ABSTRACT
Antibodies against the neuraminidase (NA) of influenza virus correlate with resistance against disease, but the effectiveness of antibodies against different NA epitopes has not been compared. In the present study, we evaluated the in vitro and in vivo efficacies of four monoclonal antibodies (MAbs): HF5 and CD6, which are specific to two different epitopes in the NA of 2009 pandemic H1N1 (pH1N1) virus, and 4E9 and 1H5, which are specific to a conserved epitope in the NA of both H1N1 and H5N1 viruses. In the in vitro assays, HF5 and CD6 inhibited virus spread and growth more effectively than 4E9 and 1H5, with HF5 being the most effective inhibitor. When administered prophylactically at 5 mg/kg of body weight, HF5 and CD6 protected ∼90 to 100% of DBA/2 mice against lethal wild-type pH1N1 virus challenge; however, at a lower dose (1 mg/kg), HF5 protected ∼90% of mice, whereas CD6 protected only 25% of mice. 4E9 and 1H5 were less effective than HF5 and CD6, as indicated by the partial protection achieved even at doses as high as 15 mg/kg. When administered therapeutically, HF5 protected a greater proportion of mice against lethal pH1N1 challenge than CD6. However, HF5 quickly selected pH1N1 virus escape mutants in both prophylactic and therapeutic treatments, while CD6 did not. Our findings confirm the important role of NA-specific antibodies in immunity to influenza virus and provide insight into the properties of NA antibodies that may serve as good candidates for therapeutics against influenza.
IMPORTANCE Neuraminidase (NA) is one of the major surface proteins of influenza virus, serving as an important target for antivirals and therapeutic antibodies. The impact of NA-specific antibodies on NA activity and virus replication is likely to depend on where the antibody binds. Using in vitro assays and the mouse model, we compared the inhibitory/protective efficacy of four mouse monoclonal antibodies (MAbs) that bind to different sites within the 2009 pandemic H1N1 (pH1N1) virus NA. The ability of each MAb to protect mice against lethal pH1N1 infection corresponded to its ability to inhibit NA activity in vitro; however, the MAb that was the most effective inhibitor of NA activity selected pH1N1 escape variants in vivo. One of the tested MAbs, which binds to a conserved region in the NA of pH1N1 virus, inhibited NA activity but did not result in escape variants, highlighting its suitability for development as a therapeutic agent.
INTRODUCTION
Neuraminidase (NA) is the second most abundant surface glycoprotein of influenza virus (1, 2). It plays an important role in virus replication; i.e., it cleaves sialic acid from cellular receptors and viral glycoproteins to facilitate the spread and disaggregation of newly formed viral particles (3–5). The licensure of NA inhibitors, e.g., oseltamivir and zanamivir, provides evidence that inhibition of NA activity impacts the duration of infection and clinical disease. Resistance to oseltamivir has possibly occurred as a result of selection during treatment with this antiviral, but it has also occurred in the absence of antiviral treatment (6–8). This has limited the options available for the control of influenza, and consequently, researchers are examining the prospect of new prophylactic and therapeutic approaches against influenza, including antibody therapy.
Immunity to NA contributes to resistance against influenza virus (9–11). NA-inhibiting antibodies do not inhibit virus attachment and entry but, rather, reduce virus replication by preventing the release of newly formed viral particles from infected cells, resulting in mild disease symptoms (12–15). Immunity to the NA of H2N2 virus may have mitigated the impact of the H3N2 pandemic in 1968 (16). Similarly, preexisting immunity to conserved regions of the NA of the previous seasonal H1N1 viruses may have provided some protection against the 2009 pandemic H1N1 (pH1N1) virus (17).
Within each NA subtype, there are genetically heterologous lineages that are serologically distinct (18, 19), suggesting the presence of strain-specific epitopes; however, analyses with polyclonal sera and monoclonal antibodies (MAbs) also demonstrate that NA contains conserved epitopes, in addition to immunodominant strain-specific epitopes (20–24). A universal epitope in NA has been reported, and a rabbit MAb against this epitope was shown to inhibit multiple subtypes of NA (25, 26), although it remains to be investigated whether this epitope elicits an immune response in vivo. We recently defined the NA antigenic domains of a seasonal H1N1 virus, A/Brisbane/59/2007 (BR/07), by identifying amino acids in NA that are critical for binding of a panel of MAbs (27). These included a group of MAbs that are highly specific for the NA of BR/07-like viruses (referred to as group A) and a group of MAbs that are broadly reactive with N1 subtypes (referred to as group B), including the NA of a 2009 pH1N1 virus, A/California/7/2009 (CA/09). The amino acids essential for binding group B antibodies are highly conserved in the N1 subtype of viruses, including H5N1. In a separate study, we characterized two MAbs, HF5 and CD6, that are specific to the NA of pH1N1 virus (28). When tested in vivo, CD6 and MAbs from group B protected DBA/2 mice against challenge with a lethal dose of reassortant vaccine candidate CA/09-X179A (27, 28). DBA/2 mice are sensitive to many subtypes of influenza viruses (29), permitting challenge experiments to be conducted with viruses that have a range of virulences. The CA/09-X179A virus was generated as a vaccine candidate by classical reassortment and was demonstrated to be less virulent than wild-type (wt) CA/09 in ferrets (30).
Although NA-specific antibodies have the potential to be used as prophylactic and therapeutic treatments, additional testing is needed to determine the efficacy of these antibodies against highly pathogenic viruses and to define the attributes of the most effective MAbs. In the present study, the in vitro functional properties of NA-specific antibodies HF5, CD6, 4E9, and 1H5 were compared, and their in vivo efficacies against wt CA/09 were tested. Our results demonstrate that the antibody which is most effective at inhibiting enzyme activity and plaque formation in vitro is also the most effective in protecting mice against lethal infection in vivo. However, selection of virus escape variants is a potential problem that would render some antibodies ineffective. Our current data suggest that NA-inhibiting MAbs with specificity for conserved epitopes are good candidates for further therapeutic development.
MATERIALS AND METHODS
Viruses and antibodies.
The wt pH1N1 virus A/California/07/2009 (CA/09) and vaccine candidate virus CA/09-X179A were propagated in 9- to 11-day-old embryonated chicken eggs and titrated in eggs or Madin-Darby canine kidney (MDCK) cells. H6N1BR/07 and H6N1CA/09 reassortant viruses, which contain the hemagglutinin (HA) gene of H6N2 virus A/turkey/Massachusetts/3740/1965 and the NA gene of CA/09 or a seasonal H1N1 virus, A/Brisbane/59/2007 (BR/07), were rescued using reverse genetics (31) and grown in 9- to 11-day-old embryonated chicken eggs for determination of the inhibitory effect of antibodies. The viruses used for enzyme-linked immunosorbent assay (ELISA) were inactivated with β-propiolactone (Sigma-Aldrich, St. Louis, MO) and purified by sucrose gradient centrifugation. Virus counting was performed as reported previously (32) using a Virus Counter 2100 system (ViroCyt, Boulder, CO). Hybridomas that secrete MAbs were generated as previously described (27, 28) and cultured in a CELLine device (BD Biosciences, San Jose, CA). MAbs were purified using protein G columns (GE Healthcare, Uppsala, Sweden) according to the instructions provided by the manufacturer.
Site-directed mutagenesis.
Nucleotide changes corresponding to single mutations (S339A, S364N, N369K, N397K, and N449D) were introduced into the CA/09 NA gene in the pCAGGS-CA/09NA plasmid (23) with a QuikChange multisite-directed mutagenesis kit (Stratagene, La Jolla, CA). The resulting plasmids were sequenced to verify the presence of the introduced mutations and the absence of additional, unwanted mutations.
Cell-based ELISA.
A cell-based ELISA was performed as previously described (27). NA was expressed on 293T cells by transfection with wt or mutant plasmids using the Lipofectamine 2000 reagent (Invitrogen Inc., Grand Island, NY). Briefly, 293T cells at approximately 90% confluence in 96-well plates were incubated with a mixture of plasmid (0.2 μg/well) and Lipofectamine 2000 (0.5 μl/well) in Opti-MEM I medium (50 μl/well) (Invitrogen, Grand Island, NY) for 6 h. The mixture was removed, and fresh medium was added. At 48 h after the addition of the transfection mixture, the cells were fixed with 0.05% glutaraldehyde for 15 min at room temperature. The cells were washed and then blocked with 3% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for 1 h at 37°C. The cells were then incubated with MAbs (1 μg/ml), followed by incubation with peroxidase-conjugated goat anti-mouse IgG (Sigma-Aldrich, St. Louis, MO). Both incubations were for 1 h at 37°C. The signal was developed using o-phenylenediamine dihydrochloride (OPD) as the substrate. The reaction was stopped with 1 N H2SO4, and the values of the optical density (OD) at 490 nm (OD490) were read. Expression of NA was confirmed with hyperimmune mouse serum against CA/09 (hemagglutination inhibition [HI] titer, ≥320). The signals generated by MAb binding to mutant NAs were normalized to the signal generated by hyperimmune mouse serum and are therefore expressed as relative binding. A >20% decrease in relative binding compared to that to wt NA was arbitrarily considered significant for the purpose of epitope identification.
