Abstract
The cell cycle of neurons remains suppressed to maintain the state of differentiation and aberrant cell cycle reentry results in loss of neurons, which is a feature in neurodegenerative disorders like Alzheimer's disease (AD). Present studies revealed that the expression of microRNA 34a (miR-34a) needs to be optimal in neurons, as an aberrant increase or decrease in its expression causes apoptosis. miR-34a keeps the neuronal cell cycle under check by preventing the expression of cyclin D1 and promotes cell cycle arrest. Neurotoxic amyloid β1–42 peptide (Aβ42) treatment of cortical neurons suppressed miR-34a, resulting in unscheduled cell cycle reentry, which resulted in apoptosis. The repression of miR-34a was a result of degradation of TAp73, which was mediated by aberrant activation of the MEK extracellular signal-regulated kinase (ERK) pathway by Aβ42. A significant decrease in miR-34a and TAp73 was observed in the cortex of a transgenic (Tg) mouse model of AD, which correlated well with cell cycle reentry observed in the neurons of these animals. Importantly, the overexpression of TAp73α and miR-34a reversed cell cycle-related neuronal apoptosis (CRNA). These studies provide novel insights into how modulation of neuronal cell cycle machinery may lead to neurodegeneration and may contribute to the understanding of disorders like AD.
INTRODUCTION
Neurons exit the cell cycle upon terminal differentiation, and their cell cycle remains suppressed to maintain the state of differentiation. There is substantial evidence which indicates that neurons can reenter the cell cycle and also undergo DNA replication. However, this attempt, which typically may occur in response to neurotoxic insults like beta amyloid peptide Aβ42 and DNA-damaging agents, leads to neuronal death but not mitosis (1–4). The role of several cell cycle proteins seems to be distinct from the cell cycle in neurons. G1-S transition in the mammalian cell cycle is initiated as a result of production of D-type cyclins, which complex with their cognate partner cdk4/6. Cyclin D-cdk4/6-mediated phosphorylation of Rb causes its dissociation from E2F, resulting in its transcriptional activation (5). E2F transcribes genes like the cyclin E gene, which are important for S-phase progression. In terminally differentiated neurons, this pathway remains largely shut down. The process of neuronal differentiation is dependent on Rb, as its knockout results in dedifferentiation and cell cycle reentry (6) in an E2F-1-dependent manner (7), indicating that it is critical for the cell cycle machinery to remain suppressed for the health of the neuron.
Although the link between neuronal cell cycle reentry has been reported for several neurodegenerative disorders (8), it is best studied for Alzheimer's disease (AD) (8, 9). Several lines of evidence implicate aberrant cell cycle reentry of neurons in the brains of AD patients, which may contribute to neuronal loss (10, 11). Studies on various in vitro and animal models of neurodegenerative disorders like AD indicate that the enhanced expression of cell cycle genes and DNA replication may precede cell death (12–14). In response to neurotoxic insults like amyloid peptide Aβ42, the aberrant expression of cell cycle proteins like cyclins facilitates cell cycle reentry (15–17). In this context, cyclin D1 seems to play an important role in cell cycle-related neuronal apoptosis (CRNA), as its increased expression by Aβ42 or DNA-damaging agents triggers this process (12, 16, 18). In addition, cyclin D1 is highly expressed in neurons of patients with mild cognitive impairment (MCI) and AD, which undergo CRNA (11). Recently, we have demonstrated that Aβ42 causes aberrant activation of the MEK–extracellular signal-regulated kinase (ERK) pathway, which leads to cyclin D1 overproduction, resulting in CRNA. Cyclin D1 overproduction abrogates interaction between p35 and cdk5, causing decreased p35-cdk5 activity, which may contribute to CRNA (12).
MicroRNAs (miRNAs), which are a subclass of noncoding RNAs, regulate the expression of genes in most mammalian cells, including neurons. miRNAs contribute significantly to nervous system development, and several miRNAs are dysregulated in neurodegenerative disorders like AD (19–21). However, it is largely unclear how miRNAs regulate or suppress the cell cycle of neurons and thereby prevent their reentry into the cell cycle. While cell cycle regulators seem to perform dual functions in neuronal differentiation and neurodegeneration, how their expression may be modulated in the two situations is poorly understood. We investigated the involvement of miRNA 34a (miR-34a), which is expressed at high levels in the brain (22), in these processes. miR-34a is regulated by p53, and its ectopic expression mimics several p53 effects in a cell-dependent manner (23, 24). In neurons, miR-34a is regulated by p73 isoform TAp73 during neuronal differentiation, and its inhibition prevents neurite outgrowth (25, 26). However, the role of miR-34a in the neuronal cell cycle is poorly understood. The balance between the TAp73 and ΔNp73 regulates cell fate. TAp73−/− (27) as well as ΔNp73−/− (28) mice exhibit brain defects like hippocampal dysgenesis, genomic instability, and neurodegeneration. Despite this information, how TAp73 isoforms regulate neuronal functions is also largely unclear.
We demonstrate that miR-34a prevents cell cycle reentry and suppresses the neuronal cell cycle by targeting cyclin D1. Furthermore, miR-34a expression was significantly altered in cortical neurons treated with neurotoxic Aβ42 and neurons from a transgenic mouse model for AD, which resulted in an aberrant increase in cyclin D1 levels, leading to cell cycle reentry and apoptosis of neurons. We have also elucidated the mechanism via which TAp73 and miR-34a are deregulated, which results in cell cycle reentry and apoptosis.
MATERIALS AND METHODS
Materials. (i) Antibodies.
Antibodies against the following were used for the described studies: cyclin D1 (sc-753; 1:500), PCNA (sc-56; 1:500), phospho-ERK (sc-7383; 1:500), ERK (sc-94; 1:1,000), p73 (Sc-9651, 1:500), ubiquitin (sc-8017; 1:1,000), actin (sc-47778; 1:1,000), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; sc-25778; 1:1,000) (all from Santa Cruz Biotechnology, Santa Cruz, CA) and cleaved caspase 3 (9661; 1:500; Cell Signaling, Beverly, MA), green fluorescent protein (GFP; 2555; 1:1,000; Cell Signaling), and 5-bromo-2′-deoxyuridine (BrdU; RPN202; GE Healthcare Biosciences, Piscataway, NJ).
(ii) miRNA, anti-miR, and small interfering RNA (siRNA). (a) miR-34a mimic and antagomir.
A miR-34a mimic (mirVana miRNA mimic; catalog no. 4464066) or antagomir34a (mirVana miRNA inhibitor; catalog no. 4464084) were from Ambion, USA. miRNA Control miRNA (Ctrl_miR) for miR-34a (mirVana miRNA mimic, negative control no. 1; catalog no. 4464058) and for anti-miR-34a (Ctrl_anti) (mirVana miRNA inhibitor, negative control no. 1; catalog no. 4464076) was used as a control.
(b) siRNA against cyclin D1.
For cyclin D1 knockdown, siRNA duplexes were designed and custom synthesized from Dharmacon, USA. Sequences were as follows: cyclin D1 siRNA, 5′ GCGAGGAGCAGAAGUGCGAUG 3′, and scrambled siRNA (ctrl_siRNA), 5′ GCGAGGAGAAGCAGUGCGAUG 3′.
(iii) Plasmid DNA constructs. (a) Cyclin D1 3′ UTR.
Using three computer-aided algorithms, including Target Scan, miRanda, and PicTar, cyclin D1 was identified as a possible target of miR-34a in humans, rats, and mice. The miR-34a target sites were predicted based on base-pairing seed sequence matches. Briefly, a 1,143-bp fragment of the cyclin D1 3′ untranslated region (UTR) containing the putative miR-34a binding site was amplified from rat mRNA by reverse transcription-PCR (RT-PCR) and cloned into the XhoI and NotI sites of the psiCheck2 vector upstream of the Renilla luciferase gene using the following primers: Cyclin D1 UTR_F, ctcgagGAGAGCCATCCAAACTGAGG, and Cyclin D1 UTR_R, gcggccgcCCAGAATACACAAAGCCAACC. Restriction sites for XhoI and NotI are indicated by lowercase letters.
The mutant of the miR-34a binding site was generated by overlapping PCR, which was performed using the following primers: Cyclin D1 Mut UTR_F, CAATGTCATAAGTACCTCTGTTCAAGTTTTAATTTCCTCGTAG, and Cyclin D1 Mut UTR_R, CTACGAGGAAATTAAAACTTGAACAGAGGTACTTATGACATTG (sequences corresponding to the mutation are underlined).
