Abstract
Transcription of herpesviral late genes is stimulated after the onset of viral DNA replication but otherwise restricted. Late gene expression in gammaherpesviruses requires the coordination of six early viral proteins, termed viral transactivation factors (vTFs). Here, we mapped the organization of this protein complex for Kaposi's sarcoma-associated herpesvirus. Disruption of this complex via point mutation of the interaction interface between the open reading frame 24 (ORF24) and ORF34 vTFs ablated both late gene expression and viral replication.
TEXT
In betaherpesviruses and gammaherpesviruses, mapped promoters driving late gene transcription are strikingly minimal and their activation is dependent on a set of conserved viral transcription regulatory proteins (1–8). Studies of the gammaherpesviruses murine gammaherpesvirus 68 (MHV68), Epstein-Barr virus (EBV), and Kaposi's sarcoma-associated herpesvirus (KSHV), as well as the betaherpesvirus human cytomegalovirus (HCMV), identified six viral transactivation factors (vTFs), open reading frame 18 (ORF18), ORF24, ORF30, ORF31, ORF34, and ORF66 (gammaherpesvirus nomenclature) necessary for late gene expression (1–4, 6, 7, 9, 10). ORF24 is a TATA box-binding protein (TBP) homolog that recruits RNA polymerase II (RNAP II) (2, 11), and HCMV UL79 (ORF18 ortholog) contributes to RNAP II elongation (10). However, little is known about the roles of the other individual vTFs or how they are organized to form the late gene transcription preinitiation complex (vPIC).
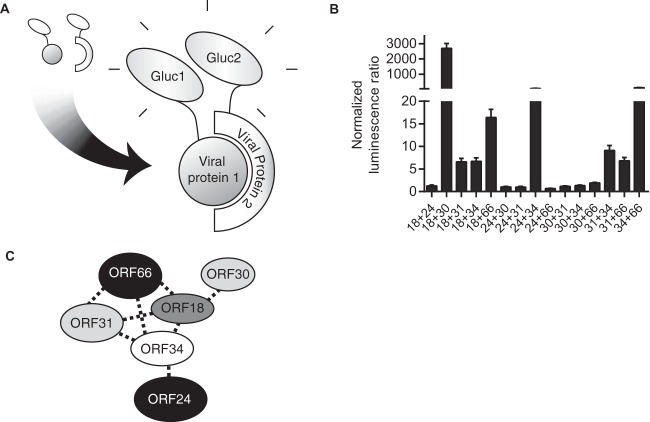
In EBV, the six vTFs have been shown to form a complex (1). To dissect the organization of the vTF complex in KSHV, we applied a high-throughput Gaussia princeps complementation assay (HT-GPCA) (Fig. 1A) (12). This HT-GPCA is based on the reconstitution of a split luciferase, and its power lies in the ability to confirm that a set of interactions occurs within an intact cell, as opposed to postlysis. Each of the pairs of vTFs being examined for interaction was fused to Gaussia luciferase fragment 2 (Gluc2) and Gluc1 by using the pSPICA-N1 and pSPICA-N2 Gateway destination vectors (13). All pairwise combinations of vTFs were transfected at a 1:1 ratio into HEK293T cells in a 24-well plate for a total of 800 ng of DNA. Individual protein-protein interactions were assessed by measuring luciferase activity 24 h later. The pairwise interaction profiles that emerged are consistent with a complex comprising all six KSHV vTFs (Fig. 1B), the connectivity of which is illustrated in Fig. 1C. They are also in agreement with a recent report demonstrating an interaction between KSHV ORF31 and ORF34 (14).
FIG 1.
vTFs interact to form the vPIC. (A) The Gaussia split luciferase assay provides a quantitative measurement of protein interactions. The two halves of the Gaussia luciferase protein (Gluc1 and Gluc2) are fused to the viral proteins to be assessed for interaction. If the two viral proteins interact, they bring the two inactive Gaussia fragments into physical proximity, allowing the formation of a functional luciferase enzyme that facilitates luminescence on the addition of luciferin substrate. (B) Results of all pairwise vTF interactions obtained by the split luciferase assay. Luminescence levels were measured and normalized to the signal of the Gluc1- and Gluc2-tagged proteins transfected on their own. (C) Summary of interaction data from panel B. Note that only ORF34 interacts with ORF24.