Conventional ELISA.
Native and denatured preparations of purified CA/09-X179A virus were coated onto ELISA plates (10 μg/ml, 100 μl/well). The purified virus was denatured by suspension in 1% sodium dodecyl sulfate (SDS) and 50 mM dithiothreitol (DTT), followed by heating at 95°C for 5 min before dilution in coating buffer. The virus-coated plates were blocked and then incubated with MAb HF5 (1 μg/ml for native virus and 2 μg/ml for denatured virus) and controls. Mouse serum against CA/09 virus (HI titer, ≥320) and rabbit MAb clone 001 (Sino Biological, Beijing, China), which binds to CA/09 NA under both reducing and nonreducing conditions, were used as positive controls, while MAb 3A2, which is specific to the NA of the BR/07-like H1N1 virus, was included as a negative control. After an additional incubation with peroxidase-conjugated goat anti-mouse IgG or peroxidase-conjugated goat anti-rabbit IgG (Sigma-Aldrich, St. Louis, MO), the signal was developed using OPD as the substrate. The reaction was stopped with 1 N H2SO4, and OD490 values were read.
Plaque assay.
MDCK cells growing in 6-well or 12-well plates were infected with 20 to 30 PFU of virus for 1 h at 37°C. After the removal of the viral inoculum, the cells were washed with PBS and overlaid with agar supplemented with MAbs and 1 μg/ml of trypsin. The cells were incubated at 37°C in 5% CO2 for 3 days and then fixed with methanol and stained with crystal violet solution to visualize the plaques. Infected cells that did not contain antibody in the agar overlay were set up as a control. To measure the sizes (diameters) of the plaques, plaque images were magnified in Photoshop software (CS6 extended); 20 plaques were randomly selected for each antibody concentration; and the diameters were determined using the measurement tool within the software, converted to real sizes (in millimeters), and compared to those of the control plaques.
Virus growth kinetics.
MDCK cells in 12-well plates were inoculated with CA/09 virus at a multiplicity of infection (MOI) of 0.001 for 1 h (run in duplicate wells). The inoculum was removed by extensive washing with PBS, and the cells were maintained in Opti-MEM I medium (Invitrogen, Grand Island, NY) supplemented with 1 μg/ml of trypsin and various concentrations of each MAb. Supernatants were sampled at time points of 0, 8, 10, 24, 36, and 48 h postinfection (p.i.). Virus in the supernatant was titrated by plaque assay.
ELLA.
The inhibition of NA activity was measured by an enzyme-linked lectin assay (ELLA) as described previously (33). Briefly, serial dilutions of mouse sera or MAbs were mixed with a predetermined amount of virus diluted in PBS containing 1% BSA and 0.05% Tween 20 (PBST). The mixture was transferred to 96-well plates coated with fetuin (Sigma-Aldrich, St. Louis, MO) and incubated at 37°C overnight. The plates were washed with PBST, followed by the addition of peanut agglutinin conjugated to peroxidase (Sigma-Aldrich, St. Louis, MO). The plates were incubated at room temperature for 2 h in the dark and washed with PBST before the addition of OPD substrate. The reaction was stopped by adding 1 N H2SO4, and OD490 values were read. The NA inhibition (NI) titer was expressed as the reciprocal of the highest dilution that exhibited ≥50% inhibition of NA activity or the 50% inhibition concentration (IC50), which was determined by nonlinear regression analysis (GraphPad Prism software, version 5).
Absorption of passively administered MAbs.
Female DBA/2 mice (8 weeks old; The Jackson Laboratory) were used in animal studies performed under protocols approved by the Center for Biologics Evaluation and Research (CBER) Animal Care and Use Committee. Antibody or PBS was administered intraperitoneally (i.p.) to groups of mice (n = 3) at a dose of 5 mg/kg of body weight. Mouse sera were collected at 1, 3, 6, 9, and 14 days after antibody or PBS injection and stored at −80°C until the NI titers were measured by ELLA.
Prophylactic and therapeutic studies.
Female DBA/2 mice (8 weeks old; The Jackson Laboratory) were used in prophylactic and therapeutic studies. To determine the impact of MAbs administered before infection (prophylactic efficacy), groups of mice (n = 14 or 20) were treated with antibodies HF5 and CD6 at doses of 0.2, 1, and 5 mg/kg, with antibodies 4E9 and 1H5 at doses of 5, 10, and 15 mg/kg, or with the control antibody, 3A2, at 5 or 15 mg/kg. Each dose was administered i.p. in a volume of 200 μl. Twelve hours later, the mice were challenged intranasally (i.n.) with 10 50% mouse lethal doses (MLD50) of CA/09. On days 6 and 8 postchallenge (p.c.), 3 mice from each group were euthanized, and the lungs were collected for viral titration in MDCK cells. In the study to evaluate the therapeutic benefit of MAbs HF5, CD6, and 1H5, groups of mice (n = 14 or 20) were infected i.n. with 10 MLD50 of CA/09 before MAb treatments: each MAb was delivered i.p. once (on day 1 p.c.), twice (on days 1 and 5 p.c.), or three times (on days 1, 2, and 3 p.c.). HF5 and CD6 were each administered at 5 mg/kg, and 1H5 was administered at 10 mg/kg, while the control antibody, 3A2, was administered at either 5 or 10 mg/kg. On days 6 and 8 p.c., 3 mice from each group were euthanized for titration of lung viral titers. For antibodies HF5 and CD6 administered prophylactically (5 mg/kg) or therapeutically (3 doses of 5 mg/kg), an additional 6 mice were included in order to determine the viral titers in lungs collected on days 10 and 12 p.c. In all groups, the remaining mice (8 per group) were weighed on day 0 or day 1 before virus challenge and monitored daily for 14 days for weight loss and survival. Mice that lost ≥25% of their body weight were euthanized.
Identification of CA/09 escape mutants in MAb-treated mice.
Lung tissue samples from mice in the prophylactic and therapeutic studies were examined for the presence of escape variants in plaque assays by supplementing the agar overlay with the selecting MAb (2 μg/ml). Plaques that were larger than the control plaques (those of wt CA/09 in the presence of each antibody) were picked and expanded in 9- to 11-day-old embryonated chicken eggs. Allantoic fluid was collected for the sequencing of the NA gene.
Reverse transcription-PCR and gene sequencing.
Viral RNA was extracted from allantoic fluid with an RNeasy minikit (Qiagen, Valencia, CA). cDNA synthesis and PCR were performed as previously described (34) to amplify the NA gene. PCR products were sequenced at the Core Facility, CBER, FDA.
Enzyme activity.
The NA activities of the MAb escape mutants and wt CA/09 obtained by ELLA and a 4-methylumbelliferyl-N-acetyl-α-d-neuraminic acid (MU-NANA) assay were compared. The latter assay was performed using MU-NANA (Sigma, St. Louis, MO) as the substrate. Serial dilutions of virus were mixed with an equal volume (50 μl) of 100 μM MU-NANA in PBS (pH 7.4), and the mixture was incubated at 37°C for 1 h. The reaction was stopped by the addition of 0.1 M glycine (pH 10.7)–25% ethanol, and the fluorescence was read (excitation, 355 nm; emission, 460 nm).
Statistical analysis.
Data were analyzed by one-way analysis of variance (ANOVA; GraphPad Prism software, version 5) using Dunnett's multiple-comparison test. P values of <0.05 were considered statistically significant.
RESULTS
The MAbs used in the study recognize different epitopes of the pH1N1 virus NA.
This study used four N1-specific MAbs that were previously identified to have potential therapeutic value. Two of the MAbs, HF5 and CD6, bind to the NA of CA/09. NA residues 95, 449, and 451 are key for the binding of CD6. Antibodies 4E9 and 1H5 are broadly reactive with the NA of H1N1 (including seasonal and pandemic H1N1 viruses) and H5N1 viruses, with NA residues 273, 338, and 339 being the critical contacts (27, 28).
