ABSTRACT
Rabies, one of the oldest infectious diseases, still presents a public health threat in most parts of the world today. Its pathogen, rabies virus (RABV), can utilize its viral proteins, such as the nucleoprotein and phosphorylation protein, to subvert the host innate immune system. For a long time, the large (L) protein was believed to be essential for RABV transcription and replication, but its role in viral pathogenicity and immune evasion was not known. Recent studies have found that the conserved K-D-K-E tetrad motif in the L protein is related to the methyltransferase (MTase) activity in the viral mRNA process. In the present study, a series of RABV mutations in this motif was constructed with the recombinant CVS-B2c (rB2c) virus. Two of these mutants, rB2c-K1685A and rB2c-K1829A, were found to be stable and displayed an attenuated phenotype in both in vitro growth and in vivo pathogenicity in adult and suckling mice. Further studies demonstrated that these two mutants were more sensitive to the expression of the interferon-stimulated gene product IFIT2 than the parent virus. Taken together, our results suggest that K1685 and K1829 in the L protein play important roles in pathogenicity and immune evasion during RABV infection.
IMPORTANCE Rabies continues to present a public health threat in most areas of the world, especially in the developing countries of Asia and Africa. The pathogenic mechanisms for rabies are not well understood. In the present study, it was found that the recombinant rabies viruses rB2c-K1685A and rB2c-K1829A, carrying mutations at the predicted MTase catalytic sites in the L protein, were highly attenuated both in vitro and in vivo. Further studies showed that these mutants were more sensitive to the expression of the interferon-stimulated gene product IFIT2 than the parent virus. These findings improve our understanding of rabies pathogenesis, which may help in developing potential therapeutics and an avirulent rabies vaccine.
INTRODUCTION
Rabies is an ancient zoonotic disease that still causes more than 55,000 human deaths around the world every year (1). Rabies virus (RABV) belongs to the family Rhabdoviridae, genus Lyssavirus. Its genome is a nonsegmented negative-strand (NNS) RNA encoding five structural proteins: the nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and large protein (L; also termed RNA-dependent RNA polymerase [RdRp]) (2). The ribonucleoprotein complex contains the RNA genome tightly encapsidated by N, P, and L. Both L and P are involved in the process of viral transcription and replication. Laboratory-adapted RABVs stimulate the host innate immune responses, while street viruses evade them (3), resulting in restricted enhancement of the blood-brain barrier (BBB) (4, 5), mild inflammation, and little to no neuronal destruction in the central nervous systems (CNS) of rabies patients (6, 7).
Type I interferon (IFN) is the first line of the host innate defense against viral infections (8, 9). Type I IFN activates the JAK/STAT intracellular signaling pathway, leading to transcriptional induction of hundreds of IFN-stimulated genes (ISGs), many of which have antiviral activities against a wide range of viruses (10). RABV infection activates the innate immune sensor RIG-I/MDA5, inducing the production of classical type I IFN in a few specific types of cells (11). However, RABV has also evolved different strategies to counteract the antiviral effects of the type I IFN responses (12–15). RABV P inhibits IFN production by impairing IFN-regulatory factor 3 (IRF-3) phosphorylation (16, 17), and it also subverts downstream signaling by blocking the nuclear transport of STAT1 (18) and alters promyelocytic leukemia protein (PML) nuclear bodies by retaining PML in the cytoplasm (19). RABV N has been reported to play an important role in the evasion of host RIG-I-mediated antiviral responses and thereby to allow the virus to propagate efficiently and spread quickly (12, 20).
Studies of other NNS RNA viruses, especially vesicular stomatitis virus (VSV), have revealed a highly conserved K-D-K-E motif within the L protein (21, 22). A set of clear biochemical assays with VSV and Sendai virus (SeV) proved that the K-D-K-E motif was related to methyltransferase (MTase) activities during the viral mRNA-capping process (21, 23). The electron cryomicroscopy structure of the full-length VSV L protein was recently resolved at 3.8 Å, providing insight into this MTase domain (24). Moreover, the structure-based alignments of some NNS viruses, including RABV, VSV, Ebola virus, respiratory syncytial virus, and measles virus, demonstrated that the K-D-K-E motif is conserved among these viruses and would function as the N-7 and 2′-O MTases during the viral mRNA-capping process (24). N-7 MTase activity has been proven to be required for recognition of the cap by the translation initiation factor eIF-4E (eukaryotic initiation factor 4E) and to facilitate the translation of viral mRNA (25). Recently, some studies showed that 2′-O MTase activity could facilitate evasion of the recognition of human and mouse coronaviruses by one of the innate sensors, MDA5, or of the restriction of West Nile virus (WNV), vaccinia virus (VACV), and poxvirus by the IFN-responsive IFIT1 and -2 genes (encoding IFN-induced proteins with tetratricopeptide repeats) (26, 27). Most recent studies have shown that VSV with defective MTase activity is highly attenuated both in vitro and in vivo, indicating that MTases play an important role in VSV pathogenesis (28).
In the present study, the roles of the K-D-K-E motif in RABV pathogenicity and immune evasion were investigated by constructing a series of recombinant RABVs (rRABVs) with amino acid substitutions throughout the predicted MTase region in the RABV L protein. It was found that two of the mutants, rB2c-K1685A and rB2c-K1829A, were stable during at least the first 15 passages in cell culture. Compared with the parent virus, rB2c, rB2c-K1685A and rB2c-K1829A exhibited highly attenuated phenotypes both in vitro and in vivo. Further studies revealed that the K1685A and K1829A mutants did not enhance the production of IFN but were more sensitive to the expression of IFIT2 than the parent virus. These findings highlight the importance of K1685 and K1829 in rabies pathogenesis and provide a rational design for novel vaccines and therapeutics.
MATERIALS AND METHODS
Cells, viruses, antibodies, and animals.
Mouse neuroblastoma cells (NA), human neuroblastoma cells (SK-N-SH), RAW 264.7 cells (a mouse macrophage cell line), HEK-293T cells, and HEK-293T-IFIT1/2 cells, which express a human IFIT1 or IFIT2 gene (29) (a gift from Jutao Guo, Drexel University, Philadelphia, PA), were maintained in RPMI 1640 medium (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY). BSR cells, a cloned cell line derived from BHK-21 cells, were maintained in Dulbecco's modified Eagle's medium (DMEM; Mediatech) containing 10% FBS. VSV labeled with green fluorescent protein (GFP) (a gift from Mingzhou Chen, Wuhan University, Wuhan, China) was propagated in BHK-21 cells. A fluorescein isothiocyanate (FITC)-conjugated antibody against RABV N was purchased from Fujirebio (Malvern, PA). Anti-RABV P monoclonal antibody 1E3 was made in our laboratory. FITC-conjugated secondary antibodies were purchased from Invitrogen (Waltham, MA, USA). Female BALB/c mice at the age of 5 weeks and 1-day-old suckling Kunming mice were purchased from the Center for Disease Control and Prevention of Hubei Province, China. All animal experiments were carried out as approved by the Laboratory Animal Monitoring Committee of Hubei Province, China.
Construction of an anticipated MTase-defective RABV genome.