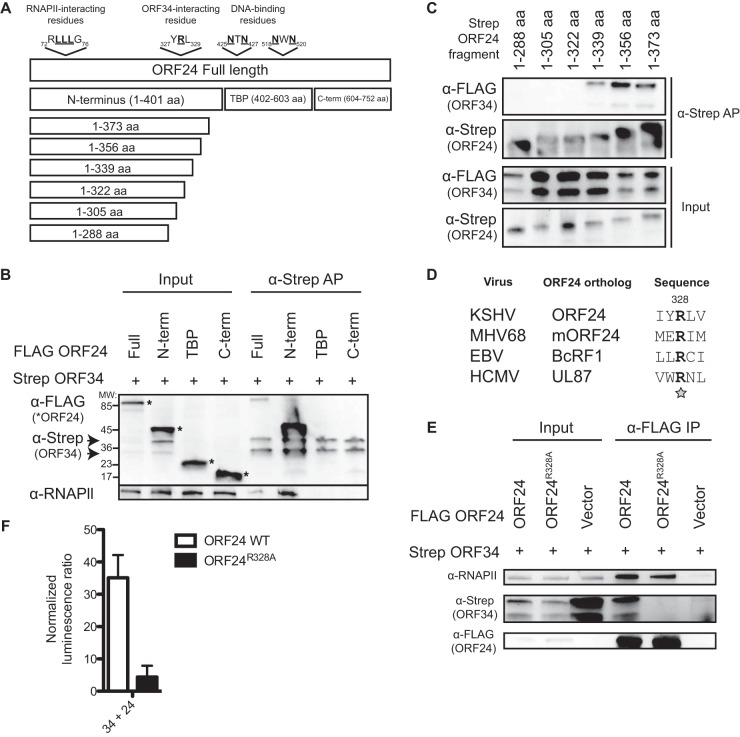
To begin to explore the importance of this vTF interaction network, we characterized the interaction between ORF34, which interacts with multiple vTFs and thus may serve as a scaffold, and ORF24, which bridges the late gene promoter DNA to RNAP II (2, 11). Affinity purification (AP) of streptavidin-tagged ORF34 from cells expressing either full-length FLAG-tagged ORF24 or portions thereof (2) revealed that ORF34 bound to the N terminus of ORF24, which is the same domain of ORF24 that binds RNAP II (Fig. 2A and B). Endogenous RNAP II copurified with ORF34 only in the presence of the ORF24 N terminus, confirming that this RNAP II interaction is indirect and bridged by ORF24 (Fig. 2B). We next constructed a panel of streptavidin-tagged successive 17-amino-acid (aa) truncation mutant forms of the ORF24 N terminus in pCDNA4/TO (Invitrogen; the cloning primers used are listed in Table 1) to better resolve the site of interaction with ORF34. Using streptavidin-AP assays, we mapped the smallest ORF34-interacting fragment of ORF24 to aa 1 to 339, with aa 1 to 322 being insufficient for ORF34 binding (Fig. 2C). We therefore hypothesized that the 17 residues between aa 323 and 339 contained residues essential for ORF34 binding. To identify such residues, we searched for those conserved across ORF24 orthologs. Sequence alignments of the N terminus of KSHV ORF24 with its orthologs in HCMV, EBV, and MHV68 revealed a single conserved residue (R328) within these 17 aa (Fig. 2D). Mutation of this residue to an alanine (ORF24R328A) abrogated the interaction with ORF34 (Fig. 2E and F) but did not impair the ability of ORF24 to bind RNAP II (Fig. 2E). Thus, ORF24 uses distinct regions within its N terminus to coordinate interactions with ORF34 and RNAP II.
FIG 2.