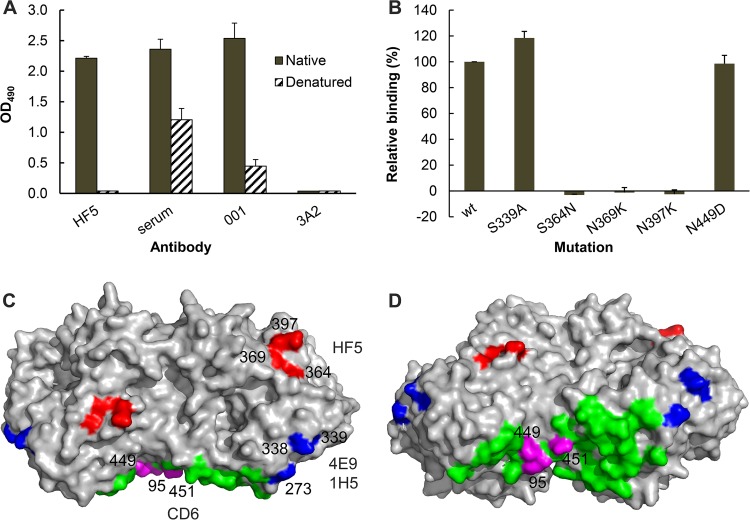
The epitope recognized by HF5 was not fully elucidated. In an ELISA using CA/09-X179A virus-coated plates, HF5 bound to native virus but not denatured virus, indicating that the HF5 epitope is dependent on the native conformation (Fig. 1A). The results of our previous NI assays suggest that HF5 may bind to an epitope involving residue 369 (28). In the present study, we examined the binding of HF5 to various mutant CA/09 NAs transiently expressed on 293T cells. A cell-based ELISA with these mutant NAs clearly demonstrated that an N-to-K mutation at residue 369 abolished the binding of NA by HF5 (Fig. 1B). This finding confirms that residue 369 is a key contact in the HF5 epitope. In addition, the S364N and N397K mutations in NA resulted in the complete loss of HF5 binding in the cell-based ELISA. Residues 364, 369, and 397 are in close proximity to one another and are clearly important contacts for HF5. The HF5 epitope is therefore distinct from the CD6 epitope as well as the epitope recognized by antibodies 4E9 and 1H5. Thus, the antigenic domains recognized by these four MAbs represent three different epitopes in N1 of the pH1N1 virus (Fig. 1C and D).
FIG 1.
MAbs HF5, CD6, 4E9, and 1H5 bind different epitopes in the NA of pH1N1 virus. (A) MAb HF5 binds to native but not denatured CA/09-X179A. Data from ELISA show the binding of HF5 to purified whole virus (solid bars) but not CA/09-X179A that has been dissociated and heat denatured (hatched bars). Assay controls are labeled serum (mouse serum against CA/09 virus), 001 (rabbit MAb against CA/09 NA), and 3A2 (MAb specific to the NA of BR/07-like H1N1 virus). Shown are mean OD490 values from two independent assays run in duplicate wells; standard deviations (SDs) are shown with error bars. (B) Binding of MAb HF5 to a panel of mutant CA/09 NAs expressed on 293T cells. The binding was measured by a cell-based ELISA, with the signals being normalized to those obtained with mouse serum against CA/09, which are therefore expressed as relative binding. Shown are the data from two independent assays run in duplicate wells. (C) A model that depicts the residues in an NA dimer (Protein Data Bank accession number 3NSS) that are critical for the binding of MAbs HF5 (red), CD6 (magenta), as well as 4E9 and 1H5 (blue). The remaining 27 residues within the CD6 epitope (Protein Data Bank accession number 4QNP) are highlighted in green. The image was generated with PyMOL software (Delano Scientific). (D) The same model in panel C shown from a different view.
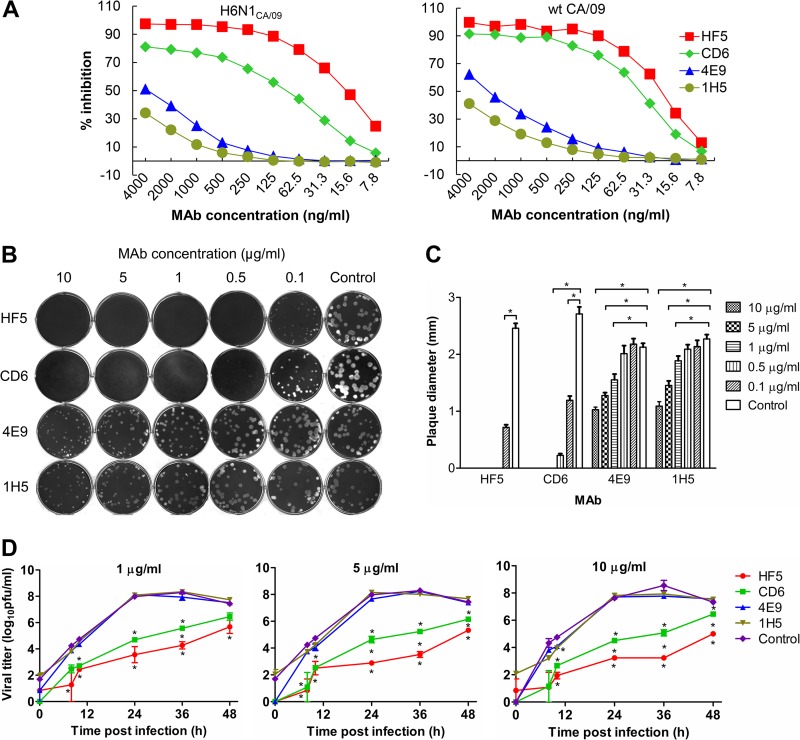
MAbs HF5 and CD6 inhibit NA activity and virus growth more efficiently than MAbs 4E9 and 1H5.
Our previous studies using H6N1CA/09 virus in an ELLA demonstrated that MAbs HF5 and CD6 are more effective in inhibiting the NA activity of pH1N1 virus than MAbs 4E9 and 1H5 (27, 28). This difference in inhibitory efficacy was also observed with wt CA/09 in ELLA: the median (50%) inhibition concentrations (IC50s) of CD6 and HF5 were <100 ng/ml, while those of 4E9 and 1H5 were >2 μg/ml (Fig. 2A). In this study, we compared the effects of the four MAbs on the growth of wt CA/09 in MDCK cells. Antibody 3A2, which is reactive only to BR/07-like H1N1 viruses, was included as a negative control. In the plaque assay, both HF5 and CD6 inhibited CA/09 plaque formation or resulted in smaller plaques when the agar overlay was supplemented with either MAb. Only pinpoint plaques were formed at MAb concentrations of ≥1 μg/ml. When applied at 0.5 μg/ml, HF5 still efficiently inhibited CA/09 plaque formation, while clear, small plaques were observed in the presence of CD6. The superior inhibitory effect of HF5 was also observed at the lowest concentration of the MAbs tested: CA/09 plaques were observed at 0.1 μg/ml of either antibody, but the plaques that formed in the presence of HF5 were slightly smaller than those that formed in the presence of CD6 (Fig. 2B and C). In comparison to the inhibition of plaque formation by MAbs HF5 and CD6, the inhibition of plaque formation by MAbs 4E9 and 1H5 was poor. There was no reduction in plaque size at 0.1 or 0.5 μg/ml MAb, but when the agar overlay was supplemented with ≥1 μg/ml MAb, both 4E9 and 1H5 reduced the size of the CA/09 plaques. Compared to the average size of the CA/09 plaques formed in the presence of 10 μg/ml of the control antibody, 3A2 (2.1 mm), plaques formed in the presence of 4E9 were significantly smaller (P < 0.001), with average diameters of 1.6, 1.3, and 1.0 mm at antibody concentrations of 1, 5, and 10 μg/ml, respectively. Similar results were observed with MAb 1H5 (Fig. 2B and C). Thus, the plaque assays revealed that HF5 and CD6 are more effective than 4E9 and 1H5 in inhibiting CA/09 spread, and this result is consistent with the ability of each MAb to inhibit NA activity.
FIG 2.
Different functional properties of MAbs HF5, CD6, 4E9, and 1H5 in vitro. (A) Inhibition of CA/09 NA activity by each MAb measured by ELLA using H6N1CA/09 and wt CA/09 viruses. (B) CA/09 virus plaques formed in the presence of various concentrations of MAbs HF5, CD6, 4E9, and 1H5 (0.1 to 10 μg/ml) or the control MAb, 3A2 (10 μg/ml), in the overlay agar. (C) Diameters of the CA/09 virus plaques shown in panel B. Plaques from each treatment were randomly chosen, and the diameters were measured and compared to the diameter of the control. Shown are mean diameters (n = 20); SDs are shown with error bars. Diameters that were significantly different from those of the control (P < 0.05) are indicated by lines and asterisks. (D) Growth kinetics of CA/09 in MDCK cells in the presence of MAb HF5, CD6, 4E9, 1H5, or 3A2 (1, 5, or 10 μg/ml). Cells growing in 12-well plates were infected with CA/09 at an MOI of 0.001, and the viral titers in the supernatant at the indicated time points were measured by plaque assay. Shown are the average titers of duplicate wells; SDs are shown with error bars. *, P < 0.05. The asterisks above the green line or below the red line indicate a significant difference between the viral titers generated in the presence of the tested MAbs (CD6 or HF5) and the control MAb 3A2, asterisks between the green and red lines indicate significant differences between the viral titers generated in the presence of MAbs CD6 and HF5, and asterisks below the blue line in the right panel indicate significant differences between groups receiving 4E9 and 1H5 and the control group (which received 3A2).
