ABSTRACT
Proteolytic maturation drives the conversion of stable, immature virus particles to a mature, metastable state primed for cell infection. In the case of human adenovirus, this proteolytic cleavage is mediated by the virally encoded protease AVP. Protein VI, an internal capsid cement protein and substrate for AVP, is cleaved at two sites, one of which is near the N terminus of the protein. In mature capsids, the 33 residues at the N terminus of protein VI (pVIn) are sequestered inside the cavity formed by peripentonal hexon trimers at the 5-fold vertex. Here, we describe a glycine-to-alanine mutation in the N-terminal cleavage site of protein VI that profoundly impacts proteolytic processing, the generation of infectious particles, and cell entry. The phenotypic effects associated with this mutant provide a mechanistic framework for understanding the multifunctional nature of protein VI. Based on our findings, we propose that the primary function of the pVIn peptide is to mediate interactions between protein VI and hexon during virus replication, driving hexon nuclear accumulation and particle assembly. Once particles are assembled, AVP-mediated cleavage facilitates the release of the membrane lytic region at the amino terminus of mature VI, allowing it to lyse the endosome during cell infection. These findings highlight the importance of a single maturation cleavage site for both infectious particle production and cell entry and emphasize the exquisite spatiotemporal regulation governing adenovirus assembly and disassembly.
IMPORTANCE Postassembly virus maturation is a cornerstone principle in virology. However, a mechanistic understanding of how icosahedral viruses utilize this process to transform immature capsids into infection-competent particles is largely lacking. Adenovirus maturation involves proteolytic processing of seven precursor proteins. There is currently no information for the role of each independent cleavage event in the generation of infectious virions. To address this, we investigated the proteolytic maturation of one adenovirus precursor molecule, protein VI. Structurally, protein VI cements the outer capsid shell and links it to the viral core. Functionally, protein VI is involved in endosome disruption, subcellular trafficking, transcription activation, and virus assembly. Our studies demonstrate that the multifunctional nature of protein VI is largely linked to its maturation. Through mutational analysis, we show that disrupting the N-terminal cleavage of preprotein VI has major deleterious effects on the assembly of infectious virions and their subsequent ability to infect host cells.
INTRODUCTION
Human adenovirus (HAdV) assembly and particle maturation are among the most poorly understood steps in the virus life cycle. Like many other viruses and bacteriophages, immature AdV particles undergo extensive proteolytic maturation, likely following assembly, to generate mature virions. Cleavage of seven preproteins (pIIIa, pVI, pVII, pVIIII, pTP, pX, and the L52/55K scaffolding protein) in the capsid is mediated by the virus-encoded adenovirus protease (AVP) (reviewed in reference 1). AVP recognizes two consensus motifs present in these precursor proteins, (M/I/L)XGX-G and (M/I/L)XGG-X (2).
A temperature-sensitive mutant of HAdV-C2 (HAdV2-ts1) has been instrumental in broadly defining the role of proteolytic processing in adenovirus maturation (3). The ts1 mutant harbors a point mutation in AVP that prevents normal protease incorporation at the nonpermissive temperature, leading to the accumulation of immature virions comprised of precursor proteins (4, 5). These ts1 particles are highly stable, rendering them noninfectious due to a failure to undergo capsid disassembly in the endosome during cell entry.
One of the substrates for AVP is preprotein VI (pVI), an internal structural protein involved in virus assembly, endosome lysis, subcellular trafficking, and the initiation of early viral gene expression (6–9). During virus replication, protein VI functions as an adaptor molecule to transport the major capsid protein, hexon, from the cytoplasm, where it is expressed, to the nucleus, where particle assembly occurs. AVP cleaves pVI near the N and C termini to generate the mature protein, VI (Fig. 1A). C-terminal cleavage liberates an 11-amino-acid (aa) peptide (pVIc) that functions as a cofactor for AVP, significantly enhancing enzyme processivity (10, 11). However, the requirement for N-terminal cleavage of pVI has not been investigated independently of ts1 particles that contain six different precursor proteins, thus making it difficult to pinpoint a specific role for pVI as well as the other precursor proteins in capsid maturation.
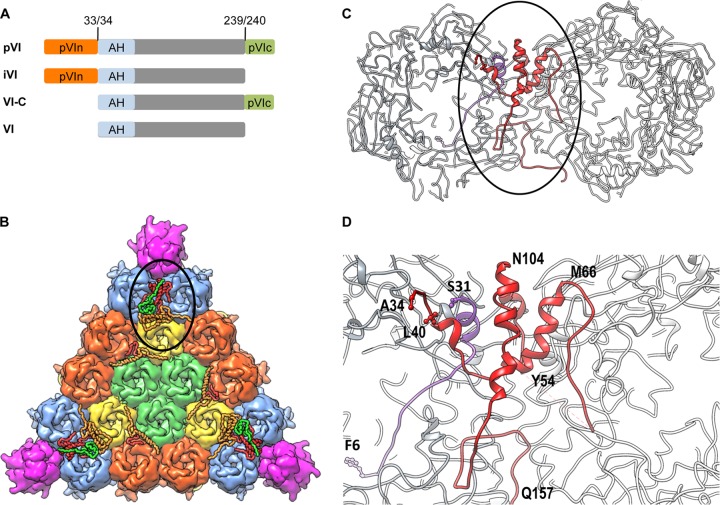
FIG 1.
Protein VI maturation and location within AdV particles. (A) Schematic of protein VI. Preprotein VI (pVI) is cleaved by the adenoviral cysteine protease (AVP) at both termini (numbered residues) to generate mature protein VI (gray). Two intermediate species cleaved at one of the two termini (iVI and VI-C) are also illustrated. The N-terminal pVIn peptide (orange) binds the internal cavity of hexon trimers. The pVIc peptide (green) is a cofactor for AVP. The membrane lytic amphipathic helix (AH), present at the N terminus of mature VI, is highlighted in light blue. (B) Crystal structure of one interior facet of the HAdV5 capsid. Color schemes are as follows: hexons, blue, orange, yellow, and light green; penton base, magenta; protein VI, red; protein V, green; protein VIII, light orange. The location of the complex formed by proteins VI, V, and VIII beneath the peripentonal hexons is circled. (C) Zoom-in view of the circled area from panel B, with the peripentonal hexons shown in gray, pVIn shown in purple, and mature VI shown in red. (D) Zoom-in view of the circled area from panel C, using the same color scheme. Selected residues from pVIn and mature VI are designated using the single-amino-acid code. PDB identifier, 4CWU.
A series of proteomics studies (12, 13), as well as the most recent crystal structure of HAdV5-35F (14), revealed that the N-terminal peptide (pVIn, residues 1 to 33) remains associated with mature virions following cleavage. Furthermore, these studies demonstrated that the pVIn peptide is buried within the internal cavity of peripentonal hexons (Fig. 1B to D) (12–14). The intimate association of hexon and pVIn within the capsid supports the known role of pVI in transporting hexon from the nucleus to the cytoplasm during virus assembly (6). The AdV crystal structure also indicates that a significant structural rearrangement takes place following cleavage at the N terminus of pVI, as ∼24 Å separates the last visible residue at the C terminus of pVIn (serine 31) from the N terminus of mature VI (alanine 34) (Fig. 1D) (14). Interestingly, native mass spectrometry analysis further showed that upon thermal perturbation that mimics virus uncoating in the endosome, the pVIn peptide remains in complex with the hexon trimers, while the majority of mature VI protein does not (12). There is also a distinct pH dependence governing hexon-pVIn association in vitro. pVIn binds hexon weakly (1 pVIn per hexon trimer) at physiologic pH and more avidly at low (endosomal) pH (3 pVIn per hexon trimer) (12). These findings suggest a new model of AdV disassembly and cell entry in which the proteolytic maturation of pVI is required for proper dissociation of mature VI from the hexon trimer within the acidic endosome.
Thus, the present work was initiated to determine what role the N-terminal cleavage of preprotein VI has in virus assembly, maturation, and infection. To that end, we describe the differential interaction of the immature and mature forms of protein VI with hexon, as well as the effect that these interactions have on the membrane lytic activity of protein VI. We identified a glycine-to-alanine mutant within the pVI N-terminal cleavage site that profoundly alters the cleavage of protein VI in vitro as revealed by proteolysis assays with recombinant proteins. Furthermore, a virus harboring this mutation shows significantly altered cell entry and virus assembly properties. Together, these studies provide evidence that proteolytic maturation at a single position within an adenovirus capsid protein can impact multiple steps in the virus life cycle.
