Abstract
The neuroendocrine regulation of reproduction is an intricate process requiring the exquisite coordination of an assortment of cellular networks, all converging on the GnRH neurons. These neurons have a complex life history, migrating mainly from the olfactory placode into the hypothalamus, where GnRH is secreted and acts as the master regulator of the hypothalamic-pituitary-gonadal axis. Much of what we know about the biology of the GnRH neurons has been aided by discoveries made using the human disease model of isolated GnRH deficiency (IGD), a family of rare Mendelian disorders that share a common failure of secretion and/or action of GnRH causing hypogonadotropic hypogonadism. Over the last 30 years, research groups around the world have been investigating the genetic basis of IGD using different strategies based on complex cases that harbor structural abnormalities or single pleiotropic genes, endogamous pedigrees, candidate gene approaches as well as pathway gene analyses. Although such traditional approaches, based on well-validated tools, have been critical to establish the field, new strategies, such as next-generation sequencing, are now providing speed and robustness, but also revealing a surprising number of variants in known IGD genes in both patients and healthy controls. Thus, before the field moves forward with new genetic tools and continues discovery efforts, we must reassess what we know about IGD genetics and prepare to hold our work to a different standard. The purpose of this review is to: 1) look back at the strategies used to discover the “known” genes implicated in the rare forms of IGD; 2) examine the strengths and weaknesses of the methodologies used to validate genetic variation; 3) substantiate the role of known genes in the pathophysiology of the disease; and 4) project forward as we embark upon a widening use of these new and powerful technologies for gene discovery.
Introduction
-
The Past: Previous Strategies Used to Discover Genes in IGD
Complex phenotypes, contiguous gene deletion syndromes, and karyotypic abnormalities point to IGD genes
Phenotypic complexity identifies pleiotropic genes in syndromic cases
Endogamous pedigrees lead to the discovery of recessive IGD genes
Candidate gene approaches reveal novel genetic causes of IGD
Summary
-
The Present
Identifying IGD genes in the “-omics” era
Limitations of current methodology
Establishing criteria for novel gene validation
A critical reappraisal of known IGD genes
The complex biology of the GnRH genetic network
The challenge of functional testing
Testing of genetic burden in IGD
-
The Future
New tools and approaches for the discovery of IGD genes
Conclusions
I. Introduction
The hypothalamic regulation of reproduction is an intricate developmental process requiring exquisite coordination of an assortment of cellular networks. During embryonic development, precursors of GnRH neurons originate from the neuroectoderm and neural crest, populate the olfactory placode, then rapidly expand their population and migrate along the vomeronasal and olfactory nerves to their final anatomic home within the medio-basal hypothalamus where they fully differentiate (1–6). Within the hypothalamus, these neurons integrate various physiologic signals and coordinately secrete GnRH in a distinctive pulsatile pattern into the hypophyseal-portal blood supply (7). This release, in turn, initiates and maintains the biosynthesis and release of LH and FSH from the anterior pituitary gonadotropes.
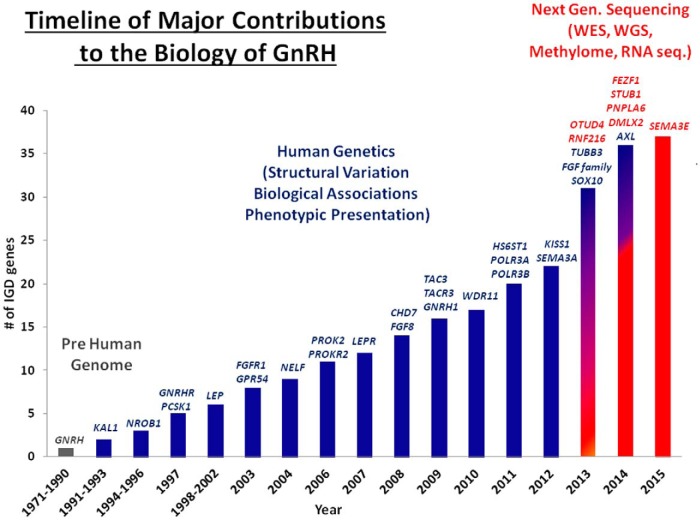
Figure 1 shows a timeline of scientific findings that have contributed to our understanding of this complex GnRH neuronal biology. These contributions were initiated and substantiated in animal models and, once the human genome was sequenced, continued to expand via genetic discoveries using the human disease model of isolated GnRH deficiency (IGD) (Online Mendelian Inheritance in Man phenotypic series: PS147950) (8–13). IGD is a family of rare Mendelian disorders (14) that have also been called isolated or idiopathic/congenital hypogonadotropic hypogonadism. The term congenital, which is often used, might well apply where evidence of the disorder exists at birth such as microphallus, cryptorchidism or hypogonadotropism. However, in the vast majority of cases, such definitive evidence is typically lacking making the use of this term somewhat inaccurate. Thus, the term IGD is preferable to these other terms because this disease occurs in the absence of any anatomical hypothalamic or pituitary lesion or other anterior pituitary hormone deficiency (ie, is isolated) and is caused by a failure of secretion and/or action of GnRH. The phenotypic result of this GnRH deficiency is hypogonadotropic hypogonadism and “downstream failure” of sexual maturation that typically manifests itself with a lack of pubertal development (8, 12, 15).
Figure 1.
Major contributions to our understanding of the ontogeny of GnRH neurons: from prehuman genome to human genetics and NGS.
The intimate topographical and developmental intertwining of the olfactory system and GnRH neurons explains the etiology of the “neurodevelopmental” form of IGD known as Kallmann syndrome (KS), which is also characterized by complete or partial loss of the sense of smell (anosmia/hyposmia) (16, 17) that accompanies the hypogonadotropism. However, despite the common feature of a reduction in (or absence of) GnRH function, IGD is clinically heterogeneous encompassing not only KS, but also normosmic idiopathic hypogonadotropic hypogonadism (nIHH) (18), the adult onset form of acquired hypogonadotropic hypogonadism (19), the more common transient loss of reproductive function seen in hypothalamic amenorrhea, and delayed gain of reproductive function seen in constitutional delay of puberty (20–22). Although these reproductive phenotypes typically dominate an IGD patient's clinical presentation, in addition to anosmia there is also a wide variety of associated nonreproductive phenotypes, including unilateral renal agenesis and other kidney malformations, cranio-facial defects (eg, cleft lip/palate), skeletal and digital defects, sensorineural hearing loss, eye and oculomotor defects, cardiac defects, and dental agenesis, among others (13, 23, 24).
Mirroring this phenotypic diversity, the underlying genetic architecture of IGD is equally heterogeneous with mutations in several genes causing the developmental loss of GnRH neurons and/or their abnormal neuroendocrine secretory function. Since the initial description of the anosmic form of this disease by Franz Kallmann in 1944, the genetic complexity underlying this disorder has puzzled physicians caring for IGD patients. In 1944, Kallmann observed that, “there is no reason to assume that it must always be the same mechanism …. or the same gene that causes the disease … ” (17). As the catalogue of candidate genes for IGD continues to expand, this prescience of Franz Kallmann is confirmed. The genetic complexity of IGD is also reflected in its different inheritance patterns. Whereas most IGD patients initially present as isolated or sporadic cases, closer reviews of their pedigrees often reveal families with X-linked, autosomal recessive, and autosomal dominant inheritance (12, 15, 23, 25). In addition to these Mendelian modes of inheritance, an even more complex genetic architecture for IGD (occurring in 10%–15% of IGD cases) has been documented wherein mutations in 2 or more IGD genes are found in a single case. These coexisting mutations could therefore play the role of “second hits” or act as “modifier” genes involved in this syndrome, perhaps accounting for some of the variable expressivity and incomplete penetrance often observed in IGD (26–30).
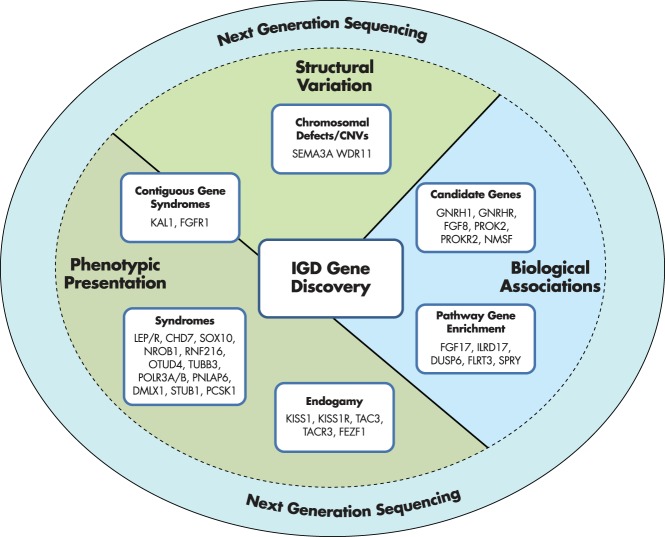
This clinical and genetic heterogeneity characterizing IGD has led to some of the most important contributions in the field of genetic discovery in reproductive endocrinology since the description of Turner and Klinefelter's syndromes (31, 32). Over the last 30 years, several research groups around the world have used a combination of clinical investigational strategies and genetic approaches to unravel the growing genetic complexity underlying IGD and link its phenotypic variation to specific genetic and biological causes (Figures 1 and 2). These approaches have been based on well-validated genetic and molecular tools and been critical to establishing this field. However, they have often lacked 2 major characteristics: speed and robustness of scale. New strategies, such as next-generation sequencing (NGS), are now empowering a global revolution in genetic discovery.
Figure 2.
Different strategies for gene discovery in IGD.
The ability of NGS to query the entire genome with increasing speed and accuracy is surfacing a surprising number of rare sequence variants (RSVs) of unknown significance in both known and novel IGD genes. Perplexingly, these RSVs are sometimes shared by both affected individuals and controls. Moreover, because many of the known IGD genes seem to act synergistically while individually displaying features of incomplete penetrance or variable expressivity, it is becoming ever more difficult to label a single genetic change in any individual IGD patient as “causative.” Therefore, before this field moves forward with new genetic tools and itsever-expanding genetic framework, it is important to pause and reassess what we know about the GnRH network and IGD genetics; how has this gene discovery evolved, and what does it mean for genes implicated in IGD? Therefore, this review will: 1) look back at the strategies used to discover the “known” IGD genes implicated in the rare forms of KS and nIHH; 2) examine the strengths and weaknesses of the methodologies used to validate their genetic variation; 3) substantiate the role of these now known genes in the pathophysiology of the disease according to the evolving standards in the field; and 4) project forward as we embark upon a widening use of these new and powerful technologies for gene discovery.
II. The Past: Previous Strategies Used to Discover Genes in IGD
A. Complex phenotypes, contiguous gene deletion syndromes, and karyotypic abnormalities point to IGD genes
The first gene associated with IGD, Kallmann 1 (KAL1), was surfaced during the investigation of brothers with complex clinical phenotypes suggestive of a large contiguous gene deletion syndrome on the X chromosome (33–35). In such syndromes, the multiorgan system defects seen in these patients represented a combination of phenotypes associated with each of the individual genes deleted within the contiguous region. As a case in point, KAL1 was identified in a male infant who displayed abnormal genitalia, hypogonadotropic hypogonadism, agenesis of the olfactory bulbs and tracts (ie, KS) associated with chondrodysplasia punctata and ichthyosis (34). Karyotype analysis and positional cloning revealed a large deletion in the Xp22.31 region that included the genes KAL1, ARSE, and STS (that together caused KS, chondrodysplasia punctata, and ichthyosis, respectively) and thus established KAL1 as an X-linked KS gene (34–38). Several years later, overlapping interstitial deletions in chromosome 8p11 were discovered in patients suffering from KS and hereditary spherocytosis and were mapped by fluorescence in situ hybridization and bacterial artificial chromosome cloning (39). Another contiguous gene deletion syndrome was verified and fibroblast growth factor receptor 1 (FGFR1) surfaced as an autosomal dominant KS gene (40).
