Abstract
Small ruminant lentivirus (SRLV) infection causes losses in the small ruminant industry due to reduced animal production and increased replacement rates. Infection of wild ruminants in close contact with infected domestic animals has been proposed to play a role in SRLV epidemiology, but studies are limited and mostly involve hybrids between wild and domestic animals. In this study, SRLV seropositive red deer, roe deer and mouflon were detected through modified ELISA tests, but virus was not successfully amplified using a set of different PCRs. Apparent restriction of SRLV infection in cervids was not related to the presence of neutralizing antibodies. In vitro cultured skin fibroblastic cells from red deer and fallow deer were permissive to the SRLV entry and integration, but produced low quantities of virus. SRLV got rapidly adapted in vitro to blood-derived macrophages and skin fibroblastic cells from red deer but not from fallow deer. Thus, although direct detection of virus was not successfully achieved in vivo, these findings show the potential susceptibility of wild ruminants to SRLV infection in the case of red deer and, on the other hand, an in vivo SRLV restriction in fallow deer. Altogether these results may highlight the importance of surveilling and controlling SRLV infection in domestic as well as in wild ruminants sharing pasture areas, and may provide new natural tools to control SRLV spread in sheep and goats.
Introduction
In the last century we have witnessed the emergence of acquired immunodeficiency syndrome, multidrug-resistant tuberculosis and tick-borne related diseases as a result of the interactions between humans and zoonotic pathogens in a pathway including wildlife and domestic animals [1].
Small ruminant lentiviruses (SRLV) infection is present in sheep and goats from Europe [2], America [3–5], Australia [6], Africa [7] and Asia [8, 9]. Economic impact of SRLV infection, highly dependent on environmental factors, breed/individual susceptibility, production system, farming practices and age of culling is often underestimated and still under study [10]. The premature removal of infected animals and the consequent increased replacement rate is a major consequence of SRLV infection. SRLV infected sheep have shown decreased fertility and number of lambs per birth, as well as a reduction of birth weight and weight gain from birth to weaning [11, 12]. Animals with advanced disease present a significantly reduced body weight at slaughter, and their carcass may not qualify for human consumption [13]. The most obvious positive result observed following the eradication of SRLV infections in goats’ herds is the disappearance of clinical cases of carpal arthritis and the improved health of the flocks [10]. This combined with the elimination of a viral infection showing a negative impact on milk production [14–16] may well explain the financial success of a combined eradication campaign comprising SRLV, such as “The Norwegian Healthier Goats program” [17].
SRLV are able to cross inter-species barrier thereby infecting sheep and goats through horizontal and lactogenic routes [10]. Since the first descriptions of natural transmissions of Visna Maedi virus (VMV) to goats, or Caprine arthritis encephalitis virus (CAEV) to sheep [18], many research groups have reported cross-species transmission in different countries [19–21]. Recently, new genotypes, subtypes and recombinant SRLV viruses have been described widening significantly their genetic and antigenic heterogeneity, likely conferring them a wider spectrum of cell and host tropism. Env and LTR genomic regions have been related to cell tropism by modifying the receptor usage [22] or by enhancing the promoter activity depending on the transcription factors present in a particular cell type, respectively [23]. Typically, the virus exists in the infected host as a continuum of related but divergent genetic variants called quasispecies that compartmentalize in different tissues or body fluids [24, 25] potentially favoring cross-species transmission.
The transmission of infectious agents from reservoir animal populations, often from domesticated species to wildlife in shared pastures or breeding areas (spill-over), may lead to the emergence of a range of infectious diseases in the wildlife. Spill-over is particularly important for endangered species and may also occur from wildlife to domestic animals (spill-back) affecting animal production [1]. A well-known example of adaptation to a new host is the human immunodeficiency virus (HIV) that successfully overcame the intrinsic restriction factors constitutive of the species-specific barrier, to successfully infect humans [26]. HIV-2 is a human adapted variant of the simian immunodeficiency virus (SIV) from Sootey mangabeys (Cercocebus atys) [27], and HIV-1 may be similarly derived from a chimpanzee (Pan troglodytes) SIV [28]. There are different examples of inter-species transmission of lentiviruses displaying more attenuated/virulent phenotype in the new host. In general, SIV cause no observable disease in their natural hosts but may become severe pathogens in novel host species [29]. In the case of BIV, which is non-pathogenic for Bos taurus, the transmission to Bos javanicus resulted in lethal Jembrana disease [30].
During the last decades many species of wild ruminants have been reintroduced and others have expanded their population across Europe, both in density and geographical range. Transmission of pathogens from or to domestic ruminants poses serious problems since infected wildlife and domestic ruminants may represent a pathogen reservoir to each other [31]. So far, SRLV have been found in Alpine ibexes (Capra ibex) from French Alps in contact with domestic goats as well as in hybrids derived from this contact, both of which showed proviral sequences related to CAEV present in local domestic goats [32]. In Rocky Mountain goats (Oreamnos americanus), CAEV is able to be transmitted by lactogenic and horizontal routes causing a severe multisystemic disease involving lungs, central nervous system and joints [33]. Mouflon (Ovis aries musimon) and domestic sheep hybrids are also susceptible to CAEV infection [34]; however, among a 101 wild mouflon population from Spain, none showed positive serology against VMV [35]. Similar results were obtained when analyzing sera from red deer in California [36–38], suggesting lack of susceptibility of these wild ruminants to SRLV.
This study aims at providing further evidence of the potential susceptibility of wild ruminants to SRLV infection. We present serological evidence that different wild ruminants may have mounted an antibody response to SRLV. Additionally, we explored the susceptibility of red- and fallow deer cells to different SRLV strains for entry, integration, replication and production of infectious viral particles.
Materials and methods
Animals and samples
259 red deer (Cervus elaphus), 36 roe deer (Capreolus capreolus), 40 fallow deer (Dama dama) and 16 mouflon (Ovis aries musimon) from different origins as specified in Figure 1, were sampled to obtain whole blood in EDTA-3 K as anticoagulant. Deer were from zoos (4%), national parks (4%), production farms (61%) and hunting campaigns (31%). Roe deer were all from hunts, fallow deer from zoos (37.5%) and national parks (62.5%) and mouflon were from zoos (62.5%) or national parks (37.5%).
Figure 1.
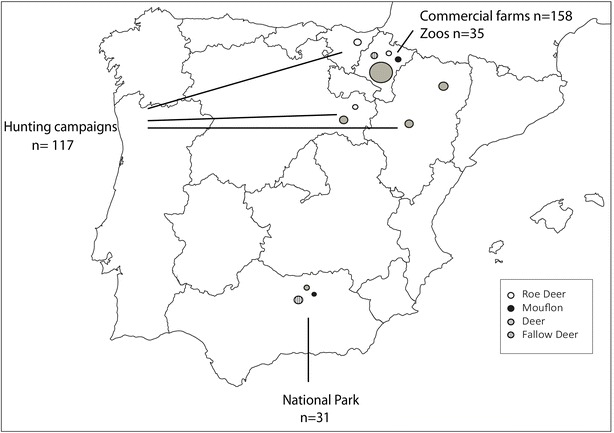
Bubble chart of the sampling distribution. Blood samples from red deer, fallow deer, roe deer and mouflon were obtained from commercial farms, zoological or national parks or hunting campaigns in different Spanish regions. The bubbles are sized according to the number of sampled animals.
Plasma was obtained and stored at −20 °C for serological determinations and buffy coat for PCR analyses. Peripheral blood leucocytes (PBLs) were isolated from buffy coat samples by density gradient centrifugation and resuspended in PBS for DNA extraction using QIAamp® DNA Blood Mini Kit (Qiagen).
