ABSTRACT
Human gastrointestinal tract research is limited by the paucity of in vitro intestinal cell models that recapitulate the cellular diversity and complex functions of human physiology and disease pathology. Human intestinal enteroid (HIE) cultures contain multiple intestinal epithelial cell types that comprise the intestinal epithelium (enterocytes and goblet, enteroendocrine, and Paneth cells) and are physiologically active based on responses to agonists. We evaluated these nontransformed, three-dimensional HIE cultures as models for pathogenic infections in the small intestine by examining whether HIEs from different regions of the small intestine from different patients are susceptible to human rotavirus (HRV) infection. Little is known about HRVs, as they generally replicate poorly in transformed cell lines, and host range restriction prevents their replication in many animal models, whereas many animal rotaviruses (ARVs) exhibit a broader host range and replicate in mice. Using HRVs, including the Rotarix RV1 vaccine strain, and ARVs, we evaluated host susceptibility, virus production, and cellular responses of HIEs. HRVs infect at higher rates and grow to higher titers than do ARVs. HRVs infect differentiated enterocytes and enteroendocrine cells, and viroplasms and lipid droplets are induced. Heterogeneity in replication was seen in HIEs from different patients. HRV infection and RV enterotoxin treatment of HIEs caused physiological lumenal expansion detected by time-lapse microscopy, recapitulating one of the hallmarks of rotavirus-induced diarrhea. These results demonstrate that HIEs are a novel pathophysiological model that will allow the study of HRV biology, including host restriction, cell type restriction, and virus-induced fluid secretion.
IMPORTANCE Our research establishes HIEs as nontransformed cell culture models to understand human intestinal physiology and pathophysiology and the epithelial response, including host restriction of gastrointestinal infections such as HRV infection. HRVs remain a major worldwide cause of diarrhea-associated morbidity and mortality in children ≤5 years of age. Current in vitro models of rotavirus infection rely primarily on the use of animal rotaviruses because HRV growth is limited in most transformed cell lines and animal models. We demonstrate that HIEs are novel, cellularly diverse, and physiologically relevant epithelial cell cultures that recapitulate in vivo properties of HRV infection. HIEs will allow the study of HRV biology, including human host-pathogen and live, attenuated vaccine interactions; host and cell type restriction; virus-induced fluid secretion; cell-cell communication within the epithelium; and the epithelial response to infection in cultures from genetically diverse individuals. Finally, drug therapies to prevent/treat diarrheal disease can be tested in these physiologically active cultures.
INTRODUCTION
Knowledge of the human small intestine has been limited by the lack of in vitro systems that recapitulate its complex nature and functions. In recent years, human intestinal enteroids (HIEs) that exhibit a similar cellular composition to and many functional, region-specific aspects of the human gastrointestinal epithelium have been established (1–4). HIEs are produced from small intestinal tissues donated by consenting individuals. The epithelial crypt domains are isolated and cultured in Wnt3A-rich growth medium ex vivo, resulting in three-dimensional cultures that contain a stem cell niche and all the differentiated epithelial cell types surrounding a single lumenal compartment (2). These HIEs offer advantages over existing cell lines because they are human, are not transformed, and demonstrate many of the biological and physiological properties of the small intestinal epithelium (1, 5). In addition, unlike many primary tissue models, HIEs are long-lived and can be passaged as well as frozen for later use (6). HIEs have been used to study intestinal stem cell behavior and epithelial responses to injury, and we have pioneered the use of HIEs to understand human gastrointestinal viral host-pathogen interactions (1, 4, 7, 8). HIEs exhibit advantages over human intestinal organoids (HIOs) derived from approved stem cell lines, which we studied previously. HIOs contain epithelial and mesenchymal cells, both of which were infected by rotaviruses (RVs), including human RV (HRV), but only a few cells were infected, possibly due to the more fetal phenotype of HIOs (9, 10). HIOs also required significantly more time to be established, maintained, and infected. Therefore, we have now examined HIE cultures because they are easier to establish from different intestinal regions from multiple patients and are reportedly more differentiated (4).
HRVs remain the major cause of severe diarrheal illness worldwide, accounting for an estimated 453,000 annual deaths in children under the age of 5 years (11). With the recent introduction of two licensed rotavirus vaccines (pentavalent RotaTeq [RV5]; Merck, Whitehouse Station, NJ, and monovalent Rotarix [RV1]; GlaxoSmithKline, Brentwood, UK), it is predicted that the global death toll attributable to rotaviruses will decrease substantially (12, 13). However, vaccine efficacy remains suboptimal in low-income settings, where the burden of disease is greatest (13–15). Recent studies have distinguished HRVs from animal rotaviruses (ARVs) based on their respective receptor usages for initial infection, with most HRVs binding human histo-blood group antigens (HBGAs) and ARVs binding sialylated glycans (16–19). HBGAs, namely, ABH and Lewis antigens, have been suggested to be genetic factors that determine host susceptibility (20), and both secretor status and Lewis status (regulated by the fucosyltransferase 2 [FUT2] and fucosyltransferase 3 enzymes, respectively) have been proposed to mediate susceptibility to infection and possibly vaccination in a rotavirus genotype-dependent manner (21). Further studies on HRV-host interactions at the cellular level are needed to advance the understanding of host susceptibility and disease pathophysiology as well as aid in the development of more efficient modes of treatment and vaccination.
Most in vitro studies of rotavirus pathogenesis in cultured cells have been performed by using simian rotavirus (rhesus rotavirus [RRV] or simian agent 11 [SA11]) to infect either homologous monkey kidney cell lines or heterologous human colonic adenocarcinoma cell lines (e.g., HT-29 and Caco-2), partially due to the limited repertoire of nontransformed human small intestinal cell culture lines (22–24). Homologous infection generally results in increased infection and disease in vivo and is the predominant form of infection seen in nature (25, 26). Many human rotaviruses do not infect or poorly infect small animal models, are attenuated in gnotobiotic large animal models (27–30), and typically grow to low titers in current in vitro models, even after cell culture adaptation, compared to animal rotaviruses (31, 32). Thus, there are few robust, biologically relevant models for studying HRV infection, as the property of host range restriction requires the study of HRV infection in human-derived cells. An in vitro model that better recapitulates in vivo infection of humans would ideally consist of a human rotavirus infecting nontransformed human small intestinal cell cultures, the natural tissue tropism of human rotaviruses.
In this study, we evaluated whether HIEs represent a robust, new, biologically relevant in vitro culture model that can be used to study aspects of human rotavirus biology and pathophysiology that have not been fully assessed previously. We examined the host range restriction of simian and human rotaviruses, including the RV1 vaccine strain; cell type restriction; and the contribution of cell differentiation to infectivity and measured enterotoxin/virus-induced fluid secretion. In addition, because HIEs can be generated from tissue donated by different people, we evaluated whether properties of HRV infection are conserved among HIEs from different individuals. We demonstrate that HIEs are a novel, genetically diverse, in vitro human model to study the replication and pathophysiology of human rotavirus infection.
MATERIALS AND METHODS
Cell lines and viruses.
African green monkey kidney (MA104) cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA). Rhesus rotavirus (G3P[3]) and human rotavirus strains Ito (G3P[8]) and Wa (G1P[8]) were propagated in MA104 cells, and for some experiments, triple-layered particles were purified by using CsCl gradient centrifugation as previously described (33). Five different preparations of RRV and three different preparations of strain Ito were utilized in this study. Lyophilized formulations of the Rotarix vaccine (G1P[8]; GlaxoSmithKline, Brentford, United Kingdom) were suspended in TNC buffer (10 mM Tris-HCl, 120 mM NaCl, 10 mM CaCl2 [pH 7.4]) (33) prior to use. The titer of virus preparations was determined by a plaque assay or a fluorescent-focus assay (FFA) in MA104 cells (34).
Human intestinal enteroid culture and media.
Duodenal and ileal biopsy specimens were obtained from adults during routine endoscopy at Baylor College of Medicine through the Texas Medical Center Digestive Diseases Center Study Design and Clinical Research Core. Jejunal tissue was obtained from patients undergoing bariatric surgery. The Baylor College of Medicine Institutional Review Board approved the study protocol (protocol numbers H-13793 and H-31910). HIEs were prepared from the tissue samples as previously described (2).
Three different types of media were used to establish, maintain, or differentiate HIEs. Complete medium without growth factors [CMGF(−) medium], also known as basal culture medium, consisted of advanced DMEM–F-12 medium (Invitrogen) supplemented with 100 U/ml penicillin-streptomycin (Invitrogen), 10 mM HEPES buffer (Invitrogen), and 1× GlutaMAX (Invitrogen) (2).
Complete medium with growth factors (CMGF+ medium), also known as stem cell culture-conditioned medium (2), consisted of CMGF(−) medium supplemented with 50 ng/ml epidermal growth factor (EGF) (Invitrogen), 10% Noggin-conditioned medium (made from Noggin-producing cells; kindly provided by G. R. van den Brink, Amsterdam, The Netherlands) (described in reference 35), 20% R-spondin-conditioned medium (R-spondin-producing cells; kindly provided by Calvin Kuo, Palo Alto, CA), 50% Wnt3A-conditioned medium produced from ATCC CRL-2647 cells (ATCC, Manassas, VA), 10 mM nicotinamide (Sigma-Aldrich, St. Louis, MO), 10 nM gastrin I (Sigma-Aldrich), 500 nM A-83-01 (Tocris Bioscience, Bristol, United Kingdom), 10 μM SB202190 (Sigma-Aldrich), 1× B27 supplement (Invitrogen), 1× N2 supplement (Invitrogen), and 1 mM N-acetylcysteine (Sigma-Aldrich).