In addition to such contiguous gene syndromes, chromosomal rearrangements have also provided powerful insights into the genetic underpinnings of IGD. Just a few years after the initial discovery of FGFR1, another analysis of a balanced chromosomal translocation (using fluorescence in situ hybridization and bacterial artificial chromosome cloning) in a patient with nIHH and cleft lip/palate led to the identification of FGFR1 as the genetic cause of that patient's phenotype, and established that FGFR1 mutations could also cause the normosmic form of IGD, nIHH (41, 42). Additional IGD cases harboring structural variations have uncovered novel genes such as the WD repeat domain 11 (WDR11) gene, which was found in a KS patient near a breakpoint of a balanced translocation between chromosomes 10 and 12 (43). Similarly, the semaphorin 3A (SEMA3A) gene was truncated in 2 seconds KS siblings harboring a large heterozygous deletion of the gene that removed 7 out of 11 of its exons and was inherited from their affected father (44). Subsequently, another group found pathogenic point mutations in SEMA3A in a cohort of KS patients (45). Importantly, the SEMA3A deletion was missed by traditional karyotyping whereas an oligonucleotide array detected it thus highlighting the importance of the constantly improving resolution of genetic technologies and their ability to contribute new insights.
Each of these genetic findings illustrate how both low-resolution karyotypes and high-resolution gene arrays have played key roles in the detection of causative candidate genes in IGD patients with complex phenotypes, implying the presence of an underlying structural event. In addition, as will be discussed, these discoveries have provided fundamental information about the underlying developmental biology of GnRH neurons. Thus, contiguous gene syndromes and complex chromosomal rearrangements represent a rich source of new gene discovery both in the field of human genetics generally, and, specifically, in the identification of genes controlling human reproduction.
B. Phenotypic complexity identifies pleiotropic genes in syndromic cases
It is important to note that the phenotypic richness found in “syndromic” cases does not always point to a contiguous gene interruption. In fact, Sanger sequencing and NGS in patients with equally complex phenotypes have yielded another large crop of novel IGD genes. These patients underscore another genetic feature of importance: the role of the pleiotropy. This phenomenon occurs when a single genetic cause results in the development of both a reproductive defect as well as additional clinical features, ie, when IGD occurs alone or accompanied by these syndromic associations.
The nuclear receptor subfamily 0, group B, member 1 (NR0B1) gene is an early example of a pleiotropic IGD gene. The discovery of this gene explained the paradox of the well-established association between the X-linked form of congenital adrenal hypoplasia and hypogonadotropic hypogonadism via its previously undescribed role in the development of the hypothalamic-pituitary axis (46, 47). Similarly, although IGD is only occasionally associated with morbid obesity, genetic defects in leptin (LEP) (48) and the leptin receptor (LEPR) (49–51) genes, as well as mutations in the proprotein convertase subtilisin/kexin type 1 (PCSK1) gene (52), have been found in IGD patients with an obesity phenotype.
There are additional pleiotropic genes that cause IGD associated with neurological phenotypes. For example, whole-exome sequencing analysis in patients with Gordon Holmes syndrome (IGD and cerebellar ataxia) led to the discovery of ovarian tumor deubiquitinase 4 gene (OTUD4) and ring finger protein 216 (RNF216) (53), as well as mutations in stress-induced phosphoprotein 1 homology and U-box containing protein 1, E3 ubiquitin protein ligase (STUB1) and patatin-like phospholipase domain-containing protein 6 (PNPLA6) (54) that were found in IGD patients who also displayed ataxia (55). There are also several other genes causing 4H syndrome (hypomyelination, hypodontia and hypogonadotropic hypogonadism), including polymerase III, RNA subunit A and B (POLR3A and POLR3B) (56, 57) Additionally, a novel clinical syndrome defined by a specific genetic mutation in the neuronal tubulin-β class 3 (TUBB3) gene was recently described having the characteristics of Moebius syndrome (facial hypoplasia/paralysis and oculomotor defects) accompanied by KS, intellectual disabilities, and in some cases, vocal cord paralysis, tracheomalacia, and cyclical vomiting (58–60). In the future, it is likely that such pleiotropic genes will be associated with the nonsyndromic form of IGD as well.
So far, 2 pleiotropic IGD genes have been found in patients with IGD alone. Coloboma of the eye, heart defects, choanal atresia, growth retardation, genital anomalies, and ear defects (CHARGE) syndrome is a disorder with multiple affected organ systems. Hypogonadotropic hypogonadism is typically a minor feature of most CHARGE cases, but this reproductive feature can be entirely absent or missed, especially if these cases are reported in childhood before the pubertal defects become manifest (61). Conversely, upon closer phenotypic examination, many IGD cases with CHARGE features (for example, hearing loss) harbor mutations in the CHARGE syndrome gene, a chromodomain helicase DNA binding protein 7 (CHD7) (61, 62). We now know that patients with KS and hearing loss are more likely to present with mutations in the CHD7 gene than any other IGD gene (63), a phenotypic association extremely useful for the prioritization of the genetic screening (24). Interestingly, rare deleterious mutations in CHD7 can also be associated with IGD in the absence of any of the other phenotypic hallmarks of CHARGE (63, 64). This finding indicates a potential differential sensitivity of developing GnRH neurons to milder deleterious mutations in CHD7. Following the same lesson of pleiotropy as seen in CHD7, mutations in the sex-determining region of Y-box 10 (SOX10) gene, previously associated with Waardenburg syndrome, were recently identified in patients with IGD alone as well as IGD patients with milder Waardenburg features, including hearing loss, pigmentation defects, intellectual disability, or psychomotor delay (65). These examples show that cases of many syndromic IGD genes may result from mutations in single genes that have pleiotropic roles with differential effect(s) on various organ systems resulting in a wide range of syndromic and nonsyndromic phenotypes. This phenomenon may be particularly true of other genes which, like CHD7 and SOX10, regulate the expression of many downstream genetic targets involved in GnRH maturation, migration, and hypothalamic function (66–68).
Taken together, these findings in patients with complex phenotypic syndromes that include hypogonadotropic hypogonadism suggest the existence of a panoply of genes whose genetic pleiotropy is a clue to their contribution to the development or regulation of GnRH neurons. In the future, there will likely be much to gain with the pursuit of other similar genetic and biological leads in piecing together the complex biology of GnRH neuronal ontogeny. In addition, syndromic cases in other nonreproductive fields may well yield a similar crop of new genes involved in isolated disease phenotypes.
C. Endogamous pedigrees lead to the discovery of recessive IGD genes
The study of IGD in multiplex families from geographically delineated populations with high rates of endogamy has been another highly fruitful method of IGD gene discovery (69). The gene coding for kisspeptin receptor 1 (KISS1R) was discovered using linkage analysis in an endogamous pedigrees with multiple affected Bedouin family members displaying nIHH (70, 71). Similarly, the discovery of tachykinin 3 (TAC3) and its receptor (TACR3), was based on haplotype analysis of endogamous nIHH Turkish and Kurdish pedigrees (72). Homozygosity mapping (73), candidate gene screening (74), and whole-exome sequencing prompted the discovery of kisspeptin (KISS1) mutations in a consanguineous nIHH family (73), whereas mutations in the fasciculation and elongation protein zeta family zinc finger 1 (FEZF1) were newly identified in consanguineous KS families of Kurdish origin (75). Most recently, linkage analysis revealed a homozygous deletion in Drosophila melanogaster X-gene like-2 (DMXL2) gene carried by 3 brothers of a Senegalese consanguineous pedigree affected with a syndromic form of IGD that includes hypothyroidism, diabetes mellitus, and various neurological phenotypes (76). These examples suggest that analysis of other endogamous pedigrees will allow researchers to uncover novel recessive genes, giving additional insights into IGD disease pathology.
D. Candidate gene approaches reveal novel genetic causes of IGD
During the more than 25 years of successful genetic studies on IGD, there have been many attempts to screen candidate genes in patients suffering from this disease. Most were based either on data from animal studies or our previously limited knowledge of the biology of GnRH neurons. Examples of excellent candidate genes surfaced based on mouse model phenotypes include the GnRH receptor (GNRHR) (77) and its ligand, GnRH (GNRH1) (78), N-Methyl-D-aspartate receptor synaptonuclear signaling and neuronal migration factor (NSMF) (79, 80), and prokineticin 2 (PROK2) and its receptor (PROKR2) (81, 82). Previous findings of FGFR1 mutations in IGD patients led to the discovery of mutations in the candidate gene FGF8 (83), a ligand of FGFR1 in the central nervous system, whereas interactions of anosmin with heparin sulfate proteoglycans led to the validation of heparan sulfate 6-O-sulfotransferase 1 (HS6ST1) as an IGD gene (84). More recently, analysis of differential gene expression in 2 different SV40 transformed mouse GnRH cell lines has led to the discovery of mutations in AXL receptor tyrosine kinase (AXL) (85, 86).
However, many other candidate gene efforts have not been similarly successful (10), due to various limiting factors. First, small numbers of patients are generally screened meaning rare mutations are often missed. Second, incomplete penetrance of the candidate genes being investigated can make segregation of variants within and across pedigrees challenging. Finally, we still have a limited knowledge about the full systems biology involved in the disease, including the roles (and redundancy) of other protein family members and/or potential modifier genes/mutations that contribute to the defective phenotypes of loss of function (LoF) in man and mice (ie, the effect of background and strain genetics) (26).
Bioinformatic tools are rapidly expanding our knowledge of the underlying biology of GnRH neurons that was previously limited to in vitro studies using mouse GnRH cell lines (87, 88), in vivo and in vitro slice preparations (89, 90), and physiological studies in knockout models (91–93). These types of bioinformatically informed approaches have made substantial progress in revealing the role of protein interactions and coordinate signaling pathways in GnRH neuronal development and are thus leading to new and better informed candidate gene discoveries. For example, a combination of proteomic gene-set enrichment analysis and more selective candidate screening has led to the identification of several additional FGF signaling pathway genes causing IGD, including an additional ligand encoded by FGF17, IL-17 receptor D (IL17RD), dual specificity phosphatase 6 (DUSP6), sprouty homolog 4 (SPRY4), and fibronectin leucine-rich transmembrane protein 3 (FLRT3) (29). Recently, a combination of whole-exome sequencing with bioinformatics, in vitro functional studies, and studies of genetically modified mice led to the description of SEMA3E as the second semaphorin family member to be implicated in KS (94).
E. Summary
Figure 2 provides a summary of the various strategies used to identify IGD genes. Past approaches have mounted evidence from phenotypic presentations, gross chromosomal and structural abnormalities, and the underlying GnRH biology to support genetic discoveries. As will be discussed in the next section, NGS approaches will require researchers to use similar approaches while also adding further stringency to prove causality of genetic candidates.