Since the four MAbs inhibited NA activity and virus spread with different efficiencies, we next tested whether the inhibitory potential could be extended to multiple cycles of virus replication. MDCK cells were infected with CA/09 at an MOI of 0.001 and maintained in medium supplemented with each MAb. HF5 and CD6 inhibited the growth of CA/09 at all of the concentrations tested (1, 5, or 10 μg/ml), with the viral titers being lower than those observed with the control antibody, 3A2, at each of the time points at which the titers were examined (Fig. 2D). The presence of HF5 and CD6 retarded virus growth: peak viral titers were not attained in the presence of these MAbs until 48 h, when most of the cells had died, whereas peak titers were measured at 36 h in the presence of control MAb 3A2. The peak titers in control culture supernatants were significantly (∼100-fold, P < 0.01 or 0.001) higher than those in supernatants containing HF5 and CD6. The absence of a dose-dependent effect with HF5 and CD6 suggests that maximal inhibition was obtained at the lowest concentration examined, 1 μg/ml. This is consistent with the fact that this concentration is ∼20 to 50 times greater than the IC50s of the two antibodies measured by ELLA. MAbs 4E9 and 1H5 did not inhibit virus growth at any of the concentrations tested (except for 10 μg/ml at 10 h p.i.), as reflected by viral titers similar to those detected in the presence of control antibody 3A2. This is also consistent with the fact that these two antibodies failed to effectively inhibit the NA activity of CA/09. HF5 and CD6 inhibited virus replication to a similar extent at early time points (8 and 10 h); however, all viral titers measured in culture supernatants containing HF5 were lower than those in samples containing CD6 at 24, 36, and 48 h (Fig. 2D), suggesting that HF5 is more effective than CD6 in inhibiting multiple cycles of CA/09 replication.
HF5 and CD6 are superior to 4E9 and 1H5 in protecting mice against wt CA/09 when administered before challenge.
Since the in vitro assays demonstrated that MAbs HF5 and CD6 inhibited CA/09 more efficiently than 4E9 and 1H5, we next investigated whether this corresponds to differences in protection against virus challenge in vivo. We previously demonstrated that CD6 and two N1 broadly reactive antibodies, 1H5 and 3H10, all protected mice against lethal challenge with a vaccine candidate virus, CA/09-X179A (27, 28), although doses of the last two MAbs higher than the dose of CD6 were required to achieve full protection. It is not known whether similar amounts of antibody would protect mice against a more virulent pH1N1 virus challenge.
CA/09-X179A is a reassortant virus that contains the HA, NA, and PB1 genes of CA/09 and 5 additional gene segments from A/Puerto Rico/8/1934 (PR8) (30). The CA/09-X179A stock had titers of 3.2 × 108 50% egg infective doses (EID50)/ml and 1.85 × 105 MLD50/ml (measured with DBA/2 mice) (Table 1); thus ∼1,730 EID50 of this virus equal 1 MLD50. In comparison, the wt CA/09 stock had titers of 5.9 × 108 EID50/ml and 3.3 × 107 MLD50/ml; therefore, ∼18 EID50 correspond to 1 MLD50. The virus count confirmed that the concentration of viral particles in each stock was similar: (7.1 ± 1.8) × 108 and (9.8 ± 2.9) × 108 viral particles/ml, respectively, indicating that ∼3,900 CA/09-X179A or ∼30 CA/09 viral particles equal 1 MLD50. These data show that wt CA/09 is more virulent than CA/09-X179A, with ∼100-fold less wt CA/09 constituting 1 MLD50 in DBA/2 mice. This study tested the efficacies of MAbs HF5, CD6, 4E9, and 1H5 against the more virulent wt CA/09 virus in the DBA/2 mouse model.
TABLE 1.
Infectious titers and viral particle count of CA/09-X179A and wt CA/09 stocks
| Virus | Titera |
Virus countb (no. of viral particles/ml) | |
|---|---|---|---|
| MLD50/ml | EID50/ml | ||
| CA/09-X179A | 1.85 × 105 | 3.2 × 108 | (7.1 ± 1.8) × 108 |
| wt CA/09 | 3.3 × 107 | 5.9 × 108 | (9.8 ± 2.9) × 108 |
MLD50 were measured in DBA/2 mice. Viral titers were determined in mice and eggs rather than MDCK cells because the CA/09-X179A virus induced opaque, small plaques in MDCK cells that were difficult to count accurately.
The number of viral particles was counted with a Virus Counter 2100 system (ViroCyt, Boulder, CO). Shown are the averages of three counts that used 1:5, 1:10, and 1:20 dilutions of each viral stock.
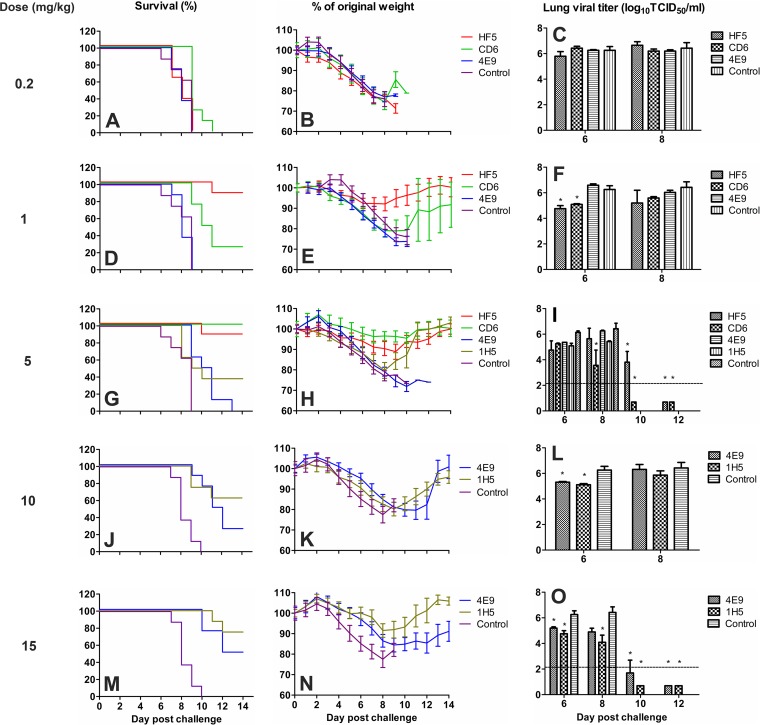
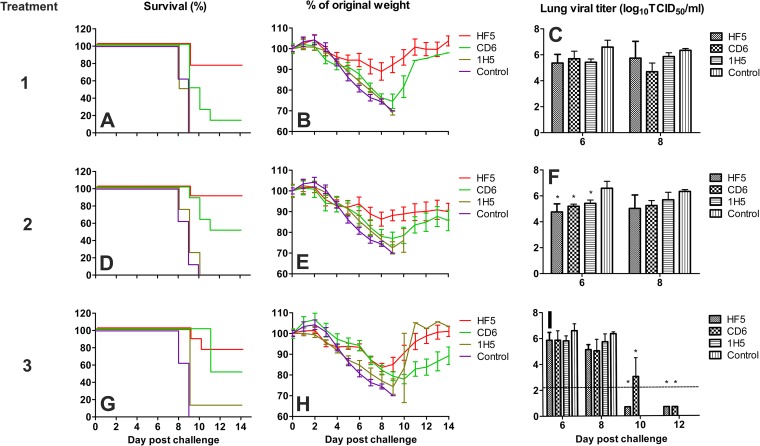
To examine the prophylactic efficacy, various doses of each MAb were administered i.p. to DBA/2 mice 12 h before challenge with 10 MLD50 of wt CA/09. The doses of MAbs used in this study were guided by the results of previous studies with the less virulent virus. HF5 and CD6 were used at doses of 0.2, 1, and 5 mg/kg, while 4E9 and 1H5 were used at doses of 5, 10, and 15 mg/kg. Survival (Fig. 3A, D, G, J, and M) and weight loss (Fig. 3B, E, H, K, and N) were monitored daily. As shown in Fig. 3, the prophylactic effect of MAbs HF5 and CD6 was dose dependent, with the greatest survival occurring at the highest dose. HF5 was more effective than CD6, as either 1 or 5 mg/kg of HF5 resulted in maximum protection (∼90% survival), whereas a 5-mg/kg dose of CD6 protected all mice from death, and 1 mg/kg of CD6 resulted in 25% survival. No mice that received 0.2 mg/kg of antibody survived. Consistent with the in vitro observations, higher doses of antibodies 4E9 and 1H5 were needed to protect against lethal wt CA/09 infection. 4E9 at 5 mg/kg did not protect any mice from death, while 1H5 at 5 mg/kg protected ∼40% of mice. At 10 and 15 mg/kg, 4E9 resulted in partial protection (∼20% and 50%, respectively) but was not as effective as 1H5, which protected ∼60% and ∼80% of mice at those doses, respectively (Fig. 3J and M). All mice in the control groups (injected with antibody 3A2 at 5 or 15 mg/kg) died or had to be euthanized within 9 to 10 days p.c. In all groups, mice began to lose weight on day 3 p.c.; however, mice that received 1 or 5 mg/kg of HF5 or 5 mg/kg of CD6 lost less weight than the mice in the other groups (Fig. 3B, E, H, K, and N).