(b) miR-34a promoter.
The pGL3Basic vector harboring the 2-kb promoter region of hsa-miR-34a (miR-34a-Prom-Luc) and its mutant were kindly provided by M. Oren (The Weizmann Institute, Israel). The putative p53/p73 binding site (5′-GGGCTTGCCTGGGCTTGTTC-3′) was mutated (to 5′-GGGtTaaCCTGGGtTTGTTC-3′) to abolish p53/p73 binding. Mutated nucleotides are indicated by lowercase letters.
(iv) Alzheimer's disease mouse model.
The amyloid-β precursor protein/presenilin 1 (APP/PS1) transgenic mouse model (APP/PS1 Tg) for AD [strain name B6C3-Tg(APPswe,PSEN1dE9)85Dbo/J; stock number 004462] maintained by The Jackson Laboratory was gifted by National Brain Research Centre, Manesar, India, to National Institute of Immunology, New Delhi, India. These mice are doubly transgenic for APP and PS1, as they express a chimeric mouse/human amyloid-β precursor protein containing the K595N/M596L Swedish mutations and a mutant human presenilin 1 carrying the exon 9 deletion variant under the control of mouse prion promoter elements, directing transgene expression predominantly to central nervous system neurons (29, 30). The transgenic mice were genotyped by PCR analysis of genomic DNA from tail biopsy specimens.
Cell culture.
For cortical neuronal culture from Sprague-Dawley rats or APP/PS1 transgenic AD mice, a previously published protocol (12) was followed. Briefly, after cortical regions of embryonic day 18 (E18) rat or E16 mouse embryos were dissected, the tissue was treated with trypsin-DNase solution until the cell mass dissociated and the solution became turbid. Excess serum-containing medium (SCM) was quickly added to the cells to stop trypsinization, and the cells were centrifuged at 500 × g for 5 min at 25°C. Subsequently, cells were plated on poly-l-lysine-coated 6-well tissue culture plates. After 12 h, cells were washed with prewarmed Tyrode's calcium- and magnesium-free (CMF) phosphate buffered saline (PBS) supplemented with glucose and NaHCO3 and were maintained in serum-free medium (SFM) containing B27 and N2 supplements, 1× penicillin-streptomycin, l-glutamine, and glucose in a CO2 incubator maintaining 5% CO2 levels.
SH-SY5Y cells were maintained in Dulbecco modified Eagle medium (DMEM) plus 10% fetal bovine serum (FBS). For differentiation, cells were cultured in DMEM plus 3% FBS and treated with 10 μM trans-retinoic acid (RA) for the desired time.
All experiments were performed in accordance with the guidelines for animal experiments of the Institutional Animal Ethical Committee of the National Institute of Immunology.
miRNA and siRNA transfections and treatment.
Typically, cortical neurons or SH-SY5Y cells were transfected with 100 nM miR-34a mimic or anti-miR-34a or 100 pmol of siRNA by using Lipofectamine 2000 (Invitrogen, USA) as per the manufacturer's instructions for siRNA transfection. Three hours after transfection, medium was changed to serum-free growth medium supplemented with B27 and N2, and cells were allowed to remain in this medium for 12 h. Subsequently, cultures were moved to serum-free medium supplemented with B27 and N2, and desired treatments were started for 48 h unless indicated otherwise. Adenovirus for GFP (control), cyclin D1 (12), or human TAp73α (gifted by Sanjeev Das, NII) was used to overexpress these proteins. Aβ1–42 (R-peptide) was used to prepare soluble oligomers of Aβ42 as described previously (12, 31). Unless indicated otherwise, cortical neurons were typically treated with a 0.5 μM concentration of the soluble oligomeric form of Aβ42 for 48 h to induce CRNA.
Real-time PCR for microRNA expression.
RNA isolation was performed by using an miRNeasy minikit (Qiagen) or by using TRIzol. Real-time PCR was performed in a Mastercycler RealPlex machine (Eppendorf). Quantitative RT-PCR (qRT-PCR) for microRNA was performed by using a TaqMan microRNA assay kit. Typically, ∼500 ng of total RNA was reverse transcribed using a TaqMan microRNA RT kit (Applied Biosystems) and microRNA-specific stem-loop RT primers provided in the TaqMan microRNA assay kit. Briefly, following the RT step, 1.33 μl of the RT reaction product was combined with 1 μl of a TaqMan microRNA assay (20×; forward primer, reverse primer, and probe, Applied Biosystems, catalog no. 4427975, hsa-miR-34a assay no. 000426, or RNU6B assay no. 001093) and 10 μl of TaqMan universal PCR master mix, No AmpErase UNG, in a 20-μl final volume. The expression of miRNA was defined on the basis of threshold cycle (CT), and the relative expression levels were determined by ΔΔCT after normalization with RNU6B.
Immunoblotting.
Cells were washed with PBS and scraped in cold lysis buffer (10% glycerol, 50 mM Tris HCl [pH 7.6], 150 mM NaCl, 1 mM MgCl2, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM sodium orthovanadate, 20 mM β-glycerophosphate, and 1× protease inhibitor cocktail) on ice. Immunoblotting was performed as described previously (12) using primary antibodies and horseradish peroxidase (HRP)-labeled secondary antibodies. The immunoreactive proteins were detected using an ECL Super Signal west Pico or west Dura kit (Pierce) on X-ray film.
Immunoprecipitation.
Fifty to 70 μg of protein lysate was used to immunoprecipitate TAp73 and incubated with ∼1 μg of anti TAp73 antibody for 12 h at 4°C. Subsequently, 50 μl of protein A+G-Sepharose (Amersham Biosciences) was incubated with the antibody-protein complex for 4 to 6 h at 4°C. The beads were washed and resuspended in cold lysis buffer, and immunoblotting was performed by using antiubiquitin antibody.
Luciferase reporter assays for the cyclin D1 3′ UTR and miR-34a promoter.
The cyclin D1 3′ UTR was cloned downstream of the Renilla luciferase gene in psiCheck2 plasmid, which also contains a synthetic firefly luciferase gene that serves as a transfection control. A miR-34a promoter-luciferase construct (see above for details) or its mutant plasmid was cotransfected with expression plasmid for β-galactosidase, which was used for normalization. The transfections were performed using Lipofectamine 2000, cells were harvested after 48 h, and luciferase activity was measured with a dual-luciferase reporter assay kit (Promega, Madison, WI) in the case of cyclin D1 3′ UTR transfection. β-Galactosidase activity was estimated by using o-nitrophenyl-β-d-galactopyranoside (ONPG) as the substrate, and the formation of a chromogenic substance was determined by measuring absorbance at 420 nm. The luciferase activity driven by miR-34a promoter was normalized with respect to β-galactosidase activity.
BrdU incorporation and TUNEL assay.
5-Bromo-2′-deoxyuridine (BrdU) labeling and terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assay were performed simultaneously to detect DNA replication and cell death as described previously (12). Anti-BrdU antibody (GE) and Hoechst 33342 were used to detect incorporated BrdU and stain the nuclei, respectively. Cells were visualized using a Zeiss AxioImager microscope equipped with an MRm camera, Axiovision software was used for image acquisition and manipulation, and Adobe Photoshop was used for preparing images for illustrations. The BrdU- and TUNEL-stained cells were counted, and the percentage of total cells that were identified by 4′,6-diamidino-2-phenylindole (DAPI) staining was determined.
Image and statistical analysis.
The densitometry analysis of desired bands in Western blots was performed by using ImageJ (NIH) software. The signal intensity of the loading control (GAPDH/actin) was used for normalization. Unless indicated otherwise, one-way analysis of variance (ANOVA) followed by the Student-Newman-Keuls (SNK) test was used for statistical analysis using Prism software (GraphPad Software Inc., USA). Data are presented as means ± standard errors of the means (SEM) from three independent experiments unless indicated otherwise. A P value of <0.05 was taken as statistically significant.
RESULTS
miR-34a suppresses the neuronal cell cycle and may promote differentiation by targeting cyclin D1.
