Abstract
Estrogens are effective in the treatment of prostate cancer; however, the effects of estrogens on prostate cancer are enigmatic. In this study, we demonstrated that estrogen (17β-estradiol [E2]) has biphasic effects on prostate tumor growth. A lower dose of E2 increased tumor growth in mouse xenograft models using DU145 and PC-3 human prostate cancer cells, whereas a higher dose significantly decreased tumor growth. We found that anchorage-independent apoptosis in these cells was inhibited by E2 treatment. Similarly, in vivo angiogenesis was suppressed by E2. Interestingly, these effects of E2 were abolished by knockdown of either estrogen receptor β (ERβ) or Krüppel-like zinc finger transcription factor 5 (KLF5). Ιn addition, E2 suppressed KLF5-mediated transcription through ERβ, which inhibits proapoptotic FOXO1 and proangiogenic PDGFA expression. Furthermore, we revealed that a nonagonistic ER ligand GS-1405 inhibited FOXO1 and PDGFA expression through the ERβ-KLF5 pathway and regulated prostate tumor growth without ERβ transactivation. Therefore, these results suggest that E2 biphasically modulates prostate tumor formation by regulating KLF5-dependent transcription through ERβ and provide a new strategy for designing ER modulators, which will be able to regulate prostate cancer progression with minimal adverse effects due to ER transactivation.
INTRODUCTION
Prostate cancer is the most frequently diagnosed cancer and the second leading cause of cancer death in the United States and other industrialized countries (1). Prostate cancer progression is initially driven by androgens through the androgen receptor (AR). Thus, androgen ablation therapy is the primary treatment approach for prostate cancer (2, 3). However, almost all patients eventually develop resistance to antiandrogen therapy, which is extremely hard to cure (4). Therefore, new molecular targets for devising novel therapies are required.
Estrogens are known to play a role in the development of the male reproductive system and prostate cancer (5, 6). The administration of estrogens has previously been extensively used in prostate cancer treatment. Early research demonstrated that estrogens exert an indirect antiandrogen action mediated through feedback inhibition of luteinizing hormone-releasing hormone and pituitary luteinizing hormone release, thereby decreasing testicular androgen levels and release (7). However, it is currently considered that estrogens modulate prostate cancer through nonandrogenic pathways (7, 8). In fact, estrogen (17β-estradiol [E2]) inhibits the development of androgen-insensitive prostate cancer xenografts in mice (9, 10). Moreover, clinical studies indicated that estrogenic therapies are useful for advanced and androgen-insensitive prostate cancer (11, 12). Despite these beneficial effects, E2 has also been revealed to be a risk factor of prostate carcinogenesis. For example, several animal studies suggested that E2 could enhance prostate cancer growth (13, 14). In addition, a recent clinicopathological study indicated that circulating E2 levels were significantly elevated in patients with prostate cancer compared with those in healthy age-matched patients (15). Thus, the molecular mechanisms underlying the contradictory effects of E2 on prostate cancer development are not well understood.
E2 acts as a physiological ligand for two nuclear receptor isoforms, estrogen receptor α (ERα) and ERβ (16, 17). Synthetic compounds also regulate gene expression in prostate cancer cells through ERβ, which is the predominant ER subtype in those cells (18–20). Being dependent on agonistic ligands such as E2, ER directly binds to estrogen response elements (EREs) within genomic DNA to induce gene expression (classical pathway) (21). However, recent studies revealed that ERs can also regulate gene expression by interacting with other DNA-binding transcription factors, such as c-Fos/c-Jun, Sp1, and NF-κB, but not by binding directly to DNA (nonclassical pathway) (22, 23). Recent reports suggested that ER ligands regulate gene expression through ERβ-dependent nonclassical pathways in prostate tissues and cancer cells (23–25). We previously reported that prostate tumor growth is regulated through the ERβ-dependent nonclassical pathway with Krüppel-like zinc finger transcription factor 5 (KLF5) (25). KLF5 (also known as BTEB2 or IKLF) is a transcription factor that possesses both tumor-suppressing and tumor-promoting activities (26–28). Analysis of the associated pathway revealed that in the absence of E2, ERβ induces the KLF5-mediated expression of FOXO1 and increases anoikis, thereby suppressing prostate tumor growth in mouse xenograft models. Conversely, E2 suppresses KLF5 transactivation through ERβ, which enhances tumor growth. However, it is unclear whether E2 regulates prostate cancer progression through ERβ and KLF5 and, if so, by which mechanism.
In this study, we demonstrated the mechanism underlying the modulation of prostate tumor formation by E2. We revealed that E2 biphasically modulates prostate tumor growth in mouse xenograft models. Our results using the nonagonistic ER ligand GS-1405 further indicated that the effects of E2 are exerted via the comprehensive regulation of FOXO1-mediated anoikis and PDGFA-mediated angiogenesis through the ERβ-KLF5 pathway. These findings may lead to the development of new therapeutic strategies for designing next-generation ER modulators.
MATERIALS AND METHODS
Cell culture and ligand treatment.
Human prostate cancer DU145 and PC-3 and human embryonic kidney HEK293 cells were obtained from the Cell Resource Center for Biomedical Research, Institute of Development, Aging and Cancer (Tohoku University, Sendai, Japan). Human prostate cancer LNCaP cells were obtained from American Type Culture Collection. DU145, PC-3, and LNCaP cells were maintained in RPMI 1640 (Nacalai Tesque), and HEK293 cells were maintained in Dulbecco modified Eagle medium (DMEM; Sigma-Aldrich). All media were supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin (Nacalai Tesque). The medium was exchanged to phenol red-free medium containing 10% charcoal-stripped FBS, and cells were cultured for 48 h before treatment with ligands. 17β-Estradiol (E2), Fulvestrant (ICI 182,780 [ICI]), 4-hydroxytamoxifen (OH-Tam), and raloxifene (Ral) were purchased from Sigma-Aldrich. 4-(6-Methyl-1,3-benzothiazol-2-yl)phenol (GS-1405 [GS]; code LTBB000265) was purchased from Labtest.
Tumor xenograft models.
All animal experiments were performed in accordance with the guidelines for the care and use of laboratory animals at University of Tsukuba. Methods for keeping mice and tumor xenograft models have been described previously (25). Each 5- to 6-week-old BALB/cA-nu castrated male mouse was injected subcutaneously with 100 μl of cell suspension (6 × 106 to 8 × 106 cells) in both flanks. Mice were subcutaneously implanted with 17β-estradiol (E2) pellets (Innovative Research of America) at 0.18 mg (E2+) or 3.4 mg (E2++) with a 60-day release, generating serum E2 concentrations from 50 to 180 pg/ml or 550 to 1,900 pg/ml, which were measured using the estradiol enzyme immunoassay (EIA) kit (Cayman). GS was subcutaneously injected in the scruff of the neck. Tumor growth was monitored by measuring the tumor size using calipers; tumor volume (V) was determined using the formula V = 1/2 × larger diameter × (smaller diameter)2. Twenty-five to 35 days after implantation, tumors were excised, weighed, and fixed or stored in liquid nitrogen for later analysis.
Expression plasmids and antibodies.
The pCMV5-FLAG-ERβ (wild-type [WT]) plasmid has been previously described (25). To generate an expression plasmid for ERβ (E305A), site-directed mutagenesis of the ERβ sequence in pCMV5-FLAG-ERβ (WT) was performed by PCR using the primers (altered sequence elements are underlined) 5′-GTTGGCCGACAAGGCGTTGGTACACATG-3′ and 5′-CATGTGTACCAACGCCTTGTCGGCCAAC-3′. cDNAs encoding full-length PDGFA were amplified by PCR and subcloned into the pcDNA3 plasmid (Invitrogen) containing sequences encoding a 6× myc sequence. Mouse anti-platelet-derived growth factor alpha (anti-PDGFA) (E-10; Santa Cruz) and anti-β-actin (A5316; Sigma-Aldrich) monoclonal antibodies and rabbit anti-ERβ (CT; Millipore) and anti-CD31 (PECAM-1) (sc-1506; Santa Cruz) polyclonal antibodies were used according to the manufacturers' instructions. The rabbit polyclonal antibodies against KLF5 and ERβ were previously generated (25).
RNA interference.
Methods for stable RNA interference and small interfering RNA (siRNA) transfection followed those described by Nakajima et al. (25). To generate the short hairpin RNA (shRNA) retroviral supernatant, GP2-293 cells (Clontech) were cotransfected with the pVSV-G vector (Clontech) encoding envelope protein and pRETRO-SUPER (OligoEngine) vector containing the ERβ, KLF5, or luciferase (control) target sequence (25). DU145 or PC-3 cells were incubated with the retroviral supernatant in the presence of 8 μg/ml of Polybrene. The infected cells were selected with 1 μg/ml of puromycin.
qRT-PCR assay.