III. The Present
A. Identifying IGD genes in the “-omics” era
Together, all of the aforementioned approaches have yielded a genetic cause in approximately 30%–35% of IGD patients (95), whereas the genetic etiology in the remaining 2/3 of patients is currently unknown. Genetic defects may have been missed due to the limited depth and range of existing DNA sequencing that, to date, has only screened coding sequences or short genome segments with low resolution techniques. Thus, mutations may still exist in noncoding areas of the “known IGD genes” or there may be unappreciated cryptic, structural defects in the genome, or epigenetic factors contributing to the disease. However, recent advances in NGS and its derivative analytic technologies (including genomics, epigenomics, proteomics, and metabolomics) have renewed efforts to unravel the genetic mystery of these “genetically unsolved” IGD cases. One key difference with these contemporary approaches is that, whereas most of the past gene discovery in IGD has been based upon limited Sanger sequencing of the genome due to cost restraints, NGS discovery tools have taken advantage of lowered costs and rapid turnaround times to base all their genetic inquiries in IGD patients on a genome-wide scale. These enhanced efforts allow for a more complete picture of the genetics underlying the disease in each patient to emerge. As high-throughput sequencing strategies become increasingly mainstream, the number of sequenced IGD patients and controls have increased, resulting in the generation of a burgeoning number of RSVs in “candidate IGD genes.” With this ever-growing list of unbiased candidates emerging from NGS, there is a pressing need for standardization of filtering based on sound statistical, biological, and genetic criteria to prioritize them and distinguish between the true genetic causes and false-positive associations.
B. Limitations of current methodology
Because IGD has been studied as a Mendelian disease, the segregation of variants within pedigrees and assessment of their penetrance have been key initial steps in the validation of putatively causative mutations. In addition to segregation, another important validation step is the replication of variants in other IGD subjects. This practice, assumes that: 1) sufficiently large cohorts of IGD patients are available for testing, and 2) large numbers of control individuals lack an association between the mutation and phenotype. However, these validation measures have been further complicated by incomplete penetrance of mutations and the fact that LoF in certain IGD candidate genes have been documented to occur in otherwise normal populations (96).
Consequently, once a rare variant is found in an affected IGD patient, variant-level experimental evidence is also required to address the functional impact of specific mutations on the level of gene/protein expression, splicing, and/or biochemical function. To address the question of how a candidate variant influences the gene function or the expression of the phenotype, several factors should be taken into consideration. First, the genetic nature of each identified variant must be assessed at the experimental level. The detection of severe, obviously “high impact” variation (intragenic deletions, frameshift deletions and insertions, stop-gain, and essential splice site mutations) may sufficiently explain the patient's phenotype without further need for experimental validation, because such mutations are a priori expected to cause LoF, particularly when occurring in the homozygous state. Conversely, although mutations in noncoding sequence and synonymous mutations are expected to be benign, this assumption has yet to be rigorously tested.
However, missense variation in a gene generally requires further experimental validation by examining its effect on mRNA expression and/or protein function. When assessing either LoF or gain of function in new candidate IGD genes, it is crucial to employ the appropriate experimental method for the functional validation of each candidate. The nature of the experimental system used to validate changes in function will vary according to the putative biology of the candidate under examination. For example, G protein-coupled receptors (97), transcription factors, and growth factors, etc, occupy special biologic niches that must be reflected in the careful choice of appropriate functional assay system selected for their validation. A corollary of this issue is that studying the gene and/or protein expression and its molecular activity in cell lines directly derived from patients may be more informative than performing such experiments in artificially constructed experimental cell lines. On the other hand, IGD is a complex disorder that ultimately affects a very small population of highly specialized neurons. Thus, simple cultured cell models may not be appropriate, whereas obtaining appropriate cell-lines, mammalian model systems, and/or human neuronal tissues can be challenging and expensive.
As an alternative to experimental validation, many researchers make use of bioinformatics tools that attempt to predict the functional effect of missense mutations (98). These programs make functional predictions based upon: 1) the biochemical role of the domain and specific amino acids within its protein; 2) the evolutionary conservation of the particular amino acid sequence across species; 3) the impact of the mutations in the different protein transcripts; and 4) the biochemical differences between the reference sequence and the proposed amino-acid substitution. One additional method that can be used to assess the causative nature of mutations is to examine the amount of observed functional variation in a gene as compared with what is expected based on evolutionary constraints (99). However, this type of analysis requires sophisticated statistical genetics.
Although the aforementioned recommendations are for assessing the variant-level implications of mutations, the remainder of this review will focus on analysis of the gene-level validation of the known IGD genes, trying to set the stage for the novel genes arising from NGS studies in IGD as well as in other rare Mendelian diseases and disorders. Lists of specific variants found in each of the IGD genes can be found in publically available databases, such as ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) along with supporting information on their pathogenicity. It is important to note that databases such as these are constantly changing and updating as new data are provided by researchers. Thus, at this time, there are relatively few RSVs that are “bona fide” pathogenic mutations (ie, have experimental validation of LoF), even in known IGD genes.
C. Establishing criteria for novel gene validation
NGS tools not only provide the opportunity for new gene discovery on a genome-wide basis but also create important statistical challenges in their interpretation and validation. The need for standards of validation for the growing number of candidate genes surfaced by NGS was thoroughly addressed in a recent publication by MacArthur et al (100). The establishment of such standards is critical, because false assignment of pathogenicity could have implications in genetic counseling and medical decision making (100).
Consequently, before the field fully embraces these novel technologies for new gene discovery, it seems reasonable to pause and examine the genes already reported in the literature that have already been associated with IGD. Our analysis will focus on 25 genes that have been found to be disrupted in human cases with pure IGD and thus are generally thought to be causative (see Table 1), even though, as previously mentioned, other genes derived from syndromic cases are also implicated in GnRH biology. We will apply the 7 criteria proposed by MacArthur et al (100) as a retrospective validation of these causative IGD genes. These criteria include:
Criterion 1: proof that the gene and/or gene product function is demonstrably altered in patients carrying these candidate mutations;
Criterion 2: evidence that there is tissue-specific expression of the protein that the gene encodes;
Criterion 3: demonstration that the biochemical function of the gene product is shared with other known genes in the disease or consistent with the phenotype;
Criterion 4: confirmation that the gene product interacts with other proteins already implicated in the disease;
Criterion 5: generation of nonhuman animal or cell-culture models with a similarly disrupted copy of the affected gene shows a phenotype consistent with human disease state;
Criterion 6: proof that the cellular phenotype in patient-derived cells or engineered equivalents can be rescued by addition of the wild-type gene product;
Criterion 7: statistical evidence that there is an increased genetic burden of variation in the gene in disease vs normal populations.
Table 1.
Assessment of the 25 IGD Genes Based on the MacArthur Criteria
| Gene | Criterion 1: Gene Disruption | Criterion 2: Expression | Criterion 3: Biochemical Function | Criterion 4: Protein Interactions | Criterion 5: Model Systems | Criterion 6: Rescue | Criterion 7: Gene Burden | Number of Criteria Met |
|---|---|---|---|---|---|---|---|---|
| KAL1 | + | + | + | The first gene discovered | + | + | 6 | |
| GNRHR | + | + | + | + | + | + | 6 | |
| GNRH1 | + | + | + | + | + | + | 6 | |
| KISS1 | + | + | + | + | + | + | 6 | |
| FGFR1 | + | + | + | + | + | 5 | ||
| FGF8 | + | + | + | + | + | 5 | ||
| KISS1R | + | + | + | + | + | 5 | ||
| TAC3 | + | + | + | + | + | 5 | ||
| TACR3 | + | + | + | + | + | 5 | ||
| PROKR2 | + | + | + | + | + | 5 | ||
| PROK2 | + | + | + | + | + | 5 | ||
| SEMA3A | + | + | + | + | + | 5 | ||
| SOX10 | + | + | + | + | + | 5 | ||
| AXL | + | + | + | + | + | 5 | ||
| SEMA3E | + | + | + | + | + | 5 | ||
| NSMF (NELF) | + | + | + | + | 4 | |||
| WDR11 | + | + | + | + | 4 | |||
| CHD7 | + | + | + | + | 4 | |||
| FGF17 | + | + | + | + | 4 | |||
| IL17RD | + | + | + | + | 4 | |||
| HS6ST1 | + | + | + | + | 4 | |||
| FEZF1 | + | + | + | + | 4 | |||
| SPRY4 | + | + | + | 3 | ||||
| FLRT3 | + | + | + | 3 | ||||
| DUSP6 | + | + | + | 3 |
The 25 IGD genes were assessed for the next criteria. Gene disruption: the gene and/or gene product function is demonstrably altered in patients carrying candidate mutations. Expression: the gene is expressed in tissues relevant to the disease of interest and/or is altered in expression in patients who have the disease. Biochemical function: the gene product performs a biochemical function shared with other known genes in the disease of interest, or consistent with the phenotype. Protein interactions: the gene product interacts with proteins previously implicated (genetically or biochemically) in the disease of interest. Model systems: nonhuman animal or cell-culture models with a similarly disrupted copy of the affected gene show a phenotype consistent with human disease state. Rescue: the cellular phenotype in patient-derived cells or engineered equivalents can be rescued by addition of the wild-type gene product. Gene burden: the affected gene shows statistical excess of rare (or de novo) probably damaging variants segregating in cases compared with control cohorts or null models.
D. A critical reappraisal of known IGD genes
Importantly, as shown in Table 1, all of the “established” IGD genes meet criteria 1 and 2 (gene disruption and expression) reflecting the careful research by different scientific groups and the dedicated efforts each has put into validation of their newly discovered genes. Most of these genes also pass at least one of the criteria assessing the biological impact of the gene(s) to the disease, ie, criteria 3, 4, and/or 5 (biochemical function, protein interactions, and model systems), because after establishing a gene disruption in human subjects, considerable efforts were made to assess these genes for an association with GnRH biology. Table 2 shows publically available gene expression data in various human tissue (101–107). Although each of the public databases have their limitations, according to existing data and current methodology, all of the genes do pass criterion 2 even when held to a standard of relevant expression in human tissues.
Table 2.
