ABSTRACT
The changing epidemiology of group A rotavirus (RV) strains in humans and swine, including emerging G9 strains, poses new challenges to current vaccines. In this study, we comparatively assessed the pathogenesis of porcine RV (PRV) G9P[13] and evaluated the short-term cross-protection between this strain and human RV (HRV) Wa G1P[8] in gnotobiotic pigs. Complete genome sequencing demonstrated that PRV G9P[13] possessed a human-like G9 VP7 genotype but shared higher overall nucleotide identity with historic PRV strains. PRV G9P[13] induced longer rectal virus shedding and RV RNAemia in pigs than HRV Wa G1P[8] and generated complete short-term cross-protection in pigs challenged with HRV or PRV, whereas HRV Wa G1P[8] induced only partial protection against PRV challenge. Moreover, PRV G9P[13] replicated more extensively in porcine monocyte-derived dendritic cells (MoDCs) than did HRV Wa G1P[8]. Cross-protection was likely not dependent on serum virus-neutralizing (VN) antibodies, as the heterologous VN antibody titers in the sera of G9P[13]-inoculated pigs were low. Thus, our results suggest that heterologous protection by the current monovalent G1P[8] HRV vaccine against emerging G9 strains should be evaluated in clinical and experimental studies to prevent further dissemination of G9 strains. Differences in the pathogenesis of these two strains may be partially attributable to their variable abilities to replicate and persist in porcine immune cells, including dendritic cells (DCs). Additional studies are needed to evaluate the emerging G9 strains as potential vaccine candidates and to test the susceptibility of various immune cells to infection by G9 and other common HRV/PRV genotypes.
IMPORTANCE The changing epidemiology of porcine and human group A rotaviruses (RVs), including emerging G9 strains, may compromise the efficacy of current vaccines. An understanding of the pathogenesis and genetic, immunological, and biological features of the new emerging RV strains will contribute to the development of new surveillance and prevention tools. Additionally, studies of cross-protection between the newly identified emerging G9 porcine RV strains and a human G1 RV vaccine strain in a susceptible host (swine) will allow evaluation of G9 strains as potential novel vaccine candidates to be included in porcine or human vaccines.
INTRODUCTION
Rotavirus (RV), a member of the Reoviridae family, has a double-stranded RNA genome with 11 segments (1). It is the most common pathogen in cases of acute gastroenteritis in children under 5 years of age (1, 2). In the United States, it causes approximately $1 billion in annual costs due to RV-associated physician visits, emergency department visits, and hospitalizations (3–5). Annually, RV causes 440,000 deaths in children under 5 years of age worldwide, with most occurring in developing countries (4). RVs also infect young domestic animals, including calves and piglets (1). RV is responsible for annual mortality rates of 7 to 20% and 3 to 15% in nursing and weaned piglets, respectively (6). The high prevalence of RV in swine results in large economic losses to the pork industry (6). Treatment of RV infection is possible only by replacing fluids and electrolyte losses, because no specific antiviral therapy is available. Therefore, effective RV vaccines are crucial to prevent morbidity and mortality in both young children and animals (7, 8).
RVs are classified into 8 groups, groups A to H, as determined by viral structural protein 6 (VP6) (9–11). Based on the outer capsid VP4 (P genotype)- and VP7 (G genotype)-encoding genes, a binary classification system has been established for RVs (12). Overall, there are at least 26 G genotypes and 33 P genotypes of group A RVs (RVAs) (13, 14). Globally, the G1 to G4, P[4], P[6], and P[8] genotypes are the most prevalent human RVAs (15). RVA G1P[8] is a common human strain worldwide and constitutes >70% of prevalent strains in North America, Australia, and Europe but only 20 to 35% of circulating strains in South America, Asia, and Africa (5, 15–17). G5 and P[7] are historically considered the most prevalent G and P RVA genotypes in swine, respectively (18). However, recent studies have shown that RVA G9 and G12 genotypes are emerging worldwide in humans and swine (2, 19–23).
Originally reported in studies of human cases in the early to mid-1980s, G9 RV strains spread quickly to all continents in the mid-1990s (24). We recently found that G9 strains are also prevalent in Ohio swine (23). Genetic analyses of the emerging G9 RVs in humans have confirmed that some G9 RV strains are phylogenetically more similar to porcine RV (PRV) than the earlier human G9 genotypes (22, 25). The emergence of PRV-like G9 RVs in children in developing countries, in addition to evidence that G1P[8] may be of swine origin, illustrates the zoonotic potential of animal RVs and collectively suggests that PRVs are a potential source of heterologous RV infections in humans (25).
Recently, a classification system encompassing all 11 RV genome segments was developed by using standardized nucleotide identity cutoff values and the notation Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx, which refers to the VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5/6 gene segments, respectively (9, 12). This system allows international standardization for analyzing RVA interspecies evolutionary relationships, gene reassortment events, emerging RVA strains, and RVA host range restriction (9, 12). Thus, full genomic sequencing and characterization of G9P[13] viruses allow whole-genome characterization and suggest their zoonotic potential by defining which gene segments are likely of swine or human origin. Knowledge of the pathogenesis and cross-protection potential of G9 RV strains will help us to understand mechanisms for the rapid global spread of G9 strains and their ability to infect diverse host species.
Because VP7 and VP4 are targets for neutralizing antibodies which elicit serotype-specific protection, they are critical for vaccine development (8, 26). Currently, two RV vaccines are licensed for humans, Rotarix and RotaTeq. Rotarix is a live attenuated human RV (HRV) vaccine that has high efficacy in preventing G1, G3, and G4 serotype-induced RV gastroenteritis, while RotaTeq is a live human-animal pentavalent vaccine that has been shown to be highly efficacious in preventing G1 to G4 serotype-induced RVA gastroenteritis in developed countries (8). However, the emergence of the G9 and G12 strains may compromise RV vaccine efficacy, as available vaccines do not provide homotypic protection against these emerging strains (26, 27). Therefore, it is critical to assess the ability of historic and emerging RV strains to elicit heterotypic immune responses against emerging and less frequent genotypes of RV to identify potential candidate vaccine strains (15, 27).
There is increasing evidence suggesting that RV can spread extraintestinally (28, 29). RVs have been shown to cause persistent infection in immunodeficient mice as well as immunodeficient young children, resulting in diseases such as encephalitis in children (30, 31). This may be due to the ability of certain RVs to infect nonepithelial cells, leading to extraintestinal spread. Our laboratory previously confirmed that gnotobiotic (Gn) piglets are susceptible to infection and disease with the virulent HRV Wa G1P[8] strain and that infected piglets have transient viremia (32). Others have suggested that in rhesus macaques, rhesus RV (RRV) escapes the intestine via a lymphatic route, with the viral nonstructural proteins (NSPs) being sequentially detected in Peyer's patches, mesenteric lymph nodes (MLNs), spleen, and liver (33). RV NSPs have been observed in mouse macrophages as well as human macrophages, dendritic cells (DCs), and B cells (34–36). These results suggest that immune cells may be permissive to RV infection and may play a role in RV persistence and extraintestinal dissemination. In this study, we compared the abilities of PRV G9P[13] and HRV Wa G1P[8] strains to infect porcine B cells, T cells, and monocyte-derived DCs (MoDCs).
Our previous pathogenesis studies demonstrated that HRV Wa G1P[8] infection of Gn piglets results in similar clinical disease and comparable levels of pathogenesis, including similar levels of intestinal lesions, to those observed in homologous PRV infections with the OSU and SB1A strains (37); however, only limited pathogenesis studies of G9 PRV strains have been conducted with neonatal piglets. In this study, we inoculated Gn pigs with PRV G9P[13] or HRV Wa G1P[8] to comparatively assess their pathogenesis and cross-protection.
MATERIALS AND METHODS
Viruses for inoculation and challenge.
The Gn pig-adapted (passage 23) HRV Wa G1P[8] strain (38) and the Gn pig-passaged (passage 2) PRV G9P[13] strain (23) were used at a dose of 105 fluorescence-forming units (FFU) for Gn pig inoculation and challenge, as described previously (38).
Complete genomic sequencing of PRV G9P[13].
Stool samples from Gn piglets infected with the PRV G9P[13] strain were processed as described previously (23). Viral RNA was then extracted by using the RNeasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Complete genome sequencing of PRV G9P[13] was conducted as previously described (39).
Animals and experimental design.
Gn pigs were hysterectomy derived and maintained in sterile isolation units as described previously (40, 41). One-week-old piglets (derived from 3 Landrace × Yorkshire sows) (postinoculation day [PID] 0) were inoculated with 105 FFU each of PRV G9P[13] (group 1 [n = 5] and group 2 [n = 4]) or HRV Wa G1P[8] (group 3 [n = 7]) in 3 ml of minimum essential medium (MEM) (Life Technologies, Grand Island, NY, USA), immediately after administration of 3 ml of 100 mM sodium bicarbonate to reduce gastric acidity. A mock group (n = 5) was inoculated with 3 ml of MEM after the sodium bicarbonate inoculation. Two pigs, from groups 1 and 3, were euthanized 1 day after diarrhea onset. At 3 weeks postinoculation (PID 21 and postchallenge day [PCD] 0), pigs of group 2 were challenged with 105 FFU HRV Wa G1P[8], and pigs of groups 1, 3, and 4 were challenged with 105 FFU PRV G9P[13], immediately after the administration of 3 ml of 100 mM sodium bicarbonate. All pigs were euthanized at 10 days postchallenge. All animal experiments were conducted according to protocol 2010A00000088 approved by the Animal Care and Use Committee of The Ohio State University.