ORF24R328 mediates the interaction with ORF34. (A) Map showing the defined functional regions of ORF24, as well as the truncation mutant forms tested. (B) FLAG-tagged full-length ORF24 and fragments thereof (N terminus [N-term], TBP-like domain, or C terminus [C-term]) were cotransfected with streptavidin-tagged ORF34 into 293T cells. Lysates were affinity purified over Strep-Tactin beads and assayed by Western blotting for FLAG (ORF24), the streptavidin (Strep) tag (ORF34), and RNAP II. The input is 5% of the lysate used for purification. (C) The N-terminal domain of ORF24 (aa 1 to 402) was further truncated to identify the site of interaction with ORF34. Streptavidin-tagged ORF24 fragments (indicated by asterisks in panel B) were cotransfected with FLAG ORF34 into 293T cells. Lysates were affinity purified as described for panel B. (D) Alignment showing a conserved amino acid (R328 in KSHV ORF24) between ORF24 orthologs from betaherpesviruses and gammaherpesviruses. This is the only conserved amino acid found in the 323-to-339-aa region. (E) FLAG-tagged WT ORF24 or ORF24R328A was cotransfected with streptavidin-tagged ORF34 into 293T cells. Lysate was immunoprecipitated with FLAG beads and assayed by Western blotting for streptavidin (ORF34) and RNAP II. (F) Gaussia split luciferase assay assessing interaction between ORF34 and WT ORF24 or ORF24R328A.
TABLE 1.
Primer sequences used in this study
| Primer | Sequence (5′-3′) | Orientationa |
|---|---|---|
| ORF18 Gateway | GGGGACAACTTTGTACAAAAAAGTTGGCATGCTCGGAAAATACGTGTGTGAGA | F |
| ORF18 Gateway | GGGGACAACTTTGTACAAGAAAGTTGGGTATTAAACCGCGTTGTTGTTAAACGCA | R |
| ORF24 Gateway | GGGGACAACTTTGTACAAAAAAGTTGGCATGGCAGCGCTCGAGGG | F |
| ORF24 Gateway | GGGGACAACTTTGTACAAGAAAGTTGGGTATTAGACCAGCGGACGGACGC | R |
| ORF30 Gateway | GGGGACAACTTTGTACAAAAAAGTTGGCATGGGTGAGCCAGTGGATCC | F |
| ORF30 Gateway | GGGGACAACTTTGTACAAGAAAGTTGGGTATCATTTCGCACCGGTGTCTAGG | R |
| ORF31 Gateway | GGGGACAACTTTGTACAAAAAAGTTGGCATGTCACAAAACAGAAAGACTCTGCC | F |
| ORF31 Gateway | GGGGACAACTTTGTACAAGAAAGTTGGGTACTACGTATCTTTCGTTGATAGCATGCG | R |
| ORF34 Gateway | GGGGACAACTTTGTACAAAAAAGTTGGCATGTTTGCTTTGAGCTCGCTCG | F |
| ORF34 Gateway | GGGGACAACTTTGTACAAGAAAGTTGGGTATTAGAGTTGGTTGAGTCCATTCTCCTT | R |
| ORF66 Gateway | GGGGACAACTTTGTACAAAAAAGTTGGCATGGCCCTGGATCAGCGC | F |
| ORF66 Gateway | GGGGACAACTTTGTACAAGAAAGTTGGGTATCAGGAGGAACACTTCCCGC | R |
| ORF24R328A SDM | CCATACACATGACGAGTGCGTAGATGGCCGGTGTGC | F |
| ORF24R328A SDM | GCACACCGGCCATCTACGCACTCGTCATGTGTATGG | R |
| ORF24R328A BAC mutant | TACCTACTATCTAGTGTATCCGGGCACACCGGCCATCTACGCACTCGTCATGTGTATGGCAGAGGATGACGACGATAAGTAGGG | F |