Cyclin D1, which is expressed in very small amounts in terminally differentiated neurons, is elevated in response to neurotoxic stimuli, contributing to the process of CRNA (12, 18). In addition, cyclin D1 is aberrantly expressed in patients with mild cognitive impairment and AD (11). However, it has remained unclear how cyclin D1 levels remain suppressed in differentiated neurons. We investigated the possibility of the involvement of miRNA-mediated regulation of cyclin D1 in the neuronal cell cycle. In silico analysis was performed using Targetscan, Miranda, and Pictar to identify putative miRNAs that may target the 3′ UTR of cyclin D1. While several candidate miRNAs emerged from these studies as putative effectors of cyclin D1 (P. K. Modi and P. Sharma, unpublished results), miR-34a was an attractive putative candidate, as it is expressed in the brain and is elevated during neuronal differentiation (26).
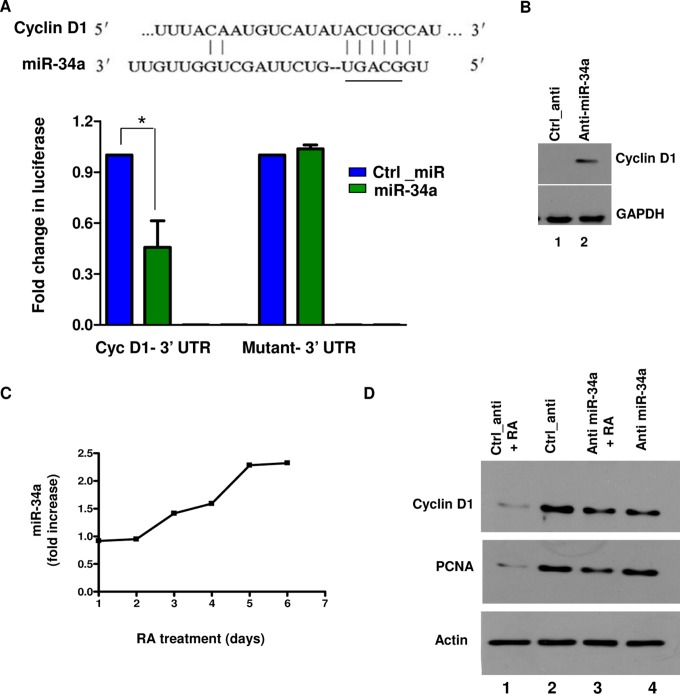
We first tested if miR-34a targets cyclin D1 in neurons. A luciferase reporter plasmid which contained the 3′ UTR of cyclin D1 was constructed. Overexpression of miR-34a suppressed expression of luciferase fused to the cyclin D1 3′ UTR in cortical neurons (Fig. 1A). In contrast, in the case of a mutant 3′ UTR in which the miR-34a target site was altered, luciferase activity remained almost unchanged. Importantly, an antagomir of miR-34a (anti-miR-34a) caused a significant increase in cyclin D1 (Fig. 1B; see also Fig. S1B in the supplemental material). Since cyclin D1 is expressed at very low levels in terminally differentiated neurons, this result suggests that miR-34a may be critical in repressing cyclin D1 expression. Collectively, these data suggested that miR-34a targets cyclin D1 in neurons.
FIG 1.
miR-34a targets cyclin D1 in neurons and promotes neuronal differentiation. (A) Rat (shown here), mouse, and human cyclin D1 3′ UTR has a region which exhibits complementarity to miR-34a seed sequence (underlined). Rat cortical neurons were transfected with the cyclin D1 3′ UTR or its mutant reporter plasmid along with miR-34a or a control miRNA (Ctrl_miR). Cell lysates were prepared after 48 h, and luciferase activity assays were performed. Fold changes in activity upon transfection with miR-34a are illustrated (*, P < 0.05, ANOVA; n = 3). (B) Rat cortical neurons were transfected with anti-miR-34a or control antagomir (Ctrl_anti). Cell lysates were prepared, and Western blotting for cyclin D1 was performed. GAPDH was used as a loading control. (C) SH-SY5Y cells were allowed to differentiate for 6 days using RA, and RNA was isolated at the indicated time points. Expression of miR-34a was quantified by qRT-PCR and is represented as fold change in miR-34a expression with respect to its levels at day 0 of RA treatment. (D) SH-SY5Y cells were transfected with anti-miR-34a or Ctrl_anti and were either left untreated (lanes 2 and 4) or treated with RA. After 72 h, lysates were prepared, followed by Western blotting with the indicated antibodies.
Next, we investigated the role of miR-34a in the neuronal cell cycle and CRNA in light of the above-described findings. The expression of miR-34a significantly increased during differentiation of cortical neurons derived from E18 rat embryos, reaching its peak 5 to 7 days in vitro (DIV) (25; P. K. Modi, S. Jaiswal, and P. Sharma, unpublished results). Retinoic acid-mediated differentiation of SH-SY5Y neuroblastoma cells caused a similar increase in miR-34a (Fig. 1C), which was consistent with previous studies (26). RA caused a significant decrease in cyclin D1 (Fig. 1D, lane 1 versus lane 2). Transfection of anti-miR-34a prevented this decrease in cyclin D1 mediated by RA, suggesting that miR-34a may target it during neuronal differentiation (lane 3 versus lane 1). Furthermore, RA treatment resulted in a significant decrease in PCNA, a cell cycle marker. Anti-miR-34a significantly reversed the decrease in PCNA levels (Fig. 1D, lane 3 versus lane 1). Collectively, these findings suggested that miR-34a is important for suppression of the neuronal cell cycle, and it may achieve this by preventing cyclin D1 expression.
miR-34a promotes suppression of the neuronal cell cycle.
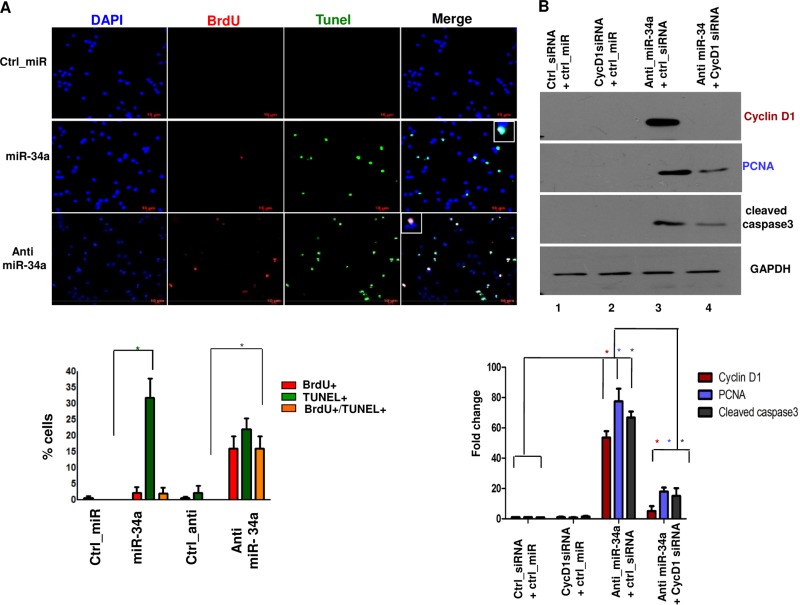
To investigate the role of miR-34a in terminally differentiated neurons, cortical neurons were transfected with miR-34a/anti-miR-34a, followed by BrdU incorporation and TUNEL assays to detect DNA replication and apoptosis, respectively. A significant increase in BrdU+/TUNEL+ neurons was observed upon anti-miR-34a treatment, which was indicative of cell cycle reentry and apoptosis (Fig. 2A). Anti-miR-34a caused an aberrant increase in cyclin D1 expression, which was accompanied by an increase in PCNA as well as cleaved caspase 3 levels (Fig. 2B; see also Fig. S1B in the supplemental material). Importantly, when cyclin D1 siRNA was cotransfected with anti-miR-34a, the increase in PCNA and caspase 3 was reversed. These data strongly indicated that miR-34a may suppress the neuronal cell cycle by preventing cyclin D1 expression. In contrast, when miR-34a was overexpressed, an increase was mainly observed in TUNEL+ and not BrdU+ cells (Fig. 2A). These data highlight the importance of optimal expression of miR-34a in neurons, as either an increase or decrease in its levels seems to cause apoptosis, albeit via different mechanisms.
