Abstract
Objective:
Age at menarche (AAM) is determined by the overall duration of endocrine-tissue sex hormone exposure levels. Osteoporosis, the most common metabolic bone disease, is characterized primarily by reduced bone mineral density (BMD) and an increased risk of low trauma fractures. Bone was an endocrine organ regulating the synthesis and secretion of sex steroid hormones. The mutual dependence between bone and gonads underscore the importance of genetic approaches to identify novel pleiotropic genetic factors coregulating BMD and AAM. In this study, we performed a bivariate genome-wide association study (GWAS) to explore novel ethnic common loci and/or genes that may influence both AAM and BMD.
Methods:
We analyzed genotyping data available for 826 unrelated Chinese subjects using genome-wide human genotyping arrays. After quality control, a total of 702 413 single-nucleotide polymorphisms (SNPs) were tested for association using a bivariate linear regression model. The interesting SNPs were replicated in three independent cohorts including 1728 unrelated Caucasians, 709 African-Americans, and 408 Hispanic-Americans.
Results:
We found four SNPs (rs10817638, rs7851259, rs10982287, and rs4979427), located upstream of the ATP6V1G1 gene, were bivariately associated with hip BMD-AAM (P = 4.90 × 10−7, P = 1.07 × 10−6, P = 1.28 × 10−5, and P = 5.42 × 10−5, respectively). These four SNPs were replicated in African-Americans, with corresponding values of P = 1.95 × 10−2, P = 3.18 × 10−2, P = 2.57 × 10−2, and P = 3.64 × 10−2, respectively. rs10817638 and rs10982287 were further replicated in Caucasians (P = 1.76 × 10−2 and P = 9.42 × 10−3, respectively) and Hispanic-Americans (P = 8.37 × 10−3 and P = 1.52 × 10−3, respectively). Meta-analyses yielded stronger association signals for rs10817638 and rs10982287 with combined values of P = 3.02 × 10−9 and P = 3.49 × 10−9, respectively.
Conclusions:
Our study implicated ATP6V1G1 as a novel pleiotropic gene underlying variation of both BMD and AAM. The findings enhance our knowledge of genetic associations between BMD and AAM and provide a rationale for subsequent functional studies of these implicated genes in the pathophysiology of diseases/traits, such as osteoporosis and AAM.
Recent genome-wide association studies (GWASs) have been highly successful at identifying novel genetic factors for many common diseases. A general practice for current GWASs is to collect multiple phenotypes of interest and analyze these phenotypes separately in a univariate framework. However, the univariate GWASs do not consider potential genetic correlation between different traits analyzed, and hence, is difficult to detect pleiotropic genes that are important to the pathogenesis of many correlated human diseases. The bivariate GWAS approach is an effective approach to tackle the challenge of detecting pleiotropic genes because it uses additional information for the correlation between correlated traits.
Osteoporosis, the most common metabolic bone disease in humans, is characterized primarily by reduced bone mineral density (BMD) and increased risk of low trauma fractures (1). BMD is the single best predictor of osteoporotic fractures and is a valuable tool for evaluating the risk of fractures (1). Osteoporosis and its related fractures confer substantive morbidity and mortality, reduction in quality of life, and the enormous financial burden to the affected families and society (2). Annually, more than 9 million osteoporotic fractures occur worldwide (3), and it is estimated that half of the world's fractures will occur in Asia by 2050 (4). The incidence of hip fractures has risen 2- to 3-fold in China over the past 15 years (5).
Age at menarche (AAM) is known to affect the health of women in the latter portion of their lives and is determined by the overall duration of endocrine-tissue sex hormone exposure levels (6). Late AAM is related to higher risk of low BMD and osteoporotic fractures in postmenopausal women (7, 8). A late onset of menarche for women can result in a shorter period of exposure to estrogen, which may partially explain the association between late AAM and low BMD or higher risk of fractures. Because estrogen increases osteoblastogenesis and apoptosis of osteoclasts, it is a pivotal hormonal factor for bone formation and growth in women (9). Recent observations found that bone via osteocalcin, an osteoblast-secreted molecule, was an endocrine organ in turn regulating the synthesis and secretion of sex steroid hormones in one or both genders and energy metabolism (10, 11). The mutual dependence between bone and gonads underscore the importance of genetic approaches to identify novel genetic factors coregulating BMD and AAM.
Previous study demonstrated that AAM is significantly related to BMD (12). Guo et al (13) reported significant phenotypic correlations between AAM and BMD at different skeletal sites and further classified these correlations into genetic vs environmental correlations. Genetic correlations between AAM and BMD at the lumbar spine and the femoral neck of the hip were −0.132 and −0.142, respectively. In contrast, environmental correlations were not significant (13), implying that the phenotypic correlation was due primarily to shared genetic factors rather than to environmental factors. A bivariate genome linkage study discovered two important regions, 22q13 and 3p25, which may harbor promising candidate genes influencing both AAM and BMD (14). It has also been recently reported that carriers of a rare nonsense mutation (c, 376C>T) in the leucine-rich-repeat-containing G-protein-coupled receptor 4 (LGR4) gene were associated with a higher risk of low BMD, osteoporosis fractures, and late AAM (15). However, despite these findings of new pleotropic loci affecting both BMD and AAM, they explain only a small component of the genetic correlation between BMD and AAM, indicating that many additional shared genetic factors affecting these two traits await identification.
Both AAM and BMD are highly heritable traits with heritability exceeding 0.5 (6, 16). Thus far, a total of 62 genetic loci were found to be robustly associated with osteoporosis-related traits (17) and 106 genomic loci were associated with AAM in multiple populations (18, 19) using a univariate GWAS. Bivariate GWASs are advantageous for detecting pleiotropic genes; however, we are not aware of any published bivariate GWASs for BMD and AAM. In this study, we report the first bivariate GWAS for BMD and AAM in a sample of 826 unrelated Chinese female subjects followed by replication analyses in three independent cohorts including 1728 unrelated Caucasians, 709 African-Americans, and 408 Hispanic-Americans. We identified gene ATP6V1G1 (ATPase, H+ transporting, lysosomal 13 kDa, V1 subunit G1) that was associated with both BMD and AAM in multiple ethnic populations, suggesting their roles in coregulating BMD and AAM.