The recombinant plasmid pcDNA3.1-B2c was constructed by inserting the genome of CVS-B2c into the mammalian expression vector pcDNA3.1 as described previously (30, 31). Key amino acids (K1685, D1797, K1829, and E1867) were mutated to alanine or other amino acids by overlapping PCR. The primers used are listed in Table 1. The PCR products were first digested with ClaI and KpnI (New England BioLabs, Beverly, MA) and then ligated into pcDNA3.1-B2c, which had been digested with ClaI and KpnI.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| ClaI-F | CAACAGATCGA CTTGTGTTACCTCCAAC |
| KpnI-R | CTCTAGACTCGAGGGTACCCGCCCTCCCTTAG |
| K1685A-F | GGTGCCCATTATGCGCTTAAGCCTATT |
| K1685A-R | AATAGGCTTAAGCGCATAATGGGCACC |
| D1797A-F | CTCATTATTTGTGCTGCAGAAGTTACTG |
| D1797A-R | CAGTAACTTCTGCAGCACAAATAATGAG |
| K1829A-F | TATCTGGTCTTCGCAACTTACGGGACT |
| K1829A-R | AGTCCCGTAAGTTGCGAAGACCAGATA |
| E1867A-F | TCCTTTTCTTCTGCGCTGTACCTCCGG |
| E1867A-R | CCGGAGGTACAGCGCAGAAGAAAAGGA |
| D1797L-F | CTCATTATTTGTCTTGCAGAAGTTACTG |
| D1797L-R | CAGTAACTTCTGCAAGACAAATAATGAG |
| D1797T-F | CTCATTATTTGTACTGCAGAAGTTACTG |
| D1797T-R | CAGTAACTTCTGCAGTACAAATAATGAG |
| E1867Q-F | TCCTTTTCTTCTCAGCTGTACCTCCGG |
| E1867Q-R | CCGGAGGTACAGCTGAGAAGAAAAGGA |
| E1867T-F | TCCTTTTCTTCTACGCTGTACCTCCGG |
| E1867T-R | CCGGAGGTACAGCGTAGAAGAAAAGGA |
| IFN-β-F | TCTTTCCATGAGCTACAACTTGCT |
| IFN-β-R | GCAGTATTCAAGCCTCCCATTC |
| hIFIT2-F | AATTCCTTGGAGAGCAGCCT |
| hIFIT2-R | AGACCCAGGCATAGTTTCCC |
| vRNA-F | CTCCACAACGAGATGCTCAA |
| vRNA-R | CATCCAACGGGAACAAGACT |
| mRNA-F | GATCGTGGAACACCATACCC |
| mRNA-R | TTCATAAGCGGTGACGACTG |
Rescue and propagation of rRABVs.
All the recombinant RABVs (rRABVs) were rescued as described previously (30) and were then propagated in BSR cells or in newborn mice. Briefly, to propagate viruses in BSR cells, BSR cells in monolayer culture were infected with RABVs at a multiplicity of infection (MOI) of 0.1 and were incubated at 34°C under 5% CO2 for 4 days. To propagate viruses in suckling mice, 1-day-old suckling Kunming mice were inoculated intracranially (i.c.) with 500 focus-forming units (FFU) of rB2c, rB2c-K1685A, or rB2c-K1829A. When paralysis developed, the mice were euthanized with CO2. The brains were harvested and were homogenized in DMEM, resulting in a 50% brain suspension. All the rRABVs used in the pathogenicity study were propagated in suckling Kunming mice.
Replication kinetics of rRABVs in vitro.
BSR cells cultured in six-well plates were infected with rRABVs at an MOI of 5 for one-step growth curves and at an MOI of 0.01 for multistep growth curves. After 1 h of incubation, supernatants were removed, the cells were washed three times with DMEM, and fresh DMEM containing 2% FBS was added. The supernatant was harvested at 1, 2, 3, 4, and 5 days postinfection (dpi). Virus titration was carried out by a direct fluorescent-antibody assay with BSR cells as described previously (32).
qRT-PCR.
The samples (tissues or cells) were collected on ice at various time points and were homogenized with TRIzol (Invitrogen). Total RNA was extracted and was used for quantitative real-time RT-PCR (qRT-PCR) as described below. First-strand cDNA was synthesized by a First-Strand cDNA synthesis kit (Toyobo) using oligo(dT) primers (TaKaRa). Each qPCR was carried out in duplicate with approximately 100 ng DNase-treated RNA and 5 nM primer pairs by using a one-step SYBR green qRT-PCR mix kit (Toyobo). The primers used are listed in Table 1. For absolute quantification of viral mRNA and viral genomic RNA (vRNA), a plasmid expressing RABV N was used as a standard (33). The copy numbers of mRNA and vRNA were normalized to 1 μg of total RNA. For IFN expression, mRNA copy numbers were normalized to those of the β-actin housekeeping gene.
Histopathology and IHC.
Animals were anesthetized with ether and were perfused by intracardiac injection of phosphate-buffered saline (PBS) as described previously (34). Brains were fixed with paraformaldehyde and were embedded with paraffin. For the immunohistochemistry (IHC) study, the sections were deparaffinized and rehydrated in xylene and ethanol, respectively. Endogenous peroxidase was quenched by incubation in 3% hydrogen peroxide. Sections were heated in 0.01 M citrate buffer at 90°C for 5 min and were naturally cooled down to room temperature. Monoclonal antibody 1E3 against RABV P was used as the primary antibody, and FITC-conjugated anti-mouse IgG was used as the secondary antibody. The sections were observed under an Olympus IX51 fluorescence microscope.
IFN pretreatment.
BSR cells were pretreated with increasing doses of human universal IFN-α (Merck Millipore) for 16 h and were then either mock infected or infected with rB2c, rB2c-K1685A, or rB2c-K1829A at an MOI of 1. At 48 h postinfection (hpi), the virus titer in the supernatant was determined by a direct fluorescent-antibody assay in BSR cells.
IFN bioassay.
To detect the active type I IFN induced by RABV, an indirect method was used as described previously (11). Briefly, RAW 264.7 cells were either mock infected or infected with rB2c, rB2c-K1685A, or rB2c-K1829A at an MOI of 1, and supernatants were collected at 12, 24, and 48 hpi. The supernatants were UV inactivated with a 254-nm UV light source for 30 min. UV-inactivated viral supernatants were then diluted 10 times in RPMI 1640 medium and were added to NA cells for 24 h. Then the NA cells were infected with VSV-GFP at an MOI of 5 for 9 h. Cells were examined for GFP expression under an Olympus IX51 fluorescence microscope.
Flow cytometry.
Infected cells in plates were digested with 0.25% EDTA-trypsin. After one wash with DMEM containing 2% FBS, cells were fixed with 4% paraformaldehyde for 1 h at 4°C. Cells were stained with an FITC-conjugated antibody against RABV N for 45 min at 37°C. After staining, cells were washed three times with PBS, flow cytometry was performed on a BD-C6 flow cytometer (BD Pharmingen), and a minimum of 100,000 events were counted. Data were analyzed by BD FACSDiva (BD Pharmingen) and FlowJo (Tree Star) software.
VNA test.