MATERIALS AND METHODS
Cell lines and viruses.
Tissue culture reagents were obtained from Life Technologies. A549 (ATCC) and 293β5 (15) cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 10 mM HEPES, 2 mM l-glutamine, 0.1 mM nonessential amino acids, 100 units/ml penicillin, and 100 μg/ml streptomycin (Strep). The replication-defective HAdV-C5 bacterial artificial chromosome (BAC) vector pAd5-GFPn1 has been previously described and harbors E1/E3 gene deletions replaced by a cytomegalovirus (CMV)-driven enhanced green fluorescent protein (EGFP) reporter cassette (8). Point mutations within the vector were made using standard recombineering technology (16). Following PacI linearization, the pAd5-GFPn1 wild-type and mutant genomes were transfected into E1/E3-complementing 293β5 cells. Lysates were serially passaged until a large virus stock was generated, at which point purified virus was isolated from cellular lysates by double banding in cesium chloride (CsCl2) gradients. Viruses were quantified with a NanoDrop 2000 spectrophotometer using the absorbance at 260 nm (A260) (particles/ml = A260 × 1.1E12). Viruses were dialyzed into A195 buffer (17), flash frozen in liquid nitrogen, and stored at −80°C until use.
Hexon purification.
Hexon was purified from virus-infected cells using a protocol adapted from the work of Rux and Burnett (18). Briefly, the hexon-containing top fraction from a HAdV-C5 CsCl2 gradient was collected and dialyzed into 10 mM Tris, pH 7.5, and purified on an AKTA purifier (GE Healthcare) equipped with a Resource Q column. The protein (>95% pure as judged by SDS-PAGE) was buffer exchanged into 20 mM Tris (pH 7.5), concentrated to 3 to 20 mg/ml, and stored in aliquots at −80°C.
Plasmids.
All plasmids encode adenovirus proteins from HAdV-C5 (AC_000008.1). Several vector backgrounds were used for expression of protein VI variants. Full-length preprotein VI (pVI) (residues 1 to 250) and mature (processed) VI (residues 34 to 239) were cloned into the pET21a backbone with a C-terminal 6×His tag (HisC) separated by a tobacco etch virus (TEV) protease cleavage site (pVI-HisC, VI-HisC). For expression of Strep-tagged pVI and VI variants, the HisC vectors were further modified via the insertion of a C-terminal double Strep tag (19) immediately upstream of the 6×His tag. The Strep tag insertion includes a stop codon such that the 6×His tag is not expressed. Point mutations in the vector were made using standard site-directed mutagenesis protocols. AVP was cloned into pET46 (Novagen) modified to include a TEV cleavage site between the N-terminal 6×His tag and the AVP open reading frame (ORF).
Recombinant protein purification.
All recombinant proteins were expressed in BL21(DE3)-RIPL cells (Agilent Technologies) cultured in LB medium supplemented with 100 μg/ml ampicillin and 30 μg/ml chloramphenicol. Cultures for protein VI expression also included 1% glucose to prevent leaky expression prior to induction. Bacteria expressing protein VI were grown at 37°C to an optical density at 600 nm (OD600) of 0.9 to 1.0, induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 1 h, pelleted, and frozen at −20°C. The purification scheme for recombinant protein VI with a C-terminal 6×His tag was adapted from reference 20. Bacterial cell pellets were resuspended in lysis buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 10 mM imidazole, 5 mM beta-mercaptoethanol [βME], 1% n-dodecyl-β-d-maltoside [DDM], 1 mM benzamidine-HCl, 1 mM phenylmethylsulfonyl fluoride [PMSF], Roche protease inhibitor cocktail, 0.1 mg/ml lysozyme, 25 U/ml Benzonase), rocked for 60 min at room temperature, and then further disrupted via sonication. Cellular debris was removed by centrifugation at 30,000 × g for 30 min. Lysates were applied to Talon cobalt resin (Clontech) equilibrated with buffer WH (50 mM Tris, pH 7.5, 100 mM NaCl, 5 mM βME, 0.025% DDM, 10 mM imidazole) and washed with buffer WH to remove unbound protein. His-tagged proteins were eluted with buffer WH supplemented with 300 mM imidazole. The sample was diluted to reduce the NaCl concentration to 50 mM and applied to a Resource S column attached to an AKTA purifier (GE Healthcare). The protein was eluted with a linear NaCl gradient from 50 to 1,000 mM. Protein VI elution fractions were concentrated to 1.5 mg/ml, and aliquots were stored at −80°C until use.
For Strep-tagged recombinant protein VI, cell pellets were resuspended in lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, 5 mM βME, 1% DDM, 1 mM benzamidine-HCl, 1 mM PMSF, Roche protease inhibitor cocktail, 0.25 mg/ml lysozyme, 20 U/ml Benzonase), incubated on ice for 30 min, and disrupted further via sonication. Cellular debris was removed by centrifugation at 30,000 × g for 30 min. Strep-tagged protein VI was bound to Strep-Tactin resin (IBA) equilibrated with buffer WS (50 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, 5 mM βME, 0.025% DDM). Unbound protein was removed by washing with buffer WS, and protein VI was eluted with buffer WS supplemented with 2.5 mM desthiobiotin. The elution was diluted to reduce the NaCl concentration to 25 mM and injected onto a Resource S column (GE Healthcare). Protein VI was eluted with a linear NaCl gradient from 25 to 500 mM in 25 mM Tris, pH 8.0. The protein VI peak was concentrated to 2 mg/ml and buffer exchanged into storage buffer (25 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, 5 mM βME, 0.025% DDM). Aliquots of protein were flash frozen and stored at −80°C.
Bacteria expressing His-AVP were grown at 28°C to an OD600 of 0.5 to 0.6, induced with IPTG to 0.5 mM, and incubated overnight for 16 to 20 h. The cells were pelleted and frozen at −20°C. Bacterial pellets were resuspended in lysis buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 5 mM βME, Roche protease inhibitor cocktail, 0.25 mg/ml lysozyme, 25 U/ml Benzonase), incubated on ice for 30 min, and sonicated. Cell debris was removed via centrifugation at 20,000 × g for 20 min. His-AVP was bound to nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) equilibrated with buffer CB (50 mM Tris, pH 8.0, 150 mM NaCl, 5 mM βME, 10 mM imidazole), washed with buffer CB containing 20 mM imidazole, and eluted with buffer CB supplemented with 300 mM imidazole. Imidazole was removed with a PD10 desalting column (GE Healthcare), and the protein was incubated with TEV protease overnight at room temperature to cleave off the C-terminal 6×His tag. Uncleaved protein and His-tagged TEV were removed by passage over Ni-NTA resin. AVP was dialyzed into 50 mM Bis-Tris, pH 6.5, 15 mM NaCl, 5 mM βME and then further purified on a Resource S column. The AVP peak was pooled, dialyzed into AVP storage buffer (20 mM Tris, pH 8.0, 25 mM NaCl, 1 mM EDTA), and concentrated to 5 to 10 mg/ml. The protein was stored in aliquots at −80°C until use.
ELISAs.
All enzyme-linked immunosorbent assays (ELISAs) were performed in high-binding enzyme immunoassay (EIA)/radioimmunoassay (RIA) plates (Corning) with proteins diluted in phosphate-buffered saline (PBS), pH 7.4. Following coating, wells were blocked with Superblock reagent (Thermo Fisher). To compare pVI and VI hexon binding, plates were coated with 1 μg/well of pVI-HisC or VI-HisC, blocked, and incubated with serial dilutions of hexon in duplicate for 2 h at room temperature. To compare the binding of wild-type pVI and that of G33A mutant pVI to hexon, ELISA plates were coated with 1 μg/well of recombinant pVI-Strep (wild type or G33A mutant) and incubated with 500 ng of hexon for 2 h at room temperature. For the competition ELISA, plates coated with 1 μg/well of pVI-Strep or VI-Strep were incubated with 500 ng of hexon or 500 ng of hexon preincubated overnight at 4°C with a 100-fold molar excess of pVIn peptide (12). Samples were prepared in triplicate. For all ELISAs, triplicate data points represent the mean ± standard deviation (SD). Hexon binding was detected with the monoclonal antihexon antibody 9C12 (21) followed by an anti-mouse secondary antibody conjugated to horseradish peroxidase (HRP). 1-Step ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] reagent (Thermo Fisher) was used as the HRP substrate.
