ABSTRACT
Human respiratory syncytial virus (RSV) is an important pathogen causing acute lower respiratory tract disease in children. The RSV attachment glycoprotein (G) is not required for infection, as G-null RSV replicates efficiently in several cell lines. Our laboratory previously reported that the viral fusion (F) protein is a determinant of strain-dependent pathogenesis. Here, we hypothesized that virus dependence on G is determined by the strain specificity of F. We generated recombinant viruses expressing G and F, or null for G, from the laboratory A2 strain (Katushka RSV-A2GA2F [kRSV-A2GA2F] and kRSV-GstopA2F) or the clinical isolate A2001/2-20 (kRSV-2-20G2-20F and kRSV-Gstop2-20F). We quantified the virus cell binding, entry kinetics, infectivity, and growth kinetics of these four recombinant viruses in vitro. RSV expressing the 2-20 G protein exhibited the greatest binding activity. Compared to the parental viruses expressing G and F, removal of 2-20 G had more deleterious effects on binding, entry, infectivity, and growth than removal of A2 G. Overall, RSV expressing 2-20 F had a high dependence on G for binding, entry, and infection.
IMPORTANCE RSV is the leading cause of childhood acute respiratory disease requiring hospitalization. As with other paramyxoviruses, two major RSV surface viral glycoproteins, the G attachment protein and the F fusion protein, mediate virus binding and subsequent membrane fusion, respectively. Previous work on the RSV A2 prototypical strain demonstrated that the G protein is functionally dispensable for in vitro replication. This is in contrast to other paramyxoviruses that require attachment protein function as a prerequisite for fusion. We reevaluated this requirement for RSV using G and F proteins from clinical isolate 2-20. Compared to the laboratory A2 strain, the G protein from 2-20 had greater contributions to virus binding, entry, infectivity, and in vitro growth kinetics. Thus, the clinical isolate 2-20 F protein function depended more on its G protein, suggesting that RSV has a higher dependence on G than previously thought.
INTRODUCTION
Human respiratory syncytial virus (hRSV or RSV) causes an annual global 3.4 million estimated severe acute lower respiratory tract infections (ALRI) in children younger than 5 years of age (1). In the United States, about 132,000 to 172,000 children younger than 5 years of age are hospitalized due to RSV every year (2). Thus far, there are no licensed vaccines, although there are multiple vaccine candidates undergoing clinical trials (3). Development of antivirals against RSV is also an active field of research and clinical development (4–6).
RSV is a member of the Paramyxoviridae family, Pneumovirus genus. Members of the paramyxovirus family encode two major glycoproteins important early during infection for attachment to the host cell and the subsequent entry process. Paramyxovirus fusion mediated by the viral fusion (F) protein is generally initiated by interaction with the homologous attachment protein upon receptor engagement (reviewed in references 7 and 8). Several studies on RSV subgroup A and B strains indicated that G is not functionally required for efficient in vitro replication in certain cell lines but is needed for optimal growth in vivo (9–11). Although not required for in vitro replication, G was shown to enhance passage of a RSV minigenome (12), and in a later study, viruses lacking G required more passages in cell culture to reach titers similar to those of viruses expressing G (10). RSV G was also shown to enhance cell-to-cell fusion, in an apparently strain-specific manner (10, 13). Similarly, human metapneumovirus (HMPV), another pneumovirus, does not require its G protein for infection (reviewed in reference 14). For both HMPV and RSV, the attachment function of G can be substituted by the F protein (15, 16). The RSV G protein has long been thought to mediate the majority of virus binding to host cells via interaction with glycosaminoglycans (GAGs) (17–19), while F is reported to bind a protein receptor (20). Considering that previous studies regarding the requirement for G during RSV infection were done with prototypical strains of this virus, we set out to reevaluate the functions of this major attachment protein using protein from a clinical isolate strain (A2001/2-20) compared to the prototypical A2 strain. We generated recombinant RSV strains harboring different combinations of the G and F proteins (GF viruses; Katushka RSV-A2GA2F [kRSV-A2GA2F] and kRSV-2-20G2-20F), along with viruses that do not express the G gene but maintain an almost identical genomic sequence composition in the G gene region (Gstop viruses; kRSV-GstopA2F and kRSV-Gstop2-20F). By comparing the G functions of each GF and Gstop virus pair, we found that there are greater contributions of 2-20 G than of A2 G to aspects of the RSV life cycle, including enhanced binding to the cell, viral entry, infectivity, and overall in vitro growth rate. Our study results show that the F protein from a clinical RSV strain has a greater dependence on its homologous G protein than the F protein of the prototypical A2 strain.
MATERIALS AND METHODS
Cell lines.
HEp-2 (ATCC CCL-23) and BEAS-2B cells were maintained as described previously (21). BSR T7/5 cells (a gift from Ursula Buchholz, National Institutes of Health, Bethesda, MD) were cultured in Glasgow's minimal essential medium (GMEM) containing 10% fetal bovine serum (FBS) and 1 μg/ml porcine serum albumin (PSA), and during every other passage, these cells were selected with Geneticin at 1 mg/ml. Chinese hamster ovary (CHO-K1) (ATCC, CCL-61) cells and a heparin sulfate-deficient derivative of that cell line, pgsD-677 (ATCC, CRL-2244), were cultured in Kaighn's modified F-12K medium (plus l-glutamine) supplemented with 10% FBS and 1 μg/ml PSA, according to ATCC instructions.
Generation of recombinant RSV strains.
The F glycoprotein genes of RSV A2 (GenBank accession number FJ614814) and A2001/2-20 (RSV 2-20 G [GenBank accession number JF279545] and RSV 2-20 F [GenBank accession number JF279544]) (21) were synthesized by GeneArt gene synthesis (Life Technologies) and cloned into a bacterial artificial chromosome (BAC) containing the antigenomic cDNA of RSV A2-K-line19F, described previously (22). All recombinant RSV strains generated in this study express a far-red fluorescent gene (monomeric Katushka-2 [mKate2]) in the first gene position—hence the kRSV designation throughout. To generate the recombinant RSV strains without G protein expression (Gstop viruses), both of the Met codons (Met1 and Met48) in the G open reading frame (ORF) were changed to Ile, with the first Met or Ile followed by a stop codon (11). Recombinant viruses were recovered by cotransfection of the RSV antigenomic BAC and four human codon bias-optimized RSV helper plasmids (N, P, L, and M2-1) into BSR T7/5 cells as described previously (22). The viruses were propagated in HEp-2 cells and the sequences of the glycoprotein ORFs confirmed. Viruses used in this study were prepared by harvesting infected HEp-2 cells followed by sonication, as described previously (23). For binding assays, virus stocks were purified by sucrose gradient centrifugation to remove the majority of cellular proteins from the virus fraction (24). Briefly, the infected HEp-2 cells were frozen at −80°C and later thawed at 37°C. Cells were scraped down and were transferred along with the medium to 50-ml conical tubes. After centrifugation at 2,000 rpm for 10 min at 4°C, supernatants were pooled and layered onto 20% sucrose-containing MEM for subsequent ultracentrifugation at 16,000 × g for 3 h at 4°C (SW32 rotor; Beckman Coulter). The resulting pellets were resuspended in MEM, and aliquots were frozen in liquid nitrogen before being stored at −80°C until use. These sucrose-purified virus stocks had infectious titers comparable to those measured for the starting material, accounting for volume change.
