Abstract
Influenza A virus infection can arrest autophagy, as evidenced by autophagosome accumulation in infected cells. Here, we report that this autophagosome accumulation can be inhibited by amantadine, an antiviral proton channel inhibitor, in amantadine-sensitive virus infected cells or cells expressing influenza A virus matrix protein 2 (M2). Thus, M2 proton channel activity plays a role in blocking the fusion of autophagosomes with lysosomes, which might be a key mechanism for arresting autophagy.
TEXT
Influenza viruses cause significant morbidity and mortality in humans (1, 2). An outstanding feature of the virus is its ability to regulate host cellular pathways for its benefit (3). Recent studies showed that influenza A virus perturbs the autophagy process in infected cells (4–6). Autophagy is an intracellular degradation process that can be divided into two stages. The first is the formation of autophagosomes in which the cytoplasmic materials, including cellular organelles, protein aggregates, and pathogens, are directed to the double-membrane vesicles. In the second, the matured autophagosomes fuse with lysosomes to form autolysosomes to degrade their contents (7). Autophagy is involved in both the sensing of and resistance to viruses invading the host. As a result, the viruses appear to have evolved mechanisms to subvert the autophagy response for their own benefit (8–10). Although influenza A virus has been shown to modulate the autophagy process (4, 5, 11–13), the underlying mechanism has remained not well defined.
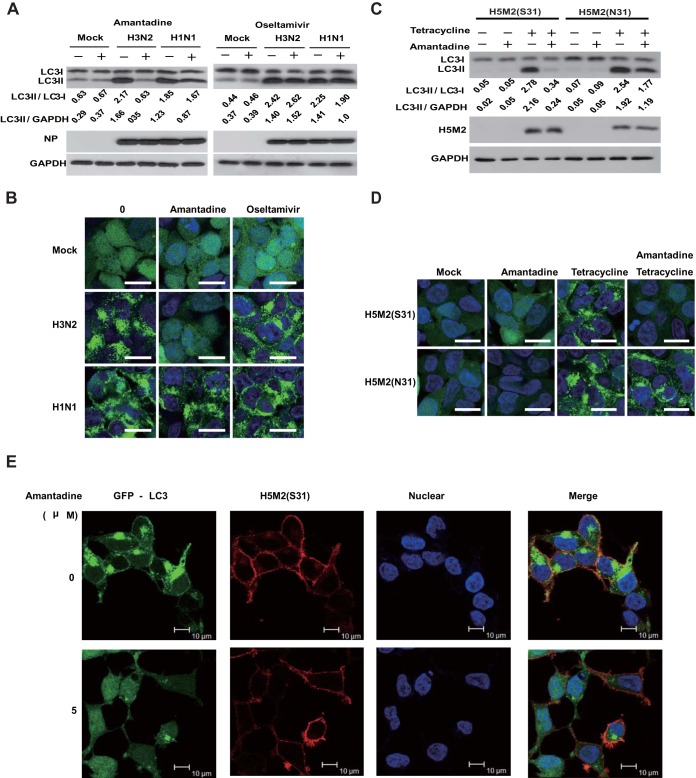
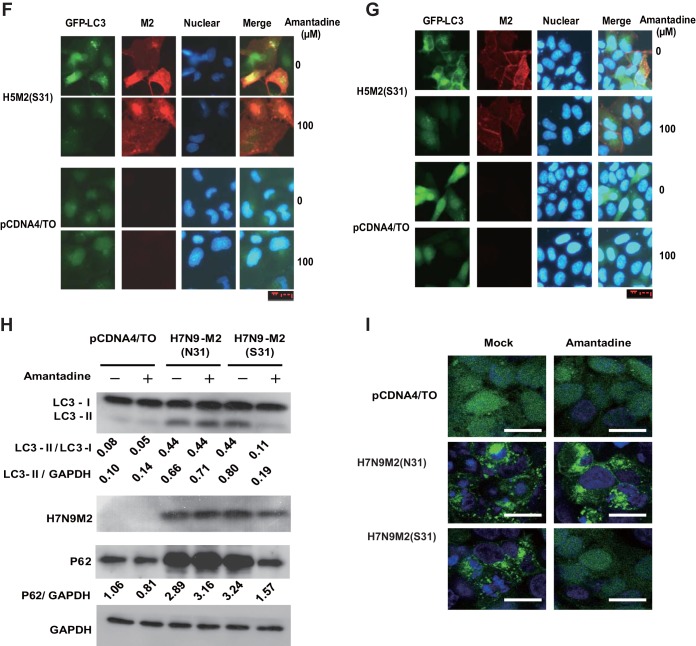
To observe the effect of influenza A virus on autophagy, we used influenza viruses A/Hong Kong/8/68 (H3N2) and A/Wisconsin/33(H1N1) to infect HEK293 cells at a multiplicity of infection (MOI) of 5. We measured the relative amounts of autophagosome marker light chain 3 (LC3) proteins (lipidated LC3-II [16 kDa] and nonlipidated LC3-I [18 kDa]) in the cells at 12 h after infection. At that time point, the cells appeared healthy, without obvious signs of cell death. We also used a cell line stably expressing a green fluorescent protein (GFP)-LC3 fusion protein for easy observation of autophagosomes (14). Both influenza viruses A/Hong Kong/8/68 (H3N2) and A/Wisconsin/33 (H1N1) increased the autophagosome marker LC3-II level (Fig. 1A) and induced punctate LC3 perinuclear localization (Fig. 1B). These results demonstrated that influenza A virus infection can induce autophagosome accumulation, which was consistent with a previous report (4). We then used amantadine, an influenza A virus M2 proton channel blocker, and oseltamivir, an influenza A virus neuraminidase inhibitor, to assess if these two antiviral agents have any effect on autophagosome accumulation at the early stage of infection. Amantadine or oseltamivir was added at 3 h, and cells were harvested at 12 h postinfection for analysis. Surprisingly, amantadine can significantly block autophagosome accumulation in influenza virus A/Hong Kong/8/68(H3N2)-infected cells but not in influenza virus A/Wisconsin/33(H1N1)-infected cells (Fig. 1A and B). It is worth noting that influenza virus A/Hong Kong/8/68(H3N2) is sensitive to amantadine and influenza virus A/WS/33 (H1N1) is