Abstract
In most human cancer cells, cellular immortalization relies on the activation and recruitment of telomerase to telomeres. The telomere-binding protein TPP1 and the TEN domain of the telomerase catalytic subunit TERT regulate telomerase recruitment. TERT contains a unique domain, called the insertion in fingers domain (IFD), located within the conserved reverse transcriptase domain. We report the role of specific hTERT IFD residues in the regulation of telomerase activity and processivity, recruitment to telomeres, and cell survival. One hTERT IFD variant, hTERT-L805A, with reduced activity and processivity showed impaired telomere association, which could be partially rescued by overexpression of TPP1-POT1. Another previously reported hTERT IFD mutant enzyme with similarly low levels of activity and processivity, hTERT-V791Y, displayed defects in telomere binding and was insensitive to TPP1-POT1 overexpression. Our results provide the first evidence that the IFD can mediate enzyme processivity and telomerase recruitment to telomeres in a TPP1-dependent manner. Moreover, unlike hTERT-V791Y, hTERT-V763S, a variant with reduced activity but increased processivity, and hTERT-L805A, could both immortalize limited-life-span cells, but cells expressing these two mutant enzymes displayed growth defects, increased apoptosis, DNA damage at telomeres, and short telomeres. Our results highlight the importance of the IFD in maintaining short telomeres and in cell survival.
INTRODUCTION
Telomeres are the protective nucleoprotein structures that cap the ends of linear eukaryotic chromosomes, thus preventing the aberrant and fatal activation of the DNA damage repair machinery. During normal somatic cell division, the end replication problem arising from the inability of DNA polymerase to completely replicate telomeres leads to progressive telomere loss and, over time, triggers cellular senescence to prevent carcinogenesis. The renewal capacity of germ cells, stem cells, and cancer cells is limited by telomere erosion and relies on the activation of a telomere maintenance mechanism for cellular survival. In over 85% of human cancers, detectable expression of telomerase, a specialized reverse transcriptase, is a requirement for cellular immortalization (1).
In humans, telomerase is minimally composed of the core catalytic subunit human telomerase reverse transcriptase (hTERT) and an intrinsic RNA moiety, human telomerase RNA (hTR), to dictate the de novo synthesis of tandem TTAGGG repeats. Telomerase has the unique ability to synthesize long stretches of telomeric sequence repeats using its short RNA template through reiterative rounds of DNA synthesis, partial dissociation, translocation, and realignment with the newly synthesized telomere end. In human cells, this unique property, termed “repeat addition processivity” (RAP), is a determinant of telomere maintenance and cellular survival (2). The reverse transcriptase region of the TERT subunit contains seven motifs (1, 2, A, B′, C, D, and E) that are also conserved in other nucleic acid polymerases. Importantly, TERT distinguishes itself from other conventional reverse transcriptases by the presence of a large insertion within the fingers subdomain between the conserved motifs A and B′, referred to as the insertion in fingers domain (IFD). The Tribolium castaneum TERT crystal structure reveals that the IFD is located on the periphery of the TERT ring (3). In Saccharomyces cerevisiae, specific residues in the IFD were found to be critical for RAP (4). More recently, we demonstrated that an hTERT variant with a mutation in the IFD, hTERT-V791Y, is defective in DNA synthesis and in localization to the telomeres and is not competent to immortalize telomerase-negative limited-life-span cells (5). This led us to speculate that the valine at position 791 and the IFD domain are important for telomerase recruitment to the telomeres and for cell survival.