FIG 3.
Prophylactic efficacy of MAbs HF5, CD6, 4E9, and 1H5 against lethal pH1N1 virus challenge in mice. DBA/2 mice (n = 14 or 20 per group) were treated i.p. with HF5, CD6, 4E9, or 1H5 at the indicated doses, followed by challenge i.n. with 10 MLD50 CA/09 12 h later. MAb 3A2, which is specific to the NA of seasonal H1N1 virus BR/07, was used as a negative control. Survival (A, D, G, J, and M) and weight loss (B, E, H, K, and N) (n = 8 per group) were monitored for up to 14 days. Lungs were collected on different days, and viral titers (C, F, I, L, and O) were determined by titration in MDCK cells. Titers are expressed as the log10 50% tissue culture infective dose (TCID50) per milliliter (n = 3); SDs are shown with error bars. The dotted lines denote the detection limit of 2.2 log10 TCID50/ml. A titer of 0.7 log10 TCID50/ml was arbitrarily assigned to samples with titers below the detection limit. The viral titers of the control groups at day 8 p.c. were used for analysis of the titers measured on days 10 and 12 p.c. because none of the mice in the control groups survived at these time points. Significant differences between the titers measured in each group and the MAb 3A2-treated control groups are shown (*, P < 0.05).
To determine whether MAb treatment reduced the viral load in mice, 3 animals from each group were sacrificed on days 6 and 8 p.c. (and also on days 10 and 12 for mice treated with HF5 and CD6 at 5 mg/kg or with 4E9 and 1H5 at 15 mg/kg), and the viral load in the lungs was measured (Fig. 3C, F, I, L, and O). High viral titers were detected in all groups of mice on days 6 and 8 p.c. For mice that received the lowest dose of either HF5 or CD6 (0.2 mg/kg), the lung viral titers were similar to those detected in the control group (in which mice received 5 mg/kg of MAb 3A2). Increasing the HF5 and CD6 doses to 1 or 5 mg/kg resulted in an ∼10-fold reduction in lung viral titers, except that a more dramatic reduction (∼300-fold) was observed on day 8 p.c. in mice that received 5 mg/kg of CD6. As the virus was not cleared by day 8 p.c., the viral titers in the lungs of additional mice in the groups receiving 5 mg/kg were examined on days 10 and 12 p.c. Viral titers were below the detection limit on both days in mice treated with CD6. Virus took longer to clear from the lungs of mice treated with HF5; there was an ∼100-fold reduction in viral titers on day 10 p.c. compared to that on day 8 p.c., but it was not until day 12 p.c. that the viral load was reduced to below the detection limit (Fig. 3I). For mice that received MAb 4E9 or 1H5, a reduction of the viral load was evident in the group given 15 mg/kg antibody; at this dose, virus was cleared by day 12 in mice treated with 4E9 and by day 10 in those treated with 1H5 (Fig. 3O). Taken together, these results indicate that NA antibodies are able to protect mice from challenge with a lethal dose of a more virulent virus and that the effective dose reflects the in vitro functional properties of the MAbs.
To confirm that the differences in protection were not due to differences in the absorption of the tested MAbs by the circulatory system, we measured the NI titers in mouse sera after i.p. injection of 5 mg/kg of each MAb. Table 2 demonstrates that the MAbs were effectively absorbed into the circulatory system. High NI titers against the homologous NA were measured on day 1 after administration of the MAbs. These levels were maintained throughout the testing period for CD6 and HF5, but serum NI titers declined at later time points for mice treated with MAbs 4E9 and 1H5 (8- to 16-fold reductions in titers compared to the titers on day 1). These data confirm that the differences in MAb protective efficacy were likely due to differences in the functional attributes of the MAbs and not an effect of MAb absorption.
TABLE 2.
Serum NI titers after passive transfer of N1-specific MAbs
| Day of serum collectiona | Serum NI titer against homologous viral NAb |
|||||
|---|---|---|---|---|---|---|
| HF5 | CD6 | 4E9 | 1H5 | 3A2 | PBS | |
| 1 | 5,120 | 2,560 | 5,120 | 640 | >5120 | <10 |
| 3 | 2,560 | 5,120 | 2,560 | 640 | >5120 | <10 |
| 6 | 2,560 | 1,280 | 1,280 | 320 | >5120 | <10 |
| 9 | 2,560 | 1,280 | 640 | 80 | >5120 | <10 |
| 14 | 1,280 | 640 | 80 | 40 | >5120 | <10 |
Mouse sera were collected on days 1, 3, 6, 9, and 14 after i.p. injection of each MAb (5 mg/kg) or PBS.
NI titers were measured by ELLA with H6N1 reassortant viruses that contained the HA gene of H6N2 virus A/turkey/Massachusetts/3740/1965 and the NA gene of CA/09 virus (H6N1CA/09) or the NA gene of BR/07 virus (H6N1BR/07). Shown are the titers from sera pooled from 3 mice at each time point. Sera from the groups receiving HF5 and CD6 were titrated against H6N1CA/09; sera from the groups receiving 4E9, 1H5, and 3A2 were titrated against H6N1BR/07; sera from the group receiving PBS were titrated against both viruses.
HF5 is superior to CD6 in protecting mice against lethal CA/09 challenge when administered therapeutically.
To examine the therapeutic efficacy of these MAbs, DBA/2 mice were infected with 10 MLD50 of wt CA/09 before each antibody was administered. 4E9 was not included in this study due to its poorer effectiveness than the other three MAbs in the prophylactic study. After challenge with wt CA/09, each antibody was administered i.p. at a dose of 5 mg/kg (HF5 and CD6) or 10 mg/kg (1H5) once (on day 1 p.c.), twice (on days 1 and 5 p.c.), or three times (on days 1, 2, and 3 p.c.). As shown in Fig. 4A, D, and G, all treatments with HF5 protected ≥75% of the challenged mice. For antibody CD6, 50% of mice that received two or three doses survived the challenge, while a single 5-mg/kg dose protected only 25% of the mice. Administration of 1H5 did not result in significant protection; only 1 out of the 8 mice that received three 10-mg/kg doses of this antibody survived.
FIG 4.
Therapeutic efficacy of MAbs HF5, CD6, and 1H5 against lethal pH1N1 virus challenge in mice. DBA/2 mice (n = 14 or 20 per group) were infected i.n. with 10 MLD50 of CA/09, followed by injection i.p. with a single dose of each MAb on day 1 p.c., two doses on days 1 and 5 p.c., or three doses (5 mg/kg per dose) on days 1, 3, and 5 p.c (shown as treatments 1, 2, and 3, respectively). MAb 3A2, which is specific to the seasonal H1N1 BR/07 virus, was used as a negative control. Survival (A, D, and G) and weight loss (B, E, and H) (n = 8 per group) were monitored for up to 14 days. Lungs were collected on different days, and viral titers (C, F, and I) were determined by titration in MDCK cells. Titers are expressed as the log10 50% tissue culture infective dose (TCID50) per milliliter; SDs are shown with error bars. The dotted line denotes the detection limit of 2.2 log10 TCID50/ml. A titer of 0.7 log10 TCID50/ml was assigned to samples with titers below the detection limit. The viral titers of the control groups at day 8 p.c. were used for analysis of the titers measured on days 10 and 12 p.c. because none of the mice in the control groups survived at these time points. Significant differences between titers measured in each group and the MAb 3A2-treated control groups are shown (*, P < 0.05).