resistant to amantadine because of an S31N mutation on M2 (15). Oseltamivir showed no effect on autophagosome accumulation in response to both influenza A viruses, indicating that neuraminidase plays no role in autophagosome accumulation, at least in the early stage of infection (Fig. 1A and B). Our study suggested that the proton channel activity of M2 might play a role in modulation of the autophagy process, in contrast to a previous report that showed that its proton channel activity was not involved in autophagy arrest (4). To further investigate whether M2 proton channel activity is sufficient to induce autophagosome accumulation and because the constitutive expression of M2 is lethal to the cells, we generated TREx-293 cell lines carrying an amantadine-sensitive (S31) or -resistant (N31) mutant form of avian influenza A virus H5N1/Vietnam/1194/2004 M2 under the control of a tetracycline-inducible promoter (15, 16). These inducible M2-expressing cell lines enabled us to observe the sole effect of M2 without the interference of influenza A virus replication or other viral proteins. Upon the expression of either H5M2 (S31) or H5M2 (N31) with induction of tetracycline, we observed remarkable autophagosome accumulation in the cells, as shown by the increase in LC3-II in a Western blot assay (Fig. 1C). Consistent with influenza A virus infection, amantadine could inhibit the increase in LC3-II and perinuclear localization induced by amantadine-sensitive H5M2 (S31) (Fig. 1C and D). In GFP-LC3-transfected cells expressing H5M2(S31), both punctate LC3 in the perinuclear region and LC3 distribution to the plasma membrane could be observed if cells were weakly permeabilized for immunostaining of M2 protein. The observation of LC3 distribution to the plasma membrane concurred with an earlier report (11). In amantadine-sensitive H5M2(S31)-expressing cells, amantadine treatment diminished both perinuclear and plasma accumulation of LC3, although the M2 protein remained at the plasma membrane (Fig. 1E). We also tested the expression of H5N1-M2(S31) on autophagy in MDCK (Fig. 1F) and MCF-7 (Fig. 1G) cells. Autophagosome accumulation could be observed in M2-expressing cells and could be inhibited by amantadine. The recent outbreak of the novel avian influenza A virus H7N9 in Asia and the high mortality rate due to it have caused global concerns (17, 18). The H7M2 protein contains asparagine (N) at amino acid position 31, conferring resistance to amantadine (17). To see whether H7M2 can also affect the autophagy process, we constructed two DNA plasmids, pCDNA4-H7N9-M2 (N31) and pCDNA4-H7N9-M2 (S31), with the asparagine at amino acid position 31 replaced with serine to restore amantadine sensitivity. Transfection of either H7M2 (N31) or H7M2 (S31) could induce substantial autophagosome accumulation in HEK293 cells. Again, amantadine could inhibit autophagosome accumulation induced by H7M2 (S31) but not that induced by amantadine-resistant H7M2 (N31) (Fig. 1H and I). We analyzed autophagosome turnover in the M2-expressing cells by using the marker SQSTM1/P62, an adaptor protein that binds to LC3 and then is degraded through the lysosomal pathway (19, 20). Both H7M2 (S31) and H7M2(N31) could induce accumulation of SQSTM1/P62, suggesting autophagy arrest, as indicated by the failure of autophagic substrate degradation. Amantadine treatment could reverse the accumulation of SQSTM1/P62 in cells transfected with H7M2 (S31) but not H7M2 (N31) (Fig. 1H). Therefore, data from different subtypes of influenza A virus M2 and multiple cell lines demonstrated that M2 proton channel activity is involved in the induction of autophagy arrest.
