ABSTRACT
Despite the recent development of highly effective anti-hepatitis C virus (HCV) drugs, the global burden of this pathogen remains immense. Control or eradication of HCV will likely require the broad application of antiviral drugs and development of an effective vaccine. A precise molecular identification of transmitted/founder (T/F) HCV genomes that lead to productive clinical infection could play a critical role in vaccine research, as it has for HIV-1. However, the replication schema of these two RNA viruses differ substantially, as do viral responses to innate and adaptive host defenses. These differences raise questions as to the certainty of T/F HCV genome inferences, particularly in cases where multiple closely related sequence lineages have been observed. To clarify these issues and distinguish between competing models of early HCV diversification, we examined seven cases of acute HCV infection in humans and chimpanzees, including three examples of virus transmission between linked donors and recipients. Using single-genome sequencing (SGS) of plasma vRNA, we found that inferred T/F sequences in recipients were identical to viral sequences in their respective donors. Early in infection, HCV genomes generally evolved according to a simple model of random evolution where the coalescent corresponded to the T/F sequence. Closely related sequence lineages could be explained by high multiplicity infection from a donor whose viral sequences had undergone a pretransmission bottleneck due to treatment, immune selection, or recent infection. These findings validate SGS, together with mathematical modeling and phylogenetic analysis, as a novel strategy to infer T/F HCV genome sequences.
IMPORTANCE Despite the recent development of highly effective, interferon-sparing anti-hepatitis C virus (HCV) drugs, the global burden of this pathogen remains immense. Control or eradication of HCV will likely require the broad application of antiviral drugs and the development of an effective vaccine, which could be facilitated by a precise molecular identification of transmitted/founder (T/F) viral genomes and their progeny. We used single-genome sequencing to show that inferred HCV T/F sequences in recipients were identical to viral sequences in their respective donors and that viral genomes generally evolved early in infection according to a simple model of random sequence evolution. Altogether, the findings validate T/F genome inferences and illustrate how T/F sequence identification can illuminate studies of HCV transmission, immunopathogenesis, drug resistance development, and vaccine protection, including sieving effects on breakthrough virus strains.
INTRODUCTION
Hepatitis C virus (HCV) infects 175,000,000 people worldwide and is a major cause of morbidity and mortality (1, 2). In the United States, HCV now exceeds human immunodeficiency virus (HIV-1) as a cause of death. Recent advances in the development of interferon sparing, direct-acting antiviral agents (DAA) suggest that most treated patients can be cured, but given the limited access and high cost of DAA on a global scale, prevention of infection by effective vaccination remains the best hope for virus eradication. However, in contrast to drug development, progress in vaccine development has been slow, in large part due to the extraordinary genetic diversity and rapid sequence evolution of the virus (3–7).
Globally, HCV is represented by seven major genotypes (1 to 7) that exhibit nucleotide sequence diversity of as much as 30% (8). Within individual infected subjects, HCV exists as a mixture of innumerable genetically distinct variants (6). It is estimated that up to 1012 virions are produced daily in a typical infected individual (9), and based on an RNA-dependent RNA polymerase error rate of ∼2.5 × 10−5 per nucleotide per replication cycle (10), most of these are expected to be unique. In fact, every possible single point mutation, as well as every possible combination of two mutations, across the 10-kb viral genome are predicted to be generated every day (11). This explains the rapid appearance of resistance mutations to DAAs and the virus's capacity to evade host adaptive immune responses.
Against this backdrop of nearly unfathomable viral diversity is substantial evidence indicating that HCV exhibits a relatively stringent population bottleneck at the moment of transmission from one individual to the next (12–14). Such a transmission bottleneck is of clinical importance because at this point in viral natural history the infection is most vulnerable to treatment and prevention measures, including, potentially, vaccination (5, 15–17). A virus bottleneck at transmission was initially inferred from clinical epidemiological studies that correlated risk of infection to the volume of blood exposure and the route of exposure (e.g., blood transfusion, injection drug use, needlestick injury, or mucosal inoculation, especially in HIV-1-positive men who have sex with men [1, 18–23]). More refined estimates of the transmission bottleneck came from studies using a variety of increasingly sensitive and specific molecular techniques to characterize viral diversity, including oligonucleotide heteroduplex gel shift, vRNA/cDNA population sequencing, and molecular cloning, followed by Sanger sequencing and next-generation deep sequencing (12, 13, 24–32). These reports described the HCV transmission bottleneck in both qualitative and quantitative terms, but none allowed for a precise identification of transmitted or founder virus genomes. The most exacting quantitative estimates of the transmission bottleneck thus far came from studies based on single-genome sequencing (SGS), mathematical modeling, and phylogenetic inference (10, 14, 33–35). This approach, which was adapted from HIV-1 infection studies (36–39), theoretically allows for an unambiguous inference and quantitative estimation of transmitted/founder (T/F) viral genomes that are responsible for establishing productive clinical infection. The conceptual and mathematical models underlying this strategy assume a virus population emanating from each T/F genome that expands exponentially with no recombination, no selection pressure on variable sites, and a constant mutation rate across lineages (36, 40, 41). Under these conditions, the frequency distribution of the Hamming distances (i.e., the pairwise distance between genetic strains) is given by a Poisson distribution whose mean depends linearly on the number of generations since the founder strain (36, 40, 41). If the expansion is sufficiently rapid, small samples of sequences exhibit a star-like phylogeny whose coalescent corresponds to the T/F genome. For this situation, an analysis by forward simulation suffices, and a Bayesian inference (e.g., BEAST [42, 43]), which is a backward simulation of genealogies, provides no advantages.
Previously, we used the forward simulation model to analyze HCV sequences from 17 subjects in the very earliest stages of acute infection (weeks before HCV antibody seroconversion). Sequences from 13 subjects conformed well to model predictions, showing a star-like phylogeny and a Poisson distribution of mutations within one or more distinct and well-separated lineages, each with low diversity (14). We interpreted these low-diversity sequence lineages to represent the progeny of discrete T/F genomes, thereby allowing for the identification and enumeration of the T/F genomes that resulted in productive clinical infection. In these 13 study subjects, the estimates of numbers of T/F genomes ranged from 1 to 13 per individual with a mean of 4 and a median of 3 (14). In the same study, however, viral sequences from four other acutely infected subjects (subjects 10003, 10016, 10020, and 106889) appeared to violate this forward simulation model by exhibiting multiple closely related lineages that failed to conform to a Poisson distribution of mutations or star-like phylogeny. Some lineages differed from others by as few as one to three informative sites (shared mutations) out of the 5,000 bases sequenced. For these patients, our estimates for the numbers of T/F genomes were as high as 30 or more. We considered two possible explanations for these outlier results: either early HCV diversification in these subjects failed to follow a simple pattern of random diversification, creating multiple distinct but closely related lineages early on because of features unique to the HCV replication schema confounded by other unknown virus-host factors, or viral sequences in the donors to these four cases had undergone a population diversity bottleneck event prior to the transmission of multiple closely related variants, which then evolved as expected by the forward simulation model. Such a bottlenecking event in the donors could occur as a result of antiviral treatment, immunological selection or very recent acquisition of virus (i.e., the donors were themselves acutely infected).
To explore these possibilities, we developed a different forward simulation model of early virus diversification that accounts for essential differences in replication dynamics between HCV and HIV-1. Based on the patterns of shared mutations observed in these forward simulations, we developed a phylogenetically based clustering method that counts the number of putative T/F genomes (14; http://www.santafe.edu/~tanmoy/programs/HCV/). This approach thus incorporates the important features of HCV's unique cytosolic life cycle, where many replication complexes continuously produce virus from long-lived hepatocytes (10, 44, 45). Because of the sequential creation of as many as 40 replication complexes per cell (46), those at the same generation depth have widely varying numbers of descendants, unlike the situation in HIV-1 infection. This leads to two predictions that differ substantially from the HIV-1 case: (i) sequence diversity of HCV saturates around the same time during primary infection that viral load stabilizes, instead of growing linearly in time as is the case for HIV-1, and (ii) there is an expectation of about three times as many stochastically shared mutations in HCV compared to HIV-1 infection when sampled in this stable phase, leading to more common deviations from a star-like phylogeny and Poisson distribution of mutations (10, 14).
When the second model was applied to sequences from subjects 10003, 10016, 10020, and 106889, it still failed to account fully for the atypical sequence diversification patterns and large numbers of potential T/F lineages observed in these subjects (14). We thus sought alternative explanations for the atypical early viral diversity, including the possibility of a pretransmission population bottleneck, followed by high-multiplicity virus transmission. In one case (subject 106889), we found evidence for such a bottleneck in the donor virus population due to prior treatment with a protease inhibitor (14). For subjects 10003, 10016, and 10020, however, there was no evidence of a drug-induced bottleneck, so we instead hypothesized that the donors for these subjects might themselves have been acutely infected or their viruses subjected to stringent immunological selection (32; unpublished data). In this scenario, the transmission of multiple viruses would be expected to yield a diversity pattern in the recipient comprised of very closely related viral lineages in addition to more distantly related lineages. In Africa, it is estimated that “acute-to-acute” infection is responsible for as many as 25% of incident HIV-1 cases (47–49). This can result in a pretransmission virus population bottleneck in the donor and acquisition of closely related viruses by the recipient (36, 38). Unfortunately, in our previous HCV studies (14), we had no documented cases of “acute-to-acute” virus transmission to evaluate this scenario. So, in the present study, we sought new examples of epidemiologically linked, sequence-confirmed, acute-to-acute HCV transmission.
Here, we analyze early patterns of HCV sequence diversity in epidemiologically linked, viral sequence confirmed, donor-recipient, acute-to-acute transmission pairs, including human-to-human and human-to-chimpanzee infections. We tested two hypotheses: first, that HCV genomes responsible for transmission to a naive host can be identified unambiguously in paired donor-recipient plasma samples and that these sequences will be identical or nearly so, and second, that high-multiplicity infection of a subject with plasma from a donor who is acutely infected (and thus harbors sequences exhibiting a pretransmission population bottleneck) will recapitulate patterns of virus diversity observed in subjects 10003, 10016, and 10020. The results we obtained affirm both hypotheses. In so doing, they validate the strategy of T/F genome identification by SGS, mathematical modeling, and phylogenetic inference for HCV, and they provide a plausible explanation for what had previously been a confusing set of HCV sequence data in a subset of acutely infected subjects. Our findings indicate that simple models of random virus diversification can generally explain early HCV evolution, although models that account for unique features of the HCV replication strategy offer a more conservative estimate of T/F genome numbers in samples that fail to conform to a star-like phylogeny and a Poisson distribution of mutations. Our results illustrate how T/F analysis in HCV infection, much like T/F analysis in HIV-1 infection, represents a novel experimental strategy that can be used to molecularly anchor and uniquely inform studies of HCV natural history, immunopathogenesis, treatment, and prevention.
