Abstract
Objective:
Epigenetic variation may contribute to the development of complex metabolic diseases such as type 2 diabetes (T2D). Hepatic insulin resistance is a hallmark of T2D. However, it remains unknown whether epigenetic alterations take place in the liver from diabetic subjects. Therefore, we investigated the genome-wide DNA methylation pattern in the liver from subjects with T2D and nondiabetic controls and related epigenetic alterations to gene expression and circulating folate levels.
Research Design and Methods:
Liver biopsies were obtained from 35 diabetic and 60 nondiabetic subjects, which are part of the Kuopio Obesity Surgery Study. The genome-wide DNA methylation pattern was analyzed in the liver using the HumanMethylation450 BeadChip. RNA expression was analyzed from a subset of subjects using the HumanHT-12 Expression BeadChip.
Results:
After correction for multiple testing, we identified 251 individual CpG sites that exhibit differential DNA methylation in liver obtained from T2D compared with nondiabetic subjects (Q < .05). These include CpG sites annotated to genes that are biologically relevant to the development of T2D such as GRB10, ABCC3, MOGAT1, and PRDM16. The vast majority of the significant CpG sites (94%) displayed decreased DNA methylation in liver from subjects with T2D. The hypomethylation found in liver from diabetic subjects may be explained by reduced folate levels. Indeed, subjects with T2D had significantly reduced erythrocyte folate levels compared with nondiabetic subjects. We further identified 29 genes that displayed both differential DNA methylation and gene expression in human T2D liver including the imprinted gene H19.
Conclusions:
Our study highlights the importance of epigenetic and transcriptional changes in the liver from subjects with T2D. Reduced circulating folate levels may provide an explanation for hypomethylation in the human diabetic liver.
The prevalence of type 2 diabetes (T2D) is rapidly increasing worldwide, and it is well established that combinations of genetic and nongenetic factors such as obesity, physical inactivity, and aging increase the susceptibility of this complex metabolic disorder. Recent genome-wide association studies (GWAS) were expected to uncover disease-causing mechanisms in T2D. However, the identified genetic variants explain only a modest proportion of the estimated heritability of the disease (1). Additional studies, going beyond the genome, are therefore needed to dissect the molecular mechanisms that contribute to T2D. These investigations may include epigenetic mechanisms such as DNA methylation and histone modifications. Indeed, recent studies from our group and others have identified altered DNA methylation patterns in some of the primary tissues for T2D including pancreatic islets, adipose tissue, and skeletal muscle from diabetic vs nondiabetic subjects (2–6). However, whether the DNA methylation pattern is altered in the liver from subjects with T2D remains unknown.
The liver plays an important role in maintaining glucose homeostasis during both the fed state, when it synthesizes and stores glycogen in response to insulin stimulation, and the fasted state, when gluconeogenesis and the release of glucose take place in response to glucagon stimulation. This fine-tuned balance is lost in subjects with T2D, in whom hepatic insulin resistance contributes to hyperglycemia. Epigenetic changes may potentially alter gene expression in the liver from subjects with T2D and thereby contribute to an impaired metabolism and hyperglycemia. Such epigenetic changes could potentially be induced by a methyl donor supply-consumption imbalance. Dietary components, including folate, serve to modulate the availability of methyl donors for methylation reactions in vivo. Folate can thereby influence the levels of DNA methylation and consequently affect gene expression and cell functions (7).
Although epigenetic studies in livers from subjects with T2D are lacking, DNA methylation changes have been identified in the liver from subjects with obesity and nonalcoholic fatty liver disease (8, 9). Interestingly, obesity has been shown to accelerate epigenetic ageing of human liver, which may partly explain why obese subjects often suffer from an early onset of age-related diseases such as T2D (8). Also, animal studies support a role for epigenetic modifications in hepatic insulin resistance (10, 11).
To identify epigenetic alterations in liver from subjects with T2D, we used a genome-wide approach in which DNA methylation of approximately 455 000 sites was analyzed in the liver from 35 diabetic and 60 nondiabetic subjects. We further tested whether epigenetic changes in the liver were associated with a differential gene expression and altered folate levels in humans.
Research Design and Methods
Study participants and analyses of clinical and metabolic parameters
Liver biopsies obtained from 35 subjects with T2D and 60 nondiabetic controls as a wedge biopsy during a Roux-en-Y gastric bypass operation were included in this study. The subjects are part of the Kuopio Obesity Surgery Study (12–14), and their characteristics are described in Table 1. Diabetes was defined according to the World Health Organization's criteria (15). Plasma glucose and serum insulin were analyzed as described before (12–14). Overall histological assessment of liver biopsy samples was performed by one pathologist according to the standard criteria (16, 17). Histological diagnosis was divided into the following three categories: 1) normal liver without any steatosis, inflammation, ballooning, or fibrosis, 2) simple steatosis (steatosis > 5%) without evidence of hepatocellular ballooning, inflammation, or fibrosis, and 3) nonalcoholic steatohepatitis (NASH). Steatosis was graded into four categories (<5%, 5%–33%, 33%–66%, and > 66%). Of the 60 nondiabetic subjects, 27 (45%) had a normal liver phenotype, 21 (35%) had simple steatosis, and 12 (20%) had NASH. Of the 35 T2D subjects, eight (23%) had a normal liver phenotype, 13 (37%) had simple steatosis, and 14 (40%) had NASH (Pχ2 < .05 comparing liver phenotype distribution between T2D and nondiabetic subjects). The level of lobular inflammation was assessed by histology as previously described (12). Erythrocyte folate levels were measured in 20 of the subjects in the fasted state using an electrochemiluminescense immunoassay (Roche Diagnostics). One measurement was in a normal probability plot defined as an outlier (2.5 SD from the mean) and discarded from the analysis. Vitamin B12 levels were measured in 16 of the subjects using an electrochemiluminescense immunoassay.
Table 1.
Clinical Characteristics of Subjects Included in the Study
| Nondiabetics | Type 2 Diabeticsa | P Valueb | |
|---|---|---|---|
| n (men/women) | 60 (17/43) | 35 (17/18) | |
| Age, y | 48.9 ± 7.9 | 50.5 ± 7.3 | ns |
| BMI, kg/m2 | 43.6 ± 5.5 | 42.2 ± 6.0 | ns |
| fP-glucose, mmol/L | 5.8 ± 0.8 | 7.7 ± 2.9 | 4.1 × 10−6 |
| fS-insulin, mU/L | 16.4 ± 8.7 (n = 42) | 57.8 ± 113.0 (n = 30) | .052 |
Abbreviation: fP, fasting plasma; fS, fasting serum; ns, not significant. Data are shown as mean ± SD.