Serological analysis
Plasma samples were analyzed for SRLV antibodies by four different ELISA methods. Commercial ELISAs for detection of SRLV antibodies were slightly modified in order to detect IgG from wild ruminant species. This was achieved in a first round, by using protein Pierce Purified Recomb® Protein G Peroxidase conjugated (Recombinant Protein G, Pierce) or secondly, with a secondary antibody able to cross-react with a wide range of ruminant species, including red deer and fallow deer (EG5, Ingenasa). ELISA plates used included commercial Chekit (AG-CHEKIT CAEV/MVV kit, IDEXX Switzerland), ELITEST (Elitest-MVV Hyphen-Biomed, France) and a previously described home-made ELISA that is based on coating with single synthetic peptides alone or in combination [39]. Procedures were carried out following manufacturer’s instructions with the exception of the conjugate antibody which, as mentioned, was substituted by a secondary antibody able to react with wild species’ IgG.
PCR
DNA samples from fifteen seropositive and four seronegative animals were subjected to amplification by described PCRs for SRLV and BIV detection (Table 1). Thermal amplification protocols were adapted to annealing temperature of each primer pair and results were analyzed in 1% agarose gels. Bands of the expected molecular weight were cut and purified using ATP Gel/PCR extraction kit (ATP Biotech Inc.). Purified products were cloned into pGEMT-easy vector (Promega) following manufacturer’s instructions and sequenced using M13 primers.
Table 1.
Primers covering gag, pol, LTR and env regions, were used for the SRLV detection by PCR
| PCR assay | Primers | Sequence 5′–3′ | References |
|---|---|---|---|
| Gag nested PCR | GAG F1 | TGG TGA RKC TAG MTA GAG ACA TGG | [64] |
| GAG F2 | CAA ACW GTR GCA ATG CAG CAT GG | ||
| POL R1 | CAT AGG RGG HGC GGA CGG CAS CA | ||
| POL R2 | GCG GAC GGC ASC ACA CG | ||
| Pol nested PCR | P28 | CAT GAA GAG GGG ACA AAT CAG CA | [18] |
| P34 | TAC ATT GGG TGC CTG GAC ATA A | ||
| P32 | TAC CTG DGT TGG TCC YWG CCA CT | ||
| P33 | CTT CCC AVA GTA CCT GDG TTG GTC | ||
| P29 | GGT GCC TGG ACA TAA AGG GAT TC | ||
| P35 | GCC ACT CTC CTG RAT GTC CTC T | ||
| Gag-Can nested PCR | CAEV F0 | AAC TGA AAC TTC GGG GAC GCC TG | [5] |
| CAEV F1 | AAG GTA AGT GAC TCT GCT GTA CGC | ||
| CAEV F2 | TGG TGA GTC TAG ATA GAG ACA TGG | ||
| CAEV R0 | GTTATCTCGTCCTAATACTTCTACTGG | ||
| CAEV R1 | TTT TTC TCC TTC TAC TAT TCC YCC | ||
| CAEV R2 | GGA CGG CAC CAC ACG TAK CCC | ||
| MVV F0 | AAG TAA GGT AAG AGA GAC ACC TAC TGG | ||
| MVV F1 | TAG ATA GAG ACA TGG CGA AGC AAG CTC | ||
| MVV F2 | GAC AGA AGG GAA CTG TCT ATG GGC | ||
| MVV R0 | GGT GGT GCT TCT GTT ACA ACA TAG G | ||
| MVV R1 | GGA CGG CAC CAC ACG TGG | ||
| MVV R2 | CCC CTC CTG YTG TTT CCC TG | ||
| BIV nested PCR | POL3 | ACA ACG GGC CGT GCT TTA CTG | [65]; [66] |
| POL4 | CCT CTT CCT CTA TTA CTG CTG C | ||
| POL5 | GAR AAT CTA TGT AAG TAT CTG GG | Unpublished data (Nadin-Davis) | |
| POL6 | CTG TTY CTT ACG TAA CAC CAC T | ||
| P09 | CAC TGG ACG AGA TGA GGT AGT | [67] | |
| P06 | TGG TAG TCT GAT AAA TGG CA | ||
| P04 | CAG GCT CTT AAG GAA ATT GT | ||
| P11 | CCA TCC TTG TGG TAG AAC ATT | ||
| Craft-Oslo | Craft-Fw | TGA CAG AAG GAA ATT GTY TRT GG | |
| Oslo-Rv | GGC ATC ATG GCT AAT ACT TCT AA | [68] | |
| LTR PCR | LTR-Fw | TGA CAC AGC AAA TGT AAC CGC AAG | [69] |
| LTR-Rv | CCA CGT TGG GCG CCA GCT GCG AGA | ||
| CO qPCR | 612-F | GAA CTC AGC CAC AAG AGG AAG AA | [70] |
| 613-R | CCT GCG GCA GCT ACT ATT GC | ||
| 614 Probe | FAM AACTAGCATAATGACCAAGCCAACGCCTCT BHQ-1 | ||
| A4 qPCR | Fs7631/Fg6221 | ACA AAC TGG ACC ACC ATG CA | [21] |
| Rg6221 | CTA GTG TTC CAT TTC CTG TTC TGT TTA | ||
| Probe g6221 | FAM GGC AAC TGT TCW CAG TGT GTT AAT GYA ACG BHQ-1 | ||
| A4-SU PCR | SU5-1F | GTA GAT GTG TAC AAA GAC CAG | [21] |
| SU5-1R | CTG CCT CTA ACA CTT GCT GC | ||
| CO-SU PCR | B1-SU5-1R | TGC CTC TAA CAC ATC CTG CTG TGC | [70] |
| B1-SU5-1F | GGT GGA ACA TAT GAC AGG AG |
Primers able to amplify bovine inmunodeficiency virus were included
Cells and SRLV in vitro infection
Red deer (DSF) and fallow deer (FSF) skin fibroblasts were obtained from skin biopsies, isolated by trypsin disruption and cultured in DMEM medium supplemented with 10% foetal bovine serum (FBS), 1% l-glutamine and 1% antibiotic/antimycotic mix (Sigma-Aldrich). Previously obtained ovine skin fibroblasts (OSF) or goat synovial membrane cells (GSM) were used as control cells as indicated throughout the experiments.
Blood derived macrophages (BDM) from deer and fallow deer were also obtained as described [40]. Briefly, buffy coat diluted in PBS was loaded onto Ficoll-Paque Premium 1.084 (GE Healthcare) gradient to isolate peripheral blood mononuclear cells. BDM were obtained by adherence and cultured in RPMI 1640 supplemented with 1% l-glutamine, 1% non-essential amino acids, 50 µM β-mercaptoethanol, 1% vitamins, 10 mM sodium pyruvate and 10% foetal bovine serum (FBS). Sheep BDM were also obtained for comparative purposes. Cultures with fibroblast-like cells overgrowth were discarded.
Established cultures were maintained and used in experimental infections with Ev1 [41], CAEV-Co [42], A4 [21] and Ov496 strains of SRLV [19]. In vitro infection was carried out at 0.1 and 1 TCID50/cell in DMEM 2% medium in the case of Ev1 and 496 strains and with 20 copies/cell in the case of CAEV-Co and A4 isolates. After 16 h, cells were washed three times with PBS and resuspended in lysis buffer AL (Qiagen) for DNA extraction and amplification of viral DNA with described primers (Table 1). For confirmatory purposes, DNA from in vitro infected fibroblastic cells was subjected to PCR with primers Craft-Oslo, A4-SU and CO-SU, for strains EV1, 496 and A4, CAEV-Co, respectively (Table 1). Amplicons were purified, cloned into pGEMT-easy vector and sequenced (StabVida). Additionally, after washing with PBS, cultures were maintained 10 days post inoculation for RT-activity determinations following manufacturer’s instructions for Ev1 and 496 strains (HS-Lenti RT Activity Kit, Cavidi). Viral quantification (viral RNA copies/µL) of A4 and CAEV-Co strains in the supernatants was done by qPCR (Qiagen QuantiTec Probe RT-PCR) using primers and TaqMan probes g6621 and 614 respectively (Table 1).