In vitro membrane lytic activity of recombinant protein VI and heat-disrupted AdV particles.
Liposome lysis assays with recombinant pVI-HisC and VI-HisC or heat-disrupted AdV particles were performed as previously described (22). Samples were prepared in triplicate, and the data represent the mean ± SD. To assess the ability of hexon to shield the membrane lytic activity of protein VI, pVI-HisC and VI-HisC (150 nM) were mixed with 2-fold serial dilutions of hexon (highest concentration was 7.5 μM, a 50-fold molar excess) before being added to liposomes. Specific liposome lysis was calculated with the following formula: % sulforhodamine B (SulfoB) released = 100 × [(Fprot − Fbkgd)/(Fdet − Fbkgd)], where Fprot is the fluorescence intensity in the presence of protein VI, Fbkgd is the background fluorescence, and Fdet is the fluorescence intensity in the presence of 0.5% Triton X-100 (full lysis). Samples were prepared in duplicate, and the mean specific lysis ± SD was plotted.
AVP cleavage assays.
The 11-amino-acid (aa) pVIc peptide (GVQSLKRRRCF) was synthesized by Eton Bioscience. In vitro cleavage reaction conditions were adapted from a previous study (23). Wild-type or G33A recombinant pVI-Strep (200 ng), 80 ng pVIc peptide, 80 ng bovine serum albumin (BSA), and 200 pM plasmid DNA were diluted in 20 mM Tris, pH 8.0, 10 mM EDTA (total reaction volume was 10 μl) and preequilibrated to room temperature for 5 min. AVP (200 ng) was added, and the cleavage reaction mixture was incubated for an additional 5 or 30 min at room temperature. The reaction was stopped via addition of SDS sample buffer, and the reaction mixture was boiled. Cleavage was visualized by SDS-PAGE and SYPRO Ruby staining.
SDS-PAGE analysis of virus composition.
Purified virus particles were analyzed by SDS-PAGE, followed by Simply Blue (Life Technologies) staining (6 μg virus/lane) or immunoblotting (3 μg virus/lane) for protein VI using a rabbit anti-VI polyclonal antibody. For determination of protein VI incorporation into virus capsids, SYPRO Ruby-stained SDS-PAGE gels were used. The gels were loaded with 5 μg of duplicate samples of two independent preparations of wild-type and G33A viruses. Following SYPRO Ruby staining, the gels were scanned on a Typhoon Fluorimager and band intensity was measured using ImageQuant software (GE Healthcare). The relative ratio of protein VI to hexon, penton base, fiber/IIIa, V, VII, or VIII/IX for each independent virus was determined. Twelve independent data points (6 from each virus preparation) were averaged together, and the mean ± standard error of the mean (SEM) is plotted in the bar graph in Fig. 4C.
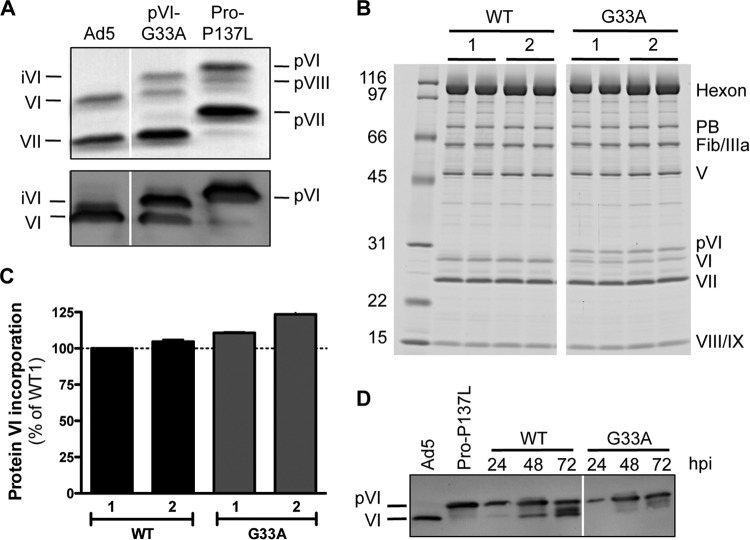
FIG 4.
The G33A mutation impacts virion composition and pVI cleavage in infected cells. (A) SDS-PAGE gel analysis of purified virus particles, including Ad5, Ad5-Pro-P137L (produces immature virions harboring precursor proteins), and Ad5-pVI-G33A. The gel in the upper panel was stained with Simply Blue and reveals various proteins from mature and immature HAdV5 that are labeled as indicated. The lower panel shows an immunoblot assay for protein VI. The additional protein VI band in the G33A virus is iVI, as it migrates differently (in between) than both pVI and mature VI. (B) Two independent preparations of wild-type (WT) or G33A virions (labeled 1 and 2) were analyzed by SDS-PAGE and SYPRO Ruby staining. Numbers at left are molecular masses in kilodaltons. (C) Quantities of the total amount of protein VI incorporated into virus particles determined by densitometry analysis of the gels shown in panel B and normalized to each of the other virus structural proteins as described in Materials and Methods. The data are the mean percentage ± SEM of protein VI incorporation shown for all viral proteins, compared to wild-type preparation 1. (D) Time course of pVI cleavage in infected cells. Lysates of infected cells were made at the indicated times (hpi) and immunoblotted for protein VI. Note the significant overall reduction and delayed processing of pVI to mature VI for the G33A point mutant.
Protein VI cleavage in infected cells.
293β5 cells were infected with 1,000 particles/cell of wild-type or G33A mutant HAdV-C5. At various times postinfection, the cells were harvested and resuspended in RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, Roche protease inhibitor cocktail, 2.5 U/ml Benzonase). Following a 1-h incubation on ice, the cell debris was pelleted at 14,000 × g for 10 min. Clarified lysates were separated by SDS-PAGE and subjected to immunoblotting with a rabbit polyclonal antibody to protein VI followed by a horseradish peroxidase-conjugated anti-rabbit secondary antibody.
Single-round infection.
Triplicate wells of A549 cells were infected with serial dilutions of wild-type HAdV-C5 or HAdV-C5 G33A mutant particles for ∼48 h. Infected cells were then fixed in 2% paraformaldehyde diluted in PBS (pH 7.4), washed twice with PBS, and resuspended in fluorescence-activated cell sorting (FACS) buffer (PBS, pH 7.4, 5% fetal calf serum, 0.2% sodium azide). Data were collected on a Novocyte flow cytometer (ACEA Biosciences) and analyzed with FlowJo to determine the percentage of GFP-positive (infected) cells. Nonlinear regression in GraphPad Prism was used to calculate the number of virus particles required to infect 50% of the cells (ID50).
Thermostability assay.
Virus thermostability assays were performed as previously described (24) with slight modification. Wild-type and G33A mutant viruses (6 μg) were diluted such that final buffer conditions were 0.05% bovine serum albumin, 50 mM NaCl, and 7.5 mM Tris, pH 7.4. Samples were incubated for 10 min at either 23°C or 50°C, before application to discontinuous HistoDenz density gradients. Supernatant and band fractions were separated by SDS-PAGE and analyzed by immunoblotting for hexon (rabbit polyclonal), fiber (4D2; NeoMarkers), and protein VI (rabbit polyclonal) using protein-specific antibodies.
Virus burst assay.
293β5 cells were infected with equivalent amounts of infectious particles (calculated from a single-round infection assay) of HAdV-C5 wild-type and G33A mutant viruses. Three independent preparations of wild-type virus were included to assess the intrinsic variability of virus production from different virus stocks. At 48 and 72 h postinfection (hpi), cells were harvested, resuspended in 200 μl fresh DMEM, and lysed by 3 rounds of freeze-thaw. The cell debris was removed by centrifugation at 5,000 × g for 10 min. To determine the virus burst size (the amount of infectious virus produced), lysates were used to infect E1-negative (noncomplementing) A549 cells in 96-well plates. The clarified lysate (100 μl) was incubated with A549 cells for 2 h at 37°C, washed once with PBS, and then overlaid with 100 μl of fresh DMEM. After 48 h, the plates were scanned with a Typhoon Fluorimager (GE Healthcare) to measure infection via expression of the GFP transgene.
