Abstract
RPE65 -associated LCA is an autosomal recessive disease that results in reduced visual acuity and night blindness beginning at birth. It is one of the few retinal degenerative disorders for which promising clinical gene transfer trials are currently underway. However, the ability to enroll patients in a gene augmentation trial is dependent upon identification of 2 bona fide disease-causing mutations and there are some patients with the phenotype of RPE65-associated disease who might benefit from gene transfer but are ineligible because 2 disease-causing genetic variations have not yet been identified. Some such patients have novel mutations in RPE65 for which pathogenicity is difficult to confirm. The goal of this study was to determine if an intronic mutation identified in a 2 year-old patient with presumed RPE65-associated disease was truly pathogenic and grounds for inclusion in a clinical gene augmentation trial. Sequencing of the RPE65 gene revealed two mutations: 1) a previously identified disease causing exonic leucine-to-proline mutation (L408P), and 2) a novel single point mutation in intron 3 (IVS3-11) resulting in an A>G change. RT-PCR analysis using RNA extracted from control human donor eye derived primary RPE, control iPSC-RPE cells and proband iPSC-RPE cells revealed that the identified IVS3-11 variation caused a splicing defect that resulted in a frame shift and insertion of a premature stop codon. In this study we demonstrate how patient-specific iPSCs can be used to confirm pathogenicity of unknown mutations, which can enable positive clinical outcomes.
Keywords: patient-specific induced pluripotent stem cells, retina, RPE, disease modeling, clinical translation
Introduction
Leber congenital amaurosis (LCA) is a rare autosomal recessive retinal degenerative disorder caused by mutations in any one of at least 20 different genes [1]. One of these genes, retinal pigment epithelium-specific 65kDa (RPE65), has been reported to account for anywhere between 6–16% of the LCA writ large [1]. The RPE65 protein functions as a retinal isomerase that catalyzes the isomerization of all-trans-retinyl to the light sensitive chromophore 11-cis-retinal, a major component of the visual cycle [2, 3]. Loss of RPE65 results in deficiency of 11-cis-retinal, inability to activate rhodopsin and failure of rod photoreceptor cells to detect light [4].
Clinically, patients with RPE65-associated LCA typically experience early-onset progressive vision loss, night blindness, nystagmus, and a profound reduction in the electroretinogram [1, 5]. Under ophthalmoscopic examination the fundus of patients with early stage disease can appear quite normal [1, 5, 6]. However, as the disease progresses, profound retinal abnormalities, including pigmentary changes, atrophic lesions, and progressive disorganization of the laminated outer retina can be detected [1, 5].
RPE65-associated LCA has gained considerable attention in recent years following the completion of three independent phase 1–2 clinical gene augmentation trials in which subretinal AAV-based delivery of the wild-type human RPE65 gene was performed [7–9]. The majority of patients that have been treated in these original studies have had significant restoration of vision without major adverse events [10]. As promising as these studies have been, some patients with phenotypic disease compatible with the RPE65 phenotype lack 2 bona fide RPE65 mutations. Over-expression of the incorrect gene in such a patient would be of no therapeutic benefit and could actually be harmful (i.e. risks associated with subretinal surgery and overexpression toxicity (e.g., CEP290 [11]). As a result, prior to enrollment into a clinical gene therapy trial, identification and molecular confirmation of both of a participant’s disease-causing alleles are required. For rare diseases such as RPE65-associated LCA, in which the disease-causing gene is expressed exclusively in a subpopulation of retinal cells, this task can be quite challenging. This is especially true when large control data sets are unavailable and the potential disease-causing mutations identified lie within noncoding regions of the gene.
With the advent of the induced pluripotent stem cell [12], and in turn the ability to generate patient-specific cell types that would be otherwise inaccessible to molecular analysis in living patients [13–15], we can now readily investigate the effect of newly identified genetic variants on cell type specific transcripts. As we have recently demonstrated, this is particularly useful for evaluation of intronic mutations that are predicted to interfere with normal splicing, i.e., cryptic splice site mutations [15].
The purpose of this study was to determine if a novel intronic RPE65 mutation, identified in a 2 year-old girl of Haitian ancestry, was pathogenic and if, in combination with a previously identified disease causing mutation, would make the patient eligible to be included in a clinical gene augmentation trial. By using a combination of Sanger sequencing and iPSC technologies we were able to demonstrate that a previously identified coding sequence mutation, L408P, was expressed in the patient’s normal length transcript. The novel IVS3-11 mutation identified was found to interfere with normal splicing, induce a frame shift and insertion of a premature stop codon in the patient’s RPE65 transcript. These findings demonstrate how patient-specific iPSCs can be used to confirm pathogenicity of unknown mutations and enable positive clinical outcomes.
Materials and Methods
Ethics statement
All experiments were conducted with the approval of the University of Iowa Internal Review Board (IRB # 200202022).
Genotyping
The proband, a 2 year-old female of Haitian ethnicity, presented with decreased visual acuity, night blindness and severly reduced electroretinogram. Bidirectional Sanger sequencing of the entire RPE65 gene was performed using DNA isolated from peripheral blood using our previously published protocols [14, 16]. The parental origin of the detected variants was determined by Sanger sequencing of a DNA sample provided by the proband’s mother. No sample was available from the proband’s biological father.