The quantitative real-time (qRT-PCR) assay was performed as described previously (25), with minor modifications. Cells were homogenized in 1 ml of Sepasol-RNA I Super G, and total RNA was extracted according to the manufacturer's instructions (Nacalai Tesque). cDNA was synthesized from total RNA using RevatraAce reverse transcriptase (Toyobo) and oligo(dT) primer. Real-time PCRs were performed to amplify fragments representing the indicated mRNAs using the Thermal Cycler Dice TP800 (TaKaRa) and SYBR Premix Ex Taq II (TaKaRa). mRNA levels were normalized to those of GAPDH. The primer sequences were as follows: FOXO1 forward primer, 5′-TCATGTCAACCTATGGCAG-3′; FOXO1 reverse primer, 5′-CATGGTGCTTACCGTGTG-3′; PDGFA forward primer, 5′-TCCACGCCACTAAGCATGTG-3′; PDGFA reverse primer, 5′-CGTAAATGACCGTCCTGGTCTT-3′; KLF5 forward primer, 5′-ATCGAGATGTTCGCTCGTGC-3′; KLF5 reverse primer, 5′-TTTAAAGGCAGACACTGAGTCAG-3′; GAPDH forward primer, 5′-ATCGTCCACCGCAAATGCTTCTA-3′; and GAPDH reverse primer, 5′-AGCCATGCCAATCTCATCTTGTT-3′.
TUNEL assay under detached conditions using poly-HEMA plates and using xenograft tissues.
One gram of poly-(2-hydroxyethyl methacrylate) (poly-HEMA) (Sigma-Aldrich) was dissolved in 25 ml of 99.5% ethanol and mixed overnight at 37°C (25). The poly-HEMA stock solution was added to each well of 12-well plates, and the plates were left to dry for a few hours. After drying, the plates were washed with phosphate-buffered saline (PBS). Cells were plated in the poly-HEMA-coated 12-well plates at a density of 60,000 (PC-3) or 200,000 (DU145)/well and incubated for 24 h. Apoptosis of the cells and xenograft tissues was analyzed by the Dead End fluorometric terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) system (Promega), and the kit was used according to the manufacturer's instructions.
Soft-agar colony formation assay.
The procedure for colony formation assay was performed as previously described (25). In total, 22,000 cells were suspended in DMEM containing 0.35% agar (Sigma-Aldrich) and layered on top of 1 ml of DMEM solidified with 0.6% agar in each well of a six-well plate. After growing at 37°C for 4 weeks, colonies with a diameter of >100 μm were observed and counted using Biozero (Keyence).
Immunohistochemical analysis.
Immunohistochemistry for KLF5 was performed as previously described (25), with the following modification for CD31 and PDGFA staining. Before incubation with anti-CD31 or anti-PDGFA antibodies, antigen retrieval was performed by microwave heating in EDTA buffer (1 mM; pH 8.0) or acid buffer (2 mM citric acid and 9 mM trisodium citrate dehydrate, pH 6.0), respectively. The antigen antibody was visualized using 3,3′-diaminobenzide.
Matrigel plug angiogenesis assay.
Matrigel angiogenesis experiments were performed for 7 days with 5- to 6-week-old castrated BALB/cA-nu mice under University of Tsukuba institutional approval. Mice were injected with 200 μl of ice-cold Matrigel (BD Biosciences) mixed with 3 × 106 cells with or without 250 ng/ml recombinant PDGFA (PeproTech). Seven days after the injection, Matrigel plugs were excised and the hemoglobin content in those plugs was determined using radioimmunoprecipitation assay (RIPA) buffer (29).
Immunoblotting.
Whole-cell lysates were extracted, and protein concentrations were quantified using bicinchoninic acid (BCA) protein assay reagent (Thermo Scientific). Cell extracts were fractionated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane using a transfer apparatus according to the manufacturer's instructions (Bio-Rad). Antibodies used were described above. Secondary antibodies were used at a concentration of 1:2,000.
Patients and tissues.
Tumor specimens were obtained from 102 patients who provided informed consent and underwent radical prostatectomy between 1987 and 2001 at Tokyo University Hospital. The mean patient age was 66.0 years (range, 52 to 75 years), the mean preoperative level of prostate-specific antigen was 16.7 ng/ml (3.2 to 136 ng/ml), and the mean follow-up period was 121 months (10 to 240 months). Thirty-seven patients were treated with surgery alone, whereas 65 patients received adjuvant antiandrogen therapy. This study was approved by the ethics committee at Graduate School of Medicine, University of Tokyo (permission number 2283).
Immunohistochemical assessment.
The immunoreactivities of KLF5 and PDGFA were evaluated in more than 1,000 carcinoma cells for each case, and subsequently, the percent immunoreactivity, i.e., labeling index, was determined. Cases with cytoplasmic staining of PDGFA in more than 10% carcinoma cells were considered high immunoreactivity in this study.
Luciferase reporter assay.
For luciferase assays, cells were cotransfected with phRG(R2.2)-Basic (Promega) and FX-luc or withERE-TATA-luc (25) with or without wild-type or mutated ERβ expression plasmids. Twenty-four hours after transfection, we replaced the culture medium with fresh medium containing ligands. Twenty-four hours after incubation with the ligands, luciferase assays were performed on cell extracts using a dual-luciferase reporter assay system (Promega), according to the manufacturer's instructions.
Structural modeling and description of the ERβ ligand-binding domain (LBD) in complex with GS.
The AutoDock Vina program (30) and AutoDock tools (31) were used for the modeling of the ligand-receptor complex. The protein structure of the hERβ LBD in complex with genistein was downloaded from the Protein Data Bank (PDB code 1QKM) (32). The exact conformation of hERβ LBD in complex with GS is unclear, in particular, the H12 configuration. Therefore, the hERβ LBD with a deletion of H12 was used for the docking simulation to avoid confusion. The model structure was described using UCSF Chimera software (33).
ChIP.
The chromatin immunoprecipitation (ChIP) assay was conducted as described previously (25). The purified DNA was analyzed to determine which DNA fragments were present in the precipitate by qRT-PCR, as described above. The primers for qRT-PCR were as follows: 5′-CCAGCCCGGCGCCCACTGGC-3′ and 5′-CAGCGGCTGCTGCGACTACC-3′ for the FOXO1 upstream region (25) and 5′-GCACTGGAGGGTGGGCAAGC-3′ and 5′-GACCCGCACCTCGGAAGCGC-3′ for the PDGFA upstream region.
Statistics.
Statistical significance was evaluated using one-way analysis of variance for multiple groups, followed by Tukey's post hoc test to evaluate differences. Cancer-specific survival rates were evaluated based on Kaplan-Meier methods, and statistical significance was determined using a log rank test.
RESULTS
E2 exerts biphasic effects on prostate tumors growth in vivo.
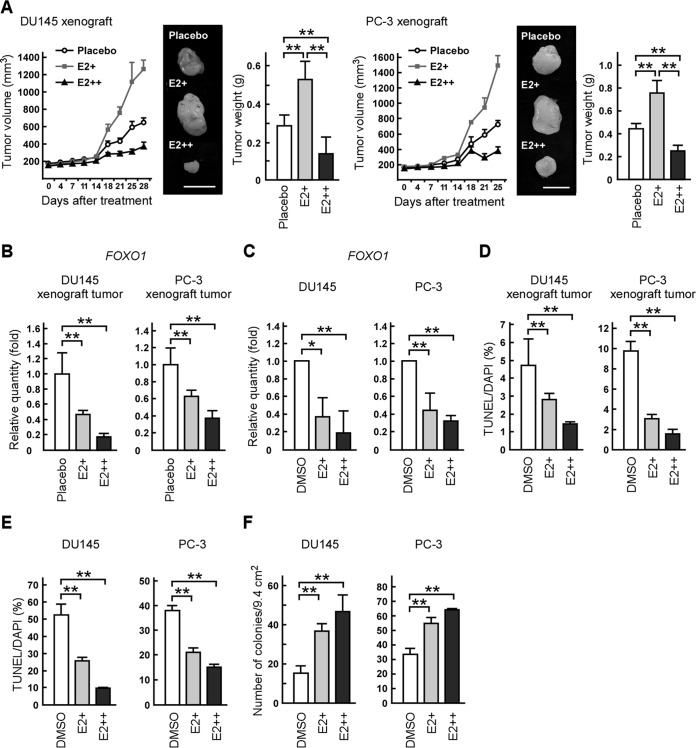
Estrogens are known to regulate prostate cancer progression, although it remains controversial whether estrogens enhance or suppress prostate cancer growth through nonandrogenic pathways (7, 8). To clarify this point, we first evaluated the dose effect of E2 on prostate tumor formation by xenograft models using AR-negative DU145 or PC-3 prostate cancer cells, which express only ERβ or both ER subtypes (25, 34, 35). Consistent with previously reported results (25), mice exposed to E2 pellets (E2+) developed larger tumors than did mice treated with placebo pellets (Fig. 1A). Surprisingly, mice exposed to pellets containing a higher dose of E2 (E2++) had smaller tumors than those treated with placebo pellets (Fig. 1A). Then, we investigated whether E2 biphasically regulated gene expression related to tumor growth. To address this, we next investigated the expression levels of FOXO1, which acts as a tumor suppressor in prostate cancer by inducing apoptosis and which is inhibited by E2 (25, 36). In cell lines and xenograft tumors, the expression levels of FOXO1 mRNA were reduced by treatment with both doses of E2 (Fig. 1B and C). The percentages of TUNEL-positive cells were also reduced by E2 treatment in xenograft tumors (Fig. 1D) and in DU145 and PC-3 cells which were cultured under anchorage-independent conditions (Fig. 1E). Moreover, an in vitro colony formation assay revealed that the anchorage-independent growth of DU145 or PC-3 cells was enhanced by E2 treatment (Fig. 1F). These results indicate that E2 has a biphasic effect on prostate cancer cell growth in vivo but not in vitro.
