Abstract
We report the crystal structure of the M2 ectodomain (M2e) in complex with a monoclonal antibody that binds the amino terminus of M2. M2e extends into the antibody binding site to form an N-terminal β-turn near the bottom of the paratope. This M2e folding differs significantly from that of M2e in complex with an antibody that binds another part of M2e. This suggests that M2e can adopt at least two conformations that can elicit protective antibodies.
TEXT
Human influenza viruses can escape antibody-mediated immunity by varying the antigenic sites in their surface antigens that are most prone to virus neutralization. Vaccines that elicit antibody responses against conserved surface-exposed antigens, such as the stem region of influenza hemagglutinin or M2e, can protect against multiple influenza virus subtypes (1). A potential clinical benefit of M2e-based vaccines can be inferred from a challenge study in human volunteers, which showed that injection of human monoclonal antibody (MAb) TCN-032 directed against M2e 1 day after inoculation of an H3N2 influenza virus resulted in reduced clinical signs and virus shedding compared to placebo treatment (2).
M2e corresponds to the surface exposed amino-terminal part of M2 and is 23 amino acid residues long (3). The first nine amino acid residues of M2 are extremely conserved across all reported influenza A viruses. Human and murine M2e-specific MAbs that recognize different parts of M2e have been described, but how these interact with M2e is largely unknown (4–6). Here, we report the crystal structure of M2e in complex with a Fab fragment from a MAb that binds to the highly conserved N-terminal part of M2.
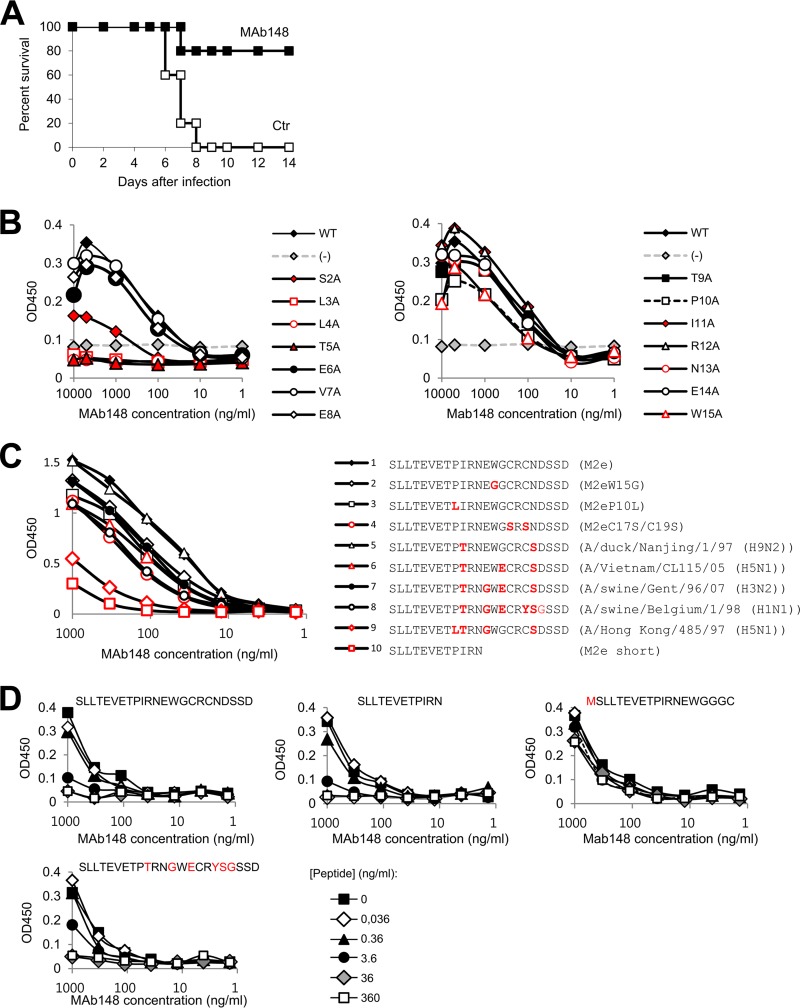
The ethical committee of Ghent University approved this mouse protection study. We isolated MAb148 from a BALB/c mouse that had been immunized with a recombinant tetrameric protein of M2e using conventional hybridoma technology (7). Intraperitoneal injection of 200 μg of this IgG1 MAb protected mice against a potentially lethal challenge with an H1N1 virus (Fig. 1A). To identify the epitope of MAb148, we transfected a series of alanine mutants of M2 into HEK293T cells. Transfected cells were subsequently fixed and used in an enzyme-linked immunosorbent assay (ELISA). This analysis revealed that M2 residues Ser2, Leu3, Leu4, and Thr5 are critical for MAb148 binding (Fig. 1B). A similar N-proximal epitope in M2e was reported for human MAb TCN-031 and -032 (8). ELISA analysis using wells coated with M2e peptide variants showed MAb148 binding was indifferent to most of the sequence variation in the M2e segment spanning residues 9 to 20 (Fig. 1C). However, we noted reduced binding to the M2e peptide derived from A/Hong Kong/485/97 (Fig. 1C). Also a shorter M2e peptide (SLLTEVETPIRN) proved less accessible for MAb148 binding (Fig. 1C), though in solution this peptide competed with MAb148 for binding the coating full-length M2e peptide as efficiently as full-length M2e (Fig. 1D), suggesting that reduced response in ELISA resulted from a coating effect.