Virus-neutralizing antibody (VNA) titers were measured using the fluorescent-antibody virus neutralization (FAVN) test. Briefly, 50-μl volumes of serial 3-fold dilutions of test serum and standard serum were prepared on 96-well microplates in 150-μl volumes. Each sample was added to four adjacent wells. A 50-μl volume of a rabies challenge virus (CVS-11) suspension, containing 50 to 200 FFU, was added to each well. The microplates were then incubated at 37°C for 1 h in an incubator with 5% CO2. Following incubation, 50 μl of the cell suspension, containing 2 × 104 BSR cells, was added to each well, and the microplates were incubated at 34°C in an incubator with 5% CO2 for 60 h. Then the plates were fixed in 80% ice-cold acetone at −20°C for 30 min and were air dried. Cells were stained with FITC-conjugated antibodies against RABV N for 45 min at 37°C. After staining, cells were washed three times with PBS. The results were assessed by using an Olympus IX51 fluorescence microscope. VNA titers were expressed in international units per milliliter by comparison with the titer of a reference serum (obtained from the National Institute for Biological Standards and Control, Herts, United Kingdom) included in each test.
Pathogenicity of MTase-defective mutants in mice. (i) Adult mice.
Five-week-old female BALB/c mice were either infected intramuscularly (i.m.) (100 μl/leg) with rB2c, rB2c-K1685A, or rB2c-K1829A at a dose of 6 × 104 FFU, mock infected with DMEM, or inoculated i.c. with the same viruses at a dose of 104 FFU in a volume of 30 μl. Five mice from each group were euthanized at 6 dpi and 10 dpi. The brains were collected for virus quantification or the IHC study; the blood was withdrawn for VNA detection. The remaining mice were monitored over 3 weeks for clinical signs of rabies, survivorship, and body weight loss as described previously (32).
(ii) Suckling mice.
One-day-old Kunming mice were either infected i.c. with rB2c, rB2c-K1685A, or rB2c-K1829A at a dose of 20 FFU or mock infected in a volume of 20 μl. The mice were observed for 21 days for clinical signs of rabies and survivorship.
Statistical analysis.
All data were analyzed with GraphPad Prism software (GraphPad Software, Inc., CA). An unpaired two-tailed t test was used to determine the statistical significance of the infection ratio and viral titers in transgenic cell lines. For experiments to determine the percentage of survival, Kaplan-Meier survival curves were analyzed by the log rank test. For all tests, the following notations were used to indicate significant differences between groups: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
RESULTS
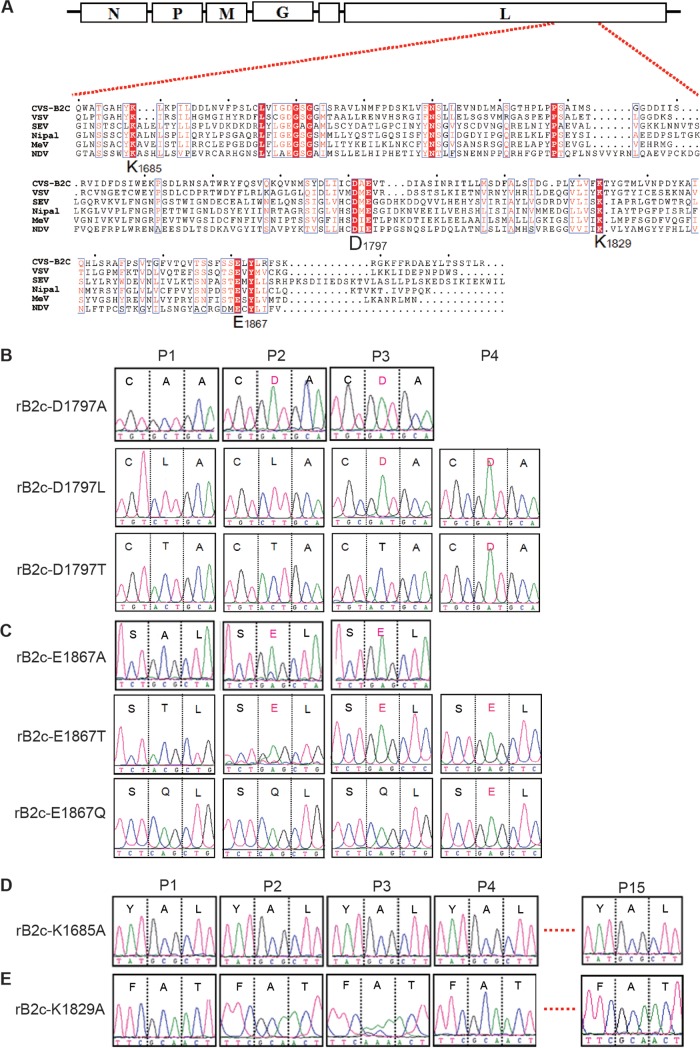
Identification of the conserved K-D-K-E tetrad in RABV L and construction of rRABVs with mutations in the K-D-K-E motif.
Multiple sequence alignments of the large (L) protein among viruses belonging to the order Mononegavirales revealed the presence of a conserved catalytic K-D-K-E tetrad in the C-terminal region of the L protein of some negative-strand RNA viruses, such as VSV, measles virus, SeV, Newcastle disease virus, and Nipah virus, as well as RABV (21, 22) (Fig. 1A). This K-D-K-E tetrad has been demonstrated to be responsible for both N-7 and 2′-O MTase activities in the capping of viral mRNA in VSV (21). To evaluate the role of MTase in the pathogenicity of RABV, the recombinant RABV strain CVS-B2c (rB2c) was chosen due to its modest virulence and easy assessment of the pathogenicity in vivo after i.c. or i.m. infection in a mouse model (30, 34). The four amino acids (Lys-1685, Asp-1797, Lys-1829, and Glu-1867) within the K-D-K-E motif of rB2c were individually mutated to alanine residues. The rRABVs were rescued as described previously (30), and the site mutation was confirmed at the nucleotide level by RT-PCR and sequencing.
FIG 1.
Construction and amino acid sequences of the recombinant RABVs with mutations in the L protein and nucleotide changes in the L gene. (A) Partial sequences of the L proteins of some NNS RNA viruses were aligned by using CLUSTAL 2 software. The conserved K-D-K-E tetrad is indicated. VSV, vesicular stomatitis virus; MeV, measles virus; NDV, Newcastle disease virus; SEV, Sendai virus; Nipal, Nipah virus. GenBank accession numbers are as follows: CVS-B2c, HQ891318.1; VSV, KF880915.1; MeV, NC_001498; NDV, EF065682.1; SEV, M30202.1; Nipal, NC_002728.1. (B to E) The recombinant RABVs with the indicated mutations in the K-D-K-E tetrad were rescued and were passaged in BSR cells. Total RNA was isolated at the indicated passages (P1 to P4). The mRNA fragment of the L gene that contained the mutation(s) in each rRABV was amplified by RT-PCR and then sequenced. Three continuous amino acids are shown in each box, with the mutated amino acid in the middle. The three letters below the large letter are the codon for the amino acid. The curves signify the four nucleotides: green, A; pink, T; blue, C; black, G. The light blue vertical lines are the default division lines per 50 nucleotides in the sequencing report.
Substitutions in the K-D-K-E motif affect the stability of rRABVs.