Detection of rectal shedding of virus by CCIF.
Rectal swabs were collected on PIDs 1 to 10, 14, and 21, as well as on PCDs 1 to 7, from all pigs for evaluation of diarrhea and rectal virus shedding, as described previously (42). The rectal swab fluid samples were tested by a cell culture immunofluorescence (CCIF) test to quantitate infectious PRV G9P[13] and HRV Wa G1P[8], as described previously (42).
Detection of PRV G9P[13] homologous/heterologous neutralizing antibody titers in convalescent-phase sera using a fluorescent-focus neutralization test.
The homologous/heterologous virus-neutralizing (VN) antibody titers against PRV G9P[13], HRV Wa G1P[8], PRV OSU G5P[7], and PRV Gottfried G4P[6] in convalescent-phase/hyperimmune sera were determined by a fluorescent-focus neutralization (FFN) test, as previously described (43). The VN titer was expressed as the reciprocal of the highest dilution of serum that reduced the number of infected-cell foci by 80%.
Detection of viral RNA in serum by real-time RT-PCR.
Blood samples were collected on PIDs 0, 3, 5, 7, 9, and 14, as well as on PCDs 3 and 5, from pigs of all treatment groups. Blood samples were centrifuged at 1,850 × g for 15 min, and sera were collected and stored at −20°C until testing. An RNeasy minikit (Qiagen, Valencia, CA, USA) was used to extract RNA from 250 μl of each serum sample according to the manufacturer's recommendations. The extracted RNA was stored at −70°C until testing. Real-time reverse transcription-PCR (RT-PCR) was used for the detection of RVA RNA by using primers NSP3F and NSP3R and a QuantiTect SYBR green RT-PCR kit (Qiagen, Valencia, CA, USA) (44). For real-time RT-PCR, the following conditions were applied: incubation for 20 min at 50°C for the reverse transcription reaction and a preheating step at 95°C for 15 min for initial denaturation, followed by 40 PCR cycles at 94°C for 15 s, 56°C for 30 s, and 72°C for 30 s. A melting-curve analysis was then performed at 95°C for 5 s and at 65°C for 1 min, slowly increasing the temperatures up to 97°C over 20 min, followed by a 40°C hold. RNA extracted from a validated RVA-positive sample was used as a positive control, while RNA-free water was used as a negative control.
Determination of RV RNA tissue distribution in vivo by real-time RT-PCR.
Two pigs, inoculated with PRV G9P[13] or HRV Wa G1P[8], were euthanized 1 day after diarrhea onset to evaluate the tissue distribution of RV RNA. One Gn pig without RVA inoculation was used as a negative control. Isolation of mononuclear cells (MNCs) from the ileum, spleen, liver, MLNs, and blood was conducted as previously described (45, 46). RNA extraction was done on 2 × 106 MNCs from each tissue sample by using an RNeasy minikit (Qiagen, Valencia, CA, USA) according to the manufacturer's recommendations. Real-time RT-PCR was conducted as described above.
Examination of PRV/HRV infection of porcine T cells and B cells in vitro.
Isolation of MNCs from the ileum, MLNs, spleen, and blood of 4-week-old uninoculated Gn piglets was conducted as previously described (45). Ileal, MLN, splenic, and blood T cells were purified by positive selection with mouse anti-porcine CD3 monoclonal antibody (MAb) (IgG1) (Southern Biotech, Birmingham, AL, USA) and goat anti-mouse IgG MicroBeads (Miltenyi Biotec, San Diego, CA, USA) according to the manufacturers' instructions. Porcine B cells were purified with mouse anti-porcine CD21 MAb (IgG1) (Southern Biotech, Birmingham, AL) and goat anti-mouse IgG MicroBeads according to the same instructions. Isolated porcine T cells and B cells were exposed to HRV Wa G1P[8] or PRV G9P[13] at a multiplicity of infection (MOI) of 0.4 for 24 h in 5% CO2 at 37°C. Untreated porcine T cells and B cells were cultured for 24 h without RV exposure as a negative control. Cells were washed twice and fixed with fixation-and-permeabilization solution (BD Bioscience, San Jose, CA, USA) for 20 min. The cells were then incubated with anti-NSP4 MAb (IgG2a) (hybridoma cell B4-2, supplied by H. B. Greenberg) for 40 min at 4°C and stained with goat anti-mouse IgG2a-R-phycoerythrin (RPE) (Life Technologies, Grand Island, NY, USA) for 20 min at 4°C. T cells were washed and stained with mouse anti-porcine CD3-fluorescein isothiocyanate (FITC) (Southern Biotech, Birmingham, AL, USA), while B cells were stained with mouse anti-porcine CD21-FITC (Southern Biotech, Birmingham, AL, USA). Mouse IgG2a-phycoerythrin (PE) and mouse IgG1-FITC (Southern Biotech, Birmingham, AL, USA) were used as isotype controls. Acquisition of 20,000 events was done by using an AccuriC6 flow cytometer (BD Bioscience, San Jose, CA, USA). Analyses were conducted by using CFlow software (BD Bioscience, San Jose, CA, USA).
Examination of PRV/HRV infection of porcine MoDCs in vitro.
Porcine blood from healthy adult pigs was collected with 30% acid citrate dextrose (ACD) anticoagulant. Blood monocytes were isolated by density centrifugation over Ficoll-Paque Premium (1.084 g/ml; GE Healthcare Life Sciences, Uppsala, Sweden), as previously described, and then suspended in RPMI 1640 medium (Life Technologies, Grand Island, NY, USA) and placed into 175-cm2 cell culture flasks (47, 48). After 3 h of incubation at 37°C, nonadherent cells were removed by washing with cold RPMI 1640 medium. The remaining adherent monocytes were cultured in Dulbecco's modified Eagle medium (DMEM) (Life Technologies, Grand Island, NY, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Atlanta Biologicals, Flowery Branch, GA, USA), 1% antibiotic-antimycotic (Life Technologies, Grand Island, NY, USA), 100 ng/ml of recombinant swine granulocyte-macrophage colony-stimulating factor (GM-CSF) (Life Technologies, Grand Island, NY, USA), and 20 ng/ml of recombinant swine interleukin-4 (IL-4) (Life Technologies, Grand Island, NY) in 5% CO2 at 37°C for 6 days to differentiate cells into MoDCs. The cells showed increased sizes and changes in morphology from round to irregular shapes with cytoplasmic projections, as previously described (48). In addition, >90% of the cells were determined to be swine workshop cluster 3 (SWC3) positive by flow cytometry. The MoDCs were exposed to HRV Wa G1P 1A[8] or PRV G9P[13] at an MOI of 0.4 for 24 h. The plates were then fixed with 80% acetone for 10 min at room temperature (RT) and air dried. Anti-NSP4 MAb was added to each well, and the wells were incubated at 4°C overnight. FITC-labeled goat anti-mouse IgG/IgM/IgA (AbD Serotec, Raleigh, NC, USA) diluted 1:500 was added to each well as a secondary antibody. The plates were incubated at 37°C for 2 h and viewed by using an Olympus IX70 fluorescence microscope (B&B Microscopes, Pittsburgh, PA, USA).
Statistical analysis.
The mean number of days to onset of virus shedding, average peak titer of virus shedding, daily fecal titer of virus shedding, mean number of days to onset of diarrhea, and mean duration (days) of diarrhea in postinoculation pigs were analyzed by a two-tailed t test. The mean number of days to onset of virus shedding, mean duration (days) of virus shedding, average peak titer of virus shedding, and mean duration (days) of diarrhea in postchallenge pigs were analyzed by one-way analysis of variance (ANOVA). Mean cumulative fecal scores in different treatment groups were compared by using the area under the curve, as previously described (49, 50). Statistical analyses were performed with GraphPad Prism 6.0c software (GraphPad Software, La Jolla, CA, USA).
Nucleotide sequence accession numbers.
The complete genomic sequences of this PRV G9P[13] strain were deposited in GenBank under the strain name RVA/Pig-hhp/USA/LS00009-RV0084/2011G9P[13] (accession numbers KR052730 through KR052740). BLAST (blastn) searches (http://www.ncbi.nlm.nih.gov/) and/or the RotaC v2.0 (http://rotac.regatools.be/) automated genotyping tool classified the full genomic constellations for this strain. The GenBank accession numbers for the genomic sequences of the HRV Wa G1P[8] strain used in this study are FJ423113 through FJ423123.
RESULTS
PRV G9P[13] has high overall identity to historic PRV strains but possesses a human-like VP7 (G9) genotype.