Differentiation medium consisted of the same components as those of CMGF+ medium without the addition of Wnt3A, SB202190, and nicotinamide as well as 50% reductions in the concentrations of Noggin and R-spondin.
HIEs were passaged in CMGF+ medium in phenol red-free, growth factor-reduced Matrigel (Corning, Corning, NY) and either (i) frozen in liquid nitrogen for later use or (ii) kept in Matrigel and used for infection experiments after being cultured in CMGF+ medium for 4 days followed by differentiation medium for 3 to 4 days.
To quantify the number of cells in HIE samples for use in calculations of multiplicity of infection (MOI), HIEs were dissociated into a single-cell suspension with Accutase cell dissociation solution (BD Biosciences) for 30 min at 37°C, and the number of cells was then quantified by using a Coulter Z1 particle counter (Beckman Coulter, Brea, CA). The MOI was calculated as amount of input virus/total number of cells in samples of HIEs. The total number of cells within a Matrigel plug of HIEs ranged from 100,000 to 200,000.
Viral infections and determination of viral infectivity.
Three to four days after culture in differentiation medium, HIEs were washed twice in cold CMGF(−) medium to remove the Matrigel. Mock-treated MA104 cell lysates and rotavirus preparations made in MA104 cells were each treated with 10 μg/ml trypsin (Worthington Biochemical Corporation, Lakewood, NJ) for 30 min at 37°C to enhance RV infectivity. HIEs were distributed equally into 5-ml round-bottom polystyrene tubes (Falcon, Corning, NY). HIEs were inoculated with different amounts of virus to achieve the desired MOI (0.5 to 20 PFU/cell or focus-forming units [FFU]/cell); mock-infected samples received a volume of trypsin-treated MA104 cell lysate equivalent to that used for viral infections. HIEs were vigorously pipetted 10 to 20 times with a P200 pipette to disperse and open the HIEs for apical exposure to the virus.
Apical exposure following pipetting was confirmed by treating HIEs with fluorescein isothiocyanate (FITC)-labeled dextran and then imaging the HIEs on a Nikon A1Rs confocal microscope. Prior to pipetting, FITC-labeled dextran (9.5 kDa) (Sigma-Aldrich) was excluded from the lumen of HIEs. After dispersion of HIEs with a P200 pipette, FITC-labeled dextran was observed within the lumenal compartment of HIEs, demonstrating apical exposure of HIEs to the surrounding medium after pipetting (data not shown).
For experiments assessing the percentage of infected cells, HIEs in polystyrene tubes were placed in an incubator at 37°C with 5% CO2 for 2 h. After 2 h of virus adsorption in the presence of 0.1 to 0.2 mg/ml of porcine pancreatin (Sigma-Aldrich) prepared in CMGF(−) medium, HIEs were washed twice with CMGF(−) medium and centrifuged at 50 × g to 70 × g to remove the inoculum and pancreatin that were present during virus adsorption. Pelleted HIEs were then suspended in differentiation medium without trypsin or pancreatin and incubated for 18 to 20 h.
To assess viral growth, virus adsorption in the presence of 0.1 to 0.2 mg/ml pancreatin occurred for 1.5 h, after which HIEs were washed 4 times with CMGF(−) medium and centrifuged at 50 × g to 70 × g to remove the inoculum. Pelleted HIEs were then suspended in differentiation medium supplemented with 0.5 mg/ml pancreatin. HIEs were either immediately frozen at −80°C to assess the amount of virus present after washing (at 1.5 h postinfection [hpi] or 2 hpi) or incubated overnight and harvested at 24 hpi. All mock- and HRV-infected samples underwent two rounds of freezing and thawing followed by 2 min of sonication. The amount of infectious virus in each sample was determined by a fluorescent-focus assay in MA104 cells (34).
Assessment of percentage of infected cells by flow cytometry.
HIEs (mock and HRV infected) were treated with Accutase cell detachment solution (BD Biosciences) for 30 min at 37°C, followed by 5 min of centrifugation at 400 × g, and the pellet was resuspended in CMGF(−) medium to create a single-cell suspension. Cells were then fixed at 4°C in 300 μl Cytofix fixation buffer (BD Biosciences) for 20 min, washed with Perm/Wash permeabilization buffer (BD Biosciences), and incubated for 30 min at room temperature (RT) with a 1:20,000 dilution of rabbit polyclonal antirotavirus antibody diluted in permeabilization buffer (36). Cells were then washed in permeabilization buffer, followed by incubation for 30 min at RT with a 1:1,000 dilution of an Alexa Fluor 488-conjugated donkey anti-rabbit antibody (Invitrogen). Cells were washed, and Alexa Fluor 488-positive cells were quantified by using an LSRII flow cytometer (BD Biosciences). Doublet discrimination was used to gate on the single-cell population. From this gate, 10,000 events were analyzed.
Quantitative reverse transcription-PCR analysis.
Total RNA was extracted from HIEs by using the RNeasy minikit and the QIAshredder kit, followed by treatment with RNase-free DNase (Qiagen, Valencia, CA) according to the manufacturer's protocol. The amount and purity of RNA were determined with a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA). Quantitative reverse transcription-PCR (qRT-PCR) was performed on a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA), utilizing One-Step RT-qPCR ToughMix with the ROX reference dye according to the manufacturer's protocol (Quanta Biosciences, Gaithersburg, MD). TaqMan primer-probe mixes were obtained from Molecular Probes (Eugene, OR), with the following assay identification numbers: Hs00356112_m1 for sucrase-isomaltase (SI), Hs00158722_m1 for lactase (LCT), Hs00357579_g1 for intestinal-type alkaline phosphatase (ALPI), Hs00900370_m1 for chromogranin A (CHGA), Hs00300531_m1 for synaptophysin (SYP), Hs03005103_g1 for mucin 2 (MUC2), Hs00902278_m1 for trefoil factor 3 (TFF3), Hs00360716_m1 for defensin alpha 5 (DEFA5), Hs00426232_m1 for lysozyme (LYZ), Hs00969422_m1 for leucine-rich-repeat-containing G-protein-coupled receptor 5 (LGR5), Hs01032443_m1 for antigen identified by monoclonal antibody Ki-67 (MKI67), and Hs02758991_g1 for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Expression levels were normalized to GAPDH levels and analyzed with StepOne v2.1 software (Applied Biosystems) by using the 2−ΔΔCT method as previously described (37).
Cytotoxicity assay.
HIEs were infected with HRV at a high multiplicity of infection to perform a one-step growth curve, as described above. Mock- and virus-treated HIEs were inoculated with an equivalent volume of MA104 lysate. At the indicated time points, HIEs were pelleted at 400 × g, and the supernatant was harvested. The cell culture supernatants from mock- and HRV-inoculated HIEs were assessed for the amount of lactate dehydrogenase (LDH) by using the CytoTox 96 Non-Radioactive cytotoxicity assay according to the manufacturer's protocols (Promega, Madison, WI). Positive controls consisted of HIEs that were lysed by freeze-thawing and assessed for LDH quantity. The percent cytotoxicity was calculated as the amount of LDH in mock-infected and infected HIE culture supernatants divided by the mean of the positive-control values.
Electron microscopy.
Pelleted mock-treated and HRV-infected HIEs (at 10 hpi) were treated with fixation buffer (2.5% glutaraldehyde, 3% formaldehyde, 0.0025% picric acid, 100 μM CaCl2, and 50 μM MgCl2 in phosphate-buffered saline [PBS] [pH 7.4]) at 4°C overnight. Fixation was quenched with 50 mM glycine in PBS, followed by postfixation with 1% OsO4 in PBS. HIEs were stained en bloc with 1% aqueous uranyl acetate and then dehydrated in serial dilutions of ethanol (50%, 70%, 90%, 95% twice, and 100% twice), transitioned with propylene oxide, and infiltrated with several changes of Spurr's resin. HIEs were placed into Beem capsules and polymerized overnight at 60°C. Thin sections were cut on an RMC Powertome XL ultramicrotome, stained with lead citrate, and visualized with a JEOL 1230 transmission electron microscope at 80 keV by using a Gatan Ultrascan-1000 charge-coupled device (CCD) and Digital Micrograph software.
Swelling assays.
To assess swelling in response to agonists, differentiated HIEs were removed from Matrigel as described above, suspended in 100 μl of CMGF(−) medium, distributed into a 96-well optical imaging plate precoated with 15 μl of Matrigel, and incubated for at least 4 h at 37°C to allow the enteroids to reseal and return to a basal state after manipulations. Stock solutions of forskolin (Sigma-Aldrich), carbachol (Sigma-Aldrich), and NSP4 peptides derived from the SA11 rotavirus strain (produced as previously described [38]) were stored at −20°C. Serial dilutions of each stock were prepared immediately prior to the experiments. HIEs were imaged in an environmental chamber on a GE Deltavision deconvolution microscope (GE, Issaquah, WA), utilizing differential interference contrast (DIC) imaging. Coordinates for the locations of HIEs were determined, and 4 to 6 images were then acquired to calculate the baseline area of each HIE. Test compounds at the indicated dilutions were then added to the chamber. An image was taken every 15 s for the duration of the experiment. The images were analyzed by using ImageJ software. The cross-sectional area of each imaged HIE was modeled as an ellipse, and perpendicular radii were measured by ImageJ at baseline and at the end of the swelling assay. The cross-sectional area was calculated as π × a × b, where “a” and “b” represent the measured radii.
Because a majority (∼80%) of HIEs expanded in response to forskolin treatment, forskolin was used as a positive control for enteroid lumenal expansion in our assays with the enterotoxin NSP4. To ensure that a lack of HIE lumenal expansion in response to a stimulus was due to the inability of the peptide to induce fluid secretion and not because the enteroid was incapable of swelling, HIEs were treated with forskolin following 45 min of NSP4 treatment. The data set that was analyzed included all HIEs that expanded in response to NSP4 or that did not expand in response to NSP4 but expanded with forskolin treatment. HIEs that did not expand in response to NSP4 or subsequent forskolin treatment were excluded, as they were considered to be incapable of swelling.