Materials and Methods
Study populations
The study was performed in two stages (discovery stage and replication stage) including multiple samples from different research and/or clinical centers. All samples were approved by the institutional review board or research administration of the institutions involved. Signed informed consent documents were obtained from all study participants before entering the study.
In the discovery stage, 826 unrelated Chinese female subjects were recruited from southwest Chinese Han adults (living in Changsha City and its neighboring areas) and northwest Chinese Han adults (living in Xi'an City and its neighboring areas). Subjects with diseases that might potentially affect bone mass, structure, or metabolism were excluded from the study as detailed previously (20).
In the replication stage, three study samples were included. The Kansas City Osteoporosis Study (KCOS) sample contained 1728 unrelated Caucasian female subjects recruited in the Midwestern United States (Kansas City, MO). The other two samples were selected from the Women's Health Initiative (WHI) observational study, a partial factorial randomized and longitudinal cohort with more than 12 000 genotyped women of African-American or Hispanic ancestry (21). The African-American ancestry (WHI-AA) and Hispanic ancestry (WHI-HIS) samples included 709 and 408 individuals, respectively, with BMD and AAM information. The WHI data sets used for the analyses described in this manuscript were obtained from the database of Genotypes and Phenotypes (dbGaP) (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap) through dbGaP accession phs000200.v6.p2.
Phenotype measurement
Areal BMD (grams per square centimeter) at the lumbar spine (L1–4, anteroposterior view) and the hip, including the femoral neck, trochanter, and intertrochanter, were measured using the Hologic QDR 4500 W bone densitometer (Hologic). The total hip BMD was a combined value at the three measured regions. The densitometer was calibrated daily, and long-term precision was monitored with the control vertebral phantom. The coefficient of variation of measured BMD values was 1.01% and 1.34% at the spine and the total hip, respectively.
AAM is defined as the age at the first menstrual period (years) and calculated as the date of menarche after the onset of menses minus the date of birth. The data on AAM were collected by a detailed nurse-administered questionnaire applied for each subject and were self-reported using a direct AAM recall method. The medical history of hormone replacement therapy and menopause age was also recorded to assess whether each subject was exposed to nonphysiological hormone concentrations. No subject had primary amenorrhea.
Genotyping and quality control
DNA was extracted using specimens collected at the time of enrollment. Genetic data of the four samples were obtained from genome-wide scans using the Genome-wide Human SNP Array 6.0 (Affymetrix, www.affymetrix.com) containing 909 622 single-nucleotide polymorphisms (SNPs) and following the manufacturer's protocol. Genotyping quality control included examination of concordance rates for blinded and unblinded duplicates. SNPs with call rates less than 95%, concordance rates less than 98%, or minor allele frequency (MAF) less than 1% were excluded. After these exclusions, 702 413 SNPs in the discovery sample, 760 794 in the KCOS sample, and 871 309 in the WHI samples were available for association analysis. For details of the genotyping method, please refer to the previous study (22).
Population stratification
To correct for a potential stratification that may lead to spurious association results, principal components analysis implement in EIGENSTART (23) was used to estimate population substructure. We applied the principal components analysis to all available genotypic data, retaining the top 10 principal components. The top 10 principal components along with height, weight, and age2 were included as covariates to adjust raw phenotypes and generate standardized residuals with SPSS linear regression analyses before subsequent association analyses.
Statistical analysis
Although previous studies have reported that BMD and AAM are the two related phenotypes (13, 14), we reestimated the phenotypic correlation between BMD and AAM in our Chinese population by the bivariate correlation analysis in SPSS.
We performed bivariate GWASs to detect associations between each SNP and two phenotypes. An additive genetic model was applied to both univariate and bivariate association analyses. Based on a linear model, bivariate regression analyses were conducted using the R software (available at http://www.r-project.org). The analysis was based on the following formula:
In this model, for an individual i, yi is a vector of the two traits; μ is the ground mean vector; Z = (Z1, Z2, …, Zn) is a vector of covariates that may include risk factors and confounding factors; βs are the effects of covariates or the SNP under test; xi is the genotype score for individual i, and ϵ is the vector of random error. We compared the likelihood of the model under the null hypothesis (the SNP effects are restricted to 0) with that under the alternative hypothesis (the SNP effects are not 0) to test the alternative hypothesis. Then the likelihood ratio can convert to an F-statistic, which follows an F-distribution under the null hypothesis. The value of P = 5 × 10−8 was used as the threshold to claim significant associations at the genome-wide level. The candidate SNPs (a total of 19 SNPs) were replicated in three independent samples. For replication, due to the prior evidence of association, we used a threshold of the significant level of P = 2.63 × 10−3 (0.05/19). The level of P = .05 was used as a threshold of nominal significance.
Individual P values achieved in the four studied samples were combined by conventional meta-analysis with weights proportional to the square root of the sample size using METAL software (http://www.sph.umich.edu/csg/abecasis/metal/, version generic-metal-2011-03-25.tar.gz) (24).
For the SNPs showing potential pleiotropic effects, we further investigated the regression coefficients (β) for each of the SNPs for the univariate and performed conditional analysis. The β-value of the SNPs was evaluated using a linear regression model implemented in PLINK (25). The conditional analysis was examined by comparing adjusted/conditional models in bivariate linear regression analyses, in which the genotype of each of the reported SNPs was adjusted as a covariate in turn.
We also performed univariate association analysis with each of the tested traits using PLINK (25) in the discovery samples to compare the results between bivariate and univariate analysis. Regional association plots of the most significant SNPs were generated using LocusZoom (26).
To calculate the statistical power of the bivariate association analyses of two continuous traits with that of univariate association analyses of each trait separately, we performed power analyses using the Generalized Estimation Equation Package implemented in the R environment (https://cran.r-project.org/web/packages/gee/index.html) for genotype-based bivariate association analyses. We used ANOVA in R and performed power analyses of genotype-based univariate association analyses. The power analyses were based on the sample sizes of 1728, 826, 709, and 408 unrelated subjects, respectively, as used in the present study for the discovery and replication analyses under additive genetic models. One thousand replicates were run in simulation to calculate the power.