BDM supernatants were collected at 3, 10 and 14 days and the CAEV-Co and A4 viral RNA was quantified by real time PCR using specific primers and Taqman probes (Table 1).
LTR promoter activity
LTR basal activity from strains A4 [21], KV1772 [43], Ov496 [19] and CAEV-Co [42] was assayed using a luciferase reporter system. Briefly, DSF, FSF and GSM cells (105 in 24-well plates) were transfected with 200 ng of each LTR construction using 4 µL of Lipofectamine-LTX Reagent and 0.2 µL of PLUS-Reagent (Life Technologies). Plasmids pGL4.13 [luc2/SV40] and “empty” p-GL4.10 [luc2] were used as positive and negative controls, respectively. At the same time, 20 ng of pGL4.73 [hRluc/SV40] Vector (Promega) were cotransfected, so that the firefly activity was standardized according to the Renilla luciferase activity. After 24 h, cells were harvested with Passive Lysis 5X Buffer (Promega), and firefly and renilla luciferase activity were measured following manufacturer’s instructions. Results were expressed as: Relative Luminiscence Units (RLU) firefly luminescence/RLU Renilla luminescence.
Entry assay
CAEV (including human alkaline phosphatase, AP) [22] virions pseudotyped with ENV proteins from strains Ev1 [41], CAEV-Co [42], 697 [44], Roccaverano [45] and Seui [46] of SRLV, as well as Vesicular Stomatitis virus protein G (VSV-G) produced in 293-T cells as described [22, 47] were used to transduce red deer and fallow deer skin fibroblasts. Briefly, pseudoviruses were incubated with target cells and after 72 h cells were washed three times with PBS and endogenous AP activity blocked by heating at 65 °C 1 h in a humidified chamber. After blocking, NCTBI reagent was added to cells for 2–24 h and finally reaction was stopped with normal water. Stained cells were visualized and counted under light microscopy by two experts.
Western blot
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was carried out using Ev1 and 496 infected DSF, FSF and OSF extracts. For Western blot analysis, monoclonal antibodies against p25 (VPM70, kindly provided by Dr B. Blacklaws, University of Cambridge, UK) was used undiluted. Anti-mouse IgG peroxidase-conjugated (Pierce) was used as secondary antibody. In addition, sera from two experimentally infected sheep with SRLV strains from genotype A and B respectively were used to reveal protein production. Reactions were developed by chemiluminescence (Amersham ECL Western blotting detection reagents; GE Healthcare, Buckinghamshire, UK).
Neutralizing antibody (NtAb) assay
Heat inactivated sera from ELISA positive red deer (n = 6) were serially diluted in DMEM 2% FBS in a 96-well plate and incubated 1:1 (vol/vol) with 100 TCID50 of strain Ev1. Six wells were used per serum dilution. After mixing and incubating overnight at 4 °C, the mixture was added to sheep fibroblasts. After 2 h at 37 °C, 5% CO2, the supernatant containing the virus and serum mixture was removed and after washing DMEM 2% FBS was added. Following a 7 day incubation at 37 °C and 5% CO2 cytopathic effect (CPE, syncitia formation) was screened by microscopy. The NtAb titer was defined as the reciprocal of the serum dilution in which 50% of the culture wells showed no signs of infection.
Results
ELISA
Using the home-made ELISA based on synthetic peptides and protein G as conjugate, SRLV antibodies were detected in 14 out of 193 (7%) red deer and one mouflon out of 10 analyzed. Using a secondary antibody able to react against wild ruminant species (EG5, Ingenasa), increased the number of seropositive red deer to 20 out of 141 (14%) and, moreover, allowed the detection of SRLV antibodies in 3 out of 36 roe deer (8%) and one mouflon, but not in fallow deer. Combining results using protein G and EG5 conjugates SRLV antibodies were detected in 26 out of 259 (10%) red deer, 3 out of 36 roe deer and one out of 16 analyzed mouflon.
Single peptide ELISA analyses indicated that peptides 126M1 and 126M2 were responsible for the seropositivity in most of the animals (Figure 2).
Figure 2.
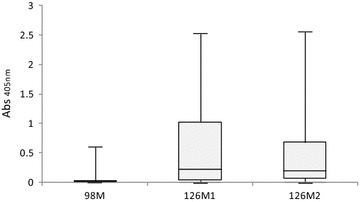
Peptide ELISA. Absorbance distribution (Abs405nm) of seropositive red deer (n = 11) obtained with individual peptides 98 M, 126M2 and 126M1 derived from Env protein of SRLV.
In order to confirm specificity of these antibody reactions, some sera were also tested for anti-SRLV antibodies using commercial ELISA (Chekit and Elitest). None of the animals that were seropositive to the peptide ELISA were seropositive to the Elitest with protein G or commercial secondary antibody conjugates. In contrast, 2 red deer that were seronegative to the peptide ELISA were seropositive with the modified Chekit test using EG5 conjugate.
Seropositive deer were mainly found (15 out of 20 seropositive deer) in commercial farms which represents the largest sample source, followed by hunting campaigns with 4 seropositive animals. Seropositive roe deer were from hunts and the seropositive mouflon was from a zoo.
Seropositive animals were tested for the presence of neutralizing antibodies obtaining titres ranging from 3 to 40.
Screening PCRs
Some bands of the expected size were obtained following PCR amplification using different sets of primers (Table 1). However, after purification, cloning and sequencing none corresponded to known SRLV proviral sequences, instead they matched bovine genomic sequences. Therefore, PCRs for the bovine immunodeficiency virus (BIV) were performed in DNA samples from seropositive and seronegative deer, but none of the amplified products presented a lentiviral-type sequence, they were all identified as genomic DNA.
In vitro infection and entry assay
After being unable to demonstrate virus presence in wild ruminants by PCR in vivo, we decided to shift to in vitro experimental infection and transduction experiments. For this, as indicated in the particular experiments, DSF, FSF and BDM cultures were obtained and infected with SRLV strains belonging to genotype A (Ev1 and Swiss A4) or genotype B (CAEV-Co and Ov496). After 16 h, CPE was evident in cells from both species (red deer and fallow deer) only in the case of Ev1 virus. Since this effect may correspond to a virus over load, viral stocks were titrated again and infection was repeated with similar results.
To clarify if these results correspond to a differential ability of SRLV in entering the cell, entry assays using CAEV-AP virions pseudotyped with envelopes from different SRLV strains were carried out. Although all the tested strains were able to enter into red deer and fallow deer cells, there were differences among them. Red deer cells were more susceptible to Seui, Ev1 and CAEV-Co strains in that order, while FSF mainly allowed the entrance of Ev1 strain from genotype A. Strains Roccaverano, and to a lesser extend 697, showed the lowest values in both cell types (Figure 3).
Figure 3.
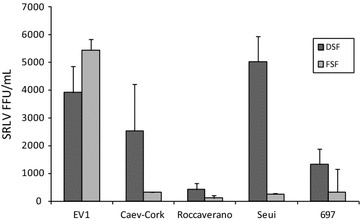
Entry assay with pseudotyped SRLV. Infectivity of CAEV-AP virions pseudotyped with Env proteins from Ev1, CAEV-Cork, Roccaverano, Seui and 697 SRLV strains on red deer skin fibroblasts (DSF) and fallow deer skin fibroblasts (FSF). Results are expressed as focus-forming units per milliliter (FFU/mL).