RSV binding assay and Western blotting.
BEAS-2B cells were seeded the prior day to be subconfluent for the experiment. Input volumes of sucrose-purified virus stocks were determined by first loading equal PFUs in SDS-PAGE gels and blotting for N and then normalizing stock dilutions (no more than 2-fold to 3-fold) to N levels. The cells were washed with cold phosphate-buffered saline (PBS), placed on ice, and inoculated with virus for 2 h. The inocula were removed by three ice-cold PBS washes, and cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich, St. Louis, MO; catalog no. R0278) supplemented with 1× protease inhibitor cocktail (Thermo Scientific, Rockford, IL; catalog no. 78430). The lysates were cleared by centrifugation at 13,200 rpm for 10 min at 4°C, and the supernatants were used for Western blotting.
Protein samples were mixed 1:1 with Laemmli sample buffer (Sigma-Aldrich) and heated at 95°C for 10 min. Samples were separated by 10% SDS-PAGE, transferred onto polyvinylidene difluoride (PVDF) membranes, and blocked with 5% nonfat dry milk–Tris-buffered saline containing 0.1% Tween 20 (TBST). Blots were probed with a mouse monoclonal antibody (clone D14; generously provided by Edward Walsh, University of Rochester, Rochester, NY) against RSV N protein followed by a horseradish peroxidase-conjugated secondary antibody. For GAPDH (glyceraldehyde-3-phosphate dehydrogenase) blots, mouse anti-GAPDH (6C5; GeneTex, Irvine, CA) was used. Chemiluminescent signal was detected with WesternBright Quantum substrate (Advansta, Menlo Park, CA). Images of Western blots were analyzed using ImageLab (v3.0.11). Relevant bands were defined manually for each group, and the total dark-pixel volume for each band was taken. The bands for the glycoproteins were then normalized by dividing their values by the respective RSV N band volumes.
FFU assay.
HEp-2 cells were seeded in 96-well plates the day before the experiment to reach 70% confluence for the assay. Fifty microliters of virus samples (serially diluted 10-fold) was inoculated onto the cells and incubated 1.5 h at room temperature with gentle rocking. After virus adsorption, 150 μl 0.75% methylcellulose (EMD, Gibbstown, NJ) in complete media was added to each well and then cells were incubated at 37°C 5% CO2 for 2 days. Wells containing 1 to 50 fluorescent-focus units (FFU) were counted and used for calculation of the virus titer in the samples. The limit of detection of this assay is 1 FFU per well, corresponding to 20 FFU/ml.
Infectivity and virus growth kinetics.
Cells were seeded into 6-well plates to be 70% confluent for infection at a multiplicity of infection (MOI) of 1.0. Cells were washed with PBS once before infection in a total volume of 500 μl per well at room temperature for 1 h with gentle rocking. The cells were then washed twice with PBS to remove the remaining inoculum. For infectivity, cells were harvested with trypsin 24 h postinfection and quantified using an LSR II flow cytometer (Becton Dickinson, Franklin Lakes, NJ) by detecting the mKate2 signal with a 532-nm-wavelength laser with a 610/20 filter. For growth kinetics, triplicate wells of infected cells were scraped in medium and resuspended at the indicated times, and aliquots were frozen at −80°C until titration by the FFU assay described above.
RSV entry assay.
This assay was performed as described previously with some modifications (25). BEAS-2B cells (70% confluent in 12-well plates) were placed on ice for 5 min and washed once with ice-cold PBS before addition of virus at an MOI of 1.0. Binding of virus to cells proceeded for 2 h on ice with gentle shaking until the inocula were removed, and cells were washed twice with ice-cold PBS. Five hundred microliters of ice-cold RPMI 1640 medium was added to each well. Plates were then warmed at 37°C for the indicated times (30 s or 1, 2, 3, 4, or 5 min). At the end of each time period, medium was removed followed by addition of 500 μl citrate buffer (400 mM sodium citrate, 10 mM potassium chloride, 135 mM sodium chloride, pH 3.0) for 2 min to inactivate any remaining extracellular virus. Cells were then washed in PBS at room temperature once before addition of complete medium, and incubation was continued at 37°C and 5% CO2 for 20 h. Infected mKate2-positive (mKate2+) cells were counted on an LSR II cytometer and analyzed using FlowJo software (Tree Star, Ashland, OR) as a quantification of the amount of virus that entered during the short warming period.
Culture and infection of primary cells.
Normal human bronchial (NHBE) cells at the air-liquid interface (NHBE/ALI cells) were obtained from Lonza (Allendale, NJ) and cultured according to the recommended protocols. For differentiation, cells were seeded onto 24-well, collagen-coated transwell supports (BD Bioscience, Bedford, MA). Cells were maintained and differentiated as described previously (26).
For binding assays, differentiated cells were cooled at 4°C and washed once with cold PBS prior to addition of virus. Equivalent amounts of virus (determined by Western blotting; see above) were used to infect the apical surface of NHBE/ALI cells for 2 h at 4°C. Following infection, the inoculum was removed and the apical surface of cells was washed three times with cold PBS. RIPA buffer supplemented with protease inhibitor cocktail was used for cell lysis. Lysates were centrifuged at 14,000 × g for 10 min for clarification prior to Western blotting.
To determine virus growth kinetics in these cells, differentiated NHBE/ALI cells were washed with PBS and apically infected at an MOI of 1.0 for 2 h at 37°C. Following infection, inocula were removed by three apical PBS washes. Virus was collected from the apical surface of cells daily by adding differential medium to the apical chamber, incubating cells in media for 10 min at 37°C, and removing media for later use in virus titration. Virus collection was performed twice per well, for a total volume of 300 μl per well for each time point. All samples were snap-frozen in liquid nitrogen until titration by FFU assay.
Statistical analysis.
Statistical analyses were performed using GraphPad Prism software version 6.0 (San Diego, CA). Data are represented as means with standard errors of the means (SEM). One-way and two-way analysis of variance (ANOVA) with Tukey's post hoc test with a P value of 0.05 was used.
RESULTS
Generation of recombinant viruses.