In type 2 diabetic subjects, 22 (63%) received oral agents (metformin, sitagliptin, glimepiride, or rosiglitazone), 10 (29%) received both insulin and oral agents, and one (3%) received insulin treatment.
Refers to Mann-Whitney statistics.
The study was performed in accordance with the Declaration of Helsinki. Written informed consent was obtained from all participants, and the study protocol was approved by the Ethics Committee of the Northern Savo Hospital District (numbers 54/2005, 104/2008, and 27/2010).
Genome-wide analysis of DNA methylation in human liver
DNA was extracted from human liver biopsies by DNeasy blood and tissue kit (QIAGEN) according to the manufacturer's protocol. Nucleic acid concentration and purity were determined using a Nano Drop 1000 spectrophotometer (NanoDrop Technologies). DNA methylation was analyzed in liver from 35 subjects with T2D and 60 nondiabetic controls using the Infinium HumanMethylation450 BeadChip (Illumina) (18). Five hundred nanograms of genomic DNA were bisulfite treated using the EZ DNA methylation kit (Zymo Research) and subsequently used for analysis of DNA methylation following the Infinium HD assay methylation protocol (Illumina). The BeadChips' images were captured using the Illumina iScan. All included samples showed a high-quality bisulfite conversion efficiency (intensity signal > 4000) and passed all GenomeStudio quality control steps based on built-in control probes for staining, hybridization, extension, and specificity. We filtered away 65 rs-probes, 14 548 cross-reactive probes based on annotation from Chen et al (19), and 10 817 X chromosome and 50 Y chromosome probes as well as 2973 non-CpG probes. Additionally, 1598 individual probes were filtered away based on mean detection values P > .01. The raw methylation data were exported from the GenomeStudio and analyzed using Bioconductor and the lumi/methylumi package, and β-values were converted to M-values (M = log2[β/(1-β)]) (20–22). Data were background corrected and normalized using quantile normalization and β-mixture quantile normalization to correct for probe design bias (23). The DNA methylation array data were corrected for batch effects using COMBAT (24). To easier interpret the results, the M-values were reconverted to β-values, which was used when describing the data and creating the figures.
Genome-wide analysis of gene expression in human liver
Total RNA was extracted from human liver tissue using the miRNeasy minikit (QIAGEN). Nucleic acid concentration and purity were determined using the Nano Drop 1000 spectrophotometer (NanoDrop Technologies). The RNA quality was determined using the Agilent 2100 bioanalyzer (Agilent Technologies). RNA expression was analyzed in the liver from a subset of subjects (19 T2D and 23 nondiabetic subjects), due to the limited size of human liver biopsies, available amounts of liver RNA, and resources. The clinical characteristics of these subjects are presented in Supplemental Table 1 and shows that the subcohort is a representative sample of the full cohort, despite that there was no significant difference in fasting insulin between cases and controls in the subcohort. RNA expression was analyzed using the HumanHT-12 Expression BeadChip (Illumina), which covers 28 688 coding transcripts, according to the manufacturer's recommendations.
Validation of expression data with real-time quantitative PCR (qPCR)
Total RNA was reverse transcribed with a high-capacity cDNA reverse transcription kit (Applied Biosystems). The qPCR was performed with the 7500 Fast real-time PCR system using SYBR Green (Applied Biosystems). The following primer sequences were used for the quantification of RIPK4 mRNA (receptor-interacting serine-threonine kinase 4): forward primer, 5′-GTTAGGCCCACCTTCCAAGA-3′, reverse primer, 5′-GGGGCTTTTCACGTCCAGAT-3′. Gene expression values were normalized to the endogenous expression of RPLP0 mRNA (ribosomal phosphoprotein large P0): forward primer, 5′-GGCGACCTGGAAGTCCAACT-3′, reverse primer, 5′-CCATCAGCACCACAGCCTTC-3′ and analyzed based on the comparative cycle threshold method (2-δδCt). RPLP0 was selected as an endogenous control based on its previous use as a control gene in liver studies and because its expression is stable between subjects with T2D and nondiabetic controls in our microarray expression data from human liver (P = .8 for probe identification 1470349) (25).
Statistical analyses
To identify the differences in DNA methylation and mRNA expression in the liver from diabetic vs nondiabetic subjects, a linear regression model was used including T2D, gender, body mass index (BMI), age, NASH diagnosis, and degree of steatosis as covariates and DNA methylation or mRNA expression as the dependent variable. Variance inflation factors (VIFs), which provides information about potential multicollinearity of studied phenotypes, were calculated (26). To account for multiple testing in the genome-wide analysis of DNA methylation, we applied false discovery rate (FDR) analysis and Q < .05 (FDR < 5%) was considered significant. Spearman correlations were used to relate folate levels and fasting glucose levels, folate levels and average degree of methylation, and RIPK4 mRNA expression and DNA methylation.
Results
Altered DNA methylation in the liver from subjects with T2D
To dissect the epigenetic basis of T2D, we analyzed DNA methylation of 455 526 CpG sites in the liver from 35 diabetic and 60 nondiabetic subjects included in the Kuopio Obesity Surgery Study. The clinical characteristics of these subjects are shown in Table 1. Although there were no significant differences in age or BMI, subjects with diabetes had elevated fasting plasma glucose levels (P = 4.1 × 10−6) and fasting insulin levels (borderline significance, P = .052) compared with nondiabetic subjects (Table 1).
A linear regression model was then used to identify differences in DNA methylation in the liver from diabetic vs nondiabetic subjects. However, to estimate the multicollinearity among the covariates included in the linear regression model, we first calculated the VIF for all covariates (T2D, gender, BMI, age, NASH diagnosis, and degree of steatosis). All calculated VIFs were close to 1 (1.06–1.41), demonstrating that the problem with multicollinearity among these variables is very limited. Based on the linear regression model and after correction for multiple testing using FDR analysis, we identified 251 CpG sites, representing 162 unique genes, with significant differences in DNA methylation in the liver from T2D vs nondiabetic subjects (Q < .05, Supplemental Table 2).