Patient specific iPSC generation
Skin biopsies were collected from patients following informed consent and were used for the generation of fibroblasts and keratinocytes as described previously [11, 13–15, 17]. iPSCs were generated via infection with 4 separate Sendai viruses (CytoTune®, Life Technologies, Carlsbad, CA) each of which were designed to drive expression of one of the transcription factors OCT3/4, SOX2, KLF4, and c-MYC. Fibroblasts plated on 6-well tissue culture plates, were infected at an MOI of 3. At 12–16hrs post-infection, cells were washed and fed with fresh growth media (DMEM media with 10% serum (Life Technologies) and 1% primocin (Invivogen, San Diego, CA)). At 5 days post-infection, cells were passaged onto 6-well Synthemax™ cell culture dishes (Corning, Corning, NY) at a density of 100,000 cells/well, and fed every day with pluripotency media (DMEM F-12 media, 15% knockout serum replacement, 1% 100× MEM NEAA, 1% penicillin/streptomycin (Life Technologies), 0.1 mM beta-mercaptoethanol (Sigma-Aldrich, St. Louis, MO), and 100ng/ml bFGF (R&D Systems, Minneapolis, MN)). Three weeks after viral transduction, iPSC colonies were picked, passaged and clonally expanded on fresh Synthemax™ plates in mTesr media (Stemcell Technologies, Vancover, BC) for further experimentation. During reprogramming and maintenance of pluripotency cells were cultured at 5% CO2 and 37°C.
RPE cell differentiation
To initiate RPE cell differentiation, iPSCs were removed from the culture substrate via manual passage and subjected to our previously described differentiation protocol [15]. As eye-cup-like structures with heavily pigmented RPE layers appeared, i.e. 60–90 days post-differentiation, pigmented clusters were isolated. Cells were then dissociated using TrypLE Express Enzyme (Life Technologies) and plated on Synthemax™-coated cell culture plates in RPE media containing DMEM (high glucose), 10% knockout serum replacement, 0.1 mM MEM NEAA, 2 mM GlutaMAX, 5% heat-inactivated fetal bovine serum (Life Technologies), and 0.1 mM beta-mercaptoethanol (Sigma-Aldrich).
Immunostaining
Cells were fixed in a 4% paraformaldehyde solution and immunostained as described previously [15]. Briefly, cells were incubated overnight at 4°C with antibodies targeted against SSEA4 (R&D Systems), Pax6 (EMD Millipore, Billerica, MA), MITF (Abcam, Cambridge, MA), or ZO1 (EMD Millipore, Billerica, MA). Subsequently, Alexafluor 488 (Life Technologies), Cy3 or Cy5-conjugated secondary antibodies (Jackson Immunochem, West Grove, PA) were used and the samples were analyzed using a Leica DM 2500 SPE confocal microscope (Leica Microsystems, Wetzlar, Germany). Microscopic analysis was performed such that exposure time, gain, and depth of field remained constant between experimental conditions.
RNA isolation, TA cloning and RT-PCR
Total RNA was extracted from undifferentiated iPSCs, differentiated iPSCs-RPE, and human primary RPE cells using the RNeasy Mini-kit (Qiagen, Germantown, MD) per the manufacturers instructions. Briefly, cells were lysed, homogenized and ethanol was added to adjust binding conditions. Samples were spun using RNeasy spin columns, washed and RNA was eluted using RNase-free water. 100ng of RNA was amplified via SuperScript III OneStep RT-PCR System with Platinum Taq DNA Polymerase (Life Technologies) using the gene specific primers indicated in supplemental table 1. Amplified PCR products were separated by electrophoresis on either 2% or 4% agarose gels (Life Technologies) and imaged. Bands were excised from the gel, purified using the QIAquick Gel Extraction kit (Qiagen) and TA cloned using the TOPO TA Cloning Kit with pCR2.1-TOPO vector (Life Technologies). Bacterial outgrowth was plated on LB/AIX plates and cultured overnight at 37°C. Clones were subsequenlty harvested and miniprepped using the QIAprep Spin Miniprep kit (Qiagen). Samples were subjected to bidirectional Sanger sequencing and analyzed using SeqMan Pro (DNASTAR, Madison, WI) and NCBI BLASTn.
Results
Molecular Genetic Analysis
A female of Haitian ethnicity first presented at 2 years-of-age with exotropia (form of strabismus causing an outturned eye), reduced visual acuity (20/60 OU) and poor night vision. Upon later ERG analysis the patient was found to have a significant reduction in B wave amplitude, under both scotopic and photopic conditions, indicating widespread photoreceptor cell dysfunction. Ophthalmoscopy revealed symmetric attenuation of the retinal vasculature and pigmentary changes consistent with an inherited retinal degenerative disease. Although the lack of nystagmus and a best-corrected visual acuity of 20/60 are not typical of LCA, i.e. relatively mild, this clinical history has been reported for patients with RPE65-associated disease [18].