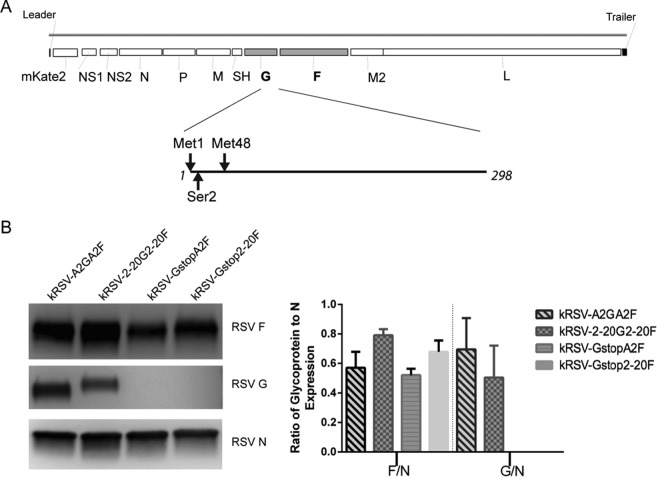
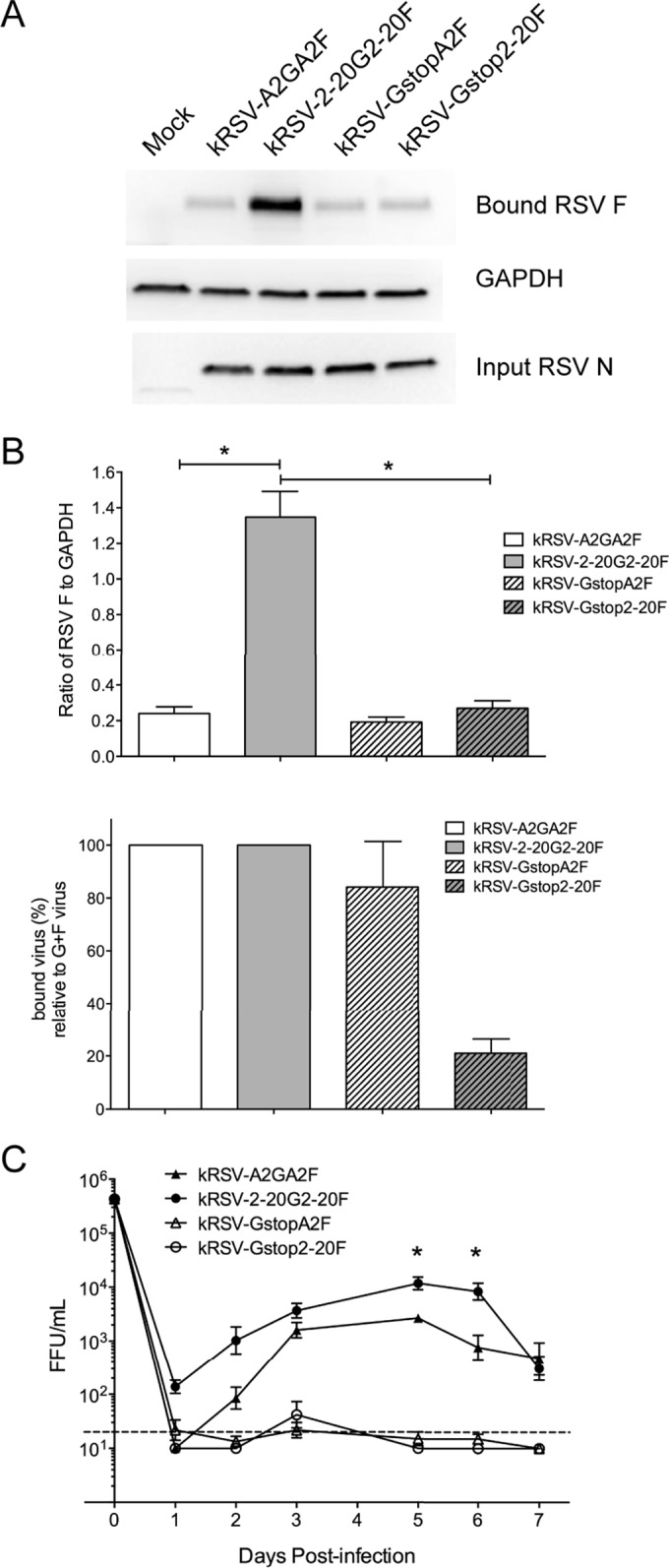
In order to assess the dependence on G protein of specific F proteins in the context of RSV infection, we used a chimeric virus approach. We generated recombinant RSV containing both G and F from either the prototypical A2 strain or the low-passage-number clinical isolate A2001/2-20 strain. We also generated G-null mutants expressing either the F protein of A2 or the F protein of 2-20. Thus, only the presence or absence of F differed between the G-null viruses. As the RSV G protein is produced both as a membrane-bound form and as a secreted form due to the presence of an alternative translation initiation site, we mutated both initiation methionines to isoleucine, as previously described (11), and we changed the second codon in the G open reading frame to a premature stop codon to abolish the expression of this protein without perturbing the gene order (Fig. 1A). We did not change the codon following the second methionine to abolish the secreted G expression because previous work showed that changing the second methionine to isoleucine is sufficient to abolish secreted G (11). As expected, Western blot analyses of sucrose-purified virus stocks showed that neither of the Gstop viruses (kRSV-GstopA2F and kRSV-Gstop2-20F) expressed detectable G protein (Fig. 1B). There was no significant difference in the levels of mature G for kRSV-A2GA2F and kRSV-2-20G2-20F (Fig. 1B). There was also no statistically significant difference in the levels of F protein abundance for kRSV-A2GA2F and kRSV-GstopA2F and for kRSV-2-20G2-20F and kRSV-Gstop2-20F (Fig. 1B), consistent with previously published data showing that the absence of G did not alter the F protein level in the virions (10). The F levels of kRSV-2-20G2-20F were 28% higher than those of kRSV-A2GA2F, normalized to N, which was not statistically significant by one-way ANOVA. Similar results were found in analyzing N, G, and F levels in HEp-2 cell lysate virus stocks that were not sucrose purified (data not shown).
FIG 1.
Schematic design of the recombinant viruses and quantification of surface glycoproteins in purified virions. (A) RSV genome with G gene open reading frame (amino acids 1 to 298) enlarged to illustrate the mutations made to generate the Gstop virus. The two methionines were changed to isoleucine, and the second codon (serine) was changed to a stop codon. (B) Western blot showing the F, G, and N protein expression levels (left) and densitometry combined from the results of four independent experiments (right). Data are represented as means ± SEM.
Differential contributions of G proteins to virus attachment to host cells.
The F protein of the prototypical A2 strain binds to host cells, possibly through interactions with heparan sulfate (16, 19, 27). We asked whether the relative contributions of G and F to cell binding differ between the A2 and 2-20 glycoproteins. To test this, we used sucrose-purified virus stocks and normalized the relative amounts of virions used as inputs for this binding assay based on the N protein expression levels determined by Western blotting. The N protein level was previously reported to correlate with radiolabeled activity in virus preparations (10). The amount of kRSV-2-20G2-20F bound to BEAS-2B cells (a human bronchial epithelial cell line) was approximately 5-fold higher than the amount of kRSV-A2GA2F (Fig. 2). Removal of the A2 G protein from the virus resulted in loss of approximately half of cell binding (comparing kRSV-GstopA2F to kRSV-A2GA2F in Fig. 2), consistent with previously published data (19). However, removal of 2-20 G protein from the virus resulted in loss of 90% of cell binding (comparing kRSV-Gstop2-20F to kRSV-2-20G2-20F in Fig. 2). We found similar virus attachment results using these viruses that were not sucrose purified (data not shown). Thus, 2-20 G displayed a greater contribution to virus binding to BEAS-2B cells than A2 G.