The vast majority of the significant CpG sites (236 sites; 94%) displayed decreased DNA methylation in subjects with T2D (Figure 1A). Because hypomethylation may be explained by a methyl donor supply-consumption imbalance, we analyzed subjects' vitamin B12 and folate levels. There was no significant difference in vitamin B12 levels between T2D (n = 6) and nondiabetic subjects (n = 10) (400.0 ± 191.3 pmol/L vs 372.6 ± 89.9 pmol/L, P = .8). Interestingly, subjects with T2D (n = 8) had significantly reduced erythrocyte folate levels compared with nondiabetic subjects (n = 11) (Figure 1B). Importantly, folate levels correlated negatively with fasting glucose levels already in nondiabetic subjects (ρ = −0.67, P = .026). Additionally, although folate levels correlated significantly with fasting glucose levels when combining nondiabetic and diabetic subjects (ρ = −0.59, P = .008), the correlation was not significant when including only subjects with T2D (ρ = −0.41, P = .32). Although there were no significant correlation between the average degree of methylation of all analyzed sites and folate levels (ρ = −0.25, P = .3), we observed a nominal significant positive correlation between the average degree of methylation of the 236 significant CpG sites with decreased methylation in T2D subjects and folate levels (ρ = 0.45, P = .051).
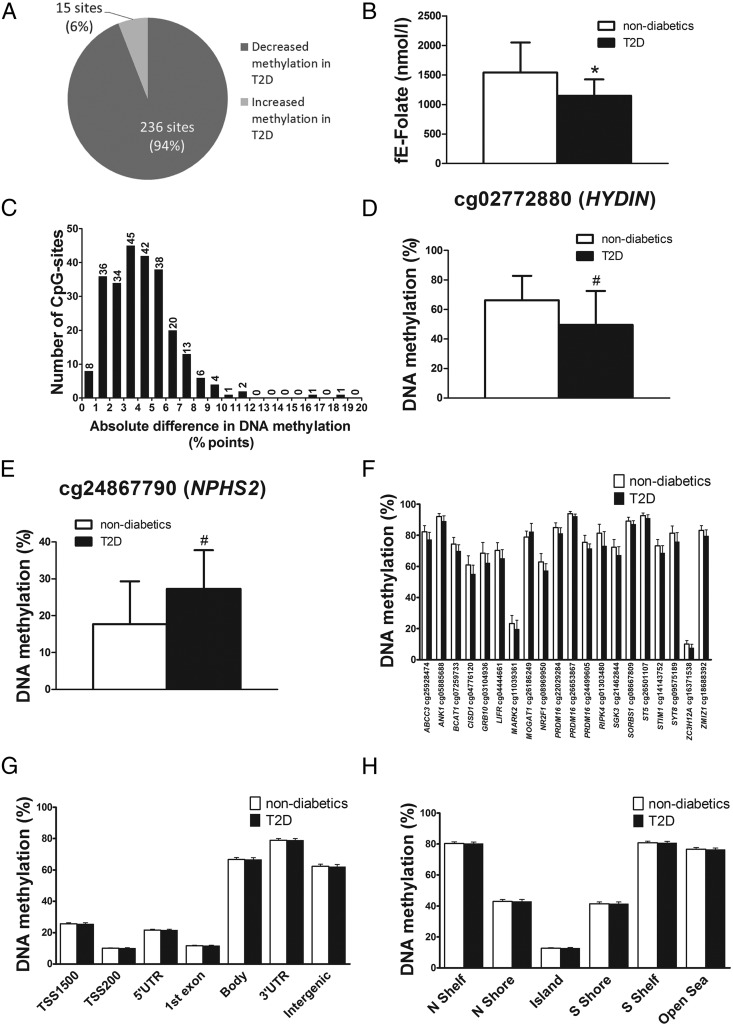
Figure 1.
The human methylome in the liver from 35 T2D and 60 nondiabetic subjects. A, Pie chart describing the number of sites that exhibit increased or decreased DNA methylation in liver from T2D compared with nondiabetic subjects (Q < .05). B, Subjects with T2D (n = 8) have significantly reduced fasting erythrocyte folate levels compared with nondiabetic subjects (n = 11). C, The absolute difference in DNA methylation in liver from T2D compared with nondiabetic subjects (Q < .05). The largest difference in methylation between T2D and nondiabetic subjects was observed for cg02772880 in the 5′UTR of HYDIN (D) and cg24867790 in the TSS1500 of NPHS2 (E). F, Genes previously associated with diabetes that exhibit differential DNA methylation in T2D compared with nondiabetic subjects. Global DNA methylation in human liver from T2D and nondiabetic subjects is shown for each gene region (G) and CpG island regions (H). Global DNA methylation is calculated as the average DNA methylation based on all CpG sites in each annotated region on the Infinium HumanMethylation450 BeadChip. Folate levels and methylation data are presented as mean ± SD. fE, fasting erythrocyte; N, northern; S, southern; Shelf, regions flanking island shores (2000–4000 bp from the CpG island); Shore, flanking region of CpG islands (0–2000 bp); TSS, proximal promoter, defined as 200 or 1500 bp upstream of the transcription start site. #, Q < .05; *, P < .05.
The distribution of the absolute differences in DNA methylation between T2D vs nondiabetic subjects is shown in Figure 1C. Among the significant sites annotated to a protein coding gene, the largest difference in methylation between T2D and nondiabetic subjects was observed for cg02772880 in the 5′-untranslated region (UTR) of HYDIN (encoding hydrocephalus-inducing protein, Figure 1D) and cg24867790 in the TSS1500 of NPHS2 (encoding nephrosis 2, Figure 1E). We next performed a literature search, in which diabetes and each gene name of the 162 identified genes were used as search terms. Here 18 genes previously associated with diabetes were found among the genes that showed differential DNA methylation in the T2D liver (Figure 1F) (27–43). These include GRB10, BCAT1, ANK1, and ZMIZ1 identified through GWAS (32, 35, 38).