Mice in all groups began to lose weight on day 3 p.c. (Fig. 4B, E, and H), and the surviving mice began gaining weight by days 9 to 11 p.c. The HF5-treated mice generally lost less weight than the CD6-treated ones: the maximum weight loss observed in HF5-treated mice was ∼17%, while the maximum weight loss observed in CD6-treated mice was 24% (Fig. 4B).
Similar to the observation in the prophylactic study, high viral titers were measured in the lungs of mice on days 6 and 8 p.c. in all groups, although the titers in the lungs of mice that received HF5 or CD6 were slightly lower than those in the lungs of mice in the control group (mice that received antibody 3A2) (Fig. 4C, F, and I). To further investigate virus clearance in mice treated with each MAb, we measured the viral load at later time points in the lungs of mice that received three doses (on days 1, 3, and 5 p.c.) of HF5 or CD6. By day 10, virus was cleared from mice that received HF5, while virus was not cleared in CD6-treated mice until day 12 p.c.
HF5 selects escape mutants in vivo.
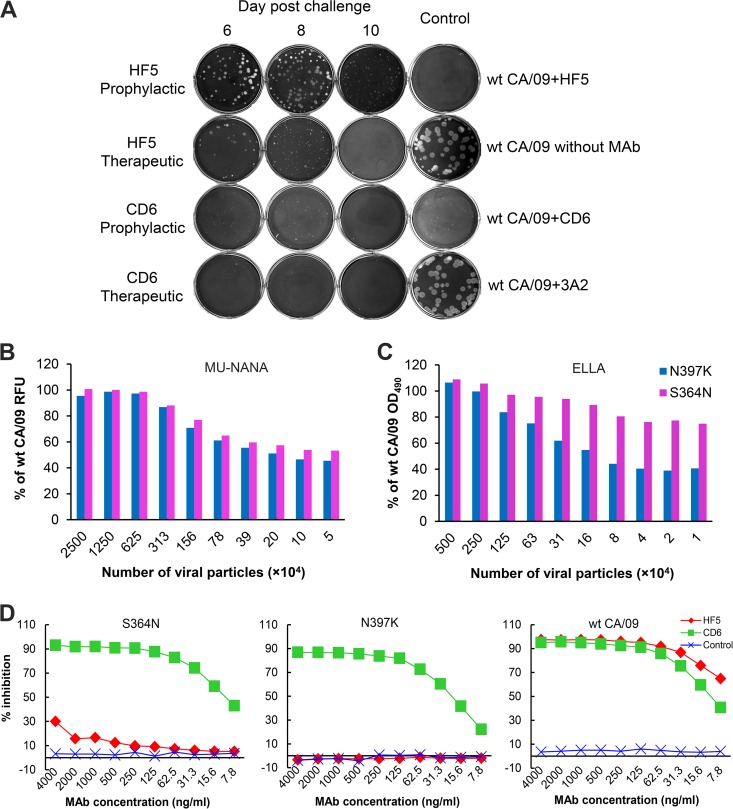
To further evaluate the potential of MAbs HF5 and CD6 as prophylactic and therapeutic agents, we examined whether these two MAbs selected escape mutants of CA/09 in vivo. Lungs collected on days 6, 8, and 10 p.c. from mice in the prophylactic study (in which 5 mg/kg of antibody was administered 12 h before challenge with wt CA/09) and the therapeutic study (in which 3 doses were administered at 5 mg/kg on days 1, 2, and 3 p.c. following challenge with wt CA/09) were tested for the presence of escape mutants. This was examined by isolating viral plaques from the supernatants of lung tissue homogenates with MAb (2 μg/ml) in the agar overlay. Despite the presence of antibody HF5 in the agar overlay, clear, big plaques were observed with lung tissue samples from mice treated with HF5 either prophylactically or therapeutically (Fig. 5A), indicating that escape mutants of CA/09 had emerged in both treatments. However, in the presence of CD6, no plaques that were bigger than those of wt CA/09 were observed with supernatants of homogenates of lung tissue from CD6-treated mice, demonstrating the absence of escape mutants that are resistant to inhibition by CD6. HF5-selected mutant viruses were expanded by picking plaques and culturing them in eggs. Sequencing of the mutant viruses revealed the S364N or N397K mutation in the NA of viruses selected in the prophylactic treatment and the S364N mutation in the NA of mutant viruses from the therapeutic treatment. wt CA/09 virus, which was used for challenge (egg passage 6, MDCK passage 1), was grown in eggs for the same amount of time (48 h) as virus plaques isolated from the lungs of MAb-treated mice; no NA sequence change was detected in the resultant virus (egg passage 7, MDCK passage 1) (data not shown), confirming that the mutations observed in the NA of HF5 escape mutants were not the result of passaging of the virus in eggs.
FIG 5.
MAb HF5 selected escape mutants of pH1N1 virus in mice. (A) Lung samples collected on days 6, 8, and 10 p.c. from mice in the prophylactic study (5 mg/kg of MAb before CA/09 challenge) and the therapeutic study (three doses of 5 mg/kg MAb after CA/09 challenge) were homogenized, and the supernatant was tested in a plaque assay, in which MAb HF5 or CD6 was supplemented in the agar overlay. (B and C) Enzyme activity of CA/09 escape variants selected with HF5. The NA activities of CA/09 variants with the S364N or N397K mutations in the NA were compared with the NA activity of the wt parent virus in an MU-NANA assay, which uses a small substrate, MU-NANA (B), and ELLA, which uses a large substrate, fetuin (C). The assay mixtures were adjusted so that they contained the same number of viral particles per milliliter, and the three viruses were tested in duplicate. The relative fluorescence units (RFUs) and OD values generated with the mutants are expressed as a percentage of the signals obtained with wt CA/09. (D) CA/09 mutants with the S364N mutation (left) or the N397K mutation (middle) in the NA were examined for the sensitivities of their NAs to MAbs HF5, CD6, and 3A2 in an ELLA. (Right) Results obtained with wt CA/09 NA were included for comparison.
The NA activity of the mutant viruses, especially the virus with the N397K mutation in the NA, was less than that of wt CA/09 when it was measured using either a small substrate (MU-NANA; Fig. 5B) or a large substrate (fetuin; Fig. 5C). NI assays were performed to confirm the resistance of mutant viruses (with the S364N or N397K mutation in the NA) to antibody HF5; while HF5 effectively inhibited wt CA/09 NA, it failed to inhibit either mutant virus (Fig. 5D). This is consistent with the observation in the cell-based ELISA, which indicates that the S364N and N397K mutations result in a dramatic reduction in the binding of HF5 to NA (Fig. 1A). As shown in Fig. 5D, both of the mutant viruses retained sensitivity to inhibition by CD6.
DISCUSSION
There is abundant evidence that NA-inhibiting antibodies contribute to resistance against influenza virus and that NA-specific MAbs have the potential to be used as prophylactic and therapeutic agents. Previous studies have also indicated the presence of multiple epitopes in NA (21, 27, 35, 36). However, the relative effectiveness of antibodies that recognize different NA epitopes is not known, and protection against wt virus infections has not been established. Since MAbs have varied specificities and functional attributes, in vitro and in vivo data are needed to guide the choice of MAbs for use for prophylactic or therapeutic treatments. In this study we compared the in vitro properties of four NA-specific MAbs, HF5, CD6, 4E9, and 1H5, and tested the efficacies of these MAbs in vivo against wt pH1N1 virus CA/09.
It is clear that the HF5, CD6, and group B MAbs (e.g., 4E9 and 1H5) recognize different epitopes, although we cannot exclude the possibility that these epitopes may overlap to some extent. Our previous investigations suggested that the CD6 epitope and the epitope recognized by group B MAbs are located relatively laterally in the NA head in relation to the enzyme active site (27, 28). Residues 364, 369, and 397, identified in the present study to be critical for the binding by MAb HF5, are located in a region of a polypeptide chain which encircles the enzyme active-site pocket. Therefore, the HF5 epitope is likely in closer proximity to the NA active center than epitopes recognized by CD6 as well as those recognized by 4E9 and 1H5.
The in vitro properties of these NA MAbs are consistent with the relative distance of the epitopes to the NA active center. For instance, in the lectin-based NI assay, HF5 inhibited CA/09 NA more efficiently than the other three MAbs (27, 28). HF5 also inhibited plaque formation and the growth kinetics of CA/09 virus to a greater extent than CD6 and the group B antibodies. It was therefore not surprising to find that HF5 exhibited higher efficacy than 4E9, 1H5, and even CD6 in protecting mice from lethal challenge with CA/09. These findings indicate that for NA MAbs, the in vitro properties (reduction in plaque size, inhibition of virus growth in cell culture, and inhibition of NA activity, as measured by ELLA) are generally consistent with each other and correlate with the in vivo effectiveness in protecting against influenza virus.