To assess swelling following virus infection, differentiated HIEs were infected with HRV as described above, and after 1 h of virus adsorption, HIEs were washed and suspended in differentiation medium. HIEs were placed into a 96-well optical imaging plate coated with 30 μl of a 20% solution of Matrigel in CMGF(−) medium and allowed to reseal before imaging began at 2 hpi. HIEs were imaged by utilizing the same procedure as the one described above for the swelling agonists. Spherical HIEs were used and analyzed in these assays. Because cross-sectional areas of these HIEs were more circular than elliptical, only one radius was measured at each time point instead of two.
Fucosyltransferase 2 genotyping of human intestinal enteroids.
DNA was extracted from HIEs by using the QIAamp DNA minikit (Qiagen) according to the manufacturer's protocol. A 546-bp fragment of the FUT2 gene was amplified by using the following primers: 5′-AGCCTCAACATCAAAGGCACTGGGA-3′ (forward) and 5′-AACCAGTCCAGGGCCTGCTGTA-3′ (reverse). Fragments were sequenced and examined for the G428A and C571T nonsense mutations and the A385T mutation, which results in an unstable enzyme (39, 40). HIEs that were homozygous for one of these mutations were designated secretor-negative HIEs.
Immunohistochemistry.
HIEs were placed into Matrigel and fixed in 10% neutral buffered formalin overnight at 4°C. The HIE-containing Matrigel was then dehydrated and embedded in paraffin. For staining, 3-μm paraffin sections of the HIEs were used. Heat-mediated antigen retrieval was performed on HIE sections in 10 mM citrate buffer (pH 6.0). The following primary antibodies were prepared in PBS and incubated overnight at 4°C: rabbit polyclonal antirotavirus (1:500 dilution) (36), E-cadherin (1:100; BD Biosciences), mucin 2 (1:500; Santa Cruz Biotechnology, Dallas, TX), sucrase-isomaltase (1:100; Santa Cruz), chromogranin A (1:100; Novus Biologicals, Littleton, CO), and lysozyme (1:100; Dako, Carpinteria, CA) antibodies. Sections were washed three times in PBS containing 0.05% Tween 20 (PBS-T). Fluorescence was visualized by using Alexa Fluor-conjugated secondary antibodies.
HRV infection of enteroendocrine cells was detected with the rabbit antirotavirus antibody, followed by secondary staining with an Alexa Fluor 488-conjugated mouse anti-rabbit antibody. The rabbit anti-human chromogranin A antibody (Novus Biologicals) was directly labeled by using the Zenon rabbit IgG labeling kit (Life Technologies, Carlsbad, CA). The sections were incubated with antibodies according to the manufacturer's instructions.
All of the sections were mounted in ProLong Gold Antifade reagent (Invitrogen). Images were obtained on a Nikon A1Rs confocal laser scanning microscope, and the images were prepared for publication by using Nikon Elements software version 3.4. The confocal stacks were processed by using Image J, and final images were prepared by using Adobe Photoshop. Routine hematoxylin and eosin (H&E) staining was done to visualize the overall morphology of the HIEs, and periodic acid-Schiff (PAS) staining was performed to detect goblet cells in the HIEs.
Statistics.
Statistical analyses were performed with Microsoft Excel software using the unpaired Student t test and one-way analysis of variance (ANOVA) with post hoc testing of significant ANOVA results using the Tukey honestly significant difference (HSD) test. Figures depict representative results from experiments that were repeated two to three times unless otherwise specified. Results were considered significant when the P value was <0.05.
RESULTS
Enteroids are a novel in vitro model of the human small intestinal epithelium.
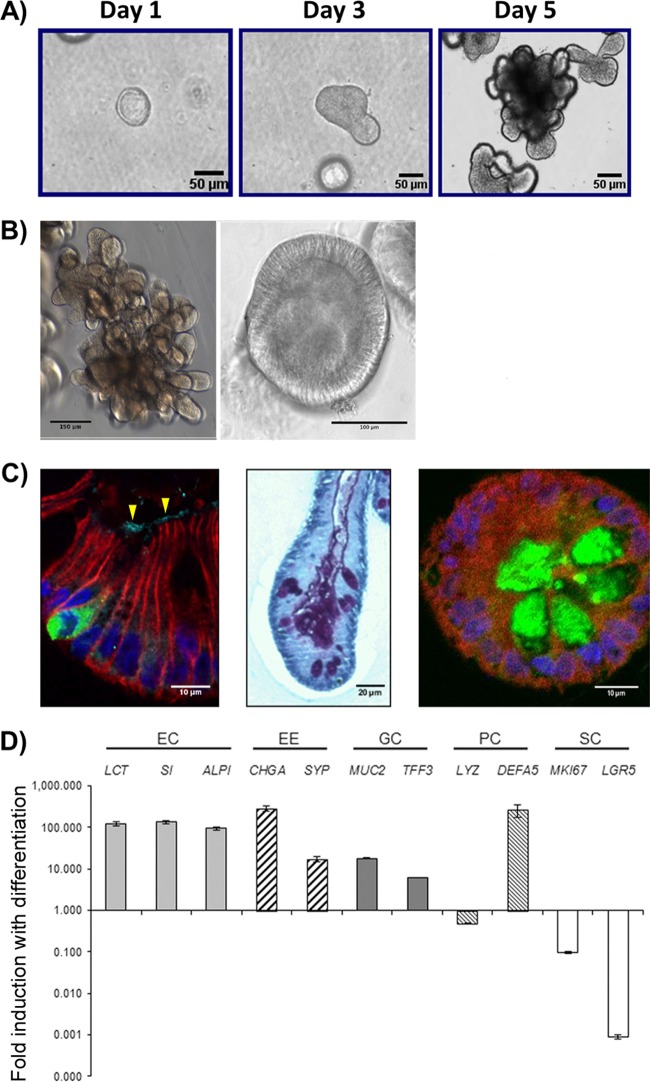
Prior to evaluation of HIE cultures as a model for HRV infection, we established a bank of cultures and characterized their structure and cell type composition as background information relevant for studying virus-host interactions. We successfully established with 100% efficiency HIEs from tissue samples of the duodenum (n = 17), jejunum (n = 13), and ileum (n = 24) from patients undergoing biopsy or gastric bypass surgery (jejunum only). Our HIE bank contains cultures from individuals who express different histo-blood group antigens; to date, we have focused on characterizing cultures from 10 secretor-positive and 3 secretor-negative individuals, all 3 of whom express the G428A nonsense mutation, which results in a premature stop codon in the FUT2 sequence. After ∼5 days in complete medium with growth factors (CMGF+ medium) (Fig. 1A), HIEs typically adopted either a multilobular (Fig. 1B, left) or a cystic (Fig. 1B, right) morphology. HIEs with both morphologies consisted of a single continuous lumen surrounded by epithelial cells. After cultivation in differentiation medium, HIEs contained differentiated epithelial cells, including absorptive enterocytes, enteroendocrine cells, and goblet cells, which increased in numbers as demonstrated by immunofluorescence analysis for protein expression and qRT-PCR for expression of cell type-specific transcripts, respectively, compared to those in undifferentiated HIEs (Fig. 1C and D). Differentiated HIEs from all segments of the small intestine showed the presence of each of these differentiated cell types (data not shown). Paneth cells were present in both undifferentiated and differentiated HIEs, as they are integral to maintaining the stem cell niche in both states (3).
FIG 1.
Characterization of differentiated human jejunal enteroids. (A) Representative images of jejunal enteroids grown over 5 days from intestinal crypts (bar = 50 μm). (B) After 5 days of growth in complete medium with growth factors (CMGF+ medium), enteroids typically result in two major morphologies, multilobular (left) (bar = 150 μm) and cystic (right) (bar = 100 μm). (C) Upon differentiation, enteroids contain the four major mature cell types of the small intestinal epithelium. (Left) Chromogranin A-containing enteroendocrine cells (green) and sucrase-isomaltase-expressing enterocytes (arrowheads) (bar = 10 μm). (Middle) Periodic acid-Schiff stain-reacting goblet cells (purple) (bar = 20 μm). (Right) Lysozyme-containing Paneth cells (green) (bar = 10 μm). E-cadherin (red) and DAPI (4′,6-diamidino-2-phenylindole) nuclear staining (blue) are shown in the left and right panels. (D) qRT-PCR results showing the fold change in levels of transcripts in differentiated enteroids relative to the transcript levels in undifferentiated enteroids. Transcript levels were first normalized to GAPDH levels prior to obtaining the relative fold change by using the 2−ΔΔCT method. Shown are markers for enterocytes (EC), enteroendocrine cells (EE), goblet cells (GC), Paneth cells (PC), and stem cells (SC). Gene symbols represent lactase (LCT), sucrase-isomaltase (SI), alkaline phosphatase (ALPI), chromogranin A (CHGA), synaptophysin (SYP), mucin 2 (MUC2), trefoil factor 3 (TFF3), lysozyme (LYZ), defensin alpha 5 (DEFA5), antigen identified by monoclonal antibody Ki-67 (MKI67), and leucine-rich-repeat-containing G-protein-coupled receptor 5 (LGR5) genes. Error bars indicate standard errors of the means (n = 3).
Human small intestinal enteroids are more susceptible to infection by homologous human rotavirus than heterologous animal rotavirus.