FIG 2.
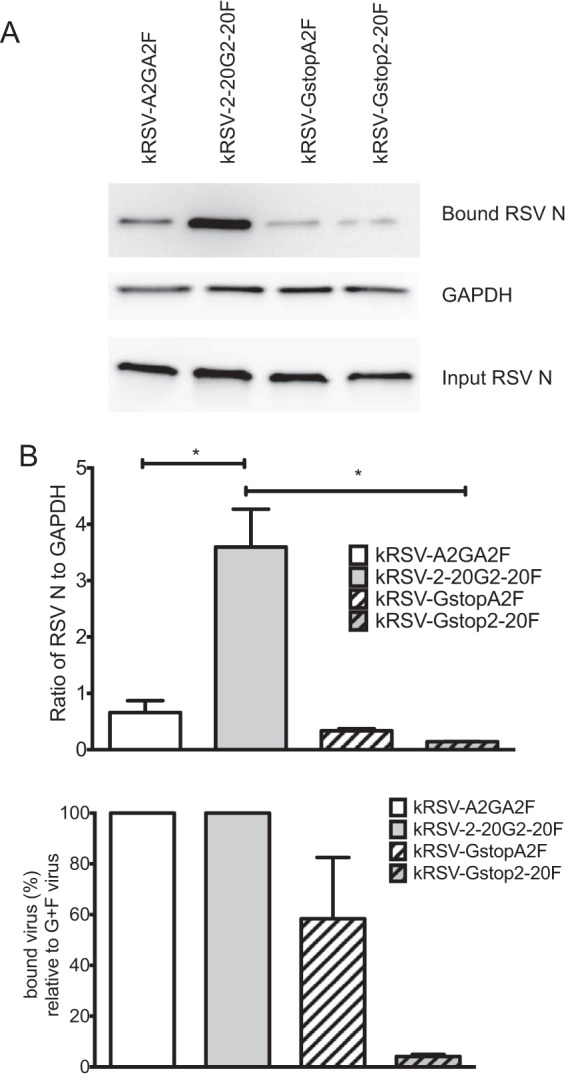
Greater contribution of 2-20 G than A2 G to binding BEAS-2B cells. (A) Western blots showing results of a representative binding assay. Input virus inocula were normalized based on the N protein expression levels as shown on the bottom blot. (B) Densitometry analysis of bound virus. Virus binding activity (top panel) was calculated by normalizing the N level (bound RSV N from the top blot) to the GAPDH level of the sample. This value for the Gstop viruses was also normalized to that of its parental virus containing both F and G and is represented as the percentage of bound virus compared to the percentage of G+F virus (bottom panel). *, P < 0.05 (comparing the bracketed groups using one-way ANOVA). Data are shown as means ± SEM and represent the combined results of three replicate experiments.
Greater contribution of 2-20 G than A2 G to virus entry kinetics.
To further explore the functional differences between 2-20 G and A2 G, we tested whether virus entry into host cells would be differentially affected by the removal of A2 or 2-20 G protein. We quantified entry kinetics of all four viruses in BEAS-2B cells by the use of a citric acid wash entry assay (25, 28). kRSV-A2GA2F had the fastest entry kinetics, followed by kRSV-GstopA2F (Fig. 3A). Removal of A2 G resulted in 3-fold-lower entry efficiency of the virus (Fig. 3B). Removal of 2-20 G from kRSV-2-20G2-20F resulted in approximately 8-fold-lower entry efficiency (Fig. 3C). These data demonstrate that although kRSV-2-20G2-20F had lower entry efficiency than kRSV-A2GA2F, 2-20 G played a relatively greater role in cell entry than A2 G. Additionally, the A2 F protein was significantly more efficient than the 2-20 F protein at mediating entry in BEAS-2B cells.
FIG 3.
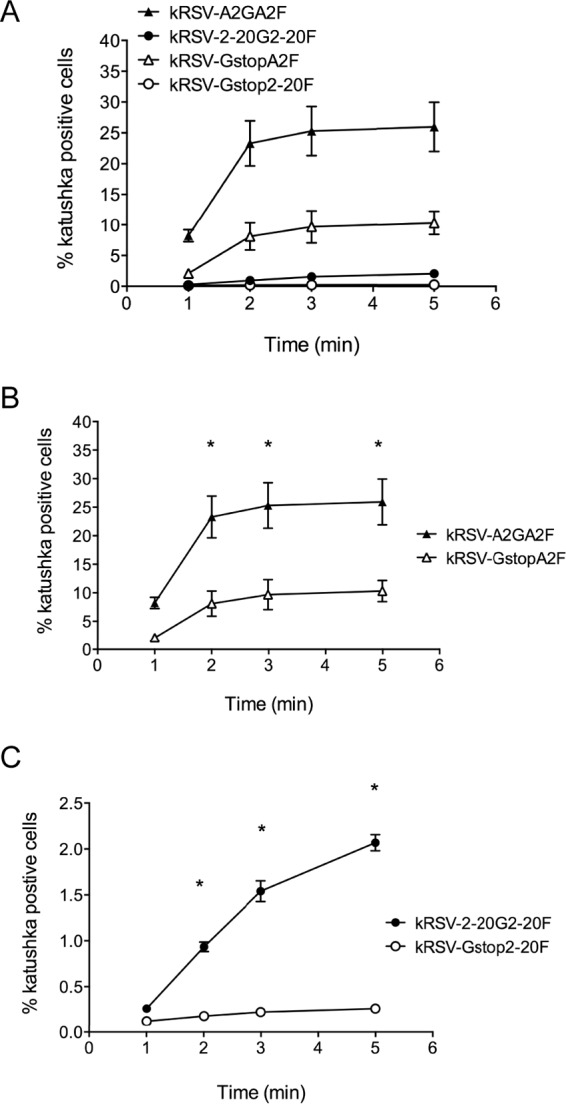
Entry kinetics in BEAS-2B cells. (A) Entry kinetics of kRSV-A2GA2F, kRSV-2-20G2-20F, kRSV-GstopA2F, and kRSV-Gstop2-20F in BEAS-2B cells using an acid wash protocol as described in Materials and Methods. (B) Contribution of A2 G protein to the entry kinetics of kRSV-A2GA2F compared to kRSV-GstopA2F. (C) Contribution of 2-20 G protein to the entry kinetics of kRSV-2-20G2-20F compared to kRSV-Gstop2-20F. *, P < 0.05 (comparing Gstop virus to GF virus at the same time point using one-way ANOVA). Data are represented as means ± SEM. The graphs in panels B and C were constructed using the data in panel A. The data represent the combined results from three independent experiments.
In vitro infectivity.