FIG 1.
17β-Estradiol (E2) has a biphasic effect on prostate tumor growth. (A) E2 biphasically regulates tumor formation in nude mice. Mice were injected with DU145 or PC-3 cells in both flanks and implanted with a control pellet (placebo) or a pellet containing 0.18 (E2+) or 3.4 mg (E2++) of E2 (released for 60 days). Tumor growth curves are presented in the left portions. After 25 or 28 days, the xenografts were removed and weighed (right portions). The middle portions show representative photographs of the tumors (scale bars, 1 cm). (B and C) E2 treatment reduces FOXO1 mRNA levels in xenografts and prostate cancer cells. (B) FOXO1 mRNA levels in the indicated xenograft tumors were determined by qRT-PCR. (C) DU145 or PC-3 cells were cultured in the absence (DMSO) or presence of E2 (E2+, 10 nM; E2++, 1 μM). Twelve hours after treatment, FOXO1 mRNA levels were determined by qRT-PCR. (D and E) E2 inhibits apoptosis in xenografts and prostate cancer cells. (D) DU145 and PC-3 xenograft tumors were examined in TUNEL assays. (E) DU145 or PC-3 cells were seeded on poly-HEMA-coated plates in the presence of DMSO or E2 (E2+, 10 nM; E2++, 1 μM). After 24 h, the cells were examined in TUNEL assays. DAPI, 4′,6-diamidino-2-phenylindole. (F) E2 enhances the anchorage-independent growth of prostate cancer cells in soft agar. DU145 or PC-3 cells were plated on 0.35% soft-agar plates in the presence of DMSO or E2 (E2+, 10 nM; E2++, 1 μM). Colonies with a diameter of more than 100 μm were counted. Values are means ± SDs. n = 4 to 6 for panels A, B, and D and 3 for panels C, E, and F. *, P < 0.05; **, P < 0.01.
E2 suppresses in vivo angiogenesis and regulates tumor growth through ERβ and KLF5.
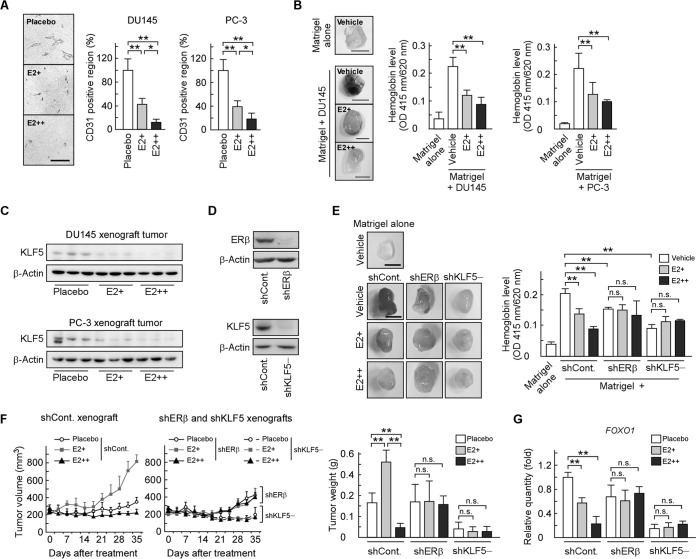
Angiogenesis plays an essential role during in vivo tumor growth (37, 38). Thus, we investigated whether angiogenesis is involved in the molecular mechanism underlying the biphasic effect of E2 on prostate tumor growth. We assessed vascular density in xenograft tumors via immunohistochemical staining for the endothelial cell marker CD31 and observed that the CD31-positive area was reduced in an E2 concentration-dependent manner (Fig. 2A). Then, we investigated the antiangiogenic activity of E2 using an in vivo Matrigel plug angiogenesis assay. DU145 or PC-3 cells were mixed with Matrigel and subcutaneously injected into mice, which were treated with E2 or vehicle. Compared with Matrigel alone, Matrigel plugs containing DU145 or PC-3 cells had a higher hemoglobin concentration (Fig. 2B). When Matrigel-implanted mice were treated with E2, hemoglobin levels in Matrigel-containing prostate cancer cells were reduced. These results indicate that E2 inhibits in vivo angiogenesis induced by prostate cancer cells.
FIG 2.
E2 modulates angiogenesis and tumor growth through ERβ and KLF5. (A) E2 inhibits angiogenesis in DU145 and PC-3 xenograft tumors. Paraffin sections of the indicated xenograft tumors were stained with antibodies for the blood vessel marker CD31, and the CD31 expression level was quantified by image analysis and expressed as a percentage of the control. Scale bar, 100 μm. (B and E) E2 inhibits angiogenesis induced by prostate cancer cells through ERβ and KLF5. Nude mice were injected subcutaneously with Matrigel, with or without the indicated cells, and the vehicle (DMSO) or E2 (E2+, 21 μg/week; E2++, 210 μg/week). Seven days after the injection, the Matrigel plugs were removed from the mice and homogenized. The supernatant was analyzed for hemoglobin content. The left portions show representative photographs of Matrigel plugs (scale bars, 0.5 cm). (C) KLF5 protein levels are lower in tumors from E2-treated mice. KLF5 protein levels in the indicated xenograft tumors were examined by immunoblotting. (D) Endogenous ERβ or KLF5 expression was stably suppressed in DU145 cells following the introduction of ERβ shRNA (shERβ) or KLF5 shRNA (shKLF5). Those protein levels were determined by immunoblotting. (F) E2 biphasically regulates tumor formation through ERβ and KLF5. Mice were injected with the indicated knockdown DU145 cells in both flanks and implanted with a placebo, E2+, or E2++ pellet. Tumor growth curves are presented in left portions. After 35 days, the xenografts were removed and weighed (right portion). (G) E2 reduces FOXO1 mRNA levels in xenografts through ERβ and KLF5. FOXO1 mRNA levels in the indicated xenograft tumors were determined by qRT-PCR. Values are means ± SDs. n = 4 to 8 for panels A, B, and E to G. *, P < 0.05; **, P < 0.01; n.s., not significant.
We previously showed that E2 reduces KLF5 protein levels and inhibits KLF5-mediated anoikis in DU145 and PC-3 cells through ERβ (25). We confirmed that KLF5 protein levels were reduced by E2 treatment in xenograft tumors (Fig. 2C). To further investigate whether ERβ and KLF5 are responsible for the E2-dependent modulation, we first performed a Matrigel plug assay using DU145 cells in which either ERβ or KLF5 was stably knocked down by shRNA (Fig. 2D). Knockdown of ERβ or KLF5 decreased hemoglobin levels and abolished the effects of E2 on angiogenesis (Fig. 2E), indicating that both ERβ and KLF5 are necessary for the promotion and E2-mediated inhibition of in vivo angiogenesis. Next, we investigated the possibility that the ERβ and KLF5 pathway contributes to the biphasic effect of E2 on prostate tumor growth using xenograft models of shERβ and shKLF5 cells. The effect of E2 on xenograft tumor growth was abolished by ERβ or KLF5 knockdown (Fig. 2F). In addition, the reduction in FOXO1 mRNA levels by E2 treatment was not observed in shERβ or shKLF5 xenografts (Fig. 2G). These data indicate that E2 modulates prostate tumor growth through the ERβ and KLF5 pathway.
KLF5 knockdown inhibits both anoikis and angiogenesis and exhibits biphasic effects on prostate tumor growth.
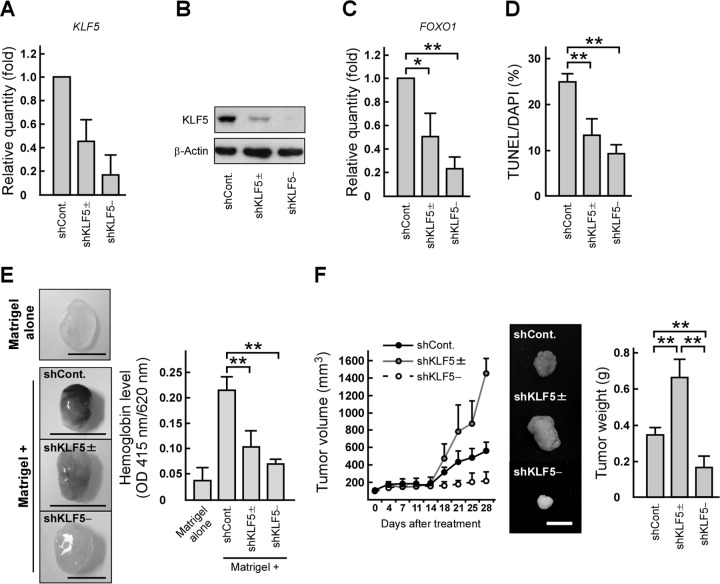
To assess the in vitro and in vivo effects of KLF5 reduction on prostate tumor growth, we generated DU145 cell lines, shKLF5± and shKLF5−, in which KLF5 expression was reduced by approximately 50% and 90%, respectively (Fig. 3A and B). The levels of FOXO1 mRNA and the number of anchorage-independent apoptotic cells were decreased in shKLF5± and shKLF5− cells (Fig. 3C and D). Interestingly, the vascularization in Matrigel plugs containing those cells was decreased by both levels of KLF5 knockdown (Fig. 3E). In contrast, xenograft tumor growth was biphasically altered (Fig. 3F). Similar results were obtained from experiments using cell lines in which KLF5 expression was reduced by other shRNA target sequences (data not shown). Taken together, our observations suggest that KLF5 exerts opposing functions on prostate tumor formation through inhibiting anoikis and angiogenesis.