MATERIALS AND METHODS
Ethics statement.
The present study was conducted according to the principles expressed in the Declaration of Helsinki. It was approved by the Institutional Review Boards of the University of Pennsylvania and Mt. Sinai Medical Center, New York, NY. Subjects provided written informed consent for the collection of blood samples and subsequent analyses. Chimpanzee specimens were collected and stored as part of a previously conducted study (50), and thus the present research is in full compliance with National Institutes of Health (NIH) policies (NOT-OD-12-025) concerning biomedical research involving chimpanzees.
Study subjects.
Plasma samples were obtained from human subjects with acute HCV infection. The subjects either were regular source plasma donors (ZeptoMetrix, Inc.; SeraCare, Inc.) who were HCV and HIV-1 antibody negative and became HCV infected sometime in the course of their twice-weekly plasma donations, as evidenced by the development of HCV viremia on sequential viral RNA testing, or they were HCV antibody-negative patients who became acutely infected with HCV and sought medical consultation. Chimpanzee inoculations with HCV-infected human plasma, subsequent specimen collections, and analysis of plasma virus load and liver specific transaminase levels were previously reported (50).
HCV RNA and antibody assays.
Plasma samples were tested for HCV RNA and antibodies by a battery of commercial tests. These included the Roche Molecular Systems COBAS Amplicor HCV Monitor (v2.0) assay, the Abbott Anti-HCV 3.0 assay, and the Ortho Anti-HCV 3.0 enzyme-linked immunosorbent assay. Additional vRNA analyses using the transcription-mediated amplification (TMA) method were performed as described previously (50).
Viral RNA extraction and cDNA synthesis.
For each plasma sample, approximately 100,000 viral RNA copies were extracted using the Qiagen BioRobot EZ1 Workstation with an EZ1 virus minikit v2.0 (Qiagen, Valencia, CA). RNA was eluted and immediately subjected to cDNA synthesis. Reverse transcription of RNA to single-stranded cDNA was performed using MuLV (SuperScript III) reverse transcriptase (RT; Invitrogen Life Technologies, Carlsbad, CA) according to the manufacturer's instructions. Briefly, each cDNA reaction included 1× RT buffer, 0.5 mM concentrations of each deoxynucleoside triphosphate, 5 mM dithiothreitol, 2 U of RNaseOUT (an RNase inhibitor)/μl, 10 U of SuperScript III reverse transcriptase/μl, and 0.25 μM antisense primer. The cDNA primer used for amplifying each genome fragment corresponded to the first-round antisense primer listed in the single-genome amplification section below. The reverse transcription reaction was carried out at 50°C for 60 min, followed by an increase in temperature to 55°C for an additional 60 min. The reaction was then heat inactivated at 70°C for 15 min and then treated with 0.1 U of RNase H/μl at 37°C for 20 min. The newly synthesized cDNA was used immediately or kept frozen at −80°C.
Single-genome amplification.
The sequences from all of the subjects were generated using the SGS method previously described (14). Nearly full length, 5′ half, 3′ half, or partial NS2, NS3, and NS4A genomes were amplified for each subject by nested or seminested PCR, respectively. cDNA was serially diluted and distributed among wells of replicate 96-well plates (Applied Biosystems, Foster City, CA) so as to identify a dilution where PCR-positive wells constituted less than 30% of the total number of reactions. At this dilution, most wells contain amplicons derived from a single cDNA molecule. This was confirmed in every positive well by direct sequencing of the amplicon and inspection of the sequence for mixed bases (double peaks), which would be evidence of priming from more than one original template or the introduction of PCR error in early cycles. Any sequence with evidence of mixed bases was excluded from further analysis. PCR amplification was carried out in the presence of 1× high-fidelity Platinum PCR buffer, 2 mM MgSO4, 0.2 mM concentrations of each deoxynucleoside triphosphate, 0.2 μM concentrations of each primer, and 0.025 U of Platinum Taq high-fidelity polymerase/μl in a 20-μl reaction (Invitrogen, Carlsbad, CA). Nearly full length, 5′ half or partial NS2, NS3, and NS4A genomes were amplified. The primers used were as follows: (i) subject 110069, first-round sense primer 1a.Core.F1 (5′-GCACGAATCCTAAACCTCAAAGAAAAA-3′; nucleotides [nt] 346 to 372, H77), first-round antisense primer 1aNS5Bend.R1a (5′-CCGGAGTGTTTATCCCAACCTTCAT-3′; nt 9374 to 9398, H77), second-round sense primer 1a.Core.F2 (5′-CGGGTGGCGGTCAGATCGTTGGTGGAGTTTA-3′; nt 415 to 445, H77), and second-round antisense primer 110069.NS5B.R2a (5′-GCGGCCGCTATTGGAGTGAGTTTGAG-3′; nt 9201 to 9226, H77); (ii) BGI and CCI, first-round sense primer 1b.Core.F1 (5′-ATGAGCACGAATCCTAAACCTCAAAGA-3′; nt 342 to 368, H77), first-round antisense primer NS4A_R1 (5′-GCACTCTTCCATCTCATCGAACTC-3′; nt 5451 to 5474, H77), second-round sense primer 1b.Core.F2 (5′-TCAAAGAAAAACCAAACGTAACACCAACCG-3′; nt 362 to 391, H77) and second-round antisense primer NS4A_R2 (5′-AGGTGCTCGTGACGACCTCCAGG-3′; nt 5297 to 5319, H77); (iii) subjects 10081 and X355, first-round sense primer 10081NS2_F2 (5′-ACCCGACCCTGATATTTGATATCACC-3′; nt 2983 to 3008, H77), first-round antisense primer 1aNS4B.R3 (5′-TATTGTATCCCACTGATGAAGTTCCACAT-3′; nt 5634 to 5662, H77), second-round sense primer 10081NS2_F3 (5′-AAAGTACCCTACTTTGTGCGCGT-3′; nt 3063 to 3085, H77), and second-round antisense primer 1aNS4B.R4 (5′-AGGGCCTTCTGCTTGAACTGCTC-3′; nt 5517 to 5539, H77); and (iv) subjects 10083 and X331, first-round sense primer 10083STF1 (5′-GACATCACTAAGCTGCTGATAG-3′, first-round antisense primer 3aV2R2 (5′-TTACTTCCAGATCAGCTGACA-3′), second-round sense primer new10083STF2 (5′-CCCGTTATATTTAATACAGGCTA-3′), and second-round antisense primer 3aV2R2 (5′-TTACTTCCAGATCAGCTGACA-3′).
DNA sequencing.
PCR amplicons were directly sequenced by cycle-sequencing using BigDye Terminator chemistry and protocols recommended by the manufacturer (Applied Biosystems). Sequencing reaction products were analyzed with an ABI 3730xl genetic analyzer (Applied Biosystems). Both DNA strands were sequenced using partially overlapping fragments. Individual sequence fragments for each amplicon were assembled and edited using the Sequencher program 5.0 (Gene Codes; Ann Arbor, MI). Inspection of individual chromatograms allowed for the identification of amplicons derived from single versus multiple templates. The absence of mixed bases at each nucleotide position was taken as evidence of amplification from a single viral RNA/cDNA template. This quality control measure enabled us to exclude from the analysis amplicons that resulted from PCR-generated in vitro recombination events or Taq polymerase errors and to obtain multiple individual sequences that proportionately represented those circulating HCV virions.
Sequence alignments.
All of the sequences alignments were initially made with CLUSTAL W and then hand-checked using Geneious 7.1.9 to improve the alignments according to the codon translation.
Sequence diversity analysis.
Eight hundred three quarter genomes, 103 5′ half genomes and 114 near full-length genomes were generated by SGS from seven human and chimpanzee subjects for the diversity analysis. Among the 1020 amplicons generated, the sequences of 928 were unambiguous at every position. Nine near-full length genomes had mixed bases at 1 to 2 positions per sequence. We inferred that these mixed bases resulted from Taq polymerase errors in the early PCR cycles because the mixed bases represented only a subset of the polymorphisms in any one sequence. These sequences were retained in the analysis and mixed bases interpreted as lineage consensus nucleotides. The other 83 amplicons contained “double peaks,” clearly resulting from amplification from more than one template, and these sequences were excluded from the analysis. Sequences were analyzed using phylogenetic tree analysis together with a sequence visualization tool, Highlighter (www.HIV.lanl.gov), that allows tracing of common ancestry between sequences based on individual nucleotide polymorphisms. Phylogenetic trees were generated by maximum-likelihood methods using PhyML (51). For subjects productively infected by more than one T/F virus, lineages containing 3 or more closely related sequences were included in the lineage diversity analyses.
Mathematical models and algorithms for estimating numbers of T/F variants.
Two mathematical models were used to analyze early HCV sequence diversity. One was a forward simulation of early viral expansion (36, 40, 41), and the second, a phylogenetic-based clustering method that accounts for essential differences in replication dynamics and replication mechanisms HCV and HIV-1 (14; http://www.santafe.edu/~tanmoy/programs/HCV/).
Statistical analysis.
Lineage-specific sequences were analyzed by the Poisson Fitter program (www.HIV.lanl.gov), which computes the best fitting Poisson distribution through Maximum Likelihood, performs a Goodness of Fit test, and tests for star-phylogeny. Power calculations to estimate the likelihood of detecting rare sequence variants based on sampling depth were performed using methods previously described (see Fig. S9 and the associated description in reference 36).
GenBank accession numbers.
All sequences were deposited in GenBank under accession numbers KT733674 to KT734610.