FIG 2.
miR-34a suppresses the neuronal cell cycle and prevents apoptosis by targeting cyclin D1. (A) Rat cortical neurons were transfected with miR-34a, anti miR-34a, or Ctrl_miR, followed by incubation with BrdU. Immunofluorescence and TUNEL assays were performed to detect BrdU incorporation (red) and apoptosis (green), respectively. (Lower panel) Percentages of cells that were BrdU+ (red), TUNEL+ (green), or BrdU+ TUNEL+ (yellow) were determined from the experiment described for panel A. Data are expressed as means ± SEM (*, P < 0.05 by ANOVA [n = 3]). miR-34a overexpression mainly caused an increase in TUNEL staining, whereas anti-miR-34a caused a significant increase in BrdU+ TUNEL+ cells. Scale bars, 10 μm. (B) Rat cortical neurons were transfected with cyclin D1 siRNA along with anti-miR-34a or Ctrl_anti. Cell lysate was prepared after 48 h, and Western blotting was performed with the indicated antibodies. (Lower panel) Densitometry of cyclin D1, PCNA, and cleaved caspase 3 bands was performed, and their levels were normalized with respect to GAPDH levels. The means of three independent experiments are shown, and error bars represent SEM (*, P < 0.05 by ANOVA; n = 3).
Aβ42 deregulates miR-34a in cortical neurons.
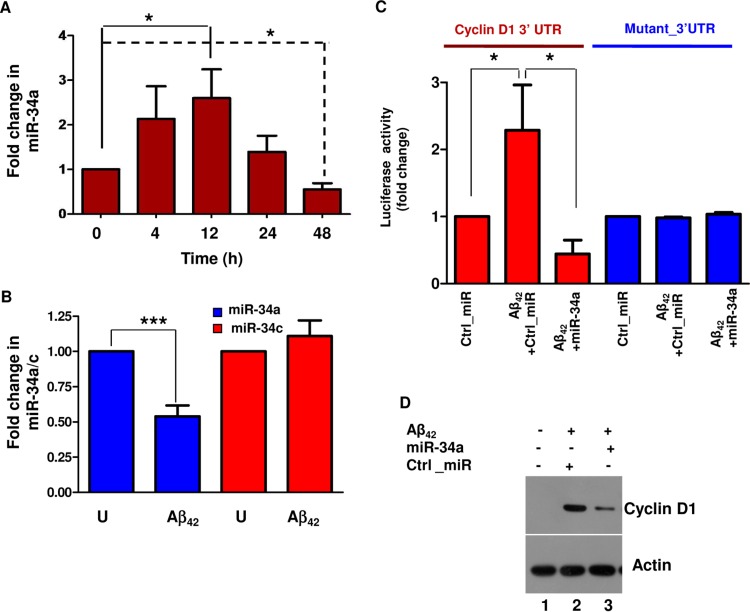
Aβ42-mediated neurotoxicity is one of the major causes for AD pathogenesis (32). Aβ42 is known to cause aberrant cell cycle reentry of cortical neurons, which is also a feature of AD animal models (13, 33) and is also detectable in the brains of AD patients (11). Since we found miR-34a to promote neuronal differentiation by preventing cell cycle entry of neurons (Fig. 2), we investigated its role in Aβ42-mediated CRNA. The treatment of cortical neurons with Aβ42 initially caused an increase in miR-34a expression, followed by a significant decline after 24 to 48 h of treatment (Fig. 3A). In contrast to miR-34a, the levels of miR-34c, which also belongs to the miR-34 family, did not change significantly (Fig. 3B). In addition, a control peptide, Aβ42–1, which possesses the sequence reverse of Aβ42 did not inhibit miR-34a expression (see Fig. S2 in the supplemental material).
FIG 3.
Aβ42 downregulates miR-34a in cortical neurons, resulting in enhanced cyclin D1 production. (A) Rat cortical neurons were treated with Aβ42 for the indicated time. Total RNA was isolated and qRT-PCR was performed to determine the levels of miR-34a; data are presented as fold change in miR-34a expression with respect to untreated neurons (0 h). Data representative of three independent experiments are shown (*, P < 0.05, ANOVA; n = 3). (B) Rat cortical neurons were treated with Aβ42 for 48 h, and qRT-PCR was performed to determine the expression of miR-34a and miR-34c. Data are means ± SEM (***, P < 0.001, t test; n = 6). However, miR-34c remained almost unchanged after Aβ42 treatment. (C) Rat cortical neurons were transfected with the cyclin D1 3′ UTR or its mutant reporter plasmid along with miR-34a or Ctrl_miR and treated with Aβ42 for 48 h. Cell lysates were prepared, luciferase activity assays were performed, and fold change in activity with respect to Ctrl_miR-transfected cells was determined (*, P < 0.05, ANOVA; n = 3). (D) Rat cortical neurons were transfected with miR-34a or Ctrl_miR and treated with Aβ42 as indicated. Cell lysates were prepared after 48 h, and Western blotting for cyclin D1 was performed; β-actin was used as a loading control.
Given the observed regulation of cyclin D1 by miR-34a, we first tested if Aβ42-mediated repression of miR-34a impacts cyclin D1 expression. Aβ42 caused a significant increase in the activity of luciferase fused to the cyclin D1 3′ UTR but not its miR-34a-resistant mutant, and miR-34a overexpression blocked this increase (Fig. 3C). Importantly, overexpression of miR-34a significantly reversed the Aβ42-mediated increase in cyclin D1 protein (Fig. 3D, lane 3 versus lane 2). Collectively, these data suggested that Aβ42-mediated downregulation of miR-34a results in aberrant cyclin D1 expression.
Deregulation of miR-34a caused by Aβ42 results in CRNA.
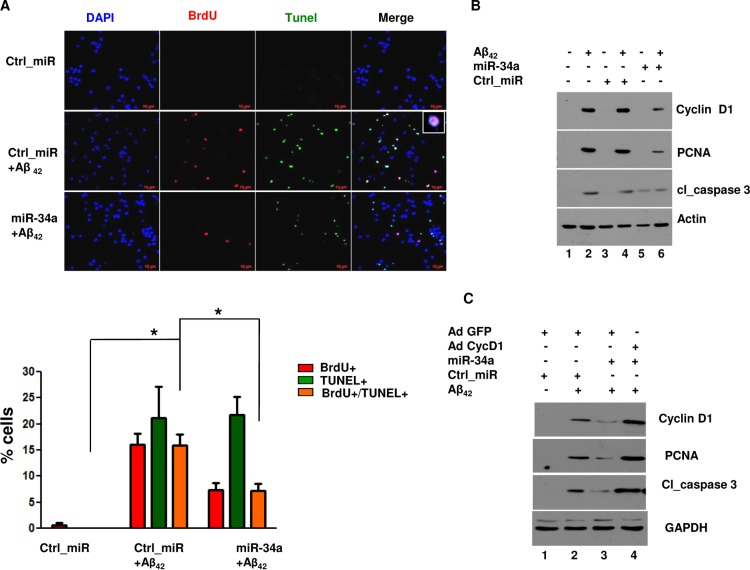
Next, we tested if downregulation of miR-34a has a bearing on neuronal cell cycle and death. While Aβ42 treatment led to an increase in BrdU+ TUNEL+ cells, miR-34a overexpression caused a significant decrease in population of these cells (Fig. 4A). These data suggested that miR-34a may protect neurons from apoptosis occurring as a result of cell cycle reentry. Furthermore, miR-34a overexpression also prevented the Aβ42-mediated increase in both PCNA and cleaved caspase 3 (Fig. 4B, lane 6 versus lane 4; see also Fig. S3A in the supplemental material) in a dose-dependent manner (see Fig. S1A in the supplemental material), which supported results from BrdU and TUNEL labeling experiments (Fig. 4A). In contrast, when miR-34a was overexpressed in the absence of Aβ42 treatment, cleaved caspase 3 and not PCNA was elevated (Fig. 4B, lane 5; see also Fig. S1A and S3A). Consistent with this, as described above, miR-34a overexpression caused an increase mainly in TUNEL+ cells (Fig. 2A). These observations highlighted the necessity of miR-34a expression to be optimal in neurons to protect them from apoptosis; its reduced levels cause apoptosis by promoting cell cycle reentry, whereas its overexpression promotes “typical” apoptosis which is largely independent of the cell cycle (see Discussion).