Telomerase recruitment to its site of action, the telomere, is essential for its function. Enzymes mutated in the N-DAT (dissociates activity of telomerase) region of the hTERT N-terminal domain (TEN) are catalytically active but fail to maintain telomeres and immortalize cells (6). A role for the N-DAT region in telomerase recruitment was later proposed (7). Several proteins are involved in the recruitment of telomerase to the telomeres, including the shelterin components TPP1 (8, 9) and TIN2 (10), the Cajal body RNA chaperone TCAB1 (11), the negative regulator of telomerase activity PinX1 (12), and the double-stranded telomeric-DNA-binding protein HOT1 (13). Extensive studies on TPP1 revealed that the TPP1 TEL patch (TPP1 glutamate [E] and leucine [L] rich) within the oligonucleotide-binding (OB) domain promotes telomerase processivity when complexed with POT1 and is required for telomerase recruitment to the telomeres (14–18). A recent study identified catalytically active TEN domain mutant enzymes defective in telomere localization and maintenance, defining a direct interaction interface between TPP1 and hTERT (19).
Here, we studied the role of the human telomerase IFD in the regulation of telomerase-specific functions, recruitment to telomeres and cellular survival. We report the implication of specific hTERT IFD residues in the regulation of telomerase activity and processivity. The hTERT IFD variant hTERT-V791Y, previously reported to display low levels of activity and processivity (5), exhibited defects in telomere binding and was insensitive to TPP1-POT1 overexpression in the present study. One hTERT IFD variant, hTERT-L805A, with reduced activity and processivity showed impaired telomere association, which could be partially rescued by TPP1-POT1 overexpression. Another hTERT IFD variant, hTERT-V763S, which had reduced activity but increased processivity, could be stimulated by overexpression of TPP1-POT1. Our results provide the first evidence that the IFD can mediate enzyme processivity and telomerase recruitment to telomeres in a TPP1-dependent manner. Moreover, unlike hTERT-V791Y, hTERT-V763S and hTERT-L805A mutant enzymes could both immortalize limited-life-span cells, but cells expressing these two variants displayed growth defects, increased apoptosis, DNA damage at telomeres, and short telomeres. Our results highlight the importance of the IFD in regulating maintenance of short telomeres and cell survival.
MATERIALS AND METHODS
Plasmid construction.
The pMSCV-puromycin-N-term-3XFLAG-hTERT was constructed by insertion of BglII- and EcoRI-digested PCR product encoding 3XFLAG-hTERT into pMSCV-puromycin. pcDNA6/myc-His C-hTERT, pVAN3XFLAG-hTERT, and pMSCV-puromycin-N-term-3XFLAG-hTERT were used as templates for the generation of pcDNA6/myc-His C-hTERT-V763S, -V791Y, and -L805A, pVAN3XFLAG-hTERT-V763S, -V791Y, and -L805A, and pMSCV-puromycin-N-term-3XFLAG-hTERT-V763S and -L805A.
Cell culture, retroviral infection, and transient transfection.
Retroviral infection of HA5 and transient transfections of HEK 293 and HeLa cells were performed as previously described (2). Retroviral infection of HeLa cells was performed using the same protocol as for HA5 cells.
Protein analysis by Western blotting.
Western blotting was performed as previously described (2). Briefly, detection of hTERT was done using anti-hTERT (C-20; Santa Cruz) with a dilution of 1:500 for 1 h 30 min at room temperature (RT), washed three times for 5 min with 1× phosphate-buffered saline–Tween (PBS-T), and blotted with rabbit anti-goat antibody (Dako) at a 1:5,000 dilution for 1 h at room temperature. Blotting of 3×FLAG-TPP1 and 3×FLAG-POT1 was performed using mouse M2 anti-FLAG antibody (F3165; Sigma) at a dilution of 1:1,500 for 30 min at room temperature. Blots were washed and blotted with horseradish peroxidase (HRP)-conjugated secondary anti-mouse antibody (Amersham) at a 1:10,000 dilution for 30 min at room temperature. Blotting of actin and tubulin was performed using mouse antiactin (Chemicon) and mouse antitubulin (Sigma) at a dilution of 1:5,000 for 30 min at room temperature, and blots were washed and probed with secondary anti-mouse HRP-conjugated antibody (Amersham) at a 1:10,000 dilution for 30 min at room temperature. After blotting with the secondary antibody, blots were washed three times in 1× PBS-T and revealed using Pierce ECL Plus Western blotting substrate (Thermo Fisher Scientific).