The rRABVs were rescued in BSR cells, and the mutant region was sequenced during each of the first 15 passages. It was noted that mutant viruses with A1797 and A1867 reverted to the wild type (D1797 and E1867) during the second passage (Fig. 1B and C). However, mutants with K1685A and K1829A were stable and retained the expected mutation even at the 15th passage (Fig. 1D and E). To make sure that the reversions did occur at D1797 and E1867, D or E was mutated to a residue with an electronic charge either similar to or different from that of the original amino acid. As a result, the mutant rRABVs rB2c-D1797L, rB2c-D1797T, rB2c-E1867T, and rB2c-E1867Q were constructed and were confirmed by RT-PCR and sequencing. Sequencing results showed that the L1797 and T1797 rRABVs began to revert to wild-type D1797 at the 3rd and 4th passages, respectively (Fig. 1B) and that the T1867 and Q1867 rRABVs reverted to wild-type E1867 at the 2nd and 4th passages, respectively (Fig. 1C). Besides the mutations at residues 1797 and 1867, no other mutation was found within the K-D-K-E tetrad coding sequence. The same phenomenon was observed when these rRABVs were amplified in the brains of suckling mice, indicating that residues D1797 and E1867 might be indispensable for the viral life cycle, while the lysines in the K-D-K-E motif, K1685 and K1829, might be important, but not necessary, for RABV reproduction.
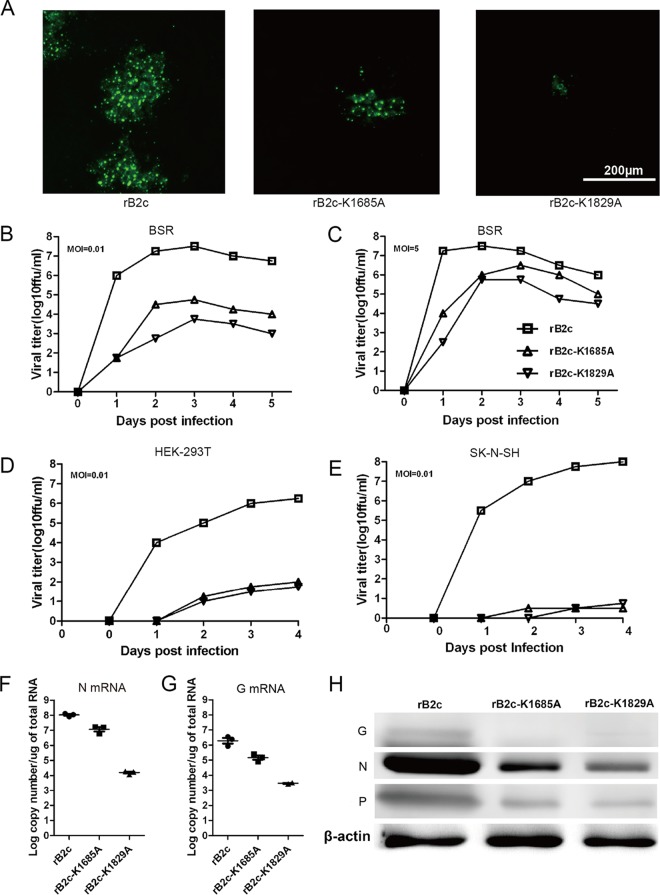
Growth kinetics of rB2c-K1685A and rB2c-K1829A in cell culture.
At 48 hpi in BSR cells, the florescence foci of rB2c-K1685A and rB2c-K1829A were much smaller than those of the parent virus, rB2c (Fig. 2A). When cells were infected at a lower MOI of 0.01, rB2c-K1685A and rB2c-K1829A attained peak titers (104.75 FFU/ml and 103.75 FFU/ml, respectively), which were much lower than that of rB2c (107.5 FFU/ml), at 3 dpi (Fig. 2B); following a high-multiplicity infection (MOI, 5), rB2c-K1685A and rB2c-K1829A reached 106.5 FFU/ml and 105.75 FFU/ml, respectively, close to the peak titer of rB2c (107.5 FFU/ml), but delayed reaching the peak by 1 day (Fig. 2C). In other IFN-competent cell lines, such as HEK-293T (Fig. 2D) and SK-N-SH (Fig. 2E) cells, rB2c-K1685A and rB2c-K1829A displayed even more-attenuated replication compared with that of rB2c. qRT-PCR and Western blot analysis demonstrated that the mutation at K1685 or K1829 decreased viral N and G mRNA transcription (Fig. 2F and G), as well as downstream N, P, and G protein expression (Fig. 2H). Taken together, these results demonstrate that the alanine mutation at K1685 or K1829 of the RABV L protein reduces viral transcription and replication in vitro.
FIG 2.
Characterization of the recombinant RABVs in vitro. (A) Comparison of the fluorescence morphologies of different rRABVs at 48 hpi in BSR cells. Bar, 200 μm. (B to E) One-step (B) or multiple-step (C) growth curves of the rRABVs in BSR cells and one-step growth curves in HEK-293T (D) and SK-N-SH (E) cells. (F and G) N (F) and G (G) mRNA transcription was measured by qRT-PCR. (H) The expression of the indicated viral proteins was determined by Western blotting.
rB2c-K1685A and rB2c-K1829A could not enhance the production of type I IFN.
It has been shown previously that 2′-O MTase-defective mutants of human and mouse coronaviruses facilitate the recognition of the innate sensor MDA5 and enhance the production of type I IFN (27). Another previous study showed that RABV induced type I IFN in only a few cell types, such as macrophages and conventional dendritic cells (cDC) (11). To investigate the production of IFN, RAW 264.7 cells were infected with rB2c, rB2c-K1685A, or rB2c-K1829A, and vRNA and IFN-β (one of the early IFNs) mRNA were measured by qRT-PCR. The kinetics of vRNA demonstrated that all the viruses were unable to replicate in RAW 264.7 cells, in agreement with the previous findings (11), and all had similar amounts of vRNA at 12, 24, and 48 hpi (Fig. 3B). VSV-GFP, which was sensitive to IFN, was utilized to monitor the production of active IFN induced by RABV. The replication of VSV-GFP suggested that rB2c-K1685A and rB2c-K1829A induced much less active IFN than rB2c, especially at 48 hpi (Fig. 3A). All these results suggest that, unlike 2′-O MTase-defective human and mouse coronaviruses, rB2c-K1685A and rB2c-K1829A did not enhance the production of type I IFN.
FIG 3.
rB2c-K1685A and rB2c-K1829A exhibit enhanced sensitivity to type I IFN. (A) RAW 264.7 cells were infected with different rRABVs at an MOI of 1. IFN produced in RAW 264.7 cells was detected by a VSV-GFP bioassay. (B) The production of viral RNA (vRNA) by different rRABVs was measured by qRT-PCR at the indicted time points. (C) BSR cells were pretreated with 1, 10, 100, or 1,000 IU/ml of human universal IFN-α for 16 h and were then infected with different rRABVs at an MOI of 0.1. At 48 hpi, the virus titers in the supernatants were determined in BSR cells. (D) The fold changes in titers were calculated.
rB2c-K1685A and rB2c-K1829A are more sensitive to IFN pretreatment than rB2c.
Pretreatment of cells with IFN can trigger the expression of hundreds of interferon-stimulated genes (ISGs), some of which can inhibit the replication of viruses by different mechanisms. To evaluate whether rB2c-K1685A and rB2c-K1829A are more sensitive to IFN pretreatment than rB2c, BSR cells pretreated with 10-fold-increased doses of universal human IFN-α were infected with each virus, and virus titers were determined at 48 hpi. As shown in Fig. 3C, rB2c was only slightly sensitive to IFN pretreatment. Even with IFN pretreatment at a dose of 1,000 IU/ml, the titer of rB2c alone decreased only 5-fold from that with mock pretreatment. In contrast, rB2c-K1685A and rB2c-K1829A showed highly upregulated sensitivity with increased concentrations of IFN (Fig. 3D). Following pretreatment with 1,000 IU/ml IFN, the titers of rB2c-K1685A and rB2c-K1829A both decreased more than 30-fold. Together, these data indicate that an amino acid substitution at K1685 or K1829 enhances the sensitivity of RABV to some (perhaps not all) specific ISGs.