Genetic analysis of the RPE65 gene revealed a previously identified disease-causing leucine-to-proline variation (L408P) in exon 11 [19], and a novel single point mutation in intron 3 (IVS3-11, A>G). Sanger sequencing of the proband’s mother revealed only the latter variant. Although RPE65-associated LCA is one of the few forms of retinal degeneration for which gene therapy is available, as indicated above to be eligible for enrolment in a clinical gene augmentation trial, confirmation of the pathogenicity of the patients’ novel intronic mutation is required. There were at least three obstacles to this confirmation: 1) introns commonly harbor non-disease causing polymorphisms, 2) a large cohort of control individuals of Haitian ethnicity was not readily available, and 3) RPE65 is expressed exclusively in retinal pigmented epithelial cells. Thus, the most direct way to confirm pathogenicity of the patient’s novel variation was to obtain RNA from the patient’s own RPE cells.
iPSC-RPE cell generation
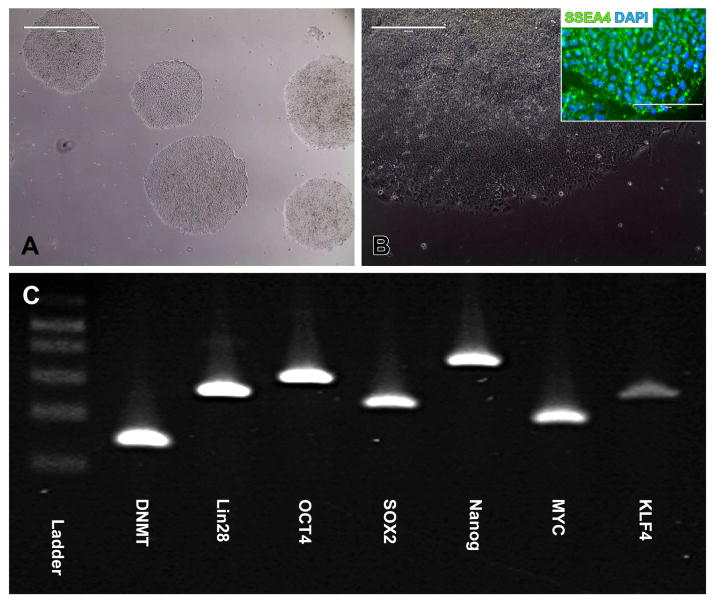
As RPE65 is exclusively expressed in the retina, i.e., predominantly in RPE, to evaluate the pathogenicity of the proband’s novel IVS3-11 mutation, generation of patient-specific RPE cells was required. iPSCs were generated from the proband as described previously [13]. Three weeks after transduction, iPSC colonies were manually isolated and clonally expanded under feeder-free conditions using Synthemax™-treated cell culture surfaces. Following expansion, well-defined, densely packed colonies consisting of cells with a high nucleus-to-cytoplasm ratio that express the pluripotency marker SSEA4 were present (Fig. 1A & B). The pluripotency markers DNMT, Lin28, OCT4, SOX2, Nanog, MYC, and KLF4 could also be detected via RT-PCR (Fig. 1C).
Figure 1. Derivation of iPSCs from a patient with clinically diagnosed LCA.
A–C: Microscopic analysis of iPSC cultures generated from a 3 year-old patient with clinically diagnosed LCA. At 2–3 weeks post-viral transduction iPSC colonies that were isolated, subcultured, and clonally expanded on SynthemaxTM cell culture surfaces maintain a pluripotent morphology (A, B), and express the pluripotency markers SSEA4 as determined by live staining (B, inset) and DNMT, LIN28, OCT4, SOX2, NANOG, C-MYC, and KLF4 as determined by RT-PCR (C). Scale bar = 1mm (A), 400um (B).
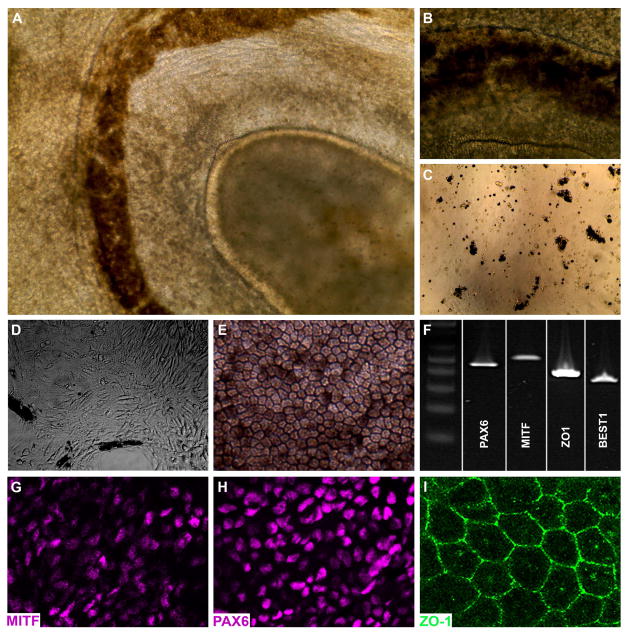
Following expansion and characterization, iPSC cultures were passaged and differentiated using our previously developed stepwise differentiation protocol [13–15]. Approximately 40 days after the onset of differentiation, iPSCs formed pigmented foci that over the next 20–50 days formed C-shaped eyecup-like structures with a clearly defined pigmented layer (Fig. 2A–B). Following Isolation, the pigmented layers were gently dissociated, sub-cultured (Fig. 2C) and expanded for subsequent analysis. By 72 hours post-plating, extensive cell spreading and a near complete loss of pigmentation was noted (Fig. 2D). Three weeks after plating, confluent cultures of densely pigmented hexagonal RPE cells were present (Fig. 2E). Expression of the retinal transcription factor PAX6, the RPE cell transcription factor, MITF, the tight junction marker, ZO1 and the RPE cell channel, BEST1, were detected via RT-PCR (Fig. 2F). These findings were corroborated via immunocytochemical analysis using antibodies targeted against MITF (Fig 2F), PAX6 (Fig 2G) and ZO1 (Fig. 2H).