Post entry restriction
Once demonstrated the SRLV ability to enter into red deer and fallow deer cells, we checked further steps of the viral cycle such as integration and RNA production. PCR at 16 h post-inoculation showed positive results in all cases confirming, the permissive entry and the presence of viral DNA within red deer and fallow deer cells. Regardless of the high transcriptional activity of the LTR in fibroblastic cells from deer and fallow deer (Figure 4), RNA production was confirmed only in the case of Ev1 infection in red deer cells 48 h after inoculation.
Figure 4.
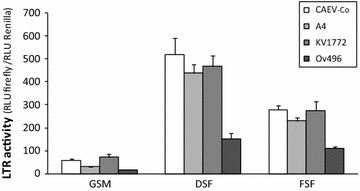
LTR promoter activity. LTR-U3-R region of SRLV strains CAEV-Co, A4, Kv1772 and Ov496 transfected in goat synovial membrane cells (GSM), red deer skin fibroblasts (DSF) and fallow deer skin fibroblasts (FSF). Results are expressed as the ratio between the relative firefly luminescence (RLU) and the Renilla luminescence.
In spite of these positive correlates of lentiviral productive infection, RT activity in the supernatants was negative in all cases at 7 and 10 days, suggesting low or no production of viral particles to the supernatants (Figures 5A and B). In order to check whether Ev1 and 496 viruses were completely absent from DSF and FSF supernatants, we transferred them to fresh ovine fibroblasts. After 7 days, RT activity from both viruses started to increase only in the case of DSF, reaching positive control values at the second supernatant transfer in OSF (Figures 5C and D).
Figure 5.
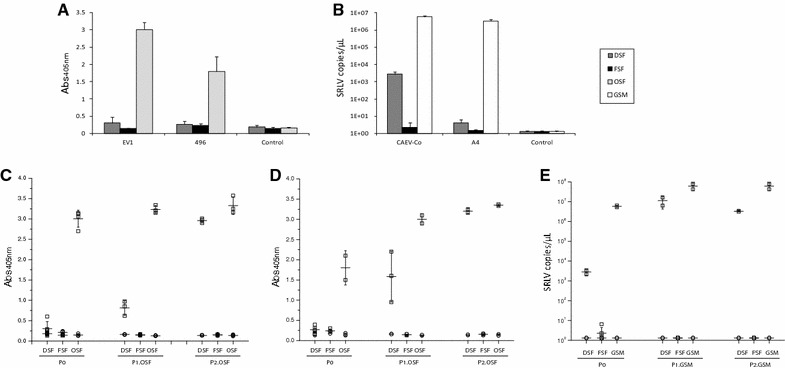
SRLV in vitro infection upon supernatant transfer. A RT activity (Abs405nm) of culture supernatants from red deer skin fibroblasts (DSF), fallow deer skin fibroblasts (FSF) and ovine fibroblasts (OSF) infected with SRLV strains Ev1 or 496. B Viral quantification of viral RNA (copies/µL) in the supernatants from DSF, FSF and goat synovial membrane cells (GSM) infected with strains A4 or CAEV-Co. C, D Culture supernatants from DSF, FSF and OSF infected with Ev1 or 496 respectively (P0) were transferred to fresh OSF twice (P1.OSF and P2.OSF) and RT values represented (Abs405nm). E SRLV RNA copies in the supernatant of DSF, FSF and GSM infected with CAEV-Co (P0) and after transfer of supernatants to fresh GSM twice (P1.GSM and P2.GSM). Squares represent values from infected cells and circles indicate non-infected cells.
In the case of strains A4 and CAEV-Co, viral RNA was detectable in the supernatants 7 days post-inoculation only in CAEV-Co infected cells. As above, supernatants from these infected-cells were used to infect GSM cells. After 7 days, viral RNA increased in the supernatant for CAEV-Co strain but remained undetectable for A4 strain (Figure 5E).
Furthermore, infected DSF and FSF were passed weekly by trypsinization and after the first passage strains Ev1, 496 and CAEV-Co showed increased production only in DSF, being the strain A4 not able to replicate. At the second passage, Ev1 was able to destroy the cellular monolayer whereas strain 496 was adapted to DSF causing persistent infection similar to ovine fibroblasts. Virus production after passages in FSF remained always negative (Figure 6).
Figure 6.
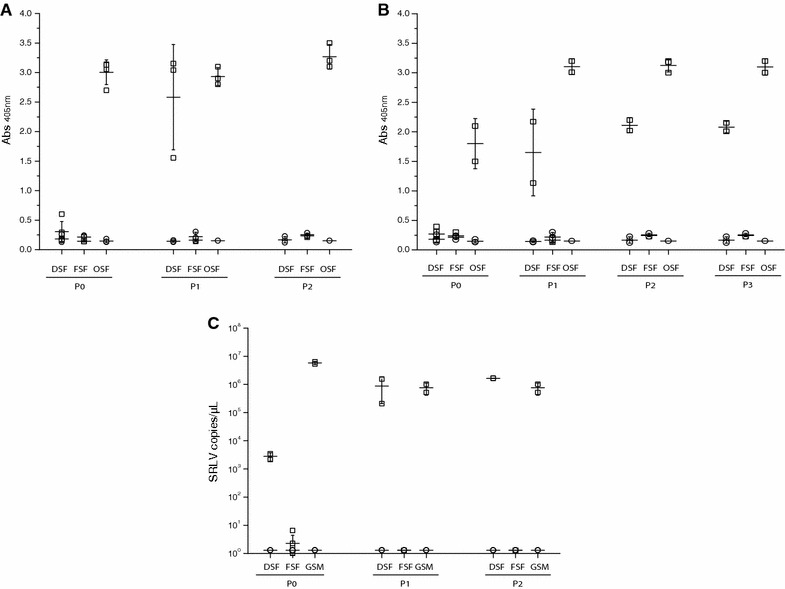
SRLV in vitro infection upon culture passages. RT activity in the supernatant from red deer (DSF), fallow deer (FSF) and ovine fibroblasts (OSF) infected with strains Ev1 (A) or 496 (B). C Viral quantification of total viral RNA (copies/µL) in the supernatants from DSF, FSF and goat synovial membrane (GSM) cells infected with strain CAEV-Co. Squares represent values from infected cells and circles indicate non-infected cells. Cell passage number is indicated (P0, P1, P2 and P3).
Production of GAG and ENV viral proteins was evaluated by western-blot using monoclonal antibodies anti-p25 and polyclonal sera from infected sheep, aiming at improving cross-reactivity. In agreement with RT activity, capsid protein production was detected in red deer cells infected with strains Ev1 and 496 at the second passage, and in OSF after inoculation with culture supernatant from infected DSF. Also in agreement was the absence of capsid protein in FSF cells neither with 496 nor Ev1 SRLV strains (Figure 7). Env protein was not specifically detected with the antisera used.
Figure 7.
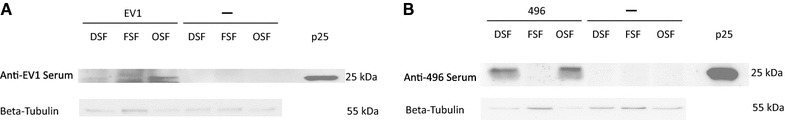
Production of P25 protein in cell culture. Western Blot of ovine fibroblasts (OSF) infected with supernatants from red deer (DSF), fallow deer (FSF) and OSF infected with strains Ev1 (A) or 496 (B) after 7 days of infection. Uninfected cells and recombinant p25 protein were also included as negative and positive controls respectively. Sera from two experimentally infected sheep with SRLV strains from genotype A and B respectively, were used to reveal protein production. Beta tubulin antibody was used as loading control.