Direct primer extension assay.
Reconstitution of wild-type (WT) telomerase and variants in HEK 293 cells and direct primer extension experiments were performed as previously described (2, 19) with the following modifications. Unless otherwise stated, reactions were performed at 30°C for 2 h and then stopped by addition of 100 μl of 3.6 M ammonium acetate (NH4OAc) and 20 μg of glycogen (20). Samples were precipitated with 450 μl of cold anhydrous ethanol at −80°C overnight. Samples were centrifuged at 4°C for 30 min. The pellets were then washed with 70% ethanol, air dried, and resuspended in 5 μl of water with 5 μl of deionized formamide loading dye (Ambion). Samples were boiled for 10 min at 95°C for denaturation prior to loading onto the 7 M urea–10% polyacrylamide–1× Tris-borate-EDTA (TBE) denaturing gel. Gels were dried at 80°C for 2 h, exposed overnight on a phosphorimager cassette and scanned using Storm 840 scanner (GE Healthcare). Data were analyzed using ImageQuant TL software (GE Healthcare). Telomerase activity was quantified by determining the number of counts in each lane relative to the number of WT counts. Telomerase processivity quantitation was done based on the “>15 repeats” method as described previously (15). In our study, the lower limit of high processivity is represented by products with 10 or more telomeric repeats added. Values are expressed relative to that for the WT, which was set to 100% RAP.
Pulse-chase time course experiment.
The pulse-chase experiment was performed and quantified as previously described (2). Reactions were stopped and processed as described above.
Immunofluorescence experiment.
The immunofluorescence experiment was performed as previously described with a few modifications (21). Briefly, HeLa cells overexpressing hTERT-WT or hTERT-variants and hTR were extracted with 0.1% Triton X-100 for 5 min on ice, followed by fixation in 4% formaldehyde–PBS for 15 min at room temperature (RT). Cells were permeabilized again for 10 min on ice and blocked in 5% bovine serum albumin (BSA) for 1 h at RT. Cells were probed using goat anti-TERT (Santa Cruz) and mouse anticoilin (Michael Terns) at 1:50 and 1:10,000 dilutions in PBG (1% cold fish water gelatin, 0.5% BSA, 1× PBS), respectively. Coverslips were washed with PBG and immunostained with secondary antibodies (1:100) conjugated to fluorescein isothiocyanate (FITC) (donkey anti-goat IgG; Jackson ImmunoResearch Lab, Inc.) or Cy3 (donkey anti-mouse; Jackson ImmunoResearch Lab, Inc.). Coverslips were washed with PBG and mounted in Vectashield with DAPI (Vector Laboratories). Images were captured using an Axio Imager M1 (63×; Carl Zeiss, Jena, Germany).
FISH.
hTR-telomere fluorescence in situ hybridization (FISH) was performed as previously described (5), using HeLa cells coexpressing hTERT-WT or hTERT-variants and hTR (22), three different Cy3-conjugated hTR probes (23), and an Oregon green-conjugated telomeric probe (8). Cy3 monoreactive dye was from GE Healthcare (Piscataway, NJ), Oregon green 488 from Invitrogen, and probes from Operon (Huntsville, AL). Images were captured using an Axio Imager M1 microscope (63×; Carl Zeiss, Jena, Germany).
ChIP.
Chromatin immunoprecipitation (ChIP) was performed using HeLa cells overexpressing 3×FLAG-tagged mutant and WT hTERTs as previously described (24) with the following modification. Ten picomoles of Alu and telomeric (T2AG3)3 probes were end labeled with 10 pmol of [γ-32P]ATP (PerkinElmer) and purified using G-25 columns (GE Healthcare). Quantitation of telomere binding was done using the formula (telo IP/telo input)/(Alu IP/Alu input) (25), and values are expressed relative to WT telomerase binding to telomeres.
Quantitative fluorescence in situ hybridization analysis and signal free ends.