| ORF24R328A BAC mutant | GTGGCCGATGCAGTCTGCCACTGCCATACACATGACGAGTGCGTAGATGGCCGGTGTGCCCGAACCAATTAACCAATTCTGATTAG | R |
| ORF24_R328A_MR_BAC | TACCTACTATCTAGTGTATCCGGGCACACCGGCCATCTACAGACTCGTCATGTGTATGGCAGAGGATGACGACGATAAGTAGGG | F |
| ORF24_R328A_MR_BAC | GTGGCCGATGCAGTCTGCCACTGCCATACACATGACGAGTCTGTAGATGGCCGGTGTGCCCGAACCAATTAACCAATTCTGATTAG | R |
| K8.1 pr pGL4.16 (KpnI-HindIII) | CCCGGCAGCAATATTAAAGGGACCA | F |
| K8.1 pr pGL4.16 (KpnI-HindIII) | AGCTTGGTCCCTTTAATATTGCTGCCGGGGTAC | R |
| ORF57_pr_KpnI_HindIII_pGL4.16 | CGGCCAGTTAATCCCACTATATAACCTGGCTGCCAGGA | F |
| ORF57_pr_KpnI_HindIII_pGL4.16 | AGCTTCCTGGCAGCCAGGTTATATAGTGGGATTAACTGGCCGGTAC | R |
| ORF24 1-288aa (NotI) | TCGAGCGGCCGCTTCACGAGCTGGTTTAGAGCCAG | R |
| ORF24 1-305aa (NotI) | TCGAGCGGCCGCTTGTATGGCTTTGGTCCGGGG | R |
| ORF24 1-322aa (NotI) | TCGAGCGGCCGCTTGCCCGGATACACTAGATAGTAGGTACAA | R |
| ORF24 1-339aa (NotI) | TCGAGCGGCCGCTTGATGCAGTCTGCCACTGCC | R |
| ORF24 1-356aa (NotI) | TCGAGCGGCCGCTTGTGGGTGCCTAAAAAGTTTGCG | R |
| ORF24 1-373aa (NotI) | TCGAGCGGCCGCTTCGGCGCGTACCGGATTC | R |
| ORF25 qPCRb | CCACCCTCGAATGCACAAC | F |
| ORF25 qPCR | GTCGGGATCGGGAAAAGCT | R |
| ORF50 qPCR | CGCAATGCGTTACGTTGTTG | F |
| ORF50 qPCR | GCCCGGACTGTTGAATCG | R |
F, forward; R, reverse.
qPCR, quantitative PCR.
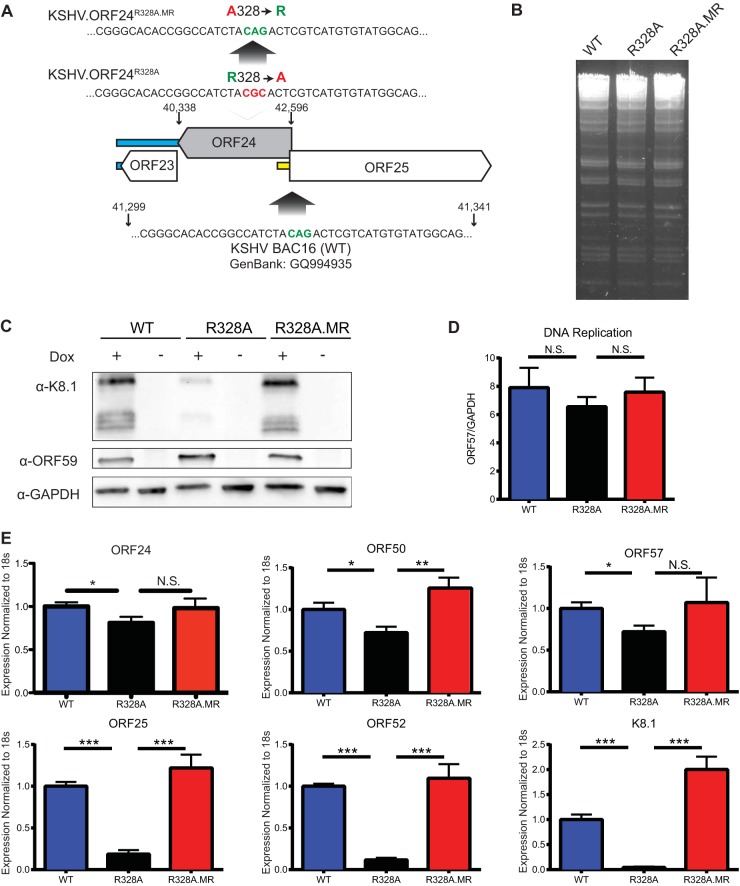
To determine the importance of the ORF24-ORF34 interaction in viral late gene expression, we engineered the ORF24R328A mutation into the KSHV bacterial artificial chromosome (BAC), as well as a repaired mutant rescue (MR) BAC (Fig. 3A). Gel electrophoresis of the NheI-digested wild-type (WT), ORF24R328A, and ORF24R328A.MR BAC16 DNAs showed identical banding patterns, indicating that no gross rearrangements or deletions occurred during BAC16 mutagenesis (Fig. 3B). We then generated KSHV.ORF24R328A and KSHV.ORF24R328A.MR stable cell lines in the iSLK background, which contains a doxycycline-inducible version of the viral lytic transactivator protein, ORF50 (RTA transcription factor) (15). iSLK cells containing WT KSHV, KSHV.ORF24R328A, or KSHV.ORF24R328A.MR were lytically reactivated with doxycycline (1 μg/ml) and sodium butyrate (1 mM) for 72 h, whereupon we measured the expression of the K8.1 (late) and ORF59 (delayed early) proteins (Fig. 3C), viral genome amplification (Fig. 3D), and the expression of early and late viral mRNAs (Fig. 3E). KSHV.ORF24R328A displayed a slight reduction in early gene expression, had no defect in viral genome amplification, but had marked defects in late gene expression relative to the WT and MR viruses (Fig. 3C to E). Collectively, these data indicate that the ability of ORF24 to bind ORF34 is critical for viral late gene expression.