FIG 1.
Inhibition of proton channel activity attenuates influenza A virus M2-induced autophagosome accumulation. (A) HEK293 cells were infected with influenza virus A/Hong Kong/8/68 (H3N2) or A/Wisconsin/33(H1N1) at an MOI of 5. Amantadine (5 μM) or oseltamivir (200 nM) was added at 3 h after infection. Cell were collected at 12 h after infection and subjected to Western blot analysis with the antibodies indicated. NP, nucleoprotein; GAPDH, glyceraldehyde 3-phosphate dehydrogenase. (B) HEK293 cells with stable expression of GFP-LC3 were treated as described for panel A and observed by confocal microscopy. Scale bars, 20 μm. (C) TREx-293 cells carrying tetracycline-inducible amantadine-sensitive H5N1-M2 (S31) or amantadine-resistant H5N1-M2 (N31) were treated with tetracycline (1 μg/ml) with or without amantadine. Cells were collected at 24 h and subjected to Western blot analysis with the antibodies indicated. (D) GFP-LC3-transfected TREx-293 cells carrying tetracycline-inducible H5N1-M2 (S31) or H5N1-M2 (N31) were treated with tetracycline with or without amantadine. Cells were observed 24 h later by confocal microscopy. Scale bars, 20 μm. (E) GFP-LC3-transfected TREx-293 cells carrying tetracycline-inducible amantadine-sensitive H5N1-M2 (S31) were treated with tetracycline with or without amantadine for 24 h. Cells were weakly permeabilized for M2 immunostaining and observed by confocal microscopy. Scale bars, 10 μm. (F, G) MDCK (F) or MCF-7 (G) cells stably expressing GFP-LC3 were transfected, respectively, with plasmids expressing H5M2(S31). Amantadine was added at 6 h after transfection. Cells were collected at 24 h and subjected to immunofluorescence staining with anti-M2 antibodies and a DyLight 549-labeled goat anti-mouse secondary antibody. The cells were observed by fluorescence microscopy. Scale bars, 20 μm. (H) HEK293 cells were transfected with plasmid pcDNA4, pcDNA4-H7N9-M2 (S31), or pcDNA4-H7N9-M2 (N31) and then treated with or without amantadine at 6 h after transfection. Cell were collected at 24 h after amantadine treatment and subjected to Western blot analysis with the antibodies indicated. (I) HEK293 cells with stable expression of GFP-LC3 were transfected with plasmid pcDNA4, pcDNA4-H7N9-M2 (S31), or pcDNA4-H7N9-M2 (N31) and then treated with amantadine at 6 h after transfection. Cells were observed by confocal microscopy at 24 h after amantadine treatment. Scale bars, 20 μm. Representative data from one of three separate experiments are shown. The relative LC3-II/LC3-I, LC3-II/GADPH, and P62/GADPH ratios were analyzed with ImageJ (National Institutes of Health) in the same Western blot assay. The anti-M2 mouse serum was generated in our lab, the anti-influenza A virus nucleoprotein antibody was Abcam Ab139361, the anti-LC3-II antibody was Sigma-Aldrich L7543, and the anti-P62 antibody was Sigma-Aldrich P0067.