Complete genomic sequencing of PRV G9P[13] was conducted. By using BLAST (blastn) searches and the RotaC v2.0 automated genotyping tool, the complete genomic constellation for this PRV G9P[13] strain was identified as G9-P13-I5-R1-C1-M1-A8-N1-T1-E1-H1. Analyses of the complete genomic nucleotide sequences (except for VP7 and VP4) of PRV G9P[13] demonstrated that this strain shared the highest overall nucleotide identity to the OSU strain (RVA/Pig-tc/USA/OSU/1977/G5P7) (89.5% to 95.6%) and the Mexican YM strain (RVA/Pig-tc/MEX/YM/1983/G11P7) (89.9% to 97.7%), followed by the Gottfried strain (RVA/Pig-tc/USA/Gottfried/1983/G4P6) (91.3% to 92%). A previously reported phylogenetic tree of the partial (nucleotides [nt] 73 to 388) sequence of the G9P[13] VP7 gene compared with available VP7 gene sequences for human and porcine RVA G genotypes indicated that this PRV G9P[13] strain shared higher identity with human RVA G9 strains (23). The nucleotide sequence of the PRV G9P[13] VP7 segment shared the highest nucleotide identity with G9-RVA/Human-wt/BEL/B3458/2003/G9P8 (92.5%).
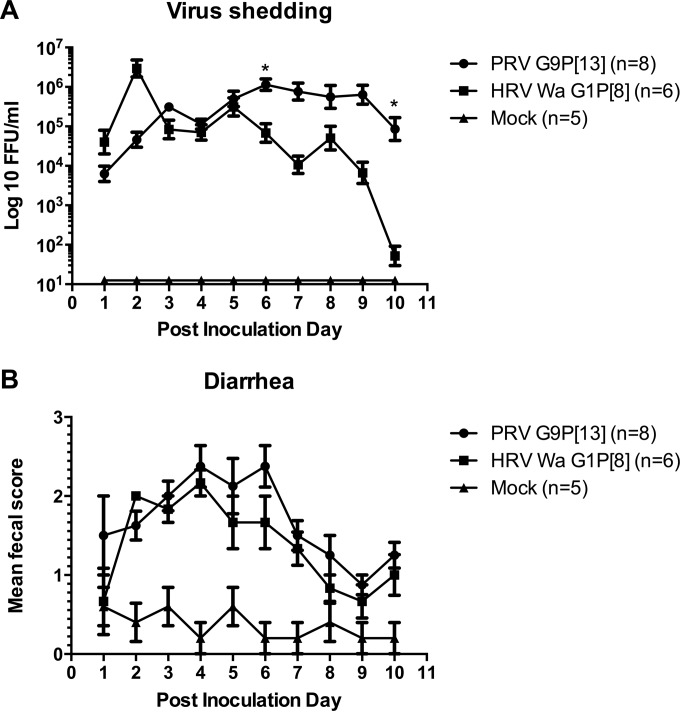
PRV G9P[13] induces increased fecal virus shedding (longer duration and higher virus loads) compared with HRV Wa G1P[8] but induces similar diarrhea severity.
Fecal virus shedding and diarrhea in PRV G9P[13]- and HRV Wa G1P[8]-inoculated pigs from PIDs 1 to 10 are summarized in Table 1 and shown in Fig. 1A and B. PRV G9P[13]- and HRV Wa G1P[8]-inoculated pigs had similar mean numbers of days to onset of virus shedding and diarrhea. Interestingly, HRV Wa G1P[8] induced a slightly higher average peak titer (at PID 2) of virus shedding in pigs than did PRV G9P[13], while PRV G9P[13] induced a numerically higher mean cumulative fecal diarrhea score in pigs than did HRV Wa G1P[8] (Table 1).
TABLE 1.
Virus shedding and diarrhea in pigs inoculated with PRV G9[13] and HRV Wa G1P[8]
| Inoculation | No. of pigs | Virus shedding |
Diarrhea |
|||||
|---|---|---|---|---|---|---|---|---|
| % of pigs with shedding | Mean no. of days to onset | Avg peak titer (FFU/ml) | % of pigs with diarrhea | Mean no. of days to onset | Mean duration (days)a | Mean cumulative fecal scoreb | ||
| PRV G9P[13] | 8 | 100 | 1.4 | 1.66E+06 | 100 | 1.9 | 5.5 | 16.9 |
| HRV Wa G1P[8] | 6 | 100 | 1.8 | 3.30E+06 | 100 | 1.7 | 4.7 | 13.8 |
Duration of diarrhea is defined as the number of days that the diarrhea score was ≥2. Fecal diarrhea were scored as follows: 0, normal; 1, pasty; 2, semiliquid; 3, liquid.
Mean cumulative fecal score = [(sum of fecal consistency score for 10 days postinoculation)/N], where N is the number of pigs receiving the inoculation.
FIG 1.
Fecal virus shedding (A) and diarrhea scores (B) for PRV G9P[13]- and HRV Wa G1P[8]-inoculated pigs from PID 1 to PID 10. Gnotobiotic pigs were orally inoculated with 105 FFU PRV G9P[13] or HRV Wa G1P[8]. Mock-infected pigs were inoculated with virus suspension medium (MEM). Rectal swabs were collected daily. Fecal consistency was scored as 0 for normal, 1 for pasty, 2 for semiliquid, and 3 for liquid, with scores of ≥2 being considered diarrhea. Virus shedding was determined by a CCIF assay. *, P ≤ 0.05.
Fecal virus shedding peaked at PID 2 (2.56E+06 FFU/ml) and then again at PID 5 in the HRV Wa G1P[8]-inoculated pigs (as observed in previous studies [51, 52]), decreasing overall in the following days (PIDs 3 to 10), whereas fecal virus shedding of the PRV G9P[13]-inoculated pigs peaked at PID 3 (6.73E+05 FFU/ml), which was then maintained at higher and relatively constant levels (1.02E+05 to 1.14E+06 FFU/ml) through PID 10 (Fig. 1A). The PRV G9P[13]-inoculated pigs had significantly higher titers of fecal virus shedding on PID 6 and PID 10 than did the HRV Wa G1P[8]-inoculated pigs (P = 0.019 and P = 0.021, respectively) (Fig. 1A). On PID 14, two (out of eight) PRV G9P[13]-inoculated pigs still had detectable levels of fecal virus shedding, while none of the pigs in the HRV Wa G1P[8]-inoculated group still shed virus (data not shown). On PID 21, neither PRV G9P[13]- nor HRV Wa G1P[8]-inoculated pigs had detectable fecal virus shedding (data not shown).
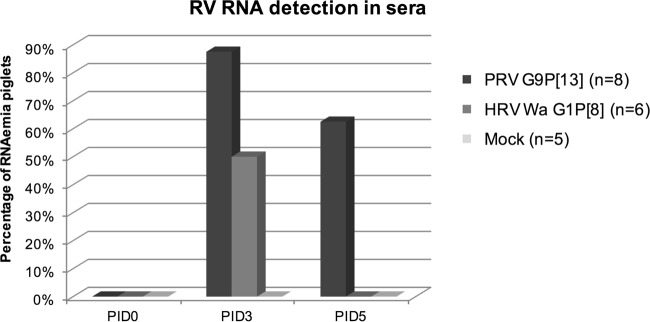
PRV G9P[13] causes higher frequencies of and more prolonged RV RNAemia in piglets, whereas HRV Wa G1P[8] causes only transient RV RNAemia.
In PRV G9P[13]-inoculated pigs, 87.5% and 62.5% of the serum samples were positive for RV RNA on PIDs 3 and 5, respectively (Fig. 2). In HRV Wa G1P[8]-inoculated pigs, viral RNA was detected in only 50% of serum samples on PID 3 and in 0% of serum samples on PID 5 (Fig. 2). None of the serum samples on PID 7, 9, or 14 or on PCDs 3 and 5 had detectable levels of RV RNA (data not shown). PRV G9P[13] caused a higher frequency of RV RNAemia in pigs on PID 3 than did HRV Wa G1P[8] (87.5% versus 50%), which then persisted for a longer time (PID 5) than for HRV Wa G1P[8] (PID 5) (Fig. 2).
FIG 2.
Detection of RV RNA in sera from pigs inoculated with PRV G9P[13] or HRV Wa G1P[8] on PID 3 and PID 5. Serum was extracted from blood collected from piglets on PIDs 0, 3, and 5. Viral RNA was extracted and detected by real-time RT-PCR.
Detection of PRV G9P[13] and HRV Wa G1P[8] RNAs in extraintestinal tissues of inoculated Gn pigs in vivo.
To validate if prolonged RV RNAemia and increased fecal RV shedding were associated with increased extraintestinal spread of PRV G9P[13], two pigs were euthanized 1 day following diarrhea onset after PRV/HRV inoculation. PRV G9P[13]- and HRV Wa G1P[8]-inoculated pigs developed diarrhea on PIDs 1 and 2, respectively, and were euthanized on PIDs 2 and 3, respectively. Results of real-time RT-PCR detection of RV RNA in MNCs of extraintestinal tissues are summarized in Table 2. PRV G9P[13] and HRV Wa G1P[8] RNAs were detected in MNCs of the ileum and in MNCs from extraintestinal tissues, including the MLNs, spleen, and liver. Only PRV G9P[13] RNA was detected in blood MNCs. These data suggest that PRV G9P[13] and HRV Wa G1P[8] may induce systemic dissemination that is possibly dependent on circulating MNCs.