FIG 3.
KLF5 knockdown suppresses anoikis and angiogenesis and exerts opposing functions on prostate tumor growth. (A and B) KLF5 expression levels in shKLF5± and shKLF5− cells. DU145 cells were transfected with luciferase shRNA (shCont) or KLF5 shRNA (shKLF5± or shKLF5−). KLF5 mRNA (A) or protein (B) levels were determined by qRT-PCR or immunoblotting, respectively. (C) KLF5 knockdown reduces FOXO1 mRNA levels in prostate cancer cells. FOXO1 mRNA levels in the indicated cells were examined by qRT-PCR. (D) KLF5 knockdown inhibits anoikis in prostate cancer cells. The indicated cells were seeded on poly-HEMA-coated plates and subjected to TUNEL assays. (E) KLF5 knockdown inhibits angiogenesis induced by prostate cancer cells. Hemoglobin content in plugs with or without the indicated cells was examined using a Matrigel plug assay (scale bars, 0.5 cm). (F) KLF5 knockdown modulates prostate tumor growth in mice. Nude mice were injected with the indicated cells in both flanks. Tumor growth curves are presented in the left portion. After 28 days, the tumors were removed and weighed (right portion). The middle portion shows representative photographs of the tumors (scale bar, 1 cm). Values are means ± SDs. n = 3 for panels A, C, and D and 4 to 6 for panels E and F. *, P < 0.05; **, P < 0.01.
PDGFA is involved in the inhibitory effect of KLF5 on prostate tumor growth through angiogenesis.
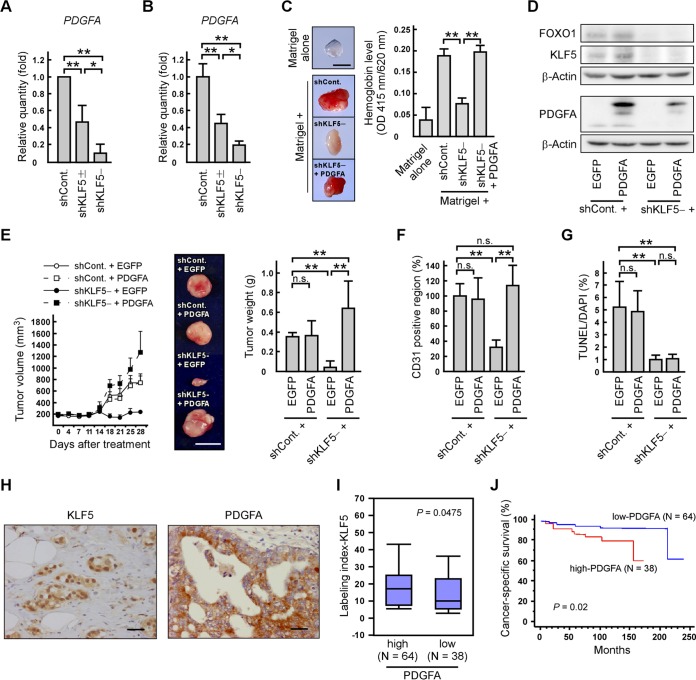
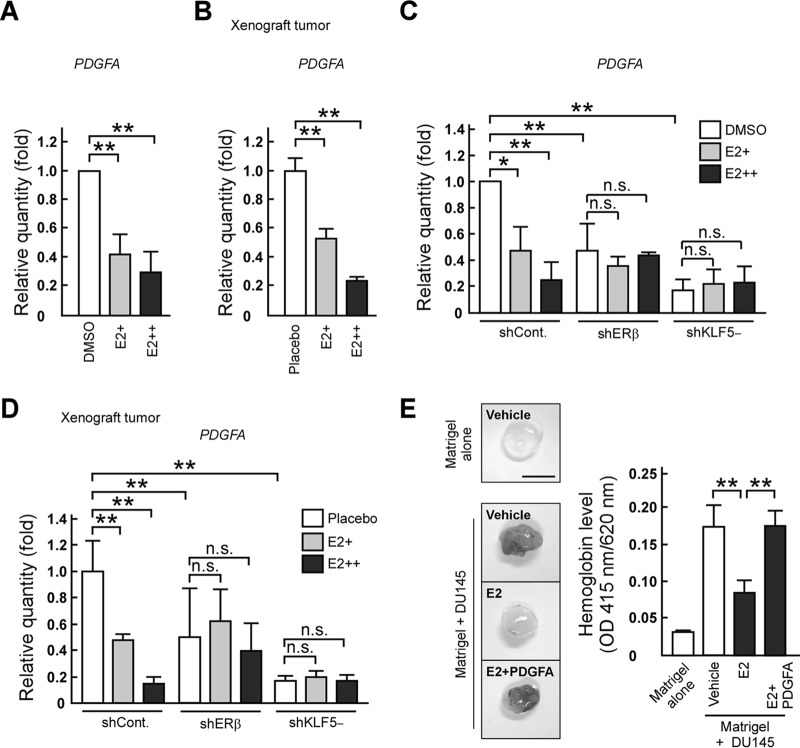
To identify a KLF5 target gene that promotes angiogenesis induced by prostate cancer cells, we focused on PDGFA because this gene is regulated by KLF5, which plays a significant role in angiogenesis (39, 40). We first revealed that PDGFA mRNA levels were decreased together with a reduction of KLF5 expression in DU145 cells and tumors (Fig. 4A and B). Next, we validated the effect of PDGFA on in vivo angiogenesis through KLF5. To address this point, we injected Matrigel containing shKLF5− cells mixed with or without PDGFA protein into mice and observed that PDGFA recovered hemoglobin levels suppressed by KLF5 depletion (Fig. 4C). Alternatively, we restored PDGFA levels in shCont. or shKLF5− cells by introducing myc-tagged PDGFA expression vectors (Fig. 4D) and injected these cells into mice. PDGFA expression in shCont. cells (shCont. + PDGFA) did not markedly modulate xenograft tumor growth compared with the growth of control tumors (shCont. + EGFP) (Fig. 4E). On the other hand, PDGFA expression in shKLF5− cells (shKLF5− + PDGFA) promoted tumor formation compared with those of shKLF5− + EGFP tumors. In shKLF5− tumors, the ratio of the CD31-positive region was recovered by PDGFA expression, but the ratio of TUNEL-positive cells was not significantly changed (Fig. 4F and G). Therefore, these results suggest that PDGFA is important for the inhibitory effect of KLF5 on prostate tumor growth through angiogenesis.
FIG 4.
PDGFA mediates the inhibitory effect of KLF5 on prostate tumor growth through angiogenesis. (A and B) PDGFA mRNA levels are reduced by KLF5 knockdown. PDGFA mRNA levels in the indicated cells (A) or xenograft tumors (B) were determined by qRT-PCR. (C) PDGFA recovers angiogenesis suppressed by KLF5 knockdown. The indicated cells were mixed with Matrigel and the vehicle or PDGFA (500 ng/plug), and the mixture was subcutaneously injected into nude mice. The quantification of hemoglobin levels within Matrigel plugs is shown in the right portion. The left portion shows representative photographs of Matrigel plugs (scale bar, 0.5 cm). (D) PDGFA, KLF5, and FOXO1 expression levels in control or shKLF5− cells expressing EGFP or PDGFA. DU145 cells were transfected with a combination of luciferase shRNA and EGFP expression plasmids (shCont + EGFP), luciferase shRNA and myc-tagged PDGFA expression plasmids (shCont. + PDGFA), KLF5 shRNA and EGFP expression plasmids (shKLF5− + EGFP), or KLF5 shRNA and myc-tagged PDGFA expression plasmids (shKLF5− + PDGFA). PDGFA, KLF5, and FOXO1 protein levels were determined by immunoblotting. (E) PDGFA expression promotes tumor formation inhibited by KLF5 knockdown. Nude mice were subcutaneously inoculated in both flanks with the indicated cells. Tumor growth curves are presented in the left portion. After 28 days, the xenografts were removed and weighed (right portion). The middle portion shows representative photographs of the tumors (scale bar, 1 cm). (F and G) PDGFA expression recovers angiogenesis but does not recover the changes of apoptosis ratios in KLF5 knockdown xenograft tumors. The indicated xenograft tumors were examined in immunostaining of CD31 (F) or TUNEL assays (G). Values are means ± SDs. n = 3 for panels A and B and 4 to 9 for panels C and E to G. *, P < 0.05; **, P < 0.01; n.s., not significant. (H) Representative prostate cancer tissues labeled with anti-KLF5 and anti-PDGFA antibodies (scale bars, 50 μm). (I) Association between the KLF5 labeling index and PDGFA expression levels in prostate cancer tissues. Prostate cancer tissues were labeled with anti-KLF5 or anti-PDGFA antibodies. “High” and “Low” indicate samples with either high (>10% positive carcinoma cells) or low (≤10% positive carcinoma cells) PDGFA immunoreactivity. (J) Clinical association of PDGFA with cancer-specific survival. Cancer-specific survival rates were analyzed using the Kaplan-Meier method for high- or low-level PDGFA-expressing samples.