FIG 1.
MAb148 binds to the N terminus of M2e. (A) MAb148 protects mice against challenge with influenza A virus. Groups of 5 female BALB/c mice were injected intraperitoneally with 200 μg of MAb148 (black squares) or phosphate-buffered saline (PBS) (white squares). Twenty-four hours later, the mice were challenged with two 50% lethal doses (LD50) of A/Swine Belgium/1/98 (H1N1) and monitored for survival. (B) HEK293T cells were transfected with pCAGGS vector (−) or with pCAGGS-M2 wt or Ala-scan mutants of the M2e domain. Twenty-four hours after transfection, the cells were fixed with 4% paraformaldehyde and used to probe binding with a dilution series of MAb148 by cellular ELISA. (C) M2e peptide ELISA using a diverse set of high-performance liquid chromatography (HPLC)-purified M2e peptides for coating to assess binding of MAb148. Residues that differ from the consensus human H3N2 virus M2e sequence (SLLTEVETPIRNEWGCRCNDSSD) are in red. (D) ELISA plates were coated with a peptide that represents the M2e sequence of most human H3N2 viruses (SLLTEVETPIRNEWGCRCNDSSD). MAb148 was preincubated with a dilution series of M2e peptide variants (the sequence of the competing peptide in solution is shown above each graph; residues in red differ from the corresponding residues in the coating peptide), and residual MAb148 binding to the coated wells was determined using a secondary horseradish peroxidase (HRP)-conjugated anti-mouse antibody.
To characterize the interactions between MAb148 and M2e in more detail, we determined the crystal structure of the M2e peptide SLLTEVETPIRNEGGCRCNDSSD (M2eW15G) in complex with Fab148 (obtained by papain digestion of MAb148). Peptide and Fab148 were mixed at a 10:1 molar ratio in 20 mM Tris-HCl (pH 8.0) and 50 mM NaCl buffer, and M2e-Fab148 complexes were purified on a Superdex 200 HR column. Needle-like crystals of M2eW15G-Fab148 complex were obtained by vapor diffusion, using 10 mg/ml of M2eW15G-Fab148 in 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, and 5 mM dithiothreitol (DTT) and a crystallization solution containing 100 mM sodium acetate (pH 5.0) and 2 M ammonium sulfate. Diffraction data to a 2.2-Å resolution were collected from a cryocooled (100 K) M2eW15G-Fab148 crystal at beamline I24 of the Diamond Light Source (Oxfordshire, United Kingdom). Diffraction data were processed and scaled using the XDS package (9), and the M2eW15G-Fab148 structure was solved by molecular replacement using Phaser (10) and refined by using Refmac5 (11) (Table 1).
TABLE 1.