We used the F-SNP program (http://compbio.cs.queensu.ca/F-SNP/) to explore potential functions of the reported SNPs.
Results
The basic characteristics of the samples are summarized in Table 1. Hip and spine BMD were lower in Chinese than in Caucasians and African Americans. Chinese females had later AAM compared with Caucasians, Hispanics, and African Americans.
Table 1.
Basic Characteristics of the Studied Samples
| Traits | Chinese Sample (n = 826) | KCOS (n = 1728) | WHI-AA (n = 709) | WHI-HIS (n = 408) |
|---|---|---|---|---|
| Age, y | 37.64 (13.84) | 51.26 (12.83) | 60.90 (6.9) | 60.70 (7.2) |
| Height, m | 1.58 (0.05) | 1.63 (0.06) | 1.62 (0.06) | 1.57 (0.06) |
| Weight, kg | 54.66 (8.18) | 71.36 (15.89) | 83.15 (17.72) | 73.87 (15.62) |
| AAM, y | 13.91 (1.61) | 12.92 (1.59) | 12.56 (1.64) | 12.60 (1.67) |
| Hip BMD, g/cm2 | 0.86 (0.11) | 0.94 (0.14) | 0.95 (0.15) | 0.86 (0.13) |
| Spine BMD, g/cm2 | 0.92 (0.12) | 1.01 (0.15) | 1.05 (0.17) | 0.97 (0.16) |
Data were presented as mean (SD).
A correlation analysis of our phenotype data showed that AAM was significantly correlated with hip and spine BMD in the Chinese sample, with correlation coefficients −0.112 and −0.157 (P < .001) for hip BMD-AAM and spine BMD-AAM, respectively. The significant correlations observed here are consistent with previous findings (13).
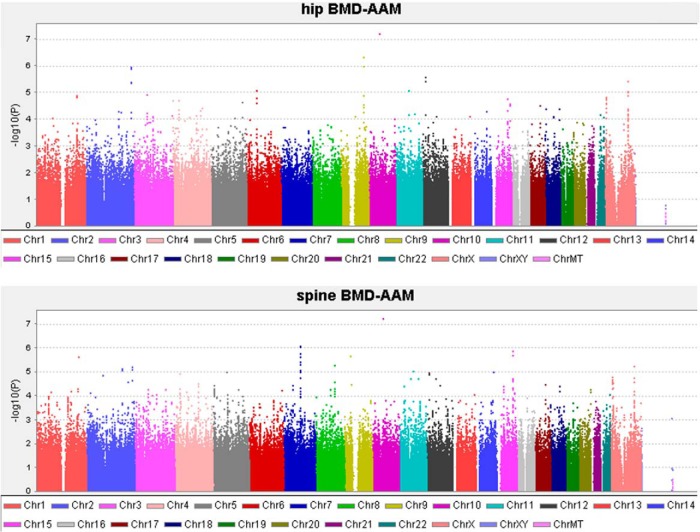
Figure 1 shows the distribution of P values of bivariate GWASs across the genome in the Chinese sample. We did not find any significant SNPs with P < 5 × 10−8. Therefore, we listed the top 10 SNPs for hip BMD-AAM and the top 10 SNPs for spine BMD-AAM in Table 2. The strongest association was found at SNP rs1930170 located in the intron of the PCDH15 (protocadherin related 15) gene, with bivariate values of P = 6.12 × 10−8 and P = 6.09 × 10−8 for spine BMD-AAM and hip BMD-AAM, respectively. However, this SNP failed to replicate in the three independent samples. Among the top 10 SNPs for spine BMD-AAM, four SNPs (rs10264856, rs7795817, rs4728709, and rs2157926) located in the ABCB1 gene were nominally associated with spine BMD-AAM. Among the top 10 SNPs for hip BMD-AAM, two SNPs (rs10817638 and rs7851259) upstream from the ATP6V1G1 gene and three SNPs on chromosome 2 (without a gene annotation) were nominally associated with hip BMD-AAM.
Figure 1.
Distribution of P values across the genome for bivariate association analyses in the Chinese sample. P values are plotted according to their physical positions on successive chromosomes.
Table 2.
Results of Bivariate Association for AAM and BMD in the Discovery Sample (Top 10 SNPs for Each Traits Pair)
| Traits Pair | SNP | Chromosome | Position | Gene | MAF | P Value |
|---|---|---|---|---|---|---|
| Hip BMD-AAM | rs1930170 | 10 | 56217752 | PCDH15 | 0.36 | 6.09E-08 |
| rs10817638 | 9 | 117322542 | ATP6V1G1 | 0.41 | 4.90E-07 | |
| rs7851259 | 9 | 117325336 | ATP6V1G1 | 0.43 | 1.07E-06 | |
| rs7598908 | 2 | 229297832 | 0.16 | 1.13E-06 | ||
| rs1469181 | 2 | 229269825 | 0.16 | 1.18E-06 | ||
| rs1863073 | 2 | 229277943 | 0.16 | 1.21E-06 | ||
| rs10846260 | 12 | 16124142 | DERA | 0.17 | 2.76E-06 | |
| rs3782545 | 12 | 16124793 | DERA | 0.17 | 2.76E-06 | |
| rs2429676 | X | 120850321 | 0.06 | 3.79E-06 | ||
| rs802075 | 6 | 49654166 | CRISP2 | 0.22 | 8.59E-06 | |
| Spine BMD-AAM | rs1930170 | 10 | 56217752 | PCDH15 | 0.28 | 6.12E-08 |
| rs10264856 | 7 | 87100517 | ABCB1 | 0.43 | 8.44E-07 | |
| rs7795817 | 7 | 87006973 | ABCB1 | 0.21 | 9.03E-07 | |
| rs10232457 | 7 | 87300451 | RUNDC3B | 0.50 | 9.41E-07 | |
| rs3893384 | 15 | 77760110 | 0.37 | 1.35E-06 | ||
| rs4728709 | 7 | 87071538 | ABCB1 | 0.14 | 1.79E-06 | |
| rs8041943 | 15 | 79966600 | 0.14 | 2.12E-06 | ||
| rs10738889 | 9 | 32458053 | DDX58 | 0.38 | 2.16E-06 | |
| rs2157926 | 7 | 87270500 | ABCB1 | 0.14 | 2.38E-06 | |
| rs1697621 | 1 | 211081181 | KCNH1 | 0.13 | 2.44E-06 |
Next, we focused on the confirmation of the significance of the three genes/regions (ATP6V1G1, ABCB1, and chromosome 2). We selected nine ABCB1 SNPs (rs7779562, rs2235047, rs4148745, rs7787082, rs10236274, rs7795817, rs4728709, rs10264856, and rs2157926), six ATP6V1G1 SNPs (rs10817638, rs7851259, rs10982287, rs4979427, rs4979428, and rs4979429), and four SNPs on chromosome 2 (rs1469181, rs1863073, rs7598908, and rs11673816) with values of P < 1 × 10−4 (ranging from 4.90 × 10−7 to 8.51 × 10−5) and replicated them in the three independent samples.