To assess if HIEs recapitulate in vivo observations of rotavirus host range restriction, differentiated HIEs were inoculated with ARV strain RRV (G3P[3]) or HRV strain Ito (G3P[8]) at a high multiplicity of infection (MOI) of ∼20. The percentage of cells infected was evaluated by detection of intracellular rotavirus antigen by flow cytometry. At 20 hpi, HRV Ito infected more cells (51.6% ± 10.8%) than did RRV (13.5% ± 2.4%) in HIEs from a single patient and resulted in a change in HIE morphology, with many more detached cells being present in the surrounding medium than in mock-infected HIEs (Fig. 2A). This observation was not dependent on specific virus preparations, as the same results were obtained after infections with multiple independent stocks of RRV (not serial passages of the same parental stock) and a different preparation of HRV Ito (data not shown). In addition, HRV Ito infected significantly more cells than did RRV in HIEs from multiple patients (data not shown), suggesting that the observation of host range restriction was not specific to HIE cultures from a single patient. These results demonstrate that jejunal HIEs are more susceptible to HRV than to RRV infection.
FIG 2.
Human rotavirus infection and replication properties in human intestinal enteroids. (A) Jejunal enteroids from one patient (patient j11) were either mock infected or infected with RV at an MOI of 20 PFU/cell. At 20 hpi, enteroids were visualized by light microscopy (left) (bar = 50 μm) for cytopathic effect. Enteroids were also assessed for the percentage of infected cells by flow cytometry. Infected cells were defined as cells containing intracellular rotavirus antigen as detected by rabbit polyclonal antirotavirus serum. Examples of individual infection results (left) are accompanied by composite results from the experiment (right). (B) Enteroids generated from 11 different patients across the three sections of the small intestine were infected with HRV Ito at an MOI of 10 FFU/cell and assessed for the percentage of infected cells as described above for panel A. (C) A one-step growth curve for HRV Ito replication was performed over 30 h. Enteroids were infected at an MOI of 10 FFU/cell. At each of the 6 time points, enteroids and the surrounding supernatant were harvested, and the amount of infectious virus was quantified by a fluorescent-focus assay. Viral titer is displayed on the left y axis and is represented by black circles on the graph. Cytotoxicity was also measured at 2 hpi and 14 hpi and is represented as bars, with values displayed on the right y axis. (D) Jejunal enteroids from 5 different patients were infected with HRV Ito at an MOI of 0.5 FFU/cell, and the amount of infectious virus was quantified as described above for panel C at 2 hpi and 24 hpi. (E) Jejunal enteroids from one patient (patient j11) were infected with either RRV or HRV at an MOI of 0.5 FFU/cell, and the amount of infectious virus was quantified at 1.5 hpi and 24 hpi. (F) Jejunal enteroids from three patients were infected with either RV1 (Rotarix) or Wa, and the amount of infectious virus was quantified at 2 hpi and 24 hpi. In panels D to F, numbers above the bars show fold increases from 1.5 or 2 hpi (light gray bars) to 24 hpi (dark gray bars). (G) Secretor-negative enteroids from 3 different patients were infected with HRV strains Ito (left), Wa (middle), and RV1 (right) at an MOI of 0.5 FFU/cell. The amount of infectious virus at 1.5 hpi and 24 hpi was quantified as described above for panel C. Each bar represents the fold increase in viral growth from 1.5 hpi to 24 hpi. Statistical analysis was performed by using one-way ANOVA, followed by post hoc analysis with a Tukey HSD test. (H) Electron micrograph of an infected cell within an enteroid. Strain Ito particles (RV) adjacent to a lipid droplet (LD) and viroplasm (V) are shown (bar = 250 nm). Results are representative of data from duplicate (B, D, and F) or triplicate (A, C, and E) independent experiments. Each data bar represents means ± standard deviations for 3 samples within each independent experiment. Statistical analyses were performed by using Student's t test unless otherwise specified.
To assess whether HRV infects different regions of the small intestine, we examined HRV infection in HIEs generated from the duodenum, jejunum, or ileum from 11 different patients (10 secretor positive and 1 secretor negative [patient j4]) by flow cytometry. HIEs from all small intestinal segments were susceptible to HRV Ito infection (Fig. 2B).
Enteroids support robust replication of human rotavirus G3P[8] and G1[P8] but not an attenuated G1[P8] human rotavirus vaccine strain.
The flow cytometry data indicated that rotavirus proteins are made in infected HIE cells but not whether a complete replication cycle of infectious particles had occurred. To determine if HIEs support replication of infectious virus and to examine the kinetics of replication, a one-step growth curve was performed by inoculating HIEs with HRV Ito at a high MOI (∼10) (Fig. 2C). Following 1 h of adsorption and washing to remove unbound residual virus, the amount of infectious virus present at each time point in the HIEs and medium was quantified by a fluorescent-focus assay (FFA) in MA104 cells. The total amount of infectious virus decreased from 1.5 to 5 hpi, indicative of an eclipse phase of the replication cycle during this time. A marked increase in the amount of infectious virus was observed from 5 to 11 hpi. Between 11 and 25 hpi, virus production increased by 50%, after which a plateau was reached. These findings indicate that an entire HRV replication cycle occurred by 11 hpi in HIEs. To evaluate cell viability during the one-step growth curve, we assessed cytotoxicity at 2 and 14 hpi. We found that the percentage of cell lysis (compared to the positive control) at 14 hpi was significantly higher (P < 0.001) in HRV-infected HIEs (89.9% ± 2.8%) than in mock-infected HIEs (31.6% ± 4.5%). These data suggest that the HRV replication cycle in HIEs is a rapid, lytic process and that the decreased rate of virus production between 11 and 30 hpi parallels a loss of cell viability.
We also examined the production of infectious virus in jejunal HIEs from 5 different patients when inoculated with a low MOI (∼0.5 FFU/cell) of virus. All HIEs supported productive replication of HRV Ito, and on average, jejunal HIEs supported an ∼2- to 3-log10-fold increase in the amount of infectious virus from 2 to 24 h (Fig. 2D). In addition, duodenal and ileal HIEs from a total of 6 different patients supported HRV Ito growth, with yields in virus output (100- to 1,000-fold increases) over 24 h similar to those observed in jejunal HIEs (data not shown). All HIEs supported rotavirus infection and replication.
The initial flow cytometry findings (Fig. 2A) demonstrated that HIEs were more susceptible to infection by HRV Ito than RRV. However, these studies did not assess whether RRV replication is restricted in HIEs compared to HRV replication. To address this question, HIEs were infected with equal amounts of infectious RRV or HRV at a low MOI (∼0.5). After 1.5 h of adsorption and removal of unbound virus, viral growth was evaluated by quantifying the amount of infectious virus by FFAs. The total amount of infectious virus present by 24 hpi was 158 times larger in HRV-infected samples than in RRV-infected samples. Calculation of the increases in infectious-virus production relative to the amount of infectious virus present at 1.5 hpi showed a 12-fold increase for infectious RRV particles compared to a 213-fold increase for infectious HRV particles. This finding of reduced RRV growth compared to that of HRV was consistent across multiple independent experiments with HIEs from different patients, and the yield of RRV did not increase more than 20-fold by 24 hpi.
We also evaluated HIE infection with two other human G1P[8] rotaviruses: the well-characterized Wa strain and RV1, an attenuated rotavirus strain in the monovalent Rotarix rotavirus vaccine. Despite data demonstrating that RV1 is attenuated in vivo, RV1 attenuation has not been reported for any in vitro models of human tissue. We assessed viral growth of both these G1 HRVs in HIEs from multiple patients and found that RV1 replication was attenuated in HIEs from all secretor-positive patients tested compared to Wa growth (Fig. 2F). Finally, we evaluated the replication of these strains in HIEs from 3 secretor-negative patients. One-way analysis of variance (ANOVA) was performed to compare levels of viral growth across multiple patients and to assess for a patient effect on viral replication in these secretor-negative patients. A patient effect was not seen for either strain Ito (P = 0.76) or strain Wa (P = 0.13), as all three secretor-negative patients supported robust viral growth in all replicate experiments (Fig. 2G, left and middle). Infection with both viruses typically resulted in a 100- to 1,000-fold increase over 24 h. In contrast, RV1 replication was consistently attenuated in HIEs from one of the secretor-negative patients (patient j4) (Fig. 2G, right) in comparison to the HIEs from the other two secretor-negative patients (P < 0.01) as well as in comparison to HIEs from secretor-positive patients (data not shown). RV1 repeatedly replicated poorly in HIEs from patient j4 in four independent experiments compared to RV1 growth in other HIEs, with only a 2- to 4-fold increase in viral growth in four consecutive experiments. These results demonstrate not only that HIEs from multiple patients support RV1 replication but also that it may be possible to study attenuation of vaccine virus replication in this novel in vitro model.
Finally, HIEs were evaluated for the presence of viral particles as well as classic structures involved in the replication cycle, such as viroplasms, by transmission electron microscopy. Large viroplasms adjacent to a lipid droplet and large numbers of triple-layered particles as well as budding triple-layered particles were readily observed in infected HIEs (Fig. 2H). These structures were not seen in mock-inoculated HIEs (data not shown). Overall, these results demonstrate that HIEs are readily infected by and support the full replication cycle of HRV strains and can be used to further study the restriction of both heterologous and vaccine strains.
Human rotavirus preferentially infects differentiated enterocytes and enteroendocrine cells of the human small intestinal epithelium.
In vivo, rotavirus preferentially infects differentiated cells at the villus tips, with sparing of the intestinal crypts. After demonstrating that HIEs are cell cultures that differentiate upon withdrawal of the growth factor Wnt3A (Fig. 1), we evaluated the effect of differentiation on susceptibility to HRV infection. HIEs were either kept in Wnt3A-rich CMGF+ medium (undifferentiated HIEs) or differentiated for 4 days in differentiation medium (differentiated HIEs). HIEs were then infected with HRV Ito, and the percentage of infected cells in both groups was assessed by flow cytometry. HRV Ito infected ∼3-fold more cells in differentiated cultures (37.6%) than in undifferentiated cultures (12.6%) (Fig. 3A). The ratio of the percentage of infected cells in differentiated HIEs to the percentage of infected cells in undifferentiated HIEs was highly reproducible across three patients (2.3- to 3.9-fold increase in the percentage of infected cells in differentiated HIEs; mean, 3.1-fold increase [data not shown]).