RESULTS
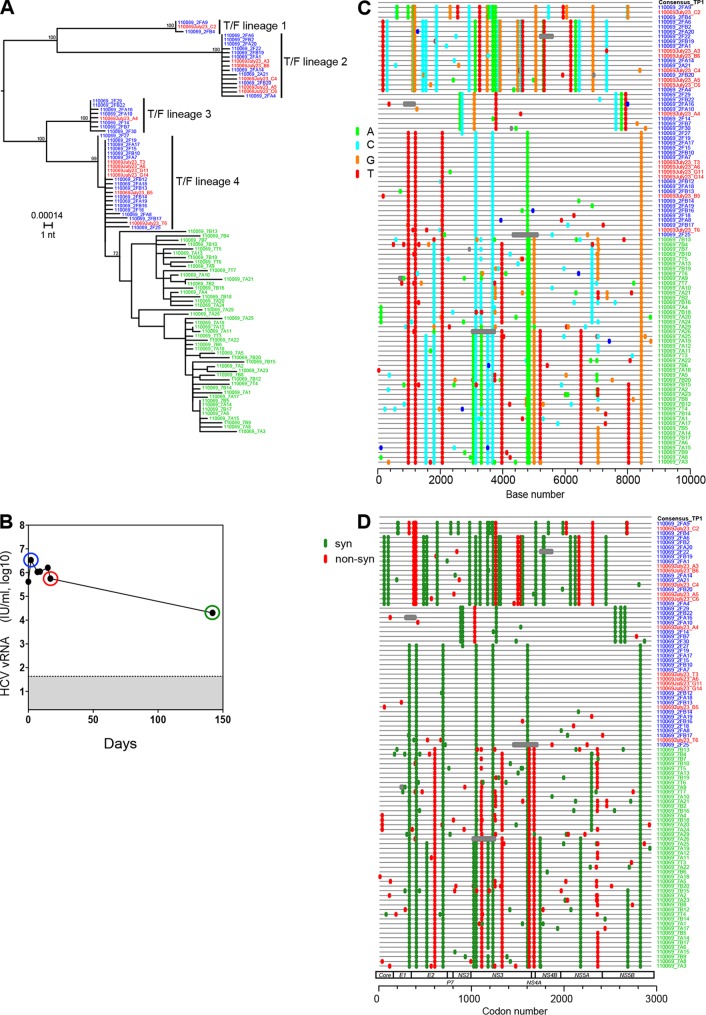
Acute HCV infection has historically been defined as the first 6 months of infection, which reflects the natural history and immunopathogenesis of disease where a minority of individuals spontaneously resolves the infection by this point, while most patients, if left untreated, exhibit persistent viremia (1, 18, 52). Whether or not an individual clears virus depends on host immunogenetics, innate and adaptive immune responses, and viral genotype, as well as other factors (1, 53–55). Figure 1 illustrates viral dynamics and diversity of plasma vRNA/cDNA sequences amplified as nearly full length (8.8 kb) genomic amplicons in a typical subject (e.g., 110069) experiencing acute (primary) HCV infection. In this case, overall viral diversity during the acute infection period was dictated far more by the number of transmitted viruses and the genetic distances among them (0.15 to 0.47%) than by early sequence evolution, which can undergo positive or negative selection, as well as population diversity bottlenecking (13, 14). Very early in acute infection prior to antibody seroconversion, sequences appeared as four readily distinguishable, low diversity lineages (T/F lineages 1 to 4). Within each lineage, sequences differed by no more than 3 nucleotides (nt) out of nearly 9,000 nt (0.03%), and exhibited a star-like phylogeny and a Poisson distribution of mutations (Table 1). Furthermore, the diversity within each lineage was roughly the same, as would be expected if all of the lineages had been diverging at about the same rate and for the same time. The pattern within each lineage conformed to a simple model of random virus evolution and is typical of most acute HCV infections (14). We inferred from these data that the coalescent of each low-diversity lineage corresponded to a distinct T/F genome in total numbering four. This is a minimum estimate because deeper sequencing could potentially detect variants present at lower abundance. In this case, 48 sequences from days 2 and 17 following the first plasma vRNA positive time point were initially analyzed. Power calculations (36) predict that at this level of sampling, there is greater than a 90% likelihood of detecting minor variants present at a frequency of 5%. However, in data not shown or reported elsewhere (33), we used other primer sets and plasma RNA samples from early time points to generate 228 additional overlapping 5′ and 3′ half genomes from this subject, giving a total of 276 sequences evaluated for diversity and phylogeny. At this level of sampling, power calculations indicate a >90% likelihood of detecting minor variant lineages present at a frequency of 1%. No additional sequence lineages were detected. Still, these are minimum estimates for the number of T/F genomes, and we cannot exclude the possibility that a sequence such as 110069_2F27 (situated between variants 3 and 4 in panel A) represents a distinct T/F genome or is an example of homoplasy. Nor can we exclude the possibility that next-generation deep sequencing could detect additional T/F lineages present at exceedingly low frequencies, although this has generally not been seen in acute HIV-1 infection (56, 57).
FIG 1.
HCV sequence diversity in acutely infected human subject 110069. A maximum-likelihood (ML) mid-point rooted phylogenetic tree (A) and Highlighter plots (C and D) of nearly full length plasma vRNA sequences spanning the initial 6-month infection period are depicted. Plasma viral load kinetics (B) are also depicted, and the sequences are color coded to correspond to sampling time points determined from the first vRNA-positive/antibody-negative time point (blue, day 2; red, day 17; green, day 142). One month earlier, the HCV vRNA and antibody tests were negative. HCV antibody tests were negative through day 17 but were positive at the next sampled time point on day 146. The tree and the Highlighter plot reveal productive clinical infection by four T/F viruses (T/F lineages 1 to 4). Viral diversity is indicated by the scale bar and bootstrap values are shown. In the Highlighter (C), tick marks are color coded to indicate nucleotide substitutions by A (green), T (red), C (blue), or G (orange) compared to the consensus sequence of the first time point (TP1). In panel D, tick marks denote synonymous (green) or nonsynonymous (red) substitutions.
TABLE 1.
Statistical analysis of Hamming distance frequency distribution and star-like phylogenya
| Sampleb | Lambda |
No. of sequences | No. of bases | HD |
Chi-squared value | DF | GOF (P)c | Star-like phylogeny | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | Maximum | |||||||
| 110069TF1 | 0.6667 | 0.2096 | 3 | 8,807 | 0.67 | 1 | 0.33520265 | 1 | 0.5626 | Yes |
| 110069TF2 | 2.533 | 0.3348 | 15 | 8,807 | 2.5 | 5 | 3.15626799 | 3 | 0.3682 | Yes |
| 110069TF3 | 1.75 | 0.3164 | 8 | 8,807 | 1.8 | 3 | 2.56564491 | 2 | 0.2773 | Yes |
| 110069TF4 | 1.81 | 0.2995 | 22 | 8,807 | 1.8 | 6 | 2.48810961 | 3 | 0.4774 | Yes |
| 110069 (late) | 11.86 | 0.5566 | 47 | 8,807 | 12 | 26 | 134,011.48 | 18 | 2.00E–16 | No |
| Pt1 (BGI) | 2.849 | 0.3567 | 28 | 4,905 | 2.8 | 8 | 5.42062263 | 5 | 0.3667 | Yes |
| Pt2 (CCI) | 2.497 | 0.2119 | 52 | 5,059 | 2.5 | 6 | 3.35842836 | 3 | 0.3396 | Yes |
| X355 (all) | 0.6286 | 0.04752 | 140 | 2,398 | 0.63 | 4 | 0.94155315 | 2 | 0.6245 | Yes |
| 10083 (all) | 4.577 | 0.2096 | 146 | 2,230 | 4.6 | 19 | 7,600.11339 | 13 | 2.00E–16 | No |
| X331 (all) | 4.276 | 0.113 | 292 | 2,230 | 4.3 | 18 | 2,270 | 13 | 2.00E–16 | No |
Poisson-Fitter (www.hiv.lanl.gov) computes the Poisson distribution by maximum-likelihood and Chi-square goodness-of-fit analyses and tests for star-like phylogeny. HD, Hamming distance; DF, degrees of freedom.
The sequences from subject 10081 exceeded the diversity limits of the Poisson Fitter and thus did not conform to a Poisson distribution or a star-like phylogeny. Pt1, patient 1; Pt2, patient 2.
A goodness-of-fit (GOF) P value of >0.05 indicates a nonsignificant divergence from a Poisson distribution.
Approximately 5 months into infection, the pattern of sequence diversity in subject 110069 plasma was much different than at earlier time points (Fig. 1). There was evidence of a stringent population genetic bottleneck between the second and third sampling time points (days 17 and 142 in Fig. 1B), as described previously in other acutely infected subjects (13). The closer similarity of the 5-month sequences to T/F lineage 4 sequences (0.1% mean diversity) compared to T/F lineages 1 (0.5% mean diversity), 2 (0.6% mean diversity), or 3 (0.3% mean diversity) suggested that the 5 month sequences had all evolved from the T/F 4 lineage, with the descendants of T/F lineages 1 to 3 lost in a population/diversity bottleneck. Later sequences emanating from the T/F 4 lineage contained many shared mutations violating both a star-like phylogeny and a Poisson distribution (Table 1). Figure 1 (panel D) suggests that the population bottleneck likely resulted from immune selection since the highly selected nonsynonymous mutations in E2 (see vertical red stripe in Fig. 1D) map to a previously identified cytotoxic T lymphocyte (CTL) epitope (58) and are adjacent to a contact residue of the broadly neutralizing antibody AR3C (59). In addition, the three strongly selected nonsynonymous mutations in NS3 map to previously identified CTL epitopes (58, 60, 61). Unfortunately, peripheral blood mononuclear cells were not available from this subject for human leukocyte antigen analysis or phenotypic testing to confirm phenotypic T cell recognition and escape of these and other potential epitopes. The patterns of sequence evolution observed here for subject 110069 illustrate how high-multiplicity transmission of very early acute infection viruses or viruses arising following an immune-mediated population bottleneck could lead to sequence patterns in a recipient that resemble what was seen in the earlier study's acutely infected outlier subjects 10003, 10016, and 10020 (14).