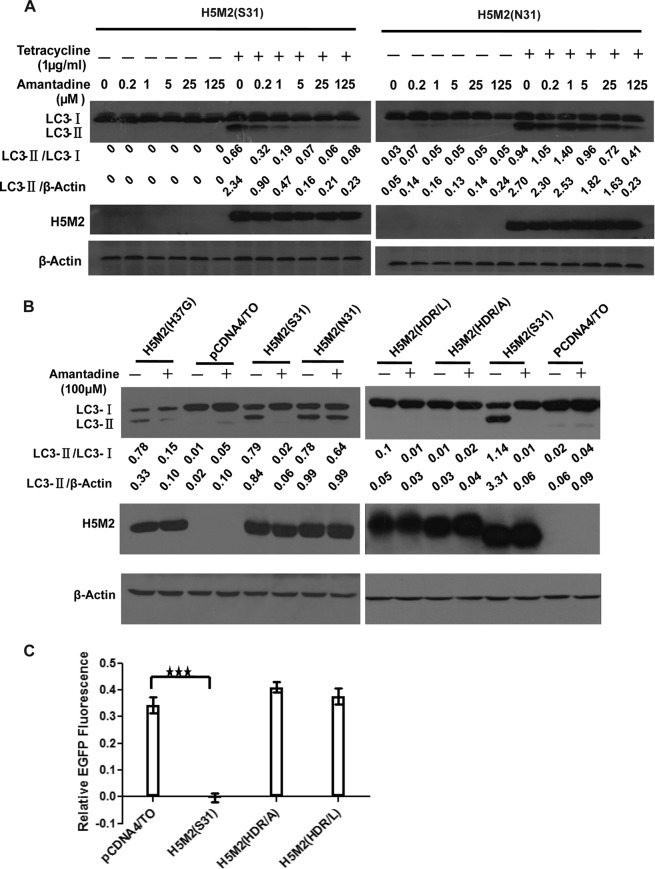
To verify that the M2 proton channel activity is involved in the induction of autophagosome accumulation, we tested the dose-dependent effect of amantadine on the inhibition of M2-induced LC3-II accumulation in TREx-293 cells expressing tetracycline-inducible M2 proteins. Amantadine showed a dose-dependent ability, starting at 0.2 μM, to block LC3-II accumulation in cells expressing amantadine-sensitive H5M2 (S31), whereas amantadine showed only a weak inhibitory effect at 125 μM in cells expressing amantadine-resistant H5M2 (N31) (Fig. 2A). To further study the role of M2 proton channel activity on autophagosome accumulation, mutations at the M2 transmembrane region were made to eliminate proton channel activity. We first constructed an M2-H37G mutation as reported previously (4). However, the M2-H37G mutant protein could still induce LC3-II accumulation, which could be inhibited by amantadine (Fig. 2B). Indeed, earlier studies have reported that the H37G mutation does not decrease ion channel activity but only eliminates its stringent ion selectivity (21, 22). We subsequently constructed proteins with more severe mutations by changing three amino acids, His37, Asp44, and Arg45, in the M2 transmembrane region to abolish proton channel activity. M2(HDR/A) has His37Ala, Asp44Ala, and Arg45Ala mutations, while M2(HDR/L) has His37Leu, Asp44Leu, and Arg45Leu mutations. Expression of the M2(HDR/A) and M2(HRD/L) proteins was verified in plasmid-transfected cells (Fig. 2B). These two mutant proteins were confirmed to have no proton channel activity by using pH-sensitive enhanced GFP (EGFP) coexpressed in the cells (23). EGFP showed a significant decrease in fluorescence intensity in H5M2(S31)-transfected cells under acidic conditions due to M2 proton channel-mediated acidification but not in M2(HDR/A)- or M2(HRD/L)-transfected cells (Fig. 2C). Neither the M2(HDR/A) nor the M2(HRD/L) mutant protein could induce autophagosome accumulation, as indicated by no increase in LC3-II (Fig. 2B).
FIG 2.