Metaphase spread analysis for detection of signal free ends (SFE) was performed as described previously (2, 5). Imaging was performed using an Axio Imager M1 microscope (63×; Carl Zeiss, Jena, Germany). Quantitative analysis of telomere length and SFE was performed with TFL-Telo (Peter Lansdorp).
Apoptosis analysis by fluorescence-activated cell sorting (FACS).
Retrovirally infected hTERT-HA5 cells were grown to confluence in a 10-cm dish. Cell medium was collected and combined with trypsinized cells from the plate. Cells were treated with propidium iodide (Sigma-Aldrich, St. Louis, MO) and annexin V-fluorescein isothiocyanate (BD Bioscience) using a BD LSRFortessa analyzer at the Lady Davis Institute Flow Cytometry Facility. Data were analyzed using BD FACSDiva dongle software.
Immunofluorescence combined with FISH for TIF detection.
For visualization of telomere dysfunction-induced foci (TIF), HA5 cells were grown on coverslips for 24 h, and then the previously described protocol (23) for detection of telomeres by FISH was carried out using the Cy3-labeled telomeric peptide nucleic acid (PNA) probe (Cy3-TelC) (Panagene). Following the last wash from the FISH protocol, coverslips were washed three times with 1× PBS, 10 min per wash, to remove any residual deionized formamide. Coverslips were preblocked in 3% BSA at RT for 1 h and then incubated with mouse anti-γH2AX (Upstate Cell Signaling Solutions) at 1:200 in 3% BSA–1× PBS-T for 1 h at RT. Coverslips were washed three times with 1× PBS–3% BSA, 5 min per wash, and blotted with FITC-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch Lab, Inc.) at a 1:100 dilution for 30 min at RT in 1× PBS-T, washed three times for 5 min in 1× PBS, and mounted in Vectashield with DAPI (Vector Laboratories).
Telomere restriction fragment length (TRF) analysis.
Analysis of telomere length was performed as previously described (2), with a few modifications. Briefly, genomic DNA was extracted from HeLa cells using 10 mM Tris (pH 8), 0.1 M NaCl, 2.5 mM EDTA, and 0.5% SDS), followed by overnight digestion with 0.1 mg/ml proteinase K. A 10% sample volume of 3 M sodium acetate (pH 5.2) was added to samples, samples were mixed, and 1 sample volume of isopropanol was added. DNA was acquired using a pipette tip and transferred to a new tube containing 75% ethanol (the process was repeated three times). DNA was washed in 100% ethanol, air dried, resuspended in 1× buffer 4 (NEB), and digested overnight with HinfI/RsaI (NEB). The next day, samples were supplemented with fresh enzymes and buffer and digested for 8 h. Electrophoresis of the digested DNA was carried out in a 0.7% agarose gel in 0.5× TBE at 70 V for 16 h. The gel then was denatured for 30 min in 0.5 N NaOH and 1.5 M NaCl and neutralized for 30 min in 1 M Tris-HCl (pH 7.5) and 1.5 M NaCl. The gel was dried at RT for 30 min and then at 50°C for 1 h. Hybridization was carried out at 37°C overnight, in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's solution, and 0.1× P-wash, in the presence of [γ-32P]ATP-labeled (C3TA2)3 probe. The gel was washed at RT for 15 min in 2× SSC, followed by 3 washes for 10 min in 0.1% SSC and 0.1% SDS. The gel was autoradiographed on X-ray film (Kodak) or on a phosphorimager cassette for 24 h and scanned using the Storm 840 imager (GE Healthcare). A [γ-32P]ATP-labeled λ-HindIII molecular weight marker was used to determine telomere length. Using ImageQuant TL software (GE Healthcare), 40 boxes were created for each sample lane. Mean telomere length was obtained using the formula Σ(ODi)/Σ(ODi/Li), where OD is the band intensity (volume) in each box (or position i) and L is the DNA size in the box (or position i).
Statistical analysis.