Although we adjusted our statistical analysis for NASH, we cannot fully exclude that some of the 251 CpG sites showing altered DNA methylation in the liver from subjects with T2D compared with nondiabetic controls reflect NASH rather than T2D. We therefore looked for an overlap between our 251 CpG sites significantly associated with T2D in liver and CpG sites significantly associated with NASH in human liver in a previous study by Ahrens et al (44). Only 4 of our 251 significant CpG sites were found to also be significant in liver from subjects with NASH compared with controls in this study (marked in Supplemental Table 2). We also looked for an overlap between our 251 CpG sites significantly associated with T2D and CpG sites significantly associated with NASH in the liver samples included in our study. Here 41 of our 251 significant sites were found to be significantly associated with NASH (marked in Supplemental Table 2). Additionally, we performed the analysis restricted to those without NASH (21 T2D and 48 nondiabetic subjects) and restricted to those without NASH or simple steatosis (eight T2D and 27 nondiabetic subjects). Importantly, although reducing the statistical power, all of the 251 CpG sites were differentially methylated in the T2D compared with the nondiabetic subjects when excluding those with NASH, and 151 of the CpG sites were still significant when excluding those with NASH and simple steatosis (Q < .05, Supplemental Table 2). These analyses suggest that most our significant CpG sites are specific for T2D and not linked to NASH. Additionally, because an altered cell composition potentially could affect DNA methylation, we further tested whether the level of lobular inflammation based on histology affects DNA methylation of our 251 CpG sites significantly associated with T2D in the liver. However, none of these CpG sites were differentially methylated based on the level of lobular inflammation in the human liver.
We proceeded to examine the average DNA methylation levels of different genomic regions in human liver from T2D and nondiabetic subjects, either based on their relation to the nearest gene and functional genome distribution (Figure 1G) or in relation to the CpG content (Figure 1H). There were no significant differences in average DNA methylation for these regions between diabetic and nondiabetic subjects. Although the average methylation level was high within the gene body, 3′UTR, and intergenic regions, it was low in the proximal promoter regions, defined as regions 200 or 1500 bp upstream of the transcription start site (TSS200 and TSS1500, respectively), 5′UTR, and the first exon (Figure 1G). Moreover, the average methylation level was low within the CpG islands and intermediate within shores (flanking regions of CpG islands [0–2000 bp]), whereas shelves (regions flanking island shores [2000–4000 bp from the CpG islands]) and open sea showed the highest average methylation levels (Figure 1H).
Altered mRNA expression of genes with differential DNA methylation in the liver from T2D subjects
Epigenetic modifications have been associated with transcriptional regulation, eg, whereas methylation close to transcription sites often correlates negatively with expression, methylation in the gene body may enhance gene elongation and be positively associated with gene expression (45). Therefore, we investigated expression of nearby genes of the 251 CpG sites significantly differentially methylated between T2D and nondiabetic subjects (transcripts with significant CpG sites within the cis distance 500 kb upstream and 100 kb downstream of the gene). We found that 26 CpG sites with significant association between T2D and DNA methylation (Q < .05) had one or more nearby gene(s) in which the liver mRNA expression differed between T2D and nondiabetics (in total 29 genes, P < .05, Table 2). Among these CpG sites, 52% had methylation and expression differences going in the opposite direction, whereas 48% had differences going in the same direction.
Table 2.
Altered mRNA Expression (P < .05) of Genes With Differential DNA Methylation (Q < .05) in the Liver From Subjects With T2D Compared With Nondiabetic Subjects
| Gene Symbol | Probe Identification | Non-T2D, Mean Expression ± SD | T2D, Mean Expression ± SD | Difference in Expression T2D vs non-T2D, % | P Value | CpG Site | Absolute Difference in DNA Methylation T2D vs non-T2D, % Points | Q Value |
|---|---|---|---|---|---|---|---|---|
| KCTD12 | 5560500 | 38.2 ± 14.4 | 51.3 ± 24.6 | 34.3 | .005 | cg01665118 | −4.1 | .025 |
| NUDT15 | 3450370 | 27.3 ± 6.3 | 23.2 ± 8.1 | −14.9 | .006 | cg18026416 | −2.9 | .018 |
| RNF144 | 3450136 | 32.0 ± 12.3 | 43.9 ± 17.3 | 37.1 | .009 | cg23335736 | −2.4 | .023 |
| AP3M2 | 3420128 | 28.8 ± 6.2 | 35.1 ± 8.6 | 21.8 | .013 | cg05885688 | −3.1 | .018 |
| MYH10 | 1770685 | 251.5 ± 63.9 | 275.2 ± 68.1 | 9.4 | .014 | cg24655262 | −4.3 | .018 |