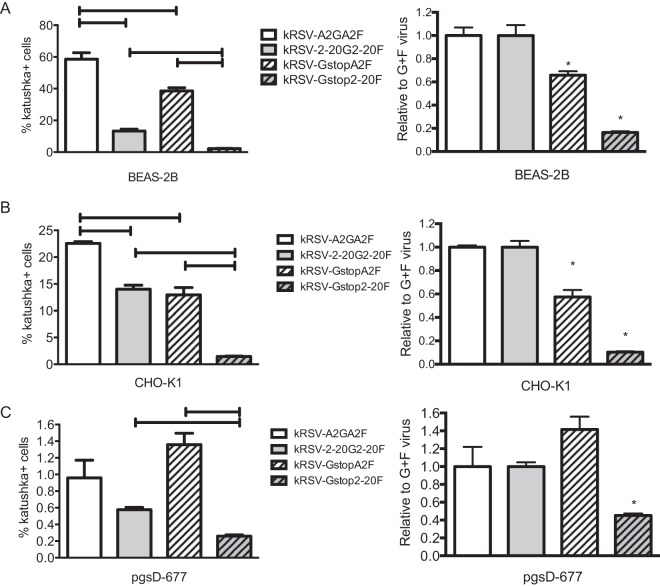
As 2-20 G contributed relatively more than A2 G to virus binding to host cells and entry kinetics, we compared the roles of A2 G and 2-20 G in infectivity in vitro. Infectivity of kRSV-A2GA2F, kRSV-2-20G2-20F, kRSV-GstopA2F, and kRSV-Gstop2-20F was assayed in three cell lines, BEAS-2B, CHO-K1, and pgsD-677. CHO-K1 and pgsD-677 were used for comparing the effects of the presence and absence of GAGs (pgsD-677 is a heparan sulfate-deficient CHO-K1 derivative). In BEAS-2B and CHO-K1 cells, kRSV-A2GA2F and kRSV-2-20G2-20F exhibited greater infectivity than kRSV-GstopA2F and kRSV-Gstop2-20F, respectively (Fig. 4). In pgsD-677 cells, kRSV-A2GA2F and kRSV-GstopA2F exhibited similar levels of infectivity, while kRSV-2-20G2-20F displayed greater infectivity than kRSV-Gstop2-20F (Fig. 4). To compare the relative contributions of the G proteins to virus infectivity, we normalized the percentage of cells infected by the Gstop viruses to the percentage of cells infected by the viruses containing both G and F proteins (kRSV-GstopA2F normalized to kRSV-A2GA2F and kRSV-Gstop2-20F normalized to kRSV-2-20G2-20F; Fig. 4). In BEAS-2B and CHO-K1 cells (both expressing heparan sulfate), A2 G contributed to 30% to 40% infectivity whereas 2-20 G contributed to more than 80% (Fig. 4A and B). Moreover, the contribution of 2-20 G protein was evident in the heparan sulfate-deficient pgsD-677 cell line whereas A2 G exhibited no significant contribution to infection in these cells (Fig. 4C). These data are consistent with the established role of A2 G binding to GAGs (10, 19) and suggest that the 2-20 G protein utilizes an additional host factor(s) in vitro.
FIG 4.
Infectivity in BEAS-2B, CHO-K1, and pgsD-677 cell lines. BEAS-2B (A), CHO-K1 (B), and pgsD-677 (C) cells were infected with kRSV-A2GA2F, kRSV-2-20G2-20F, kRSV-GstopA2F, or kRSV-Gstop2-20F at an MOI of 1. Twenty-four hours later, infected cells were quantified using flow cytometry. The left panels show the percentages of RSV-infected cells. Brackets represent a P value of <0.05 for the indicated group comparison. The right panels show the levels of infectivity of the virus without G protein normalized to that of the virus containing both G and F proteins of the same strain (kRSV-GstopA2F normalized to kRSV-A2GA2F and kRSV-Gstop2-20F normalized to kRSV-2-20G2-20F) expressed as values relative to the levels seen with G+F virus on the y axis. *, P < 0.05 (comparing each Gstop virus to the corresponding parental GF virus using one-way ANOVA). Data are represented as means ± SEM. Each graph depicts data from the combined results of three independent experiments.
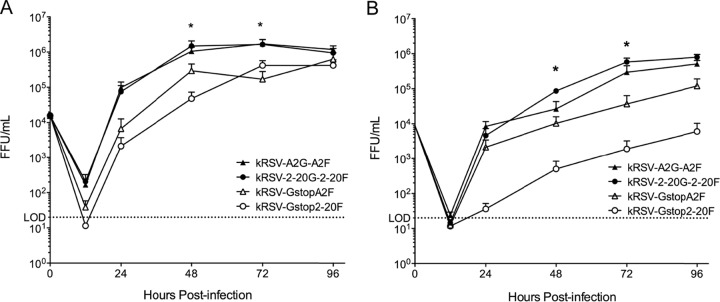
In vitro growth kinetics in HEp-2 and BEAS-2B cell lines.
We compared the growth kinetics of kRSV-A2GA2F, kRSV-2-20G2-20F, kRSV-GstopA2F, and kRSV-Gstop2-20F in the HEp-2 and BEAS-2B cell lines using an MOI of 0.01. As reported previously, the absence of G protein from A2 reduced the infectious yield of the virus in HEp-2 cells (Fig. 5A) (11). The origin of the G and F glycoprotein did not affect the growth of the virus, as kRSV-A2GA2F and kRSV-2-20G2-20F grew to similar levels in both cell lines (Fig. 5A). However, the absence of 2-20 G from the virus resulted in reduced infectious yield in HEp-2 cells and, to a greater degree, in BEAS-2B cells (Fig. 5B). Compared to kRSV-2-20G2-20F, kRSV-Gstop2-20F consistently had a titer in the BEAS-2B cell line that was more than 2 log10 lower. This growth deficiency was greater than that of kRSV-GstopA2F relative to kRSV-A2GA2F (≤1 log10 difference), suggesting that the 2-20 G protein contributes more to in vitro growth than the A2 G protein.
FIG 5.
Contribution of G protein to virus in vitro growth kinetics. HEp-2 cells (A) and BEAS-2B cells (B) were infected with kRSV-A2GA2F, kRSV-2-20G2-20F, kRSV-GstopA2F, or kRSV-Gstop2-20F using an MOI of 0.01. Time zero represents the calculated input titer. Virus titers at each time point thereafter were assayed by FFU assay. *, P < 0.05 (comparing the Gstop virus and parental GF virus at the same time point using one-way ANOVA). Data are represented as means ± SEM. LOD, limit of detection. FFU, fluorescent-focus unit. Each graph depicts data from the combined results of four independent experiments in each cell line.
Contribution of G proteins to binding and growth kinetics in primary airway epithelial cells.