FIG 3.
The interaction between ORF24 and ORF34 is required for late gene expression. (A) Schematic of the BAC mutagenesis strategy used to create the KSHV.ORF24R328A and KSHV.ORF24R328A.MR mutant forms (B) NheI-digested KSHV.WT, KSHV.ORF24R328A, and KSHV.ORF24R328A.MR BAC DNAs were electrophoresed on a 0.7% agarose gel for 24 h. (C) KSHV.WT-, KSHV.ORF24R328A-, and KSHV.ORF24R328A.MR-infected iSLK cells were lytically reactivated for 72 h. Protein lysates were collected and analyzed by Western blotting for the K8.1 (late) and ORF59 (delayed early) proteins with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a loading control. Dox, doxycycline. (D) iSLK cells infected with the viruses indicated were lytically reactivated, and viral genome levels were assessed at 72 h postreactivation by comparing the ratios of ORF57 to GAPDH DNA in induced and uninduced samples. N.S., not statistically significantly different. (E) RNA was collected, and levels were determined for early genes (ORF50, ORF57) at 24 h postreactivation and late genes (K8.1, ORF25, ORF52) at 72 h postreactivation by reverse transcription-quantitative PCR with the primers in Table 1 and previously described primers (2). ORF24 transcript levels were also measured at 72 h postreactivation. All values were normalized to the 18S rRNA level, with the viral mRNA levels present during WT infection set to 1. P values determined by Student's t test: *, <0.05; **, <0.01; ***, <0.0001.
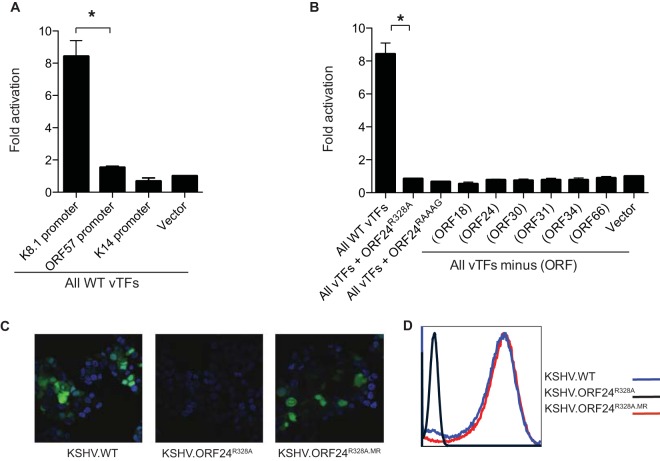
Recently, a plasmid-based late gene promoter activation assay was established for EBV, enabling the study of vTF contributions to late transcription in a manner separable from viral DNA replication (1). To establish an analogous system for KSHV, we cloned the KSHV K8.1 late gene promoter or, as controls, the K14 and ORF57 early gene promoters upstream of a firefly luciferase reporter gene (5). Cotransfection of the six KSHV vTFs with the luciferase reporter plasmid led to specific activation of the late K8.1 promoter, whereas the six vTFs had no effect on the KSHV K14 or ORF57 early promoter (Fig. 4A). Furthermore, removal of any one of the vTFs from the transfection prevented activation of the K8.1 promoter (Fig. 4B). We then used this assay to evaluate the role of the ORF24-ORF34 interaction in late gene promoter activation and observed that substitution of ORF24R328A for ORF24WT in the vPIC resulted in a complete loss of luciferase activity (Fig. 4B). Similarly, the RNAP II interaction mutant form ORF24RAAAG (2) was unable to activate the K8.1 promoter.