Replication analyses confirmed the association of four ATP6V1G1 SNPs with hip BMD-AAM, one ABCB1 SNP, and one SNP on chromosome 2 with spine BMD-AAM (Table 3). The four ATP6V1G1 SNPs (rs10817638, rs7851259, rs10982287, and rs4979427) were in strong linkage disequilibrium (LD) (r2 > 0.85) with each other. Among them, rs10817638 and rs10982287 showed consistent associations with hip BMD-AAM in all samples. Bivariate values for rs10817638 and rs10982287 were P = 4.90 × 10−7 and P = 1.28 × 10−5 in the discovery sample, P = 1.76 × 10−2 and P = 9.42 × 10−3 in Caucasians, P = 8.37 × 10−3 and P = 2.57 × 10−2 in Hispanic-Americans, and P = 1.95 × 10−2 and P = 1.52 × 10−3 in African-Americans, respectively. Meta-analyses yielded stronger associations, with combined values of P = 3.02 × 10−9 and P = 3.49 × 10−9 for rs10817638 and rs10982287, respectively. The other two SNPs (rs7851259 and rs4979427) were replicated only in Hispanic-Americans with replication values of P = 3.18 × 10−2 and P = 3.64 × 10−2, respectively. The four ATP6V1G1 SNPs showed no association signals with the spine BMD-AAM in the Chinese, suggesting that the associations of these SNPs were skeletal site specific.
Table 3.
Results of Bivariate Association for BMD and AAM for the Six Replicated SNPs
| Traits Pair | SNP | Chromosome | Position | Gene | Allele | Chinese Sample P/MAF | KCOS P/MAF | WHI-AA P/MAF | WHI-HIS P/MAF | Combined P Value |
|---|---|---|---|---|---|---|---|---|---|---|
| Spine BMD-AAM | rs10817638 | 9 | 117322542 | ATP6V1G1 | G/A | 1.97E-01/0.41 | 7.65E-02/0.35 | 7.65E-02/0.46 | 8.85E-03/0.36 | 1.71E-05 |
| rs7851259 | 9 | 117325336 | ATP6V1G1 | G/A | 5.18E-02/0.43 | 1.93E-01/0.46 | 1.93E-01/0.50 | 9.01E-02/0.50 | 2.88E-03 | |
| rs10982287 | 9 | 117314215 | ATP6V1G1 | A/G | 2.14E-01/0.42 | 8.23E-02/0.33 | 8.23E-02/0.34 | 1.64E-03/0.45 | 8.68E-06 | |
| rs4979427 | 9 | 117323426 | ATP6V1G1 | C/T | 1.78E-01/0.47 | 1.06E-01/0.29 | 1.06E-01/0.37 | 9.76E-01/0.29 | 3.93E-02 | |
| rs7787082 | 7 | 86994987 | ABCB1 | A/G | 6.38E-05/0.31 | 7.00E-03/0.42 | 6.40E-01/0.54 | 7.28E-01/0.23 | 6.05E-05 | |
| rs1469181 | 2 | 229269825 | T/G | 6.13E-06/0.38 | 3.24E-01/0.16 | 6.52E-02/0.30 | 5.45E-01/0.40 | 1.29E-04 | ||
| Hip BMD-AAM | rs10817638 | 9 | 117322542 | ATP6V1G1 | G/A | 4.90E-07/0.41 | 1.76E-02/0.35 | 1.95E-02/0.46 | 8.37E-03/0.36 | 3.02E-09 |
| rs7851259 | 9 | 117325336 | ATP6V1G1 | G/A | 1.07E-06/0.43 | 4.76E-01/0.46 | 3.18E-02/0.50 | 8.93E-02/0.50 | 1.48E-05 | |
| rs10982287 | 9 | 117314215 | ATP6V1G1 | A/G | 1.28E-05/0.42 | 9.42E-03/0.33 | 2.57E-02/0.34 | 1.52E-03/0.45 | 3.49E-09 | |
| rs4979427 | 9 | 117323426 | ATP6V1G1 | C/T | 5.42E-05/0.47 | 7.65E-01/0.29 | 3.64E-02/0.37 | 9.16E-01/0.29 | 4.56E-04 | |
| rs7787082 | 7 | 86994987 | ABCB1 | A/G | 9.63E-01/0.31 | 1.02E-02/0.42 | 4.61E-01/0.54 | 6.91E-01/0.23 | 2.63E-02 | |
| rs1469181 | 2 | 229269825 | T/G | 1.18E-06/0.38 | 3.04E-02/0.16 | 2.12E-01/0.30 | 5.52E-01/0.40 | 6.81E-06 |
rs7787082 in the ABCB1 gene showed moderate association with spine BMD-AAM in the discovery sample (P = 6.38 × 10−5) and was replicated in Caucasians with a value of P = 7.00 × 10−3. However, rs7787082 was not associated with hip BMD-AAM. SNP rs1469181 on chromosome 2 showed nominal association signals with spine BMD-AAM in the discovery sample (P = 1.18 × 10−6) and Caucasians (P = 3.04 × 10−2). rs1469181 also showed suggestive association with hip BMD-AAM (P = 6.13 × 10−6) in the discovery sample.