Because RSV primarily infects ciliated airway epithelial cells, we investigated the effects of the absence of G on virus binding and growth in normal human bronchial epithelial (NHBE) cells cultured at the air-liquid interface (ALI). In a binding assay using well-differentiated NHBE/ALI cells, we found that kRSV-2-20G2-20F bound more efficiently than kRSV-A2GA2F (Fig. 6A and B). Additionally, when we normalized binding of the Gstop viruses with binding of the parental viruses to NHBE/ALI cells, we demonstrated that A2 G contributed very little to binding by kRSV-A2GA2F (Fig. 6B). This was in contrast to removal of 2-20 G from kRSV-2-20G2-20F, which resulted in loss of approximately 70% of binding (Fig. 6B). These results imply that most of the binding difference between kRSV-A2GA2F and kRSV-2-20G2-20F in these cells is due to the G protein and that 2-20 G serves a greater role than A2 G in binding primary airway epithelial cells.
FIG 6.
Contribution of A2 and 2-20 G to binding and growth kinetics in primary airway epithelial cells. (A) Representative Western blots showing binding assays in normal human bronchial epithelial/air-liquid interface (NHBE/ALI) cells. Input virus inocula were normalized based on the N protein expression levels as shown in the bottom blot. (B) Densitometry analysis of bound virus. Virus binding (top panel) was calculated by normalizing the F level to the GAPDH level of the sample. This value for the Gstop viruses was also normalized to that of the corresponding parental virus with both F and G and is represented as the percentage of bound virus compared to that of the G+F virus (bottom panel). Graphs depict results from two independent experiments performed in triplicate wells. (C) NHBE/ALI cells were infected with kRSV-A2GA2F, kRSV-2-20G2-20F, kRSV-Gstop-A2F, or kRSV-Gstop-2-20F at an MOI of 1.0. Time zero represents the calculated input titer. Virus titers at each time point thereafter were quantified by FFU assay. *, P < 0.05 (comparing the bracketed groups by one-way ANOVA in panel B and comparing kRSV-A2GA2F and kRSV-2-20G2-20F in panel C). Data from the results of two independent experiments performed in triplicate wells are represented as means ± SEM.
We also monitored the ability of kRSV-A2GA2F, kRSV-2-20G2-20F, kRSV-Gstop-A2F, and kRSV-Gstop-2-20F to infect NHBE/ALI cells. By days 5 and 6 postinfection, kRSV-2-20G2-20F exhibited greater infectious yield from NHBE/ALI cells than kRSV-A2GA2F (Fig. 6C). Neither Gstop virus was able to replicate to detectable levels in NHBE/ALI cells. While we did not detect a difference in the G-specific contributions of these viruses to infectivity in NHBE/ALI cells, our data indicate that, together, the 2-20 G and F proteins provide an advantage to kRSV-2-20G2-20F relative to kRSV-A2GA2F in primary human bronchial epithelial cells.
DISCUSSION
Previous studies analyzing functions of the RSV G protein used prototypical strains such as A2 (10, 11) or a subgroup B strain (9). In the present studies, deletion of the attachment protein from the virus attenuated virus replication in HEp-2 but not Vero cell lines. We reevaluated the requirement for G in the context of a recent clinical isolate strain of RSV and expanded our study to include a human bronchial epithelial cell line (BEAS-2B) as well as primary normal human bronchial epithelial cells cultured at the air-liquid interface (NHBE/ALI cells). We generated recombinant viruses containing G and F glycoproteins of either A2 or the 2-20 clinical isolate and compared the functional contributions of G from the A2 and 2-20 strains.
Congruent with previously published reports, removal of G from the A2 strain resulted in a decrease in virus-to-cell binding and infectivity compared to parental A2 results (10, 11). In our study, Gstop-A2F was approximately one-third less efficient at binding and one-third less infectious in BEAS-2B cells than kRSV-A2GA2F. We found that the 2-20 G protein showed greater contributions to virus attachment to host cells, virus entry, infectivity, and overall growth kinetics than the A2 G protein. In contrast to the A2 G protein, the 2-20 G protein played a role in infection of heparan sulfate-deficient cells, suggesting that the ability of 2-20 G to bind to a receptor(s) other than GAGs may contribute to the high level of attachment. The level of entry of BEAS-2B cells by virus expressing 2-20 G and 2-20 F was reduced approximately 10-fold compared to that seen with virus expressing A2 G and A2 F. It is likely that the lower entry efficiency of the virus expressing 2-20 G and 2-20 F is due to the 2-20 F protein, because the entry of the G-null virus expressing 2-20 F was approximately 40-fold lower than the entry of the G-null virus expressing A2F.
Earlier reports determined that A2 G is involved in binding of RSV to glycosaminoglycans (GAGs) on the surface of cells but that this GAG dependence varies depending on the cell line used to produce the virus (19, 29, 30). Using our Gstop mutant virus, we replicated this result with A2 (Fig. 4). kRSV-Gstop-2-20F, however, still exhibited reduced infectivity compared to the GF virus when heparan sulfate-containing GAGs were not present on cells. This implies that the 2-20 G protein likely binds other types of GAGs or a host cell protein that is not bound by A2 G. Our data from NHBE/ALI cells further support this, as kRSV-2-20G2-20F replicated to higher titers than kRSV-A2GA2F in these cells (Fig. 6C), which do not express heparan sulfate-containing GAGs (31).
Together, our data suggest that the F protein from a recent clinical isolate of RSV depends more on its attachment protein for efficient viral infection than a prototypical laboratory strain. Our results contrast with those of a recent study using clinical isolate 98-25147-X (RSV-X). In that study, deletion of G from the virus had little effect on replication in HEp-2 cells or Vero cells (32). We postulate one main reason for the divergent results. In that study, the G gene was completely deleted in the virus, resulting in a shift forward of the downstream genes along the transcriptional gradient (32). It is likely that the complete removal of the G gene resulted in greater F glycoprotein expression in this recombinant strain and that greater F levels potentially compensated more for the missing G function than was the case in the wild-type strain. Our study avoided this potential confounding factor by utilizing Gstop viruses, which still contain the complete G gene but do not express the G protein. On the other hand, our results may be confounded by using a chimeric virus approach because G and F expression and/or incorporation may be modulated by viral sequences in clinical isolates outside the G and F open reading frames.
In vitro culture-adapted strains may lose or gain biological functions during passage and selection in cell culture. This has been noted for a variety of paramyxoviruses, such as Newcastle disease virus (NDV). The fusion protein of NDV has a very stringent requirement for its HN protein, but there have been reports that changes of certain amino acids (amino acids 211, 289, and 463) in the F1 domain can completely abolish the requirement for HN for fusion (33, 34). It is possible that similar mutations can arise in RSV strains during serial passage in tissue culture, thus somehow changing the requirement for the RSV attachment protein function for infection. The RSV 2-20 clinical isolate used in the current study was generated as a low-passage-number (passage 12) stock, and both this stock and the initial viral stock (passage 2) were sequenced. There was one nucleotide change in the 2-20 F protein, resulting in an amino acid change at position 76. No nucleotide changes were seen in the G protein sequence (21).