Statistical significance was calculated using the program GraphPad Prism. Parameters were analyzed using one-way analysis of variance (ANOVA) and the Dunnett posttest, which compares all samples to the control sample (WT). Significance level was determined using 95% confidence intervals. A P value of ≥0.05 is considered not significant, a value of 0.01 to 0.05 is significant, a value of 0.001 to 0.01 is very significant, and a value of 0.0001 to 0.001 is extremely significant.
RESULTS
hTERT-V763S and hTERT-L805A expressed in cells reconstitute lower levels of telomerase activity and altered levels of repeat addition processivity compared with wild-type telomerase.
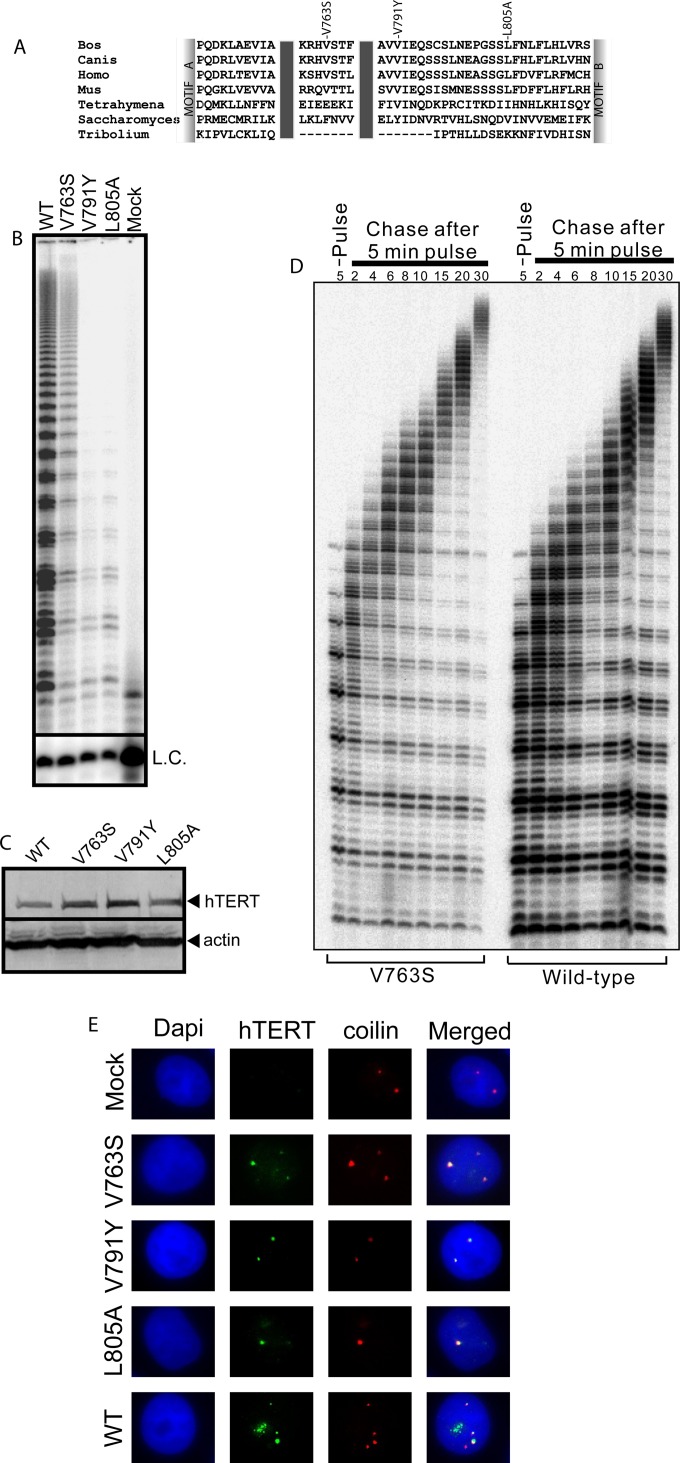
Sequence alignment of the human telomerase IFD with the corresponding region from various organisms using Clustal W (Fig. 1A) illustrates the high degree of sequence conservation of this domain. In order to identify IFD residues involved in the regulation of telomerase-specific functions, we mutated two well-conserved hydrophobic residues, valine 763 and leucine 805, to serine and alanine, respectively, in an effort to reduce enzyme activity and/or processivity. Valine 763 was mutated to serine based on an earlier published sequence alignment (4) in which the corresponding Saccharomyces cerevisiae TERT amino acid is a serine residue. In vitro, yeast telomerase is less processive than human telomerase (26), and we predicted that this substitution might negatively impact telomerase catalytic functions. The S. cerevisiae amino acid corresponding to hTERT L805 is a valine, which is similar to a leucine. We thus mutated L805 to alanine. We also included, as a control, another IFD variant, hTERT-V791Y, previously reported to display reduced enzyme activity and processivity (5). We overexpressed hTERT-V763S, hTERT-V791Y, hTERT-L805A, or wild-type hTERT (hTERT-WT), and hTR, in HEK 293 cells (often referred to as supertelomerase cells) (5, 27, 28). Next, using the direct primer extension assay, we assessed the ability of the hTERT variant complexes to reconstitute enzymatic functions (Fig. 1B) and quantified levels of DNA synthesis and RAP relative to levels reconstituted by hTERT-WT (set at 100%). Western analysis confirmed that the hTERT variants and hTERT-WT were expressed at similar levels (Fig. 1C). The hTERT-V791Y mutant enzyme displayed defects in DNA synthesis and RAP (DNA synthesis, 9.63% ± 3.31% [mean ± standard deviation]; RAP, 58.66% ± 8.82%), as previously reported (5). Similarly to the hTERT-V791Y mutant enzyme, hTERT-L805A also displayed defects in DNA synthesis (33.18% ± 9.08%) and RAP (46.72% ± 0.82%). Interestingly, the hTERT-V763S mutant enzyme, despite showing defects in DNA synthesis (37.80% ± 11.62%), had 1.51-fold- ± 0.07-fold-higher levels of RAP than the WT enzyme. Using a pulse-chase time course experiment, we observed that hTERT-V763S displays a 1.76- ± 1.20-fold increase in the rate of RAP compared to the WT enzyme, thus confirming that this mutant enzyme exhibits true processivity rather than extending the telomeric substrate in a distributive manner (Fig. 1D).