IGD Gene Expression in Human Tissues
| Tissue Expression Database |
||||||
|---|---|---|---|---|---|---|
| TiGER | BioGPS | Human Protein Atlas (n = 3, 1 Child, 2 Adults) | Riken FANTOM5 (Adult n = 1) | Riken FANTOM5 (Fetal n = 1) | GTEx (n = 22–30 Adults) | |
| Total # Tissue Types | 30 | 78 | 45 | 56 | 20 | 46 |
| Relevant tissue samples | Whole brain | Whole brain fetal brain olfactory bulb prefrontal cortex pituitary hypothalamus | Cerebral cortex lateral ventricle | Diencephalons olfactory apparatus pituitary | Occipital lobe parietal lobe temporal lobe | Frontal cortex hypothalamus pituitary |
| Gene | Expression pattern | |||||
| KAL1 | + | All | Cerebral cortex (RNA) | All | All | Frontal cortex, hypothalamus |
| GNRHR | + | All | − | Pituitary | − | Pituitary |
| GNRH1 | + | All | All | Diencephalon (low) | Parietal (low) | All |
| KISS1 | − | − | − | − | Temporal (low) | − |
| FGFR1 | + | All | All | All low | All low | All |
| FGF8 | Not found | All | − | − | − | Frontal cortex, hypothalamus |
| KISS1R | Not found | All | − | Pituitary (low) | − | Hypothalamus, pituitary |
| TAC3 | + | − | All | All (pituitary low) | All | Frontal cortex, hypothalamus |
| TACR3 | Not found | All | All | Olfactory apparatus (low) | All low | Hypothalamus |
| PROKR2 | Not found | − | All | − | − | All |
| PROK2 | Not found | − | Lateral ventricle (protein) | Diencephalon (low) | Temporal (low) | − |
| SEMA3A | + | All | All | Pituitary (low) | − | All |
| SOX10 | + | Whole brain, olfactory bulb, prefrontal cortex, hypothalamus | All | All low | All | Frontal cortex, hypothalamus |
| AXL | + | All | Cerebral cortex (RNA and protein) | All low | Occipital (low), Temporal | All (low) |
| SEMA3E | + | All | All | All low | All low | Frontal cortex, hypothalamus |
| NSMF (NELF) | Not found | Whole brain, prefrontal cortex | All | All (pituitary low) | All | All |
| WDR11 | Not found | Prefrontal cortex, hypothalamus | All | All low | Occipital, parietal, temporal (low) | All |
| CHD7 | + | All | All | All low | All | All |
| FGF17 | Not found | All | Cerebral cortex (RNA) | − | − | All |
| IL17RD | + | Prefrontal cortex, hypothalamus, olfactory bulb | All | All | All | All |
| HS6ST1 | + | All | Cerebral cortex (RNA) | − | − | All |
| FEZF1 | Not found | − | − | − | − | Hypothalamus |
| SPRY4 | + | All | All | − | Occipital (low), temporal (low) | All |
| FLRT3 | + | − | Cerebral cortex (RNA and protein) | All | All | Frontal cortex, hypothalamus |
| DUSP6 | + | Olfactory bulb | All | All low | Occipital (low), temporal (low) | All |
Expression of the 25 IGD genes according to publically available databases. Only relevant tissue types were examined. TiGER (Tissue-specific Gene Expression and Regulation) (http://bioinfo.wilmer.jhu.edu/tiger/) (107) and BioGPS (http://biogps.org/#goto=welcome) (101) databases integrate data from multiple sources. The Human Protein Atlas (http://www.proteinatlas.org/) (106), FANTOM5 (Riken consortium Functional Annotation of the Mammalian Genome) (103), and GTEx (Genotype-Tissue Expression) (http://www.gtexportal.org/home/) (105) databases contain expression data from multiple tissues taken from human subjects. Data from the FANTOM5 project were obtained indirectly from the EMBL-EBI expression atlas, which compiles expression data from many of these databases (https://www.ebi.ac.uk/gxa/home) (102). +, expressed; −, not expressed; not found, expression not examined; (low), expression was just detectable.
E. The complex biology of the GnRH genetic network
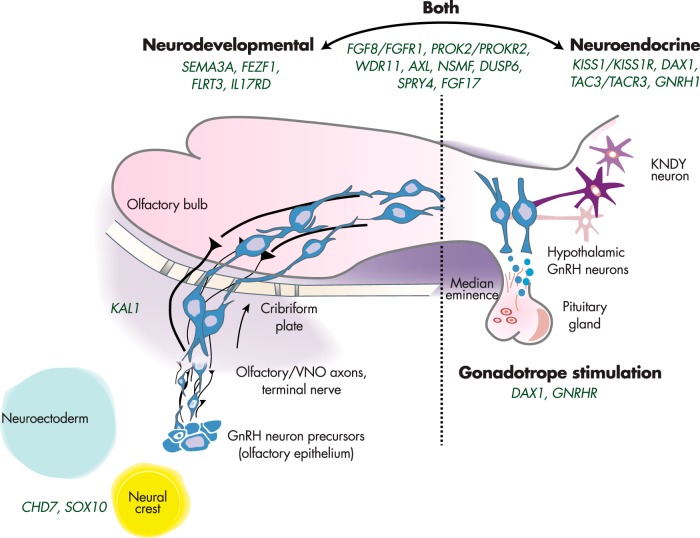
We now know that most IGD genes can either disrupt the development and/or migration of the GnRH neurons and cause KS (neurodevelopmental pathway) or alter the secretion and/or action of GnRH neurons after this migratory journey has been completed, in which case they cause nIHH (neuroendocrine pathway). In addition, several IGD genes are considered “overlap genes,” because they are found to be disrupted in both KS and nIHH carriers. Presumably, these overlap genes have multiple roles in GnRH biology although for many genes this overlap remains to be examined. Figure 3 portrays the summary of all of these genes and their roles in each of these pathways. Although it is clear that the complete biology of GnRH neurons will ultimately be far more complex than this relatively simplistic formulation, this schema represents at least a temporary map onto which each new IGD candidate gene can be placed.
Figure 3.
Complex neurodevelopmental and neuroendocrine regulation of GnRH neurons. Genes to the left are critical to GnRH neuronal migration, whereas those on the right are involved in proper GnRH function. A number of IGD genes (center) are involved in both processes.
Several KS genes affect the formation of the olfactory bulbs and the migration of the GnRH neurons from the olfactory placode (8, 12). Significant protein interactions between some of these gene products, such as FGFR1 and its ligands with the heparin sulfate proteoglycans and anosmin (the protein encoded by KAL1), explain the effects of mutations in these proteins on the expression of the KS phenotype (84, 108–110). In contrast, some more recently discovered genes (eg, SOX10 and CHD7), are not associated with olfactory development, but rather cause a disruption of neural crest cell migration indicating their potential role in a possible alternate GnRH neuronal migratory pathway. These observations suggest and that the primary origin of some GnRH neurons might exist in other central nervous system sites distinct from the olfactory placode within the brain (5). Importantly, IGD patients carrying mutations in these genes display other phenotypic characteristics that could be attributed to the defective development of neural crest cells in other organ systems such as craniofacial nerve defects, midline and retinal abnormalities, cardiac defects, and inner ear maldevelopment (61, 65).
Genes that control the neuroendocrine regulation of GnRH cause the nIHH phenotype. Apart from the obvious biological significance of GNRH1 and its receptor GNRHR, a more complex neuronal network forms a multilevel neuroendocrine regulatory system that controls GnRH secretion. This complex neuroendocrine network appears to be “supervised” by the kisspeptin-neurokinin B-dynorphin neurons that reside in the arcuate nucleus and regulate GnRH neuronal function (111–113). Although the kisspeptin signaling system is highly sensitive to the feedback of sex steroid hormones and acts directly on a large proportion of the GnRH neurons (114–117), there are findings suggesting a more modulatory role of the neurokin B/neurokinin B receptor 3 feedback (118–120). However, kisspeptin-neurokinin B-dynorphin neurons project their axons directly to GnRH neuron terminals that express the neurokinin B receptor 3 (TACR3) in the median eminence, suggesting a direct interaction of neurokin B with kisspeptin neurons (121, 122).
F. The challenge of functional testing
One of the most compelling ways to substantiate the causative nature of a novel gene is to model the genetic mutation in an animal or cell system to recapitulate the disease phenotype. Although there are many examples highlighting the successful use of animal models in IGD gene validation (Table 1, criterion 5) (92), knockout and mutant mouse models are slow to develop and expensive to maintain. In addition, phenotypic readouts from animal models are often difficult to interpret and, due to different genetic background strains, do not always produce a human disease phenocopy nor provide an accurate readout of the reproductive defects caused by specific RSVs in these genes. Even though recessive LoF mutations in genes can be validated by the use of knockout mice and other knockdown models, missense mutations require further individual evaluation because of unpredictable effects on protein function. Most studies have used in vitro modeling to assess individual missense mutations. However, such experiments are technically demanding, time consuming, and costly. Importantly, because most IGD mutations are either heterozygous, compound heterozygous, or arise in a digenic or oligogenic manner, accurate modeling necessitates the development of a model system, such as zebrafish, worm, or modified stem cells, that can tolerate the simultaneous insertion of multiple hits and use the emerging CRISPR/Cas9 system of gene perturbation (123). Alternatively, it may be possible to use patient-derived samples to generate IPS cell lines for in vitro studies. These may be particularly useful for fulfilling criterion 7, the rescue of specific mutations. Currently, with the exception of a few mutations in 4 IGD genes, KAL1, KISS1, GNRH1, and GNRHR, most IGD mutations have not been tested in rescue experiments to prove that they are a causative (Table 1, criterion 7) (124–128). Although rescue of specific mutations is currently not the “gold-standard” for validating a gene candidate, it may well be critical to assigning pathogenicity to missense mutations in known genes.
Moving forward, it will be crucial to consider how to select the right functional assays for gene validation, particularly as some mutations may only alter specific second-messenger pathways or imbue neomorphic protein functions. The development of alternative reproductive model systems that are more scalable, cost effective, robust, and high throughput is a highly desirable goal and may, in fact, be critical to the future of the field.
G. Testing of genetic burden in IGD
Although a few previous studies have used case-control association tests to establish the potential burden of pathogenicity of individual IGD gene variants based on their frequency in IGD vs control populations (Table 3) (14, 28–30, 41, 44, 45, 61, 62, 71–75, 77, 78, 80, 83–85, 94, 129–146), few have addressed the overall genetic burden for each individual IGD gene. Past pursuit of this particular line of investigation has been limited for several reasons including: 1) the rarity of most genes causing IGD, which argues for large numbers of control individuals to be screened to obtain a proper grasp of any potential genetic burden in the disease cohort; 2) the lack of such data from appropriate ethnically matched controls; 3) the economic burden of screening full coding sequences of large genes for all allelic RSVs when compared with the more limited costs of targeted screening of specific mutations; and 4) the known limitations of bioinformatic programs based upon protein structure, evolutionary conservation, etc, to predict the functional impact of RSVs accurately.
Table 3.
Properties of Statistical Association Tests Performed on Identified Variants in IGD Genes
| Gene | Total Number of IGD Probands Screened | Diagnosisa | Total Number of Controls | Citation |
|---|---|---|---|---|
| KAL1 | 1 | KS | 100 | 129 |
| 1 | KS | 150 | 130 | |
| 28 | KS | 100 | 131 | |
| 30 | KS | 100 and dbSNP | 14 | |
| GNRHR | 2 | nIHH | 20 | 77 |
| 46 | nIHH | 75 | 132 | |
| GNRH1 | 310 | nIHH | 192 | 78 |
| FGFR1 | 28 | KS | 100 | 131 |
| 80 | nIHH and KS | 100 | 133 | |
| 7 | nIHH | 235 | 41 | |
| 141 | KS | 150 | 134 | |
| 150 | KS | 250 | 135 | |
| 134 | nIHH | 270 | 136 | |
| 2 | nIHH and KS | 200 | 28 | |
| 30 | KS | 100 and dbSNP | 14 | |
| FGF8 | 451 | nIHH, KS and AHH | 180 | 83 |
| KISS1R | 64a | nIHH | 130 | 71 |
| 30 | nIHH and CDP | 185 | 137 | |
| 166 | nIHH | 276 | 138 | |
| 69 | nIHH | 120 | 139 | |
| KISS1 | 103b | nIHH | 100 | 73 |
| 1025 | nIHH, KS, CDP, HA, AHH | 703 (178 and 1kG) | 74c | |
| TAC3/TACR3 | 56b | nIHH | 100 | 72 |
| 154b | nIHH | 100 | 140 | |
| 345 | nIHH | 292 | 141 | |
| PROK2/PROKR2 | 192 | KS | 250 | 30c |
| 107 | nIHH and KS | 100 | 142 | |
| SEMA3A | 386 | KS | 386 and EVS | 45 |
| 48 screened for deletions | KS | 520 | 44 | |
| 34 | KS | 200 and EVS | 143 | |
| SOX10 | 103 | KS +− WS | 50, 1kG and EVS | 65 |
| 18 | KS | 90, dbSNP, 1kG, EVS, and LOVD | 144 | |
| AXL | 104 | nIHH and KS | 150, NHLBI, 1kG, and EVS | 85 |
| NSMF (NELF) | 33 | KS | 100 | 80 |
| 168 | nIHH and KS | 372 | 145 | |
| 1 | KS | 384 | 28 | |
| WDR11 | 201 | nIHH and KS | 880 | 146 |
| CHD7 | 101 (+96 exons 6–10) | nIHH and KS | >180 | 61 |
| 36 | KS | 300 | 62 | |
| IL17RD | 386 | KS | 155 and 1kG | 29 |
| DUSP6 | KS | |||
| SPRY4 | nIHH and KS | |||
| FLRT3 | nIHH and KS | |||
| FGF17 | nIHH and KS | |||
| HS6ST1 | 338 | nIHH and KS | 500 and EVS | 84 |
| FEZF1 | 30 | KS | 136, dbSNP, 1kG, and EVS | 75 |
| SEMA3E | 121 | nIHH and KS | 1kG and EVS | 94 |
Size of patient and control cohorts used for statistical association analyses for variants found in each of the 25 genes. dbSNP, The Single Nucleotide Polymorphism Database (http://www.ncbi.nlm.nih.gov/SNP/); 1kG, 1000 genomes database (http://www.1000genomes.org/); EVS, Exome variant server (http://evs.gs.washington.edu/EVS/); LOVD, Leiden Open Variation Database (http://www.lovd.nl/3.0/home); HA, hypothalamic amenorrhea.