Crystallographic X-ray diffraction and refinement statistics
| Statistic | Value for M2e peptide + Fab148 |
|---|---|
| Data statistics | |
| Wavelength (Å) | 1.000 |
| No. of reflections (unique reflections) | 326,286 (95,371) |
| Resolution range (Å) | 50.0∼2.20 (2.26∼2.20) |
| Completeness (%) | 97.1 (84.1) |
| Rmerge (%)a | 6.9 (33.6) |
| I/σ | 11.8 (3.5) |
| Space group | C2221 |
| Unit cell parameters (Å) | a = 92.6, b = 101.4, c = 212.5 |
| α = β = γ = 90° | |
| Refinement statistics | |
| Resolution range (Å) | 49.2∼2.2 |
| No. of reflections | 47,639 |
| R/Rfree (%)b | 18.7/24.0 |
| RMS deviationc | |
| Bonds (Å) | 0.024 |
| Angles (°) | 2.26 |
| No. of water molecules | 309 |
| Average B (Å2) | 29.4 |
| Ramachandran statistics (%)d | |
| Favored | 97.6 |
| Allowed | 2.4 |
| Outlier | 0.0 |
Rmerge = Σ|I − <I>|/Σ<I>, where I and <I> are the measured and averaged intensities of multiple measurements of the same reflection, respectively. The summation is over all the observed reflections.
R = Σ|Fo − Fc|/Σ|Fo|, calculated for all observed data. Rfree = Σ|Fo − Fc|/Σ|Fo|, calculated for a specified number of randomly chosen reflections that were excluded from the refinement.
RMS, root mean square.
Calculated using RAMPAGE (23).
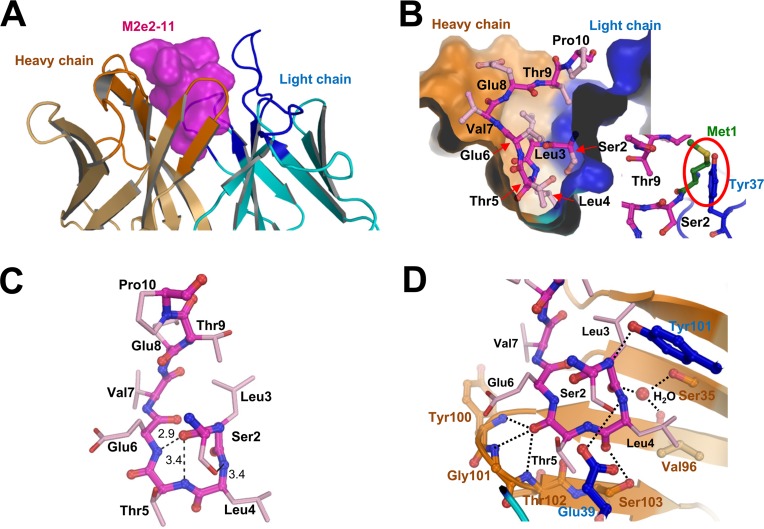
The electron density maps allowed unambiguous modeling of MAb148, as well as M2eW15G residues Ser2 to Pro10. The complement-determining regions (CDRs) form a deep and narrow binding pocket that accommodates the N-terminal part of M2e (Fig. 2A and B). M2ePro10 and Ile11 (the latter was not modeled due to weak electron density) emerge from the MAb148 binding pocket, where Pro10 kinks the M2e peptide such that its C-terminal segment is projected away from the MAb (Fig. 2B). The antigen's binding epitope resembles a fishing hook, with residues Ser2-Leu3-Leu4-Thr5-Glu6 forming a β-turn that is complementary in shape to the MAb148 paratope (Fig. 2B and C). Five H-bonds are directed toward the main chain atoms of M2e's N-terminal β-turn (Fig. 2D). Involvement of a β-turn in several peptide-antibody complexes has been reported in other X-ray structures (12–18) and in different antigenic peptides (19–22). Such a structure enhances the number of interactions that can be made by an antibody to an antigen with a limited number of residues. Our solution binding studies also show that the presence of an N-terminal Met on M2e, which is naturally removed posttranslationally (3), precludes binding of MAb148 (Fig. 1D). This is consistent with the structural data, which show that Met1 would be sterically hindered by Tyr37 in the light chain CDR3 of MAb148 (Fig. 2B, inset).
FIG 2.