TABLE 2.
RVA detection in MNCs from ileum, MLNs, spleen, liver, and blood of PRV G9P[13]- or HRV Wa G1P[8]-inoculated pigsa
| Virus | Detection of MNCs from: |
||||
|---|---|---|---|---|---|
| Ileum | MLNs | Spleen | Liver | Blood | |
| PRV G9P[13] | + | + | + | + | + |
| HRV WA G1P[8] | + | + | + | + | − |
+, rotavirus RNA detected in MNCs; −, rotavirus RNA not detected in MNCs.
PRV G9P[13] confers complete short-term (PCDs 1 to 7) protection against homologous/heterologous RV infection and diarrhea.
Virus shedding and diarrhea postchallenge are summarized for all groups in Table 3. No pigs from group 1 (PRV G9P[13]-inoculated pigs) and group 2 (HRV Wa G1P[8]-inoculated pigs) shed virus or had diarrhea after PRV G9P[13] (homologous strain) or HRV Wa G1P[8] (heterologous strain) challenge. Hence, PRV G9P[13] successfully protected against homologous and heterologous RV infection. Five of six (83.3%) group 3 (HRV Wa G1P[8]-inoculated) pigs and five of five (100%) group 4 (mock) pigs shed virus after PRV G9P[13] challenge. HRV Wa G1P[8] did not completely prevent PRV G9P[13] infection but significantly shortened the duration of virus shedding (P < 0.005) and decreased the titer of peak shedding (P < 0.05) compared to the mock group. None of 4 (0%) group 1 (PRV G9[13]/PRV G9P[13]) pigs, 0 of 4 (0%) group 2 (PRV G9P[13]/HRV Wa G1P[8]) pigs, 2 of 6 (33.3%) group 3 (HRV Wa G1P[8]/PRV G9P[13]) pigs, and 3 of 5 (60%) group 4 (control) pigs developed diarrhea. PRV G9P[13] completely protected against homologous/heterologous RV diarrhea, whereas HRV Wa G1P[8] resulted in 66.7% protection against diarrhea. Therefore, PRV G9P[13] conferred 100% short-term protection against homologous/heterologous RV challenge and diarrhea, whereas HRV Wa G1P[8] induced less heterologous protection.
TABLE 3.
Diarrhea and rectal virus shedding in postchallenge pigs of different treatment groups from PCD 1 to PCD 7
| Group | Treatment (inoculation/challenge) | No. of pigs | Virus shedding |
Diarrhea |
Rate of protection (%) against shedding | Rate of protection (%) against diarrhea | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| % of pigs with shedding | Mean no. of days to onset | Mean duration (days)a | Avg peak titer (FFU/ml) | % of pigs with diarrhea | Mean duration (days)b | Mean cumulative fecal scorec | |||||
| 1 | PRV G9P[13]/PRV G9P[13] | 4 | 0 | 0 | ≤12.5 | 0 | 0 | 4.3 | 100 | 100 | |
| 2 | PRV G9P[13]/HRV Wa G1P[8] | 4 | 0 | 0 | ≤12.5 | 0 | 0 | 5 | 100 | 100 | |
| 3 | HRV Wa G1P[8]/PRV G9P[13] | 6 | 83.3 | 2.8 | 1.3 | 3.33E+03 | 33.3 | 0.7 | 7 | 16.7 | 66.7 |
| 4 | Mock/PRV G9P[13] | 5 | 100 | 1.2 | 5.4 | 4.26E+05 | 60 | 1.2 | 6.2 | ||
The mean duration of virus shedding (days) is defined as the number of days with detectable fecal virus shedding.
Duration of diarrhea is defined as the number of days that the diarrhea score was ≥2. Diarrhea severity was scored as follows: 0, normal; 1, pasty; 2, semiliquid; 3, liquid.
Mean cumulative fecal score = [(sum of fecal consistency score for 10 days postinoculation)/N], where N is the number of pigs receiving the inoculation.
PRV G9P[13] induces low heterologous VN antibody titers against selected porcine and human RVs.
To determine if PRV G9P[13]-induced protection/cross-protection was mediated by systemic (serum) VN antibody against homologous or heterologous HRV Wa G1P[8] challenge, we used FFN to measure the levels of cross-neutralizing antibodies against PRV G9P[13] and other selected RVs (HRV Wa G1P[8], PRV OSU G5P[7], and PRV Gottfried G4P[6]) isolated in our laboratory (53) in convalescent-phase/hyperimmune sera. The VN antibody titers and the percent relatedness (R%) values against selected RVs for convalescent-phase or hyperimmune sera are summarized in Table 4. The PRV G9P[13] convalescent-phase sera showed weak reactivity with other historic porcine or human RVs, suggesting that heterologous protection against HRV Wa G1P[8] was not dependent on the heterotypic serum VN antibody titers. Similarly, HRV Wa G1P[8] hyperimmune sera had low titers of heterologous VN antibody that varied for the different PRVs.
TABLE 4.
Virus-neutralizing antibody titers (and relatedness values) against selected RVsa
| Virus | VN antibody titer (R% value) |
|||
|---|---|---|---|---|
| C serum |
HI serum |
|||
| Anti-PRV G9P[13] | Anti-HRV Wa G1P[8] | Anti-PRV OSU G5P[7] | Anti-PRV Gottfried G4P[6] | |
| PRV G9P[13] | 4,096 (100) | 5 (0.12) | 1,024 (12) | 64 (3.5) |
| HRV Wa G1P[8] | 8 (0.12) | 6,856 (100) | 37 (4.2) | 71 (15) |
| PRV OSU G5P[7] | 256 (12) | 1,437 (4.2) | 4,400 (100) | 8 (1.5) |
| PRV Gottfried G4P[6] | 156 (3.5) | 4,436 (15) | 254 (1.5) | 2,000 (100) |
R% values are indicated in parentheses and were calculated as R% = 100(√r1 × r2), where r1 is the heterologous titer of strain 2 divided by the homologous titer of strain 1 and r2 is the heterologous titer of strain 1 divided by the homologous titer of strain 2. C, convalescent phase; HI, hyperimmune. Boldface type identifies homologous virus neutralizing antibody titers.
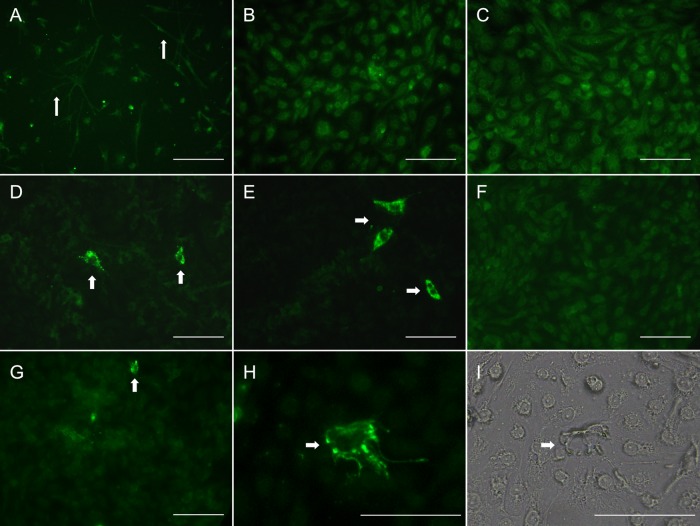
In vitro detection of both PRV G9P[13] and HRV Wa G1P[8] NSP4 antigens in porcine MoDCs at an MOI of 0.4 but not in porcine T cells and B cells.
MoDCs were used in this study to determine if PRV G9P[13] and HRV Wa G1P[8] replicate in porcine DCs based on the detection of RVA NSP4 antigen by an immunofluorescence assay. Untreated MoDCs had distinct branched projections, which are typical morphological characteristics of DCs (Fig. 3A) (48, 54). Four hours after RV exposure, no distinct immunofluorescence was observed in PRV G9P[13]- or HRV Wa G1P[8]-exposed MoDCs (Fig. 3B and C). Twenty-four hours after RV exposure, granular immunofluorescence was observed in both PRV G9P[13]-exposed and HRV Wa G1P[8]-exposed MoDCs (Fig. 3D to G). A population of PRV G9P[13]-exposed MoDCs exhibited distinct granular immunofluorescence with a perinuclear cytoplasmic distribution (Fig. 3D and E), while HRV Wa G1P[8]-exposed MoDCs exhibited less immunofluorescence in the cytoplasm of infected cells, and there were lower frequencies of HRV Wa G1P[8]-infected MoDCs than PRV G9P[13]-infected MoDCs (Fig. 3F and G). In the PRV G9P[13]-positive MoDCs and HRV G1P[8]-positive MoDCs, the immunofluorescence was concentrated in cytoplasmic granules that might correspond to viroplasmic inclusions (Fig. 3D and E). These results suggested that PRV G9P[13] replicated in MoDCs to a greater extent than did HRV Wa G1P[8]. After RV exposure, no NSP4-positive (NSP4+) T cells were detected in RV-exposed T cells or T cells of the untreated group (data not shown). A similar result was observed for porcine B cells (data not shown), suggesting that porcine T cells and B cells were not susceptible to infection by PRV G9P[13] or HRV Wa G1P[8] in vitro.