Diagnosis of probands with mutations in the target gene.
The studies included both endogamous pedigrees and sporadic cases.
Additional genetic burden test performed.
The first attempt at establishing the genetic burden of an IGD gene was reported by Dode et al (30), who sequenced 192 unrelated KS patients and 250 ethnically matched control subjects for the genes PROK2 and PROKR2. Burden testing demonstrated that the total proportion of PROKR2 alleles was significantly higher in KS patients than in controls, strongly suggesting the involvement of this gene in IGD (30). This observation of an enhanced genetic burden in IGD vs controls was also supported by the validating presence of deleterious (nonsense) LoF mutations in their IGD cohort. Although this study was rigorous by the standards at the time, the number of healthy controls used is now considered inadequate. Employing similar statistical methods, Chan et al (74) found no statistically significant difference in the prevalence of the total number of KISS1 RSVs in 1025 patients (displaying both rare and common forms of IGD) vs 703 controls, mainly due to lack of controls.
Apart from these efforts, the cumulative burden of a subset of IGD genes was first tested by Sykiotis et al (26), who analyzed a cohort of 397 IGD patients and 179 healthy controls that underwent Sanger sequencing for 8 known IGD genes (KAL1, FGFR1, PROKR2, PROK2, NSMF, FGF8, GNRHR, and KISSR1). Even though detailed burden analysis for each separate gene was not described, the overall proportion of altered alleles in these 8 genes in the IGD cohort exceeded those found in the controls, showing that the IGD patients were significantly more likely to harbor rare protein-altering variants (26). This finding of the cumulative enrichment of RSVs in a subset of IGD genes remained consistent in a subsequent study by Miraoui et al, who investigated an expanded list of 17 genes (29), and in a study of GnRH-deficient females screened for 11 genes (147). Although these studies also had small numbers of controls available for comparisons, taken together, it is clear that there is a cumulative burden of variation in these genes in IGD patients.
More recently, a large control cohort became available to assert the causality of RNF216 in syndromic patients with Gordon Holmes syndrome (cerebellar ataxia and IGD). Although this gene is not yet a proven IGD gene, lessons can be learned from the approach taken to assess its burden. In comparison with the burden test done for PROKR2, this gene was screened in a patient population of 8 probands contrasted with more than 13 000 control chromosomes assembled from patients with apparently unrelated conditions assembled by the National Heart, Lung, and Blood Institute's Exome Sequencing Project as well as 672 chromosomes from ethnically matched control subjects. Using such large control populations, these investigators were able to conclude that the percentage of deleterious mutations in RNF216 in IGD patients exceeded what is expected by chance (53).
Importantly, although using such large cohorts robustly increases the number of control subjects, false positive findings may still occur due to the intrinsic imbalance of contrasting large-scale control datasets obtained from loosely phenotyped “normals” to smaller, more densely phenotyped and sequenced rare IGD patient cohorts. Additionally, to reduce the “noise” in these complex comparisons as much as possible, the screening of healthy individuals should ideally use identical sequencing pipelines and variant calling algorithms as the one that surfaced the candidate mutations in the disease population, a criterion that is often difficult to fulfill. Moreover, the depth of detailed phenotyping almost always varies considerably in subjects recruited for large-scale control cohorts, indicating the need for both careful clinical assessments and expansion of the number of individuals needed for recruitment to these datasets.
Another important consideration is that studies in isolated populations have now revealed significant enrichment for rare LoF mutations in known IGD genes (148). For example, the frequency of mutations in the PROKR2 gene is far higher in the geographically limited Maghrebian (North African) KS population than in European KS patients (149, 150). Additionally, 77% of familial nIHH cases of Turkish origin have been shown to be enriched for variants in a subset of neuroendocrine IGD genes, including GNRHR, TACR3, TAC3, KISS1R, and KISS1 (69). These examples suggest that different selection pressures operating within isolated populations may cause some variants to look common when in other contexts they are actually quite rare.
In summary, although the biological relevance of known IGD genes is fairly well established, in the past, these genes have been indicted largely owing to the considerable weight of LoF mutations, chromosomal structural abnormalities, and evidence from human genetics (such as large pedigrees with endogamy). Although deleterious variants and other evidence may have been sufficient to substantiate a gene in the past, moving forward, as NGS technology continues to improve, we will soon be faced with such a large number of novel genes and an even larger number of missense RSVs, which cannot be eliminated by pedigree analyses or other traditional approaches. Hence, when sequencing is done on a cohort of patients, rather than a family, it will ultimately become crucial to calculate the enrichment for the RSVs in a single gene (ie, their “genetic burden”) in the disease population compared with an ethnically matched control population with much greater accuracy. Although there may be reasons (such as gene size, sequence coverage, domain-specific location of RSVs, etc) that a valid candidate gene may not pass a genetic burden test, when there is a sufficiently large, well-phenotyped, ethnic-specific control population to compare with, the assessment of genetic burden should really occur as an important step to reduce the number of genes/RSVs requiring detailed functional analyses. In addition, although this review is only a discussion of “gene-level” burden, it will become increasingly important for this analysis to be done at the “variant level,” even after a gene has been sufficiently indicted as causative in IGD.
IV. The Future
A. New tools and approaches for the discovery of IGD genes
Although the clinical presentation of IGD patients has remained constant, newly developed genetic discovery tools are continually improving and there is constant effort to optimize these new techniques for genetic discovery. Thus, even though the theory behind the genetic analysis of IGD cases remains the same, NGS is highlighting the complexity of the underlying human genetic architecture of this disease and the need for methods to analyze it further. Several of the most recent successes in gene discovery in IGD are now derived from genome-wide searches, whereas in the past they had been quite targeted to shorter DNA segments based on the available approaches. It seems clear that NGS methods will continue to improve and as the numbers of IGD patients tested increases, the yield in new gene discovery will similarly rise. Consequently, we anticipate the rapid accrual of several helpful new tools that should improve the current ambiguities of determining the pathogenicity of RSVs in candidate genes. These include: 1) improved accuracy and calling programs for NGS; 2) increasing size and robustness of control cohorts from appropriate ethnic population studies; 3) rapid improvements in the understanding of the structural biology of proteins and the potential functional implications of specific mutations from crystallographic structural modeling; 4) increased availability of gene expression data from both diseased and normal human tissues, with more tissue subtype specificity; and 5) improved bioinformatics programs using all of the known interactions between proteins, pathway analyses, evolutionary data, and genetic associations of related diseases, etc.
Clearly, whole-genome sequencing (WGS) will increasingly become the genetic standard for mutational screening as its costs decline and supporting bioinformatics capabilities become more widely available and standardized. The data generated from WGS will provide additional insights into the genomic complexity of IGD patients and help optimize the data already collected using already standardized tools. For example, even though traditional karyotyping can identify chromosomal abnormalities, novel techniques like jumping libraries-based WGS that focus on deeper genomic resolution allow for a more precise definition of breakpoints and the detection of cryptic structural events, balanced chromosomal rearrangements, and deletions/duplications of various sizes (151, 152).
In addition to WGS to identify rare variants, genome-wide association studies to assess common variation are increasingly large, available, and able to identify genetic loci associated with more quantitative traits such as the onset of puberty (153) and menopause (154–157), thereby offering new insights in the genetic complexity of reproductive disorders. Importantly, few loci associated with the age of menarche were mapped close to known IGD genes, and most genome-wide associations are either 1) localized at some distance from coding regions revealing possible long-range DNA interactions; 2) interrupt transcription factor binding with possible effects on downstream targets; or 3) interrupt noncoding RNA, which may play a role in the regulation of transcription and translation of target genes (158, 159). New approaches will need to be used to assist in the interpretation of such variation found in noncoding regions. This will be a particularly challenging endeavor, because there is likely exquisite tissue specificity of expression of many genes. To prioritize the gene candidates, publicly available online tools (such as GENets, the ENCODE database, etc) should be used to construct gene-regulatory networks that map the new candidate in gene ontology pathways, assess DNA methylation, histone marks, DNase-I hypersensitivity sites, as well as RNA sequencing data. Additional prioritization of the target genes will also be based on tissue-specific expression and their potential to play a role in the development of the GnRH network.
All of these new types of data generation and analyses will also introduce powerful new ways to investigate environmental impacts on known genes in the initiation of puberty and the biology of pubertal development. In fact, both DNA methylation and histone modifications are now being investigated for several of the IGD genes (160, 161). Thus, apart from the traditional approaches already used for detection of known genes, new tools are likely to provide greater insight in the genomic and epigenomic complexity of the both known IGD genes and future gene candidates, while pointing the field towards genomic regulation and the detection of rare cryptic events.
V. Conclusions
The discovery and validation of the known 25 IGD genes has led to a remarkable improvement in our basis of understanding of the development of the GnRH neuronal network, the neuroendocrine regulation of these unique neurons, and the role these genes play in several reproductive disorders. With the field moving forward using NGS, we assessed the known genes as held to the current standards in the field. Notably, most the genes lack rescue experiments that provide support for genetic causality and are missing well-constructed gene burden testing (criteria 6 and 7). Although it may not be necessary to meet these 2 criteria when the other (particularly genetic) support is overwhelming, in the future, we expect that the rescue experiments will be particularly critical for variant-level analyses, and that the statistical analysis will become an important proxy for genetic information in sporadic cases or small families. As we expected, most the criteria were met by most of the older and more established genes; however, further analysis is required for the determination of the causality of the most recently reported candidates (see Table 1). Such efforts should be focused on mapping the role of the new candidates in the known neurodevelopmental or neuroendocrine pathways of IGD, using expression experiments and bioinformatic tools that assess the interactions between candidate proteins, leveraging those already known to cause IGD. Further studies using the proper control cohorts should also be undertaken for a more precise assessment of the genetic burden for each of the associated genes. In addition, functional studies should be conducted for testing the pathogenicity of the detected variants both in vitro and in vivo. Although the focus of this review has been assessing the candidates on the gene level, variant level analysis will also be critical to ensure that reporting of “causal” genetic defects is confined to RSVs that have sufficient experimental validation. In addition, although we have applied criteria and analyzed genetic discoveries in a specific disease model, many of the same methodologies and considerations apply in the analyses of other Mendelian disorders.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant U54 HD028138. M.I.S. was supported by the Daland Fellowship/American Philosophical Society. K.H.C. was supported by NIH T32 DK007028.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- CHARGE
- coloboma of the eye, heart defects, choanal atresia, growth retardation, genital anomalies, and ear defects
- CHD7
- chromodomain helicase DNA binding protein 7
- DUSP6
- dual specificity phosphatase 6
- FEZF1
- fasciculation and elongation protein zeta family zinc finger 1
- FGFR1
- fibroblast growth factor receptor 1
- FLRT3
- fibronectin leucine-rich transmembrane protein 3
- GNRHR
- GnRH receptor
- HS6ST1
- heparan sulfate 6-O-sulfotransferase 1
- IGD
- isolated GnRH deficiency
- IL17RD
- IL-17 receptor D
- KAL1
- Kallmann 1
- KISS1R
- kisspeptin receptor 1
- KS
- Kallmann syndrome
- LoF
- loss of function
- NGS
- next-generation sequencing
- nIHH
- normosmic idiopathic hypogonadotropic hypogonadism
- NSMF
- NMDA receptor synaptonuclear signaling and neuronal migration factor
- PROK2
- prokineticin 2
- RNF216
- ring finger protein 216
- RSV
- rare sequence variant
- SEMA3A
- semaphorin 3A
- SOX10
- SRY-box 10
- SPRY4
- sprouty homolog 4
- TAC3
- tachykinin 3
- WDR11
- WD repeat domain 11
- WGS
- whole-genome sequencing.