Red deer BDM allowed Swiss A4 infection but few viral RNA copies were amplified from the supernatant. Instead, CAEV-Co replicated more efficiently reaching values obtained with ovine BDM. Fallow deer BDMs showed a more restricted phenotype, similar to that observed in skin fibroblasts showing undetectable virus production. Sheep BDM differentially allowed the replication of SRLV A4 and CAEV-Co strains. While A4 replicated at high titres, CAEV-Co RNA was produced at low levels in the supernatant, confirming the impaired replication of this virus in ovine cells (data not shown).
Discussion
The emergence of the devastating HIV epidemics in the human population following cross species transmission from the natural non-human primate host, in which these lentiviruses cause non-pathogenic persistent infections [48], highlights the ability of this group of viruses to cross the species barrier, adapt to a new host and dramatically increase their virulence.
Transgression of the species-specific barrier by SRLV has been explored in cattle through experimental infection of cows with CAEV-Co strain [49]. SRLV infection induced a persistent antibody production, similar to that observed in sheep or goats, suggesting viral protein production. However, this was not accompanied by productive long-term infection; indeed although authors demonstrated proviral integration into leukocytes and tissues, infection did not persist more than 4 months, and virus was not recovered even after attempts to reactivate viral replication. Virus clearance was unlikely due to the humoral response, since antibodies were not neutralizing.
Here, we show positive serological reaction against different SRLV antigen preparations in red deer, roe deer and mouflon, being fallow deer negative. However, this serological reaction was not accompanied by the detection of SRLV sequences by PCR in vivo. In vitro, skin cell cultures from red deer and fallow deer did not produce detectable levels of SRLV Gag or RT proteins in spite of the presence of syncytia and viral DNA and RNA. Infection was rescued by transferring supernatants from red deer skin fibroblasts infected in vitro, to fully permissive ovine fibroblasts suggesting the production of low levels of virus. Although highly dependent on the infecting strain, virus adaptation could be induced in DSF through periodical cell passages in vitro. In contrast, fallow deer cells did not support SRLV infection despite supernatant transfer or cell passages in agreement with serological findings. These results may suggest that SRLV are at different steps in the viral adaptation to red deer and fallow deer. In the last case, host may have evolved to mount an effective long-lasting restriction mechanism of lentiviral replication in vivo. Neutralizing antibodies were detected in ELISA positive sera from red deer however, they unlikely play an important role in the viral infection restrictive pathway as described in caprine and ovine species [21, 50]. Serological reaction was mostly detected by home-made ELISA in comparison with commercial tests as described for ovine and caprine infections likely due to the inclusion of novel epitopes providing wider cross-reactivity [39, 51]. Reaction was mainly directed against peptides 126M1 and 126M2 suggesting a potential SRLV infection involving multiple genotypes. Synthetic peptide ELISA performance was better using secondary antibody than protein G whose specificity against IgG from sheep and goats is moderate and unknown in red deer [52]. Contact with sheep or goats cannot be excluded in animals coming from hunts or national parks, however seropositive animals were mainly found in commercial farms and zoos in which these contacts are at present highly restricted.
Intra-individual SRLV viral reservoir is represented by proviral integration into monocytes that after maturation to tissue macrophages become permissive to viral replication. Wild ruminants could act as inter-individual reservoir potentially responsible for the re-emergence of infection in domestic small ruminant flocks [32]. The high SRLV genome plasticity is translated into a wide tropism that allows the generation of immune escape mutants and the colonization of new target cells and hosts [53]. Accordingly, cells from humans, monkey, hamster, mice, quail, cows [49, 54, 55] and, as shown in this study, red deer and fallow deer are permissive to SRLV entry since viral DNA was detected in vitro upon infection of DSF and FSF. Consequently, amplification and adaptation to a broad spectrum of host species could be expected. Indeed, SRLV are causing natural cross-species infection in wild ruminants following contact during the free grassing season in wilderness areas, potentially generating uncontrollable reservoir of viruses [32]. This could eventually represent a major obstacle in SRLV eradication programs in domestic sheep and goat flocks.
Endogenous retroviruses from cervids (CERVs) have been described in Mule deer (Odocoileus hemionus) genome [56], confirming previous contacts with retroviruses. Strong humoral and T cell responses are elicited against human retroviruses, especially to HERV-K10 in healthy but mostly in cancer patients due to higher viral expression [57, 58]. However, small ruminants do not mount a strong humoral or cellular immune response against exogenous JSRV, likely due to the presence of a related endogenous counterpart that induce tolerance [59]. Interestingly, sequences from CERV gag region were compared with VMV-like isolates and preliminary analyses indicate some degree of structural similarity between both sets of sequences that might explain the presence of serological reaction in the absence of an exogenous lentivirus in vivo. Unfortunately, it is unknown whether red deer develop a detectable antibody production against CERV.
Antigenic cross-reactivity with CAEV in humans has been reported likely due to the consumption of contaminated caprine dairy products [60] potentially favoring SRLV adaptation to humans. In fact, numerous emerging infectious diseases including zoonosis have been originated from wildlife [1, 61]. Domestic animals have been selected for centuries towards a specific production (milk, meat, wool, etc.) and have not been subjected to the same natural selection pressure as their wildlife counterparts and as a result, they are less resistant to a high number of pathogens [62]. However, repeated contacts of potentially susceptible wildlife with domestic SRLVs may lead to the emergence of new adapted lentivirus variants [32].
Lack of viral proteins in cultured cells from wild ruminants could be a consequence of the abnormal processing of the Env protein in these cell types. This has been previously described in sheep choroid plexus cells infected with CAEV-like virus [63]. However, if this was the case no CPE would be expected in DSF cells. Moreover, in vitro findings in this study clearly show the presence of newly synthesized infectious particles in DSF and the ability of SRLV to adapt rapidly to cells from wild ruminants in vitro. However, since in vivo lentivirus cross-species transmission is a rare event that only occurs under specific circumstances other factors related to the host immune response may hamper the progression of infection.
The host innate immunity may play an important role in counteracting infection independent of the presence of antibodies which are generated following antigenic presentation in the presence or absence of viral replication. Remarkably, cell factors involved in innate and adaptive immunity may evolve much quicker than those required for cell survival likely contributing to lentiviral resistance. Intracellular restriction mechanisms described in small ruminants include tripartite motif containing (TRIM5α) and APOBEC3, both leading to proteasomal degradation of viral proteins and therefore sharing the MHC-I antigenic presentation pathway that could result in antibody production in the absence of viral replication. On the other hand, tetherin, which impedes normal virus burden at final steps of the infection cycle, may interfere virus release without affecting proviral load or RNA transcripts. Therefore a future study on SRLV restriction due to intrinsic factors from wild ruminants is warranted.
In summary, our results suggest that cells from the Cervidae family are susceptible to SRLV infection in vitro but factors involved at post-entry steps are controlling infection spread in vivo. These results may indicate that cervids have a bystander role in SRLV epidemiology.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LS carried out the serological screening, PCR and in vitro experiments. HC carried out entry assays and in vitro infections. IG and LBC were involved in LTR promoter activity determinations, macrophage culture and SRLV infection. CMC and EB were involved in the sampling of tissues and in the writing of the manuscript. BA and DA searched for funding resources, coordinated the sampling process and were involved in the writing of the manuscript. GB and RR designed this study and were involved in work supervision and writing of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Funded by CICYT (AGL2010-22341-C04-01 and AGL2013-49137-C3-1-R) and Navarra’s Government (IIQ010449.RI1 and IIQ14064.RI1). L. Sanjosé was a FPI fellow of the Spanish MINECO and R. Reina had contracts from the Public University of Navarra and CSIC. We acknowledge Marta Gil Antona for sampling in hunting expeditions and Hunting Associations (FEDEMCA, Tragsa, INTIA, etc.) for their collaboration in the obtention of samples. We acknowledge support in the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Contributor Information
Leticia Sanjosé, Email: Leticia.sanjose@unavarra.es.