Proton channel activity contributes to influenza A virus M2-induced autophagosome accumulation in a dose-dependent manner. (A) TREx-293 cells carrying tetracycline-inducible amantadine-sensitive H5N1-M2 (S31) or amantadine-resistant H5N1-M2 (N31) were treated and with increasing concentrations of amantadine with or without tetracycline as indicated. Cells were collected at 24 h and subjected to Western blot analysis with the antibodies indicated. (B) HEK293 cells were transfected with plasmids expressing H5M2 (S31), H5M2 (N31), H5M2 (H37G), H5M2 (HDR/A), or H5M2 (HDR/L). Amantadine was added at 6 h after transfection. Cells were collected at 24 h and subjected to Western blot analysis with the antibodies indicated. Representative data from one of three separate experiments are shown. (C) MDCK cells were transfected with plasmids expressing H5M2 (S31), H5M2 (HDR/A), or H5M2 (HDR/L) with a pH-sensitive EGFP protein in the wells of a 96-well plate in six replicates. At 16 h after transfection, the culture medium was replaced with pH 7.4 phosphate-buffered saline containing 0.3% bovine serum albumin for detection of fluorescence intensity at neutral pH. The change in fluorescence intensity was detected at 1 min after the medium was changed to pH 5.0. The relative fluorescence was the ratio of fluorescence intensity at pH 5.0 normalized to that at pH 7.4. Statistical analyses were performed by two-tailed Mann-Whitney test. ***, P < 0.001. Representative data from one of three separate experiments are shown. The relative LC3-II/LC3-I and LC3-II/β-actin ratios were analyzed by using ImageJ (National Institutes of Health) in the same Western blot assay. M2(HDR/A) has His37Ala, Asp44Ala, and Arg45Ala mutations. M2(HDR/L) has His37Leu, Asp44 Leu, and Arg45Leu mutations.
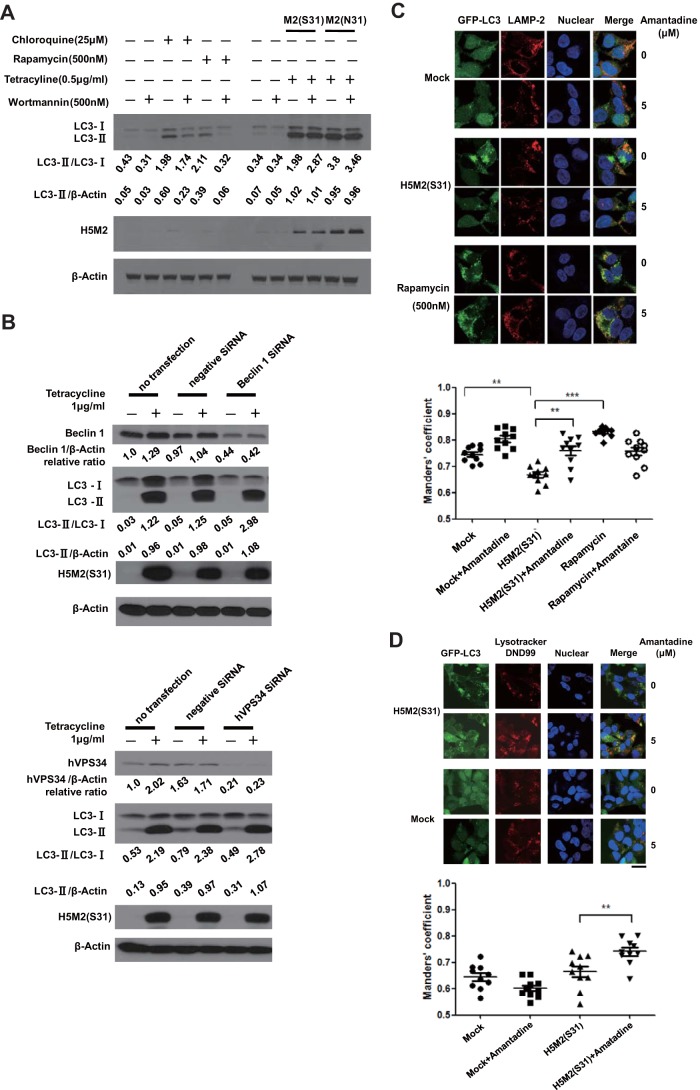
Previous studies have shown that the autophagy process can be activated by the mTOR inhibitor rapamycin, which can be inhibited by wortmannin, a general inhibitor of phosphatidylinositol-3-phosphate kinases (24–27). We compared the effect of wortmannin on rapamycin- or chloroquine-treated cells with that on cells that express H5M2 (S31) or H5M2 (N31). Rapamycin or chloroquine could induce an LC3-II level increase in treated cells. Wortmannin could significantly inhibit LC3-II accumulation in rapamycin-treated cells and modestly inhibit LC3-II accumulation in chloroquine-treated cells. Interestingly, wortmannin had no inhibitory effect on M2-induced LC3-II accumulation (Fig. 3A). When the expression of Becline 1 (Atg6), and phosphatidylinositol 3-kinase (PI3K) VPS34 was knocked down with small interfering RNA (siRNA), the M2-mediated increase in LC3-II was not affected by the silencing of either Beclin1 (Atg6) or VPS34 (Fig. 3B), indicating that M2 may induce LC3-II accumulation via a VPS34/Beclin 1-indepedent pathway. In M2(S31)-expressing cells, there was no obvious colocalization of GFP-LC3-labeled autophagosomes with lysosomal marker LAMP2-labeled lysosomes (Fig. 3C), indicating little fusion