FIG 1.
Specific IFD residues regulate telomerase activity and processivity. (A) Sequence alignment of TERT IFDs from selected organisms (Bos taurus, Canis lupus familiaris, Homo sapiens, Mus musculus, Tetrahymena thermophila, Saccharomyces cerevisiae, and Tribolium castaneum) was performed using Clustal W2. Labels above the alignment indicate the valines at amino acids 763 and 791 as well as the leucine at 805 in hTERT and the corresponding residues at equivalent positions in the other TERTs. (B) Representative gel from a direct primer extension experiment using mutant and WT telomerases expressed and reconstituted with coexpressed hTR in HEK 293 cells (supertelomerase system) for assessment of telomerase total DNA synthesis (activity) and repeat addition processivity levels using the telomeric substrate (T2AG3)3. L.C., loading and recovery control (T2AG3)2. Results are representative of three experiments. (C) Representative immunoblot of hTERT using 50 μg of total protein from HEK 293 supertelomerase cells used for panel B to confirm equal hTERT protein expression. Blots were probed using antibodies against hTERT or the loading control actin. Results are representative of three experiments. (D) A representative pulse-chase time course experiment for the hTERT-V763S mutant enzyme and hTERT-WT was conducted to confirm processive telomeric substrate extension and to measure the relative rate of repeat addition processivity. During the 5-min pulse reaction, hTERT-V763S and WT telomerases reconstituted in HEK 293 cells were incubated with the telomeric substrate (T2AG3)3 to incorporate [α-32P]dGTP into any new telomeric repeats synthesized. During the chase reaction, excess nonradioactive dGTP was added to initiate several chase reactions. Reactions were stopped at the indicated time points (in minutes). L.C., loading and recovery control [(T2AG3)2]. Results are representative of three experiments. (E) Representative coimmunofluorescence experiment using mutant and WT telomerases overexpressed and reconstituted with coexpressed hTR in HeLa cells (supertelomerase system). An untransfected sample (mock) was used as a negative control to assess nonspecific signals. DAPI was used for detection of the nucleus. Fixed cells were probed using antibodies against hTERT and coilin to visualize telomerase and Cajal bodies, respectively. Results are representative of two experiments.