FIG 4.
miR-34a prevents Aβ42-induced CRNA in cortical neurons.(A) Rat cortical neurons were transfected with miR-34a or Ctrl_miR and treated with Aβ42, followed by incubation with BrdU. Immunofluorescence assay and TUNEL assay were performed as described for Fig. 2A. (Lower panel) The percentage of cells that exhibited BrdU and/or TUNEL staining was determined. Data are means ± SEM from 3 independent experiments. (*, P < 0.05, ANOVA; n = 3). Scale bars, 10 μm. (B) Rat cortical neurons were transfected with miR-34a or Ctrl_miR and treated with Aβ42 for 48 h. Cell lysates were prepared, followed by Western blotting with the indicated antibodies. (C) Rat cortical neurons were transfected with miR-34a or Ctrl_miR and infected with recombinant adenovirus to express cyclin D1 (Ad-cyc D1) or GFP alone (Ad-GFP), followed by treatment with Aβ42 for 48 h. Western blotting was performed using antibodies against cyclin D1, PCNA, and cleaved caspase 3. GAPDH was used as a loading control. The change in expression of cyclin D1, PCNA, and cl_caspase 3 was quantified by densitometry (see Fig. S3B in the supplemental material).
We further tested if the observed neuroprotective effects of miR-34a were a result of its ability to target cyclin D1. Aβ42 caused an expected increase in PCNA and caspase 3 levels in the presence of a control GFP adenovirus, which was prevented upon overexpression of miR-34a (Fig. 4C, lane 3 versus lane 2; see also Fig. S3B in the supplemental material). When cyclin D1 adenovirus was used, miR-34a failed to prevent CRNA caused by Aβ42 treatment (Fig. 4C, lane 4 versus lane 3; see also Fig. S3B), which confirmed that the observed reversal of CRNA may be a result of miR-34a-mediated downregulation of cyclin D1. Collectively, these data suggested that downregulation of miR-34a by Aβ42 may lead to enhanced expression of cyclin D1, which triggers cell cycle reentry, resulting in neuronal apoptosis.
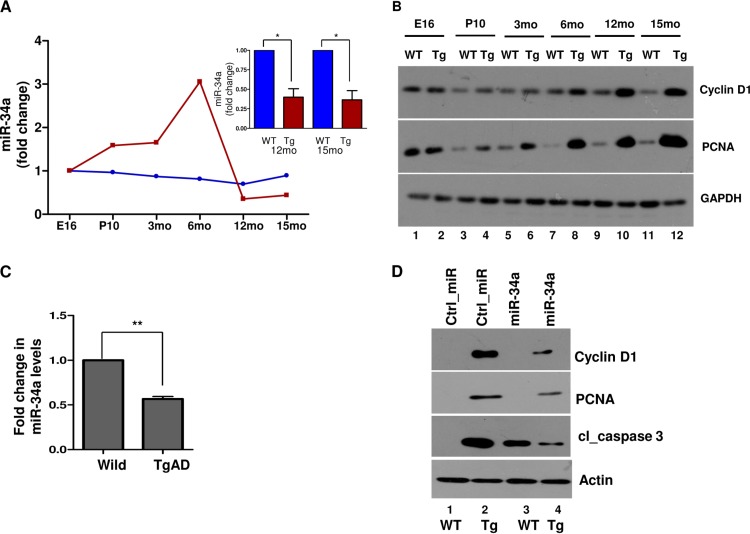
miR-34a is deregulated in APP/PS1 Tg mice, which may result in cell cycle reentry and neuronal apoptosis.
Cell cycle reentry of neurons has been observed in various AD mouse models and postmortem brains of MCI and AD patients (11, 33). Therefore, we investigated the regulation of miR-34a in a transgenic mouse model for AD. Since APP and PS1 mutants associated with AD are expressed in these animals (APP/PS1 Tg), it results in enhanced Aβ42 production (29) and causes aberrant cell cycle reentry in aging or adult animals (13, 33). First, the expression of miR-34a in mouse cortex was determined at various stages of development. The levels of miR-34a in wild-type (WT) mouse cortex altered only modestly during development. In contrast, miR-34a expression increased significantly until 6 months, followed by a decrease, which was significantly lower than in the WT animals aged more than 12 months (Fig. 5A). The levels of expression of cyclin D1 and PCNA were also compared during development: while cyclin D1 and PCNA were detected at E16, there was almost no difference in the expression of cell cycle proteins at this stage in the cortices of WT and APP/PS1 Tg mice (Fig. 5B). However, an increase was observed in the expression of these proteins at postnatal day 10 (P10), which further enhanced during development of the adult Tg animal. It was interesting to note that the maximum increase in the PCNA in Tg cortex was observed in the adult brain (>6 months), which correlates well with reduced miR-34a expression in the adult animal.
FIG 5.
miR-34a expression is altered in the cortices of APP/PS1 Tg mice and cortical neurons, which may lead to aberrant cyclin D1 expression. (A and B) Cerebral cortices were isolated from wild-type (WT) or APP/PS1 transgenic (Tg) mice, and the expression of miR-34a (A) or the indicated proteins (B) was evaluated at the indicated developmental stage by qRT-PCR (A) and Western blotting (B). Fold change in miR-34a expression in WT and APP/PS1 Tg mice at the indicated developmental stage in comparison to E16 is indicated. The inset in panel A shows the fold difference in miR-34a expression in 12- and 15-month-old APP/PS1 Tg mice with respect to WT mice (*, P < 0.05, ANOVA; n = 2). (C) Cortical neurons were cultured from WT and Tg mice for 7 DIV. RNA was isolated and miR-34a expression was quantified by qRT-PCR. Fold change in miR-34a levels with respect to that in WT mice is illustrated. Data are expressed as means ± SEM (**, P < 0.01, t test; n = 4). (D) Cortical neurons were cultured from WT or Tg mice and transfected with miR-34a or Ctrl_miR. After 48 h, cell lysate was prepared and Western blotting was performed for cyclin D1, PCNA, and cleaved caspase 3. A significant increase in cyclin D1, PCNA, and cleaved caspase 3 in Tg neurons was observed, which was significantly reversed upon miR-34a overexpression.
Since miR-34a is likely to be expressed in nonneuronal brain cells, in order to work specifically with neuronal populations, cortical neurons from the AD mouse model were used to assess miR-34a expression. qRT-PCR data suggested that miR-34a expression was significantly lower in APP/PS1 Tg neurons (Fig. 5C). In addition, cyclin D1 expression was also higher in APP/PS1 Tg neurons, as was the case with PCNA and cleaved caspase 3, suggesting that cortical neurons of APP/PS1 Tg mice have a significantly higher propensity to undergo CRNA (Fig. 5D, lane 2). Upon transfection of miR-34a in APP/PS1 Tg neurons, a significant reduction in cyclin D1 was observed, which was accompanied by a decrease in PCNA and cleaved caspase 3 and was indicative of reversal of CRNA (Fig. 5D). It is important to note that overexpression of miR-34a caused elevation in cleaved caspase 3 in WT neurons but PCNA was not detectable in this case (Fig. 5D, lane 3), which was consistent with results of the TUNEL labeling experiment whose results are shown in Fig. 2A. These data indicate that miR-34a expression may be critical for preventing aberrant cell cycle reentry of neurons of APP/PS1 Tg mice and may help explain cell cycle reentry and neuronal apoptosis in the brains of AD patients.
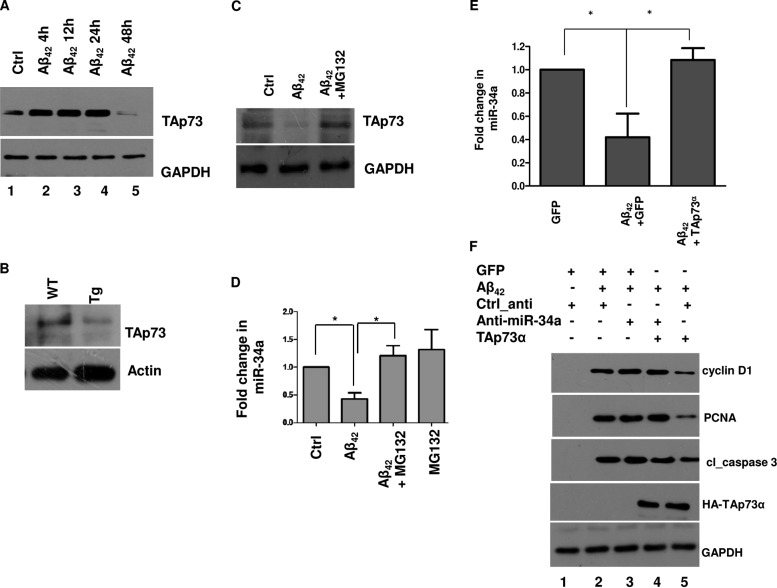
The loss of TAp73 deregulates miR-34a expression in neurons.