IFIT2 attenuates the replication of B2c-K1685A and B2c-K1829A viruses in vitro.
Recent data show that some 2′-O MTase-defective viruses, such as mouse hepatitis virus (MHV), West Nile virus (WNV), poxvirus, and vaccinia virus, can be restricted by the expression of the IFIT1 and/or IFIT2 interferon-stimulated gene (26, 27, 35). To evaluate whether rB2c-K1685A and rB2c-K1829A might be suppressed by IFIT1 and/or IFIT2, IFIT1/IFIT2-expressing cells were infected with each of the viruses, and the virus growth kinetics and the percentage of virus-infected cells were evaluated. Overexpression of IFIT1 did not affect the replication of rB2c-K1685A, rB2c-K1829A, or rB2c in 293T cells (Fig. 4A). In contrast, IFIT2 expression significantly reduced the rate of virus replication for rB2c-K1685A and rB2c-K1829A by 103 and 105 FFU/ml, respectively, reductions much greater than that for rB2c (102 FFU/ml), at 3 dpi (Fig. 4B). Analysis of virus-infected cells by flow cytometry confirmed that IFIT2 overexpression reduced the rate of virus replication by 60% for rB2c-K1685A and rB2c-K1829A, a reduction much greater than that for rB2c (30%), at 3 dpi (Fig. 4C). Interestingly, the basal mRNA level of IFIT2 in SK-N-SH and 293T cells is higher than that in BSR cells (Fig. 4D), inversely correlating with more-attenuated replication of mutant viruses in these cells (Fig. 2B, D, and E). Taken together, these data illustrate that overexpression of IFIT2, but not that of IFIT1, reduced the rate of replication for rB2c-K1685A and rB2c-K1829A in cell cultures.
FIG 4.
Overexpression of IFIT2 restricts the replication of B2c-K1685A and B2c-K1829A in vitro. HEK-293T, HEK-293T-IFIT1, or HEK-293T-IFIT2 cells were infected with different viruses at an MOI of 0.5. (A and B) Virus titers were determined at the indicated time points. d, day. (C) Infected 293T-IFIT2 cells were sorted and quantified by fluorescence-activated cell sorting at the indicated time points. (D) Basal IFIT2 mRNA levels in the indicated cells were analyzed by qRT-PCR. ND, not detectable. All data were obtained from 4 samples in each group and are given as means ± standard errors. Asterisks indicate significant differences between the experimental groups indicated.
rB2c-K1685A and rB2c-K1829A exhibit attenuated pathogenicity in vivo.
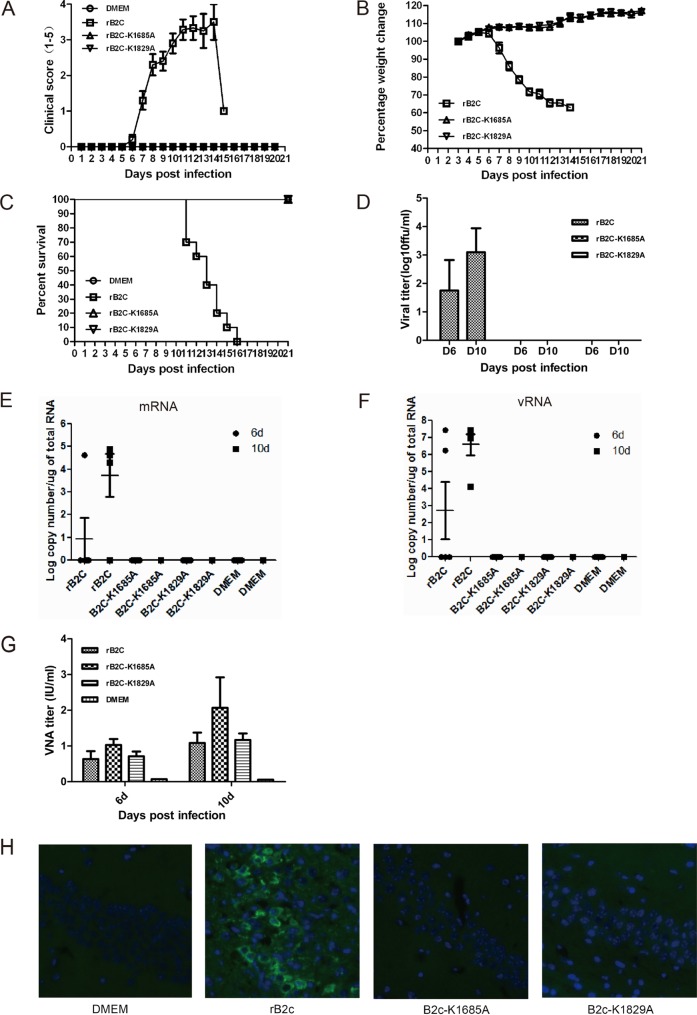
To investigate whether the mutations at K1685 and K1829 within the K-D-K-E tetrad motif affect the pathogenicity of RABV in vivo, 5- to 6-week-old BALB/c mice were either mock infected or infected i.m. with 6 × 104 FFU of each rRABV. Mice infected with rB2c exhibited severe clinical signs of rabies from 8 dpi (Fig. 5A) and lost >30% of body weight at 10 dpi (Fig. 5B), and all (n = 10) died within 3 weeks (Fig. 5C). However, rB2c-K1685A- or rB2c-K1829A-infected mice showed no clinical signs of rabies and no body weight loss. The virus titer (Fig. 5D), viral mRNA (Fig. 5E) or vRNA (Fig. 5F), and viral antigen detected by IHC (Fig. 5H) all demonstrated that rB2c, but neither rB2c-K1685A nor rB2c-K1829A, could cause infection of mouse brains. Together, these results indicated that no virulence of rB2c-K1685A or rB2c-K1829A was detectable in the mouse CNS by i.m. infection. Interestingly, measurement of the VNA titers in serum at 6 and 10 dpi demonstrated that rB2c-K1685A and rB2c-K1829A induced levels of humoral immunity comparable to that with rB2c (Fig. 5G).
FIG 5.
Pathogenicity of rB2c-K1685A and rB2c-K1829A in adult mice following i.m. infection. (A to G) Five-week-old female BALB/c mice (n = 10) were infected i.m. with 6 × 104 FFU of wild-type or mutant RABV. (A to G) The clinical score (A), body weight loss (B), percentage of survival (C), virus titers (D), viral mRNA (E), and viral genomic RNA (vRNA) (F) in the brains and VNA titers in the sera (G) were measured. (H) Brain sections were costained for the RABV antigen (green) and with the nuclear stain 4′,6-diamidino-2-phenylindole (blue). Images are representative of 2 sections from each of 3 animals from each group. Bar, 100 μm.