Helena Crespo, Email: Helena.crespo@unavarra.es.
Laure Blatti-Cardinaux, Email: laure.cardinaux@vetsuisse.unibe.ch.
Idoia Glaria, Email: idoia.g@unavarra.es.
Carlos Martínez-Carrasco, Email: cmcpleit@um.es.
Eduardo Berriatua, Email: berriatu@um.es.
Beatriz Amorena, Email: bamorena@unavarra.es.
Damián De Andrés, Email: ancad@unavarra.es.
Giuseppe Bertoni, Email: bertoni@vetsuisse.unibe.ch.
Ramses Reina, Email: ramses.reina@unavarra.es.
References
- 1.Daszak P, Cunningham AA, Hyatt AD. Emerging infectious diseases of wildlife–threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- 2.Ramirez H, Reina R, Amorena B, de Andres D, Martinez HA. Small ruminant lentiviruses: genetic variability, tropism and diagnosis. Viruses. 2013;5:1175–1207. doi: 10.3390/v5041175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cutlip RC, Lehmkuhl HD, Sacks JM, Weaver AL. Prevalence of antibody to caprine arthritis-encephalitis virus in goats in the United States. J Am Vet Med Assoc. 1992;200:802–805. [PubMed] [Google Scholar]
- 4.Ravazzolo AP, Reischak D, Peterhans E, Zanoni R. Phylogenetic analysis of small ruminant lentiviruses from Southern Brazil. Virus Res. 2001;79:117–123. doi: 10.1016/S0168-1702(01)00339-2. [DOI] [PubMed] [Google Scholar]
- 5.L’Homme Y, Ouardani M, Levesque V, Bertoni G, Simard C, Pisoni G. Molecular characterization and phylogenetic analysis of small ruminant lentiviruses isolated from Canadian sheep and goats. Virol J. 2011;8:271. doi: 10.1186/1743-422X-8-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis TM, Robinson WF, Wilcox GE. The pathology and aetiology of lung lesions in goats infected with caprine arthritis-encephalitis virus. Aust Vet J. 1988;65:69–73. doi: 10.1111/j.1751-0813.1988.tb07361.x. [DOI] [PubMed] [Google Scholar]
- 7.Querat G, Audoly G, Sonigo P, Vigne R. Nucleotide sequence analysis of SA-OMVV, a visna-related ovine lentivirus: phylogenetic history of lentiviruses. Virology. 1990;175:434–447. doi: 10.1016/0042-6822(90)90428-T. [DOI] [PubMed] [Google Scholar]
- 8.Tolari F, Al-Ramadneh W, Mazzei M, Carrozza ML, Forzan M, Bandecchi P, Grego E, Rosati S. Small ruminant lentiviruses in Jordan: evaluation of sheep and goat serological response using recombinant and peptide antigens. Trop Anim Health Prod. 2013;45:1335–1340. doi: 10.1007/s11250-013-0366-7. [DOI] [PubMed] [Google Scholar]
- 9.Tageldin MH, Johnson EH, Al-Busaidi RM, Al-Habsi KR, Al-Habsi SS. Serological evidence of caprine arthritis-encephalitis virus (CAEV) infection in indigenous goats in the Sultanate of Oman. Trop Anim Health Prod. 2012;44:1–3. doi: 10.1007/s11250-011-9883-4. [DOI] [PubMed] [Google Scholar]
- 10.Peterhans E, Greenland T, Badiola J, Harkiss G, Bertoni G, Amorena B, Eliaszewicz M, Juste RA, Krassnig R, Lafont JP, Lenihan P, Petursson G, Pritchard G, Thorley J, Vitu C, Mornex JF, Pépin M. Routes of transmission and consequences of small ruminant lentiviruses (SRLVs) infection and eradication schemes. Vet Res. 2004;35:257–274. doi: 10.1051/vetres:2004014. [DOI] [PubMed] [Google Scholar]
- 11.Keen JE, Hungerford LL, Littledike ET, Wittum TE, Kwang J. Effect of ewe ovine lentivirus infection on ewe and lamb productivity. Prev Vet Med. 1997;30:155–169. doi: 10.1016/S0167-5877(96)01101-4. [DOI] [PubMed] [Google Scholar]
- 12.Pekelder JJ, Veenink GJ, Akkermans JP, Van Eldik P, Elving L, Houwers DJ. Ovine lentivirus induced indurative lymphocytic mastitis and its effect on the growth of lambs. Vet Rec. 1994;134:348–350. doi: 10.1136/vr.134.14.348. [DOI] [PubMed] [Google Scholar]
- 13.Arsenault J, Dubreuil P, Girard C, Simard C, Belanger D. Maedi-visna impact on productivity in Quebec sheep flocks (Canada) Prev Vet Med. 2003;59:125–137. doi: 10.1016/S0167-5877(03)00086-2. [DOI] [PubMed] [Google Scholar]
- 14.Martinez-Navalon B, Peris C, Gomez EA, Peris B, Roche ML, Caballero C, Goyena E, Berriatua E. Quantitative estimation of the impact of caprine arthritis encephalitis virus infection on milk production by dairy goats. Vet J. 2013;197:311–317. doi: 10.1016/j.tvjl.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 15.Ryan DP, Greenwood PL, Nicholls PJ. Effect of caprine arthritis-encephalitis virus-infection on milk cell count and N-acetyl-beta-glucosaminidase activity in dairy goats. J Dairy Res. 1993;60:299–306. doi: 10.1017/S0022029900027643. [DOI] [PubMed] [Google Scholar]
- 16.Leitner G, Silanikove N, Merin U. Estimate of milk and curd yield loss of sheep and goats with intrammamary infection and its relation to somatic cell count. Small Rumin Res. 2008;74:221–225. doi: 10.1016/j.smallrumres.2007.02.009. [DOI] [Google Scholar]
- 17.Nagel-Alne GE, Asheim LJ, Hardaker JB, Solverod L, Lindheim D, Valle PS. The Norwegian Healthier Goats programme–a financial cost-benefit analysis. Prev Vet Med. 2014;114:96–105. doi: 10.1016/j.prevetmed.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Shah C, Huder JB, Boni J, Schonmann M, Muhlherr J, Lutz H, Schupbach J. Direct evidence for natural transmission of small-ruminant lentiviruses of subtype A4 from goats to sheep and vice versa. J Virol. 2004;78:7518–7522. doi: 10.1128/JVI.78.14.7518-7522.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glaria I, Reina R, Crespo H, de Andres X, Ramirez H, Biescas E, Perez MM, Badiola J, Lujan L, Amorena B, de Andres D. Phylogenetic analysis of SRLV sequences from an arthritic sheep outbreak demonstrates the introduction of CAEV-like viruses among Spanish sheep. Vet Microbiol. 2009;138:156–162. doi: 10.1016/j.vetmic.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Fras M, Leboeuf A, Labrie FM, Laurin MA, Singh Sohal J, L’Homme Y. Phylogenetic analysis of small ruminant lentiviruses in mixed flocks: multiple evidence of dual infection and natural transmission of types A2 and B1 between sheep and goats. Infect Genet Evol. 2013;19:97–104. doi: 10.1016/j.meegid.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 21.Cardinaux L, Zahno ML, Deubelbeiss M, Zanoni R, Vogt HR, Bertoni G. Virological and phylogenetic characterization of attenuated small ruminant lentivirus isolates eluding efficient serological detection. Vet Microbiol. 2013;162:572–581. doi: 10.1016/j.vetmic.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 22.Hotzel I, Cheevers W. Differential receptor usage of small ruminant lentiviruses in ovine and caprine cells: host range but not cytopathic phenotype is determined by receptor usage. Virology. 2002;301:21–31. doi: 10.1006/viro.2002.1575. [DOI] [PubMed] [Google Scholar]