FIG 3.
Detection of RVA anti-NSP4 antigen in untreated or PRV G9P[13]- or HRV Wa G1P[8]-exposed MoDCs. (A) MoDCs were incubated without RV for 24 h. (B) MoDCs were exposed to PRV G9P[13] for 4 h. (C) MoDCs were exposed to HRV G1P[8] for 4 h. (D and E) MoDCs were exposed to PRV G9P[13] for 24 h. (F and G) MoDCs were exposed to HRV G1P[8] for 24 h. (H and I) NSP4-positive PRV G9P[13]-exposed MoDCs visualized by fluorescence microscopy (H) and light microscopy (I). Protracting dendrites (A) and NSP4-positive cells (D, E, and G to I) are labeled with white arrows. The condensed green fluorescent granular particles in the cytoplasm surrounding the nucleus may represent viroplasmic inclusions. Bars, 200 μm.
DISCUSSION
The increasing RV group A strain diversity, including the worldwide emergence of G9 strains, poses new challenges to existing RV vaccines for humans and swine (19, 55). Therefore, it is necessary to genetically and biologically characterize the emerging G9 strains and to investigate whether the current human RV vaccines could elicit sufficient heterotypic protection against the heterologous G- and P-type emerging strains. In this study, we chose the dominant G-P combination strain in Ohio swine, PRV G9P[13], and a prevalent human strain as well as the G-P combination used in the Rotarix vaccine, HRV Wa G1P[8], to comparatively study their pathogenesis and cross-protection in Gn piglets and to examine the factors influencing PRV G9P[13] pathogenicity and spread.
The high overall nucleotide identity between PRV G9P[13] and historic PRV strains suggests the relative genetic stability of PRVs. Therefore, the HRV-like G9 VP7 genotype of this PRV G9P[13] strain likely emerged from reassortment events (56, 57). Recent studies confirmed that some emerging G9 RVs in humans were phylogenetically more similar to PRVs than to earlier human G9 genotypes, suggesting that PRVs are probable sources of heterologous RV infections in humans (22, 25). Since the majority of the serotype-specific antigenic regions of VP7 were expected to be conserved among RVs of the same G type (58), and the VP7 nucleotide of this PRV G9P[13] strain was most similar to a human G9 strain, it is plausible to extrapolate our data obtained by evaluating the cross-protection of PRV G9P[13] and HRV Wa G1P[8] in Gn pigs to predict whether current vaccines can confer heterotypic protection against emerging G9 strains in humans. We found that PRV G9P[13] inoculation conferred complete short-term protection against homologous (PRV G9P[13]) and heterologous (HRV Wa G1P[8]) RV infection and diarrhea, whereas HRV Wa G1P[8] inoculation provided partial heterologous protection against diarrhea and minimum heterologous protection against virus shedding after PRV G9P[13] challenge. Our results suggest that the current vaccine for humans (Rotarix) might not protect sufficiently against emerging G9 strains.
The introduction of RV vaccines may result in additional selective pressure on circulating RV strains and may affect their rates of evolution (20). If vaccine-induced selective pressure against G9 strains is lacking, this could facilitate the rapid global spread of G9 strains. As the commercially available PRV vaccines include only G4 and G5 PRV strains, a similar scenario may occur in swine, where the current PRV vaccines may lack effective protection against emerging G9 strains and may contribute to the dominance of the G9 strains in Ohio swine and to their being the fourth-most-dominant strain in the swine population of the Americas (18, 23). However, a recent study demonstrated that the A2 strain (previously identified as a G4 PRV strain) in a commercial PRV vaccine was in fact a G9 strain (59). If so, the prolonged administration of this vaccine could have introduced the G9 strain into the swine population that had no herd immunity against G9. This may provide a potential explanation for the emergence and subsequent spread of the G9 strains in swine in the United States and even the potential zoonotic transfer of reassortant G9 variants to humans.
The short-term cross-protection observed in pigs was likely not dependent on serum VN antibody titers. Similarly, previous studies indicated that protection against RV diarrhea was not dependent on epizootic diarrhea of infant mice (EDIM)-specific neutralizing antibody (60–62). Consistent with data from previous studies, we also observed that PRV G9P[13] induced low heterologous VN antibody titers in convalescent-phase sera. These results suggest that there are other factors associated with the high levels of cross-protection, including upregulated innate and mucosal or cellular immune responses possibly enhanced by the extended and significantly greater magnitude of PRV G9P[13] replication in vivo (61). Recent studies demonstrated that interferon lambda (IFN-λ) played a role in the intestinal epithelial antiviral responses to RV infection in neonatal mice (63, 64). Moreover, IFN-λ had a more important role in restricting the early replication of heterologous RV strains in suckling mice than in controlling homologous RV replication (65).
Mature enterocytes of the small intestine are the main target for RVs, which infect and destroy them, causing diarrhea (66, 67). Pathogenic RVs replicated faster in piglets and calves than did apathogenic RVs, with enterocyte losses from pathogenic RV infection surpassing their physiologic replacement (68–70). The similar severities of diarrhea in PRV G9P[13]- and HRV Wa G1P[8]-inoculated pigs indicated that these two RVA strains might have similar rates of replication in mature enterocytes of the small intestine, causing similar damage to the gut epithelium. In one study, RV antigen (VP6) and infectious virus (using NSP4 detection as an indicator of virus replication) were detected in multiple organs of RRV-inoculated neonatal rats (29). In a retrospective study, 34 of 353 (9.6%) children with confirmed RV gastroenteritis had extraintestinal RV infection and central nervous system complications (71). Moreover, an in vitro study indicated that RV NSPs were expressed in human peripheral blood mononuclear cells (PBMCs) after in vitro exposure to RRV, and the expression levels of the RRV NSPs varied in T cells, B cells, NK cells, monocytes, and DCs (35). Our studies indicated that PRV G9P[13] and/or HRV Wa G1P[8] may infect MNCs in the small intestine and/or adjacent lymph nodes, facilitating subsequent RV extraintestinal spread.
We observed prolonged RV RNAemia (suggestive of viremia) in PRV G9P[13]-inoculated pigs (at least on PIDs 3 and 5) and transient RV RNAemia in HRV Wa G1P[8]-inoculated pigs (only on PID 3). Our current results are consistent with data from a previous study from our laboratory showing that HRV Wa G1P1[8] caused transient RV RNAemia in Gn pigs (32). However, we detected HRV Wa G1P[8] RV RNAemia at a lower frequency than in the previous study. This may be due to the use of a lower inoculation dose (105 FFU versus 106 FFU) or the different time points selected for RV RNAemia detection (PID 3 versus PID 1). In addition to detection in sera, PRV G9P[13] and HRV Wa G1P[8] RNAs were also detected in MNCs from the MLNs, spleen, and liver. This result is in accordance with the detection of extraintestinal RRV in the neonatal mouse model after oral inoculation (33). Furthermore, we found viral RNA in blood MNCs of PRV G9P[13]-inoculated pigs but surprisingly not in blood MNCs from HRV Wa G1P[8]-inoculated pigs. Assessments of the extraintestinal spread of RV in the neonatal mouse model after oral inoculation with RRV, SA11 clone 4, and several single-segment reassortant viruses confirmed the ability of RVs to spread beyond the gut, which was primarily determined by the NSP3 phenotype and secondarily modified by VP6 (33, 72). Therefore, the different disseminating modes between the two RVA strains in our study might be due to different VP6 (I5 versus I1) genotypes and variations within the NSP3 sequences (T1).
Recent studies demonstrated that RRV and HRV Wa could infect human intestinal and blood B cells and that susceptibility was dependent on the B cell state and tissue origin (73). In our study, however, we failed to detect RV NSP4 antigen in T cells and B cells isolated from different tissues (ileum, MLNs, spleen, and blood) of uninoculated Gn pigs after exposure to PRV G9P[13] and HRV Wa G1P[8] in vitro using flow cytometry (data not shown). In the study conducted by Narváez et al., an MOI of 5 was used to infect human B cells, with primarily mature B cell subsets being infected, whereas the MOI that we used was only 0.4, and the neonatal Gn pig B cells that we used may be less mature and not in the same status (73). Conversely, we found RV NSP4 antigen in PRV G9P[13]-exposed MoDCs (same MOI of 0.4) by immunofluorescence. As a type of professional antigen-presenting cell (APC), DCs can take up and digest pathogens to present antigen to other immune cells to activate adaptive immune responses (74, 75). After phagocytosis, acidification of the phagosome and lysosomal fusion in mouse bone marrow-derived macrophages were achieved within 15 min and 2 h, respectively, suggesting rapid antigen degradation in macrophages (76). However, the antigen degradation capacity in either bone marrow-derived DCs in vitro or splenic and lymph node DCs in vivo indicates that the levels of phagosomal acidification and degradation are much lower in DCs than in macrophages and neutrophils, especially in immature DCs (77–79). Conflictingly, recent studies found that human MoDCs, in contrast to other types of human DCs, had lysosomal proteolysis levels and antigen degradation capacities similar to those of macrophages (80). Additionally, MoDCs rapidly and efficiently take up antigens from the environment (81). Therefore, we hypothesized that if RVs were unable to replicate in MoDCs, RVs would be taken up and rapidly digested by the MoDCs. In our study, we did not detect any RV NSP4 antigen in MoDCs after 4 h of exposure to PRV G9P[13] or HRV Wa G1P[8], but we found NSP4 antigen in DCs 24 h after exposure. This result suggested that the RV NSP4 antigen that we detected in DCs after 24 h of RV exposure was not accumulated by DC engulfment but was due to RV replication in DCs. In studies of NSP4 using enhanced green fluorescent protein (EGFP) in HEK 293 “Tet-on” cells, Berkova et al. found that NSP4-EGFP was expressed and distributed in novel vesicular structures throughout the cytoplasm and associated with viroplasms (82). This evidence supports our findings that PRV G9P[13] likely replicated and formed viroplasms in MoDCs. However, we do not know if RV infectious particles can be efficiently released from DCs to facilitate viral dissemination. More studies are required to verify this hypothesis.
