ABSTRACT
Antiviral CD8+ T cells are a key component of the adaptive immune response against HCV, but their impact on viral control is influenced by preexisting viral variants in important target epitopes and the development of viral escape mutations. Immunodominant epitopes highly conserved across genotypes therefore are attractive for T cell based prophylactic vaccines. Here, we characterized the CD8+ T cell response against the highly conserved HLA-B*51-restricted epitope IPFYGKAI1373–1380 located in the helicase domain of NS3 in people who inject drugs (PWID) exposed predominantly to HCV genotypes 1a and 3a. Despite this epitope being conserved in both genotypes, the corresponding CD8+ T cell response was detected only in PWID infected with genotype 3a and HCV-RNA negative PWID, but not in PWID infected with genotype 1a. In genotype 3a, the detection of strong CD8+ T cell responses was associated with epitope variants in the autologous virus consistent with immune escape. Analysis of viral sequences from multiple cohorts confirmed HLA-B*51-associated escape mutations inside the epitope in genotype 3a, but not in genotype 1a. Here, a distinct substitution in the N-terminal flanking region located 5 residues upstream of the epitope (S1368P; P = 0.00002) was selected in HLA-B*51-positive individuals. Functional assays revealed that the S1368P substitution impaired recognition of target cells presenting the endogenously processed epitope. The results highlight that, despite an epitope being highly conserved between two genotypes, there are major differences in the selected viral escape pathways and the corresponding T cell responses.
IMPORTANCE HCV is able to evolutionary adapt to CD8+ T cell immune pressure in multiple ways. Beyond selection of mutations inside targeted epitopes, this study demonstrates that HCV inhibits epitope processing by modification of the epitope flanking region under T cell immune pressure. Selection of a substitution five amino acids upstream of the epitope underlines that efficient antigen presentation strongly depends on its larger sequence context and that blocking of the multistep process of antigen processing by mutation is exploited also by HCV. The pathways to mutational escape of HCV are to some extent predictable but are distinct in different genotypes. Importantly, the selected escape pathway of HCV may have consequences for the destiny of antigen-specific CD8+ T cells.
INTRODUCTION
Based on phylogenetic analysis, hepatitis C virus (HCV) can be classified into at least seven genotypes and multiple subtypes that differ up to 20% at the amino acid level (1). The HCV genotypes have distinct epidemiological characteristics, they are associated with different transmission risk factors, and their frequencies in a population are regionally different. In Europe and North America the HCV genotypes 1 and 3 are most common (2, 3). Since routine screening via nucleic acid amplification for HCV eliminated the risk for infection through blood products, the most important risk group for incident HCV infection are people who inject drugs (PWID). The high prevalence of HCV infection with seroprevalence rates up to 80% paired with frequent risk practices for HCV transmissions in PWID results in incidence rates between 8 and 25% per year in young adult injectors (4, 5), and there is strong evidence that multiple exposures are common in this risk group (6, 7). The degree of sequence diversity between genotypes and subtypes precludes broad protection against reinfection. Accordingly, multiple infections of the same individual with different viruses have been reported (7).
Even at the subtype level HCV isolates typically differ between hosts. Lack of a proof reading function of the virus encoded RNA-dependent RNA polymerase results in a high error rate during RNA replication. As a consequence, HCV exists in chronically infected patients as a quasispecies of closely related but genetically distinct viral variants. There is now strong evidence that viral genetic variation between hosts is the product of continuous selection of mutations by host immune pressure (8–14). Collectively, this inherent sequence diversity of HCV at the genotype, subtype, and quasispecies level is a major obstacle to vaccine design (15). Strategies that aim to develop prophylactic vaccines against HCV have to cope with this genetic heterogeneity either by inducing immune responses with a high degree of cross-reactivity (16, 17), by inducing multiple responses against different sequence variants (18), or by focusing the immune response on highly conserved regions of the virus.
Dominant CD8+ T cell epitopes that are conserved across different HCV genotypes are rare (19, 20). Here, we characterized a highly conserved dominant HLA-B*51-restricted CD8+ T cell epitope (IPFYGKAI1373–1380) in HCV NS3 in PWID predominantly exposed to genotype 1a and 3a. Vigorous responses were detected in PWID with spontaneous immune control of HCV and in PWID with genotype 3a infection, but not in PWID infected with genotype 1a. Although selection of mutations inside the epitope was overall rare, there was evidence for mutational escape in HCV genotype 1b and 3a by population sequence analysis. Interestingly, HCV genotype 1a followed a distinct escape pathway by selecting a substitution (S1368P) located five amino acids upstream of the epitope. Further analysis of the functional relevance revealed that the S1368P substitution altered epitope processing. The results demonstrate that beyond selection of mutations inside CD8+ epitopes HCV adapts to immune pressure by selecting mutations in the epitope flanking region. The evolutionary escape pathways differ between HCV genotypes, indicating distinct genetic plasticity.
MATERIALS AND METHODS
Patients.
Blood samples from patients with a history of injection drug use were collected from the ward for inpatient detoxification treatment of drug addicts or the clinic for opioid maintenance treatment at the Department of Addictive Behavior and Addiction Medicine, Rhine State Hospital Essen, Hospital of the University of Duisburg-Essen. Written informed consent was obtained from all study participants and the study was approved by the ethics committee of the Medical Faculty of the University of Duisburg-Essen in accordance with the Declaration of Helsinki. In total 43 HLA-B*51-positive treatment-naive subjects who were HCV antibody positive (by anti-HCV chemiluminescent microparticle immunoassay from Abbott) were analyzed, including 15 HCV-RNA-negative patients and 28 subjects with detectable HCV-RNA. Peripheral blood mononuclear cells (PBMCs) were isolated from blood via Ficoll gradient centrifugation and subsequently cryopreserved.
Analysis and alignment of HCV sequences.
The frequency of HLA-B*51-associated sequence polymorphisms was analyzed in 405 HCV genotype 1a sequences and 145 HCV genotype 1b sequences from a multicenter cohort (21) and in a large HCV genotype 1b outbreak (22). A region covering the epitope IPFYGKAI1373–1380 was amplified and sequenced from additional 37 HCV genotype 1a isolates, as well as 102 HCV genotype 3a isolates collected in Germany. All sequences have been submitted to GenBank (accession no. FJ864775 to FJ864816, KJ130249 to KJ130317, and KJ668232 to KJ668268).
Analysis of the CD8+ T cell response.
Antigen-specific T cells were expanded from cryopreserved HLA-B*51-positive PBMCs utilizing synthetic peptides (>70% purity) purchased from EMC, Tübingen, Germany. After thawing, the PBMCs were cultured in RPMI 1640 medium containing 10% fetal calf serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, 10 mM HEPES buffer, and 25 U of recombinant interleukin-2 (IL-2)/ml and then stimulated with HLA-B*51 peptide 1373 (1 μg/ml) and 0.1 μg of anti-CD28 and anti-CD49d/ml. After 7 days medium containing IL-2 was added. On day 10 the cells were restimulated with the same peptide (10 μg/ml) in the presence of brefeldin A (100 ng/ml) for 4 h and then analyzed for their CD4+, CD8+ and gamma interferon (IFN-γ) expression via flow cytometry. To determine the degree of cross-reactivity between different B*51-1373 variants, PBMCs were cultivated in the presence of the HLA-B*51-1373 prototype or the variant peptide. After 10 days, both cultures were restimulated with the prototype and the variant peptide at different concentrations before intracellular IFN-γ staining. For the detection of HCV-specific cells ex vivo and after peptide-specific in vitro expansion, thawed PBMCs were stained with phycoerythrin-labeled IPFYGKAI-specific HLA-B*5101 dextramer (Immudex, Denmark), followed by surface staining with CD8+ PerCP-Cy5.5 (eBioscience). All samples were acquired using a FACSCanto (BD), and the data were analyzed by using FlowJo software (Tree Star, Inc.).
Expression plasmids for HCV-GFP fusion proteins.