Virus assembly assay.
Eight 15-cm plates of 293β5 cells were infected with equivalent infectious units of HAdV-C5 wild-type or G33A mutant viruses (500 or 900 particles/cell, respectively) for 53 h. The cells were harvested and divided into two aliquots (94% for cesium chloride gradients and 6% for Virabind columns) and lysed by three rounds of freeze-thaw, and cellular debris was removed via centrifugation at 4,000 × g for 10 min. To purify virus by cesium chloride density gradient (15 to 40%) ultracentrifugation, lysate was applied to the top of the gradients and centrifuged at 111,000 × g for 1.5 h. The immature and mature bands were syringe extracted from the gradients. HAdV particles were also purified on Virabind columns (Cell Biolabs) as suggested by the manufacturer's instructions. All virus samples were dialyzed into A195 buffer. Mature virus concentrations were determined based on the absorbance at 260 nm as described above. These values were then used to establish a ratio between A260 and A280 (A260/A280 = 2.6) for mature virus, which was subsequently used to determine virus concentrations based on the A280 for the immature and mixed samples that are lacking (in whole or in part) viral DNA. Infectivity of the various purified virus samples was assessed in a single-round infection assay as described above.
RESULTS
Proteolytic maturation of protein VI impacts hexon binding.
To better understand how protein VI proteolytic cleavage affects the early events in the HAdV infection cycle, we first determined the relationship between hexon-protein VI binding and the maturation state of protein VI. During virus replication, pVI binds hexon in the cytoplasm and escorts it into the nucleus for particle assembly. In subsequent rounds of infection by newly assembled virions, mature VI dissociates from the hexon in the endosome to mediate membrane lysis. These multiple functions of protein VI suggest that its maturation may alter hexon interaction. While the crystal structure of the HAdV capsid reveals an intimate association between the pVIn peptide and hexon (14), it is unclear whether the binding of these two proteins depends upon the pVIn peptide specifically. Recent studies have demonstrated a high-affinity interaction between pVI and hexon (20) but did not investigate the association of mature VI with hexon. We therefore compared the hexon binding properties of recombinant immature pVI and mature VI using an ELISA. Our results clearly indicate that unprocessed pVI, but not mature VI, binds specifically to hexon (Fig. 2A). Additionally, hexon-pVI association is competed by an excess of the pVIn peptide (Fig. 2B), indicating that residues 1 to 33 in the pVIn peptide, which are normally buried in the internal hexon cavity of the mature virus capsid, regulate these protein-protein interactions.
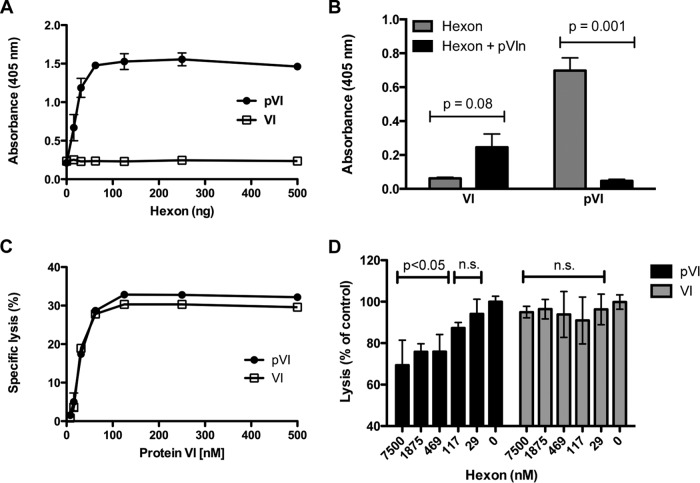
FIG 2.
Preprotein VI, but not mature VI, associates with hexon. (A) Hexon binding by recombinant protein VI. ELISA plates, coated with recombinant pVI or VI, were incubated with serial dilutions of hexon, and then hexon was detected with an antihexon antibody. (B) Competition ELISA between different recombinant protein VI molecules and pVIn peptide. ELISA plates coated with pVI or VI were incubated with hexon, hexon preincubated with a 100-fold molar excess of pVIn, or buffer. The pVIn peptide competes only with pVI for hexon binding. An unpaired two-tailed t test was used to calculate statistical significance. (C) The membrane lytic activities of pVI and VI are similar. Serial dilutions of recombinant protein VI were incubated with SulfoB-entrapped liposomes, and specific lysis was measured using a Fluorimager. (D) Hexon attenuates the membrane lytic activity of pVI but not VI. pVI or VI (150 nM) was mixed with serial dilutions of hexon and then added to liposomes. Data are the percentage of specific lysis compared to the lytic activity of pVI or VI in the absence of hexon. P values were calculated using a one-way analysis of variance and Dunnett's multiple-comparison test (compared to the 0 nM hexon control). n.s., not significant (P > 0.05).
Hexon shields the membrane lytic activity of pVI.
In further studies, we sought to determine the functional consequences of hexon-pVIn association. The membrane lytic activity of protein VI primarily lies within the N-terminal amphipathic helix (residues 34 to 54) of mature VI (7, 25). When we compared recombinant pVI with VI in an in vitro fluorescence-based liposome lysis assay, we observed very similar dose-dependent lytic activities for the two forms of the protein (Fig. 2C). This finding is consistent with previous studies (7, 26) indicating that maturational processing is not a prerequisite for protein VI lytic activity. However, the new knowledge that pVI interacts primarily with hexon via the pVIn peptide suggests a requirement for maturational processing to fully expose the amphipathic helix and allow membrane access. In support of this, we found that coincubation of pVI with hexon decreased the membrane lytic activity of pVI but had little effect on the lytic activity of mature VI (Fig. 2D). Combined with the structural information and the finding that the pVIn peptide is the critical determinant for hexon binding, these results suggest that hexons may shield the membrane lytic activity of pVI in the later stages of virus replication when production of late gene products and new particle assembly are occurring.
A glycine-to-alanine mutation within the N-terminal cleavage site alters proteolytic processing.
As our hexon binding studies and previous structural analyses suggested an important role for the pVIn region, we next sought to test this by performing mutagenesis studies. There are two consensus recognition motifs for AVP, (M/I/L)XGX-G and (M/I/L)XGG-X, where X is any amino acid. The latter site is highly conserved among HAdVs at the N terminus of protein VI, particularly the critical double glycine residues immediately preceding the cleavage site (Fig. 3A). To determine how a mutation within the AVP recognition motif would affect AVP-mediated cleavage, we engineered a glycine-to-alanine mutation at residue 33 (G33A) in a protein VI bacterial expression vector. We found that in vitro cleavage of pVI by AVP is profoundly altered by the G33A mutation (Fig. 3B). Native pVI is almost fully cleaved to mature protein VI after 30 min (see Materials and Methods for details); however, the G33A mutant was processed solely at the C terminus and thus trapped in a partially cleaved intermediate (iVI) state (Fig. 1A and 3B). Interestingly, proteolytic processing of the G33A mutant proceeded through an alternative pathway compared to wild-type pVI, which cleaves first at the N terminus (Fig. 3B; compare 5-min time points) (27). In contrast, the G33A mutant is cleaved by AVP only at the C terminus, indicating that while the N terminus is the preferred initial cleavage site in vitro, AVP adopts an alternative processing scheme when the N-terminal cleavage site is disrupted.
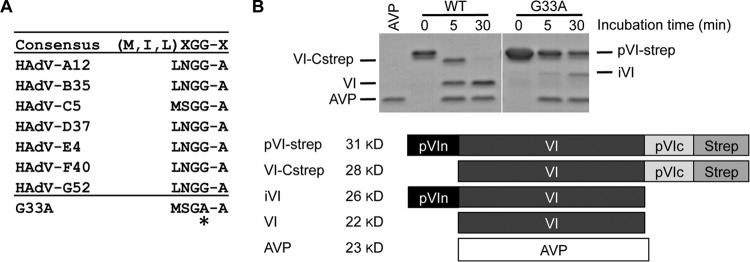
FIG 3.