Figure 2. Generation of iPSC-derived RPE cells from a patient with clinically diagnosed LCA.
A–B: iPSC-derived eyecup-like structures generated from a patient with clinically diagnosed LCA (A, low magnification – B, high magnification). C–E: Following isolation, dissociation and subculture (C), pigmented cells adhere to the cell culture surface, spread, lose pigmentation, and assume a fibroblastic morphology (D). By 3 weeks post-plating, a confluent monolayer of RPE cells are present that have taken on the typical cuboidal RPE morphology and regained pigmentation (E). F: RT-PCR analysis using primer pairs targeted against the RPE transcription factors PAX6 and MITF, the tight junction marker ZO1, and the RPE-specific channel BEST1. G–I: Immunocytochemical analysis using antibodies targeted against the RPE transcription factors PAX6 (G) and MITF (H), and the tight junction marker ZO1 (I).
Transcriptional analysis of RPE65 mutations in patient-specific iPSC-RPE cells
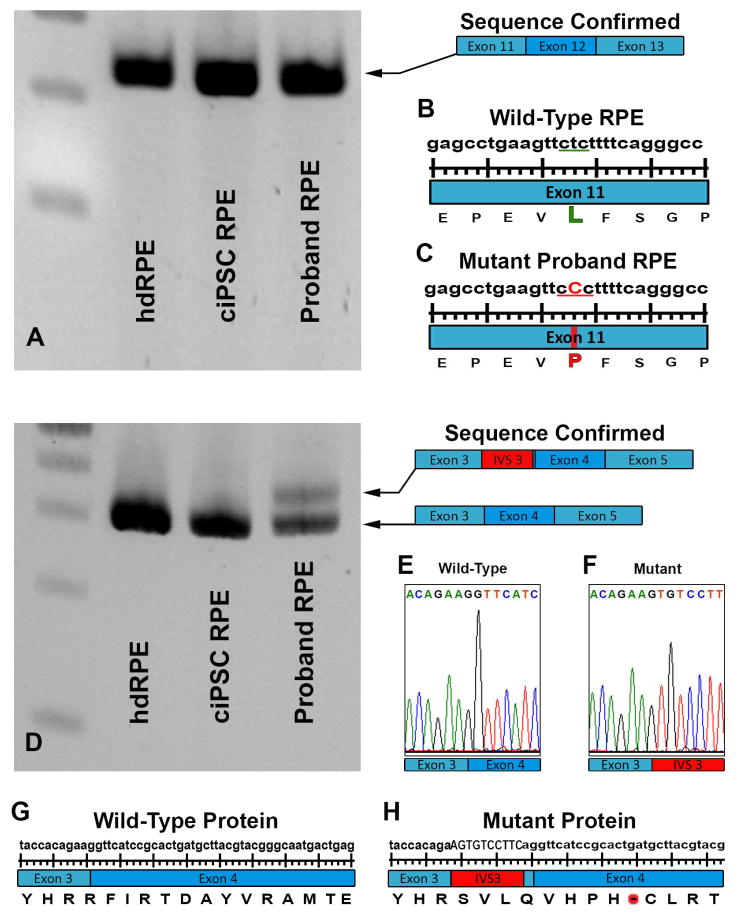
To determine if the RPE65 mutations identified were truly pathogenic, i.e. that they alter the RPE65 transcript, mRNA isolated from RPE cells derived from human donor eyes, control iPSC-RPE cells (Fig. 3) and proband iPSC-RPE cells was subjected to RT-PCR analysis. As expected, Sanger sequencing of the RT-PCR product generated using primers spanning Exons 11 through 13 confirmed the presence of the previously reported disease causing leucine-to-proline variation (L408P) (Fig. 4A, C). This variation was not detected in the transcript of either human donor eye derived primary RPE or control iPSC-RPE cells (Fig. 4A, B).