We confirmed that the observed defects were not due to inefficient assembly of the mutant proteins with hTR. Since localization of hTERT to Cajal bodies is dependent on its assembly with hTR (19, 29, 30), we performed coimmunofluorescence experiments using antibodies against hTERT and coilin, a marker of Cajal bodies (Fig. 1E). We observed colocalization of WT and mutant hTERTs with coilin (Fig. 1E), confirming that the hTERT-V763S, hTERT-V791Y, and hTERT-L805A are competent in holoenzyme assembly. We also assessed the translocation efficiency (31) of the mutant hTERTs and confirmed that the observed changes in activity and processivity were not due to differences in the rate-limiting translocation step (data not shown).
Defects in DNA synthesis and processivity of hTERT-L805A but not hTERT-V791Y can be rescued by overexpression of TPP1-POT.
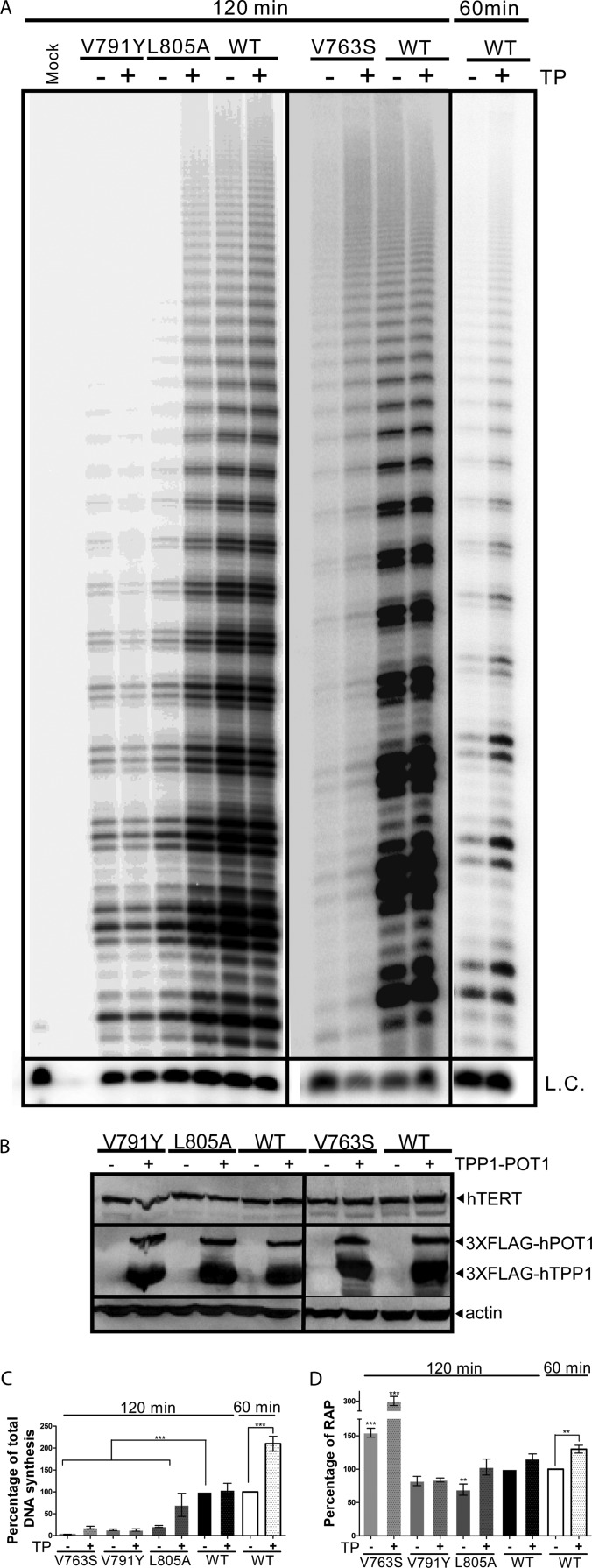
The role of TPP1-POT1 in the stimulation of telomerase activity and RAP is well established (15–17, 31–33). Stimulation of telomerase by TPP1-POT1 depends on the association of telomerase with TPP1-POT1 and is used as a biochemical indicator for this interaction (19). Recently, the hTERT TEN domain was identified as directly interacting with TPP1 to mediate telomerase recruitment to telomeres (19). Based on our previous observations that hTERT-V791Y is defective in associating with telomeres (5), we investigated the possibility that TPP1-POT1 stimulates the function of hTERT-V791Y, hTERT-L805A, or hTERT-V763S. Stimulation of telomerase requires both TPP1 and POT1 (31); we therefore coexpressed TPP1 and POT1 in HEK293 cells expressing the various supertelomerase enzymes and performed a direct primer extension assay (Fig. 2A). Protein levels were assessed by Western blotting to confirm equal expression of TPP1, POT1, and the hTERT variants in the different cells (Fig. 2B). We observed a partial rescue of the levels of DNA synthesis (70.19% ± 26.29% versus 20.84% ± 2.08%) and complete rescue in RAP (103.24% ± 11.99% versus 69.34% ± 8.41%) reconstituted by hTERT-L805A in the presence of TPP1-POT1 (Fig. 2C). However, levels