Immunohistochemical staining of human prostate cancer tissues revealed that FOXO1 expression levels were positively correlated with KLF5 positivity and favorable cancer-specific survival in patients with prostate cancer (25). We first immunohistochemically tested (Fig. 4H) the correlation between KLF5 immunoreactivity and PDGFA expression levels in prostate cancer tissues. KLF5 immunoreactivity was higher in tumor samples expressing high levels of PDGFA than in samples expressing low levels of PDGFA (Fig. 4I) (P = 0.0475), suggesting a positive correlation between the abundance of KLF5 and the expression levels of PDGFA. Next, we investigated the relationships between PDGFA immunoreactivity and the cancer-specific survival rate of patients with prostate cancer using the Kaplan-Meier method. Patients with low PDGFA-expressing tumors had higher cancer-specific survival rates than patients with high PDGFA-expressing tumors (Fig. 4J) (P = 0.02), indicating that PDGFA expression is negatively correlated with the prognosis of patients with prostate cancer.
E2 suppresses angiogenesis by inhibiting PDGFA expression through ERβ and KLF5.
We next examined the inhibitory effect of E2 on angiogenesis that is mediated through PDGFA expression. E2 treatment decreased PDGFA mRNA levels in DU145 cells and its xenograft tumors (Fig. 5A and B). Then, we investigated whether ERβ and KLF5 are also responsible for the E2-dependent suppression of PDGFA expression. The E2-dependent reduction of PDGFA mRNA levels was abrogated by knockdown of ERβ or KLF5 (Fig. 5C and D). In the absence of E2, PDGFA mRNA levels were reduced by ERβ knockdown (Fig. 5C and D), supporting a role for unliganded ERβ as a coactivator of KLF5 (25). To confirm the participation of PDGFA in angiogenesis inhibition by E2, we injected Matrigel containing DU145 cells mixed or not with PDGFA protein into mice and observed that E2-dependent reduction of hemoglobin levels was restored by PDGFA protein (Fig. 5E). Thus, our results suggest that E2 suppresses angiogenesis by inhibiting the ERβ- and KLF5-mediated expression of PDGFA.
FIG 5.
E2 inhibits angiogenesis through the suppression of PDGFA expression. (A to D) E2 treatment reduces PDGFA mRNA levels through ERβ and KLF5. DU145 (A) or the indicated knockdown cells (C) were cultured in the absence (DMSO) or presence of E2 (E2+, 10 nM; E2++, 1 μM). PDGFA mRNA levels in the indicated cells (A and C) or tumors (B and D) were determined by qRT-PCR. (E) PDGFA counteracts the inhibition of angiogenesis induced by E2. Nude mice were injected subcutaneously with Matrigel, with or without DU145 cells and proteins (PDGFA; 500 ng/plug), and the vehicle or E2 (210 μg/week). Quantification of hemoglobin levels within Matrigel plugs is shown in the right portion. Representative photographs are displayed in the left portion (scale bar, 0.5 cm). Values are means ± SDs. n = 3 to 6. *, P < 0.05; **, P < 0.01; n.s., not significant.
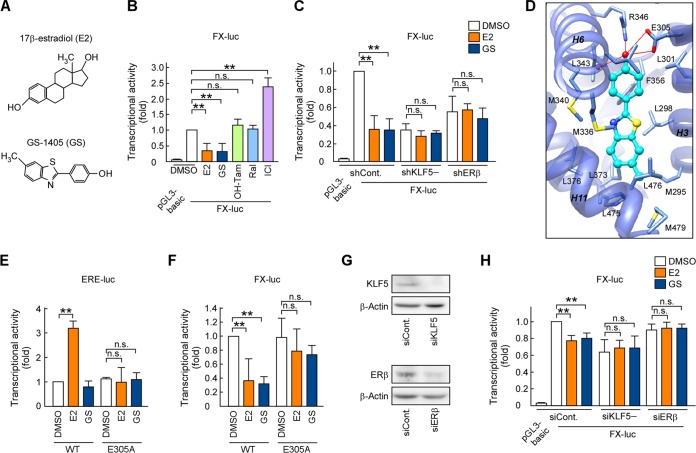
The nonagonistic ER ligand GS inhibits the KLF5 pathway through ERβ.
Previously, we identified GS as a nonagonistic ER ligand (Fig. 6A) (25). We next investigated whether GS inhibits the ERβ and KLF5 pathway without enhancing the transactivation of ERβ.
FIG 6.
The nonagonistic ER ligand GS inhibits KLF5-mediated transcription through ERβ. (A) Chemical structures of E2 and GS. (B, C, and H) E2 and GS inhibit FOXO1 promoter activity through ERβ and KLF5. A luciferase reporter plasmid containing the FOXO1 promoter (−83 to +56; FX-luc) was transfected into DU145 (B and C) or LNCaP (H) cells. Cell extracts derived from cultures containing E2, GS, 4-hydroxytamoxifen (OH-Tam), raloxifene (Ral), or ICI 182,780 (ICI) (1 μM) were examined using luciferase assays. (D) GS forms the hydrogen bond with the hERβ LBD in a docking model. GS is represented as a ball-and-stick model (cyan), whereas ligand-interacting residues are represented as sticks (light blue). Hydrogen bonds between GS and the hERβ LBD are indicated as red lines. The main chain of the hERβ LBD (PDB code 1QKM) is represented with a cartoon model (transparent blue). (E) E2, but not GS, enhances ERE-mediated transcription. ER-negative HEK293 cells were transfected with ERE-TATA-luc and the indicated ERβ expression plasmid. Transfected cells were then treated with E2 or GS (10 nM) for 24 h before the preparation of extracts. Cell extracts derived from cultures were examined using luciferase assays. (F) E305A mutation of ERβ abolishes the inhibition of FOXO1 expression by GS. FX-luc and the indicated ERβ expression plasmid were transfected into HEK293 cells. Cell extracts derived from cultures containing the indicated ER ligands (1 μM) were examined using luciferase assays. (G) Endogenous KLF5 or ERβ expression was suppressed in LNCaP cells following the introduction of KLF5 siRNA (siKLF5) or ERβ siRNA (siERβ). Those protein levels were determined by immunoblotting. Values are means ± SDs. n = 3 or 4. **, P < 0.01; n.s., not significant.
First, we compared the effects of GS and anti-estrogens on KLF5-mediated transcription using a luciferase assay with a FOXO1-promoter reporter construct containing KLF5-binding sites (FX-luc) (25). As antiestrogens, we used two selective estrogen receptor modulators, 4-hydroxytamoxifen (OH-Tam) and raloxifene (Ral), and one pure ER antagonist, ICI 182,780 (ICI). Consistent with the findings of our previous study (25), E2 inhibited KLF5-mediated transcription through ERβ, whereas ICI enhanced FOXO1 promoter activity in DU145 cells (Fig. 6B). We also observed that GS inhibited the activity in a manner similar to that of E2. On the other hand, OH-Tam and Ral did not affect the activity. To validate whether GS functions through ERβ and KLF5, we additionally performed the FX-luc assay using shERβ and shKLF5 cells and showed that the inhibitory effect of GS was abolished by ERβ or KLF5 knockdown (Fig. 6C). Then, we performed docking simulation between GS and the LBD of human ERβ (hERβ LBD). In the model structure, GS formed a hydrogen bond network involving Glu305, Arg346, and a water molecule in the LBD (Fig. 6D). Because these ligand-LBD interactions are important for the ERE-mediated transcription of ERβ induced by E2 (Fig. 6E) (41, 42), we introduced a point mutation in Glu305. We confirmed that in contrast to E2, GS did not enhance ERE-mediated transcription (Fig. 6E). The E305A mutation reduced the E2- and GS-induced transcriptional inhibition of FOXO1 promoter activity (Fig. 6F), confirming the inhibitory effects of these ligands on KLF5-mediated transcription through ERβ.
Emerging studies have demonstrated that AR plays a critical role in prostate cancer development and progression, even after castration (4, 43). Therefore, we investigated whether E2 and GS suppress KLF5-mediated transcription in the presence of AR using AR-positive LNCaP cells, which express KLF5 and ERβ (Fig. 6G). In these cells, E2 and GS inhibited FOXO1 promoter activity, whereas the inhibitory effects were abolished by KLF5 or ERβ reduction (Fig. 6H). These results suggest the possibility that E2 and GS also inhibit the KLF5 pathway through ERβ in the presence of AR.