A single G33A mutation in the N-terminal AVP cleavage site of protein VI reduces in vitro processing. (A) Amino acid sequence alignment of the N-terminal regions of protein VI in different human AdVs. The consensus sequence for recognition by AVP is given at the top of the table. Directly below is an alignment of the pVI N-terminal cleavage site sequences for different species (A to G). The HAdV5 protein VI cleavage site sequence spans residues 30 to 34. The highly conserved glycine residue at position 33 was mutated to alanine, and the mutation is indicated by an asterisk. (B) In vitro cleavage of pVI by AVP. Recombinant pVI-Strep was incubated with recombinant AVP for the indicated number of minutes at room temperature. Labeled bands to the left of the gel correspond with the labeling scheme in the schematic below, which describes the different recombinant proteins and indicates the expected molecular mass of each following processing at the N and/or C terminus by AVP. A nonspecific degradation product copurifies with the full-length proteins (slightly lower band at the 0-min point).
The G33A mutation restricts protein VI processing and incorporation into AdV particles.
As our initial experiments indicated that the G33A mutation impedes proteolytic processing of protein VI in vitro, we next wished to ascertain how the G33A mutation impacts the composition of AdV particles with respect to protein VI maturation and incorporation. Thus, we engineered the G33A mutation in protein VI harbored in a HAdV-C5 vector. The resultant stock of purified virus particles was first analyzed by SDS-PAGE and immunoblotting to determine the extent of protein VI proteolysis (Fig. 4A). We included two control viruses in the SDS gels and immunoblot analyses as markers for the migration patterns of immature (pVI) and mature (VI) protein. Wild-type HAdV-C5 possesses mature VI in its capsid, while the Ad5-Pro-P137L virus (28), which harbors the same AVP point mutation as HAdV2-ts1, contains unprocessed precursor proteins, including pVI. Interestingly, capsids harboring the G33A mutant VI contained an ∼50:50 mixture of intermediate (iVI, cleaved only at C terminus) and mature VI (Fig. 4A). We found that while cleavage was impacted by the G33A mutation, the total levels of protein VI (iVI plus VI) in the capsid were very similar to those for wild-type virus (Fig. 4B and C). These results suggest that the virus is able to package the G33A mutant protein VI normally but fails to cleave all ∼360 copies of protein VI postassembly. We further noted that cleavage of other capsid precursor cement proteins (pIIIa, pVII, and pVIII) occurred normally for multiple preparations of the G33A virus (Fig. 4B), indicating that there were no additional effects on capsid maturation beyond the decreased cleavage of G33A protein VI at the N terminus. This result indicates that AVP was activated properly by the C-terminal peptide cofactor of protein VI (VI-C).
The kinetics of maturation processing by AVP are altered in mutant-virus-infected cells.
Based on the differences in protein VI cleavage in wild-type and mutant virus particles, we next assessed whether the G33A mutation also affected proteolytic maturation of pVI during the later stages of virus infection when progeny virions are being assembled. A time course of infection revealed that precursor maturation was significantly delayed by the G33A point mutation within pVIn (Fig. 4D). At the 24-hpi point, cells infected with wild-type virus harbor a small portion of mature VI, although the majority of total protein VI exists in the precursor form. In contrast, G33A mutant-infected cells show no detectable mature VI. At 48 and 72 hpi, a much higher proportion of protein VI molecules is in the fully cleaved form in wild-type-virus-infected cells. However, the cells infected with the G33A mutant virus exhibit much less processing of the immature pVI, even at later time points. Overall, the percentage of total cellular protein VI processed to mature VI was reduced for the G33A mutant compared to the wild-type virus (Fig. 4D), and this finding confirms that precursor proteolysis of pVI is delayed and/or reduced in infected cells.
The G33A mutant virus exhibits reduced infection and membrane lysis.
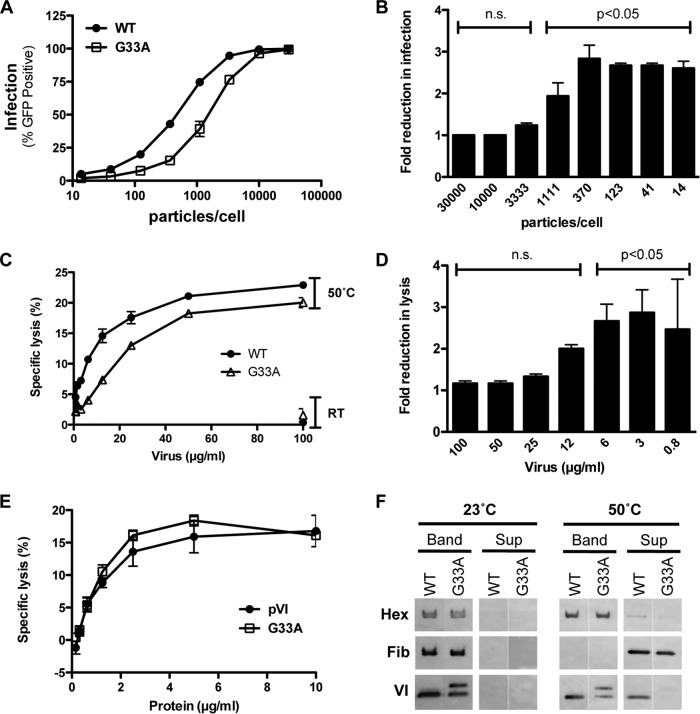
During AdV cell entry, incoming capsids undergo controlled, stepwise disassembly that is characterized by the release of proteins at the vertex region, including protein VI (29). Partial capsid disassembly and exposure of the membrane lytic protein VI (30) are critical for productive endosome escape (7). The presence of ∼180 copies of iVI in G33A virus particles (Fig. 4A) suggests that this mutant may have a reduced capacity to mediate endosome lysis during cell entry if iVI molecules cannot be effectively released from virus particles in the endosome. Thus, we evaluated the infectivity of the G33A mutant virus in a single-round infection assay, using GFP transgene expression as an indicator of successful cell entry. Noncomplementing A549 cells that lack the AdV E1 genes required for replication were infected with serial dilutions of the same number of physical particles of wild-type and G33A AdVs (Fig. 5A). While both viruses showed a dose-dependent increase in virus infection, the overall infectivity of the G33A virus was diminished with respect to wild-type virus. Additionally, we observed an inverse relationship between the virus input and the fold reduction in infection compared to wild-type virus for the G33A mutant (Fig. 5B). At very low particle-per-cell ratios, the mutant virus showed greater reductions in infectivity than at higher doses. We used nonlinear regression analysis to calculate the number of input particles required to infect 50% of the cells (ID50) for independent experiments with separate virus preparations. The mean fold reduction in ID50 compared to wild-type virus was 4.0 ± 1.5 for the G33A virus (4 experiments, 3 virus preparations). This indicates that the G33A mutant requires ∼4-fold more virus particles than the wild-type virus to productively infect the same number of cells.
FIG 5.
The G33A mutation attenuates infection and membrane lysis. (A) Single-round infection. Noncomplementing (E1-negative) A549 cells were infected with the indicated virus for 48 h, and the percentage of infected (GFP+) cells was determined by flow cytometry. WT, wild type. (B) Fold reduction in G33A mutant virus infection as a function of input particles/cell compared to wild-type virus. The data are derived from the samples depicted in panel A. P values are calculated with a one-way analysis of variance and Dunnett's multiple-comparison test. n.s., not significant. (C) Liposome lysis by heat-disrupted virus. Various amounts of viruses were incubated at room temperature (RT) (control) or heated to 50°C for 10 min to liberate protein VI and then mixed with liposomes. The percent specific lysis for each concentration is indicated. (D) Fold reduction in liposome lysis as a function of virus concentration compared to wild-type virus. The data are a subset of samples depicted in panel C. P values are calculated with a one-way analysis of variance and Dunnett's multiple-comparison test. (E) Liposome lysis by recombinant protein VI. Serial dilutions of wild-type and G33A mutant proteins were incubated with fluorescent dye-entrapped liposomes, and specific lysis was measured with a Fluorimager. (F) Thermostability assay to compare the dissociations of capsid proteins in mutant and wild-type viruses. Wild-type or mutant viruses were incubated at the indicated temperature and then subjected to density gradient ultracentrifugation to separate core (Band) and released (Sup) proteins. Proteins (hexon, fiber, and VI) were visualized via immunoblotting with the indicated antibodies.