References
- 1. Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. [DOI] [PubMed] [Google Scholar]
- 2. Schlosser G. Making senses development of vertebrate cranial placodes. Int Rev Cell Mol Biol. 2010;283:129–234. [DOI] [PubMed] [Google Scholar]
- 3. Forni PE, Taylor-Burds C, Melvin VS, Williams T, Wray S. Neural crest and ectodermal cells intermix in the nasal placode to give rise to GnRH-1 neurons, sensory neurons, and olfactory ensheathing cells. J Neurosci. 2011;31:6915–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci USA. 1989;86:8132–8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Whitlock KE, Illing N, Brideau NJ, Smith KM, Twomey S. Development of GnRH cells: setting the stage for puberty. Mol Cell Endocrinol. 2006;254–255:39–50. [DOI] [PubMed] [Google Scholar]
- 6. Schwanzel-Fukuda M, Crossin KL, Pfaff DW, Bouloux PM, Hardelin JP, Petit C. Migration of luteinizing hormone-releasing hormone (LHRH) neurons in early human embryos. J Comp Neurol. 1996;366:547–557. [DOI] [PubMed] [Google Scholar]
- 7. Belchetz PE, Plant TM, Nakai Y, Keogh EJ, Knobil E. Hypophysial responses to continuous and intermittent delivery of hypopthalamic gonadotropin-releasing hormone. Science. 1978;202:631–633. [DOI] [PubMed] [Google Scholar]
- 8. Balasubramanian R, Crowley WF., Jr Isolated GnRH deficiency: a disease model serving as a unique prism into the systems biology of the GnRH neuronal network. Mol Cell Endocrinol. 2011;346:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sykiotis GP, Pitteloud N, Seminara SB, Kaiser UB, Crowley WF., Jr Deciphering genetic disease in the genomic era: the model of GnRH deficiency. Sci Transl Med. 2010;2:32rv32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bonomi M, Libri DV, Guizzardi F, et al. New understandings of the genetic basis of isolated idiopathic central hypogonadism. Asian J Androl. 2012;14:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Valdes-Socin H, Rubio Almanza M, Tome Fernandez-Ladreda M, Debray FG, Bours V, Beckers A. Reproduction, smell, and neurodevelopmental disorders: genetic defects in different hypogonadotropic hypogonadal syndromes. Front Endocrinol (Lausanne). 2014;5:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Balasubramanian R, Dwyer A, Seminara SB, Pitteloud N, Kaiser UB, Crowley WF., Jr Human GnRH deficiency: a unique disease model to unravel the ontogeny of GnRH neurons. Neuroendocrinology. 2010;92:81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dodé C, Hardelin JP. Kallmann syndrome. Eur J Hum Genet. 2009;17:139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Laitinen EM, Vaaralahti K, Tommiska J, et al. Incidence, phenotypic features and molecular genetics of Kallmann syndrome in Finland. Orphanet J Rare Dis. 2011;6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crowley WF. The developmental biology of the GnRH neurons. Mol Cell Endocrinol. 2011;346:1–3. [DOI] [PubMed] [Google Scholar]
- 16. Bouloux PM, Munroe P, Kirk J, Besser GM. Sex and smell–an enigma resolved. J Endocrinol. 1992;133:323–326. [DOI] [PubMed] [Google Scholar]
- 17. Kallmann FJ, Schoenfeld W, Barrera S. The genetic aspects of primary eunuchoidism. Am J Ment Defic. 1944;48:203–236. [Google Scholar]
- 18. Quinton R, Duke VM, de Zoysa PA, et al. The neuroradiology of Kallmann's syndrome: a genotypic and phenotypic analysis. J Clin Endocrinol Metab. 1996;81:3010–3017. [DOI] [PubMed] [Google Scholar]
- 19. Nachtigall LB, Boepple PA, Pralong FP, Crowley WF., Jr Adult-onset idiopathic hypogonadotropic hypogonadism–a treatable form of male infertility. N Engl J Med. 1997;336:410–415. [DOI] [PubMed] [Google Scholar]
- 20. Santoro N, Filicori M, Crowley WF., Jr Hypogonadotropic disorders in men and women: diagnosis and therapy with pulsatile gonadotropin-releasing hormone. Endocr Rev. 1986;7:11–23. [DOI] [PubMed] [Google Scholar]
- 21. Caronia LM, Martin C, Welt CK, et al. A genetic basis for functional hypothalamic amenorrhea. N Engl J Med. 2011;364:215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu J, Choa RE, Guo MH, et al. A shared genetic basis for self-limited delayed puberty and idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2015;100:E646–E654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quinton R, Duke VM, Robertson A, et al. Idiopathic gonadotrophin deficiency: genetic questions addressed through phenotypic characterization. Clin Endocrinol (Oxf). 2001;55:163–174. [DOI] [PubMed] [Google Scholar]
- 24. Costa-Barbosa FA, Balasubramanian R, Keefe KW, et al. Prioritizing genetic testing in patients with Kallmann syndrome using clinical phenotypes. J Clin Endocrinol Metab. 2013;98:E943–E953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seminara SB, Hayes FJ, Crowley WF., Jr Gonadotropin-releasing hormone deficiency in the human (idiopathic hypogonadotropic hypogonadism and Kallmann's syndrome): pathophysiological and genetic considerations. Endocr Rev. 1998;19:521–539. [DOI] [PubMed] [Google Scholar]
- 26. Sykiotis GP, Plummer L, Hughes VA, et al. Oligogenic basis of isolated gonadotropin-releasing hormone deficiency. Proc Natl Acad Sci USA. 2010;107:15140–15144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quaynor SD, Kim HG, Cappello EM, et al. The prevalence of digenic mutations in patients with normosmic hypogonadotropic hypogonadism and Kallmann syndrome. Fertil Steril. 2011;96:1424–1430.e1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pitteloud N, Quinton R, Pearce S, et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miraoui H, Dwyer AA, Sykiotis GP, et al. Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 are identified in individuals with congenital hypogonadotropic hypogonadism. Am J Hum Genet. 2013;92:725–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dode C, Teixeira L, Levilliers J, et al. Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet. 2006;2:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Turner HH. A syndrome of infantilism, congenital webbed neck, and cubrtus valgus. Endocrinology. 1938;23:566–674. [PubMed] [Google Scholar]
- 32. Klinefelter HF, Reifenstein EC, Albright F. Syndrome characterized by gynecomastia, aspermatogenesis without a-leydigsm and increased secretion of follicle-stimulating hormone. J Clin Endocrinol Metab. 1942;615–622. [Google Scholar]
- 33. Schwanzel-Fukuda M, Bick D, Pfaff DW. Luteinizing hormone-releasing hormone (LHRH)-expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Brain Res Mol Brain Res. 1989;6:311–326. [DOI] [PubMed] [Google Scholar]
- 34. Bick D, Curry CJ, McGill JR, Schorderet DF, Bux RC, Moore CM. Male infant with ichthyosis, Kallmann syndrome, chondrodysplasia punctata, and an Xp chromosome deletion. Am J Med Genet. 1989;33:100–107. [DOI] [PubMed] [Google Scholar]
- 35. Ballabio A, Parenti G, Tippett P, et al. X-linked ichthyosis, due to steroid sulphatase deficiency, associated with Kallmann syndrome (hypogonadotropic hypogonadism and anosmia): linkage relationships with Xg and cloned DNA sequences from the distal short arm of the X chromosome. Hum Genet. 1986;72:237–240. [DOI] [PubMed] [Google Scholar]
- 36. Ballabio A, Zollo M, Carrozzo R, et al. Deletion of the distal short arm of the X chromosome (Xp) in a patient with short stature, chondrodysplasia punctata, and X-linked ichthyosis due to steroid sulfatase deficiency. Am J Med Genet. 1991;41:184–187. [DOI] [PubMed] [Google Scholar]
- 37. Franco B, Guioli S, Pragliola A, et al. A gene deleted in Kallmann's syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353:529–536. [DOI] [PubMed] [Google Scholar]
- 38. Petit C, Levilliers J, Weissenbach J. Long-range restriction map of the terminal part of the short arm of the human X chromosome. Proc Natl Acad Sci USA. 1990;87:3680–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vermeulen S, Messiaen L, Scheir P, De Bie S, Speleman F, De Paepe A. Kallmann syndrome in a patient with congenital spherocytosis and an interstitial 8p11.2 deletion. Am J Med Genet. 2002;108:315–318. [DOI] [PubMed] [Google Scholar]
- 40. Dodé C, Levilliers J, Dupont JM, et al. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2003;33:463–465. [DOI] [PubMed] [Google Scholar]
- 41. Pitteloud N, Acierno JS, Jr, Meysing A, et al. Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2006;103:6281–6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim HG, Herrick SR, Lemyre E, et al. Hypogonadotropic hypogonadism and cleft lip and palate caused by a balanced translocation producing haploinsufficiency for FGFR1. J Med Genet. 2005;42:666–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim HG, Ahn JW, Kurth I, et al. WDR11, a WD protein that interacts with transcription factor EMX1, is mutated in idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2010;87:465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Young J, Metay C, Bouligand J, et al. SEMA3A deletion in a family with Kallmann syndrome validates the role of semaphorin 3A in human puberty and olfactory system development. Hum Reprod. 2012;27:1460–1465. [DOI] [PubMed] [Google Scholar]
- 45. Hanchate NK, Giacobini P, Lhuillier P, et al. SEMA3A, a gene involved in axonal pathfinding, is mutated in patients with Kallmann syndrome. PLoS Genet. 2012;8:e1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Seminara SB, Achermann JC, Genel M, Jameson JL, Crowley WF., Jr X-linked adrenal hypoplasia congenita: a mutation in DAX1 expands the phenotypic spectrum in males and females. J Clin Endocrinol Metab. 1999;84:4501–4509. [DOI] [PubMed] [Google Scholar]
- 47. Muscatelli F, Strom TM, Walker AP, et al. Mutations in the DAX-1 gene give rise to both X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism. Nature. 1994;372:672–676. [DOI] [PubMed] [Google Scholar]
- 48. Strobel A, Issad T, Camoin L, Ozata M, Strosberg AD. A leptin missense mutation associated with hypogonadism and morbid obesity. Nat Genet. 1998;18:213–215. [DOI] [PubMed] [Google Scholar]
- 49. Farooqi IS, Jebb SA, Langmack G, et al. Effects of recombinant leptin therapy in a child with congenital leptin deficiency. N Engl J Med. 1999;341:879–884. [DOI] [PubMed] [Google Scholar]
- 50. Clément K, Vaisse C, Lahlou N, et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. [DOI] [PubMed] [Google Scholar]
- 51. Farooqi IS, Wangensteen T, Collins S, et al. Clinical and molecular genetic spectrum of congenital deficiency of the leptin receptor. N Engl J Med. 2007;356:237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jackson RS, Creemers JW, Ohagi S, et al. Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat Genet. 1997;16:303–306. [DOI] [PubMed] [Google Scholar]