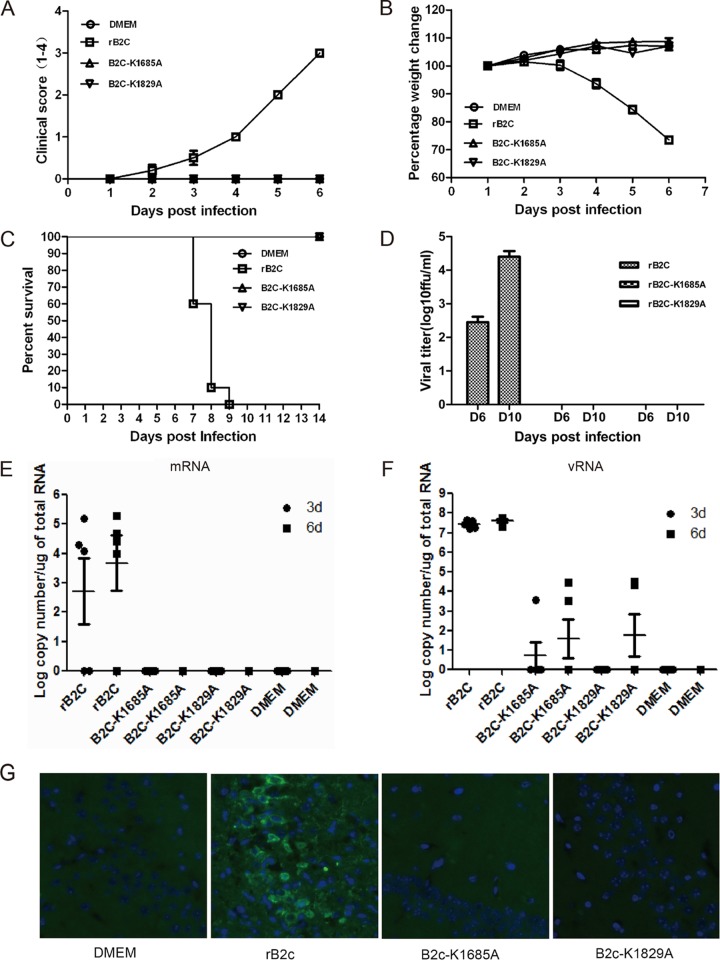
To investigate whether rB2c-K1685A and rB2c-K1829A stimulate robust innate immune responses, enabling the host to clear RABV before it invades the CNS, the pathogenicity of rB2c-K1685A and rB2c-K1829A was evaluated after direct inoculation into the mouse CNS. Strikingly, mice infected with rB2c-K1685A or rB2c-K1829A exhibited no clinical signs of rabies (Fig. 6A) and no body weight loss (Fig. 6B), while mice infected with rB2c quickly succumbed to rabies (Fig. 6C). No virus, viral antigen, or viral RNA could be detected by virus titration, qRT-PCR, or IHC in the brains of mice infected with rB2c-K1685A or rB2c-K1829A (Fig. 6D to G). Together, these studies indicate that rB2c-K1685A and rB2c-K1829A are highly attenuated and exhibit no detectable virulence in adult mice, even by i.c. infection.
FIG 6.
Virulence of rB2c-K1685A and rB2c-K1829A in adult mice infected i.c. (A to F) Five-week-old female BALB/c mice (n = 10) were inoculated i.c. with 104 FFU of wild-type or mutant RABV. The clinical score (A), weight loss (B), percentage of survival (C), virus titers (D), and viral mRNA (E) and vRNA (F) copy numbers in mouse brains are shown. (G) The distribution of viral antigen was revealed by costaining as described in the legend to Fig. 5H. Bar, 100 μm.
To investigate if rB2c-K1685A and rB2c-K1829A can infect the CNS of suckling mice, 1-day-old suckling mice were infected i.c. with different doses of rRABV. As shown in Fig. 7A to C, 40 FFU of rB2c-K1685A and 80 FFU of rB2c-K1829A could kill 90% and 70% of suckling mice, respectively. With a low virus dose (20 FFU), 40% of rB2c-K1685A-infected mice and 90% of rB2c-K1829A-infected mice survived, a survival rate significantly greater than that of rB2c-infected mice (all of which died by 6 dpi) (P < 0.001) (Fig. 7D). Together, these data demonstrate that rB2c-K1685A and rB2c-K1829A are only modestly virulent in suckling mice but are much more attenuated than rB2c.
FIG 7.
Virulence of rB2c-K1685A and rB2c-K1829A in suckling mice. (A to C) One-day-old Kunming suckling mice (n = 10) were infected i.c. with the indicated dose of rB2c (A), rB2c-K1685A (B), or rB2c-K1829A (C). The mice were monitored for 4 weeks, and the survival rate was recorded daily. (D) The percentages of survival of suckling mice (n = 10) inoculated i.c. with 20 FFU of different rRABVs were compared. Kaplan-Meier survival curves were analyzed by the log rank test. Asterisks indicate significant differences between the experimental groups indicated.
DISCUSSION
NNS RNA viruses belong to four families: Rhabdoviridae, Filoviridae, Bornaviridae, and Paramyxoviridae (28). All these viruses possess a large polymerase (L), which is a 220- to 240-kDa protein with multiple functions, including nucleotide polymerization and viral mRNA capping (36). The rhabdoviruses, including VSV and RABV, have evolved an unconventional capping mechanism (37). In the VSV L, there is a GxxT[n]HR motif (where n is 68 to 70 residues in length), which is required for the activity of GDP polyribonucleotidyltransferase (PRNTase), and a K-D-K-E tetrad, which catalyzes both guanine-N-7 (N-7) and ribose 2′-O methylation (21, 38, 39). The nascent viral mRNA is first transferred as a monophosphate RNA onto a GDP acceptor by GDP PRNTase and then sequentially methylated at the ribose 2′-O and N-7 positions, reversing the traditional methylation order (37). Apart from sequence analyses, very little is known about the RABV L protein, because there is no easy system for reconstituting transcription and replication in vitro as there is for other viruses, such as VSV and SeV (40). Thus, the details of mRNA modification for RABV are poorly understood, and most of the information comes from the study of VSV.
In the sequence alignment of the MTase domains of some typical NNS RNA viruses, a highly conserved K-D-K-E catalytic tetrad is found among the L proteins (Fig. 1A). Recent studies indicate that substitutions in the K-D-K-E tetrad motif abolish 2′-O or N-7 MTase activities, or both, for some viruses. For flaviviruses, including Japanese encephalitis virus (JEV), WNV, and dengue virus (DENV), mutation of the E in the K-D-K-E tetrad completely abolishes the 2′-O MTase activities, while the N-7 MTase activities are retained (41–44). In the severe acute respiratory syndrome coronavirus (SARS-CoV), a mutation at any of these residues ablates 2′-O but not N-7 MTase activities (45, 46). However, biochemical assays with VSV demonstrate that substitution at any residue in the K-D-K-E motif ablates both guanine-N-7 and ribose 2′-O MTase activities (21). N-7 methylation is believed to be essential for RNA translation and stability (21, 47). The role of 2′-O methylation has been elusive for a long time. Recent studies demonstrate that 2′-O methylation activity helps a virus escape host innate immune recognition. Mutants of WNV, JEV, DENV, and SARS-CoV defective in 2′-O MTase activity exhibit an attenuated viral pathogenicity phenotype (26, 35, 41, 48).
In contrast to 2′-O MTase-defective mutants of human and mouse coronaviruses, rB2c-K1685A and rB2c-K1829A fail to induce more-robust type I IFN production than rB2c, indicating that the absence of MTase activity in RABV does not contribute to recognition by the innate sensor RIG-I/MDA5. Nevertheless, like the other 2′-O MTase-defective mutant viruses (WNV-E218A, MHV-D130A, and VACV-J3-K175R) (26), rB2c-K1685A and rB2c-K1829A are more sensitive to type I IFN than rB2c, suggesting that these mutants maintain IFN-based attenuation in vitro.