We considered the possibility that the presence of ∼50% partially processed precursor protein in G33A virions was responsible for the observed reduction in virus infectivity in the single-round infection assay. We further reasoned that this decreased infection might be due to a lack of iVI release during G33A virus disassembly in the endosome. To test this possibility, we utilized an in vitro liposome lysis assay that relies on heat-disrupted virus. Gentle heating of wild-type AdV causes vertex removal and protein VI release similar to that occurring in the endosome during the cell entry process (31–34). While viruses incubated at room temperature showed no significant lytic activity, both wild-type and G33A viruses heated to 50°C exhibited a clear dose-dependent increase in membrane lytic activity due to protein VI release (Fig. 5C). The G33A mutant virus showed a modest but reproducible and statistically significant decrease in the ability to mediate membrane disruption compared to wild-type virus. In agreement with what we observed in the infectivity assay, the reduced lytic activity for the G33A mutant was greatest at lower virus particle numbers (Fig. 5D), suggesting that there is a synergistic effect for protein VI molecules that are released from multiple virus capsids to disrupt a single liposome. We also considered the possibility that the G33A mutation directly impacts membrane lysis, perhaps by reducing lytic activity of the protein overall. We therefore compared the in vitro lytic activity of recombinant protein bearing the G33A mutation to that of wild-type protein in a liposome lysis assay. However, contrary to this notion, the recombinant G33A mutant was equally as capable of disrupting model membranes as the wild-type protein (Fig. 5E). These results indicate that the decreased membrane lytic activity observed for the virus particles was not due to alterations in the intrinsic membrane disruption potential of the protein. Rather, reduced lysis was more likely linked to reduced protein VI release. To gather further support for this possibility, we explored whether the G33A mutation impacts capsid disassembly by using an in vitro uncoating assay (Fig. 5F). We noted that the G33A mutant exhibited higher capsid stability, with significantly less protein VI being exposed at 50°C. These results provide a mechanistic basis for the observed reduction in membrane lytic activity for heat-disrupted G33A particles. Overall, these experiments suggest that the decrease in infection observed for the G33A virus may be linked to a reduction in protein VI exposure.
The G33A cleavage site mutant reduces infectious particle yield.
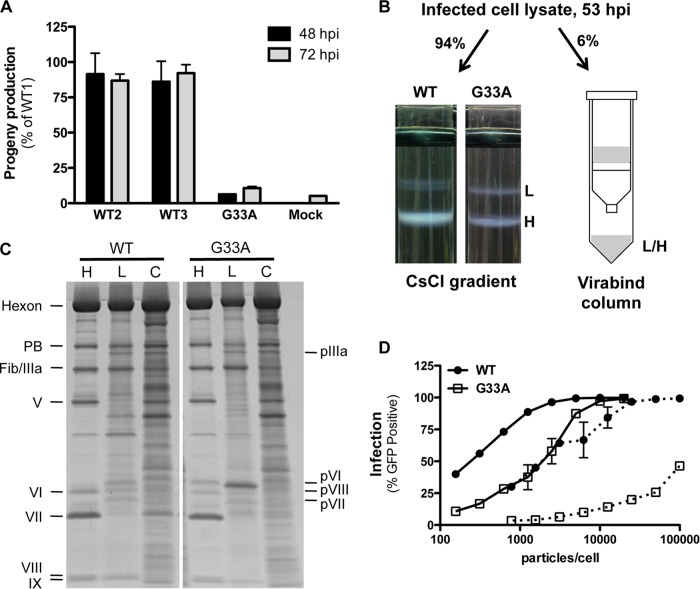
During the initial rescue (propagation) of the G33A mutant virus, we noted a distinctly lower growth rate compared to wild-type virus. While we attribute some of the slow growth kinetics to reduced infectivity, we were also interested to experimentally determine if the G33A mutation impacted virus replication. Therefore, we utilized a virus burst assay to ascertain how the G33A mutation impacted the assembly of progeny virions. For this experiment, we infected complementing 293β5 cells with an equivalent number of infectious units to control for the differences in cell entry between wild-type and G33A viruses. The equivalent infection allows for differences in infectious particle generation to be attributed to differences in assembly as opposed to infectivity. Using this assay, we found that the number of infectious particles produced by the G33A virus was significantly lower than that of wild-type virus (Fig. 6A). The reduction in infectious particles for the G33A virus was quite pronounced, with a 15.8-fold and a 9.4-fold reduction compared to wild-type virus at 48 and 72 hpi, respectively. Interestingly, infectious particle production increased with time for the G33A virus, implying that this virus mutant requires additional time to mature. Due to the fact that the G33A mutation also impacts a single-round infection, the observed decrease in infectious progeny is likely due to a combination of reduced particle production as well as reduced secondary infection.
FIG 6.
The G33A cleavage site mutant reduces infectious particle assembly. (A) Virus burst assay. Complementing 293β5 (E1-positive) cells were infected with equivalent amounts of infectious particles in triplicate. Cell lysates were prepared 48 and 72 hpi and passaged onto noncomplementing A549 cells. Virus infection (GFP transgene expression) was measured 48 hpi. Three independent preparations of wild-type (WT) Ad5 were included to assess intrinsic variations in virus burst. Progeny production is shown as a percentage of the value for wild-type preparation 1. (B) Illustration of sample preparation for studies shown in panels C and D. Infected cell lysates were divided as indicated, and virus was purified by cesium chloride density gradient ultracentrifugation or by binding and elution in a Virabind column. The light (L, genome-lacking) and heavy (H, genome-containing) virus bands were harvested separately from cesium chloride gradients, while the eluate from the Virabind column contains both heavy and light particles. (C) SDS-PAGE analysis of the virus purified by the different methods described in the legend to panel B. Note the differences in precursor capsid protein processing in the heavy and light particles. H, heavy; L, light; C, column purified. (D) The G33A cleavage mutant severely impacts particle assembly. Equivalent numbers of particles from the samples generated in panel B were used to infect A549 cells, and infection (GFP expression) was measured by flow cytometry. Mature viruses from the cesium chloride gradient are shown as solid lines, while column-purified virus is indicated with dotted lines.
Therefore, we sought to rigorously compare particle production between the wild-type virus and G33A cleavage site mutant. We utilized two independent purification methods (Fig. 6B; see Materials and Methods for full details) to assess the relative amounts of infectious units of virus particles produced following cell infection with an equivalent input of infectious units. Heavy (H, viral genome-containing) and light (L, genome-lacking) particles were harvested separately from cesium chloride density gradients. In parallel, commercially available Virabind columns, which allow purification of AdV particles based on size and charge, were used to isolate a mixture of heavy and light particles. The quality and purity of heavy, light, and mixed populations isolated by each of the methods were analyzed by SDS-PAGE (Fig. 6C). This analysis confirmed that heavy wild-type particles from cesium chloride gradients contain fully mature proteins, including protein VI. The heavy particles of the G33A mutant, in contrast, contained the same mixture of iVI and VI molecules as observed in Fig. 4A and B. As expected, the light particles for both the wild type and the G33A mutants contained the precursor proteins pIIIa, pVI, pVII, and pVIII and also lacked the DNA-associated protein V. Additionally, the gel profiles of column-purified viruses confirmed that they were a mixture of heavy and light particles, containing both processed and unprocessed proteins.
Quantification of the different virus particle samples isolated from the cesium chloride gradients revealed that the G33A mutant had a significantly higher ratio of light to heavy particles (Table 1). The wild-type virus produces nearly 4-fold more heavy than light particles, while the G33A virus produces slightly more light particles than heavy particles. Furthermore, there is a substantial decrease in the total amount of mature particles produced for the G33A mutant compared to wild-type virus (Table 1). These results indicate that virus assembly overall is significantly impacted by the G33A mutation.
TABLE 1.