There is some evidence suggesting that RSV strain differences contribute to different pathogenesis outcomes (21, 35–37). Several studies using different clinical isolates of RSV compared to the prototypical A2 strain demonstrated that some strains are more virulent in well-differentiated cell culture systems or in animal models (21, 35, 37). Thus, reevaluating the gene functions for clinical isolates of RSV may contribute to our understanding of the pathogenesis of RSV. The strain used in this study, 2-20, has been shown to induce disease that is more severe than that seen with RSV A2 in the BALB/c mouse model (21, 38, 39). The contribution of the 2-20 G protein may be linked to its pathogenesis, and it would be interesting to explore whether the relatively greater contribution of G can be extended to other RSV clinical isolates. The results from these further studies could potentially guide vaccine design and antiviral drug development.
Although many members of the Paramyxoviridae family, such as NDV and measles virus, have a strict requirement for the attachment protein (HN or H) to trigger fusion, there are some known exceptions in this virus family. The fusion protein of parainfluenza virus 5 (PIV5) has been shown to induce membrane fusion independently of the abundance of its HN protein, which is thought to provide only the binding activity necessary for the optimal distance between the fusion protein and the cellular target(s) (40). In addition to RSV, other members of the Pneumovirus genus, namely, human metapneumovirus and bovine RSV, harbor attachment glycoproteins which are dispensable for virus growth in vitro (41, 42). Previous work determined that the 2-20 G protein enhanced fusion of the 2-20 F protein (13). The G protein of RSV has been thought to simply facilitate fusion enhancement by bridging the two membranes in close proximity (7). At this point, we cannot rule out this scenario for RSV 2-20 G as it did enhance the binding activity of the virus to the host cells, as shown in this study. Future studies may determine the role of specific domains in 2-20 G as important for boosting 2-20 F fusion activity. As the virus containing only A2 F still retained more than half of the binding activity seen with the virus with only 2-20 F (Fig. 2), differences in the F binding activity could be the determining factor for their different levels of dependence on the G protein for infection. Further studies are needed to completely dissect the interaction of RSV G and F proteins in regard to fusion activity, and, as a whole, more focus on clinically relevant viruses will aid our knowledge of this evolving pathogen.
ACKNOWLEDGMENTS
We thank the Emory Children's Pediatric Research Center flow cytometry core supported by Children's Healthcare of Atlanta (CHOA).
We thank Edward Walsh for the monoclonal antibody to the RSV N protein and Ursula Buchholz and Karl-Klaus Conzelmann for the BSR-T7/5 cell line. We also thank Nancy Ulbrandt (MedImmune) for providing motavizumab antibody.
M.L.M. and Emory University are entitled to licensing fees derived from various agreements that Emory has entered into related to products used in this research described in this paper. This study could affect the personal financial status of M.L.M. The terms of this agreement have been reviewed and approved by Emory University in accordance with its conflict-of-interest policies.
Funding Statement
This study was also supported by funds from Emory University and CHOA. Additional support was provided by the NIH through Emory Vaccinology Training Grant T32AI074492 (Christopher C. Stobart), by Emory and CHOA under joint grant number 33515 (Michael G. Currier, Christopher C. Stobart, Sujin Lee, and Martin L. Moore), and by Emory through Nelson Memorial Fund R6336510 (Anne L. Hotard and Martin L. Moore).
REFERENCES
- 1.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simoes EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stockman LJ, Curns AT, Anderson LJ, Fischer-Langley G. 2012. Respiratory syncytial virus-associated hospitalizations among infants and young children in the United States, 1997–2006. Pediatr Infect Dis J 31:5–9. doi: 10.1097/INF.0b013e31822e68e6. [DOI] [PubMed] [Google Scholar]
- 3.Guvenel AK, Chiu C, Openshaw PJ. 2014. Current concepts and progress in RSV vaccine development. Expert Rev Vaccines 13:333–344. doi: 10.1586/14760584.2014.878653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeVincenzo JP, Whitley RJ, Mackman RL, Scaglioni-Weinlich C, Harrison L, Farrell E, McBride S, Lambkin-Williams R, Jordan R, Xin Y, Ramanathan S, O'Riordan T, Lewis SA, Li X, Toback SL, Lin SL, Chien JW. 2014. Oral GS-5806 activity in a respiratory syncytial virus challenge study. N Engl J Med 371:711–722. doi: 10.1056/NEJMoa1401184. [DOI] [PubMed] [Google Scholar]
- 5.Mackman RL, Sangi M, Sperandio D, Parrish JP, Eisenberg E, Perron M, Hui H, Zhang L, Siegel D, Yang H, Saunders O, Boojamra C, Lee G, Samuel D, Babaoglu K, Carey A, Gilbert BE, Piedra PA, Strickley R, Iwata Q, Hayes J, Stray K, Kinkade A, Theodore D, Jordan R, Desai M, Cihlar T. 2015. Discovery of an oral respiratory syncytial virus (RSV) fusion inhibitor (GS-5806) and clinical proof of concept in a human RSV challenge study. J Med Chem 58:1630–1643. doi: 10.1021/jm5017768. [DOI] [PubMed] [Google Scholar]
- 6.Wang G, Deval J, Hong J, Dyatkina N, Prhavc M, Taylor J, Fung A, Jin Z, Stevens SK, Serebryany V, Liu J, Zhang Q, Tam Y, Chanda SM, Smith DB, Symons JA, Blatt LM, Beigelman L. 2015. Discovery of 4′-chloromethyl-2′-deoxy-3′,5′-di-O-isobutyryl-2′-fluorocytidine (ALS-8176), a first-in-class RSV polymerase inhibitor for treatment of human respiratory syncytial virus infection. J Med Chem 58:1862–1878. doi: 10.1021/jm5017279. [DOI] [PubMed] [Google Scholar]
- 7.Chang A, Dutch RE. 2012. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses 4:613–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plattet P, Plemper RK. 2013. Envelope protein dynamics in paramyxovirus entry. mBio 4:e00413-13. doi: 10.1128/.00413-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karron RA, Buonagurio DA, Georgiu AF, Whitehead SS, Adamus JE, Clements-Mann ML, Harris DO, Randolph VB, Udem SA, Murphy BR, Sidhu MS. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci U S A 94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Techaarpornkul S, Barretto N, Peeples ME. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J Virol 75:6825–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teng MN, Whitehead SS, Collins PL. 2001. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 289:283–296. [DOI] [PubMed] [Google Scholar]