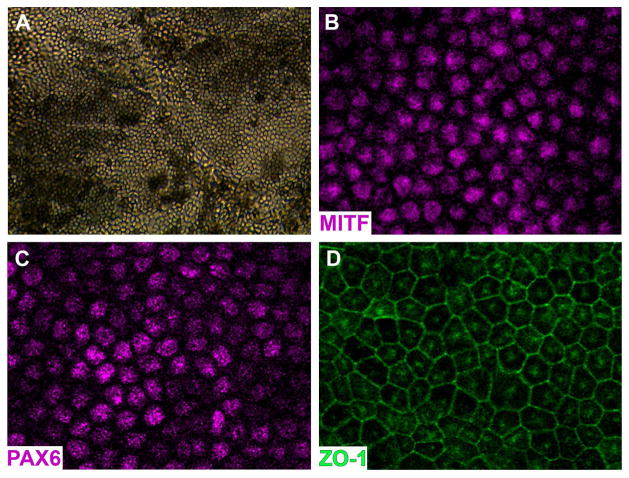
Figure 3. Generation of iPSC derived RPE cells from a control non-LCA individual.
A: Phase micrograph of a confluent monolayer of pigmented iPSC-derived RPE cells. B–D: Immunocytochemical analysis using antibodies targeted against the RPE transcription factors PAX6 (G) and MITF (H), and the tight junction marker ZO1 (I).
Figure 4.
Investigation of genomic RPE65 variants in human donor eye derived primary RPE and iPSC-derived RPE cells. A: RT-PCR analysis of RPE65 exons 11 to 13 in human control RPE/choroid (hdRPE), human control iPSC-derived RPE cells (ciPSC-RPE) and human LCA iPSC-derived RPE cells (Proband-RPE). A previously identified single heterozygous point mutation detected in exon 11 of DNA isolated from blood (L408P) was confirmed in the proband’s RPE cell transcript. D: RT-PCR analysis of RPE65 exon 3 to 5 in control human donor eye derived primary RPE (hdRPE), human control iPSC-derived RPE cells (ciPSC-RPE), and human LCA iPSC-derived RPE cells (Proband-RPE). An intronic splice site mutation in intervening sequence 3 of the RPE65 gene results in the introduction of a pseudoexon (IVS3 Red) causing a translational frameshift and a premature stop codon. B & C: Schematic diagram depicting wildtype and proband L408P transcript. E & F: Chromatogram depicting of wildtype and proband IVS3-11 transcript. G & H: Schematic diagram depicting wildtype and proband IVS3-11 protein.
If the novel IVS3-11 A>G variant identified in the proband is truly disease causing, we predicted that it would function by altering splicing of the patient’s RPE65 transcript. To determine if this was the case, a series of RT-PCR/sequencing experiments similar to those described above, using primers spanning exons 3 through 5, were performed. As compared to human donor eye derived primary RPE and control iPSC-RPE cells, iPSC-RPE generated from the proband contained two separate RPE65 transcripts (Fig. 4D). The lower molecular weight wild-type transcript was identical to that seen in donor and control iPSC-RPE cells and contained the normal exon 3 to exon 4 transition (Fig. 4E). The higher molecular weight transcript resulted from insertion of the remaining 10 nucleotides of intron 3 before transitioning into exon 4 (Fig. 4F–H). Introduction of intronic sequence between Exons 3 and 4 resulted in a frame shift and a premature stop codon just 9 amino acids downstream (Fig. 4H). To determine the degree to which this variation induces mis-splicing, and in turn loss of wild type transcript, rtPCR products, generated using primers spanning the patient’s entire RPE65 transcript, were TA cloned and bidirectionally sequenced. Of the 249 clones successfully sequenced, 11 (4.4% of the total RPE65 message) were found to contain the normal wild type transcript, 199 (79.9% of the total RPE65 message) were found to contain the L408P variant and 39 (15.7% of the total RPE65 message) were found to contain the IVS3-11 variant (importantly none of the clones contained both IVS3-11 and L408P variants). The predominance of the L408P transcript suggests that the majority of the IVS3-11 transcript is lost to nonsense-mediated decay. Note that the presence of wild type transcript, albeit low, could explain why the patient’s disease phenotype at presentation was slightly milder than that reported for patients with two null mutations [1, 5].
Discussion
Patient-specific iPSCs provide a valuable tool for the evaluation of potential disease-causing mutations. This technology has enabled scientists to explore the pathophysiology of human diseases in ways that were once only possible by the use of animal models. For rare autosomal recessive disorders, such as RPE65-associated LCA, there is often little statistical evidence for the pathogenicity of one or both of a patient’s putative disease-causing mutations. This is especially true for ethnic groups that have not been well characterized and from which control samples and large genetic data sets are unavailable.
Non-coding mutations are not an uncommon cause of recessive diseases. In fact, in diseases such as CEP290-associated LCA a single intronic variation accounts for the largest proportion of disease-causing alleles [20]. Unfortunately, unlike coding sequence mutations, for which an effect can often be readily predicted, it is frequently difficult to determine if a variation identified in an intron has an effect on the transcript that could confidently be considered as pathogenic. This is especially true when the gene of interest is expressed exclusively in a tissue that is inaccessible for molecular analysis.