When administered at effective doses (e.g., ≥1 mg/kg prophylactically or 5 mg/kg therapeutically), HF5 appeared to be a suitable treatment. However, it rapidly selected escape mutants of CA/09 virus in vivo, which diminishes its potential to serve as a therapeutic agent against pH1N1 virus infection. In fact, the N397K mutation, detected in the NA of escape mutants from the HF5-treated mice, has occurred in the current circulating pH1N1 viruses (37, 38). In addition, the N369K mutation, which abolishes the binding of HF5 to CA/09 NA, was selected soon after the introduction of pH1N1 virus into the human population and by 2011 was present in ≥99% of the circulating strains (8). This suggests that the HF5 epitope is also probably targeted by the human NA-specific antibodies, which resulted in the antigenic drift of this epitope. The prevalence of the N369K and, to a lesser extent, N397K mutations in circulating pH1N1 viruses (7, 8) indicates that antibodies elicited against the HF5 epitope in humans no longer bind to the majority of circulating pH1N1 viruses. Thus, although MAb HF5 is more effective than the other MAbs in inhibiting the NA of the prototype pH1N1 virus CA/09, it is not a suitable candidate for further development as a therapeutic. It was interesting to find that mutant viruses with the S364N and N397K mutations but not viruses with the N369K mutation in the NA were selected by MAb HF5 in mice. This phenomenon was also noted for N9-specific MAb NC41: when MAbs that bind N9 were used to select escape variants, mutations at residue 369 were selected by some MAbs but not by NC41, even though X-ray crystallography shows that both heavy and light chains of the antibody have contact with amino acid 369 (39). It is likely that the amino acid changes selected are unique to each antibody. In addition, the selective pressure exerted by the human immune response (elicited by either natural infection or vaccination) might be different from that exerted by mouse MAb HF5, which may result in different amino acid mutations in the NA. However, the reasons for selecting specific mutations are likely to be complex and, as these examples show, are sometimes not easily predicted. In contrast to the various mutations within the HF5 epitope that are observed in the NA of circulating pH1N1 viruses, phylogenetic analysis shows that the CD6 epitope has remained largely conserved. The N449D/D451G double mutation in CA/09 NA, which was selected in vitro (28), has not been identified in natural pH1N1 viruses so far, highlighting the potential of antibody CD6 to serve as an ideal target for antiviral development. It is noteworthy that the D-to-G mutation at residue 451, one of the critical contacts in the CD6 epitope, has been detected in some circulating pH1N1 strains. However, it does not appear to affect the inhibition by CD6, as additional amino acid mutations in the NA may help these strains to retain the sensitivity to CD6 (28). When these findings are taken into consideration, antibody CD6 is more ideal than HF5 as an antiviral, even though it is slightly less effective than HF5 in inhibiting CA/09 NA activity.
In our previous mouse challenge studies with the vaccine candidate virus CA/09--X179A, MAb CD6 protected all mice after a single dose of 1 mg/kg was administered prophylactically or a single dose of 5 mg/kg was administered therapeutically. High doses (15 mg/kg) of the broadly reactive group B MAbs 1H5 and 3H10 also provided full protection against lethal challenge with this virus. In the present study, however, administration of group B MAb 4E9 failed to result in similar protective efficacy against the wt CA/09, and a higher dose of CD6 was needed to obtain optimal protection. These data reflect a difference in the virulence of wt CA/09 and CA/09-X179A in the DBA/2 mouse model. The molecular basis for the greater virulence of wt CA/09 than the vaccine reassortant virus remains to be investigated. The more stringent conditions imposed by using the more virulent virus in the mouse model are an important step toward the development of antivirals that are likely to be effective in the clinic.
Antibodies vary in their properties, including specificity, affinity, and the ability to neutralize/inhibit virus. The present study highlights the importance of carefully selecting appropriate antibodies for development as antiviral therapeutics. The efficacy of NA-specific MAbs is largely due to the release of reduced amounts of newly formed virus particles from infected cells, although the clearance of virus-infected cells through complement- or antibody-dependent cytotoxicity mechanisms may also contribute to their effectiveness. Our results suggest that the rate at which virus replication is reduced by NA-specific MAbs likely depends on the virus strain. At high doses (≥2.5 mg/kg), prophylactic treatment with MAb CD6 cleared the CA/09-X179A vaccine virus from the lungs of mice by day 3 p.c. (28); however, in the current study, the same MAb applied at a higher dose (5 mg/kg) did not result in clearance of the more virulent wt CA/09 until after day 10 p.c. Regardless of their initial effectiveness, antibodies that select escape variants may quickly become obsolete as antigenic drift variants become predominant in the circulating virus population. For NA antibodies, those that bind to conserved antigenic domains and also have inhibitory properties are better candidates for antiviral development, as they are more likely to be effective against a broad range of circulating viruses and are less likely to select escape variants. Considering the various advantages of antibodies with different specificities, it may be beneficial to use a mixture of MAbs with specificity for different NA epitopes or a mixture of NA- and HA-specific MAbs to minimize the generation of escape mutants and improve the effectiveness of treatment.
ACKNOWLEDGMENTS
We thank J. Taubenberger (National Institutes of Health, Bethesda, MD) for providing the pCAGGS-CA/09NA plasmid and R. Webster (St. Jude Children's Research Hospital, Memphis, TN) for providing the plasmids used to generate influenza viruses by reverse genetics. We appreciate the technical support provided by Xiaoquan Wang. We are also indebted to staff of the Division of Veterinary Services, CBER, FDA, for excellent animal care and the CBER core facility for sequence analyses.
Funding Statement
This work was supported by intramural FDA funds. L.J. and G.F. were supported by training funds administered by the Oak Ridge Institute for Science and Education.
REFERENCES
- 1.Kilbourne ED, Johansson BE, Grajower B. 1990. Independent and disparate evolution in nature of influenza A virus hemagglutinin and neuraminidase glycoproteins. Proc Natl Acad Sci U S A 87:786–790. doi: 10.1073/pnas.87.2.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Air GM. 2012. Influenza neuraminidase. Influenza Other Respir Viruses 6:245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. 2004. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol 78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner R, Wolff T, Herwig A, Pleschka S, Klenk HD. 2000. Interdependence of hemagglutinin glycosylation and neuraminidase as regulators of influenza virus growth: a study by reverse genetics. J Virol 74:6316–6323. doi: 10.1128/JVI.74.14.6316-6323.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palese P, Compans RW. 1976. Inhibition of influenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoroacetylneuraminic acid (FANA): mechanism of action. J Gen Virol 33:159–163. doi: 10.1099/0022-1317-33-1-159. [DOI] [PubMed] [Google Scholar]
- 6.Sheu TG, Deyde VM, Okomo-Adhiambo M, Garten RJ, Xu X, Bright RA, Butler EN, Wallis TR, Klimov AI, Gubareva LV. 2008. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob Agents Chemother 52:3284–3292. doi: 10.1128/AAC.00555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okomo-Adhiambo M, Fry AM, Su S, Nguyen HT, Elal AA, Negron E, Hand J, Garten RJ, Barnes J, Xiyan X, Villanueva JM, Gubareva LV. 2015. Oseltamivir-resistant influenza A(H1N1)pdm09 viruses, United States, 2013-14. Emerg Infect Dis 21:136–141. doi: 10.3201/eid2101.141006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurt AC, Hardie K, Wilson NJ, Deng YM, Osbourn M, Leang SK, Lee RT, Iannello P, Gehrig N, Shaw R, Wark P, Caldwell N, Givney RC, Xue L, Maurer-Stroh S, Dwyer DE, Wang B, Smith DW, Levy A, Booy R, Dixit R, Merritt T, Kelso A, Dalton C, Durrheim D, Barr IG. 2012. Characteristics of a widespread community cluster of H275Y oseltamivir-resistant A(H1N1)pdm09 influenza in Australia. J Infect Dis 206:148–157. doi: 10.1093/infdis/jis337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eichelberger MC, Wan H. 2015. Influenza neuraminidase as a vaccine antigen. Curr Top Microbiol Immunol 386:275–299. doi: 10.1007/82_2014_398. [DOI] [PubMed] [Google Scholar]