To explore whether the six replicated SNPs were at increased risk for late menarche and low BMD, we calculated the β-values of the six SNPs using the univariate association analyses approach in Supplemental Table 1. For the 4 ATP6V1G1 SNPs, the minor allele is associated with high BMD value and early menarche. For the ABCB1 rs7787082 and rs1469181 on chromosome 2, the minor allele is associated with low BMD value and late menarche. For the SNP rs10817638 with most strong association signal, we calculated raw BMD and AAM values for groups that are stratified by different alleles. Subjects with the G allele of rs10817638 in ATP6V1G1 had lower mean AAM and higher hip BMD and spine BMD values than those with the A allele (raw AAM, hip BMD, and spine values were 13.91 y, 0.872 g/cm2, 0.920 g/cm2 vs 13.92 y, 0.856 g/cm2, 0.914 g/cm2 for G allele vs A allele carriers).
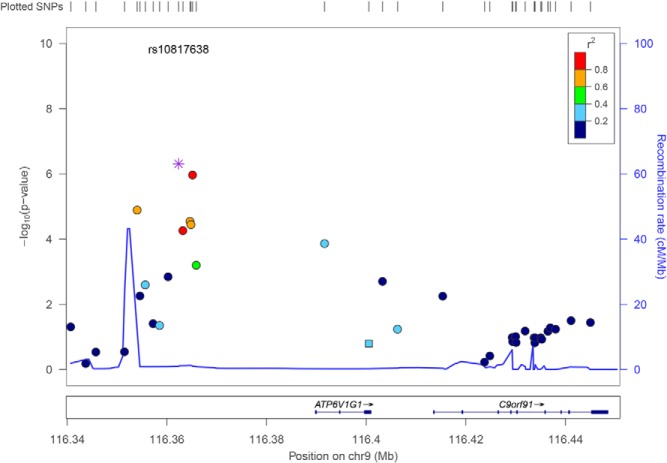
Using each SNP as a covariate, we performed conditional analyses on the remaining five SNPs (Table 4). It can be seen that for the four ATP6V1G1 SNPs, when the genotype of one SNP was used as a covariate, there is significant drop of association signals for hip BMD-AAM in the conditional analysis as compared with the regular association analysis, suggesting that the association signals between these four SNPs are highly correlated. The correlation of the association signals between these four SNPs is primarily due to the linkage disequilibrium between them (Figure 2). However, there is no ATP6V1G1 SNP that, when used as a covariate for conditional analysis, had made the association signals disappear for all the remaining three SNPs, suggesting that none of the four SNPs can explain on its own the association signals for all of the other three SNPs. When the genotypes of SNPs located in different genes were used as a covariate, the association signals remained, suggesting that ATP6V1G1, ABCB1, and chromosome 2 are independent genetic loci.
Table 4.
The Conditional Analysis of the Six SNPs for the Two Trait Pairs in the Discovery Sample
| Trait Pair | Covariate | Gene | rs10817638 | rs7851259 | rs10982287 | rs4979427 | rs7787082 | rs1469181 |
|---|---|---|---|---|---|---|---|---|
| Spine BMD-AAM | rs10817638 | ATP6V1G1 | 2.32E-01 | 4.45E-01 | 3.26E-01 | 6.43E-01 | 3.64E-01 | |
| rs7851259 | ATP6V1G1 | 4.95E-01 | 2.80E-01 | 2.13E-01 | 4.18E-01 | 1.33E-01 | ||
| rs10982287 | ATP6V1G1 | 5.70E-01 | 3.15E-01 | 4.17E-01 | 6.11E-01 | 2.99E-01 | ||
| rs4979427 | ATP6V1G1 | 5.01E-01 | 3.60E-01 | 2.95E-01 | 5.70E-01 | 3.49E-01 | ||
| rs7787082 | ABCB1 | 2.71E-01 | 8.48E-02 | 1.87E-01 | 2.34E-01 | 2.56E-04 | ||
| rs1469181 | 8.99E-05 | 6.59E-04 | 5.47E-05 | 4.99E-05 | 1.30E-03 | |||
| Hip BMD-AAM | rs10817638 | ATP6V1G1 | 3.44E-02 | 7.59E-02 | 5.92E-02 | 3.70E-05 | 2.69E-06 | |
| rs7851259 | ATP6V1G1 | 2.19E-02 | 8.66E-02 | 1.39E-01 | 4.92E-05 | 8.45E-06 | ||
| rs10982287 | ATP6V1G1 | 2.14E-03 | 8.19E-03 | 2.55E-02 | 1.74E-05 | 5.85E-05 | ||
| rs4979427 | ATP6V1G1 | 4.09E-03 | 2.21E-02 | 1.65E-02 | 3.52E-05 | 1.95E-04 | ||
| rs7787082 | ABCB1 | 7.83E-06 | 2.22E-05 | 1.30E-04 | 2.65E-04 | 9.92E-01 | ||
| rs1469181 | 3.68E-05 | 1.51E-04 | 1.09E-05 | 2.41E-05 | 5.50E-04 |
Figure 2.
Regional plots of the ATP6V1G1 gene. SNPs are plotted by position on the chromosome against bivariate association (−log10 P value). Recombination rates (from HapMap) are plotted in blue to reflect the local LD structure. The SNPs surrounding the most significant SNP (in purple) are color coded to reflect their LD with this SNP (r2 values from the 1000 Genomes Mar 2012 ASN data).
We also performed a univariate association analysis for the six confirmed SNPs in the discovery sample. As demonstrated in Supplemental Table 2, the four ATP6V1G1 SNPs (rs10817638, rs7851259, rs10982287, and rs4979427) showed nominal associations with hip BMD (P = 9.21 × 10−5 to P = 1.22 × 10−3) and no associations with AAM and spine BMD. Similarly, rs7787082 in the ABCB1 gene showed only a weak association with spine BMD (P = 1.20 × 10−2) and no associations with AAM and hip BMD. rs1469181 showed suggestive association with AAM (P = 6.81 × 10−3) and no associations with spine BMD and hip BMD. The results of the univariate association analyses were less significant than those of bivariate analyses.