Structure of the Fab148-M2eW15G complex. (A) Cartoon representation of the modeled part of M2e (surface presentation; purple) surrounded by the heavy (brown) and light (blue) chain complementarity-determining regions of MAb 148. (B) Cross section of M2e (sticks; purple) bound to MAb148. Amino acid residues of M2e are labeled. The inset is a theoretical modeling of Met1 (green) of M2eW15G. The red oval highlights the predicted steric hindrance of Met1 by Tyr37 of HCDR1. (C) Residues Ser2 to Thr5 of M2eW15G form a β-turn that is followed by a turn in M2e. Dotted lines represent hydrogen bonds, and distances between atoms are in angstroms. (D) Hydrophilic interactions between M2e (stick presentation) and the antibody are with main chain atoms of M2e except for the oxygen of the Ser2 side chain. Heavy and light chain residues are in brown and blue, respectively, and dotted lines represent hydrogen bonds. Oxygen and nitrogen atoms in this figure are in red and blue, respectively.
Apart from main chain interactions, MAb148 forms H bonds with side chain hydroxyls of Ser2, Thr5, Thr9, and the Glu8 carboxyl. Additionally, the nonpolar atoms in the M2e β-turn contribute hydrophobic interactions with the bottom of the MAb148 paratope. This suggests that hydrophobic interactions are necessary for the binding with MAb148, consistent with the loss of MAb148 binding with M2 Ala mutants at positions Leu3, Leu4, and Thr5 (Fig. 1A).
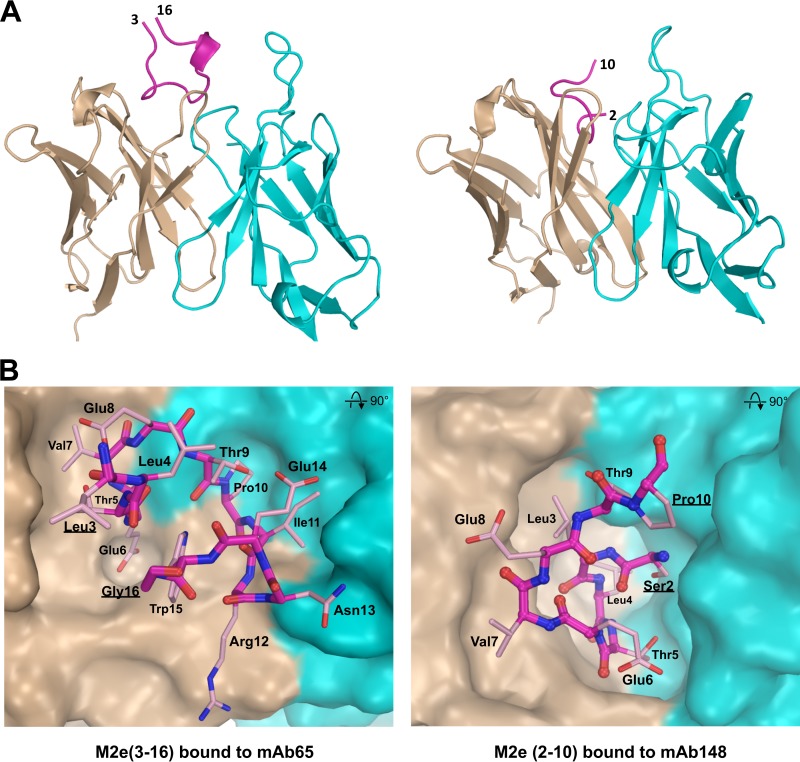
In the MAb65 complex that we resolved previously, the observed M2e conformation is quite different from that seen in the MAb148 complex (Fig. 3) (6). The N-terminal β-turn from Ser2 to Glu6 that determines M2e binding to MAb148 is absent in the MAb65 complex (Fig. 3), suggesting this segment of M2e can adopt multiple conformations on the virus coats. For practical reasons, we solved the cocomplex structure of MAb148 with the M2e peptide that has a Trp15-to-Gly15 mutation (M2eW15G) (in the presence of Trp15, no crystals were obtained, possibly due to incompatibility of the increased bulk of Trp with crystal packing). Although Trp15 is highly conserved in M2e, we assume that the structure of Fab148 complexed with M2eW15G is biologically significant, since the structural data show that the antigenic epitope comprises residues 2 to 11 only and we found MAb148 to interact equally well with both M2eWT and M2eW15G, as well as with the shorter M2e peptide SLLTEVETPIRN (Fig. 1C and D).