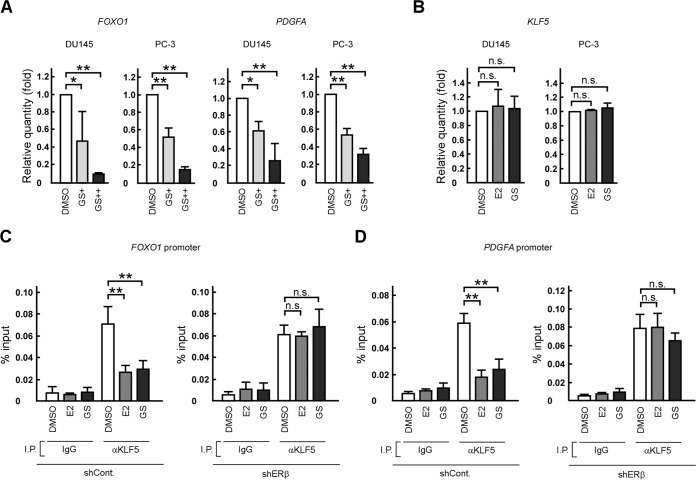
We then investigated the effect of GS on the mRNA levels of KLF5 target genes. Similar to the case with E2, GS treatment decreased FOXO1 and PDGFA mRNA levels but not those of KLF5 in DU145 and PC-3 cells (Fig. 7A and B). Furthermore, a ChIP experiment revealed that both ligands inhibited the binding of KLF5 to the FOXO1 or PDGFA promoter regions containing functional or potential KLF5 response elements (25, 44) (Fig. 7C and D). The inhibitory effects of E2 and GS were not observed in shERβ cells.
FIG 7.
E2 and GS suppress FOXO1 and PDGFA expression through inhibiting KLF5 interaction to those promoter regions. (A) GS treatment reduces FOXO1 and PDGFA mRNA levels in prostate cancer cells. DU145 or PC-3 cells were cultured in the absence (DMSO) or presence of GS (GS+, 10 nM; GS++, 1 μM), and FOXO1 or PDGFA mRNA levels were determined by qRT-PCR. (B) E2 or GS treatment does not affect KLF5 mRNA levels. DU145 or PC-3 cells were cultured in the absence (DMSO) or presence of E2 or GS (1 μM), and KLF5 mRNA levels were determined by qRT-PCR. (C and D) E2 or GS treatment inhibits the binding of KLF5 to the FOXO1 (C) and PDGFA promoter regions (D) through ERβ. Control (shCont.) and ERβ knockdown (shERβ) DU145 cells were cultured in the absence (DMSO) or presence of the indicated ER ligands (1 μM). ChIP assays were performed using anti-KLF5 antibodies. Immunoprecipitated DNA was assessed in qRT-PCR assays using primers specific for the FOXO1 or PDGFA promoter. Samples were normalized to the input DNA. Values are means ± SDs. n = 3. *, P < 0.05; **, P < 0.01; n.s., not significant.
Taken together, these results suggest that GS inhibits KLF5 recruitment to the target promoter through ERβ for the suppression of KLF5-mediated transcription without enhancing ERβ transactivation.
GS inhibits anoikis and angiogenesis and regulates prostate tumor growth through ERβ.
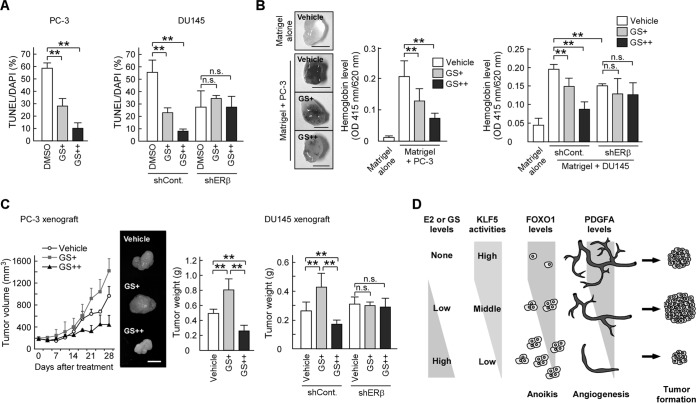
Finally, we investigated the in vitro and in vivo effects of GS on prostate tumor growth. To address this issue, we investigated whether GS affects anoikis and angiogenesis. GS treatment decreased the number of apoptotic cells in poly-HEMA-coated plates (Fig. 8A). In addition, GS inhibited angiogenesis in the Matrigel plugs containing prostate cancer cells (Fig. 8B). Then, we used DU145 and PC-3 xenograft models to evaluate the effect of GS on prostate tumor growth. Mice treated with GS (GS+) developed larger tumors than did control mice treated with dimethyl sulfoxide (DMSO), whereas those injected with a higher dose of GS (GS++) had smaller tumors than control mice (Fig. 8C). We confirmed that these effects of GS were abolished by ERβ knockdown (Fig. 8A to C). These results suggest that the nonagonistic ER ligand GS inhibits anoikis and angiogenesis through ERβ and modulates prostate tumor growth.
FIG 8.
GS regulates prostate tumor growth through the inhibition of anoikis and angiogenesis. (A) GS suppresses anoikis in prostate cancer cells. DU145 or PC-3 cells were seeded onto poly-HEMA-coated plates in the presence of DMSO or GS (GS+, 10 nM; GS++, 1 μM). After 24 h, the cells were examined by TUNEL assays. (B) GS inhibits in vivo angiogenesis through ERβ. Hemoglobin content in plugs with the indicated cells treated with GS (GS+, 5 mg/week; GS++, 25 mg/week) or left untreated was examined using a Matrigel plug assay (scale bars, 0.5 cm). (C) GS modulates prostate tumor growth. Nude mice were injected with the indicated cells followed by the vehicle (DMSO) or GS (GS+, 5 mg/week; GS++, 25 mg/week). Tumor growth curves are presented in the left portion. After 28 days, the tumors were removed and weighed (right portions). The middle portion shows representative photographs of the tumors (scale bar, 1 cm). (D) Schematic model of the mechanism by which E2 or GS biphasically regulates prostate tumor growth. Values are means ± SDs. n = 3 for panel A and 4 to 8 for panels B and C. **, P < 0.01; n.s., not significant.
DISCUSSION
In this study, our results address the molecular basis of the paradoxical effects of E2 in prostate cancer. Our previous results revealed that E2 treatment decreased KLF5-dependent FOXO1 transcription in prostate cancer cells though ERβ, thereby inhibiting apoptosis and increasing tumor weight in mouse xenograft models (25). In contrast, our present results showed that when mice were treated with higher doses of E2, prostate tumor growth was suppressed through ERβ and KLF5 in those models (Fig. 1A and 2F). We also demonstrated that E2 inhibited PDGFA transcription and suppressed angiogenesis through ERβ and KLF5 (Fig. 2E, 5C, and D). Moreover, PDGFA recovered angiogenesis inhibited by E2 (Fig. 5E). Apoptosis serves as a natural barrier for cancer development (45). Conversely, angiogenesis is indispensable for tumorigenesis (46). Considering the previous reports together with our data, angiogenesis may be sufficient for tumor growth in mice treated with lower doses of E2, which enhances xenograft tumor growth through the inhibition of apoptosis. On the other hand, when both PDGFA and FOXO1 expressions were markedly suppressed by higher doses of E2, angiogenesis may have been insufficient for prostate tumor growth, thereby suppressing tumor growth. Therefore, our previous and present results suggest that E2 biphasically regulates prostate tumor growth by suppressing FOXO1 and PDGFA expression levels through the ERβ-KLF5 pathway (Fig. 8D).
In response to ligands, ERs initiate transcription by binding directly to EREs (classical pathway) or by interacting with other transcription factors (nonclassical pathway) (22, 23). Recently, we indicated that in the absence of a ligand, ERβ acts as a coactivator of KLF5 by recruiting CBP, thereby enhancing FOXO1 expression and anchorage-independent apoptosis (25). In this study, we further found that in vivo angiogenesis was suppressed by ERβ depletion in the absence of ER ligands (Fig. 2E and 8B). ERβ depletion also reduced PDGFA mRNA levels in DU145 cells and xenograft tumors that were not treated with ER ligands (Fig. 5C and D). Moreover, PDGFA was targeted by KLF5 (Fig. 4A and B and 7D) and was involved in KLF5-mediated angiogenesis (Fig. 4C). Taken together, these results suggest that unliganded ERβ regulates PDGFA expression through KLF5 transactivation and thereby mediates angiogenesis in vivo.