According to our power analyses, analyzing two traits simultaneously using a bivariate association approach consistently achieved higher statistical power than analyzing each trait separately using a univariate association approach (Supplemental Figure 1). For example, the power to detect a genetic locus of a heritability of 0.01 is 73.5% using univariate an association analysis approach, as compared with a 91% power using bivariate association analysis approach in our discovery Chinese sample. The higher power of bivariate analyses over the univariate analyses is derived from the fact that the bivariate analyses use the information from both of the two related traits and hence can detect with higher power those significant loci missed in the univariate analyses. The power analyses also showed that our study had moderate to high power to detect a pleiotropic genetic locus. For example, the power to detect a locus of a heritability of 0.01 is 91% using a bivariate association analysis approach in our discovery Chinese sample, 99.7% in the KCOS sample, 86.4% in the WHI-AA sample, and 64.8% in the WHI-HIS sample, respectively (Supplemental Figure 1).
Using the F-SNP program, we investigated the potential functions of these six SNPs. The ABCB1 SNP, rs7787082, was located at potential transcription factor-binding sites and thus may have a role in transcription regulation. A G-A change at rs7787082 may produce a binding site of Sox-5 and HFH-2. The protein encoded by Sox-5 may play a role in chondrogenesis. The other five SNPs (ie, rs10817638, rs7851259, rs10982287, rs4979427, and rs1469181) did not show any known function according to the F-SNP program.
Discussion
In the present study, we performed a bivariate GWAS analysis for BMD and AAM and identified four SNPs (rs10817638, rs7851259, rs10982287, and rs4979427) upstream from the ATP6V1G1 gene that were associated with both BMD and AAM in multiple populations. Our study represents the first bivariate GWAS to detect shared genetic factors for the closely related BMD and AAM phenotypes.
The ATP6V1G1 gene encodes the V1G1 subunit of the vacuolar proton pump H+-ATPs (V-ATPase). The V1G1 subunit is a component of the peripheral stalk and is fundamental for correct V-ATPase assembly. V-ATPase is a proton pump, capable of acidifying intracellular compartments and the pericellular space. The complex is located on the ruffled border plasma membrane of bone-resorbing osteoclasts, mediating extracellular acidification for bone demineralization during bone resorption (27). Genetic studies in mice and humans have suggested that V-ATPase subunits play a critical role in osteoclast-related diseases including osteopetrosis (28) and osteoporosis (29). V-ATPase has also been shown to be expressed in ovarian follicles of Drosophila and played a role in follicle growth, vesicle acidification, and yolk processing (30). Therefore, V-ATPase may indirectly play an important role in the onset of menarche.
The ABCB1 (also called MDR1 or TAP1) gene encodes the intestinal efflux transporter P-glycoprotein. P-glycoprotein can transport substrates (glucocorticoids, steroids, some cytokines such as IL-1β, and a number of endogenous substances) from the inside to the outside cells using ATP as an energy source. Mutoh et al (31) reported that estrogen down-regulates P-glycoprotein expression in human breast cancer cells, and it is well documented that the onset of menarche is partly determined by the estrogen concentration (32). The documented capacity of estrogen to effect P-glycoprotein expression levels, albeit in cancer cells, suggests that estrogen may be involved in the molecular mechanism underlying the association between ABCB1 and the onset of menarche. A recent study reported that a common polymorphism in the ABCB1 gene (rs1045642) is associated with reduced bone mass and might confer susceptibility to glucocorticoid induced osteoporosis (33). Statistical evidence obtained in our study, together with previously reported biological functions, suggests that ABCB1 may play a role in both bone metabolism and onset of menarche.
The four ATP6V1G1 SNPs that were modestly associated with BMD-AAM in our Chinese discovery sample were replicated in Caucasians, African-Americans, and/or Hispanic-Americans. Thus, the relatively broad associations of these four SNPs with BMD-AAM in multiple populations suggest that the ATP6V1G1 gene has broad functional effects on people from diverse ethnic backgrounds, and the generalizability of these variants can be used in risk modeling. However, ethnic genetic heterogeneity has also been observed among different races when studying specific phenotypes or diseases (34). Our study found PCDH15 was the only gene that had SNPs near to it that were significant in both hip BMD-AAM and spine BMD-AAM, but these findings were not replicated in the three replication populations. It is possible that the failure to replicate these findings in the replication populations was attributable to differential effects of this gene or differential tagging for this gene on different ethnicities in this region, although the relatively small sample sizes provides an alternative explanation. We did not exclude the potential role of PCDH15 underlying BMD and AAM variation. Further study is needed to replicate this gene in other independent Chinese subjects.
The bone densitometer can measure the BMD of multiple sites. We test only the bivariate association of spine BMD-AAM and hip BMD-AAM because osteoporotic fracture at the spine and hip are the two most common and/or most serious fracture types and are linked with increased mortality (35). Spine and hip BMD are the two most often studied densitometer output in the osteoporosis genetic and epidemiological studies. Our study showed that the four ATP6V1G1 SNPs and one ABCB1 SNP were associated with either spine BMD-AAM or hip BMD-AAM, which suggested the site-specific effects of these SNPs. Other studies also showed that the MPP7 (36) gene was associated with spine BMD but not hip BMD, and RUNX2 (37) was associated with a site-specific fracture in postmenopausal females. However, the intergenic SNP on chromosome 2 showed consistent association with spine BMD-AAM and hip BMD-AAM. Our study supported the previous hypothesis of site-specific regulation of BMD as well as shared common genetic factors. It is well known that bone property differs between the spine and hip, with the spine comprising mostly trabecular bone and hip having 50% cortical and 50% trabecular bone. Therefore, the gene regulation of hip and spine BMD is not entirely the same.
Previous studies have implicated the LGR4 (15) and PPARG and EP300 genes (14) in both osteoporosis and onset of menarche. To confirm the findings, we checked the association results of these three genes in the Chinese sample. Our results showed that LGR4 rs2472625 and PPARG rs1175543 were associated with spine BMD-AAM with bivariate values of P = .02 and P = .047, respectively. PPARG rs1177809 and rs12490265 and EP300 rs2413637 were associated with hip BMD-AAM with bivariate values of P = .041, P = .042 and P = .047, respectively. Hence, our results confirmed the importance of the three genes in both osteoporosis and the onset of menarche.
