred–predicting nuclear localization of proteins. Bioinformatics 23, 1159–1160 (2007). [DOI] [PubMed] [Google Scholar]
- Hrabak E. M. et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132, 666–680 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J. et al. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 (2010). [DOI] [PubMed] [Google Scholar]
- Grant D., Nelson R. T., Cannon S. B. & Shoemaker R. C. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic acids res 38, D843–846 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Hettenhausen C., Meldau S. & Baldwin I. T. Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19, 1096–1122 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill N. et al. Molecular and chromosomal evidence for allopolyploidy in soybean. Plant Physiol 151, 1167–1174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschmann F., Stalder U., Piotrowski M., Oecking C. & Schaller A. LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization. Plant Physiol 129, 156–168 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon G. M., Cho H. S., Ha H. J., Liu J. R. & Lee H. S. P. Characterization of NtCDPK1, a calcium-dependent protein kinase gene in Nicotiana tabacum, and the activity of its encoded protein. Plant Mol Biol 39, 991–1001 (1999). [DOI] [PubMed] [Google Scholar]
- Diezel C., von Dahl C. C., Gaquerel E. & Baldwin I. T. Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiol 150, 1576–1586 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. & Baldwin I. T. New insights into plant responses to the attack from insect herbivores. Annu Rev Genet 44, 1–24 (2010). [DOI] [PubMed] [Google Scholar]
- Schmelz E. A., Engelberth J., Alborn H. T., Tumlinson J. H. & Teal P. E. A. Phytohormone-based activity mapping of insect herbivore-produced elicitors. P Natl Acad Sci USA 106, 653–657 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelckel C., Weisser W. W. & Baldwin I. T. An analysis of plant-aphid interactions by different microarray hybridization strategies. Mol Ecol 13, 3187–3195 (2004). [DOI] [PubMed] [Google Scholar]
- von Dahl C. C. & Baldwin I. T. Deciphering the role of ethylene in plant-herbivore interactions. J Plant Growth Regul 26, 201–209 (2007). [Google Scholar]
- Asano T., Hayashi N., Kikuchi S. & Ohsugi R. CDPK-mediated abiotic stress signaling. Plant Signal Behav 7, 817–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R. & Pandey G. K. Expressional analysis and role of Calcium regulated kinases in abiotic stress signaling. Curr Genomics 11, 2–13 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S. et al. A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol Biochem 71, 112–120 (2013). [DOI] [PubMed] [Google Scholar]
- Zhu S. Y. et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19, 3019–3036 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince V. E. & Pickett F. B. Splitting pairs: The diverging fates of duplicated genes. Nat Rev Genet 3, 827–837 (2002). [DOI] [PubMed] [Google Scholar]
- Boudsocq M. & Sheen J. CDPKs in immune and stress signaling. Trends Plant Sci 18, 30–40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T. & Herde M. From local to global: CDPKs in systemic defense signaling upon microbial and herbivore attack. Curr opinion in plant biol 20C, 1–10 (2014). [DOI] [PubMed] [Google Scholar]
- Goodstein D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic acids res 40, D1178–1186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E. et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic acids res 34, W362–365 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A. Y., Zhu Q. H., Chen X. & Luo J. C. GSDS: a gene structure display server. Yi Chuan 29, 1023–1026 (2007). [PubMed] [Google Scholar]
- Ren J. et al. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel 21, 639–644 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
- Keane T. M., Creevey C. J., Pentony M. M., Naughton T. J. & McLnerney J. O. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol 6, 29 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M. et al. PlantCARE: a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic acids res, 30 325–327 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.