In various cancers, including prostate cancer, KLF5 was inactivated by chromosomal deletion, transcriptional silencing, and excessive protein degradation, thereby suggesting that KLF5 acts as a tumor suppressor (47–50). On the contrary, in prostate cancer cells, KLF5 levels are most often decreased as a result of hemizygous deletion; KLF5 is hardly deleted homozygously (49). Thus, these observations raise the possibility that KLF5 both possesses a tumor-suppressive function and is also necessary for tumor formation. In this study, we illustrated by knockdown experiments that an approximately 50% reduction of KLF5 expression in DU145 cells inhibited apoptosis under anchorage-independent conditions (Fig. 3D, shKLF5±). The ratio of apoptosis was more strongly suppressed by a severe reduction of KLF5 expression (Fig. 3D, shKLF5−). Although these results suggest that shKLF5− cells possess the potential to form larger tumors than shKLF5± cells, we unexpectedly found that shKLF5− cells did not form tumors in mice (Fig. 3F). In contrast, Matrigel plug assays indicated that KLF5 knockdown reduced angiogenesis (Fig. 3E). Considering that angiogenesis plays an indispensable role in tumorigenesis (51, 52), our results suggest that prostate cancer cells, in which KLF5 has been homozygously deleted, may not be able to form tumors because of inhibited angiogenesis.
KLF5 is involved in cancer development in a number of human tissues, although its function remains controversial (26, 27). For instance, expression of KLF5 enhances cell proliferation in untransformed cells and transformed fibroblasts, whereas KLF5 suppresses cell growth in some cancer cells (28). Recent reports disclosed that xenograft tumor growth was suppressed by the expression of wild-type KLF5 but enhanced by the expression of a deacetylated KLF5 mutant (K369R) in prostate cancer cells, suggesting that the roles of KLF5 are regulated by posttranscriptional modifications (53). It is also known that KLF5 activity is regulated by steroid hormones in breast cancer cells (54, 55). In fact, we found in this study that ER ligands inhibited KLF5-mediated transcription in prostate cancer cells (Fig. 6) and altered xenograft tumor growth (Fig. 1A and 8C). Thus, the specific roles of KLF5 in cancer development appear to be context dependent, including posttranscriptional modifications and hormone levels. Therefore, further studies are needed to address the mechanism underlying the modulation of prostate cancer tumorigenesis by KLF5.
Estrogens, including the synthetic estrogen diethylstilbestrol, have previously been used in prostate cancer treatment; however, adverse effects limited their use (8, 56). These undesirable effects of estrogenic drugs are probably mediated in part by the transactivation of ERs (classical pathway) (57). Our previous and present results showed that E2 enhanced the transcriptional activity of ERβ and suppressed that of KLF5, whereas the nonagonistic ER ligand GS inhibited KLF5-mediate transactivation through ERβ (Fig. 6) (25). We further revealed that high-dose GS inhibited angiogenesis and prostate tumor growth in mouse xenograft models through ERβ (Fig. 8B and C). These results suggest that selective inhibition of KLF5 activity via ERβ could be useful in prostate cancer therapies that minimize adverse effects caused by ER transactivation through the classical pathway. Previous reports indicated that ERs bind to and modulate the transcriptional activity of several transcription factors, including Sp1, NF-κB, and AP1 (23, 58, 59). According to our results, it is possible to develop compounds that regulate these transcription factors separately. Therefore, our results provide a new strategy for designing next-generation ER modulators that can regulate nonclassical pathways without affecting the classical pathway.
ACKNOWLEDGMENTS
We sincerely thank A. Fukamizu for a critical reading of the manuscript and helpful scientific input. We are also grateful to N. Ohnuma for valuable discussions and technical advice.
This work was supported by a Grant-in Aid for Young Scientists (B) from the Japan Society for the Promotion of Science (23790075 to Y.N.), the research program of the Project for Development of Innovative Research on Cancer Therapeutics (P-Direct), Ministry of Education, Culture, Sports, Science and Technology of Japan (to J.Y.), and the Open Innovation Core (OIC) project (Yuka Nakajima; a member of OIC) of Life Science Center, Tsukuba Advance Research Alliance (TARA), University of Tsukuba, Japan.
Funding Statement
The Open Innovation Core (OIC) project of Life Science Center, Tsukuba Advance Research Alliance (TARA), University of Tsukuba, Japan, provides the research facility for Yuka Nakajima.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. 2013. Cancer statistics, 2013. CA Cancer J Clin 62:11–30. [DOI] [PubMed] [Google Scholar]
- 2.Ockrim J, Lalani El-N, Abel P. 2006. Therapy insight: parental estrogen treatment for prostate cancer-a new dawn for an old therapy. Nat Clin Pract Oncol 3:552–563. doi: 10.1038/ncponc0602. [DOI] [PubMed] [Google Scholar]
- 3.Shafi AA, Yen AE, Weigel NL. 2013. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther 140:223–238. doi: 10.1016/j.pharmthera.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Karantanos T, Corn PG, Thompson TC. 2013. Prostate cancer progression after androgen deprivation therapy: mechanism of castrate resistance and novel therapeutic approaches. Oncogene 32:5501–5511. doi: 10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellem SJ, Risbridger GP. 2010. Aromatase and regulating the estrogen:androgen ratio in the prostate gland. J Steroid Biochem Mol Biol 118:246–251. doi: 10.1016/j.jsbmb.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 6.Ho SM, Lee MT, Lam HM, Leung YK. 2011. Estrogens and prostate cancer: etiology, mediators, prevention, and management. Endocrinol Metab Clin North Am 40:591–614. doi: 10.1016/j.ecl.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelles JL, Hu WY, Prins GS. 2011. Estrogen action and prostate cancer. Expert Rev Endocrinol Metab 6:437–451. doi: 10.1586/eem.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Härkönen PL, Makela SI. 2004. Role of estrogens in development of prostate cancer. J Steroid Biochem Mol Biol 92:297–305. doi: 10.1016/j.jsbmb.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Corey E, Quinn JE, Emond MJ, Buhler KR, Brown LG, Vessella RL. 2002. Inhibition of androgen-independent growth of prostate cancer xenografts by 17β-estradiol. Clin Cancer Res 8:1003–1007. [PubMed] [Google Scholar]
- 10.Coleman IM, Kiefer JA, Brown LG, Pitts TE, Nelson PS, Brubaker KD, Vessella RL, Corey E. 2006. Inhibition of androgen-independent prostate cancer by estrogenic compounds is associated with increased expression of immune-related genes. Neoplasia 8:862–878. doi: 10.1593/neo.06328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ravery V, Fizazi K, Drouet L, Eymard JC, Culine S, Gravis G, Hennequin C, Zerbib M. 2011. The use of estramustine phosphate in the modern management of advanced prostate cancer. BJU Int 108:1782–1786. doi: 10.1111/j.1464-410X.2011.10201.x. [DOI] [PubMed] [Google Scholar]
- 12.Clemons J, Glodé LM, Gao D, Flaig TW. 2013. Low-dose diethylstilbestrol for the treatment of advanced prostate cancer. Urol Oncol 31:198–204. doi: 10.1016/j.urolonc.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho SM. 2004. Estrogens and anti-estrogens: key mediators of prostate carcinogenesis and new therapeutic candidates. J Cell Biochem 91:491–503. doi: 10.1002/jcb.10759. [DOI] [PubMed] [Google Scholar]
- 14.Bonkhoff H, Berges R. 2009. The evolving role of oestrogens and their receptors in the development and progression of prostate cancer. Eur Urol 55:533–542. doi: 10.1016/j.eururo.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 15.Abd Elmageed ZY, Moroz K, Srivastav SK, Fang Z, Crawford BE, Moparty K, Thomas R, Abdel-Mageed AB. 2013. High circulating estrogens and selective expression of ERβ in prostate tumors of Americans: implications for racial disparity of prostate cancer. Carcinogenesis 34:2017–2023. doi: 10.1093/carcin/bgt156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson JA. 1996. Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci U S A 93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao W, Brown M. 2004. Advances in estrogen receptor biology: prospects for improvements in targeted breast cancer therapy. Breast Cancer Res 6:39–52. doi: 10.1186/bcr742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enmark E, Pelto-Huikko M, Grandien K, Lagercrantz S, Lagercrantz J, Friend G, Nordenskjöld M, Gustafsson JÅ. 1997. Human estrogen receptor beta-gene structure, chromosomal localization, and expression pattern. J Clin Endocrinol Metab 82:4258–4265. doi: 10.1210/jcem.82.12.4470. [DOI] [PubMed] [Google Scholar]
- 19.Bosland MC. 2005. The role of estrogens in prostate carcinogenesis: a rationale for chemoprevention. Rev Urol 3:S4–S10. [PMC free article] [PubMed] [Google Scholar]