- 12.Teng MN, Collins PL. 1998. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J Virol 72:5707–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stokes KL, Currier MG, Sakamoto K, Lee S, Collins PL, Plemper RK, Moore ML. 2013. The respiratory syncytial virus fusion protein and neutrophils mediate the airway mucin response to pathogenic respiratory syncytial virus infection. J Virol 87:10070–10082. doi: 10.1128/JVI.01347-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox RG, Williams JV. 2013. Breaking in: human metapneumovirus fusion and entry. Viruses 5:192–210. doi: 10.3390/v5010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox RG, Livesay SB, Johnson M, Ohi MD, Williams JV. 2012. The human metapneumovirus fusion protein mediates entry via an interaction with RGD-binding integrins. J Virol 86:12148–12160. doi: 10.1128/JVI.01133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman SA, Audet S, Beeler JA. 2000. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J Virol 74:6442–6447. doi: 10.1128/JVI.74.14.6442-6447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldman SA, Hendry RM, Beeler JA. 1999. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J Virol 73:6610–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine S, Klaiber-Franco R, Paradiso PR. 1987. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J Gen Virol 68(Pt 9):2521–2524. doi: 10.1099/0022-1317-68-9-2521. [DOI] [PubMed] [Google Scholar]
- 19.Techaarpornkul S, Collins PL, Peeples ME. 2002. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology 294:296–304. doi: 10.1006/viro.2001.1340. [DOI] [PubMed] [Google Scholar]
- 20.Tayyari F, Marchant D, Moraes TJ, Duan W, Mastrangelo P, Hegele RG. 2011. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat Med 17:1132–1135. [DOI] [PubMed] [Google Scholar]
- 21.Stokes KL, Chi MH, Sakamoto K, Newcomb DC, Currier MG, Huckabee MM, Lee S, Goleniewska K, Pretto C, Williams JV, Hotard A, Sherrill TP, Peebles RS Jr, Moore ML. 2011. Differential pathogenesis of respiratory syncytial virus clinical isolates in BALB/c mice. J Virol 85:5782–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hotard AL, Shaikh FY, Lee S, Yan D, Teng MN, Plemper RK, Crowe JE Jr, Moore ML. 2012. A stabilized respiratory syncytial virus reverse genetics system amenable to recombination-mediated mutagenesis. Virology 434:129–136. doi: 10.1016/j.virol.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham BS, Perkins MD, Wright PF, Karzon DT. 1988. Primary respiratory syncytial virus infection in mice. J Med Virol 26:153–162. doi: 10.1002/jmv.1890260207. [DOI] [PubMed] [Google Scholar]
- 24.Boyoglu-Barnum S, Gaston KA, Todd SO, Boyoglu C, Chirkova T, Barnum TR, Jorquera P, Haynes LM, Tripp RA, Moore ML, Anderson LJ. 2013. A respiratory syncytial virus (RSV) anti-G protein F(ab′)2 monoclonal antibody suppresses mucous production and breathing effort in RSV rA2-line19F-infected BALB/c mice. J Virol 87:10955–10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White LK, Yoon JJ, Lee JK, Sun A, Du Y, Fu H, Snyder JP, Plemper RK. 2007. Nonnucleoside inhibitor of measles virus RNA-dependent RNA polymerase complex activity. Antimicrob Agents Chemother 51:2293–2303. doi: 10.1128/AAC.00289-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng J, Lee S, Hotard AL, Moore ML. 2014. Refining the balance of attenuation and immunogenicity of respiratory syncytial virus by targeted codon deoptimization of virulence genes. mBio 5:e01704-14. doi: 10.1128/mBio.01704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crim RL, Audet SA, Feldman SA, Mostowski HS, Beeler JA. 2007. Identification of linear heparin-binding peptides derived from human respiratory syncytial virus fusion glycoprotein that inhibit infectivity. J Virol 81:261–271. doi: 10.1128/JVI.01226-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan D, Lee S, Thakkar VD, Luo M, Moore ML, Plemper RK. 2014. Cross-resistance mechanism of respiratory syncytial virus against structurally diverse entry inhibitors. Proc Natl Acad Sci U S A 111:E3441–E3449. doi: 10.1073/pnas.1405198111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hallak LK, Spillmann D, Collins PL, Peeples ME. 2000. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J Virol 74:10508–10513. doi: 10.1128/JVI.74.22.10508-10513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwilas S, Liesman RM, Zhang L, Walsh E, Pickles RJ, Peeples ME. 2009. Respiratory syncytial virus grown in Vero cells contains a truncated attachment protein that alters its infectivity and dependence on glycosaminoglycans. J Virol 83:10710–10718. doi: 10.1128/JVI.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Monzon ME, Casalino-Matsuda SM, Forteza RM. 2006. Identification of glycosaminoglycans in human airway secretions. Am J Respir Cell Mol Biol 34:135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Widjojoatmodjo MN, Boes J, van Bers M, van Remmerden Y, Roholl PJ, Luytjes W. 2010. A highly attenuated recombinant human respiratory syncytial virus lacking the G protein induces long-lasting protection in cotton rats. Virol J 7:114. doi: 10.1186/1743-422X-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ayllón J, Villar E, Muñoz-Barroso I. 2010. Mutations in the ectodomain of Newcastle disease virus fusion protein confer a hemagglutinin-neuraminidase-independent phenotype. J Virol 84:1066–1075. doi: 10.1128/JVI.01473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sergel TA, McGinnes LW, Morrison TG. 2000. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J Virol 74:5101–5107. doi: 10.1128/JVI.74.11.5101-5107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villenave R, Thavagnanam S, Sarlang S, Parker J, Douglas I, Skibinski G, Heaney LG, McKaigue JP, Coyle PV, Shields MD, Power UF. 2012. In vitro modeling of respiratory syncytial virus infection of pediatric bronchial epithelium, the primary target of infection in vivo. Proc Natl Acad Sci U S A 109:5040–5045. doi: 10.1073/pnas.1110203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melero JA, Moore ML. 2013. Influence of respiratory syncytial virus strain differences on pathogenesis and immunity. Curr Top Microbiol Immunol 372:59–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derscheid RJ, van Geelen A, Gallup JM, Kienzle T, Shelly DA, Cihlar T, King RR, Ackermann MR. 2014. Human respiratory syncytial virus Memphis 37 causes acute respiratory disease in perinatal lamb lung. Biores Open Access 3:60–69. doi: 10.1089/biores.2013.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Almeida Nagata DE, Demoor T, Ptaschinski C, Ting HA, Jang S, Reed M, Mukherjee S, Lukacs NW. 2014. IL-27R-mediated regulation of IL-17 controls the development of respiratory syncytial virus-associated pathogenesis. Am J Pathol 184:1807–1818. doi: 10.1016/j.ajpath.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen BC, Dolgachev V, Rasky A, Lukacs NW. 2014. IL-17E (IL-25) and IL-17RB promote respiratory syncytial virus-induced pulmonary disease. J Leukoc Biol 95:809–815. doi: 10.1189/jlb.0913482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dutch RE, Joshi SB, Lamb RA. 1998. Membrane fusion promoted by increasing surface densities of the paramyxovirus F and HN proteins: comparison of fusion reactions mediated by simian virus 5 F, human parainfluenza virus type 3 F, and influenza virus HA. J Virol 72:7745–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biacchesi S, Skiadopoulos MH, Yang L, Lamirande EW, Tran KC, Murphy BR, Collins PL, Buchholz UJ. 2004. Recombinant human Metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. J Virol 78:12877–12887. doi: 10.1128/JVI.78.23.12877-12887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karger A, Schmidt U, Buchholz UJ. 2001. Recombinant bovine respiratory syncytial virus with deletions of the G or SH genes: G and F proteins bind heparin. J Gen Virol 82:631–640. doi: 10.1099/0022-1317-82-3-631. [DOI] [PubMed] [Google Scholar]