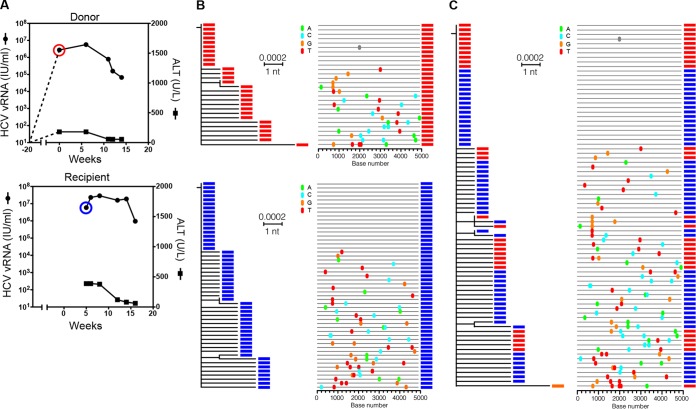
To further test the validity of HCV T/F genome inferences, we next studied epidemiologically linked human-to-human and human-to-chimpanzee donor-recipient HCV transmission pairs in an attempt to document transmission of actual virus genomes and to study their subsequent evolution. Figure 2 illustrates plasma vRNA kinetics and 80 5′ half genome sequences from two human subjects (patient 1 [BGI], donor; patient 2 [CCI], recipient), who became acutely infected with HCV at about the same time. These two individuals were HIV-1-positive men who have sex with men, who were regular sexual partners, who denied injection drug use (IDU), and who were HCV antibody negative at the time of first vRNA detection. The clinical history indicated that patient 1 first became symptomatic with acute HCV infection (fatigue, jaundice, elevated liver transaminases) leading to HCV screening of himself and his partner patient 2 by viral RNA assay. Both individuals were confirmed to be positive for HCV RNA and negative for HCV antibody. Figure 2 shows that patient 1 and patient 2 each harbored single low-diversity HCV sequence lineages that exhibited a near star-like phylogeny and a Poisson distribution of mutations (Table 1), indicating that each had been productively infected by a single T/F virus. Surprisingly, when the viral sequences from these individuals were analyzed together, they were found to be identical or nearly so. Whether the sequences were analyzed separately or together, they coalesced phylogenetically to the same T/F virus genome (Fig. 2). To be certain that a sample mix-up had not occurred, which could explain the identity or near identity of their sequences, five additional plasma samples taken from these subjects in subsequent weeks were analyzed. A total of 162 sequences from patients 1 and 2 spanning a 15-week period all coalesced to the same single T/F genome. This confirmed that donor and recipient had both been productively infected by viruses containing an identical 5′ half-genome sequence. Given the clinical history, the most plausible explanation for this result is that patient 1 became acutely infected by a single T/F virus, and very early in his acute infection period, when most of his circulating virus still contained no mutations, he transmitted a single virus to patient 2. A less likely scenario is that patient 1 and patient 2 each acquired their virus from a third individual. This interpretation was not consistent with the clinical history and would require that this third individual also be acutely infected so as to transmit identical viruses to two partners. Either way, acquisition of viruses containing identical genomes by patient 1 and patient 2 corroborates the T/F genome strategy and the conclusion that early virus evolution generally follows a simple model of random sequence diversification.
FIG 2.
HCV plasma vRNA kinetics and 5′ half genome HCV sequences from an epidemiologically linked human HCV transmission pair. (A) Viral load profiles in the presumed donor (patient 1 [BGI]) and recipient (patient 2 [CCI]) are illustrated. The sampling time points are circled and color coded red (donor) and blue (recipient). (B) ML phylogenetic trees and Highlighter plots show that each subject was productively infected by a single virus. (C) The combined tree and plot show that the T/F viral genomes infecting both subjects were identical in sequence.
We next sought to evaluate the T/F strategy in situations of low and high multiplicity infection from a transmitting donor who was acutely infected. This was done to determine whether HCV sequence patterns similar to those found in subjects 10003, 10016, and 10020 could be recapitulated and explained on the basis of a pretransmission virus population bottleneck. Because we had no access to additional epidemiologically linked human-to-human transmission pairs, we studied HCV transmission in the closely related human-to-chimpanzee infection model. Biomedical research in chimpanzees is now restricted, so we analyzed stored plasma samples from a previous study by Busch et al. (50) that was designed to evaluate the clinical infectivity of human plasma sampled from the earliest phases of infection before plasma HCV vRNA could be detected by U.S. Food and Drug Administration (FDA)-licensed diagnostic assays. In those experiments, closely spaced sequential plasma samples from individuals who subsequently became acutely infected by HCV were inoculated into virus-naive chimpanzees to determine whether they contained infectious HCV. This study plan provided an ideal experimental design and sets of well-pedigreed frozen plasma samples from linked “donors” and “recipients” to test the concept of T/F genome identification under conditions of low- and high-multiplicity infections. The human donors for these studies were HCV antibody-negative source plasma donors who had donated plasma once or twice weekly for many months until, unexpectedly, they were found to have become positive for plasma HCV RNA. In both human subjects whom we studied, plasma HCV RNA increased rapidly and plateaued at high levels, followed weeks later by HCV antibody seroconversion in patterns typical of acute (primary) HCV infection.
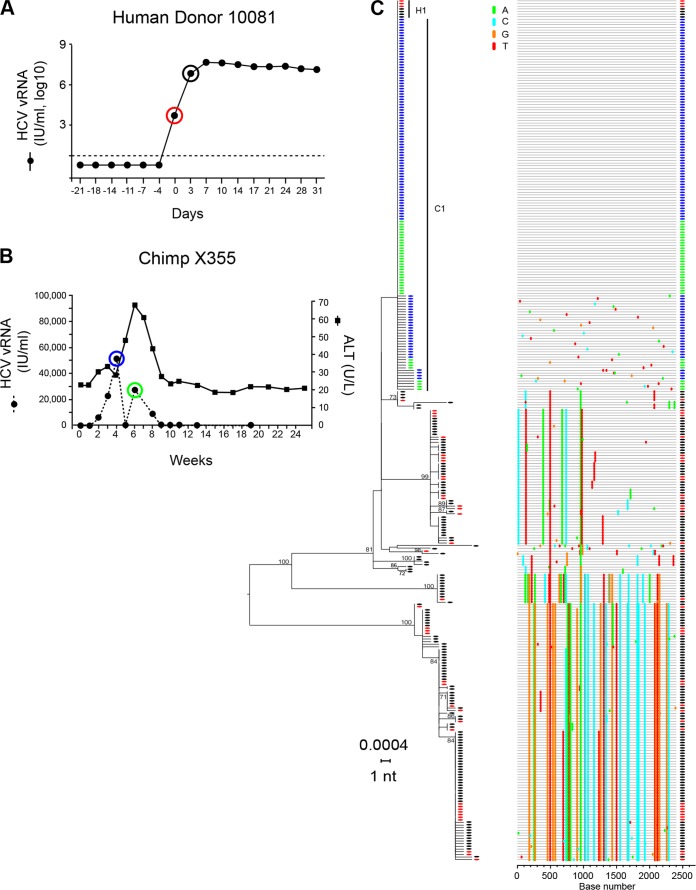
Figure 3A depicts plasma vRNA kinetics of the acutely infected human donor 10081 (50) and shows the time of sampling for our HCV sequence determinations. The first plasma sample positive for HCV RNA by an FDA-licensed assay (Roche Molecular Systems COBAS Amplicor HCV Monitor PCR assay, v2.0; detection limit of 600 IU/ml or 2,040 vRNA copies/ml) is designated as day 0 so as to maintain consistency with the previous report (50). Samples for day −21 through day −4 all tested negative for HCV RNA by this FDA-licensed viral load assay. A more sensitive transcription-mediated amplification (TMA) assay (Gen-Probe/Hologic, Inc.; detection threshold of 12 vRNA copies/ml) was then used to test replicate plasma samples. Samples from days −21, −18, −14, and −11 were determined to be negative for vRNA by this ultrasensitive test, but a subset of the replicates from days −7 and −4 was positive. Samples (50 ml) of plasma from day −18 through day −7 were infused into a virus-naive chimpanzee but did not lead to productive HCV infection. However, intravenous infusion of 50 ml of subject 10081 plasma from day −4, which was estimated to contain ∼60 (95% confidence interval, 25 to 95) vRNA copies based on the TMA assay, led to productive infection of chimpanzee X355. Plasma viremia in chimpanzee X355 was first detected 2 weeks after inoculation and remained variably positive through week 8 (Fig. 3B). This pattern of early viremia, transient elevation of alanine aminotransferase (ALT), followed by resolution of viremia is typical of chimpanzee HCV infection.
FIG 3.
HCV viral load kinetics and sequences from a human-to-chimpanzee transmission pair. (A) Viral load kinetics in acutely infected human subject 10081. Red and black circles indicate plasma samples subjected to sequence analysis. (B) Fifty milliliters of 10081 plasma from the day −4 time point was infused intravenously into chimpanzee X355 at week 0. Plasma vRNA kinetics are indicated by filled black dots, with blue and green circles indicating time points subjected to sequence analysis. Alanine aminotransferase (ALT) is a hepatic enzyme that when elevated above baseline indicates liver inflammation or injury. An ML phylogenetic tree (with sequences color coded red, black, blue, and green to correspond to time points indicated in panels A and B) and a Highlighter plot show that the human subject 10081 was acutely infected by multiple viruses, one of which, H1 (top of panel C), was transmitted to chimpanzee X355 giving rise to the C1 T/F lineage. Note that the T/F genome corresponding to H1 is identical in sequence to the T/F genome corresponding to the C1 lineage.
Plasma samples from human subject 10081 at day 0 and day 3 (Fig. 3A) and from chimpanzee X355 at weeks 4 and 6 postinoculation (Fig. 3B), were analyzed by SGS. One hundred eighty-four human HCV sequences and 140 chimpanzee HCV sequences are shown (Fig. 3C). This sequencing depth provided over 90% likelihood of detecting minor variants present at frequencies of ∼2%. Phylogenetic analysis suggested infection of the human subject 10081 by a large number of viruses. In 10081, widely divergent lineages were evident but so too were many additional lineages comprised of sequences that contained one, two, three, or more shared nucleotide polymorphisms. These multiple sets of very closely related sequences failed to conform to a simple model of random virus diversification and resembled those from subjects 10003, 10016, and 10020 (14). Using the revised models to account for differences in replication strategies of HCV and HIV-1 (14; http://www.santafe.edu/~tanmoy/programs/HCV/), the sequence pattern in subject 10081 (Fig. 3C) could be explained by the acquisition of between 9 and 14 T/F viruses. In contrast, the chimpanzee sequences shown in Fig. 3C comprise a single lineage containing 140 identical or nearly identical sequences. These sequences exhibited a star-like phylogeny and a Poisson distribution of random mutations (Table 1). This sequence pattern indicated productive infection by a single T/F virus. Importantly, the inferred T/F HCV genome in the acutely infected chimpanzee (T/F lineage C1) was found to be identical to one of the human T/F sequence lineages (H1). This result documents the transmission of an HCV genome unchanged from donor to recipient, again corroborating the T/F concept and the observation that early virus evolution generally follows a simple pattern of random sequence diversification.