FIG 3.
Effect of differentiation status on susceptibility to HRV infection. (A) Undifferentiated and differentiated jejunal enteroids from the same patient were either mock infected or infected with HRV Ito at an MOI of 10 FFU/cell. At 20 hpi, single-cell suspensions were assessed for the presence of intracellular rotavirus antigen by flow cytometry. Results are representative of data from triplicate independent experiments. Each data bar represents the mean ± standard deviation for 3 samples within each independent experiment. (B) Undifferentiated and differentiated jejunal enteroids from the same patient (patient j2) were infected with HRV Ito at an MOI of 0.5 FFU/cell, and the amount of infectious virus was quantified at 1.5 hpi (light gray bars) and 24 hpi (dark gray bars). Fold increases are displayed above the bars. Results are representative of data from replicate experiments performed with enteroids from two additional patients (patients j3 and j11). Each data bar represents the mean ± standard deviation for 4 samples within each independent experiment. Statistical analyses were performed by using Student's t test.
In addition, we found that undifferentiated cultures supported significantly less viral growth than did differentiated cultures (Fig. 3B). In undifferentiated cultures from patient j2, there was a 2-fold increase in viral growth from 1.5 hpi to 24 hpi, compared to a 291-fold increase in differentiated cultures over the same time span. These findings were verified with HIEs from two additional patients, in which the increase in viral growth in undifferentiated cultures varied from 2- to 5-fold over 24 h, whereas the increase in viral growth in differentiated cultures ranged from 291- to 1,159-fold. Together, these results indicate that differentiation status affects susceptibility to HRV infection as well as the total virus yield.
We next examined HRV-infected HIEs by immunofluorescence to identify which differentiated cell types were infected in these multicellular cultures by detecting rotavirus-positive cells with a polyclonal antirotavirus antibody. Enterocytes were identified by staining for E-cadherin expression. Mock-infected HIEs did not contain any rotavirus antigen-positive cells, while HRV Ito-infected HIEs had numerous rotavirus antigen-positive enterocytes (Fig. 4A). The predominant cell type infected by HRV in HIEs was enterocytes.
FIG 4.

Assessment of differentiated cell types infected by HRV. Ileal enteroids were either mock infected or infected with HRV Ito at an MOI of 10, fixed at 10 hpi, processed for immunofluorescence staining, and visualized by using confocal microscopy. (A) Paraffin-embedded sections of mock-infected (left) and HRV-infected (right) ileal enteroids were assessed for intracellular rotavirus antigen (green), E-cadherin (red), and nuclei (blue). Infected enterocytes (arrows) are identified as E-cadherin-expressing cells containing rotavirus antigen (bar = 20 μm). (B) Enteroendocrine cell infection was assessed by visualizing rotavirus antigen-containing cells (green) (left), chromogranin A-containing cells (magenta) (middle), and the merged image (right). Staining of nuclei (blue) and E-cadherin (red) is present in all panels (bar = 10 μm).
RRV infection of enteroendocrine cells in mice and primary human carcinoid enterochromaffin cells has been reported (41), but whether human rotaviruses infect these cells in vitro or in vivo is not known. To examine this question, jejunal and ileal HIEs infected with HRV Ito were costained for viral antigen and the enteroendocrine marker chromogranin A (CHGA). Mock-infected HIEs stained positive for CHGA but did not contain any rotavirus antigen-positive cells (Fig. 4B, top). In addition to enterocytes, a subset of CHGA-positive cells in HRV-infected HIEs also stained positive for rotavirus antigen (Fig. 4B, bottom). To quantify the frequency of enteroendocrine cell infection, five independent, randomly selected visual fields among sections of HRV-infected HIEs from two patients were visually examined. The number of cells costaining for both RV antigen and CHGA was compared to the total number of CHGA-positive cells. We observed 6/9 (67%) CHGA-positive cells in jejunal HIEs from one patient, and 6/14 (43%) CHGA-positive cells in ileal HIEs from a second patient contained RV antigen. This finding in HIEs shows that HRV can infect enteroendocrine cells of the human small intestine. We evaluated serial sections for goblet cell infection using the goblet cell marker MUC2, and we did not observe costaining of RV antigen and MUC2 (data not shown).
Human intestinal enteroids swell in response to HRV infection and the enterotoxin NSP4.
We examined whether HIEs can be used as a system to study apical fluid secretion in response to human rotavirus infection and to the rotavirus enterotoxin by measuring HIE lumenal expansion. Lumenal expansion represents a surrogate marker for the movement of fluid from the basolateral to the apical compartment, mimicking fluid secretion during diarrheal processes (42). We first examined whether forskolin, a cyclic AMP (cAMP) agonist, induced swelling in our HIEs. To evaluate forskolin-induced lumenal expansion, cross sections of duodenal HIEs were imaged in a 96-well optical plate by utilizing deconvolution microscopy. Images were captured every 15 s following forskolin (20 μM) treatment until the lumens reached their maximum cross-sectional area (Fig. 5A). A minority (∼20%) of HIEs did not expand in response to forskolin, which we interpreted as a sign that the enteroid was not completely sealed, and thus, intraluminal effects could not be observed.
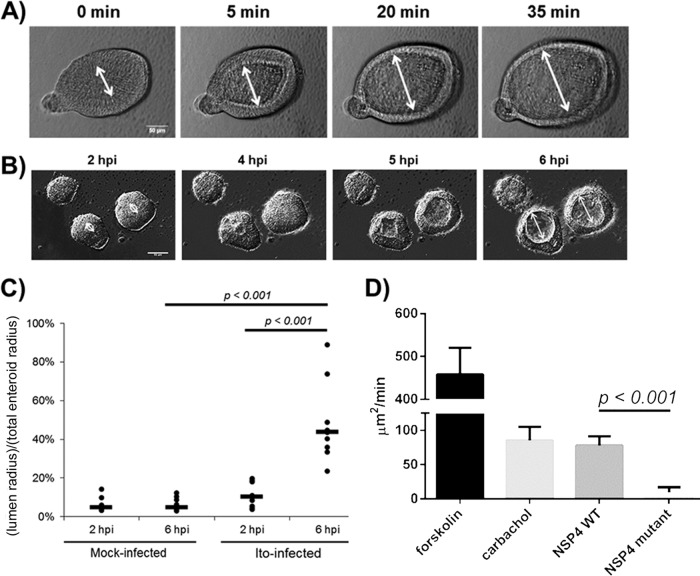
FIG 5.
Fluid dynamics of human intestinal enteroids in response to rotavirus. (A) Duodenal HIEs were treated with forskolin and imaged over 40 min. The white line marks the enteroid lumen diameter (bar = 50 μm). (B) Duodenal HIEs were infected with HRV Ito at an MOI of 20 PFU/cell. HIEs were imaged from 2 hpi to 12 hpi. Images from four representative time points are shown. White lines demarcate the lumen diameter (bar = 50 μm). (C) Duodenal HIEs were either mock infected or infected with HRV as described above for panel B. The lumenal radius and the total enteroid radius were measured at 2 hpi and 6 hpi for mock-infected (n = 10) and HRV-infected (n = 9) HIEs. The ratio of these two radii is shown as lumen radius/total enteroid radius at 2 hpi and at 6 hpi. Black bars represent median values. (D) Duodenal HIEs were treated with forskolin (20 μM), carbachol (200 μM), the NSP4 wild-type (WT) peptide spanning residues 95 to 146 (20 μM), or the E120A/Q123A mutant NSP4 peptide spanning residues 95 to 146 (20 μM). Forskolin-treated HIEs (n = 5) were imaged for 10 min. Carbachol (n = 7)-, NSP4 wild-type peptide (n = 10)-, and NSP4 mutant peptide (n = 5)-treated HIEs were imaged for 40 min, and the rate of increase in the cross-sectional area is shown in square micrometers per minute. Statistical analyses of data in panels C and D were performed by using Student's t test.
We next evaluated if spherical duodenal HIEs expand after HRV Ito infection. We measured the lumenal radius (the distance from the apical surface to the center of the lumen) and the total enteroid radius (the distance from the basolateral surface to the center of the lumen) for comparison at each time point. In contrast to the rapid expansion in response to forskolin, the lumens of HRV Ito-infected HIEs began expanding between 3 and 4 hpi, reaching a maximum lumenal radius at 6 hpi (Fig. 5B; see also Movie S1A in the supplemental material). Between 6 hpi and 7 hpi, the lumens of HRV-infected HIEs collapsed, and individual cells were observed to disassociate from the enteroids at between 7 hpi and 12 hpi (see Movie S1B in the supplemental material).
To quantify the degree of lumenal expansion, we calculated the ratio of the lumenal radius to the total enteroid radius. Only HIEs that expanded during infection were included in the data analysis. No mock-infected HIEs expanded during the 10 h of imaging, and thus, 10 randomly chosen mock-infected spherical HIEs were chosen for data analysis. The ratio in mock-infected HIEs remained between 4% and 5% at both time points (Fig. 5C). For HRV-infected compared to mock-infected HIEs, the ratio was higher at 2 hpi (P = 0.053) and 6 hpi (P = 0.0003). The ratio increased over time only in HRV-infected cells, from a median value of 10% at 2 hpi to 44% at 6 hpi (P = 0.0005). These data show that HIEs are physiologically active when treated with an agonist (forskolin) that induces fluid secretion and that HRV infection elicits a similar but slower response.