FIG 3.
Comparison of M2e bound to MAb65 and MAb148. (A) The conformation of M2e peptide bound by mAb65 (left) is entirely different from M2e bound by MAb148 (right). M2e peptide backbones are shown as purple lines. The 310 helix in M2e from Ile11 to Trp15 in the M2e-mAb65 complex is shown as a short ribbon, and the terminal residues of M2e are labeled. Antibody variable heavy and light chains are in brown and blue, respectively. (B) M2e has a β-turn from Thr5 to Glu8 and is folded in a U-shaped conformation with central Trp15 positioning when bound to MAb 65 (left) (PDB ID 4N8C [6]). M2e has an N-terminal β-turn involving Ser2 to Thr5 when M2e interacts with MAb 148 (right). The surfaces of heavy and light chains are in brown and blue, respectively.
In conclusion, MAb148 targets the conserved N-terminal residues of M2e and proves largely indifferent to known sequence variation immediately downstream of this segment.
Protein structure accession number.
The complex structure has been deposited in the Protein Data Bank under the PDB code 5DLM.
ACKNOWLEDGMENTS
K.J.C. was supported by FWO project 3G052412N, B.S. and K.M. are postdoctoral fellows of FWO-Vlaanderen, and L.D. was supported by State Scholarship Fund (file no. 2011674067) from the China Scholarship Council. We also acknowledge support from VIB, the IUAP BELVIR project p7/45, and Ghent University Special Research Grant BOF12/GOA/014 and Hercules foundation grant UABR/09/005.
We acknowledge use of beamline I24 of the Diamond Light Source under proposal MX9426 and are grateful for the help provided by the MX beamline staff.
Funding Statement
Fonds Wetenschappelijk Onderzoek (FWO) provided postdoctoral fellowships to Bert Schepens and Kristof Moonens. The Hercules Foundation funded the structural biology infrastructure under grant UABR/09/005. Access to the Diamond Light Source was supported in part by EU FP7 infrastructure grant BIOSTRUCT-X (contract 6601).
REFERENCES
- 1.Krammer F, Palese P. 2015. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov 14:167–182. doi: 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 2.Ramos EL, Mitcham JL, Koller TD, Bonavia A, Usner DW, Balaratnam G, Fredlund P, Swiderek KM. 2015. Efficacy and safety of treatment with an anti-m2e monoclonal antibody in experimental human influenza. J Infect Dis 211:1038–1044. doi: 10.1093/infdis/jiu539. [DOI] [PubMed] [Google Scholar]
- 3.Lamb RA, Zebedee SL, Richardson CD. 1985. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 40:627–633. doi: 10.1016/0092-8674(85)90211-9. [DOI] [PubMed] [Google Scholar]
- 4.Fu TM, Freed DC, Horton MS, Fan J, Citron MP, Joyce JG, Garsky VM, Casimiro DR, Zhao Q, Shiver JW, Liang X. 2009. Characterizations of four monoclonal antibodies against M2 protein ectodomain of influenza A virus. Virology 385:218–226. doi: 10.1016/j.virol.2008.11.035. [DOI] [PubMed] [Google Scholar]
- 5.Wang R, Song A, Levin J, Dennis D, Zhang NJ, Yoshida H, Koriazova L, Madura L, Shapiro L, Matsumoto A, Yoshida H, Mikayama T, Kubo RT, Sarawar S, Cheroutre H, Kato S. 2008. Therapeutic potential of a fully human monoclonal antibody against influenza A virus M2 protein. Antiviral Res 80:168–177. doi: 10.1016/j.antiviral.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Cho KJ, Schepens B, Seok JH, Kim S, Roose K, Lee JH, Gallardo R, Van Hamme E, Schymkowitz J, Rousseau F, Fiers W, Saelens X, Kim KH. 2015. Structure of the extracellular domain of matrix protein 2 of influenza A virus in complex with a protective monoclonal antibody. J Virol 89:3700–3711. doi: 10.1128/JVI.02576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Filette M, Martens W, Roose K, Deroo T, Vervalle F, Bentahir M, Vandekerckhove J, Fiers W, Saelens X. 2008. An influenza A vaccine based on tetrameric ectodomain of matrix