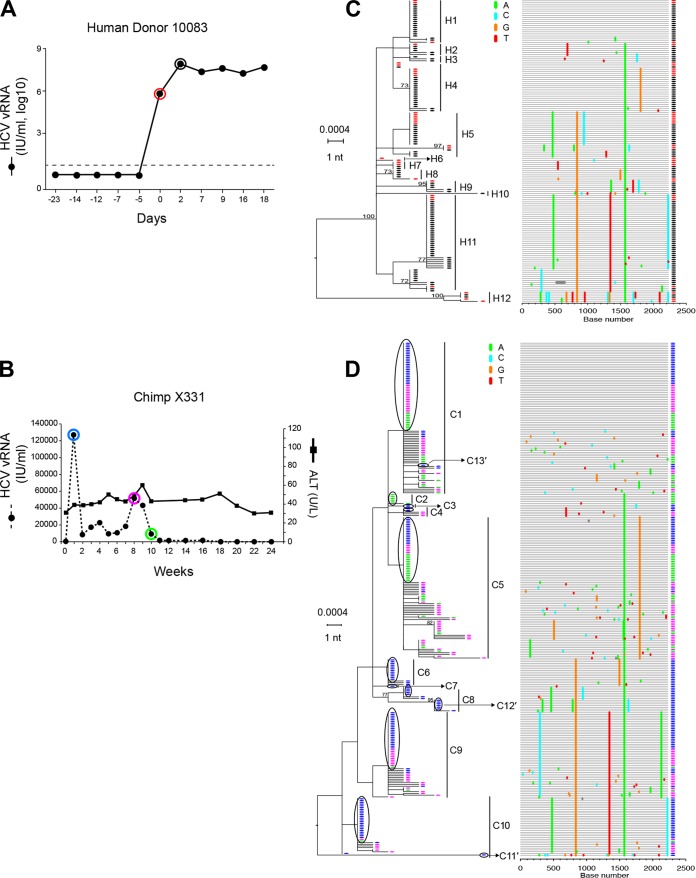
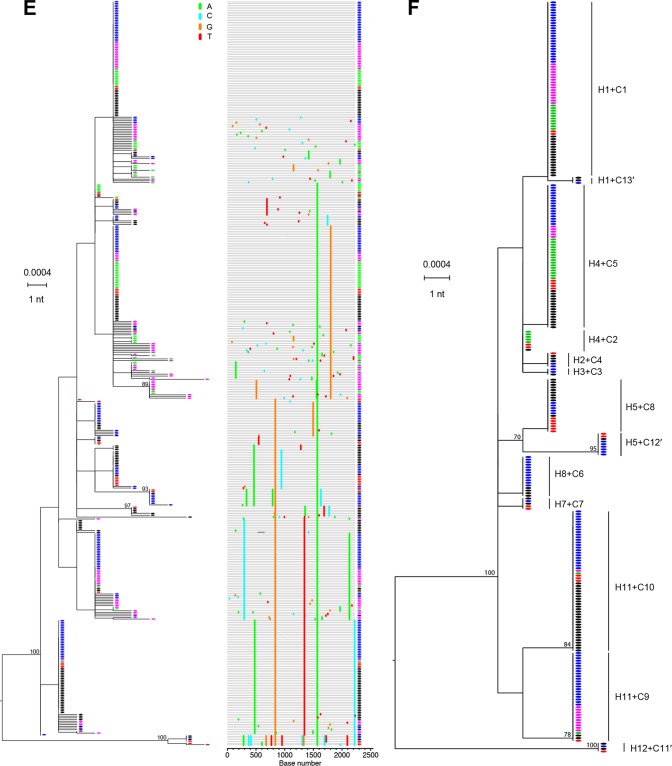
Figure 4 illustrates a second case of human-to-chimpanzee HCV transmission but at a much higher multiplicity of infection. The experimental design was similar to that for Fig. 3, except that in this case, plasma samples from human donor 10083 from days −49, −7, and −5 (the sample immediately preceding frank viremia) failed to infect chimpanzee X331 despite containing ∼784 HCV vRNA copies (50). To show that X331 was infectible by HCV, Busch et al. next inoculated this chimpanzee intravenously with 50 ml of the day 0 human plasma, estimated to contain 3.4 × 107 HCV vRNA copies (50). Productive infection of chimpanzee X331 was detected 1 week later and persisted through week 16 (Fig. 4B). We sequenced by SGS the human 10083 plasma samples from days 0 and 2 (Fig. 4A) and chimpanzee X331 plasma samples from weeks 1, 8, and 10 (Fig. 4B). A total of 146 HCV sequences from 10083 (Fig. 4C) and 292 HCV sequences from X331 (Fig. 4D) were analyzed. As for human donor 10081, phylogenetic analysis of human donor 10083 sequences revealed multivariant HCV infection resulting in multiple sets of very closely and more distantly related sequence lineages. Using our previously described clustering model (14; http://www.santafe.edu/~tanmoy/programs/HCV/), we estimated that subject 10083 had been productively infected by at least 12 T/F lineages (H1 to H12). This is a minimum estimate because of sampling limitations and because lineages differing by as few as one or two shared mutations could represent distinct transmitted variants. The diversity among the 12 inferred T/F genomes ranged from as little as 0.1% between H1 and H2 to as much as 0.6% between H11 and H12 (Fig. 4D), reflecting the sequence diversity in the individual from whom 10083 acquired HCV infection. The phylogeny of sequences from chimpanzee X331 was quite different from that observed in chimpanzee X335 where only a single T/F lineage was identified. Instead, in chimpanzee X331, we found evidence by the clustering model (14; http://www.santafe.edu/~tanmoy/programs/HCV/) of a minimum of 10 T/F lineages (C1 to C10), with an overall pattern of diversity closely resembling that of subject 10083 HCV sequences (compare Fig. 4C and D). When 10083 and X331 sequences were combined, phylogenetic analysis confirmed their close similarity (Fig. 4F). In order to analyze this combined sequence set for unequivocal evidence of transmission of sequences from donor to recipient, we selected all sequences that were identical between 10083 and X331 and analyzed them in a combined phylogenetic tree (Fig. 4E). We identified 13 discrete T/F lineages in chimpanzee X331 that had matching identical sequences in the human donor 10083. This was three more T/F genomes (C11′, C12′, and C13′) than were inferred by the clustering model (Fig. 4D). The 13 T/F genome lineages thus correspond to viral genomes that were transmitted unaltered from the human donor 10083 to chimpanzee X331. This is likely an underestimate of the numbers of T/F viruses giving rise to productive infection in chimpanzee X331, given the limitations of sampling and the very high virus inoculum. Nonetheless, the unequivocal identification of 13 T/F genomes that were identical in donor and recipient again corroborates the T/F concept for HCV and recapitulates the patterns of sequence diversity that we observed previously in subjects 10003, 10016, and 10020 (14).
FIG 4.
HCV viral load kinetics and sequences in a human-to-chimpanzee transmission pair. (A) Viral load kinetics in acutely infected human subject 10083. Red and black circles indicate plasma samples subjected to sequence analysis. (B) Fifty milliliters of 10083 plasma from the day zero time point was infused intravenously into chimpanzee X331 at week 0. Plasma vRNA kinetics are indicated by filled dots, with blue, purple, and green circles indicating time points subjected to sequence analysis. ALT values are shown. (C) An ML phylogenetic tree (with sequences color-coded red and black to correspond to time points indicated in panel A) and a Highlighter plot show that the human subject 10083 was acutely infected by multiple viruses. The model described by Bhattacharya and coworkers (14; http://www.santafe.edu/~tanmoy/programs/HCV/) suggests a minimum of 12 T/F genomes (H1-H12). Panel D depicts sequences (color coded blue, purple, and green to correspond to time points indicated in panel B) from chimpanzee X331, and the Bhattacharya model suggests a minimum of 10 T/F viruses (C1 to C10). (E) Human (red and black) and chimpanzee (blue, purple, and green) sequences are combined. (F) An ML tree of sequences found to be identical between the human donor and the chimpanzee recipient. This tree reveals 13 T/F genome lineages (C1 to C13′) that were transmitted unaltered from humans to chimpanzees. These 13 transmitted genomes are enclosed in black ovals in panel D and include three lineages (C11′, C12′, and C13′) that were identified empirically (F) but not by the Bhattacharya clustering analysis (D).
DISCUSSION
The findings of the present study provide an essential validation of the T/F strategy, originally developed to identify transmitted HIV-1 genomes (36, 38, 39, 62, 63), for HCV. The results distinguish modes and multiplicities of virus transmission (14, 35) and mathematical models of early virus diversification and evolution (10, 14, 36, 40, 41; http://www.santafe.edu/~tanmoy/programs/HCV/). The analyses included individual cases of acute human infection by HCV (Fig. 1) (14) and cases of human-to-human (Fig. 2) and human-to-chimpanzee (Fig. 3 and 4) transmission where HCV sequences in donors and recipients were found to be identical, thus proving that the inferred T/F viral genomes in these subjects corresponded to actual transmitted viruses. These findings further support the conclusion that HCV sequence evolution in the initial weeks following virus transmission generally conforms to a simple model of random virus evolution with sequences characterized by a within-lineage star-like phylogeny and a Poisson distribution of mutations. This allows for precise estimates to be made of numbers of T/F genomes associated with different clinicoepidemiological circumstances or risks of HCV transmission such as needlestick injury, blood transfusion, injection drug use, mucosal exposure, and acute-to-acute infection outbreaks (1, 14, 18–23, 28, 35, 50, 64), where the numbers of T/F viruses responsible for productive clinical infection may vary widely (14, 35). These observations are important for future studies aimed at molecularly characterizing viruses responsible for infection, coinfection, superinfection and reinfection (64), for analyzing the selective or sieving effects of neutralizing antibodies and virus specific T cells that emerge postinfection or postvaccination (65–69), and for analyzing the transmission or emergence of drug-resistant mutants (14).