| PPP1R1A | 2750563 | 470.5 ± 164.2 | 362.6 ± 120.8 | −22.9 | .015 | cg16784745 | −4.3 | .028 |
| PLA2G4C | 1990672 | 63.9 ± 11.9 | 86.5 ± 35.2 | 35.4 | .018 | cg26873392 | −1.8 | .044 |
| TMCO7 (TANGO6) | 6330437 | 25.5 ± 8.0 | 22.4 ± 7.8 | −12.2 | .021 | cg07735969 | −4.9 | .018 |
| GOLPH4 (GOLIM4) | 5870221 | 81.4 ± 37.2 | 59.6 ± 39.1 | −26.8 | .022 | cg18142906 | −5.2 | .046 |
| UPF2 | 5390603 | 187.6 ± 53.6 | 221.1 ± 66.8 | 17.9 | .023 | cg23421114 | −6.2 | .031 |
| TICAM1 | 5570730 | 61.0 ± 17.8 | 74.5 ± 19.2 | 22.2 | .024 | cg04410777 | −0.5 | .018 |
| H19 | 5340017 | 1053.5 ± 742.0 | 1267.8 ± 1237.9 | 20.3 | .025 | cg09575189 | −5.5 | .042 |
| ARFGEF1 | 3710681 | 26.0 ± 6.9 | 20.9 ± 7.0 | −19.5 | .026 | cg21462844 | −5.1 | .018 |
| SUMO1P3 | 4050358 | 204.8 ± 39.2 | 172.5 ± 42.6 | −15.8 | .028 | cg19308497 | −2.3 | .048 |
| IL23Ap19 | 6280750 | 26.7 ± 9.3 | 38.4 ± 23.4 | 43.9 | .029 | cg14940636 | 2.2 | .020 |
| ZFP90 | 5900286 | 60.0 ± 10.0 | 69.0 ± 16.0 | 15.0 | .031 | cg07735969 | −4.9 | .018 |
| SLAMF6 | 7200743 | 35.0 ± 15.8 | 46.4 ± 24.0 | 32.6 | .032 | cg19308497 | −2.3 | .048 |
| PRCC | 6520156 | 30.5 ± 9.6 | 27.0 ± 7.0 | −11.6 | .034 | cg05954120 | −3.3 | .023 |
| ELOF1 | 630709 | 37.6 ± 8.7 | 33.5 ± 8.5 | −11.1 | .035 | cg14296903 | −2.0 | .023 |
| CYB561D1 | 1990092 | 59.9 ± 12.0 | 69.5 ± 14.1 | 16.0 | .035 | cg19244300 | −6.5 | .033 |
| RAD50 | 1400253 | 30.3 ± 8.8 | 25.1 ± 8.3 | −17.0 | .037 | cg03046705 | −1.1 | .018 |
| ZNF23 | 4260221 | 31.6 ± 9.5 | 26.0 ± 6.6 | −17.7 | .038 | cg02772880 | −16.8 | .043 |
| FAM173B | 7320368 | 29.6 ± 6.5 | 26.6 ± 5.4 | −10.2 | .038 | cg07429192 | −5.0 | .043 |
| RIPK4 | 5860138 | 37.2 ± 12.7 | 51.9 ± 33.6 | 39.5 | .040 | cg01303480 | −8.3 | .048 |
| cg13520715 | −9.8 | .018 | ||||||
| CCNY | 1340291 | 73.6 ± 15.8 | 66.9 ± 13.1 | −9.1 | .040 | cg16832648 | −1.7 | .044 |
| LOC148413 | 2060541 | 168.0 ± 43.5 | 193.4 ± 31.0 | 15.1 | .043 | cg22627753 | −3.7 | .018 |
| C4orf19 | 670475 | 78.7 ± 19.3 | 67.9 ± 23.1 | −13.7 | .045 | cg23862925 | −4.7 | .018 |
| ZNF295 | 6060326 | 28.6 ± 7.7 | 32.8 ± 8.0 | 15.0 | .048 | cg01303480 | −8.3 | .048 |
| cg13520715 | −9.8 | .018 | ||||||
| VWA1 | 50682 | 36.4 ± 8.1 | 30.9 ± 7.9 | −15.1 | .048 | cg22627753 | −3.7 | .018 |
This analysis was restricted to transcripts with significantly differentially methylated CpG sites within the cis distance 500 kb upstream and 100 kb downstream of the gene.
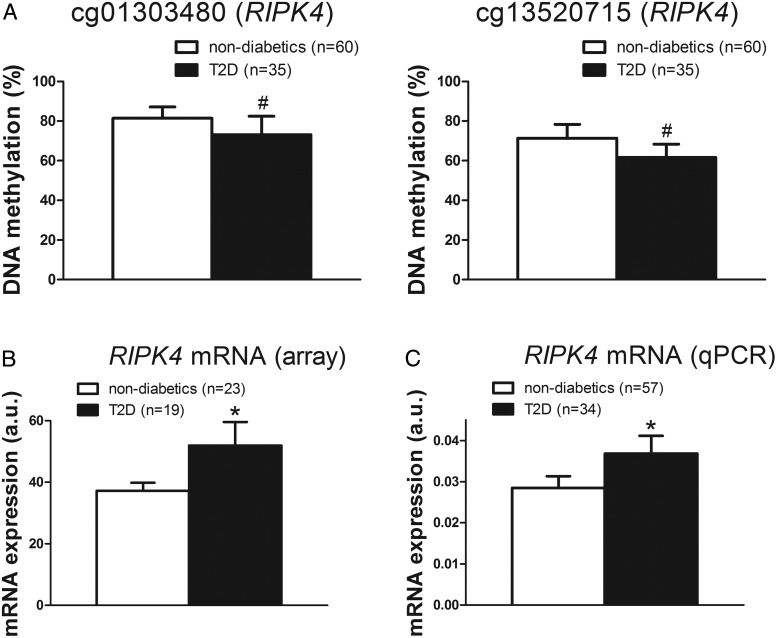
Based on a literature search in which diabetes and each name of the 29 identified genes were used as search terms, we further examined whether genes of potential importance in diabetes were among the differentially expressed transcripts. It should be noted that this second literature search includes some different genes compared with the first literature search because a cis distance was applied in the combined analysis of methylation and expression data, whereas Illumina's annotations were used for the analysis of DNA methylation only. We identified six diabetes-related genes including RIPK4, H19, TICAM1, MYH10, PPP1R1A, and RAD50 (39, 46–50) (Table 2). Among these, RIPK4 displayed the largest difference in DNA methylation and mRNA expression, respectively, in liver from the T2D compared with nondiabetic subjects (Figure 2, A and B). There were nonsignificant negative correlations between the RIPK4 mRNA expression and DNA methylation (ρ = −0.22, P = .16, and ρ = −0.21, P = .18, for cg01303480 and cg13520715, respectively).
Figure 2.
Altered mRNA expression of a gene with differential DNA methylation. For RIPK4, both DNA methylation (A) and mRNA expression (B) differed between T2D subjects and nondiabetic controls. The RIPK4 mRNA expression data were validated with qPCR in the full cohort (C). Data are presented as mean ± SD for methylation and mean ± SEM for expression. a.u., arbitrary units. #, Q < .05; *, P < .05.
To validate our expression microarray data, we further analyzed the expression of RIPK4 with qPCR in the full cohort. The RIPK4 expression could be validated in the full cohort and showed significant associations with T2D in the same direction as the array data (Figure 2C).
We then performed correlations between DNA methylation and expression for the transcripts located in the genomic region around the 251 CpG sites differentially methylated in diabetic liver (transcripts with significant CpG sites within the cis distance 500 kb upstream and 100 kb downstream of the gene). We identified 57 (32 negative and 25 positive) correlations between methylation and expression with P < .05 (Supplemental Table 3).
To assess the accuracy and reproducibility of the microarray experiments, we performed technical replicates. The correlation coefficient for pairwise comparisons of data from two liver RNA samples from the same individual analyzed on two different occasions showed a high reproducibility of Illumina's HumanHT-12 expression array data (r = 0.99, P < 2.2 × 10−16, Supplemental Figure 1).