The different biologic functions of IFN are mediated by the products of specific IFN-stimulated genes (ISGs) in the target cells. Among them, the IFN-induced proteins with tetratricopeptide (IFIT) repeats are expressed at very low basal levels, but these are the most strongly induced ISGs during many viral infections (49–51). There are three IFIT family members in mice (IFIT1 [ISG56], IFIT2 [ISG54], and IFIT3 [ISG49]) and four in humans (IFIT1 [ISG56], IFIT2 [ISG54], IFIT3 [ISG60], and IFIT5 [ISG58]) (50). Recent studies demonstrated that IFIT1 and -2 could specifically inhibit virus replication by recognizing viral mRNA lacking 2′-O methylation. IFIT1 could inhibit JEV replication by interacting with 2′-O unmethylated RNA in vitro (26, 35). However, in our present study, we found that IFIT1 overexpression could not significantly inhibit the replication of rB2c-K1685A or rB2c-K1829A in 293T cells, compared with that of rB2c (Fig. 4). In agreement with other studies with WNV, MHV, poxvirus, and vaccinia virus, IFIT2 overexpression significantly restricts MTase-defective RABV in vitro (26). Interestingly, IFIT2 also showed the ability to suppress the replication of the parent virus, rB2c, in cell cultures. A recent report showed that IFIT2 could suppress the spread of VSV in the mouse CNS (52). It will be interesting to investigate the role of IFIT2 in the pathogenicity of RABV in vivo.
As expected, rB2c-K1685A and rB2c-K1829A displayed highly attenuated phenotypes in vivo following i.m. infection. Surprisingly, these two mutants exhibited undetectable virulence in the adult mouse after i.c. infection, suggesting that substitution at K1685 or K1829 almost abrogates the virulence of rB2c in adult mice. There are at least two explanations for this attenuation mechanism. First, the attenuated replication of rB2c-K1685A and rB2c-K1829A in BSR cells relative to that of rB2c and the much smaller size of the mutants' fluorescence foci indicate that mutation of K1685 or K1829 in the L protein significantly reduces RABV replication and spreading. Second, without 2′-O MTase activity, rB2c-K1685A and rB2c-K1829A exhibit enhanced sensitivity to IFIT1 and -2 inhibitory activity. In suckling mice, which lack strong immune responses, rB2c-K1685A and rB2c-K1829A restored robust viral replication and displayed modest virulence in the CNS, suggesting that K1685 and K1829 contribute to immune evasion in vivo.
In summary, our studies demonstrate that substitutions within the K-D-K-E tetrad in RABV L highly attenuate viral pathogenicity both in vitro and vivo. The K1685A and K1829A mutant RABVs display undetectable virulence but elicit humoral immunity comparable to that with the wild type; thus, they may have the potential to be developed as avirulent RABV vaccines.
ACKNOWLEDGMENT
We thank Robert Jeffrey Hogan, University of Georgia, for critical readings of the manuscript.
Funding Statement
This work was partially supported by the National Natural Science Foundation of China (grant 31372419), the Wuhan Science and Technology Bureau of China (grant 2014020101010071), and the Huazhong Agricultural University Scientific & Technological Self-Innovation Foundation (2012RC009) (to L.Z.) and by U.S. Public Health Service grant AI-051560 from the National Institute of Allergy and Infectious Diseases, the National Natural Science Foundation of China (grant 31330078), the China Department of Sciences and Technology (863 Program grant 2011AA10A212), and the China Department of Agriculture (Special Fund for Agro-Scientific Research in the Public Interest grant 201303042) (to Z.F.F.).
REFERENCES
- 1.WHO. 2013. WHO Expert Consultation on Rabies. Second report. World Health Organ Tech Rep Ser 2013(982):1–139. [PubMed] [Google Scholar]
- 2.Wunner WH, Larson JK, Dietzschold B, Smith CL. 1988. The molecular biology of rabies viruses. Rev Infect Dis 10:771–784. [DOI] [PubMed] [Google Scholar]
- 3.Wang ZW, Sarmento L, Wang Y, Li XQ, Dhingra V, Tseggai T, Jiang B, Fu ZF. 2005. Attenuated rabies virus activates, while pathogenic rabies virus evades, the host innate immune responses in the central nervous system. J Virol 79:12554–12565. doi: 10.1128/JVI.79.19.12554-12565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roy A, Phares TW, Koprowski H, Hooper DC. 2007. Failure to open the blood-brain barrier and deliver immune effectors to central nervous system tissues leads to the lethal outcome of silver-haired bat rabies virus infection. J Virol 81:1110–1118. doi: 10.1128/JVI.01964-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy A, Hooper DC. 2008. Immune evasion by rabies viruses through the maintenance of blood-brain barrier integrity. J Neurovirol 14:401–411. doi: 10.1080/13550280802235924. [DOI] [PubMed] [Google Scholar]
- 6.Miyamoto K, Matsumoto S. 1967. Comparative studies between pathogenesis of street and fixed rabies infection. J Exp Med 125:447–456. doi: 10.1084/jem.125.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson AC. 2002. Rabies pathogenesis. J Neurovirol 8:267–269. doi: 10.1080/13550280290100725. [DOI] [PubMed] [Google Scholar]
- 8.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Honda K, Takaoka A, Taniguchi T. 2006. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faul EJ, Wanjalla CN, Suthar MS, Gale M, Wirblich C, Schnell MJ. 2010. Rabies virus infection induces type I interferon production in an IPS-1 dependent manner while dendritic cell activation relies on IFNAR signaling. PLoS Pathog 6:e1001016. doi: 10.1371/journal.ppat.1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masatani T, Ito N, Shimizu K, Ito Y, Nakagawa K, Sawaki Y, Koyama H, Sugiyama M. 2010. Rabies virus nucleoprotein functions to evade activation of the RIG-I-mediated antiviral response. J Virol 84:4002–4012. doi: 10.1128/JVI.02220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rieder M, Conzelmann KK. 2009. Rhabdovirus evasion of the interferon system. J Interferon Cytokine Res 29:499–509. doi: 10.1089/jir.2009.0068. [DOI] [PubMed] [Google Scholar]
- 14.Rieder M, Brzózka K, Pfaller CK, Cox JH, Stitz L, Conzelmann KK. 2011. Genetic dissection of interferon-antagonistic functions of rabies virus phosphoprotein: inhibition of interferon regulatory factor 3 activation is important for pathogenicity. J Virol 85:842–852. doi: 10.1128/JVI.01427-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lafon M. 2011. Evasive strategies in rabies virus infection. Adv Virus Res 79:33–53. doi: 10.1016/B978-0-12-387040-7.00003-2. [DOI] [PubMed] [Google Scholar]
- 16.Brzózka K, Finke S, Conzelmann KK. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J Virol 79:7673–7681. doi: 10.1128/JVI.79.12.7673-7681.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rieder M, Conzelmann KK. 2011. Interferon in rabies virus infection. Adv Virus Res 79:91–114. doi: 10.1016/B978-0-12-387040-7.00006-8. [DOI] [PubMed] [Google Scholar]
- 18.Vidy A, Chelbi-Alix M, Blondel D. 2005. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J Virol 79:14411–14420. doi: 10.1128/JVI.79.22.14411-14420.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blondel D, Regad T, Poisson N, Pavie B, Harper F, Pandolfi PP, de Thé H, Chelbi-Alix MK. 2002. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21:7957–7970. doi: 10.1038/sj.onc.1205931. [DOI] [PubMed] [Google Scholar]