But if identification of T/F genomes is straightforward in cases where sequences conform to a simple model of random virus diversification, how can the present results explain the atypical examples previously described where acute infection sequences show very closely related lineages that appear to violate neutral evolution (i.e., subjects 10003, 10016, and 10020 in reference 14) and subjects 10081 and 10083 in the present study? The current data show that in cases where a pretransmission virus population diversity bottleneck is present in a donor (e.g., subject 10083 in Fig. 4C) and the multiplicity of infection is high, the resulting pattern of virus diversity in the recipient (chimpanzee X331 in Fig. 4D) can mirror that in subjects 10003, 10016, and 10020 (14). A pretransmission bottleneck can result from acute infection of the donor, treatment of the donor with antiviral drugs, or, in some instances, potent immune selection by neutralizing antibodies and/or CTLs. If instead, a low-multiplicity infection occurs from a similarly bottlenecked donor (e.g., subject 10081 in Fig. 3C), sequence diversity in the recipient (chimpanzee X335) follows the predicted random pattern with star-like phylogeny and a Poisson distribution of mutations (Fig. 3C). These findings suggest that during acute infection, the majority of distinct sequence lineages reflect diversification from discrete, and in some cases, very closely related T/F genomes. A star-like topology is expected to persist, while the majority of the replication complexes present result from a rapid burst of diversification, probably up to 6 weeks or so postinfection when an acquired immune response is typically first detected. The 13 T/F genome lineages in chimpanzee X331 support this conclusion (Fig. 4D to F). However, in the absence of SGS data from linked donors and recipients, the complex patterns of closely related, low diversity sequence lineages such as shown for 10081 (Fig. 3C), 10083 (Fig. 4C), and X331 (Fig. 4D), cannot all be assumed to represent distinct T/F genomes, and it is impossible to determine with certainty which were transmitted and which evolved posttransmission as a consequence of early stochastic mutations that then persisted in the population. Such complex data sets can be evaluated more conservatively by a relaxed deterministic model of HCV diversification (14; http://www.santafe.edu/~tanmoy/programs/HCV/). In the case of chimpanzee X331, we could evaluate how closely the approximations of T/F lineages based on this mathematical model corresponded to T/F lineages documented empirically: 10 by the former, 13 by the latter (Fig. 4D and F).
An interesting question is why, given the complex replication strategy of HCV, early evolution of individual T/F genome lineages does not violate the predictions of a simple model of random diversification more frequently. The likely explanation is that in acute HCV infection, plasma virus titers, the numbers of infected hepatocytes, and the numbers of intracellular replication complexes all increase exponentially and proportionately until plateau viremia is reached. For a fast enough exponential expansion rate, at each generation the progeny of new replication complexes dominate the virus population and, as a result, the phylogenetic tree of a small sample of viruses shows few coalescent events at generations much after the common ancestor. In this situation, HCV diversification resembles that of a fast-growing HIV-1 viral population, and the long-lived nature of HCV replication complexes in untreated infection (10) does not modify the essentially random mutational patterns observed in early samples. During the plateau phase of viremia, the linear “stamping press” replication process from a relatively fixed number of infected hepatocytes (10) dominates and most replication complexes formed show evidence of this shared viral ancestry. Consistent with this picture, it was observed that the average sequence diversity in a sample grows almost linearly at very early times during the virus titer exponential growth phase and saturates later when the virus reaches the plateau phase (10, 14). Once adaptive immune responses, including neutralizing antibodies and cytotoxic T cells, emerge, the viral load decreases and nonrandom virus diversity becomes evident (Fig. 1) (10, 14; unpublished data).
There are limitations and caveats to the present study. We presented examples of nearly full length (8.8 kb), 5′ half (4.9 kb) and 5′ quarter (2.4) genomic analyses. Nearly full length single-genome amplifications are not always possible because of priming and extension inefficiencies, time and cost considerations, and other factors. However, because HCV contains a nonsegmented RNA genome, analyses of sequences of a reasonable length (∼2 to 5 kb) that contain sufficient numbers of informative sites (shared nucleotide polymorphisms) are sufficient to identify and distinguish distinct T/F genomes. In addition, for applications where the intent is to characterize particular gene products such as Env (E1E2), a 2-kb 5′ amplification product may be sufficient to identify T/F genomes, characterize population diversity bottlenecks, and identify mutational escape patterns. Where the intent is to map on a proteome-wide basis all T cell epitopes recognized or escaped, nearly full length amplification is required in order to maintain genetic linkage across all alleles, as we have described previously for HIV-1 (37, 70).
There are additional caveats to the extrapolation of the findings reported here to other cases of acute HCV infection and to different risk groups. First, we studied relatively few subjects, and for most of these individuals, risk behaviors leading to HCV infection were unknown. Future studies could benefit from an analysis of larger numbers of human subjects with more clearly defined risk factors for HCV acquisition (e.g., needlestick, blood products, IDU, sexual, and other). Second, our models of early virus replication and diversification of HCV (14), as for HIV-1 (36), cannot fully account for the possibility of extremely early mutations that occur during one or more of the initial replication cycles of the incoming virus (for HCV, plus-sense RNA to minus-sense RNA to plus-sense RNA, etc.). This can result in the early occurrence and persistence of shared mutations, deviations from a star-like phylogeny, and inference of a T/F genome one or a few virus generations subsequent to the actual transmission event. In practice, very early mutations resulting in shared polymorphisms are uncommon in acute HCV vRNA sequence data sets (14, 33), and when they do occur, they do not confound the inference of founder genomes. The paucity of very early mutations persisting to later virus generations may be due in part to a lower in vivo error rate of the HCV polymerase than that estimated from in vitro analyses (10) and because random mutations are generally neutral or deleterious and subject to early purifying selection (71). Thus, while it will not always be the case that the consensus of a low-diversity sequence lineage that conforms to model predictions represents the transmitted genome, such a sequence is nonetheless a founder sequence likely to have arisen within one or few virus generations of the transmitted genome. The phenotypic properties of such founder viruses, including their antigenicity, is likely to be equivalent to that of the corresponding transmitted viruses because of the short time interval and small numbers of virus generations that separate the two.
What are the implications of the present findings for future HCV research? Because of the exceedingly short half-life of HCV in human plasma (t1/2 ∼ 45 min) (72), plasma virus titers and genetic composition closely reflect the spectrum of viruses replicating in hepatocytes, along with the composite effects of biological, immunological and antiviral drug pressures influencing virus replication and persistence. As a consequence, quantitative analyses of plasma virus titers and genetic composition provide an extremely sensitive and precise real-time indicator of biologically relevant, and oftentimes clinically relevant, virus-host interactions. The current findings show how SGS of HCV genes and nearly full length genomes can inform important areas of HCV research. For example, with the recent development and clinical introduction of new curative treatment strategies, elucidation of treatment “failures” due to antiviral drug resistance, inadequate dosing or compliance, or virus reinfection becomes paramount. SGS, which maintains genetic linkage within genes (e.g., NS5B in patients receiving polymerase inhibitors) and across genomes (e.g., NS3-NS5B in patients receiving protease, NS5A, and polymerase inhibitors), can contribute uniquely to the elucidation of mechanisms of treatment failure, as described previously for HIV-1 infection (73). Similarly, distinguishing HCV coinfection from superinfection or reinfection can be important in explaining apparent cases of treatment failure. This can best be done by SGS, as suggested by Irving and Brown (28, 64) and by the findings reported here. In the area of vaccine development, SGS has been uniquely informative and represents a major tool for assessing virus responses to host adaptive immunity in natural HIV-1 infection and following vaccination. This has included proteome-wide analyses of T-cell responses (37, 70, 74), Env gp160-wide analyses of neutralizing antibody responses (36, 75, 76), and sieve analyses of vaccine breakthrough cases (65–68, 77). Current and future HCV vaccine trials (3, 4) are primed to adopt this methodology and the associated bioinformatic and statistical platforms to accelerate HCV vaccine development and testing. Comparable studies in other flavivirus or alphavirus systems may similarly benefit from a precise molecular identification of T/F genomes and their early molecular pathways of diversification.
ACKNOWLEDGMENTS
This study was supported by the University of Pennsylvania Centers for AIDS Research Sequencing Core Facility (NIH P30 AI 045008) and by grants from the NIH, including AI106000 (G.M.S.) and AI02433 (A.S.P.), OD011095 (A.S.P.), and AI078881 (A.S.P.).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We disavow any financial or other relationships that could represent a conflict of interest with respect to this study.