As an excellent example of this dilemma, a 2 year-old Haitian girl presented with atypical LCA. Following sequencing of the patient’s RPE65 gene, two mutations were identified. The first a single point mutation in exon 11, resulted in a leucine-to-proline amino acid substitution at position 408. This previously reported mutation had been demonstrated to cause RPE65-associated disease via reduced enzyme activity [21], induction of protein misfolding, ubiquitination and rapid proteasome-dependent degradation [19]. The proband’s biological father was not available for testing and thus it is theoretically possible that the L408P variant occurred as a de novo event on the maternal allele. The arguments against this are: 1) following PCR and cloning of the patients entire RPE65 transcript the L408P and IVS3-11 variants were never detected in the same clone, 2) the presence of a rare phenotype (LCA) in the proband that agrees with her genotype, 3) the fact that L408P has been previously observed in LCA patients, and 4) that L408P has never been reported as a recurrent mutation; that is, it does not appear to be a hyper-mutable site. The second mutation identified in the proband was a novel IVS3-11 A>G variation that was directly confirmed to have been inherited from her mother. Since this variation lies within an intron, it would not be expected to induce an amino acid change. Rather, we would predict that if this mutation were truly disease causing, it would most probably act via creation of a cryptic splice site, exonification of a portion of intron 3, generation of a frame shift and insertion of a premature stop codon. As RPE65 is expressed exclusively in RPE cells, a tissue that is for obvious reasons not amenable to biopsy for molecular analysis, in the pre-iPSC era this hypothesis would have been nearly impossible to test in a clinically relevant patient-specific fashion. In this study we have demonstrated how patient-specific iPSC-derived RPE cells can be used to rapidly investigate the pathogenicity of a novel intronic mutation. Specifically, the novel IVS3-11 A>G RPE65 variation was demonstrated to induce a splicing abnormality that resulted in exonification of 10 nucleotides of intron 3, induction of a translational frame shift and insertion of a premature stop codon just 9 amino acids downstream.
RPE65-associated LCA is one of the few forms of inherited retinal degenerative diseases for which clinical gene augmentation trials are currently underway. Although these trials have been largely successful, it has become clear that the age of the patient and in turn the stage of disease at which the treatment is administered has a significant impact on clinical outcome. As one would expect, the ability to restore visual function decreases with disease progression [22]. Maguire and co-workers demonstrated this effect in RPE65-LCA patients, in which the average change in light sensitivity of treated patients in the 8–11 year old age group was found to be approximately 2.2 log units, whereas in the 19–44 year old age group the average change in light sensitivity was significantly lower at 1.2 log units [22]. The youngest of the patients treated in this trial, an 8-year-old male, experienced an improvement in light sensitivity of as much as 3.8 log units [22]. Although the improvements detected in older individuals were still significant, these data underscore the importance of early molecular diagnosis and treatment.
As indicated above, for rare intronic variants, such as the one identified in the proband presented in this study, in the pre-iPSC era it would have been difficult, if not impossible, to obtain the degree of confidence in the pathogenicity required to allow the patient to be enrolled. Patients such as this would have likely been categorized as molecularly unconfirmed, where they would remain until a new genetic finding of convincing pathogenicity was identified, new patients with the same mutation and a large cohort of control individuals of the Haitian ancestry who lacked this mutation became available, or a convincing animal model was created and studied. This process would likely take months or even years before a conclusion regarding the pathogenicity of this novel mutation could be reached. All the while the patient’s disease would continue to progress and a gene augmentation based treatment approach would be rendered less and less effective.
As the iPSC technology continues to mature, the evaluation of RNA from iPSC-derived retinal cells may become a common step in the analysis of genetic data. That said, for this technology to become widely employed as a diagnostic tool, issues of cost and scalability will need to be addressed. At present, iPSC generation, characterization, expansion, differentiation and analysis are rather expensive and somewhat difficult to perform in a high throughput fashion. For example, iPSC generation currently requires the daily attention of a highly skilled individual, for an average of 10–12 weeks. Additionally, depending on the cell type required and differentiation protocol used [15, 23–28], the process of differentiation can take an additional 20–30 weeks to generate a sufficient number of cells for the types of analysis reported here. Development of reagents and automated systems capable of performing these tasks rapidly, inexpensively, and on a per patient basis would be advantageous. Until such time, this technology will be most appropriate for situations in which an individual’s disease course might be altered with an existing gene-specific therapy. That is, whenever solid confirmation of the pathogenicity of a patient’s novel disease-causing mutation could be used to determine their eligibility for either a clinical trial or an expensive and somewhat invasive approved treatment, this degree of functional investigation of the patient’s alleles may be warranted despite the associated expense.
Table 1.