- 20.Masatani T, Ito N, Shimizu K, Ito Y, Nakagawa K, Abe M, Yamaoka S, Sugiyama M. 2011. Amino acids at positions 273 and 394 in rabies virus nucleoprotein are important for both evasion of host RIG-I-mediated antiviral response and pathogenicity. Virus Res 155:168–174. doi: 10.1016/j.virusres.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Fontaine-Rodriguez EC, Whelan SP. 2005. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J Virol 79:13373–13384. doi: 10.1128/JVI.79.21.13373-13384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferron F, Longhi S, Henrissat B, Canard B. 2002. Viral RNA-polymerases—a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem Sci 27:222–224. doi: 10.1016/S0968-0004(02)02091-1. [DOI] [PubMed] [Google Scholar]
- 23.Bujnicki JM, Rychlewski L. 2002. In silico identification, structure prediction and phylogenetic analysis of the 2′-O-ribose (cap 1) methyltransferase domain in the large structural protein of ssRNA negative-strand viruses. Protein Eng 15:101–108. doi: 10.1093/protein/15.2.101. [DOI] [PubMed] [Google Scholar]
- 24.Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SP. 2015. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 1975. 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature 255:33–37. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- 26.Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M Jr, Shi PY, Diamond MS. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Züst R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12:137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma YM, Wei YW, Zhang XD, Zhang Y, Cai H, Zhu Y, Shilo K, Oglesbee M, Krakowka S, Whelan SPJ, Li JR. 2014. mRNA cap methylation influences pathogenesis of vesicular stomatitis virus in vivo. J Virol 88:2913–2926. doi: 10.1128/JVI.03420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang D, Weidner JM, Qing M, Pan XB, Guo H, Xu C, Zhang X, Birk A, Chang J, Shi PY, Block TM, Guo JT. 2010. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J Virol 84:8332–8341. doi: 10.1128/JVI.02199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang G, Wang H, Mahmood F, Fu ZF. 2013. Rabies virus glycoprotein is an important determinant for the induction of innate immune responses and the pathogenic mechanisms. Vet Microbiol 162:601–613. doi: 10.1016/j.vetmic.2012.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue K, Shoji Y, Kurane I, Iijima T, Sakai T, Morimoto K. 2003. An improved method for recovering rabies virus from cloned cDNA. J Virol Methods 107:229–236. doi: 10.1016/S0166-0934(02)00249-5. [DOI] [PubMed] [Google Scholar]
- 32.Zhao L, Toriumi H, Kuang Y, Chen H, Fu ZF. 2009. The roles of chemokines in rabies virus infection: overexpression may not always be beneficial. J Virol 83:11808–11818. doi: 10.1128/JVI.01346-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao L, Toriumi H, Wang H, Kuang Y, Guo X, Morimoto K, Fu ZF. 2010. Expression of MIP-1α (CCL3) by a recombinant rabies virus enhances its immunogenicity by inducing innate immunity and recruiting dendritic cells and B cells. J Virol 84:9642–9648. doi: 10.1128/JVI.00326-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuang Y, Lackay SN, Zhao L, Fu ZF. 2009. Role of chemokines in the enhancement of BBB permeability and inflammatory infiltration after rabies virus infection. Virus Res 144:18–26. doi: 10.1016/j.virusres.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura T, Katoh H, Kayama H, Saiga H, Okuyama M, Okamoto T, Umemoto E, Matsuura Y, Yamamoto M, Takeda K. 2013. Ifit1 inhibits Japanese encephalitis virus replication through binding to 5′ capped 2′-O unmethylated RNA. J Virol 87:9997–10003. doi: 10.1128/JVI.00883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poch O, Blumberg BM, Bougueleret L, Tordo N. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J Gen Virol 71(Part 5):1153–1162. [DOI] [PubMed] [Google Scholar]
- 37.Decroly E, Ferron F, Lescar J, Canard B. 2011. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol 10:51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Wang JT, Whelan SP. 2006. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc Natl Acad Sci U S A 103:8493–8498. doi: 10.1073/pnas.0509821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li J, Rahmeh A, Morelli M, Whelan SP. 2008. A conserved motif in region V of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J Virol 82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albertini AA, Ruigrok RW, Blondel D. 2011. Rabies virus transcription and replication. Adv Virus Res 79:1–22. doi: 10.1016/B978-0-12-387040-7.00001-9. [DOI] [PubMed] [Google Scholar]
- 41.Li SH, Dong H, Li XF, Xie X, Zhao H, Deng YQ, Wang XY, Ye Q, Zhu SY, Wang HJ, Zhang B, Leng QB, Zuest R, Qin ED, Qin CF, Shi PY. 2013. Rational design of a flavivirus vaccine by abolishing viral RNA 2′-O methylation. J Virol 87:5812–5819. doi: 10.1128/JVI.02806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Züst R, Dong H, Li XF, Chang DC, Zhang B, Balakrishnan T, Toh YX, Jiang T, Li SH, Deng YQ, Ellis BR, Ellis EM, Poidinger M, Zolezzi F, Qin CF, Shi PY, Fink K. 2013. Rational design of a live attenuated dengue vaccine: 2′-O-methyltransferase mutants are highly attenuated and immunogenic in mice and macaques. PLoS Pathog 9:e1003521. doi: 10.1371/journal.ppat.1003521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. 2007. Structure and function of flavivirus NS5 methyltransferase. J Virol 81:3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol 80:8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y, Su C, Ke M, Jin X, Xu L, Zhang Z, Wu A, Sun Y, Yang Z, Tien P, Ahola T, Liang Y, Liu X, Guo D. 2011. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog 7:e1002294. doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Decroly E, Debarnot C, Ferron F, Bouvet M, Coutard B, Imbert I, Gluais L, Papageorgiou N, Sharff A, Bricogne G, Ortiz-Lombardia M, Lescar J, Canard B. 2011. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog 7:e1002059. doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furuichi Y, Shatkin AJ. 2000. Viral and cellular mRNA capping: past and prospects. Adv Virus Res 55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menachery VD, Yount BL, Josset L, Gralinski LE, Scobey T, Agnihothram S, Katze MG, Baric RS. 2014. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-O-methyltransferase activity. J Virol 88:4251–4264. doi: 10.1128/JVI.03571-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diamond MS, Farzan M. 2013. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fensterl V, Sen GC. 2011. The ISG56/IFIT1 gene family. J Interferon Cytokine Res 31:71–78. doi: 10.1089/jir.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wacher C, Muller M, Hofer MJ, Getts DR, Zabaras R, Ousman SS, Terenzi F, Sen GC, King NJ, Campbell IL. 2007. Coordinated regulation and widespread cellular expression of interferon-stimulated genes (ISG) ISG-49, ISG-54, and ISG-56 in the central nervous system after infection with distinct viruses. J Virol 81:860–871. doi: 10.1128/JVI.01167-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fensterl V, Wetzel JL, Ramachandran S, Ogino T, Stohlman SA, Bergmann CC, Diamond MS, Virgin HW, Sen GC. 2012. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog 8:e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]