All constructs were based on the parental plasmids pEGFP-N1 (Clontech, Germany) featuring a cytomegalovirus promoter-controlled enhanced green fluorescent protein for N-terminal fusion. Fragments of NS3 were generated by amplifying NS3 1329-1433 by nested PCR with the primers 4b-F (CCTACGGCAAGTTCCTTGC) and 4b-R_new (GCAGTCTATCACCGAGTCG) and the primers B51-for-EcoRI (CCAAGGGAATTCTTGGCTTCGTCTTCACCCTCGGCATCGGCACYGTCCTTGACCAAG) and B51-Rev-SalI (CGGATACCGTCGACCCGCGGTARTAMGCCACGGC). Subsequently, the fragment was cloned via EcoRI/SalI into pEGFP-N1. The substitution S1368P was introduced via site-directed mutagenesis (Stratagene) using the primer B51-S1368P-for (GGAGGTTGCTCTGCCCACCACCGGAGA) and B51-S1368P-rev (TCTCCGGTGGTGGGCAGAGCAACCTCC) according to the manufacturer's instructions. All constructs were verified by DNA sequencing.
Analysis of the CD8+ T cell response against endogenously processed antigens.
HLA-B*51 PBMCs from healthy donors were obtained from buffy coats (Department of Transfusion Medicine, University Hospital Essen, Essen, Germany). For analysis of endogenously processed antigens, HLA-B*51-positive cells were electroporated with plasmids pac-NS3-S1368S-GFP or pac-NS3-S1368P-GFP encoding an NS3-GFP fusion protein utilizing the Amaxa T Cell Nucleofector kit (vpa-1002; Lonza). In brief, after thawing PBMCs were cultured in RPMI medium. After 24 h, 7 × 106 cells were resuspended in 100 μl of Nucleofector solution, mixed with 7.5 μg of DNA and pulsed with the optimized protocol for unstimulated human T cells (Program v024; Amaxa2b) with an Amaxa apparatus (Lonza). After electroporation, the PBMCs were cultured in RPMI medium for 24 h cells before GFP-positive cells were sorted using a FACSAria II (BD). Subsequently, 105 B*51-1373-specific cells from a 10-day culture were restimulated with GFP-positive cells in a 1:1 ratio for 4 h before intracellular IFN-γ staining.
In vitro proteasome digestion.
The synthetic peptides EVALSTTGEIPFYGKAIPLEAIKGG and EVALPTTGEIPFYGKAIPLEAIKGG (≥95% purity) were purchased from EMC, Tübingen, Germany, and digested using a 20S proteasome assay kit complemented with human constitutive or immunoproteasome (Boston Biochem). In brief, 20 μg of peptide was digested with 4 μg of sodium dodecyl sulfate-activated proteasome in a 500-μl reaction. The reaction was stopped on indicated time points by adding 3 volumes of ice-cold acetone. For precipitation of proteasome, the samples were subsequently frozen at −20°C for 30 min and then centrifuged for 30 min at 4°C with 15,000 rpm. The supernatants were collected, evaporated to dryness, and dissolved in 50 μl of 0.1% trifluoroacetic acid. The peptide concentration of the resulting solution was determined by amino acid analysis as previously described (23). For the subsequent liquid chromatography-tandem mass spectrometry experiment, the sample was diluted to a concentration of 0.33 pmol/μl, and 5 pmol was analyzed using an UltiMate 3000 RSLCnano system online coupled to a Velos Pro linear ion trap mass spectrometer (both from Thermo Scientific, Bremen, Germany) as described earlier (23). The acquired raw files were further analyzed with Proteome Discoverer software (Thermo Scientific, v1.3.0.339) and searched with Sequest (24) against a self-written database containing the investigated prototype sequence (pt) and S1368P peptides. Precursor and fragment ion mass tolerance was set to 0.4 Da, and the confidence level of peptide identification was set to a false discovery rate of 1%. Relative peptide quantification was carried out via spectral counting. Then, a normalized spectral index was calculated for each of the identified peptides by dividing the number of acquired peptide spectrum matches (PSMs) of a particular peptide by the number of PSMs acquired in the whole sample.
Generation and analysis of TNcc viruses.
Plasmid pTNcc encoding for the GT1a virus TNcc was kindly provided by Jens Bukh (25). Mutations S1368P, I1373V, and I1380L were introduced by site-directed mutagenesis, and all constructs were verified by DNA sequencing. In vitro transcription and electroporation of Huh-7.5 cells was performed as described before (26). At 4 h posttransfection, the medium was changed with fresh Dulbecco modified Eagle medium (2 mM l-glutamine, nonessential amino acids, 100 U of penicillin/ml, 100 μg of streptomycin/ml, 10% fetal calf serum) supplemented with or without 10 mM 2′CMA (kindly provided by T. Tellinghuisen [27]). At 72 h posttransfection, cell-free supernatant was filtered through 0.45-μm-pore-size filters and 10× concentrated through Amicon centrifugal filters (Millipore). Virus titers were determined as described elsewhere (28) with slight modifications. In brief, Huh-7.5 cells were seeded in 96-well plates at a density of 104 cells per well 24 h prior to inoculation with dilutions of filtered cell culture supernatant (at least six wells were used per dilution). After 3 days, cells were washed with phosphate-buffered saline (PBS), fixed for 20 min with ice-cold methanol at −20°C, and washed three times with PBS. HCV-infected cells were detected with anti-core C7.50 (1:300) (29) and anti-NS5A 9E10 (1:1,000) (28) antibody in PBS for 45 min at room temperature. The cells were washed as described above, and bound antibodies were detected by incubation with horseradish peroxidase-conjugated antibodies specific to murine IgG (Sigma-Aldrich, Steinheim, Germany) diluted 1:200 in PBS. After 1 h of incubation at room temperature, the cells were washed as specified above. The peroxidase activity was detected by using carbazole substrate (0.32% [wt/vol] of 3-amino-9-ethylcarazole [Sigma] in N,N-dimethyl-formamide was diluted at a ratio of 1:3.3 with 15 mM acetic acid, 35 mM sodium acetate, [pH 8.0], and 0.4% H2O2). Virus titers (50% tissue culture infective doses [TCID50]/ml) were calculated based on the method of Spearman and Kärber. HCV RNA was quantified by X-tail reverse transcriptase-PCR as described previously (22). For the detection of HCV core protein, virus-containing supernatant was inactivated by the addition of Triton X-100 to a final concentration of 1% (vol/vol) and the amount of released core protein was determined by a commercially available core enzyme-linked immunosorbent assay (Architect HCV Core AG test; Abbott, Wiesbaden, Germany).
Statistical analysis.
All statistical tests were performed using GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA).
RESULTS
CD8+ T cells directed against the epitope IPFYGKAI1373–1380 are detected after spontaneous resolution and in chronic genotype 3a infection but not in chronic genotype 1a infection.