p53 family member p73 has been implicated in neuronal differentiation and death (34). TAp73 isoforms interact with a p53 binding site on the miR-34a promoter and regulate its expression (25). We specifically evaluated the role of TAp73 in miR-34a biogenesis and CRNA by overexpressing TAp73α. miR-34a promoter activity, which is inhibited in response to Aβ42, was restored upon TAp73α overexpression (see Fig. S6 in the supplemental material). Next, the effect of Aβ42 on TAp73 was investigated. A time course experiment revealed that a slight increase in TAp73 protein levels was followed by a decrease after 24 to 48 h of Aβ42 treatment (Fig. 6A). Consistent with this, a similar decrease in TAp73 was observed in neurons of APP/PS1 Tg AD mice (Fig. 6B). In addition, TAp73 was significantly lower in >12-month-old APP/PS1 Tg mice than in age-matched WT mice (see Fig. S5B in the supplemental material). The decrease in TAp73 expression correlates well with the loss of miR-34a in Aβ42-treated neurons or neurons from APP/PS1 Tg mice (Fig. 3A and 5C). Since almost no change in the TAp73 transcript was observed (data not shown), its regulation via posttranslational mechanisms was investigated. The loss of TAp73 upon Aβ42 treatment was significantly reverted by proteasomal inhibitor MG132, indicating that Aβ42 promotes proteasomal degradation of TAp73 (Fig. 6C). A similar recovery of miR-34a expression was observed when neurons were treated with MG132 (Fig. 6D). To further ascertain if the loss of miR-34a was a result of TAp73 degradation, TAp73α was overexpressed, followed by Aβ42 treatment. The downregulation of miR-34a promoter activity caused by Aβ42 (see Fig. S6 in the supplemental material) and its expression (Fig. 6E) were significantly prevented by TAp73α overexpression. Importantly, the Aβ42-mediated increase of cyclin D1, which is targeted by miR-34a (Fig. 3D and 4C), was prevented by TAp73α expression (Fig. 6F, lane 2 versus lane 5; see also Fig. S3C in the supplemental material), and when anti-miR-34a was coexpressed with TAp73α, these effects were reversed (Fig. 6F, lane 4 versus lane 5; see also Fig. S3C). Moreover, the increases in the levels of PCNA and cleaved caspase 3 were also reversed by TAp73α overexpression in an miR-34a-dependent manner (Fig. 6F; see also Fig. S3C). Collectively, these results strongly indicate that TAp73 may be a key player in the prevention of CRNA, as it regulates miR-34a expression. TAp73 degradation triggered by Aβ42 attenuates miR-34a expression, which leads to cell cycle reentry and apoptosis.
FIG 6.
Aβ42-mediated TAp73 degradation causes deregulation of miR-34a, resulting in CRNA. (A) Rat cortical neurons were treated with Aβ42 for the indicated time. Western blotting was performed with an anti-TAp73 antibody, and GAPDH was used as a loading control. (B) Cortical neurons for WT or APP/PS1 Tg mice were cultured. At DIV 7, cell lysate was prepared and Western blotting was performed using anti-TAp73. Actin was used as a loading control. (C and D) Rat cortical neurons were treated with Aβ42 for 48 h in the presence or absence of 10 μM MG132, which was included for 12 h. Subsequently, Western blotting (C) or qRT-PCR (D) was performed for assessing the expression of TAp73 (C) or miR-34a (D), respectively (*, P < 0.05, ANOVA; n = 3). (E) Rat cortical neurons were infected with adenovirus for GFP or TAp73α-HA and treated with Aβ42 for 48 h. RNA was isolated and qRT-PCR was performed to determine miR-34a levels. Results are expressed as means ± SEM of three independent experiments (*, P < 0.05, ANOVA; n = 3). (F) Rat cortical neurons were transfected with anti-miR-34a or Ctrl_anti and infected with TAp73α or GFP adenovirus. Subsequently, neurons were treated with Aβ42 for 48 h, and Western blotting was performed by using antibodies against the indicated proteins. The change in expression of cyclin D1, PCNA, and cl-caspase3 was quantified by densitometry (see Fig. S3C in the supplemental material).
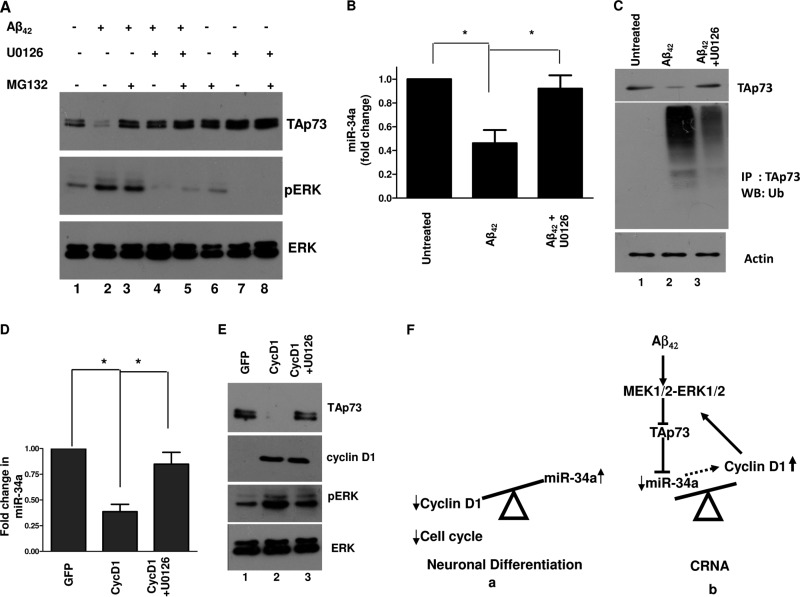
TAp73 and miR-34a are negatively regulated by the MEK-ERK pathway.
Given the observed deregulation of TAp73 and miR-34a by Aβ42, efforts were made to decipher the molecular events which may regulate this process. In this context, we investigated the role of the MEK-ERK pathway, as aberrant activation of this pathway by Aβ42 causes cyclin D1 production, which leads to CRNA (12). Prolonged Aβ42 treatment caused hyperactivation of MEK-ERK, as suggested by elevated phospho-ERK levels (Fig. 7A, lane 2) (12), which was accompanied by a marked reduction in TAp73 expression (Fig. 7A, lane 2). Strikingly, the MEK inhibitor U0126 prevented the loss of TAp73 (Fig. 7A, lane 4 versus lane 2). Since proteasomal degradation can be either ubquitination dependent or independent, we investigated if TAp73 is ubiquitinated in response to Aβ42. For this purpose, TAp73 was immunoprecipitated from cortical neurons, followed by Western blotting with ubiquitin antibody. A marked increase in ubiquitination of TAp73 was observed following Aβ42 treatment, which was significantly prevented upon addition of U0126 (Fig. 7C). These results correlated well with the observed decrease in TAp73 upon Aβ42 treatment in a MEK-ERK-dependent manner (Fig. 7A). Consistent with this, U0126 significantly restored miR-34a expression (Fig. 7B). These observations suggested that aberrant and sustained MEK-ERK pathway activation promotes TAp73 degradation, resulting in attenuated miR-34a expression.
FIG 7.
Cyclin D1 downregulates TAp73 and miR-34a via MEK-ERK signaling during CRNA in cortical neurons. (A) Cortical neurons were treated with Aβ42 peptide for 48 h with dimethyl sulfoxide (DMSO) or 10 μM U0126 in the presence or absence of 10 μM MG132, followed by Western blotting with the indicated antibodies. (B) Cortical neurons were treated with Aβ42 peptide for 48 h with DMSO or 10 μM U0126, followed by qRT-PCR for miR-34a. Aβ42 treatment led to a significant decrease in miR-34a levels, which was restored by U0126. Data represent means ± SEM of four independent experiments (*, P < 0.05, ANOVA; n = 4). (C) Rat cortical neurons were treated with Aβ42 in the presence or absence of U0126. After 48 h, protein lysates were prepared and immunoprecipitation (IP) was performed using anti-TAp73 antibody, followed by Western blotting (WB) using antiubiquitin antibody. Western blotting was also performed on total cell lysate with the indicated antibodies. (D and E) Rat cortical neurons were infected with cyclin D1 or GFP adenovirus, followed by U0126 treatment. Western blotting was performed with the indicated antibodies (E), and qRT-PCR was performed to measure miR-34a (D). Data are means ± SEM (*, P < 0.05 by ANOVA; n = 3). (F) Molecular mechanism via which miR-34a may regulate neuronal cell cycle suppression and cell cycle reentry. Present studies demonstrate that TAp73 drives the expression of miR-34a, which is necessary for neuronal differentiation and keeps the neuronal cell cycle suppressed (a), by preventing cyclin D1 expression. Neurotoxic insults like Aβ42 cause aberrant activation of the MEK-ERK pathway and result in TAp73 degradation and miR-34a repression. As a result, cyclin D1 expression is elevated, which feeds back positively into this pathway, contributing to CRNA (b).