In this study, we demonstrated that PRV G9P[13] induced prolonged fecal virus shedding and RV RNAemia compared to HRV Wa G1P[8]. This may be associated with the capacity of PRV G9P[13] to spread more efficiently beyond the gut and to replicate more in porcine immune cells (MoDCs) than HRV Wa G1P[8]. These characteristics of the G9 strain may contribute to its dominance in Ohio swine. To our knowledge, this is the first report of heterologous protection between an emerging G9 strain and an HRV vaccine-like G1 strain. Our data suggest that heterologous protection by the current monovalent HRV G1P[8] vaccine against the current worldwide-emerging RVA G9 strains should be evaluated further in relevant clinical and experimental studies. A similar scenario may be observed in swine populations, where current G4- and G5-based PRV vaccines may be ineffective against emerging porcine G9 strains, which also requires further studies. This may facilitate the increased global spread of RVA G9 strains. However, another study demonstrated that the A2 strain in the commercial PRV vaccine in the United States in fact included the G9 PRV strain and not a G4 strain (59). We hypothesize that the use of this vaccine might have contributed to the diversity of G9 strains in swine populations. Moreover, the potent short-term heterotypic protection induced by PRV G9P[13] is not dependent on the heterologous serum VN antibody titer but might rely on its potential to evoke IFN-λ responses or other mucosal or cellular immune responses. More studies on the long-term protection and immunogenicity of attenuated and virulent PRV G9P[13] using more Gn piglets and vaccination regimens allowing booster vaccinations (as done for current RV vaccines) are required to confirm and elucidate the mechanisms of heterotypic protection. Such data will aid in candidate vaccine strain selection and provide strategies for future RV vaccine design.
ACKNOWLEDGMENTS
We thank Juliette Hanson, Ronna Wood, and Jeff Ogg for animal care. We also thank Marcia Lee for conducting the fluorescent-focus neutralization test and technical support.
REFERENCES
- 1.Martella V, Bányai K, Matthijnssens J, Buonavoglia C, Ciarlet M. 2010. Zoonotic aspects of rotaviruses. Vet Microbiol 140:246–255. doi: 10.1016/j.vetmic.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 2.Midgley SE, Bányai K, Buesa J, Halaihel N, Hjulsager CK, Jakab F, Kaplon J, Larsen LE, Monini M, Poljšak-Prijatelj M, Pothier P, Ruggeri FM, Steyer A, Koopmans M, Böttiger B. 2012. Diversity and zoonotic potential of rotaviruses in swine and cattle across Europe. Vet Microbiol 156:238–245. doi: 10.1016/j.vetmic.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 3.Parashar UD, Hummelman EG, Bresee JS, Miller MA, Glass RI. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg Infect Dis 9:565–572. doi: 10.3201/eid0905.020562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parashar UD, Gibson CJ, Bresee JS, Glass RI. 2006. Rotavirus and severe childhood diarrhea. Emerg Infect Dis 12:304–306. doi: 10.3201/eid1202.050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Payne DC, Staat MA, Edwards KM, Szilagyi PG, Weinberg GA, Hall CB, Chappell J, Curns AT, Wikswo M, Tate JE, Lopman BA, Parashar UD, New Vaccine Surveillance Network. 2011. Direct and indirect effects of rotavirus vaccination upon childhood hospitalizations in 3 US counties, 2006-2009. Clin Infect Dis 53:245–253. doi: 10.1093/cid/cir307. [DOI] [PubMed] [Google Scholar]
- 6.Chang KO, Kim Y, Saif LJ. 2012. Rotaviruses and reoviruses, p 621–634. In Zimmerman JJ, Kariker LA, Ramirez A, Schwartz KJ, Stevenson GW (ed), Diseases of swine, 10th ed Wiley-Blackwell, Chichester, United Kindom. [Google Scholar]
- 7.Bernstein DI. 2009. Rotavirus overview. Pediatr Infect Dis J 28:S50–S53. doi: 10.1097/INF.0b013e3181967bee. [DOI] [PubMed] [Google Scholar]
- 8.Dennehy PH. 2008. Rotavirus vaccines: an overview. Clin Microbiol Rev 21:198–208. doi: 10.1128/CMR.00029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gómara M, Maes P, Patton JT, Rahman M, Van Ranst M. 2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol 82:3204–3219. doi: 10.1128/JVI.02257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marthaler D, Rossow K, Culhane M, Goyal S, Collins J, Matthijnssens J, Nelson M, Ciarlet M. 2014. Widespread rotavirus H in commercially raised pigs, United States. Emerg Infect Dis 20:1195–1198. doi: 10.3201/eid2007.140034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molinari BLD, Lorenzetti E, Otonel RAA, Alfieri AF, Alfieri AA. 2014. Species H rotavirus detected in piglets with diarrhea, Brazil, 2012. Emerg Infect Dis 20:1019–1022. doi: 10.3201/eid2006.130776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Bányai K, Estes MK, Gentsch JR, Iturriza-Gómara M, Kirkwood CD, Martella V, Mertens PPC, Nakagomi O, Patton JT, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Desselberger U, Van Ranst M. 2008. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol 153:1621–1629. doi: 10.1007/s00705-008-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abe M, Ito N, Masatani T, Nakagawa K, Yamaoka S, Kanamaru Y, Suzuki H, Shibano KI, Arashi Y, Sugiyama M. 2011. Whole genome characterization of new bovine rotavirus G21P[29] and G24P[33] strains provides evidence for interspecies transmission. J Gen Virol 92:952–960. doi: 10.1099/vir.0.028175-0. [DOI] [PubMed] [Google Scholar]
- 14.Miyazaki A, Kuga K, Suzuki T, Kohmoto M, Katsuda K, Tsunemitsu H. 2011. Genetic diversity of group A rotaviruses associated with repeated outbreaks of diarrhea in a farrow-to-finish farm: identification of a porcine rotavirus strain bearing a novel VP7 genotype, G26. Vet Res 42:112. doi: 10.1186/1297-9716-42-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santos N, Hoshino Y. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol 15:29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- 16.Chen S-C, Tan L-B, Huang L-M, Chen K-T. 2012. Rotavirus infection and the current status of rotavirus vaccines. J Formos Med Assoc 111:183–193. doi: 10.1016/j.jfma.2011.09.024. [DOI] [PubMed] [Google Scholar]
- 17.Iturriza-Gómara M, Dallman T, Bányai K, Böttiger B, Buesa J, Diedrich S, Fiore L, Johansen K, Korsun N, Kroneman A, Lappalainen M, László B, Maunula L, Matthinjnssens J, Midgley S, Mladenova Z, Poljsak-Prijatelj M, Pothier P, Ruggeri FM, Sanchez-Fauquier A, Schreier E, Steyer A, Sidaraviciute I, Tran AN, Usonis V, Van Ranst M, de Rougemont A, Gray J. 2009. Rotavirus surveillance in Europe, 2005-2008: Web-enabled reporting and real-time analysis of genotyping and epidemiological data. J Infect Dis 200(Suppl 1):S215–S221. doi: 10.1086/605049. [DOI] [PubMed] [Google Scholar]
- 18.Papp H, László B, Jakab F, Ganesh B, De Grazia S, Matthijnssens J, Ciarlet M, Martella V, Bányai K. 2013. Review of group A rotavirus strains reported in swine and cattle. Vet Microbiol 165:190–199. doi: 10.1016/j.vetmic.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi N, Ishino M, Wang Y-H, Chawla-Sarkar M, Krishnan T, Naik TN. 2007. Diversity of G-type and P-type of human and animal rotaviruses and its genetic background, p 847–858. In Mendez-Vulas A. (ed), Communicating current research and educational topics and trends in applied microbiology. Formatex, Badajoz, Spain. [Google Scholar]
- 20.Matthijnssens J, Heylen E, Zeller M, Rahman M, Lemey P, Van Ranst M. 2010. Phylodynamic analyses of rotavirus genotypes G9 and G12 underscore their potential for swift global spread. Mol Biol Evol 27:2431–2436. doi: 10.1093/molbev/msq137. [DOI] [PubMed] [Google Scholar]
- 21.Lachapelle V, Sohal JS, Lambert M-C, Brassard J, Fravalo P, Letellier A, L'Homme Y. 2014. Genetic diversity of group A rotavirus in swine in Canada. Arch Virol 159:1771–1779. doi: 10.1007/s00705-013-1951-9. [DOI] [PubMed] [Google Scholar]