- 23.Oskarsson T, Hreggvidsdottir HS, Agnarsdottir G, Matthiasdottir S, Ogmundsdottir MH, Jonsson SR, Georgsson G, Ingvarsson S, Andresson OS, Andresdottir V. Duplicated sequence motif in the long terminal repeat of maedi-visna virus extends cell tropism and is associated with neurovirulence. J Virol. 2007;81:4052–4057. doi: 10.1128/JVI.02319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramirez H, Reina R, Bertolotti L, Cenoz A, Hernandez MM, San Roman B, Glaria I, de Andres X, Crespo H, Jauregui P, Benavides J, Polledo L, Perez V, Garcia-Marin JF, Rosati S, Amorena B, de Andres D. Study of compartmentalization in the visna clinical form of small ruminant lentivirus infection in sheep. BMC Vet Res. 2012;8:8. doi: 10.1186/1746-6148-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pisoni G, Moroni P, Turin L, Bertoni G. Compartmentalization of small ruminant lentivirus between blood and colostrum in infected goats. Virology. 2007;369:119–130. doi: 10.1016/j.virol.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 26.Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature. 1989;339:389–392. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 28.Gao F, Bailes E, Robertson DL, Chen YL, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 29.Kaur A, Grant RM, Means RE, McClure H, Feinberg M, Johnson RP. Diverse host responses and outcomes following simian immunodeficiency virus SIVmac239 infection in sooty mangabeys and rhesus macaques. J Virol. 1998;72:9597–9611. doi: 10.1128/jvi.72.12.9597-9611.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soeharsono S, Wilcox GE, Dharma DM, Hartaningsih N, Kertayadnya G, Budiantono A. Species differences in the reaction of cattle to Jembrana disease virus infection. J Comp Pathol. 1995;112:391–402. doi: 10.1016/S0021-9975(05)80020-9. [DOI] [PubMed] [Google Scholar]
- 31.VandeWoude S, Troyer J, Poss M. Restrictions to cross-species transmission of lentiviral infection gleaned from studies of FIV. Vet Immunol Immunopathol. 2010;134:25–32. doi: 10.1016/j.vetimm.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erhouma E, Guiguen F, Chebloune Y, Gauthier D, Lakhal LM, Greenland T, Mornex JF, Leroux C, Alogninouwa T. Small ruminant lentivirus proviral sequences from wild ibexes in contact with domestic goats. J Gen Virol. 2008;89:1478–1484. doi: 10.1099/vir.0.2008/000364-0. [DOI] [PubMed] [Google Scholar]
- 33.Patton KM, Bildfell RJ, Anderson ML, Cebra CK, Valentine BA. Fatal Caprine arthritis encephalitis virus-like infection in 4 Rocky Mountain goats (Oreamnos americanus) J Vet Diagn Invest. 2012;24:392–396. doi: 10.1177/1040638711435503. [DOI] [PubMed] [Google Scholar]
- 34.Guiguen F, Mselli-Lakhal L, Durand J, Du J, Favier C, Fornazero C, Grezel D, Balleydier S, Hausmann E, Chebloune Y. Experimental infection of Mouflon-domestic sheep hybrids with caprine arthritis-encephalitis virus. Am J Vet Res. 2000;61:456–461. doi: 10.2460/ajvr.2000.61.456. [DOI] [PubMed] [Google Scholar]
- 35.Lopez-Olvera JR, Vidal D, Vicente J, Perez M, Lujan L, Gortazar C. Serological survey of selected infectious diseases in mouflon (Ovis aries musimon) from south-central Spain. Eur J Wildl Res. 2009;55:75–79. doi: 10.1007/s10344-008-0215-6. [DOI] [Google Scholar]
- 36.Clark RK, Jessup DA, Kock MD, Weaver RA. Survey of desert bighorn sheep in California for exposure to selected infectious diseases. J Am Vet Med Assoc. 1985;187:1175–1179. [PubMed] [Google Scholar]
- 37.Cutlip RC, Lehmkuhl HD, Brogden KA, Schmerr MJ. Seroprevalence of ovine progressive pneumonia virus in various domestic and wild animal species, and species susceptibility to the virus. Am J Vet Res. 1991;52:189–191. [PubMed] [Google Scholar]
- 38.Chomel BB, Carniciu ML, Kasten RW, Castelli PM, Work TM, Jessup DA. Antibody prevalence of eight ruminant infectious diseases in California mule and black-tailed deer (Odocoileus hemionus) J Wildl Dis. 1994;30:51–59. doi: 10.7589/0090-3558-30.1.51. [DOI] [PubMed] [Google Scholar]
- 39.Sanjose L, Pinczowski P, Crespo H, Perez M, Glaria I, Gimeno M, de Andres D, Amorena B, Lujan L, Reina R. Diagnosing infection with small ruminant lentiviruses of genotypes A and B by combining synthetic peptides in ELISA. Vet J. 2015;204:88–93. doi: 10.1016/j.tvjl.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Cross ML, Thomson AJ, Slobbe LJ, Griffin JF, Buchan GS. Macrophage function in deer. Vet Immunol Immunopathol. 1996;49:359–373. doi: 10.1016/0165-2427(95)05479-0. [DOI] [PubMed] [Google Scholar]
- 41.Sargan DR, Bennet ID, Cousens C, Roy DJ, Blacklaws BA, Dalziel RG, Watt NJ, McConnell I. Nucleotide-sequence of EV1, a British isolate of maedi visna virus. J Gen Virol. 1991;72:1893–1903. doi: 10.1099/0022-1317-72-8-1893. [DOI] [PubMed] [Google Scholar]
- 42.Saltarelli M, Querat G, Konings DAM, Vigne R, Clements JE. Nucleotide sequence and transcriptional analysis of molecular clones of CAEV which generate infectious virus. Virology. 1990;179:347–364. doi: 10.1016/0042-6822(90)90303-9. [DOI] [PubMed] [Google Scholar]
- 43.Andresson OS, Elser JE, Tobin GJ, Greenwood JD, Gonda MA, Georgsson G, Andresdottir V, Benediktsdottir E, Carlsdottir HM, Mantyla EO. Nucleotide sequence and biological properties of a pathogenic proviral molecular clone of neurovirulent visna virus. Virology. 1993;193:89–105. doi: 10.1006/viro.1993.1106. [DOI] [PubMed] [Google Scholar]
- 44.Glaria I, Reina R, Ramirez H, de Andres X, Crespo H, Jauregui P, Salazar E, Lujan L, Perez MM, Benavides J, Perez V, Polledo L, Garcia-Marin JF, Riezu JI, Borras F, Amorena B, de Andres D. Visna/Maedi virus genetic characterization and serological diagnosis of infection in sheep from a neurological outbreak. Vet Microbiol. 2012;155:137–146. doi: 10.1016/j.vetmic.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 45.Reina R, Grego E, Bertolotti L, De Meneghi D, Rosati S. Genome analysis of small-ruminant lentivirus genotype E: a caprine lentivirus with natural deletions of the dUTPase subunit, vpr-like accessory gene, and 70-base-pair repeat of the U3 region. J Virol. 2009;83:1152–1155. doi: 10.1128/JVI.01627-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reina R, Bertolotti L, Dei Giudici S, Puggioni G, Ponti N, Profiti M, Patta C, Rosati S. Small ruminant lentivirus genotype E is widespread in Sarda goat. Vet Microbiol. 2010;144:24–31. doi: 10.1016/j.vetmic.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 47.Juganaru M, Reina R, Bertolotti L, Stella MC, Profiti M, Armentano M, Bollo E, Amorena B, Rosati S. In vitro properties of small ruminant lentivirus genotype E. Virology. 2011;410:88–95. doi: 10.1016/j.virol.2010.10.031. [DOI] [PubMed] [Google Scholar]