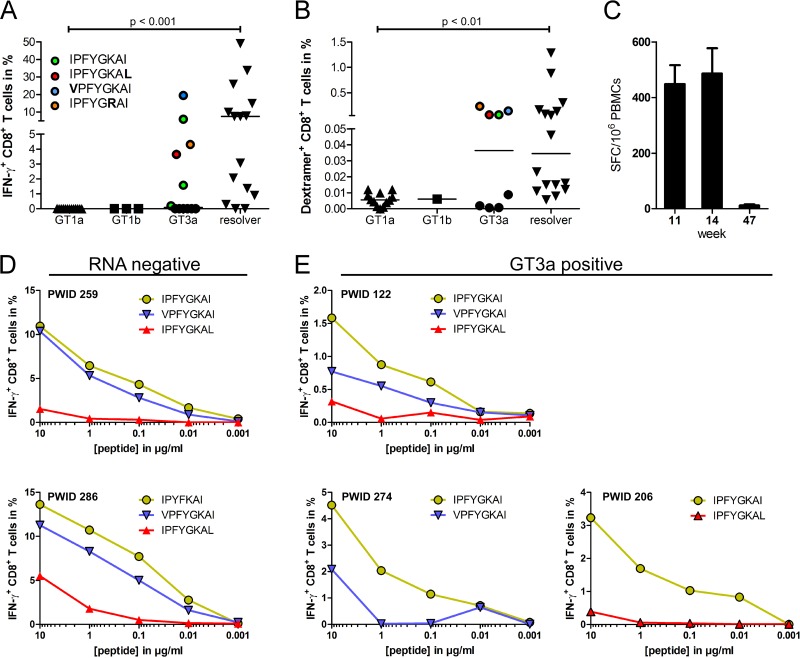
We previously reported an HLA-B*51-restricted epitope in NS3 (IPFYGKAI1373–1380) that is >90% conserved across the HCV genotypes 1a, 1b, and 3a (19). The CD8+ T cell immune response directed against the epitope IPFYGKAI1373–1380 was analyzed in a cohort of anti-HCV-positive PWID carrying the HLA-B*51 allele. A total of 43 subjects were analyzed, including 15 anti-HCV-positive PWID with undetectable HCV-RNA, 13 PWID infected with GT1a, 3 PWID infected with GT1b, and 12 PWID infected with GT3a. After 10 days of in vitro expansion IPFYGKAI-specific CD8+ T cells were detectable in 12 of 15 (80%) individuals with undetectable viremia and in 6 of 12 (50%) patients chronically infected with genotype 3a (Fig. 1A). In contrast, CD8+ T cells directed against this epitope were not detectable after antigen-specific expansion in patients with genotype 1a or 1b infection (Fig. 1A). Similar results were obtained when CD8+ T cell were directly analyzed ex vivo with HLA class I/peptide dextramers. IPFYGKAI-specific CD8+ T cells were detectable in most HCV-RNA-negative PWID and in some PWID with a genotype 3a infection. Notably, the four genotype 3a-infected patients with high frequencies were identical to those where antigen-specific CD8+ T cells were efficiently expanded in vitro (Fig. 1A). In turn, the analysis with HLA class I/peptide dextramers confirmed lack of detection of IPFYGKAI-specific CD8+ T cells in patients infected with genotype 1 (Fig. 1B). The CD8+ T cell response against the epitope IPFYGKAI1373–1380 was also studied in a patient with acute HCV genotype 1a infection. The patient was previously reported in a study on the impact of HLA-B*57 on HCV infection outcome (30) and was also HLA-B*51 positive. Interestingly, here, the response was detectable by week 11 after infection but became completely undetectable in an IFN-γ enzyme-linked immunospot (ELISpot) assay by week 47 (Fig. 1C).
FIG 1.
CD8+ T cell response against the epitope IPFYGKAI1373–1380 in PWID exposed to HCV. (A) T cells were expanded for 10 days from PBMCs in the presence of the peptide IPFYGKAI. After in vitro expansion, the cells were restimulated with the peptide before intracellular IFN-γ staining. The autologous viral epitope sequences from GT3a-infected PWID with detectable responses are indicated. (B) HLA-B*511373–1380-specific T cells were detected ex vivo via IPFYGKAI-specific HLA-B*5101 dextramer staining. (C) The HLA-B*511373–1380 specific T cell response was determined by ELISpot assay at the indicated time points in a patient with acute HCV infection. (D and E) Serial peptide dilutions of the prototype (green), I1373V (blue), or I1380L (red) sequence of the B51-1373 peptide were tested in RNA-negative (D) or GT3a-infected (E) patients. Statistical comparisons between groups in panels A and B were done with a Kruskal-Wallis test, and significant P values are indicated.
The autologous virus of 22 PWID from the total of 26 with detectable HCV-RNA was sequenced. The sequences of the epitope and the flanking region are shown in Table 1 and are indicated in Fig. 1A for PWID with genotype 3a infection with a detectable CD8+ T cell response (colored in Fig. 1A). In 3 of 10 HCV genotype 3a sequences, the virus harbored substitutions in the epitope region (I1373V, K1377R, or I1380L). Of note, all three patients mounted robust responses against the prototype sequence (Fig. 1A). In functional assays the I1380L variant impaired recognition by antigen-specific CD8+ T cells, whereas the I1373V variant was cross-reactive in HCV-RNA-negative PWID and to a lesser extent in GT3a-infected PWID (Fig. 1D and E). Notably, the three remaining GT3a-infected patients with detectable CD8+ T cell responses and no evidence for escape mutations had a history of injection drug use of less than 6 months, consistent with more recent exposure and infection with HCV, possibly at a stage prior to mutational escape. One of nine GT1a sequences and one of three GT1b sequences harbored the I1373V substitution in position 1 of the epitope. In both patients no CD8+ T cell response was detectable. One PWID with genotype 1a infection harbored a S1368P substitution. Notably, this substitution was also selected in the patient with acute HCV genotype 1a infection and with the rapid decline of the CD8+ T cell response (Fig. 1C and data not shown).
TABLE 1.
Patient characteristics
| Patient ID | Genotype | Viral load (IU/ml) | Sequencea |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | L | S | T | T | G | E | I | P | F | Y | G | K | A | I | P | |||
| 138 | 1a | 2,148,000 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 154 | 1a | 777,200 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 175 | 1a | 2,200,000 | . | . | P | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 251 | 1a | 61,790 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 278 | 1a | 3,015,000 | . | . | . | . | . | . | . | V | . | . | . | . | . | . | . | . |
| 283 | 1a | 2,422,000 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 299 | 1a | 275,700 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 324 | 1a | 2,469,000 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 348 | 1a | 83,080 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 393 | 1a | 5,282,000 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 408 | 1a | 1,562,000 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| 423 | 1a | 320,900 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 581 | 1a | 3,942,000 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 084 | 1b | 61,550 | . | . | . | N | I | . | . | . | . | . | . | . | . | . | . | . |
| 117 | 1b | 1,033,000 | . | . | . | N | I | . | . | V | . | . | . | . | . | . | . | . |
| 332 | 1b | 220,000 | . | . | . | N | . | . | . | . | . | . | . | . | . | . | . | . |
| 096 | 3a | 247,700 | . | . | G | S | E | . | . | . | . | . | . | . | . | . | . | . |
| 113 | 3a | 2258,000 | . | . | G | S | E | . | . | . | . | . | . | . | . | . | . | . |
| 122 | 3a | 87,400 | . | . | G | S | E | . | . | . | . | . | . | . | . | . | . | . |
| 137 | 3a | 1,046,000 | . | . | G | S | E | . | . | . | . | . | . | . | . | . | . | . |
| 176 | 3a | 924,700 | . | . | G | S | E | . | . | . | . | . | . | . | . | . | . | . |
| 206 | 3a | 368,000 | . | . | G | S | E | . | . | . | . | . | . | . | . | . | L | . |
| 240 | 3a | 154,500 | . | . | G | S | E | . | . | . | . | . | . | . | . | . | . | . |
| 257 | 3a | 3,642 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 274 | 3a | 146,300 | . | . | G | S | E | . | . | V | . | . | . | . | . | . | . | . |
| 292 | 3a | 80,690 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| 466 | 3a | 34,010,000 | . | . | G | S | E | . | . | . | . | . | . | . | . | . | . | . |
| 533 | 3a | 511,500 | . | . | G | S | D | . | . | . | . | . | . | . | R | . | . | . |
| 042 | RNA negb | |||||||||||||||||
| 062 | RNA neg | |||||||||||||||||
| 110 | RNA neg | |||||||||||||||||
| 161 | RNA neg | |||||||||||||||||
| 196 | RNA neg | |||||||||||||||||
| 242 | RNA neg | |||||||||||||||||
| 264 | RNA neg | |||||||||||||||||
| 286 | RNA neg | |||||||||||||||||
| 344 | RNA neg | |||||||||||||||||
| 365 | RNA neg | |||||||||||||||||
| 417 | RNA neg | |||||||||||||||||
| 418 | RNA neg | |||||||||||||||||
| 474 | RNA neg | |||||||||||||||||
| 574 | RNA neg | |||||||||||||||||
| 587 | RNA neg | |||||||||||||||||
The H77 prototype sequence is indicated in the column subheadings. Differences from the prototype sequence are specified by the appropriate letter in the table. A period indicates no difference from the prototype sequence. –, Not done.