Discussion
This study describes the methylome in human liver from a large number of obese T2D and nondiabetic subjects. Our data support a role for epigenetic alteration in human liver in T2D. The vast majority of the significantly differentially methylated CpG sites (94%) showed decreased DNA methylation in the liver from the T2D compared with the nondiabetic subjects. Intriguingly, we have previously observed a similar pattern in pancreatic islets, in which 97% of differentially methylated CpG sites showed decreased DNA methylation in diabetic compared with nondiabetic islets (2). This may be explained by changed expression and/or activity of proteins controlling DNA methylation. However, we found no significant difference in the gene expression of DNA methylation regulators such as DNA methyltransferases or ten-eleven translocation enzymes between T2D and nondiabetic subjects in this study (data not shown).
Hypomethylation may also have other explanations such as a methyl donor supply-consumption imbalance (51). Folate is a methyl donor in the methylation cycle, which maintains adequate cellular levels of S-adenosylmethionine for biological methylation reactions, including DNA methylation. Interestingly, we observed significantly reduced circulating folate levels in the T2D compared with the nondiabetic subjects. The reduced folate levels may potentially explain why most significant sites in diabetic subjects are hypomethylated. Indeed, the average DNA methylation of the 236 hypomethylated sites correlated (nominally significant, P = .051) with subjects folate levels. Our human data are in line with a previous rodent study, in which the reduced folate levels were associated with reduced global DNA methylation in the rat liver (52). In addition, folate intake and folate levels have been associated with liver lipid metabolism in animal studies (53–56). It is difficult to draw conclusions about causality in case-control cohorts, and the ideal study design would be to longitudinally assess changes in folate levels during individuals' transition into disease. Nevertheless, we found that folate levels correlated negatively with fasting glucose levels already in nondiabetic subjects, suggesting that reduced circulating folate levels may contribute to the development of T2D. It should further be noted that 89% of subjects with T2D were treated with metformin, and hence, we cannot exclude that this treatment affected the degree of methylation in the liver. Nevertheless, persons with diabetes treated with metformin had no difference in the methylation of the 251 CpG sites identified in this study compared with persons with diabetes without this treatment, suggesting a limited effect of metformin on the degree of methylation of these sites.
In complex polygenic diseases like T2D, it is believed that multiple modest changes may interact to drive a physiological response and disease development. Several affected CpG sites may in combination potentially contribute to diabetes. We identified 251 individual CpG sites that exhibit differential DNA methylation in liver obtained from diabetic compared with nondiabetic subjects. Although many of these sites display absolute differences in methylation less than 5% points between T2D and nondiabetic subjects, 86 individual CpG sites had an absolute difference in methylation of 5%–18.5% points, representing a fold change between 6% and 54% in T2D compared with nondiabetic liver. The differentially methylated sites include CpG sites in genes with previous known functions in liver and/or in the development of T2D such as GRB10, encoding the growth factor receptor-bound protein 10. Common variants in the GRB10 have in GWAS been associated with increased risk of T2D and impaired insulin secretion (38, 57). In the present study, subjects with T2D had lower methylation of GRB10 in liver compared with nondiabetic controls. Interestingly, in a previous study, we found lower DNA methylation of GRB10 in skeletal muscle from first-degree relatives of T2D compared with subjects without any family history of the disease (58).
We have also shown that a risk SNP for elevated glucose/T2D is associated with differential DNA methylation of GRB10 in human pancreatic islets (59). Together these studies support that altered DNA methylation of GRB10 in primary tissues for T2D affect the disease. GRB10 is imprinted in a parent-of-origin fashion in different tissues. Recent gene knockouts in mice have established that Grb10 acts as an inhibitor of intracellular signaling pathways regulating growth and metabolism. Ablation of Grb10 impacts on hepatic glycogen synthesis and probably on glucose homoeostasis (60). Grb10 has also been found to affect the proportions of lean and fat tissue during embryonic development, thereby influencing energy homeostasis in the adult (61).
One should be aware of that this study is performed in very obese subjects and do not necessary reflect T2D in general across the range of BMI. Also, in addition to hepatocytes, liver tissue comprises a mixture of different cell types. Differences in the cell type composition between T2D and nondiabetics could therefore potentially be responsible for some of the observed differences in the DNA methylation and mRNA expression. Based on the histological assessment of liver biopsy samples, simple steatosis or NASH could be detected in several subjects included in this study. Importantly, these phenotypes were taken into consideration when identifying differences in DNA methylation and mRNA expression in the liver from diabetic vs nondiabetic subjects in this study. Only a modest proportion of our significant CpG sites were associated with both T2D and NASH. Additionally, all of the 251 CpG sites were differentially methylated (Q < .05) in T2D compared with nondiabetic subjects when excluding those with NASH. Moreover, the level of lobular inflammation based on histology was not associated with an altered methylation of our significant CpG sites. Therefore, we believe that most our significant results represent independent effects of T2D on DNA methylation in the human liver.
By combining our genome-wide DNA methylation data with transcriptome profiles, we identified 29 genes that exhibit both differential DNA methylation and gene expression in T2D liver. These include H19 and RIPK4, which both showed a decreased methylation and an increased expression in the diabetic liver. H19 is a paternally imprinted gene, and studies in mice have shown that maternal diabetes leads to differential methylation of this gene in the offspring (48). RIPK4 encodes an activator of nuclear factor-κB. Nuclear factor-κB activation results in increased levels of proinflammatory mediators leading to decreased insulin sensitivity in liver (39). It should be noted that the expression data were nominally significant after correction for multiple testing. However, we managed to validate expression data by qPCR in a larger number of samples.
DNA methylation was initially thought to be a silencing mark and increased DNA methylation in promoter regions has been associated with decreased gene expression (45). However, recent studies suggest that DNA methylation also affects additional biological processes such as the expression of noncoding RNAs, transcriptional elongation, splicing events, and the overall genomic stability, which may depend on the genomic location of the methylated CpG sites (45). Therefore, altered DNA methylation in the diabetic liver may affect other biological processes not studied here, and it may be positively associated with gene expression. In fact, we found an altered DNA methylation of CpG sites annotated to miRNAs, including miR-548d2, miR-148b, and miR-30a.
In conclusion, we find that subjects with T2D exhibit epigenetic and transcriptional changes in the liver relevant to the development of the disease together with reduced folate levels. Our data support the role of epigenetic variation in the pathogenesis of T2D and suggest that future studies should test whether folic acid supplementation may prevent and/or treat the disease.