- 22.Teodoroff TA, Tsunemitsu H, Okamoto K, Katsuda K, Kohmoto M, Kawashima K, Nakagomi T, Nakagomi O. 2005. Predominance of porcine rotavirus G9 in Japanese piglets with diarrhea: close relationship of their VP7 genes with those of recent human G9 strains. J Clin Microbiol 43:1377–1384. doi: 10.1128/JCM.43.3.1377-1384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amimo JO, Vlasova AN, Saif LJ. 2013. Detection and genetic diversity of porcine group A rotaviruses in historic (2004) and recent (2011 and 2012) swine fecal samples in Ohio: predominance of the G9P[13] genotype in nursing piglets. J Clin Microbiol 51:1142–1151. doi: 10.1128/JCM.03193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirkwood CD. 2010. Genetic and antigenic diversity of human rotaviruses: potential impact on vaccination programs. J Infect Dis 202:S43–S48. doi: 10.1086/653548. [DOI] [PubMed] [Google Scholar]
- 25.Phan TG, Okitsu S, Maneekarn N, Ushijima H. 2007. Genetic heterogeneity, evolution and recombination in emerging G9 rotaviruses. Infect Genet Evol 7:656–663. doi: 10.1016/j.meegid.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Hoshino Y, Kapikian AZ. 2000. Rotavirus serotypes: classification and importance in epidemiology, immunity, and vaccine development. J Health Popul Nutr 18:5–14. [PubMed] [Google Scholar]
- 27.Angel J, Franco MA, Greenberg HB. 2012. Rotavirus immune responses and correlates of protection. Curr Opin Virol 2:419–425. doi: 10.1016/j.coviro.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blutt SE, Kirkwood CD, Parreno V, Warfield KL, Ciarlet M, Estes MK, Bok K, Bishop RF, Conner ME. 2003. Rotavirus antigenaemia and viraemia: a common event? Lancet 362:1445–1449. doi: 10.1016/S0140-6736(03)14687-9. [DOI] [PubMed] [Google Scholar]
- 29.Crawford SE, Patel DG, Cheng E, Berkova Z, Hyser JM, Ciarlet M, Finegold MJ, Conner ME, Estes MK. 2006. Rotavirus viremia and extraintestinal viral infection in the neonatal rat model. J Virol 80:4820–4832. doi: 10.1128/JVI.80.10.4820-4832.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riepenhoff-Talty M, Dharakul T, Kowalski E, Michalak S, Ogra PL. 1987. Persistent rotavirus infection in mice with severe combined immunodeficiency. J Virol 61:3345–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sood M, Booth IW. 1999. Is prolonged rotavirus infection a common cause of protracted diarrhoea? Arch Dis Child 80:309–310. doi: 10.1136/adc.80.4.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Azevedo MS, Yuan L, Jeong KI, Gonzalez A, Nguyen TV, Pouly S, Gochnauer M, Zhang W, Azevedo A, Saif LJ. 2005. Viremia and nasal and rectal shedding of rotavirus in gnotobiotic pigs inoculated with Wa human rotavirus. J Virol 79:5428–5436. doi: 10.1128/JVI.79.9.5428-5436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mossel EC, Ramig RF. 2003. A lymphatic mechanism of rotavirus extraintestinal spread in the neonatal mouse. J Virol 77:12352–12356. doi: 10.1128/JVI.77.22.12352-12356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohanty SK, Ivantes CAP, Mourya R, Pacheco C, Bezerra JA. 2010. Macrophages are targeted by rotavirus in experimental biliary atresia and induce neutrophil chemotaxis by Mip2/Cxcl2. Pediatr Res 67:345–351. doi: 10.1203/PDR.0b013e3181d22a73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mesa MC, Rodríguez L-S, Franco MA, Angel J. 2007. Interaction of rotavirus with human peripheral blood mononuclear cells: plasmacytoid dendritic cells play a role in stimulating memory rotavirus specific T cells in vitro. Virology 366:174–184. doi: 10.1016/j.virol.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Narváez CF, Angel J, Franco MA. 2005. Interaction of rotavirus with human myeloid dendritic cells. J Virol 79:14526–14535. doi: 10.1128/JVI.79.23.14526-14535.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H-H, Park J-G, Matthijnssens J, Kim H-J, Kwon H-J, Son K-Y, Ryu E-H, Kim D-S, Lee WS, Kang M-I, Yang D-K, Lee J-H, Park S-J, Cho K-O. 2013. Pathogenicity of porcine G9P[23] and G9P[7] rotaviruses in piglets. Vet Microbiol 166:123–137. doi: 10.1016/j.vetmic.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward LA, Rosen BI, Yuan L, Saif LJ. 1996. Pathogenesis of an attenuated and a virulent strain of group A human rotavirus in neonatal gnotobiotic pigs. J Gen Virol 77(Part 7):1431–1441. [DOI] [PubMed] [Google Scholar]
- 39.Nyaga MM, Jere KC, Esona MD, Seheri ML, Stucker KM, Halpin RA, Akopov A, Stockwell TB, Peenze I, Diop A, Ndiaye K, Boula A, Maphalala G, Berejena C, Mwenda JM, Steele AD, Wentworth DE, Mphahlele MJ. 2015. Whole genome detection of rotavirus mixed infections in human, porcine and bovine samples co-infected with various rotavirus strains collected from sub-Saharan Africa. Infect Genet Evol 31:321–334. doi: 10.1016/j.meegid.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan L, Saif LJ. 2002. Induction of mucosal immune responses and protection against enteric viruses: rotavirus infection of gnotobiotic pigs as a model. Vet Immunol Immunopathol 87:147–160. doi: 10.1016/S0165-2427(02)00046-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saif LJ, Ward LA, Yuan L, Rosen BI, To TL. 1996. The gnotobiotic piglet as a model for studies of disease pathogenesis and immunity to human rotaviruses. Arch Virol Suppl 12:153–161. [DOI] [PubMed] [Google Scholar]
- 42.Saif LJ, Yuan L, Ward L, To T. 1997. Comparative studies of the pathogenesis, antibody immune responses, and homologous protection to porcine and human rotaviruses in gnotobiotic piglets. Adv Exp Med Biol 412:397–403. doi: 10.1007/978-1-4899-1828-4_62. [DOI] [PubMed] [Google Scholar]
- 43.Zhang W, Azevedo MSP, Wen K, Gonzalez A, Saif LJ, Li G, Yousef AE, Yuan L. 2008. Probiotic Lactobacillus acidophilus enhances the immunogenicity of an oral rotavirus vaccine in gnotobiotic pigs. Vaccine 26:3655–3661. doi: 10.1016/j.vaccine.2008.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pang XL, Lee B, Boroumand N, Leblanc B, Preiksaitis JK, Yu Ip CC. 2004. Increased detection of rotavirus using a real time reverse transcription-polymerase chain reaction (RT-PCR) assay in stool specimens from children with diarrhea. J Med Virol 72:496–501. doi: 10.1002/jmv.20009. [DOI] [PubMed] [Google Scholar]
- 45.Yuan L, Ward LA, Rosen BI, To TL, Saif LJ. 1996. Systematic and intestinal antibody-secreting cell responses and correlates of protective immunity to human rotavirus in a gnotobiotic pig model of disease. J Virol 70:3075–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong Z-J, Wei H-M, Sun R, Tian Z-G, Gao B. 2004. Isolation of murine hepatic lymphocytes using mechanical dissection for phenotypic and functional analysis of NK1.1+ cells. World J Gastroenterol 10:1928–1933. doi: 10.3748/wjg.v10.i13.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCullough KC, Basta S, Knotig S, Gerber H, Schaffner R, Kim YB, Saalmuller A, Summerfield A. 1999. Intermediate stages in monocyte-macrophage differentiation modulate phenotype and susceptibility to virus infection. Immunology 98:203–212. doi: 10.1046/j.1365-2567.1999.00867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin C-M, Jeng C-R, Hsiao S-H, Lee Y, Tsai Y-C, Chia M-Y, Pang VF. 2012. Monocyte-derived dendritic cells enhance cell proliferation and porcine circovirus type 2 replication in concanavalin A-stimulated swine peripheral blood lymphocytes in vitro. Vet Immunol Immunopathol 145:368–378. doi: 10.1016/j.vetimm.2011.12.009. [DOI] [PubMed] [Google Scholar]
- 49.Perez-Cano FJ, Marin-Gallen S, Castell M, Rodriguez-Palmero M, Rivero M, Castellote C, Franch A. 2008. Supplementing suckling rats with whey protein concentrate modulates the immune response and ameliorates rat rotavirus-induced diarrhea. J Nutr 138:2392–2398. doi: 10.3945/jn.108.093856. [DOI] [PubMed] [Google Scholar]
- 50.Chattha KS, Vlasova AN, Kandasamy S, Rajashekara G, Saif LJ. 2013. Divergent immunomodulating effects of probiotics on T cell responses to oral attenuated human rotavirus vaccine and virulent human rotavirus infection in a neonatal gnotobiotic piglet disease model. J Immunol 191:2446–2456. doi: 10.4049/jimmunol.1300678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vlasova AN, Chattha KS, Kandasamy S, Liu Z, Esseili M, Shao L, Rajashekara G, Saif LJ. 2013. Lactobacilli and bifidobacteria promote immune homeostasis by modulating innate immune responses to human rotavirus in neonatal gnotobiotic pigs. PLoS One 8:e76962. doi: 10.1371/journal.pone.0076962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crouch CF, Woode GN. 1978. Serial studies of virus multiplication and intestinal damage in gnotobiotic piglets infected with rotavirus. J Med Microbiol 11:325–334. doi: 10.1099/00222615-11-3-325. [DOI] [PubMed] [Google Scholar]