We also assessed whether exposure of the HIEs to the rotaviral enterotoxin NSP4 alone would induce HIE swelling. The lumenal cross-sectional area of duodenal HIEs was measured before and after the application of forskolin, carbachol, or a purified, recombinantly expressed wild-type NSP4 fragment (amino acids [aa] 95 to 146), which includes the enterotoxin domain but not the viroporin domain (43). The cholinergic agonist carbachol was included in our assessment because, similarly to NSP4, it causes chloride secretion through a calcium-mediated pathway (44–47). In addition, to evaluate the specificity of the response, we measured changes in the HIE lumenal area after administration of a mutant enterotoxin peptide (NSP4 peptide spanning aa 95 to 146 with an E120A/Q123A mutation) that does not increase intracellular calcium concentrations in vitro or induce diarrhea in neonatal mice (48).
The HIE cross-sectional area was calculated both at baseline and at 10 min (forskolin) or 40 min (carbachol and wild-type and mutant NSP4s). Untreated HIEs did not expand during this time frame (see Movie S2A in the supplemental material). HIEs expanded fastest in response to forskolin treatment. HIEs treated with carbachol and the wild-type NSP4 peptide spanning aa 95 to 146 exhibited similar increases in the lumen cross-sectional area (Fig. 5D). As expected, the wild-type NSP4 peptide induced significantly greater lumenal expansion than did the mutant NSP4 peptide (see Movie S2B in the supplemental material). These results indicate that the rotavirus enterotoxin NSP4 peptide, but not the biologically inactive mutant peptide, results in apical fluid secretion. This swelling assay for fluid movement further documents HIEs as a physiologically relevant in vitro model for rotavirus infection with features mimicking the pathophysiological observations of diarrhea.
DISCUSSION
The development of nontransformed human intestinal epithelial cell cultures that model intestinal biology (2) has stimulated interest in new studies of host-pathogen interactions, with most recently reported studies focusing on bacterial infections (1, 7, 49–51, 64). Here, we demonstrate the utility of such novel ex vivo multicellular, physiologically active cultures to study a human enteric virus (HRV) to gain new insight into interactions with human small intestinal cells in cultures established from different patients. In spite of effective RV vaccines reducing the incidence of rotavirus diarrhea in developed countries, rotaviruses remain the leading cause of severe secretory diarrhea in infants and young children worldwide. This is attributed to limited vaccine availability, lower-level vaccine effectiveness, and possibly diverse circulating virus strains as well as the nutritional and genetic variability of children in developing countries (52). These issues emphasize the continued need to more fully understand HRV biology and pathogenesis. We demonstrate that HIEs are uniquely able to model host range restriction, cell type restriction, virus-induced fluid secretion, and the epithelial cell response to infection in cultures from genetically diverse individuals.
We utilized laboratory HRV strains representing two of the most common circulating HRV genotypes: G1P[8] (Wa) and G3P[8] (Ito). Most of our studies of HIEs used G3P[8] strain Ito as it shares the same G type as the prototype animal rotavirus RRV previously used in the majority of rotavirus studies to study and compare homologous and heterologous rotavirus infections in cultured cells or animal models. We found that RV infection of HIEs recapitulates the cell type and host range restriction of in vivo RV infection based on several parameters.
First, we found that HRV replicates in HIEs established from all three regions of the small intestine. This could be tested because HIEs maintain a transcriptional signature corresponding to their region of origin, indicating that duodenal, jejunal, and ileal HIEs each exhibit segment-specific properties (4). All three segments of the small intestine are also susceptible to infection with homologous ARVs in multiple animal models (53, 54). The regional replication that we observed is consistent with the discovery of HRV in duodenal epithelial cells (55, 56) and the subsequent detection of HRVs in the duodenum and occasionally in the upper jejunum and ileum in children (57, 58). Our results indicate that this novel ex vivo system will now allow examination of whether there are different consequences of or host responses to infection of different intestinal regions.
Second, HRVs infect up to 50% of the cells in differentiated HIEs that exhibit a “villus-like” epithelial phenotype, while undifferentiated “crypt-like” HIEs are infected at a significantly lower rate; this confirms the lack of detection of RV infection in intestinal crypts in prior studies of infected tissues from children or animals. This result also explains the lower percentage of epithelial cells (<20%) infected by HRVs in human intestinal organoid cultures derived from the differentiation of human pluripotent stem cells (10); these cultures are now known to most closely resemble the human fetal intestine (9).
Third, enteroendocrine cells were also a secondary cell type infected by HRV in differentiated HIEs. These results confirm and extend with HRV previous reports that RRV can infect human enteroendocrine cells in tumor cell lines and mouse jejunum, and the rotavirus enterotoxin NSP4 stimulates serotonin release from carcinoid enterochromaffin cells (41). This led to a proposed model of human disease that involves rotavirus stimulation of serotonin mediators from enteroendocrine cells that would lead to the activation of the enteric nervous system (59, 60) and fluid secretion as well as changes in intestinal motility leading to vomiting and diarrhea (59, 61). Our data showing that enteroendocrine cells in nontransformed, multicellular HIEs from multiple patients are susceptible to HRV infection will allow future determination of whether multiple subtypes or a particular subset of the multiple (>15) subtypes of enteroendocrine cells are infected and whether serotonin only or other secreted factors contribute to pathogenesis.
Fourth, HRVs infect more cells within differentiated HIEs than do ARVs (50% and <15%, respectively), and different strains of HRV, including the RV1 vaccine strain, show heterogeneity in infection of HIEs from different patients. HRVs replicated to titers between 100- and 1,000-fold higher than those of RRV in 24 h. This property of restriction of heterologous rotavirus infection is consistently demonstrated in animal models; however, this has not been recapitulated previously in transformed human tissue culture cell lines in vitro, where RRV and other ARVs infect as many cells as or more cells than HRVs and typically replicate to higher titers. To our knowledge, HIEs represent the first in vitro model of rotavirus infection to demonstrate host range restriction of RRV similar to in vivo findings while permitting robust infection by and growth of HRVs. Because HRV infection of HIEs mimics in vivo cell type tropism and host range restriction, HIEs will be useful to pursue the identification of both viral factors (using reassortant strains) and cell factors (including apical versus basolateral infection) that restrict RV tropism in cells within the differentiated, small intestinal epithelium.
Building off our findings that HRV Ito replicates well in all HIEs tested, we investigated the replication of HRV Wa and the attenuated HRV Rotarix vaccine strain (RV1) in HIEs that were genetically typed as secretor positive and secretor negative. Variable replication efficiencies were observed with these viruses. Using different HIEs, both Wa and RV1 replicated less well than strain Ito, suggesting that heterogeneity in host factors may affect the robustness of HRV replication. In addition, RV1 was attenuated in HIEs from multiple patients compared to laboratory HRV strain Wa. Although our data suggest that RV1 replication is attenuated in HIEs, the mechanism of RV1 attenuation in vivo is not known. To our knowledge, this is the first demonstration of detection of possible vaccine strain attenuation in a cell culture system.
This finding is also interesting from the perspective of understanding host range restriction. Both RV1 and Wa are G1P[8] HRVs, sharing the same genotype classification for their 11 gene segments (62). Thus, RV1 represents an HRV strain that is restricted in HIEs despite having the same genotype as the prototypic HRV strain Wa. Passaging of RV1 through African green monkey kidney and Vero cells of simian origin to produce the vaccine may have introduced mutations in its genome that allow it to replicate better in simian than in human cells. Although these mutations do not change the strain's genotype, they may attenuate its replication in human cells. Future potential studies of reassortants of RV1 in HIEs could allow the genetic determinants of vaccine attenuation to be defined, and this system might be useful for testing the attenuation of other candidate rotavirus vaccines. Furthermore, the observation that RV1 did not grow well in HIEs from one of the three patients with the secretor-negative genotype is of interest. This observation needs to be pursued by using HIEs derived from additional individuals to obtain data on how common host factors that contribute to this restriction might be and to determine whether these cultures represent a potential preclinical model for evaluating the effects of host genetic polymorphisms at the epithelial cell level on vaccine growth.
Finally, we examined whether HIEs could provide an in vitro model to study the core characteristic of rotavirus disease, diarrhea. Both mouse and human enteroids respond to known agonists of chloride secretion by exhibiting lumenal expansion/swelling, a marker of intestinal epithelial fluid secretion (42, 63). Treatment of our enteroids with the agonists forskolin and carbachol, which mediate chloride secretion through activation of cAMP and calcium signaling, respectively, resulted in a significant increase in the lumenal expansion of HIEs, as previously reported (42). Treatment of HIEs with HRV also resulted in increased HIE lumenal expansion. Swelling was observed between 5 and 7 hpi, at which point it appeared that tight-junction integrity failed as the HIEs rapidly collapsed. Analysis of the timing of HIE swelling and collapse together with the results from the HRV one-step growth curve demonstrate that intraluminal expansion coincides with the end of the viral replication eclipse phase. Large quantities of viral proteins, including the viral enterotoxin NSP4, are present within the cells at between 5 and 7 hpi (data not shown). The exact mechanism responsible for the HRV-induced swelling of HIEs remains to be determined, but it is likely due to autocrine or paracrine effects on noninfected cells by the viral enterotoxin NSP4 secreted from infected cells (44, 52). This was confirmed by treatment of HIEs with an NSP4 peptide fragment (aa 95 to 146) containing the enterotoxin domain (aa 114 to 135) that caused HIE swelling similar to that induced by carbachol, an agonist of calcium-induced chloride secretion. In contrast, the NSP4 mutant peptide containing mutations in the E120 and Q123 residues, which does not mobilize intracellular calcium or cause diarrhea in mice (48), failed to induce HIE swelling. These findings showing that wild-type NSP4 but not the mutant peptide can induce HIE swelling correlate with in vivo findings of NSP4 virulence and further validate this system as a functional assay to study fluid secretion and “diarrheagenic” properties of RV and its toxin. In addition, for rotavirus in particular, this assay should provide a functional assay for determining the virulence of different rotavirus strains and NSP4s by assessing their ability to cause HIE swelling, a potential in vitro correlate of rotavirus-induced secretory diarrhea. Finally, HIEs can be utilized for the development and preclinical testing of drug therapies to prevent/treat diarrheal disease.