- 48.Wolfe ND, Prosser TA, Carr JK, Tamoufe U, Mpoudi-Ngole E, Torimiro JN, LeBreton M, McCutchan FE, Birx DL, Burke DS. Exposure to nonhuman primates in rural Cameroon. Emerg Infect Dis. 2004;10:2094–2099. doi: 10.3201/eid1012.040062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morin T, Guiguen F, Bouzar BA, Villet S, Greenland T, Grezel D, Gounel F, Gallay K, Garnier C, Durand J, Alogninouwa T, Mselli-Lakhal L, Mornex JF, Chebloune Y. Clearance of a productive lentivirus infection in calves experimentally inoculated with caprine arthritis-encephalitis virus. J Virol. 2003;77:6430–6437. doi: 10.1128/JVI.77.11.6430-6437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andresdottir V, Skraban R, Matthiasdottir S, Lutley R, Agnarsdottir G, Thorsteinsdottir H. Selection of antigenic variants in maedi-visna virus infection. J Gen Virol. 2002;83:2543–2551. doi: 10.1099/0022-1317-83-10-2543. [DOI] [PubMed] [Google Scholar]
- 51.de Andres X, Ramirez H, Bertolotti L, San Roman B, Glaria I, Crespo H, Jauregui P, Minguijon E, Juste R, Leginagoikoa I, Perez M, Lujan L, Badiola JJ, Polledo L, Garcia-Marin JF, Riezu JI, Borras-Cuesta F, de Andres D, Rosati S, Reina R, Amorena B. An insight into a combination of ELISA strategies to diagnose small ruminant lentivirus infections. Vet Immunol Immunopathol. 2013;152:277–288. doi: 10.1016/j.vetimm.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 52.Kramsky JA, Manning EJ, Collins MT. Protein G binding to enriched serum immunoglobulin from nondomestic hoofstock species. J Vet Diagn Invest. 2003;15:253–261. doi: 10.1177/104063870301500306. [DOI] [PubMed] [Google Scholar]
- 53.Haflidadottir BS, Matthiasdottir S, Agnarsdottir G, Torsteinsdottir S, Petursson G, Andresson OS, Andresdottir V. Mutational analysis of a principal neutralization domain of visna/maedi virus envelope glycoprotein. J Gen Virol. 2008;89:716–721. doi: 10.1099/vir.0.83410-0. [DOI] [PubMed] [Google Scholar]
- 54.Lyall JW, Solanky N, Tiley LS. Restricted species tropism of maedi-visna virus strain EV-1 is not due to limited receptor distribution. J Gen Virol. 2000;81:2919–2927. doi: 10.1099/0022-1317-81-12-2919. [DOI] [PubMed] [Google Scholar]
- 55.Bruett L, Clements JE. Functional murine leukemia virus vectors pseudotyped with the visna virus envelope show expanded visna virus cell tropism. J Virol. 2001;75:11464–11473. doi: 10.1128/JVI.75.23.11464-11473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elleder D, Kim O, Padhi A, Bankert JG, Simeonov I, Schuster SC, Wittekindt NE, Motameny S, Poss M. Polymorphic integrations of an endogenous gammaretrovirus in the mule deer genome. J Virol. 2012;86:2787–2796. doi: 10.1128/JVI.06859-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sauter M, Roemer K, Best B, Afting M, Schommer S, Seitz G, Hartmann M, MuellerLantzsch N. Specificity of antibodies directed against Env protein of human endogenous retroviruses in patients with germ cell tumors. Cancer Res. 1996;56:4362–4365. [PubMed] [Google Scholar]
- 58.Wang-Johanning F, Radvanyi L, Rycaj K, Plummer JB, Yan P, Sastry KJ, Piyathilake CJ, Hunt KK, Johanning GL. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008;68:5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmarini M, Mura M, Spencer TE. Endogenous betaretroviruses of sheep: teaching new lessons in retroviral interference and adaptation. J Gen Virol. 2004;85:1–13. doi: 10.1099/vir.0.19547-0. [DOI] [PubMed] [Google Scholar]
- 60.Tesoro-Cruz E, Feria-Romero IA, Orozco-Suarez S, Hernandez-Gonzalez R, Silva-Garcia R, Valladares-Salgado A, Bekker-Mendez VC, Blanco-Favela F, Aguilar-Setien A. Frequency of the serological reactivity against the caprine arthritis encephalitis lentivirus gp135 in children who consume goat milk. Arch Med Res. 2009;40:204–207. doi: 10.1016/j.arcmed.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 61.Minardi da Cruz JC, Singh DK, Lamara A, Chebloune Y. Small ruminant lentiviruses (SRLVs) break the species barrier to acquire new host range. Viruses. 2013;5:1867–1884. doi: 10.3390/v5071867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mariante ADS, Egito AA. Animal genetic resources in Brazil: result of five centuries of natural selection. Theriogenology. 2002;57:223–235. doi: 10.1016/S0093-691X(01)00668-9. [DOI] [PubMed] [Google Scholar]
- 63.Chebloune Y, Sheffer D, Karr BM, Stephens E, Narayan O. Restrictive type of replication of ovine/caprine lentiviruses in ovine fibroblast cell cultures. Virology. 1996;222:21–30. doi: 10.1006/viro.1996.0394. [DOI] [PubMed] [Google Scholar]
- 64.Grego E, Bertolotti L, Quasso A, Profiti M, Lacerenza D, Muz D, Rosati S. Genetic characterization of small ruminant lentivirus in Italian mixed flocks: evidence for a novel genotype circulating in a local goat population. J Gen Virol. 2007;88:3423–3427. doi: 10.1099/vir.0.83292-0. [DOI] [PubMed] [Google Scholar]
- 65.Nadin-Davis SA, Chang SC, Smith H, Jacobs RM. Detection of bovine immunodeficiency-like virus by the polymerase chain reaction. J Virol Methods. 1993;42:323–336. doi: 10.1016/0166-0934(93)90043-Q. [DOI] [PubMed] [Google Scholar]
- 66.Lew AE, Bock RE, Miles J, Cuttell LB, Steer P, Nadin-Davis SA. Sensitive and specific detection of bovine immunodeficiency virus and bovine syncytial virus by 5′ Taq nuclease assays with fluorescent 3′ minor groove binder-DNA probes. J Virol Methods. 2004;116:1–9. doi: 10.1016/j.jviromet.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 67.Suarez DL, Whetstone CA. PCR diagnosis of the bovine immunodeficiency-like virus. Methods Mol Biol. 1998;92:67–79. doi: 10.1385/0-89603-497-6:67. [DOI] [PubMed] [Google Scholar]
- 68.Rimstad E, East NE, Torten M, Higgins J, DeRock E, Pedersen NC. Delayed seroconversion following naturally acquired caprine arthritis-encephalitis virus infection in goats. Am J Vet Res. 1993;54:1858–1862. [PubMed] [Google Scholar]
- 69.Zanoni RG, Nauta IM, Kuhnert P, Pauli U, Pohl B, Peterhans E. Genomic heterogeneity of small ruminant lentiviruses detected by PCR. Vet Microbiol. 1992;33:341–351. doi: 10.1016/0378-1135(92)90061-W. [DOI] [PubMed] [Google Scholar]
- 70.Ravazzolo AP, Nenci C, Vogt HR, Waldvogel A, Obexer-Ruff G, Peterhans E, Bertoni G. Viral load, organ distribution, histopathological lesions, and cytokine mRNA expression in goats infected with a molecular clone of the caprine arthritis encephalitis virus. Virology. 2006;350:116–127. doi: 10.1016/j.virol.2006.02.014. [DOI] [PubMed] [Google Scholar]