FIG 4.
The ORF24-ORF34 interaction is required for activation of a late gene promoter and progeny virion production. (A) Cells were transfected with all six WT vTFs or the empty vector and assessed for the ability to activate a luciferase reporter driven by the K8.1 (late), ORF57 (early), or K14 (early) promoter. An asterisk indicates a P value of <0.05, as determined by Student's t test. (B) 293T cells were transfected with all six vTFs or lacking the vTF in parentheses and assessed for the ability to activate a cotransfected late gene promoter (K8.1)-driven firefly luciferase reporter. All values were normalized to a cotransfected simian virus 40 promoter-driven Renilla luciferase expression plasmid. (C) iSLK cells infected with the viruses indicated were lytically reactivated for 72 h, whereupon supernatants were collected, mixed with Polybrene, and added to 293T cells. Twenty-four hours later, the 293T cells were monitored for GFP expression by confocal microscopy, indicating the transfer of infectious virus. DNA is stained (blue) with 4′,6-diamidino-2-phenylindole (DAPI). A minimum of 30 fields were monitored for each experiment, and a single representative field is shown. No GFP-positive cells were detected in any field of recipient cells containing supernatant from ORF24R328A-infected cells. (D) Supernatant from the reactivated iSLK cells indicated (described in panel C) was added to 293T cells and assessed for GFP expression by flow cytometry after 24 h.
Late gene products generally comprise structural components of the virion, so we assessed whether the late gene expression defect of the ORF24R328A mutant virus impacted the production of infectious virions. iSLK.BAC16 cells contain a green fluorescent protein (GFP) cassette under the control of a constitutive EF-1α promoter, allowing infectious virion production to be assessed by the presence of GFP-positive cells following a supernatant infection assay (15). Consistent with its defect in late gene expression, KSHV.ORF24R328A failed to produce infectious progeny virions, as measured by both confocal microscopy and flow cytometry (Fig. 4C and D). In contrast, transfer of the WT or MR virus resulted in comparable levels of virion production. Thus, the ORF24-ORF34 interaction is required specifically for the activation of late genes, and a point mutation that selectively disrupts the interaction prevents KSHV virion production. Collectively, our findings provide a model of the general organization of the KSHV vTF complex and highlight how the N terminus of ORF24 coordinates both intraviral and virus-host protein-protein interactions essential for late gene transcription.
ACKNOWLEDGMENTS
We thank Caroline Demeret and Yves Jacob (Institut Pasteur) for their generous sharing of the split luciferase plasmids, Mandy Muller for technical advice on HT-GPCA, and other members of the Glaunsinger lab for helpful discussions.
Funding Statement
This research was funded by a W.M. Keck Foundation Distinguished Young Scholars Award and a Burroughs Wellcome Foundation Investigators in the Pathogenesis of Infectious Diseases Award to Britt A. Glaunsinger, who is an investigator of the Howard Hughes Medical Institute. Cell imaging was performed at the CRL Molecular Imaging Center, which is supported by the Gordon and Betty Moore Foundation.