Heavy and light particle yields
| Virus | Lighta | Heavya | Heavy/lightb | Mixturea |
|---|---|---|---|---|
| WTc | 8.8 × 1011 | 3.3 × 1012 | 3.8 | 1.1 × 1012 |
| G33A | 9.4 × 1011 (0.9) | 7.7 × 1011 (4.3) | 0.8 | 1.5 × 1012 (0.7) |
Values are the total heavy and light particle yields from the cesium chloride gradients and the heavy/light particle mixture from the Virabind column. It is important to note the intrinsic variability associated with syringe extraction of virus bands from cesium chloride gradients, and total particle yields are approximations. Numbers in parentheses indicate the fold reduction in G33A mutant particle yield compared to wild-type virus for each of the categories.
Heavy/light indicates the ratio of heavy to light particles produced for each of the viruses.
WT, wild type.
We next compared the infectivity of heavy particles isolated from the G33A mutant with those obtained from wild-type AdV. In agreement with the cell-based infection burst assay (Fig. 6A), the G33A mutation substantially decreased infectious particle production compared to wild-type virus (Fig. 6D). Comparing the cesium chloride density gradient-purified heavy particles with the total virus pool isolated on the Virabind column confirms that infectious particle production is severely diminished for the column-purified mutant virus (Fig. 6D; compare solid and dotted lines for each virus). We next calculated ID50 values for each of the viruses and found that wild-type column-purified virus was ∼8-fold less infectious than cesium chloride heavy particles. In contrast, the G33A column-purified virus was nearly 100-fold less infectious than heavy particles. These findings indicate that a much greater number of immature (genome-lacking) or defective particles are made for the G33A mutant, highlighting a major defect in normal viral particle assembly. Thus, the G33A mutation in pVI reveals a dual role for processing at the N-terminal region of protein VI. Proteolytic maturation is a prerequisite for promoting the correct assembly of mature, progeny capsids as well as for the efficient release of processed VI from mature virus particles during cell entry.
DISCUSSION
HAdV maturation is a highly complex process that involves the proteolytic processing of six precursor proteins (pIIIa, pVI, pVII, pVIII, pX, and pTP), as well as the viral scaffolding protein L1 52/55K (1). When proteolytic maturation is completely blocked, as in the case of the HAdV-C2-ts1 virus, particles are locked in an immature state and comprised completely of precursor molecules. The ts1 capsids are noninfectious, attributed to a failure to partially become uncoated in the endosome and release the membrane lytic protein VI. While the ts1 mutant provides insight into proteolytic maturation overall, analysis of the key event(s) in the maturation of individual precursor protein has not been carried out. A better understanding of the molecular and functional connections underlying maturation is clearly needed. The studies presented here were designed to address this gap in our knowledge to better understand a key step in the virus life cycle. To that end, we have analyzed a rather subtle glycine-to-alanine point mutation within the N-terminal cleavage site of protein VI that nonetheless disrupts pVI cleavage in vitro and in vivo as well as reducing virus cell entry and assembly. The properties of this mutant are summarized in Table 2. To our knowledge, the VI-G33A HAdV-C5 mutant represents the first example of a single virus proteolytic maturation event linked to both entry and assembly of adenovirus.
TABLE 2.
Summary of G33A cleavage site mutant properties compared to wild typea
| Mutation | In vitro cleavage | Rescue virus? | Protein VI incorporation | Cleavage in capsid | Infectivity | Progeny |
|---|---|---|---|---|---|---|
| WT | N/C | Yes | Normal | VI | +++ | +++ |
| G33A | C | Yes | Normal | iVI/VI | + | + |
In vitro cleavage is evaluated with recombinant protein. N, cleavage at N terminus; C, cleavage at C terminus; WT, wild type. Cleavage in capsid reflects the cleavage state of protein VI in the virus, and nomenclature for the protein species is listed as depicted in Fig. 1A. Infectivity and progeny production are given on a relative scale compared to wild-type virus; + and +++ represent the relative amounts of infectivity and progeny virions produced by the wild-type and G33A viruses.
Our results indicate that residues 1 to 33 of pVI are the primary determinant controlling hexon interactions in solution (outside the viral capsid). This finding is in conflict with the previously reported regions of pVI essential for hexon binding determined by deletion mapping (residues 48 to 74 and 233 to 239) (23). While this earlier approach failed to identify the N-terminal portion of protein VI as being involved in hexon interactions, the N-terminal 6×His tag present on the recombinant pVI in this study may have interfered with pVIn-hexon interactions. Furthermore, it is now clear from both the HAdV crystal structure (14) and hydrogen-deuterium exchange mass spectrometry analyses (12) that pVIn makes extensive contacts with hexon. There is also evidence from the crystal structure that significant portions of mature VI make contacts with hexon, but these residues primarily stabilize hexon-hexon junctions, requiring an assembled capsid to provide the necessary binding sites.
Our results also expand our understanding of AVP-mediated cleavage of protein VI. We observed diminished AVP-mediated processing of immature pVI to mature VI both in our in vitro cleavage assay and in AdV-infected cells for the G33A mutant. The time frame for cleavage in infected cells is much longer than the 30 min (maximum) used for in vitro cleavage in this study. However, even an extended cleavage time of up to 3 h at both room temperature and 37°C yielded no detectable production of mature VI (data not shown). The higher local concentrations and close proximity of AVP and pVI within the virus capsid may favor productive cleavage of pVI to mature VI. Moreover, pVI interaction with viral DNA, hexon, pVIII/VIII, and V in the capsid may present pVI in a more favorable conformation for AVP cleavage than in the in vitro system.
The studies presented here also provide clues into one of the key remaining questions in understanding AdV cell entry: how many molecules of protein VI are required to escape the endosome? AdV particles harbor approximately 360 copies of mature protein VI. The G33A mutant, which harbors ∼50% iVI, is attenuated for infection (a 4-fold decrease with respect to wild-type virus). We surmise that G33A iVI proteins (∼180 copies) are not efficiently released in the endosome, which reduces the pool of membrane lytic protein VI molecules to below the threshold required to efficiently lyse the endosome. This likely traps incoming particles within endocytic compartments and ultimately decreases AdV-mediated transgene delivery.
The single-round infection assay indicates that the G33A mutant is defective for reporter transgene delivery to the nucleus. This assay cannot distinguish between defects in receptor binding, particle internalization, endosome escape, subcellular trafficking, and nuclear translocation of the viral DNA. However, based on the known binding of pVIn to hexon under the acidic conditions similar to those found in the endosome (12), the decrease in G33A virus infection may be linked to defective separation of iVI from hexons. The nearly identical lytic activity of recombinant pVI bearing the G33A mutation rules out the possibility that the mutation itself impairs membrane rupture. The G33A cleavage site mutant virus also offers insight on the necessity of pVI cleavage for particle assembly and maturation. This mutant showed delayed particle production, suggesting that this mutation has a broad-reaching impact on precursor maturation overall. Furthermore, both the virus burst assay and the assembly assay clearly show an increase in total immature particle yields, even when the primary infection is initiated with the same number of infectious particles/cell.
Overall, these results support a model in which the interaction with hexon shields pVI lytic activity during the assembly phase of the AdV life cycle. This prevents premature interaction with membranes, which would damage or even destroy the host cell prior to virus assembly. Upon capsid assembly, AVP cleaves pVI, priming it for appropriate spatiotemporal release during virus uncoating within endocytic vesicles. In the case of the G33A mutant virus, a reduction in protein VI exposure due to the maintenance of hexon binding of the iVI intermediate likely attenuates infectivity. The G33A mutant also impacts virus assembly, reducing the number of infectious particles produced by an infected cell. This is likely due to defects in preprotein maturation. Taken together, our results increase our understanding of the complicated maturational pathway that involves conversion of an immature virus capsid to a mature metastable particle that is primed for host cell infection.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants HL054352 to G.R.N. and 5T32AI007354 to C.L.M.
We thank Vijay Reddy for helpful discussions and for assistance in figure generation.
Footnotes
This is paper no. 29141 from The Scripps Research Institute.