- 20.McPherson SJ, Hussain S, Balanathan P, Hedwards SL, Niranjan B, Grant M, Chandrasiri UP, Toivanen R, Wang Y, Taylor RA, Risbridger GP. 2010. Estrogen receptor-β activated apoptosis in benign hyperplasia and cancer of the prostate is androgen independent and TNFα mediated. Proc Natl Acad Sci U S A 107:3123–3128. doi: 10.1073/pnas.0905524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll JS, Brown M. 2006. Estrogen receptor target gene: an evolving concept. Mol Endocrinol 20:1707–1714. doi: 10.1210/me.2005-0334. [DOI] [PubMed] [Google Scholar]
- 22.McDevitt MA, Glidewell-Kenney C, Jimenez MA, Ahearn PC, Weiss J, Jameson JL, Levine JE. 2008. New insights into the classical and non-classical actions of estrogen: evidence from estrogen receptor knock-out and knock-in mice. Mol Cell Endocrinol 290:24–30. doi: 10.1016/j.mce.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leung YK, Ho SM. 2011. Estrogen receptorβ: switching to a new partner and escaping from estrogen. Sci Signal 4:pe19. [DOI] [PubMed] [Google Scholar]
- 24.Leung YK, Gao Y, Lau KM, Zhang X, Ho SM. 2006. ICI 182,780-regulated gene expression in DU145 prostate cancer cells is mediated by estrogen receptor-beta/NFkappaB crosstalk. Neoplasia 8:242–249. doi: 10.1593/neo.05853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakajima Y, Akaogi K, Suzuki T, Osakabe A, Yamaguchi C, Sunahara N, Ishida J, Kako K, Ogawa S, Fujimura T, Homma Y, Fukamizu A, Murayama A, Kimura K, Inoue S, Yanagisawa J. 2011. Estrogen regulates tumor growth through a nonclassical pathway that includes the transcription factors ERβ and KLF5. Sci Signal 4:ra22. [DOI] [PubMed] [Google Scholar]
- 26.Dong JT, Chen C. 2009. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci 66:2691–2706. doi: 10.1007/s00018-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simmen RC, Pabona JM, Velarde MC, Simmons C, Rahal O, Simmen FA. 2010. The emerging role of Krüppel-like factors in endocrine-responsive cancers of female reproductive tissues. J Endocrinol 204:223–231. doi: 10.1677/JOE-09-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diakiw SM, D'Andrea RJ, Brown AL. 2013. The double life of KLF5: opposing role in regulation of gene-expression, cellular function, and transformation. IUBMB Life 65:999–1011. doi: 10.1002/iub.1233. [DOI] [PubMed] [Google Scholar]
- 29.Greenberg JI, Shields DJ, Barillas SG, Acevedo LM, Murphy E, Huang J, Scheppke L, Stockmann C, Johnson RS, Angle N, Cheresh DA. 2008. A role for VEGF as a negative regulator of pericyte function and vessel maturation. Nature 456:809–813. doi: 10.1038/nature07424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Himmel DM, Gourinath S, Reshetnikova L, Shen Y, Szent-Gyorgyi AG, Cohen C. 2002. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: further insights into the mechanics of the motor. Proc Natl Acad Sci U S A 99:12645–12650. doi: 10.1073/pnas.202476799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanner MF. 1999. Python: a programming language for software integration and development. J Mol Graph Model 17:57–61. [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Lau KM, LaSpina M, Long J, Ho SM. 2000. Expression of estrogen receptor (ER)-α and ER-β in normal and malignant prostatic epithelial cells: regulation by methylation and involvement in growth regulation. Cancer Res 60:3175–3182. [PubMed] [Google Scholar]
- 35.Dey P, Ström A, Gustafsson J̊A. 2014. Estrogen receptor β upregulates FOXO3a and causes induction of apoptosis through PUMA in prostate cancer. Oncogene 33:4213–4225. doi: 10.1038/onc.2013.384. [DOI] [PubMed] [Google Scholar]
- 36.Huang H, Tindall DJ. 2008. Regulation of FOXO protein stability via ubiquitination and proteasome degradation. Oncogene 27:2312–2319. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrae J, Gallini R, Betsholtz C. 2008. Role of platelet-derived growth factors in physiology and medicine. Genes Dev 22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heldin CH. 2013. Targeting the PDGF signaling pathway in tumor treatment. Cell Commun Signal 11:97. doi: 10.1186/1478-811X-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, Kawai-Kowase K, Moriyama N, Imai Y, Kawakami H, Nishimatsu H, Ishikawa T, Suzuki T, Morita H, Maemura K, Sata M, Hirata Y, Komukai M, Kagechika H, Kadowaki T, Kurabayashi M, Nagai R. 2002. Krüppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med 8:856–863. [DOI] [PubMed] [Google Scholar]
- 40.Pietras K, Pahler J, Bergers G, Hanahan D. 2008. Functions of paracrine PDGF signaling in the proangiogenic tumor stroma revealed by pharmacological targeting. PLoS Med 5:e19. doi: 10.1371/journal.pmed.0050019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi Y, Koh JT. 2002. Functionally orthogonal ligand-receptor pairs for the selective regulation of gene expression generated by manipulation of charged residues at the ligand-receptor interface of ER alpha and ER beta. J Am Chem Soc 124:6921–6928. doi: 10.1021/ja016897x. [DOI] [PubMed] [Google Scholar]
- 42.Bertini S, De Cupertinis A, Granchi C, Bargagli B, Tuccinardi T, Martinelli A, Macchia M, Gunther JR, Carlson KE, Katzenellenbogen JA, Minutolo F. 2011. Selective and potent agonists for estrogen receptor beta derived from molecular refinements of salicylaldoximes. Eur J Med Chem 46:2453–2462. doi: 10.1016/j.ejmech.2011.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lonergan PE, Tindall DJ. 2011. Androgen receptor signaling in prostate cancer development and progression. J Carcinog 10:20. doi: 10.4103/1477-3163.83937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aizawa K, Suzuki T, Kada N, Ishihara A, Kawai-Kowase K, Matsumura T, Sasaki K, Munemasa Y, Manabe I, Kurabayashi M, Collins T, Nagai R. 2004. Regulation of platelet-derived growth factor-A chain by Krüppel-like factor 5: new pathway of cooperative activation with nuclear factor-kappaB. J Biol Chem 279:70–76. doi: 10.1074/jbc.M306621200. [DOI] [PubMed] [Google Scholar]
- 45.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 46.Folkman J. 2003. Angiogenesis and apoptosis. Semin Cancer Biol 13:159–167. doi: 10.1016/S1044-579X(02)00133-5. [DOI] [PubMed] [Google Scholar]
- 47.Knuutila S, Aalto Y, Autio K, Björkqvist AM, El-Rifai W, Hemmer S, Huhta T, Kettunen E, Kiuru-Kuhlefelt S, Larramendy ML, Lushnikova T, Monni O, Pere H, Tapper J, Tarkkanen M, Varis A, Wasenius VM, Wolf M, Zhu Y. 1999. DNA copy number losses in human neoplasm. Am J Pathol 155:683–694. doi: 10.1016/S0002-9440(10)65166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dong JT. 2001. Chromosomal deletions and tumor suppressor genes in prostate cancer. Cancer Metastasis Rev 20:173–193. doi: 10.1023/A:1015575125780. [DOI] [PubMed] [Google Scholar]
- 49.Chen C, Bhalala HV, Vessella RL, Dong JT. 2003. KLF5 is frequently deleted and down-regulated but rarely mutated in prostate cancer. Prostate 55:81–88. doi: 10.1002/pros.10205. [DOI] [PubMed] [Google Scholar]
- 50.Chen C, Sun X, Ran Q, Wilkinson KD, Murphy TJ, Simons JW, Dong JT. 2005. Ubiquitin-proteasome degradation of KLF5 transcription factor in cancer and untransformed epithelial cells. Oncogene 24:3319–3327. doi: 10.1038/sj.onc.1208497. [DOI] [PubMed] [Google Scholar]
- 51.Bergers G, Hanahan D. 2008. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer 8:592–603. doi: 10.1038/nrc2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ribeiro AL, Okamoto OK. 2015. Combined effects of pericytes in the tumor microenvironment. Stem Cells Int 2015:868475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Zhang B, Wu Q, Ci X, Zhao R, Zhang Z, Xia S, Su D, Chen J, Ma G, Fu L, Dong JT. 2015. Interruption of KLF5 acetylation converts its function from tumor suppressor to tumor promoter in prostate cancer cells. Int J Cancer 136:536–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao KW, Sikriwal D, Dong X, Guo P, Sun X, Dong JT. 2011. Oestrogen causes degradation of KLF5 by inducing the E3 ubiquitin ligase EFP in ER-positive breast cancer cells. Biochem J 437:323–333. doi: 10.1042/BJ20101388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu R, Zhou Z, Zhao D, Chen C. 2011. The induction of KLF5 transcription factor by progesterone contributes to progesterone-induced breast cancer cell proliferation and dedifferentiation. Mol Endcrinol 25:1137–1144. doi: 10.1210/me.2010-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wibowo E, Schellhammer P, Wassersug RJ. 2011. Role of estrogen in normal male function: clinical implications for patients with prostate cancer on androgen deprivation therapy. J Urol 185:17–23. doi: 10.1016/j.juro.2010.08.094. [DOI] [PubMed] [Google Scholar]
- 57.Ho SM, Leung YK, Chung I. 2006. Estrogens and antiestrogens as etiological factors and therapeutics for prostate cancer. Ann N Y Acad Sci 1089:177–193. doi: 10.1196/annals.1386.005. [DOI] [PubMed] [Google Scholar]
- 58.Bjornström L, Sjoberg M. 2005. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol 19:833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- 59.Zhao C, Gao H, Liu Y, Papoutsi Z, Jaffrey S, Gustafsson JA, Dahlman-Wright K. 2010. Genome-wide mapping of estrogen receptor-beta-binding regions reveals extensive cross-talk with transcription factor activator protein-1. Cancer Res 70:5174–5183. doi: 10.1158/0008-5472.CAN-09-4407. [DOI] [PubMed] [Google Scholar]