We compared the 62 genetic loci associated with osteoporosis-related traits (17) and 106 genomic loci with AAM (18, 19) and found that the ESR1 gene overlapped between these two groups of loci. We then checked the association results of ESR1 with BMD-AAM in the Chinese sample. We found five SNPs (rs9322336, rs17091835, rs4458702, rs4870061, and rs7220295) that were associated with both hip BMD-AAM and spine BMD-AAM with bivariate P values ranging from P = 1.45 × 10−3 to P = 2.35 × 10−2.
The transethnic meta-analysis using Meta-Analysis of Transethnic Association studies (MANTRA) software (38) is an alternative and robust analytic approach that accounts for the transethnic nature of the various cohorts. In contrast to the traditional meta-analysis, the transethnic approach in MANTRA accommodates between-population heterogeneity of associated variants and their effect sizes by allowing for allelic effects to be most similar between the most closely related populations. However, this approach is not feasible in a bivariate association context because of the unavailability of effect sizes for two traits in our bivariate model. In our previous paper (39), we performed a conventional meta-analysis using METAL and a transethnic meta-analysis using MANTRA using the similar study populations of various ethnicities with the present study for univariate analyses, and the transethnic meta-analysis using MANTRA closely mirrored the results from the standard meta-analysis using METAL. Therefore, the results of the meta-analysis using METAL in our study are reasonable and reliable.
This study has some limitations that must be considered. First, the average age among the four cohorts differed. Second, the sample size of African-Americans and Hispanic-Americans was relatively small. These two factors may potentially contribute to the discrepant findings between the discovery and replication samples. An additional limitation is that information regarding AAM was collected through individual retrospective self-reporting. Although some minor reporting inaccuracies may exist in the very old age group (>70 y), the age of menarche, as a milestone of a woman's reproductive period, is generally remembered correctly by most individuals, regardless of their chronological age (40). In our sample, most females were younger than 70 years. Thus, we consider the retrospective acquisition of AAM data via recollection to be a reasonable and reliable method for our study.
In conclusion, we used a bivariate GWAS approach in a Chinese discovery sample, and a follow-up replication study, combined with functional evidence to identify the ATP6V1G1 gene, which appear to coregulate BMD and AAM in multiple ethnic populations. These findings enhance our knowledge of genetic associations between BMD and AAM and provide a rationale for subsequent functional studies of these implicated genes in the pathophysiology of diseases related to these phenotypes, such as osteoporosis and AAM.
Acknowledgments
We thank Dr Jian Li for his assistance in usage of the data from the database of Genotypes and Phenotypes. We also thank all the study subjects for volunteering to participate in the study.
This work was supported by Natural Science Foundation of China Grants 31271344 and 81570807, Hunan Provincial Construct Program of the Key Discipline in Ecology (0713), and the Cooperative Innovation Center of Engineering and New Products for Developmental Biology of Hunan Province (20134486). H.-W.D. was supported in part by National Institutes of Health Grants P50AR055081, R01AG026564, R01AR050496, RC2DE020756, R01AR057049, and R03TW008221, the Edward G. Schlieder Endowment and the Franklin D Dickson/Missouri Endowment. The Women's Health Initiative program is supported by the National Heart, Lung, and Blood Institute, National Institutes of Health, and US Department of Health and Human Services through Contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221.
This manuscript was not prepared in collaboration with investigators of the Women's Health Initiative, has not been reviewed and/or approved by the Women's Health Initiative, and does not necessarily reflect the opinions of the Women's Health Initiative investigators or the National Heart, Lung, and Blood Institute. The data sets used for the analyses described in this manuscript were obtained from the database of Genotypes and Phenotypes (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap) through dbGaP accession number phs000200.v6.p2.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AAM
- age at menarche
- BMD
- bone mineral density
- dbGaP
- database of Genotypes and Phenotypes
- GWAS
- genome-wide association study
- KCOS
- Kansas City Osteoporosis Study
- LD
- linkage disequilibrium
- MAF
- minor allele frequency
- MANTRA
- Meta-Analysis of Transethnic Association studies
- SNP
- single-nucleotide polymorphism
- V-ATPase
- vacuolar proton pump H+-adenosine triphosphatases
- WHI
- Women's Health Initiative
- WHI-AA
- women of African-American ancestry in WHI
- WHI-HIS
- women of Hispanic ancestry in WHI.
References
- 1. Ammann P, Rizzoli R. Bone strength and its determinants. Osteoporos Int. 2003;14(suppl 3):S13–S18. [DOI] [PubMed] [Google Scholar]
- 2. Hawker GA, Jamal SA, Ridout R, Chase C. A clinical prediction rule to identify premenopausal women with low bone mass. Osteoporos Int. 2002;13(5):400–406. [DOI] [PubMed] [Google Scholar]
- 3. Burge R, Dawson-Hughes B, Solomon DH, Wong JB, King A, Tosteson A. Incidence and economic burden of osteoporosis-related fractures in the United States, 2005–2025. J Bone Miner Res. 2007;22(3):465–475. [DOI] [PubMed] [Google Scholar]
- 4. Dhanwal DK, Dennison EM, Harvey NC, Cooper C. Epidemiology of hip fracture: worldwide geographic variation. Indian J Orthop. 2011;45(1):15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xia WB, He SL, Xu L, et al. Rapidly increasing rates of hip fracture in Beijing, China. J Bone Miner Res. 2012;27(1):125–129. [DOI] [PubMed] [Google Scholar]