RNA neg, RNA negative.
The residue under selection pressure in HLA-B*51-positive individuals depends on the viral genotype.
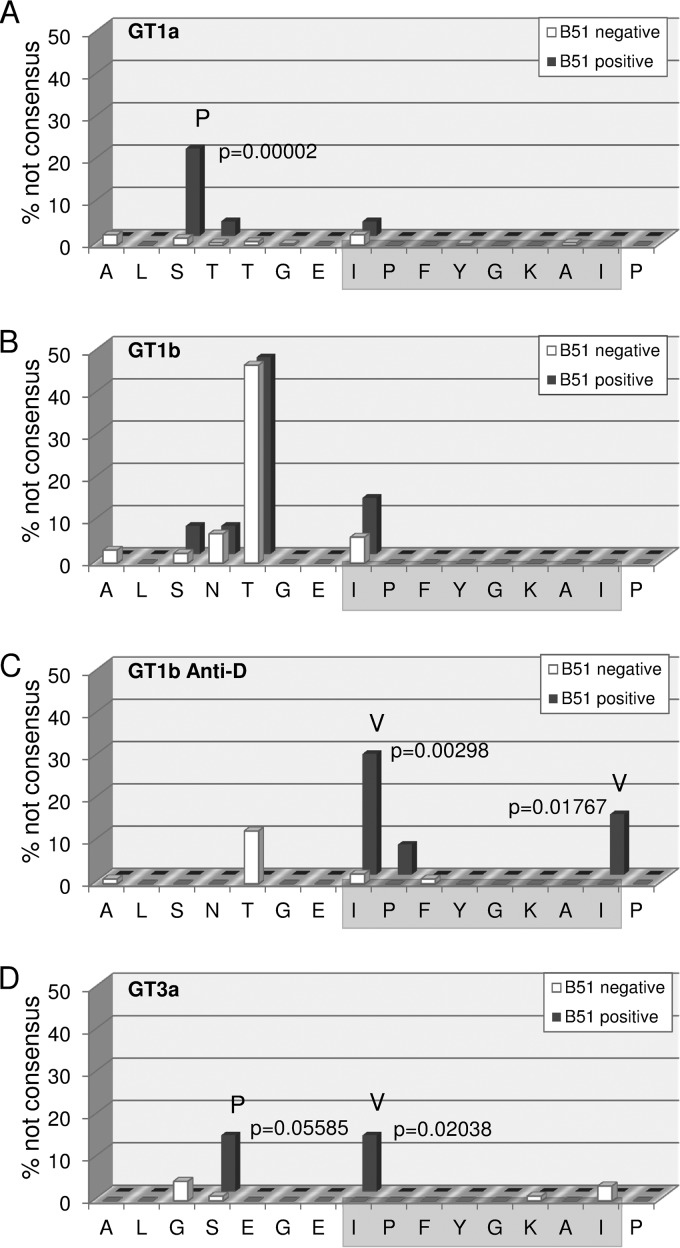
Given the relatively high reproducibility of IPFYGKAI-specific CD8+ T cell responses in HCV-RNA negative PWID, we aimed to address whether there is evidence for immune escape in patients with persistent HCV infection at a population level. We therefore analyzed the impact of HLA-B*51 expression on the frequency of sequence polymorphisms in this epitope in different cohorts. In an analysis of 442 genotype 1a sequences from a multicenter cohort (Fig. 2A), there was no evidence for mutational escape inside the epitope. Although an I1373V polymorphism was observed, this substitution was not enriched in HLA-B*51-positive patients. In contrast, the I1373V polymorphism was slightly enriched in HLA-B*51-positive patients from a multicenter cohort of 145 genotype 1b-infected patients (Fig. 2B), although this difference was not statistically significant. However, in a large genotype 1b single source outbreak there was statistical evidence for selection of polymorphisms in position 1 (I1373V) and 8 (I1380L) of the epitope in the presence of HLA-B*51 (Fig. 2C). Finally, we also analyzed a local cohort of 102 patients infected with genotype 3a (Fig. 2D). Here, there was statistical evidence for selection of the I1373V polymorphism inside the epitope. Importantly, even though in genotype 1a there was no evidence for mutational escape inside the epitope, there was strong statistical evidence for HLA-B*51-associated selection pressure on a residue located five amino acids upstream of the epitope (S1368) (Fig. 2A). In genotype 1a, 20.7% of the HLA-B*51-positive patients carried a S1368P substitution; in contrast, only 0.8% of the HLA-B*51-negative patients carried any substitutions in this position (P = 0.00002). Notably, in genotype 3a an S1369P substitution was also enriched in HLA-B*51-positive individuals; however, the difference was not statistically significant (Fig. 2D). Taken together, by analyses of different cohorts we found statistical evidence for mutational escape inside the HLA-B*51-restricted epitope (IPFYGKAI) in genotype 1b and 3a and evidence for the selection of a distinct substitution in the epitope flanking region in genotype 1a.
FIG 2.
Frequency of HLA-B*51-associated viral polymorphisms in the epitope region. The frequency of variations from the reported prototype sequence of the epitope region in GT1a (A), GT1b (B), the East-German Anti-D cohort (C), and GT3a (D) are shown for patients carrying the HLA-B*51 allele (red) and patients not carrying the HLA-B*51 allele (green). Positions with significant differences in polymorphism frequencies in the absence or presence of HLA-B*51 are marked, and the P values (Fisher exact test) and the most frequent variant amino acid are indicated.
The S1368P substitution impairs targeting of the endogenously processed HLA-B*51-restricted epitope IPFYGKAI1373–1380.
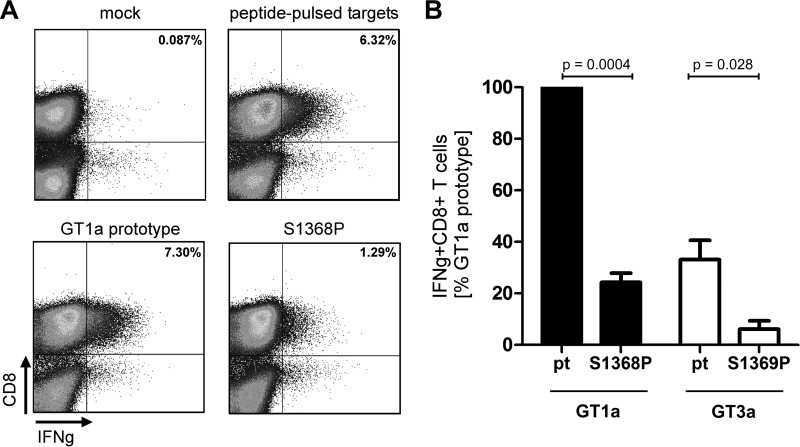
HCV genotype 1a was unique because evidence for HLA-B*51-associated selection pressure was only observed in the epitope flanking region. Of note, this region does not contain a second HLA-B*51 binding motif by analysis with prediction algorithms for HLA class I binding (www.immuneepitope.org). We therefore hypothesized that the S1368P may impair processing and presentation of the epitope. To address this experimentally, an assay that allows analysis of endogenously processed antigens was established. Effector CD8+ T cells directed against the HLA-B*51-restricted epitope IPFYGKAI were obtained by 10 days of antigen-specific expansion from different PWID with spontaneous immune control of HCV. To generate target cells that present the endogenously processed epitope HLA-B*51-positive buffy coat cells were transfected with expression plasmids encoding a fusion protein of a short fragment of NS3 containing the epitope fused to GFP. At 24 h after transfection GFP-positive cells were sorted and used as targets for specific CD8+ T. As a positive-control native target cells were pulsed with the synthetic peptide overnight. Figure 3A shows a representative result of one experiment. Upon stimulation with peptide-pulsed targets 6.3% IFN-γ+ CD8+ T cells were detectable. When targets were transfected with a genotype 1a prototype sequence 7.3% of CD8+ T cells secreted IFN-γ. In contrast, when targets were transfected with the plasmid harboring the S1368P substitution the number of IFN-γ+CD8+ T cells was reduced to 1.3%. In seven independent experiments the CD8+ T cell response against targets transfected with the S1368P variant was reproducibly reduced to levels of 24% compared to prototype 1a (Fig. 3B). This suggests that less antigen was presented on S1368P-transfected target cells consistent with impaired endogenous processing associated with this substitution. To compare the processing efficiency between genotype 1a and genotype 3a, the same fusion proteins were constructed with the 3a prototype sequence and the genotype 3a S1369P substitution. In three independent experiments the CD8+ T cell response against targets transfected with prototype 3a were reproducibly weaker (33%) compared to prototype 1a. The S1369P substitution further reduced the response to 6% compared to prototype 1a. This suggests that the epitope is less efficiently processed in a genotype 3a context compared to a genotype 1a context. Moreover, both substitutions in the epitope flanking region (S1368P in genotype 1a and S1369P in genotype 3a) further reduce the antigen processing efficiency.