Primer sequences
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Best1 | CCTGCTCTGCTACTACATCATC | GTGTTCTGCCGGAGTCATAA |
| c-Myc | GCTGCTTAGACGCTGGATTT | AGCAGCTCGAATTTCTTCCA |
| DNMT | TGTACCGAGTTGGTGATGGTGTGT | TGCTGCCTTTGATGTAGTCGGAGT |
| Klf4 | AGAAGGATCTCGGCCAATTT | AAGTCGCTTCATGTGGGAGA |
| Lin28 | AGAGTAAGCTGCACATGGAAGGGT | TATGGCTGATGCTCTGGCAGAAGT |
| Mitf | AGAAAGTAGAGGGAGGGATAGTC | CCGAGACAGGCAACGTATTT |
| Nanog | TTCTTCCACCAGTCCCAAAG | TTGCTCCACATTGGAAGGTT |
| Oct4 | AACTCGAGCAATTTGCCAAGCTCC | TTCGGGCACTGCAGGAACAAATTC |
| Pax6 | GGGCAATCGGTGGTAGTAAA | CCTCAATTTGCTCTTGGGTAAAG |
| RPE65ex3-5 | GGCCAGGACTCTTTGAAGTT | TCTCTGTGCAAGCGTAGTAATC |
| RPE65ex11-13 | GCTGACACAGGCAAGAATTTAG | ACATTCAGCTTACAGAGCCTATC |
| Sox2 | CATCACCCACAGCAAATGAC | GCAAACTTCCTGCAAAGCTC |
| ZO1 | GGAGTGGTGTGGTTAACAGAA | TCGTCTCTCTCAGAGGCATTAG |
Background
Although gene augmentation to treat molecularly confirmed RPE65-LCA has been a promising, patients with clinical disease that lack 2 disease-causing mutations exist. For rare disorders in which the disease-causing gene is expressed exclusively in a subpopulation of retinal cells, validating mutations and conferring treatment eligibility can be challenging.
Translational Significance
By using iPSCs one can now generate cells inaccessible to molecular analysis and investigate the effect of newly identified genetic variants on cell type-specific transcripts. Here we demonstrate how iPSCs can be combined with sequencing approaches to interrogate the pathophysiology of novel mutations and enable clinically meaningful outcomes.
Acknowledgments
Grant support: NIH Directors New Innovator Award 1-DP2-OD007483-01, Research to Prevent Blindness, Foundation Fighting Blindness, Howard Hughes Medical Institute.
This work was supported by funding from the NIH New Investigators award-DP2OD007483-01, the Grousbeck Family Foundation, the Foundation Fighting Blindness and the HHMI.
Footnotes
All authors have read the journal’s authorship agreement and policy on disclosure of potential conflicts of interest, and have no potential conflicts of interest to declare.
Author contributions:
BAT, CC, SS, KRA, LMS, APD, and LA, Collection and/or assembly of data
BAT, LA, RFM, and EMS, Conception and design, Financial support, Provision of study material or patients, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chacon-Camacho OF, Zenteno JC. Review and update on the molecular basis of Leber congenital amaurosis. WORLD JOURNAL OF CLINICAL CASES. 2015;3(2):112–124. doi: 10.12998/wjcc.v3.i2.112. Available at: http://www.wjgnet.com/2307-8960/full/v3/i2/112.htm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Redmond TM, Yu S, Lee E, et al. Rpe65 is necessary for production of 11-cis-vitamin A in the retinal visual cycle. NAT GENET. 1998;20(4):344–351. doi: 10.1038/3813. Available at: http://www.nature.com/doifinder/10.1038/3813. [DOI] [PubMed] [Google Scholar]
- 3.Redmond TM, Poliakov E, Yu S, et al. Mutation of key residues of RPE65 abolishes its enzymatic role as isomerohydrolase in the visual cycle. PROC NATL ACAD SCI US A. 2005;102(38):13658–13663. doi: 10.1073/pnas.0504167102. Available at: http://www.pnas.org/cgi/doi/10.1073/pnas.0504167102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns ME, Baylor DA. Activation, deactivation, and adaptation in vertebrate photoreceptor cells. ANNUAL REVIEW OF NEUROSCIENCE. 2001;24(1):779–805. doi: 10.1146/annurev.neuro.24.1.779. Available at: http://www.annualreviews.org/doi/abs/10.1146/annurev.neuro.24.1.779. [DOI] [PubMed] [Google Scholar]
- 5.Redmond TM. Focus on Molecules: RPE65, the visual cycle retinol isomerase. EXP EYE RES. 2009;88(5):846–847. doi: 10.1016/j.exer.2008.07.015. Available at: http://linkinghub.elsevier.com/retrieve/pii/S0014483508002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ripamonti C, Henning GB, Ali RR, et al. Nature of the visual loss in observers with Leber’s congenital amaurosis caused by specific mutations in RPE65. INVEST OPHTHALMOL VIS SCI. 2014;55(10):6817–6828. doi: 10.1167/iovs.14-14923. Available at: http://www.iovs.org/cgi/doi/10.1167/iovs.14-14923. [DOI] [PubMed] [Google Scholar]