Acknowledgments
We thank Päivi Turunen, Tiina Sistonen, and Matti Laitinen for their technical assistance in the KOBS study. We also thank the Swegene Center for Integrative Biology at Lund University Genomics Facility for help with the DNA methylation and mRNA expression analyses.
Author contributions include the following: E.N. researched the data and wrote/reviewed the manuscript, A.M performed the validation experiments, collected clinical data, and reviewed the manuscript, V.D.d.M. and P.K. collected the clinical data and reviewed the manuscript, A.P. analyzed the data and reviewed the manuscript, J.P. designed the study, collected the clinical data, and reviewed the manuscript, and C.L. designed the study and wrote/reviewed the manuscript. E.N. and C.L. are guarantors of this work and, as such, had full access to all of the data in the study and take responsibility for the integrity of the data.
This work was supported by grants from the Academy of Finland (Contracts 120979, 138006, and 131593), the Finnish Diabetes Research Foundation, the Finnish Cultural Foundation, and the Kuopio University Hospital EVO and VTR funding. This work was also supported by grants from the Swedish Research Council, Region Skåne, the Knut and Alice Wallenberg Foundation, the Novo Nordisk Foundation, the EFSD/Lilly Fellowship, the Söderberg Foundation, The Swedish Diabetes Foundation, the Påhlsson Foundation, EXODIAB, and a Linné grant.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- BMI
- body mass index
- FDR
- false discovery rate
- GWAS
- genome-wide association studies
- NASH
- nonalcoholic steatohepatitis
- qPCR
- quantitative PCR
- T2D
- type 2 diabetes
- UTR
- untranslated region
- VIF
- variance inflation factor.
References
- 1. Groop L, Pociot F. Genetics of diabetes—are we missing the genes or the disease? Mol Cell Endocrinol. 2014;382:726–739. [DOI] [PubMed] [Google Scholar]
- 2. Dayeh T, Volkov P, Salo S, et al. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 2014; 10:e1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nilsson E, Jansson PA, Perfilyev A, et al. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63:2962–2976. [DOI] [PubMed] [Google Scholar]
- 4. Ribel-Madsen R, Fraga MF, Jacobsen S, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS One. 2012;7:e51302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barres R, Osler ME, Yan J, et al. Non-CpG methylation of the PGC-1α promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–198. [DOI] [PubMed] [Google Scholar]
- 6. Volkmar M, Dedeurwaerder S, Cunha DA, et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012;31:1405–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crider KS, Yang TP, Berry RJ, Bailey LB. Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate's role. Adv Nutr. 2012;3:21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Horvath S, Erhart W, Brosch M, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci USA. 2014;111:15538–15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pirola CJ, Gianotti TF, Burgueno AL, et al. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62:1356–1363. [DOI] [PubMed] [Google Scholar]
- 10. Zhang H, Chu X, Huang Y, et al. Maternal vitamin D deficiency during pregnancy results in insulin resistance in rat offspring, which is associated with inflammation and Iκbα methylation. Diabetologia. 2014;57:2165–2172. [DOI] [PubMed] [Google Scholar]
- 11. Chang X, Yan H, Fei J, et al. Berberine reduces methylation of the MTTP promoter and alleviates fatty liver induced by a high-fat diet in rats. J Lipid Res. 2010;51:2504–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mannisto VT, Simonen M, Soininen P, et al. Lipoprotein subclass metabolism in nonalcoholic steatohepatitis. J Lipid Res. 2014;55:2676–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pihlajamaki J, Gronlund S, Simonen M, et al. Cholesterol absorption decreases after Roux-en-Y gastric bypass but not after gastric banding. Metabolism. 2010;59:866–872. [DOI] [PubMed] [Google Scholar]
- 14. Pihlajamaki J, Kuulasmaa T, Kaminska D, et al. Serum interleukin 1 receptor antagonist as an independent marker of non-alcoholic steatohepatitis in humans. J Hepatol. 2012;56:663–670. [DOI] [PubMed] [Google Scholar]
- 15. Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15:539–553. [DOI] [PubMed] [Google Scholar]
- 16. Brunt EM, Janney CG, Di Bisceglie AM, Neuschwander-Tetri BA, Bacon BR. Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am J Gastroenterol. 1999;94:2467–2474. [DOI] [PubMed] [Google Scholar]
- 17. Kleiner DE, Brunt EM, Van Natta M, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. [DOI] [PubMed] [Google Scholar]
- 18. Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. [DOI] [PubMed] [Google Scholar]
- 19. Chen YA, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. [DOI] [PubMed] [Google Scholar]
- 21. Du P, Zhang X, Huang CC, et al. Comparison of β-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Teschendorff AE, Marabita F, Lechner M, et al. A β-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. [DOI] [PubMed] [Google Scholar]
- 25. Bertola A, Bonnafous S, Anty R, et al. Hepatic expression patterns of inflammatory and immune response genes associated with obesity and NASH in morbidly obese patients. PLoS One. 2010;5:e13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fox J, Monette G. Generalized collinearity diagnostics. J Am Stat Assoc. 1992;87:178–183. [Google Scholar]
- 27. Broholm C, Brandt C, Schultz NS, Nielsen AR, Pedersen BK, Scheele C. Deficient leukemia inhibitory factor signaling in muscle precursor cells from patients with type 2 diabetes. Am J Physiol Endocrinol Metab. 2012;303:E283–E292. [DOI] [PubMed] [Google Scholar]
- 28. Ferecatu I, Goncalves S, Golinelli-Cohen MP, et al. The diabetes drug target MitoNEET governs a novel trafficking pathway to rebuild an Fe-S cluster into cytosolic aconitase/iron regulatory protein 1. J Biol Chem. 2014;289:28070–28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gupta RK, Gao N, Gorski RK, et al. Expansion of adult β-cell mass in response to increased metabolic demand is dependent on HNF-4α. Genes Dev. 2007;21:756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hayashi Y, Suemitsu E, Kajimoto K, et al. Hepatic monoacylglycerol O-acyltransferase 1 as a promising therapeutic target for steatosis, obesity, and type 2 diabetes. Mol Ther Nucleic Acids. 2014;3:e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hurov JB, Huang M, White LS, et al. Loss of the Par-1b/MARK2 polarity kinase leads to increased metabolic rate, decreased adiposity, and insulin hypersensitivity in vivo. Proc Natl Acad Sci USA. 2007;104:5680–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Imamura M, Maeda S, Yamauchi T, et al. A single-nucleotide polymorphism in ANK1 is associated with susceptibility to type 2 diabetes in Japanese populations. Hum Mol Genet. 2012;21:3042–3049. [DOI] [PubMed] [Google Scholar]