FIG 3.
CD8+ T cell response against the endogenously processed epitope IPFYGKAI1373–1380. Effector T cells were expanded for 10 days from PBMCs in the presence of the peptide IPFYGKAI. HLA-B*51-positive target cells were generated by electroporation with a NS3(aa1330-2420)-GFP fusion protein. GFP-positive cells were sorted and used as targets for restimulation of IPFYGKAI-specific effector CD8+ T cells in an effector/target ratio of 1:1 for 4 h, followed by an intracellular cytokine staining (ICS). Mock transfected targets and peptide-pulsed targets served as negative or positive controls, respectively. (A) Representative fluorescence-activated cell sorting results of one ICS. (B) The IFN-γ response against targets transfected with GT1a prototype NS3 was normalized to 100% and compared to the response against other targets as indicated. The data represent results from at least three independent experiments. The P values were calculated using a one-sample t test with a hypothetical value of 100 (S1368P versus pt1a) or an unpaired t test (pt GT3a versus S1369P).
Differential proteasomal cleavage of peptides with the S1368P substitution.
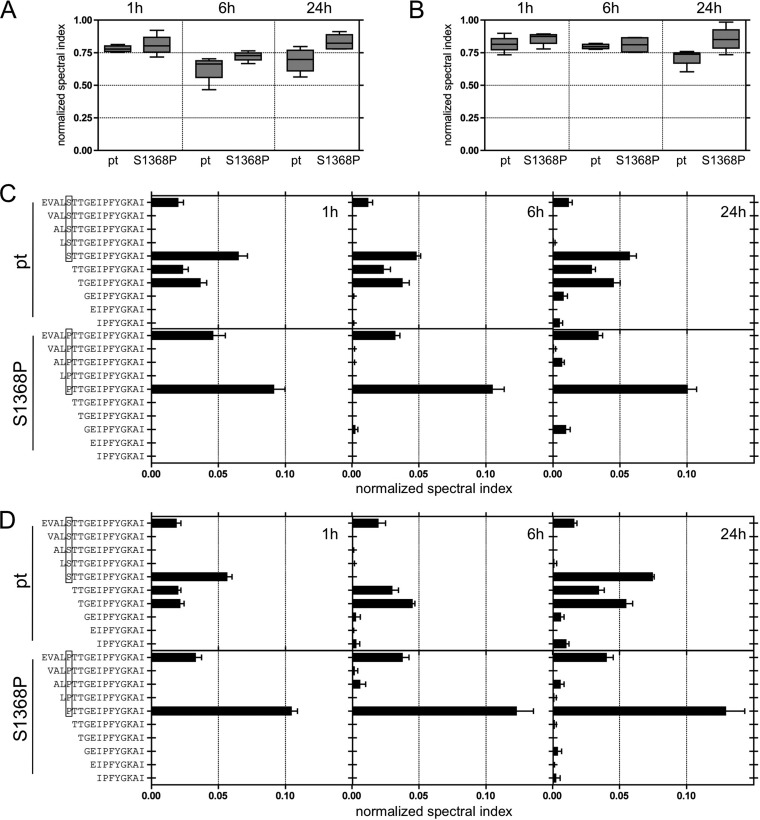
It was next addressed whether the S1368P substitution selected in genotype 1a has an impact on proteasomal cleavage consistent with altered processing. Therefore, synthetic peptides 25 amino acids in length either with the prototype sequence (pt) or harboring the S1368P substitution (S1368P) were digested with constitutive or immune proteasome, and the cleavage products were analyzed by mass spectrometry. In Fig. 4 the normalized spectral indices for cleavage products containing the full epitope sequence are shown at different time points after digestion with the constitutive proteasome (Fig. 4A) or the immune proteasome (Fig. 4B). There was no significant difference in the relative frequency of epitope containing peptides upon digestion of the prototype or the S1368P variant at any time. In fact, there was a minor trend toward higher frequencies of epitope containing peptides after 24 h when the S1368P variant was digested (Fig. 4A and B). Since carboxypeptidase activity is basically absent from the endoplasmic reticulum (ER), antigen presentation requires peptides with correct C-terminal ends after proteasomal cleavage (31). We therefore focused the analysis on peptide products ending with the epitope sequence IPFYGKAI. Figure 4C and D show the normalized spectral indices of individual peptides after digestion by the constitutive proteasome (Fig. 4C) or the immune proteasome (Fig. 4D) at different time points. After digestion of the prototype peptide the most prevalent cleavage product was STTGEIPFYGKAI carrying the residue that was under selection pressure in HLA-B*51-positive patients in genotype 1a at its N-terminal end (boxed in Fig. 4C and D). Also highly prevalent were N-extended peptides with three (TGEIPFYGKAI) or four (TTGEIPFYGKAI) additional residues. When the S1368P variant was digested the most prevalent cleavage product (PTTGEIPFYGKAI) carried the substitution selected in HLA-B*51-positive patients in genotype 1a at its N-terminal end. Importantly, shorter N-extended cleavage products with three or four additional residues were nearly absent after digestion of the S1368P variant. Similar results were obtained after digestion by the constitutive proteasome (Fig. 4C) or the immune proteasome (Fig. 4D).
FIG 4.
Impact of the S1368P substitution on proteasomal degradation. (A and B) The relative abundance of epitope-containing cleavage products after digestion of 25mer peptides with the prototype sequence (pt) or carrying the S1368P substitution (S1368P) with constitutive proteasome (A) or immune proteasome (B) is shown by normalized spectral indices. (C and D) The relative abundance of cleavage products with the correct C-terminal epitope end is shown by normalized spectral indices of individual peptides after digestion with constitutive proteasome (C) or immune proteasome (D).
The S1368P substitution does not impair viral fitness in HCV genotype 1a.
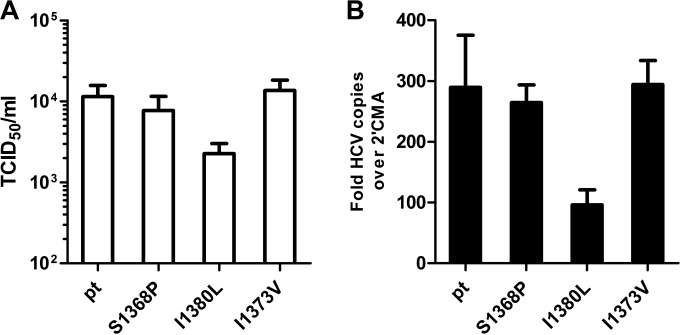
We hypothesized that fitness constraints caused preferred selection of the S1368P substitution in genotype 1a. To address this experimentally, the recently described genotype 1a full genome virus TNcc (25) was utilized to study the replication fitness and infectivity associated with the substitutions S1368P, I1373V or I1380L. At 72 h after the electroporation of virus RNA into Huh-7.5 cells, the supernatants were used for infection of naive Huh-7.5 cells (Fig. 5A). Upon transfection with the parental HCV TN genome the TCID50 was 1.1 × 104 infectious particles per ml. Approximately the same amount of infectious particles was released upon transfection with the S1368P variant (0.8 × 104 TCID50/ml) or the I1373V variant (1.3 × 104 TCID50/ml). In contrast, the I1380L substitution was associated with a decreased TCID50 of 0.2 × 104 per ml. The same hierarchy was observed at the replication level, with only a slight reduction of replication for the S1368P and I1373V substitution and a 3-fold reduced replication for the I1380L substitution. This indicates that only the substitution I1380L impairs viral fitness in genotype 1a, whereas the substitutions S1368P and I1380L do not.