between autophagosomes and lysosomes. Amantadine treatment for 3 h decreased punctate GFP-LC3, and the preformed GFP-LC3 vesicles appeared to overlap LAMP2-positive compartments in M2(S31)-expressing cells. In comparison, rapamycin induced colocalization of GFP-LC3-labeled autophagosomes with LAMP2-labeled lysosomes (Fig. 3C). We finally confirmed M2-mediated blockage of autophagosome fusion with lysosomes by using LysoTracker DND99 to label lysosomes in living cells. In mock-transfected cells, there was no punctate GFP-LC3. In M2(S31)-expressing cells, there was punctate GFP-LC3 but little colocalization with LysoTracker-labeled lysosomes. With amantadine treatment for just 1 h, punctate GFP-LC3 appeared to localize with lysosomes in M2(S31)-expressing cells (Fig. 3D). Taking these results together, we propose that M2 proton channel activity plays an important role in inducing autophagy arrest by blocking the fusion of autophagosomes with lysosomes.
FIG 3.
Proton channel activity of influenza A virus M2 is required to block the fusion of autophagosomes with lysosomes. (A)TREx-293 cells carrying tetracycline-inducible H5N1-M2 (S31) or H5N1-M2 (N31) were treated with different compounds as indicated. Cells were collected at 24 h and subjected to Western blot analysis. (B) TREx-293 cells carrying tetracycline-inducible H5N1-M2 (S31) were transfected with siRNA for Beclin 1 (Atg6), hVPS34, or negative siRNA as a control (purchased from RiboBio). Twenty-four hours later, cells were treated with 1 μg/ml tetracycline for another 24 h. Cell lysates were subjected to Western blot analysis with the antibodies indicated (anti-Beclin 1 [Sigma B6186] and anti-PI3K hVPS34 [Cell Signaling 3358] antibodies). (C) HEK293 cells stably expressing GFP-LC3 were either transfected with a plasmid encoding H5N1-M2 (S31) or treated with rapamycin for 24 h. Amantadine was then added for another 3 h of incubation. Cells were stained with an anti-LAMP2 antibody (Abcam Ab18529) and a Cy3-conjugated goat anti-rabbit secondary antibody and observed by confocal microscopy. Scale bars, 20 μm. (D) GFP-LC3-transfected TREx-293 cells with tetracycline-inducible H5N1-M2 (S31) were treated with tetracycline for 24 h and then treated with LysoTracker Red DND99 (Life Technologies L7528) for 2 h and amantadine for 1 h. The cells were observed by fluorescence microscopy. Scale bars, 20 μm. The colocalization of GFP-LC3 with the lysosome marker LAMP2 or LysoTracker Red DND99 was analyzed with Image Pro (Media Cybernetics). Overlap is indicated by Manders' overlap coefficients ranging from 0 to 1. A value of 1 implies that 100% of both selected channels is colocalized. A total of 10 cells were analyzed. Statistical analyses were performed by two-tailed Mann-Whitney test. **, P < 0.01; ***, P < 0.001. Representative data from one of three separate experiments are shown. Relative LC3-II/LC3-I, LC3-II/β-actin, Beclin 1/β-actin, and hVPS34/β-actin ratios were analyzed by using ImageJ (National Institutes of Health) in the same Western blot assay.