- 10.Murphy BR, Kasel JA, Chanock RM. 1972. Association of serum anti-neuraminidase antibody with resistance to influenza in man. N Engl J Med 286:1329–1332. doi: 10.1056/NEJM197206222862502. [DOI] [PubMed] [Google Scholar]
- 11.Ogra PL, Chow T, Beutner KR, Rubi E, Strussenberg J, DeMello S, Rizzone C. 1977. Clinical and immunologic evaluation of neuraminidase-specific influenza A virus vaccine in humans. J Infect Dis 135:499–506. doi: 10.1093/infdis/135.4.499. [DOI] [PubMed] [Google Scholar]
- 12.Couch RB, Kasel JA, Gerin JL, Schulman JL, Kilbourne ED. 1974. Induction of partial immunity to influenza by a neuraminidase-specific vaccine. J Infect Dis 129:411–420. doi: 10.1093/infdis/129.4.411. [DOI] [PubMed] [Google Scholar]
- 13.Kilbourne ED, Laver WG, Schulman JL, Webster RG. 1968. Antiviral activity of antiserum specific for an influenza virus neuraminidase. J Virol 2:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson BE, Grajower B, Kilbourne ED. 1993. Infection-permissive immunization with influenza virus neuraminidase prevents weight loss in infected mice. Vaccine 11:1037–1039. doi: 10.1016/0264-410X(93)90130-P. [DOI] [PubMed] [Google Scholar]
- 15.Brett IC, Johansson BE. 2005. Immunization against influenza A virus: comparison of conventional inactivated, live-attenuated and recombinant baculovirus produced purified hemagglutinin and neuraminidase vaccines in a murine model system. Virology 339:273–280. doi: 10.1016/j.virol.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Monto AS, Kendal AP. 1973. Effect of neuraminidase antibody on Hong Kong influenza. Lancet i:623–625. [DOI] [PubMed] [Google Scholar]
- 17.Marcelin G, DuBois R, Rubrum A, Russell CJ, McElhaney JE, Webby RJ. 2011. A contributing role for anti-neuraminidase antibodies on immunity to pandemic H1N1 2009 influenza A virus. PLoS One 6:e26335. doi: 10.1371/journal.pone.0026335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, Cox NJ, Bender CA, Regnery HL, Shaw MW. 1996. Genetic variation in neuraminidase genes of influenza A (H3N2) viruses. Virology 224:175–183. doi: 10.1006/viro.1996.0519. [DOI] [PubMed] [Google Scholar]
- 19.Sandbulte MR, Jimenez GS, Boon AC, Smith LR, Treanor JJ, Webby RJ. 2007. Cross-reactive neuraminidase antibodies afford partial protection against H5N1 in mice and are present in unexposed humans. PLoS Med 4:e59. doi: 10.1371/journal.pmed.0040059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wohlbold TJ, Nachbagauer R, Xu H, Tan GS, Hirsh A, Brokstad KA, Cox RJ, Palese P, Krammer F. 2015. Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. mBio 6:e02556. doi: 10.1128/mBio.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Webster RG, Brown LE, Laver WG. 1984. Antigenic and biological characterization of influenza virus neuraminidase (N2) with monoclonal antibodies. Virology 135:30–42. doi: 10.1016/0042-6822(84)90114-4. [DOI] [PubMed] [Google Scholar]
- 22.Chen Z, Kim L, Subbarao K, Jin H. 2012. The 2009 pandemic H1N1 virus induces anti-neuraminidase (NA) antibodies that cross-react with the NA of H5N1 viruses in ferrets. Vaccine 30:2516–2522. doi: 10.1016/j.vaccine.2012.01.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Easterbrook JD, Schwartzman LM, Gao J, Kash JC, Morens DM, Couzens L, Wan H, Eichelberger MC, Taubenberger JK. 2012. Protection against a lethal H5N1 influenza challenge by intranasal immunization with virus-like particles containing 2009 pandemic H1N1 neuraminidase in mice. Virology 432:39–44. doi: 10.1016/j.virol.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcelin G, Sandbulte MR, Webby RJ. 2012. Contribution of antibody production against neuraminidase to the protection afforded by influenza vaccines. Rev Med Virol 22:267–279. doi: 10.1002/rmv.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doyle TM, Jaentschke B, Van Domselaar G, Hashem AM, Farnsworth A, Forbes NE, Li CG, Wang JZ, He RT, Brown EG, Li XG. 2013. The universal epitope of influenza A viral neuraminidase fundamentally contributes to enzyme activity and viral replication. J Biol Chem 288:18283–18289. doi: 10.1074/jbc.M113.468884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyle TM, Hashem AM, Li C, Van Domselaar G, Larocque L, Wang J, Smith D, Cyr T, Farnsworth A, He R, Hurt AC, Brown EG, Li X. 2013. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antiviral Res 100:567–574. doi: 10.1016/j.antiviral.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Wan H, Gao J, Xu K, Chen H, Couzens LK, Rivers KH, Easterbrook JD, Yang K, Zhong L, Rajabi M, Ye J, Sultana I, Wan XF, Liu X, Perez DR, Taubenberger JK, Eichelberger MC. 2013. Molecular basis for broad neuraminidase immunity: conserved epitopes in seasonal and pandemic H1N1 as well as H5N1 influenza viruses. J Virol 87:9290–9300. doi: 10.1128/JVI.01203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan H, Yang H, Shore DA, Garten RJ, Couzens L, Gao J, Jiang L, Carney PJ, Villanueva J, Stevens J, Eichelberger MC. 2015. Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers. Nat Commun 6:6114. doi: 10.1038/ncomms7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boon AC, deBeauchamp J, Krauss S, Rubrum A, Webb AD, Webster RG, McElhaney J, Webby RJ. 2010. Cross-reactive neutralizing antibodies directed against pandemic H1N1 2009 virus are protective in a highly sensitive DBA/2 mouse influenza model. J Virol 84:7662–7667. doi: 10.1128/JVI.02444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson JS, Nicolson C, Harvey R, Johnson R, Major D, Guilfoyle K, Roseby S, Newman R, Collin R, Wallis C, Engelhardt OG, Wood JM, Le J, Manojkumar R, Pokorny BA, Silverman J, Devis R, Bucher D, Verity E, Agius C, Camuglia S, Ong C, Rockman S, Curtis A, Schoofs P, Zoueva O, Xie H, Li X, Lin Z, Ye Z, Chen LM, O'Neill E, Balish A, Lipatov AS, Guo Z, Isakova I, Davis CT, Rivailler P, Gustin KM, Belser JA, Maines TR, Tumpey TM, Xu X, Katz JM, Klimov A, Cox NJ, Donis RO. 2011. The development of vaccine viruses against pandemic A(H1N1) influenza. Vaccine 29:1836–1843. doi: 10.1016/j.vaccine.2010.12.044. [DOI] [PubMed] [Google Scholar]
- 31.Sandbulte MR, Westgeest KB, Gao J, Xu X, Klimov AI, Russell CA, Burke DF, Smith DJ, Fouchier RA, Eichelberger MC. 2011. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc Natl Acad Sci U S A 108:20748–20753. doi: 10.1073/pnas.1113801108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plant EP, Ye Z. 2015. Chimeric neuraminidase and mutant PB1 gene constellation improves growth and yield of H5N1 vaccine candidate virus. J Gen Virol 96:752–755. doi: 10.1099/jgv.0.000025. [DOI] [PubMed] [Google Scholar]
- 33.Couzens L, Gao J, Westgeest K, Sandbulte M, Lugovtsev V, Fouchier R, Eichelberger M. 2014. An optimized enzyme-linked lectin assay to measure influenza A virus neuraminidase inhibition antibody titers in human sera. J Virol Methods 210C:7–14. doi: 10.1016/j.jviromet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol 146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 35.Air GM, Els MC, Brown LE, Laver WG, Webster RG. 1985. Location of antigenic sites on the three-dimensional structure of the influenza N2 virus neuraminidase. Virology 145:237–248. doi: 10.1016/0042-6822(85)90157-6. [DOI] [PubMed] [Google Scholar]
- 36.Varghese JN, Laver WG, Colman PM. 1983. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature 303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- 37.Takashita E, Ejima M, Itoh R, Miura M, Ohnishi A, Nishimura H, Odagiri T, Tashiro M. 2014. A community cluster of influenza A(H1N1)pdm09 virus exhibiting cross-resistance to oseltamivir and peramivir in Japan, November to December 2013. Euro Surveill 19(1): pii=20666 10.2807/1560-7917.ES2014.19.1.20666. [DOI] [PubMed] [Google Scholar]
- 38.Takashita E, Kiso M, Fujisaki S, Yokoyama M, Nakamura K, Shirakura M, Sato H, Odagiri T, Kawaoka Y, Tashiro M. 2015. Characterization of a large cluster of influenza A(H1N1)pdm09 viruses cross-resistant to oseltamivir and peramivir during the 2013-2014 influenza season in Japan. Antimicrob Agents Chemother 59:2607–2617. doi: 10.1128/AAC.04836-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Air GM, Laver WG, Webster RG. 1990. Mechanism of antigenic variation in an individual epitope on influenza virus N9 neuraminidase. J Virol 64:5797–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]