FIG 5.
Infectivity and replication of wild-type and mutant TNcc strains. RNA transcripts of the parental HCV TN genome and TN mutants were transfected into Huh-7.5 cells. (A) Seventy-two hours later, cell-free supernatant was used for inoculation of naive Huh-7.5 cells. The TCID50 of the variants was determined by a limiting-dilution assay and staining with core and NS5A-specific antibodies. (B) RNA levels at 72 h posttransfection were measured by quantitative real-time reverse transcription-PCR. The RNA levels in the presence of 2′methyladenosin (2′CMA) was used for normalization. The ratio of measured HCV RNA to measured RNA levels in the presence of 2′CMA was calculated for each construct. The data from four independent experiments are shown.
DISCUSSION
Here, we characterized a highly conserved HLA-B*51-restricted CD8+ T cell epitope located in the helicase domain of HCV NS3. The high frequency of CD8+ T cells specific for this epitope in PWID who achieved spontaneous immune control of HCV infection suggests that this CD8+ T cell response is reproducibly mounted in HLA-B*51-positive patients. Given the high degree of conservation of the epitope, it was therefore surprising that in chronic infection CD8+ T cell responses against this epitope were detectable only in PWID infected with genotype 3a and were completely absent in patients infected with genotype 1a. Although viral sequence polymorphisms were overall rare, there was selection pressure on the epitope containing region, as evidenced by HLA-B*51-associated viral sequence polymorphisms in all studied genotypes. Notably, in genotype 1b there was statistical support for the selection of escape mutations inside the epitope in the single-source outbreak, whereas there was only a nonsignificant trend in the genotype 1b multicenter cohort. It is possible that escape patterns inside this epitope differ between cohorts. Alternatively, because the natural sequence variability in this epitope in the genotype 1b multicenter cohort is higher than in the single-source outbreak, the size of the multicenter cohort may be too small to unmask an existing effect of HLA-B*51-associated selection pressure.
The predominant escape mechanisms of substitutions selected by CD8+ T cell immune pressure in HCV epitopes are impaired binding of the variant epitope to the HLA class I-molecule or impaired binding of the T cell receptor to the HLA class I/peptide-complex. As an alternative mechanism, processing of the variant epitope can be affected by substitutions in the epitope region. We previously reported CD8+ T cell selection of a substitution inside the HLA-B*08-restricted epitope NS3 HSKKKCDEL1395–1403 located in position 9 of the epitope (9). Functional analyses revealed that the selected leucine to valine or phenylalanine substitution impaired presentation of the endogenously processed epitope. Similarly, Kimura et al. (32) described substitutions selected inside targeted CD8+ T cell epitopes that impaired proteasomal processing in the chimpanzee model. In contrast, CD8+ T cell selection of substitutions in the epitope flanking region has not been described yet in HCV. Seifert et al. (33) demonstrated that a tyrosine to phenylalanine polymorphism located in the C-terminal position +1 of the HLA-A*02-restricted epitope NS3 CVNGVCWTV1073–1081 influenced carboxy-terminal cleavage of the epitope by the proteasome; however, there was no conclusive evidence for immune selection pressure on this residue (22, 33). In HIV larger population studies of HLA class I-associated viral adaptation suggested that selection of processing mutations in the epitope flanking region contributes to mutational immune escape (34). This included substitutions in the C-terminal and N-terminal epitope flanking regions. Only a few studies directly addressed the functional consequences of such putative processing mutations (35, 36). The example of selection of altered processing in HLA-B*51-positive patients infected with HCV genotype 1a supports that similar immune escape strategies are utilized by HCV.
Processing and presentation of viral epitopes is a multistep process starting with production of peptide precursors by proteasomal degradation of viral proteins (reviewed in reference 37). These peptide precursors undergo N-terminal trimming by different endopeptidases and aminopeptidases in the cytosol and by the aminopeptidases ERAP1 and ERAP2 after the peptides have been transported by TAP to the ER (38, 39). Importantly, since carboxypeptidase activity is absent from the ER, production of peptide precursors with correct C-terminal ends by the proteasome is required for subsequent HLA class I presentation (31). It has been highlighted that the trimming efficiency of N-extended epitopes strongly depends on the amino acid composition of the epitope flanking region. Some amino acids, such as leucine, lysine, phenylalanine, and methionine, are cleaved with high activity, whereas amino acids such as proline and glutamic acid are only poorly cleaved (40). Accordingly, these latter amino acids are underrepresented in flanking regions of known CD8+ T cell epitopes (41). In HCV genotype 1a there was HLA-B*51-associated selection pressure on position −5 of the epitope with a substitution from serine to proline. Digestion of the N- and C-extended variant peptide by the constitutive and the immune proteasome yielded predominantly precursor peptides starting with proline, whereas digestion of the prototype yielded also shorter N-extended precursor peptides. The shorter peptides are advantageous for the transport into the ER (42), and more importantly, N-terminal trimming of the peptide precursor starting with proline is inhibited (31). This suggests that the serine to proline substitution altered proteasomal production of epitope precursors with the consequence of a very low abundance of cleavable N-extended epitopes in the ER, although the latter was not formally tested.
Selection of CD8+ T cell escape mutations is a trade-off between functional escape and viral constraints. Indeed, HCV may explore its replication space with different substitutions in targeted epitopes before some mutations reach fixation (9, 43). The example presented here underlines that even despite an epitope being conserved across HCV genotypes the selected escape pathways and the CD8+ T cell response can substantially differ. Although not fully conclusive, our data suggest that selection of the S1368P substitution represents the optimal trade-off between functional immune escape and viral fitness in genotype 1a. The I1380L substitution may be disadvantageous due to its fitness costs despite efficient immune escape. In turn, the degree of cross-reactivity of the I1373V variant may prevent fixation in genotype 1a despite its low fitness costs. Interestingly, we observed interindividual differences in the level of cross-reactivity of the I1373V substitution with high cross-reactivity in HCV-RNA-negative PWID and lower levels of cross-reactivity in PWID infected with genotype 3a (Fig. 1D and E), suggesting that the degree of cross-reactivity may influence the outcome of infection as previously reported (17). Absence of the CD8+ T cell response in PWID with chronic genotype 1a infection is unlikely caused by lack of priming. The rapid decline of the response in the patient with acute HCV genotype 1a infection rather suggests secondary failure. This patient harbored a virus with the S1368P substitution. Although the CD8+ T cell response was clearly impaired against endogenously processed antigens carrying the S1368P substitution, there was still some degree of T cell reactivity in our assays. This residual epitope presentation may cause continuous CD8+ T cell stimulation associated with progressive dysfunction or exhaustion of antigen-specific CD8+ T cells (44–46) and possibly ultimate extinction of these cells (46).
The degree of CD8+ T cell reactivity against the S1368P substitution in the context of genotype 1a was comparable to the reactivity against the endogenously processed genotype 3a antigen, suggesting that the epitope is less efficiently processed in the context of genotype 3a. Notably, in genotype 3a the substitution S1369P also impaired recognition of the endogenously processed antigen; however, here the I1373V was preferentially selected. One possibility is that in combination with the lower processing efficiency of the epitope in genotype 3a the I1373V substitution is sufficient to escape from the immune response. A second plausible explanation is that the I1373V is associated with lower fitness costs compared to S1369P and I1380L in genotype 3a. However, this cannot be addressed until robust genotype 3a infectious viruses are publicly available. In either case, at least in some individuals the corresponding CD8+ T cell response was preserved in our cohort of PWID infected with genotype 3a, a finding consistent with a memory T cell phenotype after mutational escape of the targeted antigen as previously reported (44, 45).