- 6. Anderson CA, Duffy DL, Martin NG, Visscher PM. Estimation of variance components for age at menarche in twin families. Behav Genet. 2007;37(5):668–677. [DOI] [PubMed] [Google Scholar]
- 7. Silman AJ. Risk factors for Colles' fracture in men and women: results from the European Prospective Osteoporosis Study. Osteoporos Int. 2003;14(3):213–218. [DOI] [PubMed] [Google Scholar]
- 8. Eastell R. Role of oestrogen in the regulation of bone turnover at the menarche. J Endocrinol. 2005;185(2):223–234. [DOI] [PubMed] [Google Scholar]
- 9. Riggs BL, Khosla S, Melton LJ., 3rd Sex steroids and the construction and conservation of the adult skeleton. Endocr Rev. 2002;23(3):279–302. [DOI] [PubMed] [Google Scholar]
- 10. Chamouni A, Oury F. Reciprocal interaction between bone and gonads. Arch Biochem Biophys. 2014;561:147–153. [DOI] [PubMed] [Google Scholar]
- 11. Karsenty G. The mutual dependence between bone and gonads. J Endocrinol. 2012;213(2):107–114. [DOI] [PubMed] [Google Scholar]
- 12. Gerdhem P, Obrant KJ. Bone mineral density in old age: the influence of age at menarche and menopause. J Bone Miner Metab. 2004;22(4):372–375. [DOI] [PubMed] [Google Scholar]
- 13. Guo Y, Zhao LJ, Shen H, Guo Y, Deng HW. Genetic and environmental correlations between age at menarche and bone mineral density at different skeletal sites. Calcif Tissue Int. 2005;77(6):356–360. [DOI] [PubMed] [Google Scholar]
- 14. Pan F, Xiao P, Guo Y, et al. Chromosomal regions 22q13 and 3p25 may harbor quantitative trait loci influencing both age at menarche and bone mineral density. Hum Genet. 2008;123(4):419–427. [DOI] [PubMed] [Google Scholar]
- 15. Styrkarsdottir U, Thorleifsson G, Sulem P, et al. Nonsense mutation in the LGR4 gene is associated with several human diseases and other traits. Nature. 2013;497(7450):517–520. [DOI] [PubMed] [Google Scholar]
- 16. Zofkova I. [Role of genetics in prediction of osteoporosis risk]. Vnitrni Lekarstvi. 2011;57(1):78–84. [PubMed] [Google Scholar]
- 17. Richards JB, Zheng HF, Spector TD. Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nat Rev Genet. 2012;13(8):576–588. [DOI] [PubMed] [Google Scholar]
- 18. Dvornyk V, Waqar-ul H. Genetics of age at menarche: a systematic review. Hum Reprod Update. 2012;18(2):198–210. [DOI] [PubMed] [Google Scholar]
- 19. Perry JR, Day F, Elks CE, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514(7520):92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deng HW, Shen H, Xu FH, et al. Tests of linkage and/or association of genes for vitamin D receptor, osteocalcin, and parathyroid hormone with bone mineral density. J Bone Miner Res. 2002;17(4):678–686. [DOI] [PubMed] [Google Scholar]
- 21. Design of the Women's Health Initiative clinical trial and observational study. The Women's Health Initiative Study Group. Control Clin Trials. 1998;19(1):61–109. [DOI] [PubMed] [Google Scholar]
- 22. Zhang L, Choi HJ, Estrada K, et al. Multistage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum Mol Genet. 2014;23(7):1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. [DOI] [PubMed] [Google Scholar]
- 24. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Renteria ME, Cortes A, Medland SE. Using PLINK for genome-wide association studies (GWAS) and data analysis. Methods Mol Biol. 2013;1019:193–213. [DOI] [PubMed] [Google Scholar]
- 26. Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qin A, Cheng TS, Pavlos NJ, Lin Z, Dai KR, Zheng MH. V-ATPases in osteoclasts: structure, function and potential inhibitors of bone resorption. Int J Biochem Cell Biol. 2012;44(9):1422–1435. [DOI] [PubMed] [Google Scholar]
- 28. Kornak U, Schulz A, Friedrich W, et al. Mutations in the a3 subunit of the vacuolar H(+)-ATPase cause infantile malignant osteopetrosis. Hum Mol Genet. 2000;9(13):2059–2063. [DOI] [PubMed] [Google Scholar]
- 29. Xu J, Cheng T, Feng HT, Pavlos NJ, Zheng MH. Structure and function of V-ATPases in osteoclasts: potential therapeutic targets for the treatment of osteolysis. Histol Histopathol. 2007;22(4):443–454. [DOI] [PubMed] [Google Scholar]
- 30. Bohrmann J, Braun B. Na,K-ATPase and V-ATPase in ovarian follicles of Drosophila melanogaster. Biol Cell. 1999;91(2):85–98. [DOI] [PubMed] [Google Scholar]
- 31. Mutoh K, Tsukahara S, Mitsuhashi J, Katayama K, Sugimoto Y. Estrogen-mediated post transcriptional down-regulation of P-glycoprotein in MDR1-transduced human breast cancer cells. Cancer Sci. 2006;97(11):1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bernstein L, Pike MC, Ross RK, Henderson BE. Age at menarche and estrogen concentrations of adult women. Cancer Causes Control. 1991;2(4):221–225. [DOI] [PubMed] [Google Scholar]
- 33. Lovas K, Gjesdal CG, Christensen M, et al. Glucocorticoid replacement therapy and pharmacogenetics in Addison's disease: effects on bone. Eur J Endocrinol. 2009;160(6):993–1002. [DOI] [PubMed] [Google Scholar]
- 34. Kochi Y, Suzuki A, Yamada R, Yamamoto K. Genetics of rheumatoid arthritis: underlying evidence of ethnic differences. J Autoimmun. 2009;32(3–4):158–162. [DOI] [PubMed] [Google Scholar]
- 35. Melton LJ., 3rd Adverse outcomes of osteoporotic fractures in the general population. J Bone Miner Res.. 2003;18(6):1139–1141. [DOI] [PubMed] [Google Scholar]
- 36. Xiao SM, Kung AW, Gao Y, et al. Post-genome wide association studies and functional analyses identify association of MPP7 gene variants with site-specific bone mineral density. Hum Mol Genet. 2012;21(7):1648–1657. [DOI] [PubMed] [Google Scholar]
- 37. Morrison NA, Stephens AS, Osato M, et al. Polyalanine repeat polymorphism in RUNX2 is associated with site-specific fracture in post-menopausal females. PloS One. 2013;8(9):e72740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol. 2011;35(8):809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tan LJ, Zhu H, He H, et al. Replication of 6 obesity genes in a meta-analysis of genome-wide association studies from diverse ancestries. PloS One. 2014;9(5):e96149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Must A, Phillips SM, Naumova EN, et al. Recall of early menstrual history and menarcheal body size: after 30 years, how well do women remember? Am J Epidemiol. 2002;155(7):672–679. [DOI] [PubMed] [Google Scholar]