REFERENCES
- 1.Ray SC, Thomas DL. 2010. Hepatitis C: principles and practice of infectious diseases, 7th ed Churchill Livingstone/Elsevier Publishers, New York, NY. [Google Scholar]
- 2.Averhoff FM, Glass N, Holtzman D. 2012. Global burden of hepatitis C: considerations for healthcare providers in the United States. Clin Infect Dis 55(Suppl 1):S10–S15. doi: 10.1093/cid/cis361. [DOI] [PubMed] [Google Scholar]
- 3.Law L, Landi A, Magee W, Tyrrell D, Houghton M. 2013. Progress toward a hepatitis C virus vaccine. Emerg Microbes Infect 2:e79. doi: 10.1038/emi.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang TJ. 2013. Current progress in development of hepatitis C virus vaccines. Nat Med 19:869–878. doi: 10.1038/nm.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houghton M, Abrignani S. 2005. Prospects for a vaccine against the hepatitis C virus. Nature 436:961–966. doi: 10.1038/nature04081. [DOI] [PubMed] [Google Scholar]
- 6.Farci P. 2011. New insights into the HCV quasispecies and compartmentalization. Semin Liver Dis 31:356–374. doi: 10.1055/s-0031-1297925. [DOI] [PubMed] [Google Scholar]
- 7.Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, Halfon P, Inchauspe G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin IT, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 8.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2014. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 10.Ribeiro RM, Li H, Wang S, Stoddard MB, Learn GH, Korber BT, Bhattacharya T, Guedj J, Parrish EH, Hahn BH, Shaw GM, Perelson AS. 2012. Quantifying the diversification of hepatitis C virus (HCV) during primary infection: estimates of the in vivo mutation rate. PLoS Pathog 8:e1002881. doi: 10.1371/journal.ppat.1002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rong R. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog 5:e1000594. doi: 10.1371/journal.ppat.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang GP, Sherrill-Mix SA, Chang KM, Quince C, Bushman FD. 2010. Hepatitis C virus transmission bottlenecks analyzed by deep sequencing. J Virol 84:6218–6228. doi: 10.1128/JVI.02271-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bull RA, Luciani F, McElroy K, Gaudieri S, Pham ST, Chopra A, Cameron B, Maher L, Dore GJ, White PA, Lloyd AR. 2011. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS Pathog 7:e1002243. doi: 10.1371/journal.ppat.1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Stoddard MB, Wang S, Blair LM, Giorgi EE, Parrish EH, Learn GH, Hraber P, Goepfert PA, Saag MS, Denny TN, Haynes BF, Hahn BH, Ribeiro RM, Perelson AS, Korber BT, Bhattacharya T, Shaw GM. 2012. Elucidation of hepatitis C virus transmission and early diversification by single-genome sequencing. PLoS Pathog 8:e1002880. doi: 10.1371/journal.ppat.1002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maheshwari A, Ray S, Thuluvath PJ. 2008. Acute hepatitis C Lancet 372:321–332. [DOI] [PubMed] [Google Scholar]
- 16.Liang TJ, Ghany MG. 2013. Current and future therapies for hepatitis C virus infection. N Engl J Med 368:1907–1917. doi: 10.1056/NEJMra1213651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauer GM. 2013. Immune responses to hepatitis C virus (HCV) infection and the prospects for an effective HCV vaccine or immunotherapies. J Infect Dis 207(Suppl 1):S7–S12. doi: 10.1093/infdis/jis762. [DOI] [PubMed] [Google Scholar]
- 18.Busch MP, Shafer KA. 2005. Acute-phase hepatitis C virus infection: implications for research, diagnosis, and treatment. Clin Infect Dis 40:959–961. doi: 10.1086/428583. [DOI] [PubMed] [Google Scholar]
- 19.Page-Shafer K, Pappalardo BL, Tobler LH, Phelps BH, Edlin BR, Moss AR, Wright TL, Wright DJ, O'Brien TR, Caglioti S, Busch MP. 2008. Testing strategy to identify cases of acute hepatitis C virus (HCV) infection and to project HCV incidence rates. J Clin Microbiol 46:499–506. doi: 10.1128/JCM.01229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suryaprasad AG, White JZ, Xu F, Eichler BA, Hamilton J, Patel A, Hamdounia SB, Church DR, Barton K, Fisher C, Macomber K, Stanley M, Guilfoyle SM, Sweet K, Liu S, Iqbal K, Tohme R, Sharapov U, Kupronis BA, Ward JW, Holmberg SD. 2014. Emerging epidemic of hepatitis C virus infections among young nonurban persons who inject drugs in the United States, 2006-2012. Clin Infect Dis 59:1411–1419. doi: 10.1093/cid/ciu643. [DOI] [PubMed] [Google Scholar]
- 21.van de Laar T, Pybus O, Bruisten S, Brown D, Nelson M, Bhagani S, Vogel M, Baumgarten A, Chaix ML, Fisher M, Gotz H, Matthews GV, Neifer S, White P, Rawlinson W, Pol S, Rockstroh J, Coutinho R, Dore GJ, Dusheiko GM, Danta M. 2009. Evidence of a large, international network of HCV transmission in HIV-positive men who have sex with men. Gastroenterology 136:1609–1617. doi: 10.1053/j.gastro.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zibbell J, Iqbal K, Patel R, Suryaprasad A, Sanders K, Moore-Moravian L, Serrecchia J, Blankenship S, Ward J, Holtzman D. 2015. Increases in hepatitis C virus infection related to injection drug use among persons aged ≤30 years—Kentucky, Tennessee, Virginia, and West Virginia, 2006-2012. MMWR Morb Mortal Wkly Rep 64:453–458. [PMC free article] [PubMed] [Google Scholar]
- 23.Fierer D, Factor S, Uriel A, Carriero D, Dieterich D, Mullen M, Klepper A, van Seggelen W, Childs K, Branch A, Holtzman D, Ward J, Khudyakov Y, Holmberg S. 2011. Sexual transmission of hepatitis C virus among HIV-infected men who have sex with men—New York City, 2005-2010. MMWR Morb Mortal Wkly Rep 60:945–950. [PubMed] [Google Scholar]
- 24.Cox AL, Mosbruger T, Mao Q, Liu Z, Wang XH, Yang HC, Sidney J, Sette A, Pardoll D, Thomas DL, Ray SC. 2005. Cellular immune selection with hepatitis C virus persistence in humans. J Exp Med 201:1741–1752. doi: 10.1084/jem.20050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, Strazzera A, Chien DY, Munoz SJ, Balestrieri A, Purcell RH, Alter HJ. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 26.Herring BL, Tsui R, Peddada L, Busch M, Delwart EL. 2005. Wide range of quasispecies diversity during primary hepatitis C virus infection. J Virol 79:4340–4346. doi: 10.1128/JVI.79.7.4340-4346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuntzen T, Timm J, Berical A, Lewis-Ximenez LL, Jones A, Nolan B, Schulze zur Wiesch J, Li B, Schneidewind A, Kim AY, Chung RT, Lauer GM, Allen TM. 2007. Viral sequence evolution in acute hepatitis C virus infection. J Virol 81:11658–11668. doi: 10.1128/JVI.00995-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith JA, Aberle JH, Fleming VM, Ferenci P, Thomson EC, Karayiannis P, McLean AR, Holzmann H, Klenerman P. 2010. Dynamic coinfection with multiple viral subtypes in acute hepatitis C. J Infect Dis 202:1770–1779. doi: 10.1086/657317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tester I, Smyk-Pearson S, Wang P, Wertheimer A, Yao E, Lewinsohn DM, Tavis JE, Rosen HR. 2005. Immune evasion versus recovery after acute hepatitis C virus infection from a shared source. J Exp Med 201:1725–1731. doi: 10.1084/jem.20042284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dowd KA, Netski DM, Wang XH, Cox AL, Ray SC. 2009. Selection pressure from neutralizing antibodies drives sequence evolution during acute infection with hepatitis C virus. Gastroenterology 136:2377–2386. doi: 10.1053/j.gastro.2009.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, Fisher BE, Dowd KA, Astemborski J, Cox AL, Ray SC. 2010. Acceleration of hepatitis C virus envelope evolution in humans is consistent with progressive humoral immune selection during the transition from acute to chronic infection. J Virol 84:5067–5077. doi: 10.1128/JVI.02265-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honegger JR, Kim S, Price AA, Kohout JA, McKnight KL, Prasad MR, Lemon SM, Grakoui A, Walker CM. 2013. Loss of immune escape mutations during persistent HCV infection in pregnancy enhances replication of vertically transmitted viruses. Nat Med 19:1529–1533. doi: 10.1038/nm.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoddard MB, Li H, Wang S, Saeed M, Andrus L, Ding W, Jiang X, Learn GH, von Schaewen M, Wen J, Goepfert PA, Hahn BH, Ploss A, Rice CM, Shaw GM. 2015. Identification, molecular cloning, and analysis of full-length hepatitis C virus transmitted/founder genotypes 1, 3, and 4. mBio 6:e02518. doi: 10.1128/mBio.02518-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown RJ, Hudson N, Wilson G, Rehman SU, Jabbari S, Hu K, Tarr AW, Borrow P, Joyce M, Lewis J, Zhu LF, Law M, Kneteman N, Tyrrell DL, McKeating JA, Ball JK. 2012. Hepatitis C virus envelope glycoprotein fitness defines virus population composition following transmission to a new host. J Virol 86:11956–11966. doi: 10.1128/JVI.01079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D'Arienzo V, Moreau A, D'Alteroche L, Gissot V, Blanchard E, Gaudy-Graffin C, Roch E, Dubois F, Giraudeau B, Plantier JC, Goudeau A, Roingeard P, Brand D. 2013. Sequence and functional analysis of the envelope glycoproteins of hepatitis C virus variants selectively transmitted to a new host. J Virol 87:13609–13618. doi: 10.1128/JVI.02119-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med 206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abrahams MR, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, Ping LH, Athreya GS, Treurnicht FK, Keele BF, Wood N, Salazar-Gonzalez JF, Bhattacharya T, Chu H, Hoffman I, Galvin S, Mapanje C, Kazembe P, Thebus R, Fiscus S, Hide W, Cohen MS, Karim SA, Haynes BF, Shaw GM, Hahn BH, Korber BT, Swanstrom R, Williamson C. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol 83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, Karita E, Manigart O, Mulenga J, Keele BF, Shaw GM, Hahn BH, Allen SA, Derdeyn CA, Hunter E. 2009. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog 5:e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee HY, Giorgi EE, Keele BF, Gaschen B, Athreya GS, Salazar-Gonzalez JF, Pham KT, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Hahn BH, Shaw GM, Korber BT, Bhattacharya T, Perelson AS. 2009. Modeling sequence evolution in acute HIV-1 infection. J Theor Biol 261:341–360. doi: 10.1016/j.jtbi.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giorgi EE, Funkhouser B, Athreya G, Perelson AS, Korber BT, Bhattacharya T. 2010. Estimating time since infection in early homogeneous HIV-1 samples using a Poisson model. BMC Bioinformatics 11:532. doi: 10.1186/1471-2105-11-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindenbach BD, Thiel HJ, Rice C. 2007. Flaviviridae: the viruses and their replication, 5th ed, vol 1 Lippincott William & Wilkins, Philadelphia, PA. [Google Scholar]