Taken together, HCV is able to evolutionarily adapt to CD8+ T cell immune pressure in multiple ways. The pathways to mutational escape are predictable but are distinct in different genotypes. Beyond selection of mutations inside targeted epitopes that impair HLA class I binding of the variant peptide ligand or TCR binding to the variant HLA class I/peptide complex HCV also inhibits epitope processing by modification of the epitope flanking region under T cell immune pressure. Importantly, the selected escape pathway of HCV may have consequences for the destiny of antigen-specific CD8+ T cells.
ACKNOWLEDGMENTS
We thank Jens Bukh for the TNcc strain, Darius Moradpour and Charles Rice for monoclonal antibodies, and Michael Engelmann and Lejla Timmer for technical assistance.
REFERENCES
- 1.Pybus OG, Cochrane A, Holmes EC, Simmonds P. 2005. The hepatitis C virus epidemic among injecting drug users. Infect Genet Evol 5:131–139. doi: 10.1016/j.meegid.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Esteban JI, Sauleda S, Quer J. 2008. The changing epidemiology of hepatitis C virus infection in Europe. J Hepatol 48:148–162. doi: 10.1016/j.jhep.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 3.Rustgi VK. 2007. The epidemiology of hepatitis C infection in the United States. J Gastroenterol 42:513–521. doi: 10.1007/s00535-007-2064-6. [DOI] [PubMed] [Google Scholar]
- 4.Page K, Hahn JA, Evans J, Shiboski S, Lum P, Delwart E, Tobler L, Andrews W, Avanesyan L, Cooper S, Busch MP. 2009. Acute hepatitis C virus infection in young adult injection drug users: a prospective study of incident infection, resolution, and reinfection. J Infect Dis 200:1216–1226. doi: 10.1086/605947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mehta SH, Astemborski J, Kirk GD, Strathdee SA, Nelson KE, Vlahov D, Thomas DL. 2011. Changes in blood-borne infection risk among injection drug users. J Infect Dis 203:587–594. doi: 10.1093/infdis/jiq112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Micallef JM, Macdonald V, Jauncey M, Amin J, Rawlinson W, van Beek I, Kaldor JM, White PA, Dore GJ. 2007. High incidence of hepatitis C virus reinfection within a cohort of injecting drug users. J Viral Hepat 14:413–418. doi: 10.1111/j.1365-2893.2006.00812.x. [DOI] [PubMed] [Google Scholar]
- 7.Aitken CK, Lewis J, Tracy SL, Spelman T, Bowden DS, Bharadwaj M, Drummer H, Hellard M. 2008. High incidence of hepatitis C virus reinfection in a cohort of injecting drug users. Hepatology 48:1746–1752. doi: 10.1002/hep.22534. [DOI] [PubMed] [Google Scholar]
- 8.von Hahn T, Yoon JC, Alter H, Rice CM, Rehermann B, Balfe P, McKeating JA. 2007. Hepatitis C virus continuously escapes from neutralizing antibody and T-cell responses during chronic infection in vivo. Gastroenterology 132:667–678. doi: 10.1053/j.gastro.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Timm J, Lauer GM, Kavanagh DG, Sheridan I, Kim AY, Lucas M, Pillay T, Ouchi K, Reyor LL, Schulze zur Wiesch J, Gandhi RT, Chung RT, Bhardwaj N, Klenerman P, Walker BD, Allen TM. 2004. CD8 epitope escape and reversion in acute HCV infection. J Exp Med 200:1593–1604. doi: 10.1084/jem.20041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruhl M, Knuschke T, Schewior K, Glavinic L, Neumann-Haefelin C, Chang DI, Klein M, Heinemann FM, Tenckhoff H, Wiese M, Horn PA, Viazov S, Spengler U, Roggendorf M, Scherbaum N, Nattermann J, Hoffmann D, Timm J. 2011. CD8+ T-cell response promotes evolution of hepatitis C virus nonstructural proteins. Gastroenterology 140:2064–2073. doi: 10.1053/j.gastro.2011.02.060. [DOI] [PubMed] [Google Scholar]
- 11.Erickson AL, Kimura Y, Igarashi S, Eichelberger J, Houghton M, Sidney J, McKinney D, Sette A, Hughes AL, Walker CM. 2001. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity 15:883–895. doi: 10.1016/S1074-7613(01)00245-X. [DOI] [PubMed] [Google Scholar]
- 12.Cox AL, Mosbruger T, Mao Q, Liu Z, Wang XH, Yang HC, Sidney J, Sette A, Pardoll D, Thomas DL, Ray SC. 2005. Cellular immune selection with hepatitis C virus persistence in humans. J Exp Med 201:1741–1752. doi: 10.1084/jem.20050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray SC, Fanning L, Wang XH, Netski DM, Kenny-Walsh E, Thomas DL. 2005. Divergent and convergent evolution after a common-source outbreak of hepatitis C virus. J Exp Med 201:1753–1759. doi: 10.1084/jem.20050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farci P, Alter HJ, Wong DC, Miller RH, Govindarajan S, Engle R, Shapiro M, Purcell RH. 1994. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci U S A 91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fauvelle C, Lepiller Q, Felmlee DJ, Fofana I, Habersetzer F, Stoll-Keller F, Baumert TF, Fafi-Kremer S. 2013. Hepatitis C virus vaccines: progress and perspectives. Microb Pathog 58:66–72. doi: 10.1016/j.micpath.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Ziegler S, Skibbe K, Walker A, Ke X, Heinemann FM, Heinold A, Mok JY, van Esch WJ, Yang D, Wolfl M, Timm J. 2014. Impact of sequence variation in a dominant HLA-A*02-restricted epitope in hepatitis C virus on priming and cross-reactivity of CD8+ T cells. J Virol 88:11080–11090. doi: 10.1128/JVI.01590-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yerly D, Heckerman D, Allen TM, Chisholm JV III, Faircloth K, Linde CH, Frahm N, Timm J, Pichler WJ, Cerny A, Brander C. 2008. Increased cytotoxic T-lymphocyte epitope variant cross-recognition and functional avidity are associated with hepatitis C virus clearance. J Virol 82:3147–3153. doi: 10.1128/JVI.02252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yusim K, Dilan R, Borducchi E, Stanley K, Giorgi E, Fischer W, Theiler J, Marcotrigiano J, Korber B, Barouch DH. 2013. Hepatitis C genotype 1 mosaic vaccines are immunogenic in mice and induce stronger T-cell responses than natural strains. Clin Vaccine Immunol 20:302–305. doi: 10.1128/CVI.00605-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giugliano S, Oezkan F, Bedrejowski M, Kudla M, Reiser M, Viazov S, Scherbaum N, Roggendorf M, Timm J. 2009. Degree of cross-genotype reactivity of hepatitis C virus-specific CD8+ T cells directed against NS3. Hepatology 50:707–716. doi: 10.1002/hep.23096. [DOI] [PubMed] [Google Scholar]