- 33. Kim SJ, Chae S, Kim H, et al. A protein profile of visceral adipose tissues linked to early pathogenesis of type 2 diabetes mellitus. Mol Cell Proteomics. 2014;13:811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. More VR, Wen X, Thomas PE, Aleksunes LM, Slitt AL. Severe diabetes and leptin resistance cause differential hepatic and renal transporter expression in mice. Comp Hepatol. 2012;11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARγ agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 2012;15:395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Piao G, Saito S, Sun Y, et al. A computational procedure for identifying master regulator candidates: a case study on diabetes progression in Goto-Kakizaki rats. BMC Syst Biol. 2012;6(suppl 1):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rampersaud E, Damcott CM, Fu M, et al. Identification of novel candidate genes for type 2 diabetes from a genome-wide association scan in the Old Order Amish: evidence for replication from diabetes-related quantitative traits and from independent populations. Diabetes. 2007;56:3053–3062. [DOI] [PubMed] [Google Scholar]
- 39. Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116:1793–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Skalniak L, Mizgalska D, Zarebski A, Wyrzykowska P, Koj A, Jura J. Regulatory feedback loop between NF-κB and MCP-1-induced protein 1 RNase. FEBS J. 2009;276:5892–5905. [DOI] [PubMed] [Google Scholar]
- 41. Wang Q, Zhang X, Wang Y, Deng A, Zhu Z, Feng Y. Significance and expression of serum and glucocorticoid-inducible kinase in kidney of mice with diabetic nephropathy. J Huazhong Univ Sci Technolog Med Sci. 2005;25:170–173. [DOI] [PubMed] [Google Scholar]
- 42. Xu Z, Lefevre GM, Felsenfeld G. Chromatin structure, epigenetic mechanisms and long-range interactions in the human insulin locus. Diabetes Obes Metab. 2012;14(suppl 3):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zbidi H, Lopez JJ, Amor NB, Bartegi A, Salido GM, Rosado JA. Enhanced expression of STIM1/Orai1 and TRPC3 in platelets from patients with type 2 diabetes mellitus. Blood Cells Mol Dis. 2009;43:211–213. [DOI] [PubMed] [Google Scholar]
- 44. Ahrens M, Ammerpohl O, von Schonfels W, et al. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 2013;18:296–302. [DOI] [PubMed] [Google Scholar]
- 45. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. [DOI] [PubMed] [Google Scholar]
- 46. da Costa AV, Calabria LK, de Souza Santos P, Goulart LR, Espindola FS. Glibenclamide treatment modulates the expression and localization of myosin-IIB in diabetic rat brain. J Neurol Sci. 2014;340:159–164. [DOI] [PubMed] [Google Scholar]
- 47. Dasu MR, Devaraj S, Park S, Jialal I. Increased toll-like receptor (TLR) activation and TLR ligands in recently diagnosed type 2 diabetic subjects. Diabetes Care. 2010;33:861–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ge ZJ, Liang QX, Hou Y, et al. Maternal obesity and diabetes may cause DNA methylation alteration in the spermatozoa of offspring in mice. Reprod Biol Endocrinol. 2014;12:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Khan S, Raghuram GV, Bhargava A, et al. Role and clinical significance of lymphocyte mitochondrial dysfunction in type 2 diabetes mellitus. Transl Res. 2011;158:344–359. [DOI] [PubMed] [Google Scholar]
- 50. Taneera J, Fadista J, Ahlqvist E, et al. Identification of novel genes for glucose metabolism based upon expression pattern in human islets and effect on insulin secretion and glycemia. Hum Mol Genet. 2015;24:1945–1955. [DOI] [PubMed] [Google Scholar]
- 51. Zhou SS, Zhou YM, Li D, Lun YZ. Dietary methyl-consuming compounds and metabolic syndrome. Hypertens Res. 2011;34:1239–1245. [DOI] [PubMed] [Google Scholar]
- 52. Mejos KK, Kim HW, Lim EM, Chang N. Effects of parental folate deficiency on the folate content, global DNA methylation, and expressions of FRα, IGF-2 and IGF-1R in the postnatal rat liver. Nutr Res Pract. 2013;7:281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Akesson B, Fehling C, Jagerstad M, Stenram U. Effect of experimental folate deficiency on lipid metabolism in liver and brain. Br J Nutr. 1982;47:505–520. [DOI] [PubMed] [Google Scholar]
- 54. Champier J, Claustrat F, Nazaret N, Fevre Montange M, Claustrat B. Folate depletion changes gene expression of fatty acid metabolism, DNA synthesis, and circadian cycle in male mice. Nutr Res. 2012;32:124–132. [DOI] [PubMed] [Google Scholar]
- 55. McNeil CJ, Hay SM, Rucklidge GJ, et al. Disruption of lipid metabolism in the liver of the pregnant rat fed folate-deficient and methyl donor-deficient diets. Br J Nutr. 2008;99:262–271. [DOI] [PubMed] [Google Scholar]
- 56. McNeil CJ, Hay SM, Rucklidge GJ, Reid MD, Duncan GJ, Rees WD. Maternal diets deficient in folic acid and related methyl donors modify mechanisms associated with lipid metabolism in the fetal liver of the rat. Br J Nutr. 2009;102:1445–1452. [DOI] [PubMed] [Google Scholar]
- 57. Prokopenko I, Poon W, Magi R, et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 2014;10:e1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nitert MD, Dayeh T, Volkov P, et al. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61:3322–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Olsson AH, Volkov P, Bacos K, et al. Genome-wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets. PLoS Genet. 2014;10:e1004735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Holt LJ, Siddle K. Grb10 and Grb14: enigmatic regulators of insulin action—and more? Biochem J. 2005;388:393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cowley M, Garfield AS, Madon-Simon M, et al. Developmental programming mediated by complementary roles of imprinted Grb10 in mother and pup. PLoS Biol. 2014;12:e1001799. [DOI] [PMC free article] [PubMed] [Google Scholar]