REFERENCES
- 1.Mangel WF, San Martin C. 2014. Structure, function and dynamics in adenovirus maturation. Viruses 6:4536–4570. doi: 10.3390/v6114536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Webster A, Russell S, Talbot P, Russell WC, Kemp GD. 1989. Characterization of the adenovirus proteinase: substrate specificity. J Gen Virol 70:3225–3234. doi: 10.1099/0022-1317-70-12-3225. [DOI] [PubMed] [Google Scholar]
- 3.Weber J. 1976. Genetic analysis of adenovirus type 2 III. Temperature sensitivity of processing viral proteins. J Virol 17:462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson CW. 1990. The proteinase polypeptide of adenovirus serotype 2 virions. Virology 177:259–272. doi: 10.1016/0042-6822(90)90479-B. [DOI] [PubMed] [Google Scholar]
- 5.Rancourt C, Keyvani-Amineh H, Sircar S, Labrecque P, Weber JM. 1995. Proline 137 is critical for adenovirus protease encapsidation and activation but not enzyme activity. Virology 209:167–173. doi: 10.1006/viro.1995.1240. [DOI] [PubMed] [Google Scholar]
- 6.Wodrich H, Guan T, Cingolani G, Von Seggern D, Nemerow G, Gerace L. 2003. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J 22:6245–6255. doi: 10.1093/emboj/cdg614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiethoff CM, Wodrich H, Gerace L, Nemerow GR. 2005. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J Virol 79:1992–2000. doi: 10.1128/JVI.79.4.1992-2000.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wodrich H, Henaff D, Jammart B, Segura-Morales C, Seelmeir S, Coux O, Ruzsics Z, Wiethoff CM, Kremer EJ. 2010. A capsid-encoded PPxY-motif facilitates adenovirus entry. PLoS Pathog 6:e1000808. doi: 10.1371/journal.ppat.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schreiner S, Martinez R, Groitl P, Rayne F, Vaillant R, Wimmer P, Bossis G, Sternsdorf T, Marcinowski L, Ruzsics Z, Dobner T, Wodrich H. 2012. Transcriptional activation of the adenoviral genome is mediated by capsid protein VI. PLoS Pathog 8:e1002549. doi: 10.1371/journal.ppat.1002549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webster A, Hay RT, Kemp G. 1993. The adenovirus protease is activated by a virus-coded disulphide-linked peptide. Cell 72:97–104. doi: 10.1016/0092-8674(93)90053-S. [DOI] [PubMed] [Google Scholar]
- 11.Mangel WF, McGrath WJ, Toledo DL, Anderson CW. 1993. Viral DNA and a viral peptide can act as cofactors of adenovirus virion proteinase activity. Nature 361:274–275. doi: 10.1038/361274a0. [DOI] [PubMed] [Google Scholar]
- 12.Snijder J, Benevento M, Moyer CL, Reddy V, Nemerow GR, Heck AJ. 2014. The cleaved N-terminus of pVI binds peripentonal hexons in mature adenovirus. J Mol Biol 426:1971–1979. doi: 10.1016/j.jmb.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benevento M, Di Palma S, Snijder J, Moyer CL, Reddy VS, Nemerow GR, Heck AJ. 2014. Adenovirus composition, proteolysis, and disassembly studied by in-depth qualitative and quantitative proteomics. J Biol Chem 289:11421–11430. doi: 10.1074/jbc.M113.537498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy VS, Nemerow GR. 2014. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc Natl Acad Sci U S A 111:11715–11720. doi: 10.1073/pnas.1408462111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith JG, Silvestry M, Lindert S, Lu W, Nemerow GR, Stewart PL. 2010. Insight into the mechanisms of adenovirus capsid disassembly from studies of defensin neutralization. PLoS Pathog 6:e1000959. doi: 10.1371/journal.ppat.1000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res 33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans RK, Nawrocki DK, Isopi LA, Williams DM, Casimiro DR, Chin S, Chen M, Zhu DM, Shiver JW, Volkin DB. 2004. Development of stable liquid formulations for adenovirus-based vaccines. J Pharm Sci 93:2458–2475. doi: 10.1002/jps.20157. [DOI] [PubMed] [Google Scholar]
- 18.Rux JJ, Burnett RM. 2007. Large-scale purification and crystallization of adenovirus hexon. Methods Mol Med 131:231–250. doi: 10.1007/978-1-59745-277-9_17. [DOI] [PubMed] [Google Scholar]
- 19.Krey T, d'Alayer J, Kikuti CM, Saulnier A, Damier-Piolle L, Petitpas I, Johansson DX, Tawar RG, Baron B, Robert B, England P, Persson MA, Martin A, Rey FA. 2010. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog 6:e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graziano V, McGrath WJ, Suomalainen M, Greber UF, Freimuth P, Blainey PC, Luo G, Xie XS, Mangel WF. 2013. Regulation of a viral proteinase by a peptide and DNA in one-dimensional space: I. Binding to DNA and to hexon of the precursor to protein VI, pVI, of human adenovirus. J Biol Chem 288:2059–2067. doi: 10.1074/jbc.M112.377150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varghese R, Mikyas Y, Stewart PL, Ralston R. 2004. Postentry neutralization of adenovirus type 5 by an antihexon antibody. J Virol 78:12320–12332. doi: 10.1128/JVI.78.22.12320-12332.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moyer CL, Nemerow GR. 2012. Disulfide-bond formation by a single cysteine mutation in adenovirus protein VI impairs capsid release and membrane lysis. Virology 428:41–47. doi: 10.1016/j.virol.2012.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matthews DA, Russell WC. 1995. Adenovirus protein-protein interactions: molecular parameters governing the binding of protein VI to hexon and the activation of the adenovirus 23K protease. J Gen Virol 76:1959–1969. doi: 10.1099/0022-1317-76-8-1959. [DOI] [PubMed] [Google Scholar]
- 24.Moyer CL, Wiethoff CM, Maier O, Smith JG, Nemerow GR. 2011. Functional genetic and biophysical analyses of membrane disruption by human adenovirus. J Virol 85:2631–2641. doi: 10.1128/JVI.02321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier O, Wiethoff CM. 2010. N-terminal alpha-helix-independent membrane interactions facilitate adenovirus protein VI induction of membrane tubule formation. Virology 408:31–38. doi: 10.1016/j.virol.2010.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maier O, Galan DL, Wodrich H, Wiethoff CM. 2010. An N-terminal domain of adenovirus protein VI fragments membranes by inducing positive membrane curvature. Virology 402:11–19. doi: 10.1016/j.virol.2010.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graziano V, Luo G, Blainey PC, Perez-Berna AJ, McGrath WJ, Flint SJ, San Martin C, Xie XS, Mangel WF. 2013. Regulation of a viral proteinase by a peptide and DNA in one-dimensional space: II. Adenovirus proteinase is activated in an unusual one-dimensional biochemical reaction. J Biol Chem 288:2068–2080. doi: 10.1074/jbc.M112.407312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen EK, Nemerow GR, Smith JG. 2010. Direct evidence from single-cell analysis that human alpha-defensins block adenovirus uncoating to neutralize infection. J Virol 84:4041–4049. doi: 10.1128/JVI.02471-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greber UF, Willetts M, Webster P, Helenius A. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477–486. doi: 10.1016/0092-8674(93)90382-Z. [DOI] [PubMed] [Google Scholar]
- 30.Luisoni S, Suomalainen M, Boucke K, Tanner LB, Wenk MR, Guan XL, Grzybek M, Coskun U, Greber UF. 2015. Co-option of membrane wounding enables virus penetration into cells. Cell Host Microbe 18:75–85. doi: 10.1016/j.chom.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Smith JG, Nemerow GR. 2008. Mechanism of adenovirus neutralization by human alpha-defensins. Cell Host Microbe 3:11–19. doi: 10.1016/j.chom.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 32.Rexroad J, Wiethoff CM, Green AP, Kierstead TD, Scott MO, Middaugh CR. 2003. Structural stability of adenovirus type 5. J Pharm Sci 92:665–678. doi: 10.1002/jps.10340. [DOI] [PubMed] [Google Scholar]
- 33.Russell WC, Valentine RC, Pereira HG. 1967. The effect of heat on the anatomy of the adenovirus. J Gen Virol 1:509–522. doi: 10.1099/0022-1317-1-4-509. [DOI] [PubMed] [Google Scholar]
- 34.Perez-Berna AJ, Ortega-Esteban A, Menendez-Conejero R, Winkler DC, Menendez M, Steven AC, Flint SJ, de Pablo PJ, San Martin C. 2012. The role of capsid maturation on adenovirus priming for sequential uncoating. J Biol Chem 287:31582–31595. doi: 10.1074/jbc.M112.389957. [DOI] [PMC free article] [PubMed] [Google Scholar]