protein 2. J Biol Chem 283:11382–11387. doi: 10.1074/jbc.M800650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grandea AG III, Olsen OA, Cox TC, Renshaw M, Hammond PW, Chan-Hui PY, Mitcham JL, Cieplak W, Stewart SM, Grantham ML, Pekosz A, Kiso M, Shinya K, Hatta M, Kawaoka Y, Moyle M. 2010. Human antibodies reveal a protective epitope that is highly conserved among human and nonhuman influenza A viruses. Proc Natl Acad Sci U S A 107:12658–12663. doi: 10.1073/pnas.0911806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabsch W. 2010. XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tormo J, Blaas D, Parry NR, Rowlands D, Stuart D, Fita I. 1994. Crystal structure of a human rhinovirus neutralizing antibody complexed with a peptide derived from viral capsid protein VP2. EMBO J 13:2247–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stanfield RL, Fieser TM, Lerner RA, Wilson IA. 1990. Crystal structures of an antibody to a peptide and its complex with peptide antigen at 2.8 A. Science 248:712–719. doi: 10.1126/science.2333521. [DOI] [PubMed] [Google Scholar]
- 14.Ghiara JB, Stura EA, Stanfield RL, Profy AT, Wilson IA. 1994. Crystal structure of the principal neutralization site of HIV-1. Science 264:82–85. doi: 10.1126/science.7511253. [DOI] [PubMed] [Google Scholar]
- 15.Serriere J, Dugua JM, Bossus M, Verrier B, Haser R, Gouet P, Guillon C. 2011. Fab′-induced folding of antigenic N-terminal peptides from intrinsically disordered HIV-1 Tat revealed by X-ray crystallography. J Mol Biol 405:33–42. doi: 10.1016/j.jmb.2010.10.033. [DOI] [PubMed] [Google Scholar]
- 16.Rini JM, Stanfield RL, Stura EA, Salinas PA, Profy AT, Wilson IA. 1993. Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen. Proc Natl Acad Sci U S A 90:6325–6329. doi: 10.1073/pnas.90.13.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding J, Smith AD, Geisler SC, Ma X, Arnold GF, Arnold E. 2002. Crystal structure of a human rhinovirus that displays part of the HIV-1 V3 loop and induces neutralizing antibodies against HIV-1. Structure 10:999–1011. doi: 10.1016/S0969-2126(02)00793-1. [DOI] [PubMed] [Google Scholar]
- 18.Olal D, Kuehne AI, Bale S, Halfmann P, Hashiguchi T, Fusco ML, Lee JE, King LB, Kawaoka Y, Dye JM Jr, Saphire EO. 2012. Structure of an antibody in complex with its mucin domain linear epitope that is protective against Ebola virus. J Virol 86:2809–2816. doi: 10.1128/JVI.05549-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell AP, McInnes C, Hodges RS, Sykes BD. 1995. Comparison of NMR solution structures of the receptor binding domains of Pseudomonas aeruginosa pili strains PAO, KB7, and PAK: implications for receptor binding and synthetic vaccine design. Biochemistry 34:16255–16268. doi: 10.1021/bi00050a005. [DOI] [PubMed] [Google Scholar]
- 20.Dyson HJ, Cross KJ, Houghten RA, Wilson IA, Wright PE, Lerner RA. 1985. The immunodominant site of a synthetic immunogen has a conformational preference in water for a type-II reverse turn. Nature 318:480–483. doi: 10.1038/318480a0. [DOI] [PubMed] [Google Scholar]
- 21.Blumenstein M, Matsueda GR, Timmons S, Hawiger J. 1992. A beta-turn is present in the 392-411 segment of the human fibrinogen gamma-chain. Effects of structural changes in this segment on affinity to antibody 4A5. Biochemistry 31:10692–10698. [DOI] [PubMed] [Google Scholar]
- 22.Campbell AP, Sykes BD, Norrby E, Assa-Munt N, Dyson HJ. 1996. Solution conformation of an immunogenic peptide derived from the principal neutralizing determinant of the HIV-2 envelope glycoprotein gp125. Fold Des 1:157–165. doi: 10.1016/S1359-0278(96)00024-7. [DOI] [PubMed] [Google Scholar]
- 23.Lovell SC, Davis IW, Arendall WB III, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. 2003. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]