- 53. Margolin DH, Kousi M, Chan YM, et al. Ataxia, dementia, and hypogonadotropism caused by disordered ubiquitination. N Engl J Med. 2013;368:1992–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Topaloglu AK, Lomniczi A, Kretzschmar D, et al. Loss-of-function mutations in PNPLA6 encoding neuropathy target esterase underlie pubertal failure and neurological deficits in Gordon Holmes syndrome. J Clin Endocrinol Metab. 2014;99:E2067–E2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shi CH, Schisler JC, Rubel CE, et al. Ataxia and hypogonadism caused by the loss of ubiquitin ligase activity of the U box protein CHIP. Hum Mol Genet. 2014;23:1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bernard G, Chouery E, Putorti ML, et al. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am J Hum Genet. 2011;89:415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tétreault M, Choquet K, Orcesi S, et al. Recessive mutations in POLR3B, encoding the second largest subunit of Pol III, cause a rare hypomyelinating leukodystrophy. Am J Hum Genet. 2011;89:652–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Abid F, Hall R, Hudgson P, Weiser R. Moebius syndrome, peripheral neuropathy and hypogonadotrophic hypogonadism. J Neurol Sci. 1978;35:309–315. [DOI] [PubMed] [Google Scholar]
- 59. Chew S, Balasubramanian R, Chan WM, et al. A novel syndrome caused by the E410K amino acid substitution in the neuronal β-tubulin isotype 3. Brain. 2013;136:522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Balasubramanian R, Chew S, MacKinnon SE, et al. Expanding the phenotypic spectrum and variability of endocrine abnormalities associated with TUBB3 E410K syndrome. J Clin Endocrinol Metab. 2015;100:E473–E477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kim HG, Kurth I, Lan F, et al. Mutations in CHD7, encoding a chromatin-remodeling protein, cause idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2008;83:511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jongmans MC, van Ravenswaaij-Arts CM, Pitteloud N, et al. CHD7 mutations in patients initially diagnosed with Kallmann syndrome–the clinical overlap with CHARGE syndrome. Clin Genet. 2009;75:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Marcos S, Sarfati J, Leroy C, et al. The prevalence of CHD7 missense versus truncating mutations is higher in patients with Kallmann syndrome than in typical CHARGE patients. J Clin Endocrinol Metab. 2014;99:E2138–E2143. [DOI] [PubMed] [Google Scholar]
- 64. Balasubramanian R, Choi JH, Francescatto L, et al. Functionally compromised CHD7 alleles in patients with isolated GnRH deficiency. Proc Natl Acad Sci USA. 2014;111:17953–17958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pingault V, Bodereau V, Baral V, et al. Loss-of-function mutations in SOX10 cause Kallmann syndrome with deafness. Am J Hum Genet. 2013;92:707–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lang D, Epstein JA. Sox10 and Pax3 physically interact to mediate activation of a conserved c-RET enhancer. Hum Mol Genet. 2003;12:937–945. [DOI] [PubMed] [Google Scholar]
- 67. Bondurand N, Pingault V, Goerich DE, et al. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Hum Mol Genet. 2000;9:1907–1917. [DOI] [PubMed] [Google Scholar]
- 68. Schulz Y, Wehner P, Opitz L, et al. CHD7, the gene mutated in CHARGE syndrome, regulates genes involved in neural crest cell guidance. Hum Genet. 2014;133:997–1009. [DOI] [PubMed] [Google Scholar]
- 69. Gürbüz F, Kotan LD, Mengen E, et al. Distribution of gene mutations associated with familial normosmic idiopathic hypogonadotropic hypogonadism. J Clin Res Pediatr Endocrinol. 2012;4:121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA. 2003;100:10972–10976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Seminara SB, Messager S, Chatzidaki EE, et al. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349:1614–1627. [DOI] [PubMed] [Google Scholar]
- 72. Topaloglu AK, Reimann F, Guclu M, et al. TAC3 and TACR3 mutations in familial hypogonadotropic hypogonadism reveal a key role for Neurokinin B in the central control of reproduction. Nat Genet. 2009;41:354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Topaloglu AK, Tello JA, Kotan LD, et al. Inactivating KISS1 mutation and hypogonadotropic hypogonadism. N Engl J Med. 2012;366:629–635. [DOI] [PubMed] [Google Scholar]
- 74. Chan YM, Broder-Fingert S, Paraschos S, et al. GnRH-deficient phenotypes in humans and mice with heterozygous variants in KISS1/Kiss1. J Clin Endocrinol Metab. 2011;96:E1771–E1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kotan LD, Hutchins BI, Ozkan Y, et al. Mutations in FEZF1 cause Kallmann syndrome. Am J Hum Genet. 2014;95:326–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tata B, Huijbregts L, Jacquier S, et al. Haploinsufficiency of Dmxl2, encoding a synaptic protein, causes infertility associated with a loss of GnRH neurons in mouse. PLoS Biol. 2014;12:e1001952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. de Roux N, Young J, Misrahi M, et al. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med. 1997;337:1597–1602. [DOI] [PubMed] [Google Scholar]
- 78. Chan YM, de Guillebon A, Lang-Muritano M, et al. GNRH1 mutations in patients with idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2009;106:11703–11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kramer PR, Wray S. Novel gene expressed in nasal region influences outgrowth of olfactory axons and migration of luteinizing hormone-releasing hormone (LHRH) neurons. Genes Dev. 2000;14:1824–1834. [PMC free article] [PubMed] [Google Scholar]
- 80. Miura K, Acierno JS, Jr, Seminara SB. Characterization of the human nasal embryonic LHRH factor gene, NELF, and a mutation screening among 65 patients with idiopathic hypogonadotropic hypogonadism (IHH). J Hum Genet. 2004;49:265–268. [DOI] [PubMed] [Google Scholar]
- 81. Pitteloud N, Zhang C, Pignatelli D, et al. Loss-of-function mutation in the prokineticin 2 gene causes Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA. 2007;104:17447–17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cole LW, Sidis Y, Zhang C, et al. Mutations in prokineticin 2 and prokineticin receptor 2 genes in human gonadotrophin-releasing hormone deficiency: molecular genetics and clinical spectrum. J Clin Endocrinol Metab. 2008;93:3551–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Falardeau J, Chung WC, Beenken A, et al. Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest. 2008;118:2822–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tornberg J, Sykiotis GP, Keefe K, et al. Heparan sulfate 6-O-sulfotransferase 1, a gene involved in extracellular sugar modifications, is mutated in patients with idiopathic hypogonadotrophic hypogonadism. Proc Natl Acad Sci USA. 2011;108:11524–11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Salian-Mehta S, Xu M, Knox AJ, et al. Functional consequences of AXL sequence variants in hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2014;99:1452–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pierce A, Bliesner B, Xu M, et al. Axl and Tyro3 modulate female reproduction by influencing gonadotropin-releasing hormone neuron survival and migration. Mol Endocrinol. 2008;22:2481–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Radovick S, Wray S, Lee E, et al. Migratory arrest of gonadotropin-releasing hormone neurons in transgenic mice. Proc Natl Acad Sci USA. 1991;88:3402–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mellon PL, Windle JJ, Goldsmith PC, Padula CA, Roberts JL, Weiner RI. Immortalization of hypothalamic GnRH neurons by genetically targeted tumorigenesis. Neuron. 1990;5:1–10. [DOI] [PubMed] [Google Scholar]
- 89. Moenter SM. Identified GnRH neuron electrophysiology: a decade of study. Brain Res. 2010;1364:10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fueshko S, Wray S. LHRH cells migrate on peripherin fibers in embryonic olfactory explant cultures: an in vitro model for neurophilic neuronal migration. Dev Biol. 1994;166:331–348. [DOI] [PubMed] [Google Scholar]
- 91. Herbison AE, Pape JR, Simonian SX, Skynner MJ, Sim JA. Molecular and cellular properties of GnRH neurons revealed through transgenics in the mouse. Mol Cell Endocrinol. 2001;185:185–194. [DOI] [PubMed] [Google Scholar]
- 92. Lippincott MF, True C, Seminara SB. Use of genetic models of idiopathic hypogonadotrophic hypogonadism in mice and men to understand the mechanisms of disease. Exp Physiol. 2013;98:1522–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kumar D, Boehm U. Genetic dissection of puberty in mice. Exp Physiol. 2013;98:1528–1534. [DOI] [PubMed] [Google Scholar]
- 94. Cariboni A, André V, Chauvet S, et al. Dysfunctional SEMA3E signaling underlies gonadotropin-releasing hormone neuron deficiency in Kallmann syndrome. J Clin Invest. 2015;125:2413–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Crowley WF., Jr Commentary: the year in endocrine genetics for basic scientists. Mol Endocrinol. 2011;25:1989–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Noel SD, Kaiser UB. G protein-coupled receptors involved in GnRH regulation: molecular insights from human disease. Mol Cell Endocrinol. 2011;346:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Giallourakis C, Henson C, Reich M, Xie X, Mootha VK. Disease gene discovery through integrative genomics. Annu Rev Genomics Hum Genet. 2005;6:381–406. [DOI] [PubMed] [Google Scholar]
- 99. Samocha KE, Robinson EB, Sanders SJ, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wu C, Orozco C, Boyer J, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kapushesky M, Emam I, Holloway E, et al. Gene expression atlas at the European bioinformatics institute. Nucleic Acids Res. 2010;38:D690–D698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Forrest AR, Kawaji H, Rehli M, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kapushesky M, Adamusiak T, Burdett T, et al. Gene Expression Atlas update–a value-added database of microarray and sequencing-based functional genomics experiments. Nucleic Acids Res. 2012;40:D1077–D1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Uhlen M, Fagerberg L, Hallstrom BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 107. Liu X, Yu X, Zack DJ, Zhu H, Qian J. TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinformatics. 2008;9:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Soussi-Yanicostas N, Hardelin JP, Arroyo-Jimenez MM, et al. Initial characterization of anosmin-1, a putative extracellular matrix protein synthesized by definite neuronal cell populations in the central nervous system. J Cell Sci. 1996;109(pt 7):1749–1757. [DOI] [PubMed] [Google Scholar]
- 109. Pellegrini L. Role of heparan sulfate in fibroblast growth factor signalling: a structural view. Curr Opin Struct Biol. 2001;11:629–634. [DOI] [PubMed] [Google Scholar]
- 110. Ayari B, Soussi-Yanicostas N. FGFR1 and anosmin-1 underlying genetically distinct forms of Kallmann syndrome are co-expressed and interact in olfactory bulbs. Dev Genes Evol. 2007;217:169–175. [DOI] [PubMed] [Google Scholar]