In summary, we demonstrate that HIEs provide a useful new model of the small intestinal epithelium to study enteric virus-host interactions at the cellular level. Using HRVs as a model pathogen of the human small intestine, we found that HIEs reveal several in vivo findings of rotavirus infection in an in vitro model, including host range restriction and vaccine attenuation, preferential infection of differentiated cells, infection of enteroendocrine cells, and fluid secretion in response to infection. A final intriguing possibility is that HIEs may provide a novel system with which to study and gain a mechanistic understanding of the effect of host polymorphisms on susceptibility to human rotaviruses and other human enteric pathogens (such as human noroviruses, astroviruses, and toroviruses as well as parasites and bacteria), which lack relevant and efficient ex vivo models that fully recapitulate the biology and pathophysiology of disease.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants U18-TR000552, U19-AI116497, and RO1 AI080656 and Howard Hughes Medical Institute grant 570076890. We are grateful for Core Support from the Integrated Microscopy Core, the Cellular and Molecular Morphology Core, and the Cytometry and Cell Sorting Core of Baylor College of Medicine, which are funded by grant SCCPR U54 HD-007495 (B. W. O'Malley); grant P30 DK-56338, which funds the Texas Medical Center Digestive Diseases Center (M.K.E.); grant P30 CA-125123 (C. K. Osborne); and the Dan L. Duncan Cancer Center of Baylor College of Medicine.
We declare no conflict of interest.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01930-15.
REFERENCES
- 1.Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Ettayebi K, Blutt SE, Hyser JM, Zeng XL, Crawford SE, Broughman JR, Estes MK, Donowitz M. 2014. Human enteroids as an ex-vivo model of host-pathogen interactions in the gastrointestinal tract. Exp Biol Med (Maywood) 239:1124–1134. doi: 10.1177/1535370214529398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. 2011. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 3.Sato T, Clevers H. 2013. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340:1190–1194. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 4.Middendorp S, Schneeberger K, Wiegerinck CL, Mokry M, Akkerman RD, van Wijngaarden S, Clevers H, Nieuwenhuis EE. 2014. Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells 32:1083–1091. doi: 10.1002/stem.1655. [DOI] [PubMed] [Google Scholar]
- 5.Leushacke M, Barker N. 2014. Ex vivo culture of the intestinal epithelium: strategies and applications. Gut 63:1345–1354. doi: 10.1136/gutjnl-2014-307204. [DOI] [PubMed] [Google Scholar]
- 6.Fuller MK, Faulk DM, Sundaram N, Shroyer NF, Henning SJ, Helmrath MA. 2012. Intestinal crypts reproducibly expand in culture. J Surg Res 178:48–54. doi: 10.1016/j.jss.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.VanDussen KL, Marinshaw JM, Shaikh N, Miyoshi H, Moon C, Tarr PI, Ciorba MA, Stappenbeck TS. 2015. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut 64:911–920. doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovbasnjuk O, Zachos NC, In J, Foulke-Abel J, Ettayebi K, Hyser JM, Broughman JR, Zeng XL, Middendorp S, de Jonge HR, Estes MK, Donowitz M. 2013. Human enteroids: preclinical models of non-inflammatory diarrhea. Stem Cell Res Ther 4(Suppl 1):S3. doi: 10.1186/scrt364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkbeiner SR, Hill DR, Altheim CH, Dedhia PH, Taylor MJ, Tsai YH, Chin AM, Mahe MM, Watson CL, Freeman JJ, Nattiv R, Thomson M, Klein OD, Shroyer NF, Helmrath MA, Teitelbaum DH, Dempsey PJ, Spence JR. 2015. Transcriptome-wide analysis reveals hallmarks of human intestine development and maturation in vitro and in vivo. Stem Cell Rep 4:1140–1155. doi: 10.1016/j.stemcr.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkbeiner SR, Zeng XL, Utama B, Atmar RL, Shroyer NF, Estes MK. 2012. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio 3:e00159-12. doi: 10.1128/mBio.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD, WHO-Coordinated Global Rotavirus Surveillance Network. 2012. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis 12:136–141. doi: 10.1016/S1473-3099(11)70253-5. [DOI] [PubMed] [Google Scholar]
- 12.Tate JE, Haynes A, Payne DC, Cortese MM, Lopman BA, Patel MM, Parashar UD. 2013. Trends in national rotavirus activity before and after introduction of rotavirus vaccine into the national immunization program in the United States, 2000 to 2012. Pediatr Infect Dis J 32:741–744. doi: 10.1097/INF.0b013e31828d639c. [DOI] [PubMed] [Google Scholar]
- 13.Babji S, Kang G. 2012. Rotavirus vaccination in developing countries. Curr Opin Virol 2:443–448. doi: 10.1016/j.coviro.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Zaman K, Dang DA, Victor JC, Shin S, Yunus M, Dallas MJ, Podder G, Vu DT, Le TP, Luby SP, Le HT, Coia ML, Lewis K, Rivers SB, Sack DA, Schodel F, Steele AD, Neuzil KM, Ciarlet M. 2010. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in Asia: a randomised, double-blind, placebo-controlled trial. Lancet 376:615–623. doi: 10.1016/S0140-6736(10)60755-6. [DOI] [PubMed] [Google Scholar]
- 15.Armah GE, Sow SO, Breiman RF, Dallas MJ, Tapia MD, Feikin DR, Binka FN, Steele AD, Laserson KF, Ansah NA, Levine MM, Lewis K, Coia ML, Attah-Poku M, Ojwando J, Rivers SB, Victor JC, Nyambane G, Hodgson A, Schodel F, Ciarlet M, Neuzil KM. 2010. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomised, double-blind, placebo-controlled trial. Lancet 376:606–614. doi: 10.1016/S0140-6736(10)60889-6. [DOI] [PubMed] [Google Scholar]
- 16.Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, Estes MK, Prasad BV. 2012. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 485:256–259. doi: 10.1038/nature10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Huang P, Tan M, Liu Y, Biesiada J, Meller J, Castello AA, Jiang B, Jiang X. 2012. Rotavirus VP8*: phylogeny, host range, and interaction with histo-blood group antigens. J Virol 86:9899–9910. doi: 10.1128/JVI.00979-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramani S, Cortes-Penfield NW, Hu L, Crawford SE, Czako R, Smith DF, Kang G, Ramig RF, Le Pendu J, Prasad BV, Estes MK. 2013. The VP8* domain of neonatal rotavirus strain G10P[11] binds to type II precursor glycans. J Virol 87:7255–7264. doi: 10.1128/JVI.03518-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohm R, Fleming FE, Maggioni A, Dang VT, Holloway G, Coulson BS, von Itzstein M, Haselhorst T. 2015. Revisiting the role of histo-blood group antigens in rotavirus host-cell invasion. Nat Commun 6:5907. doi: 10.1038/ncomms6907. [DOI] [PubMed] [Google Scholar]
- 20.Imbert-Marcille BM, Barbe L, Dupe M, Moullac-Vaidye B, Besse B, Peltier C, Ruvoen-Clouet N, Le Pendu J. 2014. A FUT2 gene common polymorphism determines resistance to rotavirus A of the P[8] genotype. J Infect Dis 209:1227–1230. doi: 10.1093/infdis/jit655. [DOI] [PubMed] [Google Scholar]
- 21.Nordgren J, Sharma S, Bucardo F, Nasir W, Gunaydin G, Ouermi D, Nitiema LW, Becker-Dreps S, Simpore J, Hammarstrom L, Larson G, Svensson L. 2014. Both Lewis and secretor status mediate susceptibility to rotavirus infections in a rotavirus genotype-dependent manner. Clin Infect Dis 59:1567–1573. doi: 10.1093/cid/ciu633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheth R, Anderson J, Sato T, Oh B, Hempson SJ, Rollo E, Mackow ER, Shaw RD. 1996. Rotavirus stimulates IL-8 secretion from cultured epithelial cells. Virology 221:251–259. doi: 10.1006/viro.1996.0374. [DOI] [PubMed] [Google Scholar]
- 23.Cuadras MA, Feigelstock DA, An S, Greenberg HB. 2002. Gene expression pattern in Caco-2 cells following rotavirus infection. J Virol 76:4467–4482. doi: 10.1128/JVI.76.9.4467-4482.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frias AH, Vijay-Kumar M, Gentsch JR, Crawford SE, Carvalho FA, Estes MK, Gewirtz AT. 2010. Intestinal epithelia activate anti-viral signaling via intracellular sensing of rotavirus structural components. Mucosal Immunol 3:622–632. doi: 10.1038/mi.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, Jain V, Cunliffe NA, Nakagomi O, Kirkwood CD, Fischer TK, Parashar UD, Bresee JS, Jiang B, Glass RI. 2005. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J Infect Dis 192(Suppl 1):S146–S159. doi: 10.1086/431499. [DOI] [PubMed] [Google Scholar]
- 26.Conner ME, Estes MK, Graham DY. 1988. Rabbit model of rotavirus infection. J Virol 62:1625–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramig RF. 1988. The effects of host age, virus dose, and virus strain on heterologous rotavirus infection of suckling mice. Microb Pathog 4:189–202. doi: 10.1016/0882-4010(88)90069-1. [DOI] [PubMed] [Google Scholar]