- 20.von Delft A, Humphreys IS, Brown A, Pfafferott K, Lucas M, Klenerman P, Lauer GM, Cox AL, Gaudieri S, Barnes E. 2015. The broad assessment of HCV genotypes 1 and 3 antigenic targets reveals limited cross-reactivity with implications for vaccine design. Gut doi: 10.1136/gutjnl-2014-308724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuntzen T, Timm J, Berical A, Lennon N, Berlin AM, Young SK, Lee B, Heckerman D, Carlson J, Reyor LL, Kleyman M, McMahon CM, Birch C, Schulze Zur Wiesch J, Ledlie T, Koehrsen M, Kodira C, Roberts AD, Lauer GM, Rosen HR, Bihl F, Cerny A, Spengler U, Liu Z, Kim AY, Xing Y, Schneidewind A, Madey MA, Fleckenstein JF, Park VM, Galagan JE, Nusbaum C, Walker BD, Lake-Bakaar GV, Daar ES, Jacobson IM, Gomperts ED, Edlin BR, Donfield SM, Chung RT, Talal AH, Marion T, Birren BW, Henn MR, Allen TM. 2008. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology 48:1769–1778. doi: 10.1002/hep.22549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruhl M, Chhatwal P, Strathmann H, Kuntzen T, Bankwitz D, Skibbe K, Walker A, Heinemann FM, Horn PA, Allen TM, Hoffmann D, Pietschmann T, Timm J. 2012. Escape from a dominant HLA-B*15-restricted CD8+ T cell response against hepatitis C virus requires compensatory mutations outside the epitope. J Virol 86:991–1000. doi: 10.1128/JVI.05603-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Megger DA, Bracht T, Kohl M, Ahrens M, Naboulsi W, Weber F, Hoffmann AC, Stephan C, Kuhlmann K, Eisenacher M, Schlaak JF, Baba HA, Meyer HE, Sitek B. 2013. Proteomic differences between hepatocellular carcinoma and nontumorous liver tissue investigated by a combined gel-based and label-free quantitative proteomics study. Mol Cell Proteomics 12:2006–2020. doi: 10.1074/mcp.M113.028027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borisenko IA, Viazovichenko Iu E, Gudkov VI. 1994. An improvement in information support in the interests of the epidemiological health welfare of the troops. Voenno-Meditsinskii Zhurnal 37–42:80 (In Russian.) [PubMed] [Google Scholar]
- 25.Li YP, Ramirez S, Jensen SB, Purcell RH, Gottwein JM, Bukh J. 2012. Highly efficient full-length hepatitis C virus genotype 1 (strain TN) infectious culture system. Proc Natl Acad Sci U S A 109:19757–19762. doi: 10.1073/pnas.1218260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinmann E, Brohm C, Kallis S, Bartenschlager R, Pietschmann T. 2008. Efficient trans-encapsidation of hepatitis C virus RNAs into infectious virus-like particles. J Virol 82:7034–7046. doi: 10.1128/JVI.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marukian S, Jones CT, Andrus L, Evans MJ, Ritola KD, Charles ED, Rice CM, Dustin LB. 2008. Cell culture-produced hepatitis C virus does not infect peripheral blood mononuclear cells. Hepatology 48:1843–1850. doi: 10.1002/hep.22550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 29.Moradpour D, Wakita T, Tokushige K, Carlson RI, Krawczynski K, Wands JR. 1996. Characterization of three novel monoclonal antibodies against hepatitis C virus core protein. J Med Virol 48:234–241. doi:. [DOI] [PubMed] [Google Scholar]
- 30.Kim AY, Kuntzen T, Timm J, Nolan BE, Baca MA, Reyor LL, Berical AC, Feller AJ, Johnson KL, Schulze zur Wiesch J, Robbins GK, Chung RT, Walker BD, Carrington M, Allen TM, Lauer GM. 2011. Spontaneous control of HCV is associated with expression of HLA-B 57 and preservation of targeted epitopes. Gastroenterology 140:686–696 e681. doi: 10.1053/j.gastro.2010.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Endert P. 2011. Post-proteasomal and proteasome-independent generation of MHC class I ligands. Cell Mol Life Sci 68:1553–1567. doi: 10.1007/s00018-011-0662-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura Y, Gushima T, Rawale S, Kaumaya P, Walker CM. 2005. Escape mutations alter proteasome processing of major histocompatibility complex class I-restricted epitopes in persistent hepatitis C virus infection. J Virol 79:4870–4876. doi: 10.1128/JVI.79.8.4870-4876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seifert U, Liermann H, Racanelli V, Halenius A, Wiese M, Wedemeyer H, Ruppert T, Rispeter K, Henklein P, Sijts A, Hengel H, Kloetzel PM, Rehermann B. 2004. Hepatitis C virus mutation affects proteasomal epitope processing. J Clin Invest 114:250–259. doi: 10.1172/JCI200420985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlson JM, Brumme ZL, Rousseau CM, Brumme CJ, Matthews P, Kadie C, Mullins JI, Walker BD, Harrigan PR, Goulder PJ, Heckerman D. 2008. Phylogenetic dependency networks: inferring patterns of CTL escape and codon covariation in HIV-1 Gag. PLoS Comput Biol 4:e1000225. doi: 10.1371/journal.pcbi.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen TM, Altfeld M, Yu XG, O'Sullivan KM, Lichterfeld M, Le Gall S, John M, Mothe BR, Lee PK, Kalife ET, Cohen DE, Freedberg KA, Strick DA, Johnston MN, Sette A, Rosenberg ES, Mallal SA, Goulder PJ, Brander C, Walker BD. 2004. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J Virol 78:7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milicic A, Price DA, Zimbwa P, Booth BL, Brown HL, Easterbrook PJ, Olsen K, Robinson N, Gileadi U, Sewell AK, Cerundolo V, Phillips RE. 2005. CD8+ T cell epitope-flanking mutations disrupt proteasomal processing of HIV-1 Nef. J Immunol 175:4618–4626. doi: 10.4049/jimmunol.175.7.4618. [DOI] [PubMed] [Google Scholar]
- 37.Sijts EJ, Kloetzel PM. 2011. The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell Mol Life Sci 68:1491–1502. doi: 10.1007/s00018-011-0657-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammer GE, Kanaseki T, Shastri N. 2007. The final touches make perfect the peptide-MHC class I repertoire. Immunity 26:397–406. doi: 10.1016/j.immuni.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. 2002. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature 419:480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 40.Zhang SC, Martin E, Shimada M, Godfrey SB, Fricke J, Locastro S, Lai NY, Liebesny P, Carlson JM, Brumme CJ, Ogbechie OA, Chen H, Walker BD, Brumme ZL, Kavanagh DG, Le Gall S. 2012. Aminopeptidase substrate preference affects HIV epitope presentation and predicts immune escape patterns in HIV-infected individuals. J Immunol 188:5924–5934. doi: 10.4049/jimmunol.1200219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schatz MM, Peters B, Akkad N, Ullrich N, Martinez AN, Carroll O, Bulik S, Rammensee HG, van Endert P, Holzhutter HG, Tenzer S, Schild H. 2008. Characterizing the N-terminal processing motif of MHC class I ligands. J Immunol 180:3210–3217. doi: 10.4049/jimmunol.180.5.3210. [DOI] [PubMed] [Google Scholar]
- 42.van Endert PM, Tampe R, Meyer TH, Tisch R, Bach JF, McDevitt HO. 1994. A sequential model for peptide binding and transport by the transporters associated with antigen processing. Immunity 1:491–500. doi: 10.1016/1074-7613(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 43.Uebelhoer L, Han JH, Callendret B, Mateu G, Shoukry NH, Hanson HL, Rice CM, Walker CM, Grakoui A. 2008. Stable cytotoxic T cell escape mutation in hepatitis C virus is linked to maintenance of viral fitness. PLoS Pathog 4:e1000143. doi: 10.1371/journal.ppat.1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kasprowicz V, Kang YH, Lucas M, Schulze zur Wiesch J, Kuntzen T, Fleming V, Nolan BE, Longworth S, Berical A, Bengsch B, Thimme R, Lewis-Ximenez L, Allen TM, Kim AY, Klenerman P, Lauer GM. 2010. Hepatitis C virus (HCV) sequence variation induces an HCV-specific T-cell phenotype analogous to spontaneous resolution. J Virol 84:1656–1663. doi: 10.1128/JVI.01499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bengsch B, Seigel B, Ruhl M, Timm J, Kuntz M, Blum HE, Pircher H, Thimme R. 2010. Coexpression of PD-1, 2B4, CD160 and KLRG1 on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. PLoS Pathog 6:e1000947. doi: 10.1371/journal.ppat.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wherry EJ. 2011. T cell exhaustion. Nat Immunol 12:492–499. [DOI] [PubMed] [Google Scholar]