REFERENCES
- 1.Aubry V, Mure F, Mariame B, Deschamps T, Wyrwicz LS, Manet E, Gruffat H. 2014. Epstein-Barr virus late gene transcription depends on the assembly of a virus-specific preinitiation complex. J Virol 88:12825–12838. doi: 10.1128/JVI.02139-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis ZH, Verschueren E, Jang GM, Kleffman K, Johnson JR, Park J, Von Dollen J, Maher MC, Johnson T, Newton W, Jager S, Shales M, Horner J, Hernandez RD, Krogan NJ, Glaunsinger BA. 2015. Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Mol Cell 57:349–360. doi: 10.1016/j.molcel.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isomura H, Stinski MF, Murata T, Yamashita Y, Kanda T, Toyokuni S, Tsurumi T. 2011. The human cytomegalovirus gene products essential for late viral gene expression assemble into prereplication complexes before viral DNA replication. J Virol 85:6629–6644. doi: 10.1128/JVI.00384-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia Q, Wu TT, Liao HI, Chernishof V, Sun R. 2004. Murine gammaherpesvirus 68 open reading frame 31 is required for viral replication. J Virol 78:6610–6620. doi: 10.1128/JVI.78.12.6610-6620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang S, Yamanegi K, Zheng ZM. 2004. Requirement of a 12-base-pair TATT-containing sequence and viral lytic DNA replication in activation of the Kaposi's sarcoma-associated herpesvirus K8.1 late promoter. J Virol 78:2609–2614. doi: 10.1128/JVI.78.5.2609-2614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong E, Wu TT, Reyes N, Deng H, Sun R. 2007. Murine gammaherpesvirus 68 open reading frame 24 is required for late gene expression after DNA replication. J Virol 81:6761–6764. doi: 10.1128/JVI.02726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu TT, Park T, Kim H, Tran T, Tong L, Martinez-Guzman D, Reyes N, Deng H, Sun R. 2009. ORF30 and ORF34 are essential for expression of late genes in murine gammaherpesvirus 68. J Virol 83:2265–2273. doi: 10.1128/JVI.01785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong-Ho E, Wu TT, Davis ZH, Zhang B, Huang J, Gong H, Deng H, Liu F, Glaunsinger B, Sun R. 2014. Unconventional sequence requirement for viral late gene core promoters of murine gammaherpesvirus 68. J Virol 88:3411–3422. doi: 10.1128/JVI.01374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arumugaswami V, Wu TT, Martinez-Guzman D, Jia Q, Deng H, Reyes N, Sun R. 2006. ORF18 is a transfactor that is essential for late gene transcription of a gammaherpesvirus. J Virol 80:9730–9740. doi: 10.1128/JVI.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perng YC, Campbell JA, Lenschow DJ, Yu D. 2014. Human cytomegalovirus pUL79 is an elongation factor of RNA polymerase II for viral gene transcription. PLoS Pathog 10:e1004350. doi: 10.1371/journal.ppat.1004350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruffat H, Kadjouf F, Mariame B, Manet E. 2012. The Epstein-Barr virus BcRF1 gene product is a TBP-like protein with an essential role in late gene expression. J Virol 86:6023–6032. doi: 10.1128/JVI.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cassonnet P, Rolloy C, Neveu G, Vidalain PO, Chantier T, Pellet J, Jones L, Muller M, Demeret C, Gaud G, Vuillier F, Lotteau V, Tangy F, Favre M, Jacob Y. 2011. Benchmarking a luciferase complementation assay for detecting protein complexes. Nat Methods 8:990–992. doi: 10.1038/nmeth.1773. [DOI] [PubMed] [Google Scholar]
- 13.Muller M, Jacob Y, Jones L, Weiss A, Brino L, Chantier T, Lotteau V, Favre M, Demeret C. 2012. Large scale genotype comparison of human papillomavirus E2-host interaction networks provides new insights for e2 molecular functions. PLoS Pathog 8:e1002761. doi: 10.1371/journal.ppat.1002761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brulois K, Wong LY, Lee HR, Sivadas P, Ensser A, Feng P, Gao SJ, Toth Z, Jung JU. 2015. Association of Kaposi's sarcoma-associated herpesvirus ORF31 with ORF34 and ORF24 is critical for late gene expression. J Virol 89:6148–6154. doi: 10.1128/JVI.00272-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brulois KF, Chang H, Lee AS, Ensser A, Wong LY, Toth Z, Lee SH, Lee HR, Myoung J, Ganem D, Oh TK, Kim JF, Gao SJ, Jung JU. 2012. Construction and manipulation of a new Kaposi's sarcoma-associated herpesvirus bacterial artificial chromosome clone. J Virol 86:9708–9720. doi: 10.1128/JVI.01019-12. [DOI] [PMC free article] [PubMed] [Google Scholar]