A recent study reported that the cytoplasmic tail of M2 can directly interact with autophagy protein LC3 and promote LC3 relocation to the plasma membrane, which may contribute to virion stability (11). M2 has been implicated to interact with Beclin-1, which may also modulate the autophagy process (4). We also found that wortmannin could inhibit LC3-II accumulation in rapamycin-treated cells but not in M2-expressing cells. The fusion of autophagosomes with lysosomes is driven by multiple proteins, and any impairments may disturb the completion of an autophagy process (7). Acidification of the lumenal space of endosomes, lysosomes, and autophagosomes is an important step. Basic chemical compounds such as chloroquine and ammonium chloride can raise the pH in acidic cytoplasmic compartments and block the degradation of autophagosomes by lysosomes (28). Bafilomycin A1, an inhibitor of V-ATPase, prevents autophagosome acidification and fusion with endosomes and lysosomes (29). It has been proposed that M2 protein can insert itself into the cellular membrane system to act as a proton channel (30–33), resulting in perturbation of the pH of the environment in cellular compartments. It is possible that M2 plays multiple roles in the modulation of the autophagy process. At least part of the mechanism of autophagy arrest by M2 is proton channel activity to prevent the fusion of autophagosomes with lysosomes, thereby leading to autophagosome accumulation. Further studies are needed to elucidate the mechanism in more detail.
ACKNOWLEDGMENTS
We thank Zhang Beiwu, Li Zhixia, and Chen Xiaomei for their technical assistance. We gratefully thank Zhimin Yuan (Harvard School of Public Health) for providing the GFP-LC3 plasmid.
This research was supported by the National Natural Science Foundation of China (81490534), a mega-project grant on infectious disease (2014ZX10004006), an H7N9 grant (KJYJ-2013-01-05) and an SKLRD grant from MOST and NHFPC of China, and grants from the municipal government of Guangzhou (201400000002, 2013X01).
Funding Statement
N/A
REFERENCES
- 1.Li KS, Guan Y, Wang J, Smith GJ, Xu KM, Duan L, Rahardjo AP, Puthavathana P, Buranathai C, Nguyen TD, Estoepangestie AT, Chaisingh A, Auewarakul P, Long HT, Hanh NT, Webby RJ, Poon LL, Chen H, Shortridge KF, Yuen KY, Webster RG, Peiris JS. 2004. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 2.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 3.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gannagé M, Dormann D, Albrecht R, Dengjel J, Torossi T, Ramer PC, Lee M, Strowig T, Arrey F, Conenello G, Pypaert M, Andersen J, Garcia-Sastre A, Munz C. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Z, Jiang X, Liu D, Fan Z, Hu X, Yan J, Wang M, Gao GF. 2009. Autophagy is involved in influenza A virus replication. Autophagy 5:321–328. doi: 10.4161/auto.5.3.7406. [DOI] [PubMed] [Google Scholar]
- 6.Comber JD, Robinson TM, Siciliano NA, Snook AE, Eisenlohr LC. 2011. Functional macroautophagy induction by influenza A virus without a contribution to major histocompatibility complex class II-restricted presentation. J Virol 85:6453–6463. doi: 10.1128/JVI.02122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klionsky DJ, Emr SD. 2000. Autophagy as a regulated pathway of cellular degradation. Science 290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine B, Deretic V. 2007. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol 7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson WT, Giddings TH Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol 3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belov GA, Altan-Bonnet N, Kovtunovych G, Jackson CL, Lippincott-Schwartz J, Ehrenfeld E. 2007. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J Virol 81:558–567. doi: 10.1128/JVI.01820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beale R, Wise H, Stuart A, Ravenhill BJ, Digard P, Randow F. 2014. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe 15:239–247. doi: 10.1016/j.chom.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y, Li C, Shu Y, Ju X, Zou Z, Wang H, Rao S, Guo F, Liu H, Nan W, Zhao Y, Yan Y, Tang J, Zhao C, Yang P, Liu K, Wang S, Lu H, Li X, Tan L, Gao R, Song J, Gao X, Tian X, Qin Y, Xu KF, Li D, Jin N, Jiang C. 2012. Inhibition of autophagy ameliorates acute lung injury caused by avian influenza A H5N1 infection. Sci Signal 5:ra16. doi: 10.1126/scisignal.2001931. [DOI] [PubMed] [Google Scholar]