TAp73 degradation may contribute to the cyclin D1-mediated feedback loop, which is critical for sustained MEK-ERK activation and CRNA.
Previous studies had indicated that cyclin D1 expression, which is triggered by MEK-ERK activation in response to Aβ42, engages in a positive feedback loop resulting in sustained and aberrant activation of the MEK-ERK pathway (12). Therefore, it was worth exploring if TAp73 and miR-34a participated in the feedback loop axis, which may be needed for the feedback loop to be operational. To test this hypothesis, cyclin D1 was overexpressed in neurons, which resulted in further enhancement of phospho-ERK levels, as observed previously (Fig. 7E, lane 2) (12). Importantly, a concomitant decrease in TAp73 was observed, which was prevented by U0126 (Fig. 7E, lane 2 versus lane 3). These data indicated that TAp73 is degraded as a result of cyclin D1-mediated aberrant MEK-ERK activation. Furthermore, cyclin D1 overexpression caused a significant decrease in miR-34a levels, which was prevented by U0126 and correlated well with its observed regulation by TAp73 (Fig. 7D).
These observations provide strong evidence for the role of the cyclin D1-mediated feedback loop in degradation of TAp73 and the loss of miR-34a, which causes sustained activation of the MEK-ERK pathway, promotes reentry of neurons into the cell cycle, and may lead to their death (Fig. 7F).
DISCUSSION
We demonstrate that miR-34a suppresses the neuronal cell cycle via its ability to target cyclin D1. Previously, miR-34a was shown to target cyclin D1 in A459 cells, but its role in the neuronal cell cycle was not understood (35). It is indeed possible that miR-34a targets multiple cell cycle genes in neurons, but since cyclin D1 production is one of the first steps in cell cycle reentry, its downregulation by miR-34a is crucial for prevention of CRNA. As reported previously (26), we found that the differentiation of rat cortical neurons and SH-SY5Y cells is accompanied by an increase in miR-34a expression. We found that anti-miR-34a prevents neuronal differentiation, as evidenced by an increase in S-phase cells and elevated cyclin D1 expression. Moreover, when miR-34a was knocked down in differentiated cortical neurons, a significant increase in cell cycle reentry and apoptosis was observed. Interestingly, overexpression of miR-34a (>100 nM) also caused apoptosis, as noted previously (36, 37), but it did not trigger the cell cycle. In this case, apoptosis was probably mediated by the conventional pathway, possibly by preventing the expression of apoptosis/survival-related proteins like Bcl2, which is an established target of miR-34a (36). Consistent with these studies, we detected a significant decrease in Bcl2 upon miR-34a overexpression (Modi and Sharma, unpublished). These data highlighted the necessity of tight regulation of miR-34a in neurons.
TAp73 was recently demonstrated to regulate miR-34a expression during neuronal differentiation (26), and the knockdown of miR-34a caused defects in neurite outgrowth (26). However, direct effects of miR-34a on the neuronal cell cycle were not analyzed previously. While miR-34a may regulate multiple processes in neurons via diverse targets, it is reasonable to state on the basis of present studies that the suppression of the cell cycle by deregulating cyclin D1 may be one of its major functions. Collectively, these findings suggest that miR-34a may regulate multiple processes in neurons via diverse targets. Present studies suggest that the suppression of the cell cycle by targeting cyclin D1 may be one of its key functions.
Soluble oligomers of Aβ42, which is generated as a result of aberrant cleavage of amyloid precursor protein (APP) (32), cause cell cycle reentry and neuronal apoptosis (12, 13). Since the cell cycle machinery undergoes reprogramming in response to Aβ42, we hypothesized that those molecular events which lead to neuronal differentiation may be altered when neurons encounter neurotoxic insults and may cause unscheduled cell cycle reentry and apoptosis. For instance, the expression of cyclin D1, which was suppressed during neuronal differentiation (Fig. 1D), was upregulated in response to Aβ42 (Fig. 4B) (12) and was significantly higher in 6- to 12-month-old transgenic AD mice (13). Aβ42-induced aberrant activation of the MEK-ERK pathway results in CRNA (12), which is caused by a dramatic increase in cyclin D1.
While underlying mechanisms may not be clear, p73 has also been implicated in neuronal apoptosis (38) and p73+/− and p73−/− mice exhibit severe neuronal loss as a result of apoptosis, which increases significantly with aging, and also exhibit AD-related pathological manifestations (39, 40). In the present studies, overexpression of TAp73α reversed the loss of miR-34a and the increase in cyclin D1 in cortical neurons. As a result, cell cycle reentry and neuronal apoptosis were significantly ablated. Therefore, it is reasonable to suggest that deregulation of miR-34a as a result of TAp73 degradation may make a significant contribution to this process.
TAp73 and miR-34a deregulation was observed in APP/PS1 Tg AD mice. While a previous study reported enhanced miR-34a expression in the cortices of 3- and 6-month-old APP/PS1 Tg mice (37), miR-34a levels at later stages were not measured in this work. Our studies indicate that TAp73 and miR-34a are downregulated after prolonged Aβ42 treatment of cortical neurons (48 h) and in the cortices of >12-month-old APP/PS1 Tg mice (Fig. 3 and 5). Corroboratively, cell cycle reentry and apoptosis were also observed during this period. To explore if the initial increase (∼12 h) in miR-34a upon Aβ42 treatment (Fig. 3A) has any bearing on apoptosis and/or the cell cycle, anti-miR-34a was used for this duration, but no significant changes were found in either of these processes (see Fig. S4 in the supplemental material).
Previous studies have indicated that cell cycle reentry in the brains of both AD patients and animal models is a slow process and in some instances leads to cell death (9, 11, 14, 33). Consistent with this, the expression of PCNA and cyclin D1 was maximal in the adult APP/PS1 Tg mice (Fig. 5B) and miR34a expression was the lowest during this period (Fig. 5A). Given the above-mentioned observations, it is possible that miR-34a and TAp73 deregulation, which appears to be a delayed response, contributes to the slow progress of CRNA.
Our studies highlight the importance of suppression of cell cycle for the survival of terminally differentiated neurons and reveal a novel molecular mechanism via which it is achieved. miR-34a and its regulator TAp73 emerge as key players in tight regulation of the neuronal cell cycle via their ability to suppress cyclin D1. This balance is significantly altered when neurons encounter insults like Aβ42, which aberrantly activates MEK-ERK signaling, leading to deregulation of TAp73/miR-34a and promoting cyclin D1 expression, which drives cell cycle reentry and apoptosis (Fig. 7F).
Supplementary Material
ACKNOWLEDGMENTS
P.K.M. acknowledges support through a DBT-RA fellowship from the Department of Biotechnology, India. S.J. acknowledges the Junior Research Fellowship from CSIR, India. This work was supported by funding from grants by Department of Biotechnology, India, and National Institute of Immunology core.
We acknowledge discussions with Sanjeev Das and gifts of some reagents from him.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00589-15.