- 28.Ciarlet M, Estes MK, Barone C, Ramig RF, Conner ME. 1998. Analysis of host range restriction determinants in the rabbit model: comparison of homologous and heterologous rotavirus infections. J Virol 72:2341–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward LA, Rosen BI, Yuan L, Saif LJ. 1996. Pathogenesis of an attenuated and a virulent strain of group A human rotavirus in neonatal gnotobiotic pigs. J Gen Virol 77(Part 7):1431–1441. [DOI] [PubMed] [Google Scholar]
- 30.Hoshino Y, Saif LJ, Kang SY, Sereno MM, Chen WK, Kapikian AZ. 1995. Identification of group A rotavirus genes associated with virulence of a porcine rotavirus and host range restriction of a human rotavirus in the gnotobiotic piglet model. Virology 209:274–280. doi: 10.1006/viro.1995.1255. [DOI] [PubMed] [Google Scholar]
- 31.Kitamoto N, Ramig RF, Matson DO, Estes MK. 1991. Comparative growth of different rotavirus strains in differentiated cells (MA104, HepG2, and CaCo-2). Virology 184:729–737. doi: 10.1016/0042-6822(91)90443-F. [DOI] [PubMed] [Google Scholar]
- 32.Superti F, Tinari A, Baldassarri L, Donelli G. 1991. HT-29 cells: a new substrate for rotavirus growth. Arch Virol 116:159–173. doi: 10.1007/BF01319239. [DOI] [PubMed] [Google Scholar]
- 33.Crawford SE, Mukherjee SK, Estes MK, Lawton JA, Shaw AL, Ramig RF, Prasad BV. 2001. Trypsin cleavage stabilizes the rotavirus VP4 spike. J Virol 75:6052–6061. doi: 10.1128/JVI.75.13.6052-6061.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciarlet M, Estes MK. 1999. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J Gen Virol 80(Part 4):943–948. [DOI] [PubMed] [Google Scholar]
- 35.Heijmans J, van Lidth de Jeude JF, Koo BK, Rosekrans SL, Wielenga MC, van de Wetering M, Ferrante M, Lee AS, Onderwater JJ, Paton JC, Paton AW, Mommaas AM, Kodach LL, Hardwick JC, Hommes DW, Clevers H, Muncan V, van den Brink GR. 2013. ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep 3:1128–1139. doi: 10.1016/j.celrep.2013.02.031. [DOI] [PubMed] [Google Scholar]
- 36.Blutt SE, Matson DO, Crawford SE, Staat MA, Azimi P, Bennett BL, Piedra PA, Conner ME. 2007. Rotavirus antigenemia in children is associated with viremia. PLoS Med 4:e121. doi: 10.1371/journal.pmed.0040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 38.Sastri NP, Viskovska M, Hyser JM, Tanner MR, Horton LB, Sankaran B, Prasad BV, Estes MK. 2014. Structural plasticity of the coiled-coil domain of rotavirus NSP4. J Virol 88:13602–13612. doi: 10.1128/JVI.02227-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svensson L, Petersson A, Henry SM. 2000. Secretor genotyping for A385T, G428A, C571T, C628T, 685delTGG, G849A, and other mutations from a single PCR. Transfusion 40:856–860. doi: 10.1046/j.1537-2995.2000.40070856.x. [DOI] [PubMed] [Google Scholar]
- 40.Lay MK, Atmar RL, Guix S, Bharadwaj U, He H, Neill FH, Sastry KJ, Yao Q, Estes MK. 2010. Norwalk virus does not replicate in human macrophages or dendritic cells derived from the peripheral blood of susceptible humans. Virology 406:1–11. doi: 10.1016/j.virol.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagbom M, Istrate C, Engblom D, Karlsson T, Rodriguez-Diaz J, Buesa J, Taylor JA, Loitto VM, Magnusson KE, Ahlman H, Lundgren O, Svensson L. 2011. Rotavirus stimulates release of serotonin (5-HT) from human enterochromaffin cells and activates brain structures involved in nausea and vomiting. PLoS Pathog 7:e1002115. doi: 10.1371/journal.ppat.1002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I, Janssens HM, de Winter-de Groot KM, Brandsma AM, de Jong NW, Bijvelds MJ, Scholte BJ, Nieuwenhuis EE, Van den Brink S, Clevers H, van der Ent CK, Middendorp S, Beekman JM. 2013. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med 19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 43.Hyser JM, Collinson-Pautz MR, Utama B, Estes MK. 2010. Rotavirus disrupts calcium homeostasis by NSP4 viroporin activity. mBio 1:e00265-10. doi: 10.1128/mBio.00265-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ball JM, Tian P, Zeng CQ, Morris AP, Estes MK. 1996. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272:101–104. doi: 10.1126/science.272.5258.101. [DOI] [PubMed] [Google Scholar]
- 45.Morris AP, Scott JK, Ball JM, Zeng CQ, O'Neal WK, Estes MK. 1999. NSP4 elicits age-dependent diarrhea and Ca(2+) mediated I(-) influx into intestinal crypts of CF mice. Am J Physiol 277:G431–G444. [DOI] [PubMed] [Google Scholar]
- 46.Dharmsathaphorn K, Pandol SJ. 1986. Mechanism of chloride secretion induced by carbachol in a colonic epithelial cell line. J Clin Invest 77:348–354. doi: 10.1172/JCI112311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lorrot M, Vasseur M. 2007. How do the rotavirus NSP4 and bacterial enterotoxins lead differently to diarrhea? Virol J 4:31. doi: 10.1186/1743-422X-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seo NS, Zeng CQ, Hyser JM, Utama B, Crawford SE, Kim KJ, Hook M, Estes MK. 2008. Integrins alpha1beta1 and alpha2beta1 are receptors for the rotavirus enterotoxin. Proc Natl Acad Sci U S A 105:8811–8818. doi: 10.1073/pnas.0803934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, Vries R, Peters PJ, Clevers H. 2015. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology 148:126–136. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nigro G, Rossi R, Commere PH, Jay P, Sansonetti PJ. 2014. The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe 15:792–798. doi: 10.1016/j.chom.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Yamamoto Y, Wilson LH, Zhang T, Howitt BE, Farrow MA, Kern F, Ning G, Hong Y, Khor CC, Chevalier B, Bertrand D, Wu L, Nagarajan N, Sylvester FA, Hyams JS, Devers T, Bronson R, Lacy DB, Ho KY, Crum CP, McKeon F, Xian W. 2015. Cloning and variation of ground state intestinal stem cells. Nature 522:173–178. doi: 10.1038/nature14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Estes MK, Greenberg HB. 2013. Rotaviruses, p 1347–1401. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 53.Boshuizen JA, Reimerink JH, Korteland-van Male AM, van Ham VJ, Koopmans MP, Buller HA, Dekker J, Einerhand AW. 2003. Changes in small intestinal homeostasis, morphology, and gene expression during rotavirus infection of infant mice. J Virol 77:13005–13016. doi: 10.1128/JVI.77.24.13005-13016.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conner ME, Ramig RF. 1997. Enteric viruses, p 713–743. In Nathason N. (ed), Viral pathogenesis. Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 55.Bishop RF, Davidson GP, Holmes IH, Ruck BJ. 1973. Virus particles in epithelial cells of duodenal mucosa from children with acute non-bacterial gastroenteritis. Lancet ii:1281–1283. [DOI] [PubMed] [Google Scholar]
- 56.Davidson GP, Barnes GL. 1979. Structural and functional abnormalities of the small intestine in infants and young children with rotavirus enteritis. Acta Paediatr Scand 68:181–186. doi: 10.1111/j.1651-2227.1979.tb04986.x. [DOI] [PubMed] [Google Scholar]
- 57.Lynch M, Shieh WJ, Tatti K, Gentsch JR, Ferebee-Harris T, Jiang B, Guarner J, Bresee JS, Greenwald M, Cullen S, Davies HD, Trevenen C, Zaki SR, Glass RI. 2003. The pathology of rotavirus-associated deaths, using new molecular diagnostics. Clin Infect Dis 37:1327–1333. doi: 10.1086/379322. [DOI] [PubMed] [Google Scholar]
- 58.Middleton PJ, Szymanski MT, Abbott GD, Bortolussi R, Hamilton JR. 1974. Orbivirus acute gastroenteritis of infancy. Lancet 303:1241–1244. doi: 10.1016/S0140-6736(74)90001-4. [DOI] [Google Scholar]
- 59.Hagbom M, Sharma S, Lundgren O, Svensson L. 2012. Towards a human rotavirus disease model. Curr Opin Virol 2:408–418. doi: 10.1016/j.coviro.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 60.Lundgren O, Peregrin AT, Persson K, Kordasti S, Uhnoo I, Svensson L. 2000. Role of the enteric nervous system in the fluid and electrolyte secretion of rotavirus diarrhea. Science 287:491–495. doi: 10.1126/science.287.5452.491. [DOI] [PubMed] [Google Scholar]
- 61.Istrate C, Hagbom M, Vikstrom E, Magnusson KE, Svensson L. 2014. Rotavirus infection increases intestinal motility but not permeability at the onset of diarrhea. J Virol 88:3161–3169. doi: 10.1128/JVI.02927-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gomara M, Johne R, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Parreno V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. 2011. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch Virol 156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, Nieuwenhuis EE, Beekman JM, Clevers H. 2013. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 64.In J, Foulke-Abel J, Zachos NC, Hansen A-M, Kaper JB, Bernstein HD, Halushka M, Blutt S, Estes MK, Donowitz M, Kovbasnjuk O. 14 October 2015. Enterohemorrhagic Escherichia coli reduces mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell Mol Gastroenterol Hepatol. doi: 10.1016/j.jcmgh.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.