- 45.Moradpour D, Penin F, Rice CM. 2007. Replication of hepatitis C virus. Nat Rev Microbiol 5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 46.Quinkert D, Bartenschlager R, Lohmann V. 2005. Quantitative analysis of the hepatitis C virus replication complex. J Virol 79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brenner BG, Roger M, Routy JP, Moisi D, Ntemgwa M, Matte C, Baril JG, Thomas R, Rouleau D, Bruneau J, Leblanc R, Legault M, Tremblay C, Charest H, Wainberg MA, Quebec Primary HIVISG . 2007. High rates of forward transmission events after acute/early HIV-1 infection. J Infect Dis 195:951–959. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 48.Pilcher CD, Eron JJ Jr, Vemazza PL, Battegay M, Harr T, Yerly S, Vom S, Perrin L. 2001. Sexual transmission during the incubation period of primary HIV infection. JAMA 286:1713–1714. doi: 10.1001/jama.286.14.1713. [DOI] [PubMed] [Google Scholar]
- 49.Ma ZM, Stone M, Piatak M Jr, Schweighardt B, Haigwood NL, Montefiori D, Lifson JD, Busch MP, Miller CJ. 2009. High specific infectivity of plasma virus from the pre-ramp-up and ramp-up stages of acute simian immunodeficiency virus infection. J Virol 83:3288–3297. doi: 10.1128/JVI.02423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Busch MP, Murthy KK, Kleinman SH, Hirschkorn DF, Herring BL, Delwart EL, Racanelli V, Yoon JC, Rehermann B, Alter HJ. 2012. Infectivity in chimpanzees (Pan troglodytes) of plasma collected before HCV RNA detectability by FDA-licensed assays: implications for transfusion safety and HCV infection outcomes. Blood 119:6326–6334. doi: 10.1182/blood-2011-12-393637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 52.Cox AL, Netski DM, Mosbruger T, Sherman SG, Strathdee S, Ompad D, Vlahov D, Chien D, Shyamala V, Ray SC, Thomas DL. 2005. Prospective evaluation of community-acquired acute-phase hepatitis C virus infection. Clin Infect Dis 40:951–958. doi: 10.1086/428578. [DOI] [PubMed] [Google Scholar]
- 53.Rehermann B. 2009. Hepatitis C virus versus innate and adaptive immune responses: a tale of coevolution and coexistence. J Clin Invest 119:1745–1754. doi: 10.1172/JCI39133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bowen DG, Walker CM. 2005. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 436:946–952. doi: 10.1038/nature04079. [DOI] [PubMed] [Google Scholar]
- 55.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. 2009. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fischer W, Ganusov VV, Giorgi EE, Hraber PT, Keele BF, Leitner T, Han CS, Gleasner CD, Green L, Lo CC, Nag A, Wallstrom TC, Wang S, McMichael AJ, Haynes BF, Hahn BH, Perelson AS, Borrow P, Shaw GM, Bhattacharya T, Korber BT. 2010. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS One 5:e12303. doi: 10.1371/journal.pone.0012303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, Berlin AM, Malboeuf CM, Ryan EM, Gnerre S, Zody MC, Erlich RL, Green LM, Berical A, Wang Y, Casali M, Streeck H, Bloom AK, Dudek T, Tully D, Newman R, Axten KL, Gladden AD, Battis L, Kemper M, Zeng Q, Shea TP, Gujja S, Zedlack C, Gasser O, Brander C, Hess C, Gunthard HF, Brumme ZL, Brumme CJ, Bazner S, Rychert J, Tinsley JP, Mayer KH, Rosenberg E, Pereyra F, Levin JZ, Young SK, Jessen H, Altfeld M, Birren BW, Walker BD, Allen TM. 2012. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog 8:e1002529. doi: 10.1371/journal.ppat.1002529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burke KP, Munshaw S, Osburn WO, Levine J, Liu L, Sidney J, Sette A, Ray SC, Cox AL. 2012. Immunogenicity and cross-reactivity of a representative ancestral sequence in hepatitis C virus infection. J Immunol 188:5177–5188. doi: 10.4049/jimmunol.1103008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, Dai X, Stanfield RL, Burton DR, Ward AB, Wilson IA, Law M. 2013. Hepatitis C virus E2 envelope glycoprotein core structure. Science 342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yerly D, Heckerman D, Allen T, Suscovich TJ, Jojic N, Kadie C, Pichler WJ, Cerny A, Brander C. 2008. Design, expression, and processing of epitomized hepatitis C virus-encoded CTL epitopes. J Immunol 181:6361–6370. doi: 10.4049/jimmunol.181.9.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bull RA, Leung P, Gaudieri S, Deshpande P, Cameron B, Walker M, Chopra A, Lloyd AR, Luciani F. 2015. Transmitted/founder viruses rapidly escape from CD8+ T cell responses in acute hepatitis C virus infection. J Virol 89:5478–5490. doi: 10.1128/JVI.03717-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Bar KJ, Wang S, Decker JM, Chen Y, Sun C, Salazar-Gonzalez JF, Salazar MG, Learn GH, Morgan CJ, Schumacher JE, Hraber P, Giorgi EE, Bhattacharya T, Korber BT, Perelson AS, Eron JJ, Cohen MS, Hicks CB, Haynes BF, Markowitz M, Keele BF, Hahn BH, Shaw GM. 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog 6:e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bar KJ, Li H, Chamberland A, Tremblay C, Routy JP, Grayson T, Sun C, Wang S, Learn GH, Morgan CJ, Schumacher JE, Haynes BF, Keele BF, Hahn BH, Shaw GM. 2010. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol 84:6241–6247. doi: 10.1128/JVI.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Irving WL, Brown RJ. 2010. Acute hepatitis C virus infection: a dynamic-and challenging-concept. J Infect Dis 202:1765–1767. doi: 10.1086/657318. [DOI] [PubMed] [Google Scholar]
- 65.Edlefsen PT, Rolland M, Hertz T, Tovanabutra S, Gartland AJ, deCamp AC, Magaret CA, Ahmed H, Gottardo R, Juraska M, McCoy C, Larsen BB, Sanders-Buell E, Carrico C, Menis S, Bose M, Team RVS, Arroyo MA, O'Connell RJ, Nitayaphan S, Pitisuttithum P, Kaewkungwal J, Rerks-Ngarm S, Robb ML, Kirys T, Georgiev IS, Kwong PD, Scheffler K, Pond SL, Carlson JM, Michael NL, Schief WR, Mullins JI, Kim JH, Gilbert PB. 2015. Comprehensive sieve analysis of breakthrough HIV-1 sequences in the RV144 vaccine efficacy trial. PLoS Comput Biol 11:e1003973. doi: 10.1371/journal.pcbi.1003973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edlefsen PT, Gilbert PB, Rolland M. 2013. Sieve analysis in HIV-1 vaccine efficacy trials. Curr Opin HIV AIDS 8:432–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rolland M, Tovanabutra S, deCamp AC, Frahm N, Gilbert PB, Sanders-Buell E, Heath L, Magaret CA, Bose M, Bradfield A, O'Sullivan A, Crossler J, Jones T, Nau M, Wong K, Zhao H, Raugi DN, Sorensen S, Stoddard JN, Maust BS, Deng W, Hural J, Dubey S, Michael NL, Shiver J, Corey L, Li F, Self SG, Kim J, Buchbinder S, Casimiro DR, Robertson MN, Duerr A, McElrath MJ, McCutchan FE, Mullins JI. 2011. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat Med 17:366–371. doi: 10.1038/nm.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, deCamp AC, Carrico C, Menis S, Magaret CA, Ahmed H, Juraska M, Chen L, Konopa P, Nariya S, Stoddard JN, Wong K, Zhao H, Deng W, Maust BS, Bose M, Howell S, Bates A, Lazzaro M, O'Sullivan A, Lei E, Bradfield A, Ibitamuno G, Assawadarachai V, O'Connell RJ, deSouza MS, Nitayaphan S, Rerks-Ngarm S, Robb ML, McLellan JS, Georgiev I, Kwong PD, Carlson JM, Michael NL, Schief WR, Gilbert PB, Mullins JI, Kim JH. 2012. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature 490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Colloca S, Barnes E, Folgori A, Ammendola V, Capone S, Cirillo A, Siani L, Naddeo M, Grazioli F, Esposito ML, Ambrosio M, Sparacino A, Bartiromo M, Meola A, Smith K, Kurioka A, O'Hara GA, Ewer KJ, Anagnostou N, Bliss C, Hill AV, Traboni C, Klenerman P, Cortese R, Nicosia A. 2012. Vaccine vectors derived from a large collection of simian adenoviruses induce potent cellular immunity across multiple species. Sci Transl Med 4:115ra112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, Keele BF, Learn GH, Turnbull EL, Salazar MG, Weinhold KJ, Moore S, Letvin N, Haynes BF, Cohen MS, Hraber P, Bhattacharya T, Borrow P, Perelson AS, Hahn BH, Shaw GM, Korber BT, McMichael AJ. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med 206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. 2010. Viral mutation rates. J Virol 84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guedj J, Dahari H, Rong L, Sansone ND, Nettles RE, Cotler SJ, Layden TJ, Uprichard SL, Perelson AS. 2013. Modeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-life. Proc Natl Acad Sci U S A 110:3991–3996. doi: 10.1073/pnas.1203110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Palmer S, Kearney M, Maldarelli F, Halvas EK, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT Jr, Dewar RL, Metcalf JA, Hammer S, Mellors JW, Coffin JM. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol 43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu MK, Hawkins N, Ritchie AJ, Ganusov VV, Whale V, Brackenridge S, Li H, Pavlicek JW, Cai F, Rose-Abrahams M, Treurnicht F, Hraber P, Riou C, Gray C, Ferrari G, Tanner R, Ping LH, Anderson JA, Swanstrom R, Cohen M, Karim SS, Haynes B, Borrow P, Perelson AS, Shaw GM, Hahn BH, Williamson C, Korber BT, Gao F, Self S, McMichael A, Goonetilleke N. 2013. Vertical T cell immunodominance and epitope entropy determine HIV-1 escape. J Clin Invest 123:380–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bar KJ, Tsao CY, Iyer SS, Decker JM, Yang Y, Bonsignori M, Chen X, Hwang KK, Montefiori DC, Liao HX, Hraber P, Fischer W, Li H, Wang S, Sterrett S, Keele BF, Ganusov VV, Perelson AS, Korber BT, Georgiev I, McLellan JS, Pavlicek JW, Gao F, Haynes BF, Hahn BH, Kwong PD, Shaw GM. 2012. Early low-titer neutralizing antibodies impede HIV-1 replication and select for virus escape. PLoS Pathog 8:e1002721. doi: 10.1371/journal.ppat.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, Zhu J, Shapiro L, Program NCS, Mullikin JC, Gnanakaran S, Hraber P, Wiehe K, Kelsoe G, Yang G, Xia SM, Montefiori DC, Parks R, Lloyd KE, Scearce RM, Soderberg KA, Cohen M, Kamanga G, Louder MK, Tran LM, Chen Y, Cai F, Chen S, Moquin S, Du X, Joyce MG, Srivatsan S, Zhang B, Zheng A, Shaw GM, Hahn BH, Kepler TB, Korber BT, Kwong PD, Mascola JR, Haynes BF. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roederer M, Keele BF, Schmidt SD, Mason RD, Welles HC, Fischer W, Labranche C, Foulds KE, Louder MK, Yang ZY, Todd JP, Buzby AP, Mach LV, Shen L, Seaton KE, Ward BM, Bailer RT, Gottardo R, Gu W, Ferrari G, Alam SM, Denny TN, Montefiori DC, Tomaras GD, Korber BT, Nason MC, Seder RA, Koup RA, Letvin NL, Rao SS, Nabel GJ, Mascola JR. 2014. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature 505:502–508. [DOI] [PMC free article] [PubMed] [Google Scholar]