- 13.Ma J, Sun Q, Mi R, Zhang H. 2011. Avian influenza A virus H5N1 causes autophagy-mediated cell death through suppression of mTOR signaling. J Genet Genomics 38:533–537. doi: 10.1016/j.jgg.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, et al. . 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu W, Zeng S, Li C, Jie Y, Li Z, Chen L. 2010. Identification of hits as matrix-2 protein inhibitors through the focused screening of a small primary amine library. J Med Chem 53:3831–3834. doi: 10.1021/jm901664a. [DOI] [PubMed] [Google Scholar]
- 16.Sun J, Li C, Xu W, Li Z, Liu J, Chen L. 2008. Establish stable cell line to express M2 ion channel of influenza A virus H5N1. Sheng Wu Gong Cheng Xue Bao 24:1902–1906. (In Chinese.) [PubMed] [Google Scholar]
- 17.Liu Q, Lu L, Sun Z, Chen GW, Wen Y, Jiang S. 2013. Genomic signature and protein sequence analysis of a novel influenza A (H7N9) virus that causes an outbreak in humans in China. Microbes Infect 15:432–439. doi: 10.1016/j.micinf.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Yuen KY. 2013. Solving the mystery of H7N9 by crystal balls. Cell Res 23:1335–1336. doi: 10.1038/cr.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding WX, Yin XM. 2008. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy 4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 20.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Lamb RA, Pinto LH. 1995. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys J 69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. 2008. Structural basis for the function and inhibition of an influenza virus proton channel. Nature 451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, Long Y, Lin Z, Jie Y, Xiao Y, Yang L, Sun J, Ren Y, Chen L, Li Z. 2013. New strategy for high throughput screening of anti-influenza virus M2 ion channel inhibitors. Curr Pharm Des 19:5146–5155. doi: 10.2174/13816128113199990001. [DOI] [PubMed] [Google Scholar]
- 24.Jiang H, Cheng D, Liu W, Peng J, Feng J. 2010. Protein kinase C inhibits autophagy and phosphorylates LC3. Biochem Biophys Res Commun 395:471–476. doi: 10.1016/j.bbrc.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullinane M, Gong L, Li X, Lazar-Adler N, Tra T, Wolvetang E, Prescott M, Boyce JD, Devenish RJ, Adler B. 2008. Stimulation of autophagy suppresses the intracellular survival of Burkholderia pseudomallei in mammalian cell lines. Autophagy 4:744–753. doi: 10.4161/auto.6246. [DOI] [PubMed] [Google Scholar]
- 26.Schmelzle T, Hall MN. 2000. TOR, a central controller of cell growth. Cell 103:253–262. doi: 10.1016/S0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 27.Tassa A, Roux MP, Attaix D, Bechet DM. 2003. Class III phosphoinositide 3-kinase–Beclin1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem J 376:577–586. doi: 10.1042/bj20030826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovács AL, Reith A, Seglen PO. 1982. Accumulation of autophagosomes after inhibition of hepatocytic protein degradation by vinblastine, leupeptin or a lysosomotropic amine. Exp Cell Res 137:191–201. doi: 10.1016/0014-4827(82)90020-9. [DOI] [PubMed] [Google Scholar]
- 29.Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. 2008. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy 4:849–850. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 30.Pinto LH, Lamb RA. 2006. The M2 proton channels of influenza A and B viruses. J Biol Chem 281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 31.Pinto LH, Lamb RA. 2007. Controlling influenza virus replication by inhibiting its proton channel. Mol Biosyst 3:18–23. doi: 10.1039/B611613M. [DOI] [PubMed] [Google Scholar]
- 32.Schnell JR, Chou JJ. 2008. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henkel JR, Popovich JL, Gibson GA, Watkins SC, Weisz OA. 1999. Selective perturbation of early endosome and/or trans-Golgi network pH but not lysosome pH by dose-dependent expression of influenza M2 protein. J Biol Chem 274:9854–9860. doi: 10.1074/jbc.274.14.9854. [DOI] [PubMed] [Google Scholar]