REFERENCES
- 1.Giovanni A, Wirtz-Brugger F, Keramaris E, Slack R, Park DS. 1999. Involvement of cell cycle elements, cyclin-dependent kinases, pRb, and E2F x DP, in B-amyloid-induced neuronal death. J Biol Chem 274:19011–19016. doi: 10.1074/jbc.274.27.19011. [DOI] [PubMed] [Google Scholar]
- 2.Herrup K, Yang Y. 2007. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci 8:368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- 3.Ino H, Chiba T. 2001. Cyclin-dependent kinase 4 and cyclin D1 are required for excitotoxin-induced neuronal cell death in vivo. J Neurosci 21:6086–6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park DS, Morris EJ, Stefanis L, Troy CM, Shelanski ML, Geller HM, Greene LA. 1998. Multiple pathways of neuronal death induced by DNA-damaging agents, NGF deprivation, and oxidative stress. J Neurosci 18:830–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartek J, Bartkova J, Lukas J. 1996. The retinoblastoma protein pathway and the restriction point. Curr Opin Cell Biol 8:805–814. doi: 10.1016/S0955-0674(96)80081-0. [DOI] [PubMed] [Google Scholar]
- 6.Lee EY, Hu N, Yuan SS, Cox LA, Bradley A, Lee WH, Herrup K. 1994. Dual roles of the retinoblastoma protein in cell cycle regulation and neuron differentiation. Genes Dev 8:2008–2021. doi: 10.1101/gad.8.17.2008. [DOI] [PubMed] [Google Scholar]
- 7.Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T. 1998. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell 2:293–304. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y, Herrup K. 2007. Cell division in the CNS: protective response or lethal event in post-mitotic neurons? Biochim Biophys Acta 1772:457–466. doi: 10.1016/j.bbadis.2006.10.002/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webber KM, Raina AK, Marlatt MW, Zhu X, Prat MI, Morelli L, Casadesus G, Perry G, Smith MA. 2005. The cell cycle in Alzheimer disease: a unique target for neuropharmacology. Mech Ageing Dev 126:1019–1025. [DOI] [PubMed] [Google Scholar]
- 10.Busser J, Geldmacher DS, Herrup K. 1998. Ectopic cell cycle proteins predict the sites of neuronal cell death in Alzheimer's disease brain. J Neurosci 18:2801–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Mufson EJ, Herrup K. 2003. Neuronal cell death is preceded by cell cycle events at all stages of Alzheimer's disease. J Neurosci 23:2557–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modi PK, Komaravelli N, Singh N, Sharma P. 2012. Interplay between MEK-ERK signaling, cyclin D1, and cyclin-dependent kinase 5 regulates cell cycle reentry and apoptosis of neurons. Mol Biol Cell 23:3722–3730. doi: 10.1091/mbc.E12-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varvel NH, Bhaskar K, Patil AR, Pimplikar SW, Herrup K, Lamb BT. 2008. Abeta oligomers induce neuronal cell cycle events in Alzheimer's disease. J Neurosci 28:10786–10793. doi: 10.1523/JNEUROSCI.2441-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Geldmacher DS, Herrup K. 2001. DNA replication precedes neuronal cell death in Alzheimer's disease. J Neurosci 21:2661–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giovanni A, Keramaris E, Morris EJ, Hou ST, O'Hare M, Dyson N, Robertson GS, Slack RS, Park DS. 2000. E2F1 mediates death of B-amyloid-treated cortical neurons in a manner independent of p53 and dependent on Bax and caspase 3. J Biol Chem 275:11553–11560. doi: 10.1074/jbc.275.16.11553. [DOI] [PubMed] [Google Scholar]
- 16.Kranenburg O, van der Eb AJ, Zantema A. 1996. Cyclin D1 is an essential mediator of apoptotic neuronal cell death. EMBO J 15:46–54. [PMC free article] [PubMed] [Google Scholar]
- 17.Park DS, Obeidat A, Giovanni A, Greene LA. 2000. Cell cycle regulators in neuronal death evoked by excitotoxic stress: implications for neurodegeneration and its treatment. Neurobiol Aging 21:771–781. doi: 10.1016/S0197-4580(00)00220-7. [DOI] [PubMed] [Google Scholar]
- 18.Malik B, Currais A, Andres A, Towlson C, Pitsi D, Nunes A, Niblock M, Cooper J, Hortobagyi T, Soriano S. 2008. Loss of neuronal cell cycle control as a mechanism of neurodegeneration in the presenilin-1 Alzheimer's disease brain. Cell Cycle 7:637–646. doi: 10.4161/cc.7.5.5427. [DOI] [PubMed] [Google Scholar]
- 19.Absalon S, Kochanek DM, Raghavan V, Krichevsky AM. 2013. MiR-26b, upregulated in Alzheimer's disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J Neurosci 33:14645–14659. doi: 10.1523/JNEUROSCI.1327-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiore R, Khudayberdiev S, Saba R, Schratt G. 2011. MicroRNA function in the nervous system. Prog Mol Biol Transl Sci 102:47–100. [DOI] [PubMed] [Google Scholar]
- 21.Lau P, De Strooper B. 2010. Dysregulated microRNAs in neurodegenerative disorders. Semin Cell Dev Biol 21:768–773. [DOI] [PubMed] [Google Scholar]
- 22.Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. 2008. MicroRNA expression in the adult mouse central nervous system. RNA 14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. 2007. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 25.Agostini M, Tucci P, Killick R, Candi E, Sayan BS, Rivetti di Val Cervo P, Nicotera P, McKeon F, Knight RA, Mak TW, Melino G. 2011. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc Natl Acad Sci U S A 108:21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, Forsythe ID, Young KW, Ventura A, Concepcion CP, Han YC, Candi E, Knight RA, Mak TW, Melino G. 2011. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci U S A 108:21099–21104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, Iovanna JL, Squire J, Jurisica I, Kaplan D, Melino G, Jurisicova A, Mak TW. 2008. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev 22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R, Itie-Youten A, Wakeham A, Arsenian-Henriksson M, Melino G, Kaplan DR, Miller FD, Mak TW. 2010. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev 24:549–560. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA, Copeland NG, Lee MK, Younkin LH, Wagner SL, Younkin SG, Borchelt DR. 2004. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet 13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- 30.Jankowsky JL, Slunt HH, Ratovitski T, Jenkins NA, Copeland NG, Borchelt DR. 2001. Co-expression of multiple transgenes in mouse CNS: a comparison of strategies. Biomol Eng 17:157–165. doi: 10.1016/S1389-0344(01)00067-3. [DOI] [PubMed] [Google Scholar]
- 31.Stine WB Jr, Dahlgren KN, Krafft GA, LaDu MJ. 2003. In vitro characterization of conditions for amyloid-beta peptide oligomerization and fibrillogenesis. J Biol Chem 278:11612–11622. doi: 10.1074/jbc.M210207200. [DOI] [PubMed] [Google Scholar]
- 32.Selkoe DJ. 1998. The cell biology of beta-amyloid precursor protein and presenilin in Alzheimer's disease. Trends Cell Biol 8:447–453. doi: 10.1016/S0962-8924(98)01363-4. [DOI] [PubMed] [Google Scholar]
- 33.Li L, Cheung T, Chen J, Herrup K. 2011. A comparative study of five mouse models of Alzheimer's disease: cell cycle events reveal new insights into neurons at risk for death. Int J Alzheimers Dis 2011:171464. doi: 10.4061/2011/171464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller FD, Pozniak CD, Walsh GS. 2000. Neuronal life and death: an essential role for the p53 family. Cell Death Differ 7:880–888. [DOI] [PubMed] [Google Scholar]
- 35.Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. 2008. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett 582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 36.Truettner JS, Motti D, Dietrich WD. 2013. MicroRNA overexpression increases cortical neuronal vulnerability to injury. Brain Res 1533:122–130. doi: 10.1016/j.brainres.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Liu P, Zhu H, Xu Y, Ma C, Dai X, Huang L, Liu Y, Zhang L, Qin C. 2009. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer's disease, inhibits bcl2 translation. Brain Res Bull 80:268–273. doi: 10.1016/j.brainresbull.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Ramadan S, Terrinoni A, Catani MV, Sayan AE, Knight RA, Mueller M, Krammer PH, Melino G, Candi E. 2005. p73 induces apoptosis by different mechanisms. Biochem Biophys Res Commun 331:713–717. doi: 10.1016/j.bbrc.2005.03.156. [DOI] [PubMed] [Google Scholar]
- 39.Pozniak CD, Barnabe-Heider F, Rymar VV, Lee AF, Sadikot AF, Miller FD. 2002. p73 is required for survival and maintenance of CNS neurons. J Neurosci 22:9800–9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wetzel MK, Naska S, Laliberte CL, Rymar VV, Fujitani M, Biernaskie JA, Cole CJ, Lerch JP, Spring S, Wang SH, Frankland PW, Henkelman RM, Josselyn SA, Sadikot AF, Miller FD, Kaplan DR. 2008. p73 regulates neurodegeneration and phospho-tau accumulation during aging and Alzheimer's disease. Neuron 59:708–721. doi: 10.1016/j.neuron.2008.07.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.