- 7.Bainbridge JWB, Smith AJ, Barker SS, et al. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N ENGL J MED. 2008;358(21):2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 8.Maguire AM, High KA, Auricchio A, Wright JF, Pierce EA, Testa F, Mingozzi F, Bennicelli JL, Ying GS, Rossi S, Fulton A, Marshall KA, Banfi S, Chung DC, Morgan JI, Hauck B, Zelenaia O, Zhu X, Raffini L, Coppieters F, De Baere E, Shindler KS, Volpe NJ, Surace EM, Acerra C, Lyubarsky A, Redmond TM, Stone E, Sun J, McDonnell JW, Leroy BP, Simonelli F, Bennett J. Age-dependent effects of RPE65 gene therapy for Leber’s congenital amaurosis: a phase 1 dose-escalation trial. Lancet. 2009 Nov 7;374(9701):1597–605. doi: 10.1016/S0140-6736(09)61836-5. Epub 2009 Oct 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hauswirth WW, Aleman TS, Kaushal S, et al. Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. HUM GENE THER. 2008;19(10):979–990. doi: 10.1089/hum.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobson SG, Cideciyan AV, Ratnakaram R, et al. Gene therapy for leber congenital amaurosis caused by RPE65 mutations: safety and efficacy in 15 children and adults followed up to 3 years. ARCH OPHTHALMOL. 2012;130(1):9–24. doi: 10.1001/archophthalmol.2011.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burnight ER, Wiley LA, Drack AV, et al. CEP290 gene transfer rescues Leber congenital amaurosis cellular phenotype. GENE THER. 2014 doi: 10.1038/gt.2014.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. CELL. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 13.Tucker BA, Anfinson KR, Mullins RF, et al. Use of a synthetic xeno-free culture substrate for induced pluripotent stem cell induction and retinal differentiation. STEM CELLS TRANSL MED. 2013;2(1):16–24. doi: 10.5966/sctm.2012-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tucker BA, Scheetz TE, Mullins RF, et al. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. PROC NATL ACAD SCI US A. 2011;108(34):E569–76. doi: 10.1073/pnas.1108918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tucker BA, Mullins RF, Streb LM, et al. Patient-specific iPSC-derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. ELIFE. 2013;2:e00824. doi: 10.7554/eLife.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun TA, Mullins RF, Wagner AH, et al. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. HUM MOL GENET. 2013 doi: 10.1093/hmg/ddt367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tucker BA, Solivan-Timpe F, Roos BR, et al. Duplication of TBK1 Stimulates Autophagy in iPSC-derived Retinal Cells from a Patient with Normal Tension Glaucoma. J STEM CELL RES THER. 2014;3(5):161. doi: 10.4172/2157-7633.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weleber RG, Michaelides M, Trzupek KM, et al. The phenotype of Severe Early Childhood Onset Retinal Dystrophy (SECORD) from mutation of RPE65 and differentiation from Leber congenital amaurosis. INVEST OPHTHALMOL VIS SCI. 2011;52(1):292–302. doi: 10.1167/iovs.10-6106. Available at: http://www.iovs.org/cgi/doi/10.1167/iovs.10-6106. [DOI] [PubMed] [Google Scholar]
- 19.Li S, Izumi T, Hu J, et al. Rescue of enzymatic function for disease-associated RPE65 proteins containing various missense mutations in non-active sites. J BIOL CHEM. 2014;289(27):18943–18956. doi: 10.1074/jbc.M114.552117. Available at: http://www.jbc.org/cgi/doi/10.1074/jbc.M114.552117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stone EM. Leber congenital amaurosis - a model for efficient genetic testing of heterogeneous disorders: LXIV Edward Jackson Memorial Lecture. AM J OPHTHALMOL. 2007;144(6):791–811. doi: 10.1016/j.ajo.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 21.Philp AR, Jin M, Li S, et al. Predicting the pathogenicity of RPE65 mutations. HUM MUTAT. 2009;30(8):1183–1188. doi: 10.1002/humu.21033. Available at: http://doi.wiley.com/10.1002/humu.21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maguire AM, High KA, Auricchio A, et al. Age-dependent effects of RPE65 gene therapy for Leber’s congenital amaurosis: a phase 1 dose-escalation trial. LANCET. 2009;374(9701):1597–1605. doi: 10.1016/S0140-6736(09)61836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong X, Gutierrez C, Xue T, et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. NAT COMMUN. 2014;5:4047. doi: 10.1038/ncomms5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips MJ, Wallace KA, Dickerson SJ, et al. Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. INVEST OPHTHALMOL VIS SCI. 2012;53(4):2007–2019. doi: 10.1167/iovs.11-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamba DA, McUsic A, Hirata RK, et al. Generation, purification and transplantation of photoreceptors derived from human induced pluripotent stem cells. PLOS ONE. 2010;5(1):e8763. doi: 10.1371/journal.pone.0008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowland TJ, Blaschke AJ, Buchholz DE, et al. Differentiation of human pluripotent stem cells to retinal pigmented epithelium in defined conditions using purified extracellular matrix proteins. J TISSUE ENG REGEN MED. 2013;7(8):642–653. doi: 10.1002/term.1458. [DOI] [PubMed] [Google Scholar]
- 27.Kamao H, Mandai M, Okamoto S, et al. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. STEM CELL REPORTS. 2014;2(2):205–218. doi: 10.1016/j.stemcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano T, Ando S, Takata N, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. CELL STEM CELL. 2012;10(6):771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]