- 53.Bohl EH, Theil KW, Saif LJ. 1984. Isolation and serotyping of porcine rotaviruses and antigenic comparison with other rotaviruses. J Clin Microbiol 19:105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen B, Shi Y, Smith JD, Choi D, Geiger JD, Mulé JJ. 1998. The role of tumor necrosis factor alpha in modulating the quantity of peripheral blood-derived, cytokine-driven human dendritic cells and its role in enhancing the quality of dendritic cell function in presenting soluble antigens to CD4+ T cells in vitro. Blood 91:4652–4661. [PubMed] [Google Scholar]
- 55.Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Bányai K, Ramachandran M, Jain V, Cunliffe NA, Nakagomi O, Kirkwood CD, Fischer TK, Parashar UD, Bresee JS, Jiang B, Glass RI. 2005. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J Infect Dis 192(Suppl 1):S146–S159. doi: 10.1086/431499. [DOI] [PubMed] [Google Scholar]
- 56.Theuns S, Desmarets LMB, Heylen E, Zeller M, Dedeurwaerder A, Roukaerts IDM, Van Ranst M, Matthijnssens J, Nauwynck HJ. 2014. Porcine group A rotaviruses with heterogeneous VP7 and VP4 genotype combinations can be found together with enteric bacteria on Belgian swine farms. Vet Microbiol 172:23–34. doi: 10.1016/j.vetmic.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 57.Theuns S, Heylen E, Zeller M, Roukaerts IDM, Desmarets LMB, Van Ranst M, Nauwynck HJ, Matthijnssens J. 2015. Complete genome characterization of recent and ancient Belgian pig group A rotaviruses and assessment of their evolutionary relationship with human rotaviruses. J Virol 89:1043–1057. doi: 10.1128/JVI.02513-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang J, Nagesha HS, Dyall-Smith ML, Holmes IH. 1989. Comparative sequence analysis of VP7 genes from five Australian porcine rotaviruses. Arch Virol 109:173–183. doi: 10.1007/BF01311079. [DOI] [PubMed] [Google Scholar]
- 59.Hoshino Y, Honma S, Jones RW, Ross J, Santos N, Gentsch JR, Kapikian AZ, Hesse RA. 2005. A porcine G9 rotavirus strain shares neutralization and VP7 phylogenetic sequence lineage 3 characteristics with contemporary human G9 rotavirus strains. Virology 332:177–188. doi: 10.1016/j.virol.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 60.Ward RL, Clemens JD, Knowlton DR, Rao MR, van Loon FP, Huda N, Ahmed F, Schiff GM, Sack DA. 1992. Evidence that protection against rotavirus diarrhea after natural infection is not dependent on serotype-specific neutralizing antibody. J Infect Dis 166:1251–1257. doi: 10.1093/infdis/166.6.1251. [DOI] [PubMed] [Google Scholar]
- 61.McNeal MM, Broome RL, Ward RL. 1994. Active immunity against rotavirus infection in mice is correlated with viral replication and titers of serum rotavirus IgA following vaccination. Virology 204:642–650. doi: 10.1006/viro.1994.1579. [DOI] [PubMed] [Google Scholar]
- 62.Ward RL, McNeal MM, Sheridan JF. 1992. Evidence that active protection following oral immunization of mice with live rotavirus is not dependent on neutralizing antibody. Virology 188:57–66. doi: 10.1016/0042-6822(92)90734-7. [DOI] [PubMed] [Google Scholar]
- 63.Pott J, Mahlakõiv T, Mordstein M, Duerr CU, Michiels T, Stockinger S, Staeheli P, Hornef MW. 2011. IFN-lambda determines the intestinal epithelial antiviral host defense. Proc Natl Acad Sci U S A 108:7944–7949. doi: 10.1073/pnas.1100552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hernández PP, Mahlakõiv T, Yang I, Schwierzeck V, Nguyen N, Guendel F, Gronke K, Ryffel B, Hölscher C, Dumoutier L, Renauld J-C, Suerbaum S, Staeheli P, Diefenbach A. 2015. Interferon-λ and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat Immunol 16:698–707. doi: 10.1038/ni.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arnold MM, Sen A, Greenberg HB, Patton JT. 2013. The battle between rotavirus and its host for control of the interferon signaling pathway. PLoS Pathog 9:e1003064. doi: 10.1371/journal.ppat.1003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graham DY, Sackman JW, Estes MK. 1984. Pathogenesis of rotavirus-induced diarrhea. Dig Dis Sci 29:1028–1035. doi: 10.1007/BF01311255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramig RF. 2004. Pathogenesis of intestinal and systemic rotavirus infection. J Virol 78:10213–10220. doi: 10.1128/JVI.78.19.10213-10220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burke B, Desselberger U. 1996. Rotavirus pathogenicity. Virology 218:299–305. doi: 10.1006/viro.1996.0198. [DOI] [PubMed] [Google Scholar]
- 69.Bridger JC, Burke B, Beards GM, Desselberger U. 1992. The pathogenicity of two porcine rotaviruses differing in their in vitro growth characteristics and genes 4. J Gen Virol 73(Part 11):3011–3015. [DOI] [PubMed] [Google Scholar]
- 70.Hall GA, Bridger JC, Parsons KR, Cook R. 1993. Variation in rotavirus virulence: a comparison of pathogenesis in calves between two rotaviruses of different virulence. Vet Pathol 30:223–233. doi: 10.1177/030098589303000302. [DOI] [PubMed] [Google Scholar]
- 71.Dalgic N, Haşim Ö, Pullu M, Sancar M, Kafadar I. 2010. Is rotavirus diarrhea a systemic viral infection? Cocuk Enf Derg 4:48–55. [Google Scholar]
- 72.Mossel EC, Ramig RF. 2002. Rotavirus genome segment 7 (NSP3) is a determinant of extraintestinal spread in the neonatal mouse. J Virol 76:6502–6509. doi: 10.1128/JVI.76.13.6502-6509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Narváez CF, Franco MA, Angel J, Morton JM, Greenberg HB. 2010. Rotavirus differentially infects and polyclonally stimulates human B cells depending on their differentiation state and tissue of origin. J Virol 84:4543–4555. doi: 10.1128/JVI.02550-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cella M, Sallusto F, Lanzavecchia A. 1997. Origin, maturation and antigen presenting function of dendritic cells. Curr Opin Immunol 9:10–16. doi: 10.1016/S0952-7915(97)80153-7. [DOI] [PubMed] [Google Scholar]
- 75.Banchereau J, Steinman RM. 1998. Dendritic cells and the control of immunity. Nature 392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 76.Yates RM, Hermetter A, Russell DG. 2005. The kinetics of phagosome maturation as a function of phagosome/lysosome fusion and acquisition of hydrolytic activity. Traffic 6:413–420. doi: 10.1111/j.1600-0854.2005.00284.x. [DOI] [PubMed] [Google Scholar]
- 77.Savina A, Amigorena S. 2007. Phagocytosis and antigen presentation in dendritic cells. Immunol Rev 219:143–156. doi: 10.1111/j.1600-065X.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- 78.Delamarre L, Pack M, Chang H, Mellman I, Trombetta ES. 2005. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science 307:1630–1634. doi: 10.1126/science.1108003. [DOI] [PubMed] [Google Scholar]
- 79.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. 2003. Activation of lysosomal function during dendritic cell maturation. Science 299:1400–1403. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- 80.McCurley N, Mellman I. 2010. Monocyte-derived dendritic cells exhibit increased levels of lysosomal proteolysis as compared to other human dendritic cell populations. PLoS One 5:e11949. doi: 10.1371/journal.pone.0011949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sallusto F, Cella M, Danieli C, Lanzavecchia A. 1995. Dendritic cells use macropinocytosis and the mannose receptor to concentrate macromolecules in the major histocompatibility complex class II compartment: downregulation by cytokines and bacterial products. J Exp Med 182:389–400. doi: 10.1084/jem.182.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berkova Z, Crawford SE, Trugnan G, Yoshimori T, Morris AP, Estes MK. 2006. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasms. J Virol 80:6061–6071. doi: 10.1128/JVI.02167-05. [DOI] [PMC free article] [PubMed] [Google Scholar]