- 111. Navarro VM, Gottsch ML, Chavkin C, Okamura H, Clifton DK, Steiner RA. Regulation of gonadotropin-releasing hormone secretion by kisspeptin/dynorphin/neurokinin B neurons in the arcuate nucleus of the mouse. J Neurosci. 2009;29:11859–11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wakabayashi Y, Nakada T, Murata K, et al. Neurokinin B and dynorphin A in kisspeptin neurons of the arcuate nucleus participate in generation of periodic oscillation of neural activity driving pulsatile gonadotropin-releasing hormone secretion in the goat. J Neurosci. 2010;30:3124–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Goodman RL, Lehman MN, Smith JT, et al. Kisspeptin neurons in the arcuate nucleus of the ewe express both dynorphin A and neurokinin B. Endocrinology. 2007;148:5752–5760. [DOI] [PubMed] [Google Scholar]
- 114. Smith JT, Dungan HM, Stoll EA, et al. Differential regulation of KiSS-1 mRNA expression by sex steroids in the brain of the male mouse. Endocrinology. 2005;146:2976–2984. [DOI] [PubMed] [Google Scholar]
- 115. Lehman MN, Hileman SM, Goodman RL. Neuroanatomy of the kisspeptin signaling system in mammals: comparative and developmental aspects. Adv Exp Med Biol. 2013;784:27–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Navarro VM, Tena-Sempere M. Kisspeptins and the neuroendocrine control of reproduction. Front Biosci (Schol Ed). 2011;3:267–275. [DOI] [PubMed] [Google Scholar]
- 117. García-Galiano D, Pinilla L, Tena-Sempere M. Sex steroids and the control of the Kiss1 system: developmental roles and major regulatory actions. J Neuroendocrinol. 2012;24:22–33. [DOI] [PubMed] [Google Scholar]
- 118. Li Q, Millar RP, Clarke IJ, Smith JT. Evidence that nurokinin B controls basal gonadotropin-releasing hormone secretion but is not critical for estrogen-positive feedback in sheep. Neuroendocrinology. 2015;101:161–174. [DOI] [PubMed] [Google Scholar]
- 119. Grachev P, Millar RP, O'Byrne KT. The role of neurokinin B signalling in reproductive neuroendocrinology. Neuroendocrinology. 2014;99:7–17. [DOI] [PubMed] [Google Scholar]
- 120. True C, Nasrin Alam S, Cox K, Chan YM, Seminara SB. Neurokinin B is critical for normal timing of sexual maturation but dispensable for adult reproductive function in female mice. Endocrinology. 2015;156:1386–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Krajewski SJ, Burke MC, Anderson MJ, McMullen NT, Rance NE. Forebrain projections of arcuate neurokinin B neurons demonstrated by anterograde tract-tracing and monosodium glutamate lesions in the rat. Neuroscience. 2010;166:680–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Rance NE, Krajewski SJ, Smith MA, Cholanian M, Dacks PA. Neurokinin B and the hypothalamic regulation of reproduction. Brain Res. 2010;1364:116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Mason AJ, Pitts SL, Nikolics K, et al. Gonadal development and gametogenesis in the hypogonadal mouse are restored by gene transfer. Ann NY Acad Sci. 1987;513:16–26. [DOI] [PubMed] [Google Scholar]
- 125. d'Anglemont de Tassigny X, Fagg LA, Dixon JP, et al. Hypogonadotropic hypogonadism in mice lacking a functional Kiss1 gene. Proc Natl Acad Sci USA. 2007;104:10714–10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Janovick JA, Maya-Nunez G, Conn PM. Rescue of hypogonadotropic hypogonadism-causing and manufactured GnRH receptor mutants by a specific protein-folding template: misrouted proteins as a novel disease etiology and therapeutic target. J Clin Endocrinol Metab. 2002;87:3255–3262. [DOI] [PubMed] [Google Scholar]
- 127. Yanicostas C, Herbomel E, Dipietromaria A, Soussi-Yanicostas N. Anosmin-1a is required for fasciculation and terminal targeting of olfactory sensory neuron axons in the zebrafish olfactory system. Mol Cell Endocrinol. 2009;312:53–60. [DOI] [PubMed] [Google Scholar]
- 128. Janovick JA, Stewart MD, Jacob D, et al. Restoration of testis function in hypogonadotropic hypogonadal mice harboring a misfolded GnRHR mutant by pharmacoperone drug therapy. Proc Natl Acad Sci USA. 2013;110:21030–21035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Zhang S, Wang T, Yang J, Liu Z, Wang S, Liu J. A fertile male patient with Kallmann syndrome and two missense mutations in the KAL1 gene. Fertil Steril. 2010;95:1789.e1783–1786. [DOI] [PubMed] [Google Scholar]
- 130. Zhang S, Xu H, Wang T, Liu G, Liu J. The KAL1 pVal610Ile mutation is a recessive mutation causing Kallmann syndrome. Fertil Steril. 2013;99:1720–1723. [DOI] [PubMed] [Google Scholar]
- 131. Sato N, Katsumata N, Kagami M, et al. Clinical assessment and mutation analysis of Kallmann syndrome 1 (KAL1) and fibroblast growth factor receptor 1 (FGFR1, or KAL2) in five families and 18 sporadic patients. J Clin Endocrinol Metab. 2004;89:1079–1088. [DOI] [PubMed] [Google Scholar]
- 132. Layman LC, Cohen DP, Jin M, et al. Mutations in gonadotropin-releasing hormone receptor gene cause hypogonadotropic hypogonadism. Nat Genet. 1998;18:14–15. [DOI] [PubMed] [Google Scholar]
- 133. Trarbach EB, Costa EM, Versiani B, et al. Novel fibroblast growth factor receptor 1 mutations in patients with congenital hypogonadotropic hypogonadism with and without anosmia. J Clin Endocrinol Metab. 2006;91:4006–4012. [DOI] [PubMed] [Google Scholar]
- 134. Dodé C, Fouveaut C, Mortier G, et al. Novel FGFR1 sequence variants in Kallmann syndrome, and genetic evidence that the FGFR1c isoform is required in olfactory bulb and palate morphogenesis. Hum Mutat. 2007;28:97–98. [DOI] [PubMed] [Google Scholar]
- 135. Pitteloud N, Meysing A, Quinton R, et al. Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. Mol Cell Endocrinol. 2006;254–255:60–69. [DOI] [PubMed] [Google Scholar]
- 136. Raivio T, Sidis Y, Plummer L, et al. Impaired fibroblast growth factor receptor 1 signaling as a cause of normosmic idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2009;94:4380–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Semple RK, Achermann JC, Ellery J, et al. Two novel missense mutations in g protein-coupled receptor 54 in a patient with hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2005;90:1849–1855. [DOI] [PubMed] [Google Scholar]
- 138. Cerrato F, Shagoury J, Kralickova M, et al. Coding sequence analysis of GNRHR and GPR54 in patients with congenital and adult-onset forms of hypogonadotropic hypogonadism. Eur J Endocrinol. 2006;155(suppl 1):S3–S10. [DOI] [PubMed] [Google Scholar]
- 139. Teles MG, Trarbach EB, Noel SD, et al. A novel homozygous splice acceptor site mutation of KISS1R in two siblings with normosmic isolated hypogonadotropic hypogonadism. Eur J Endocrinol. 2010;163:29–34. [DOI] [PubMed] [Google Scholar]
- 140. Young J, Bouligand J, Francou B, et al. TAC3 and TACR3 defects cause hypothalamic congenital hypogonadotropic hypogonadism in humans. J Clin Endocrinol Metab. 2010;95:2287–2295. [DOI] [PubMed] [Google Scholar]
- 141. Gianetti E, Tusset C, Noel SD, et al. TAC3/TACR3 mutations reveal preferential activation of gonadotropin-releasing hormone release by neurokinin B in neonatal life followed by reversal in adulthood. J Clin Endocrinol Metab. 2010;95:2857–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Abreu AP, Trarbach EB, de Castro M, et al. Loss-of-function mutations in the genes encoding prokineticin-2 or prokineticin receptor-2 cause autosomal recessive Kallmann syndrome. J Clin Endocrinol Metab. 2008;93:4113–4118. [DOI] [PubMed] [Google Scholar]
- 143. Känsäkoski J, Fagerholm R, Laitinen EM, et al. Mutation screening of SEMA3A and SEMA7A in patients with congenital hypogonadotropic hypogonadism. Pediatr Res. 2014;75:641–644. [DOI] [PubMed] [Google Scholar]
- 144. Vaaralahti K, Tommiska J, Tillmann V, et al. De novo SOX10 nonsense mutation in a patient with Kallmann syndrome and hearing loss. Pediatr Res. 2014;76:115–116. [DOI] [PubMed] [Google Scholar]
- 145. Xu N, Kim HG, Bhagavath B, et al. Nasal embryonic LHRH factor (NELF) mutations in patients with normosmic hypogonadotropic hypogonadism and Kallmann syndrome. Fertil Steril. 2011;95:1613–1620.e1611–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Kim HG, Ahn JW, Kurth I, et al. WDR11, a WD protein that interacts with transcription factor EMX1, is mutated in idiopathic hypogonadotropic hypogonadism and Kallmann syndrome. Am J Hum Genet. 2010;87:465–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Shaw ND, Seminara SB, Welt CK, et al. Expanding the phenotype and genotype of female GnRH deficiency. J Clin Endocrinol Metab. 2011;96:E566–E576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Lim ET, Wurtz P, Havulinna AS, et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Sarfati J, Fouveaut C, Leroy C, Jeanpierre M, Hardelin JP, Dodé C. Greater prevalence of PROKR2 mutations in Kallmann syndrome patients from the Maghreb than in European patients. Eur J Endocrinol. 2013;169:805–809. [DOI] [PubMed] [Google Scholar]
- 150. Avbelj Stefanija M, Jeanpierre M, Sykiotis GP, et al. An ancient founder mutation in PROKR2 impairs human reproduction. Hum Mol Genet. 2012;21:4314–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Talkowski ME, Rosenfeld JA, Blumenthal I, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Talkowski ME, Ordulu Z, Pillalamarri V, et al. Clinical diagnosis by whole-genome sequencing of a prenatal sample. N Engl J Med. 2012;367:2226–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Perry JR, Day F, Elks CE, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Perry JR, Corre T, Esko T, et al. A genome-wide association study of early menopause and the combined impact of identified variants. Hum Mol Genet. 2013;22:1465–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Carty CL, Spencer KL, Setiawan VW, et al. Replication of genetic loci for ages at menarche and menopause in the multi-ethnic Population Architecture using Genomics and Epidemiology (PAGE) study. Hum Reprod. 2013;28:1695–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Rahmani M, Earp MA, Ramezani Tehrani F, et al. Shared genetic factors for age at natural menopause in Iranian and European women. Hum Reprod. 2013;28:1987–1994. [DOI] [PubMed] [Google Scholar]
- 157. Chen CT, Liu CT, Chen GK, et al. Meta-analysis of loci associated with age at natural menopause in African-American women. Hum Mol Genet. 2014;23:3327–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Guil S, Esteller M. RNA-RNA interactions in gene regulation: the coding and noncoding players. Trends Biochem Sci. 2015;40:248–256. [DOI] [PubMed] [Google Scholar]
- 160. Lomniczi A, Loche A, Castellano JM, et al. Epigenetic control of female puberty. Nat Neurosci. 2013;16:281–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Lomniczi A, Wright H, Ojeda SR. Epigenetic regulation of female puberty. Front Neuroendocrinol. 2014;36C:90–107. [DOI] [PMC free article] [PubMed] [Google Scholar]