of DNA synthesis and RAP reconstituted by hTERT-V791Y were similar in the presence or absence of TPP1-POT1 overexpression (Fig. 2C). Overexpression of TPP1-POT1 and hTERT-V763S led to an enhancement of both total DNA synthesis and RAP (18.33% ± 2.86% versus 2.63% ± 0.42 and 300.34% ± 9.30% versus 154.63% ± 6.63%, respectively) (Fig. 2C and D). Furthermore, stimulation of the WT telomerase total DNA synthesis and RAP by TPP1-POT1 (210.00% ± 16.87% and 130.00% ± 5.89%, respectively, versus 100%) (Fig. 2C and D) was observed when the incubation time was reduced to 60 min, consistent with published reports (15–17, 31–33). However, due to the low levels of catalytic activity of the hTERT V791Y and L805A variants, a 120-min incubation was necessary to obtain a quantifiable signal for measurement of telomerase activity or processivity.
FIG 2.
Defects in DNA synthesis and RAP of hTERT-L805A but not hTERT-V791Y can be rescued by overexpression of TPP1-POT. (A) Representative gel of a direct primer extension experiment using mutant and WT telomerases expressed and reconstituted with coexpressed hTR in HEK 293 cells (supertelomerase system) in the presence or absence of coexpressed TPP1-POT1 (TP) for assessment of telomerase total DNA synthesis (activity) and repeat addition processivity levels using the telomeric substrate (T2AG3)3. L.C., loading and recovery control (T2AG3)2. (B) Representative immunoblot of hTERT, 3×FLAG-hTPP1, and 3×FLAG-hPOT1 using 50 μg of total protein from the HEK 293 supertelomerase cells used for panel A to confirm equal protein expression. Blots were probed using antibodies against hTERT, FLAG (to detect hTPP1 and hPOT1) or the loading control actin. (C) Quantitation of relative total DNA synthesis reconstituted by mutant and WT telomerases expressed in HEK 293 cells as for panel A was performed by measuring the total incorporation of radiolabeled dGTP into the extension products. Values are normalized to the L.C. DNA synthesis of WT telomerase in the absence of coexpressed TPP1-POT1 was set to 100%. A P value of 0.0001 to 0.001 is extremely significant (***). Values are from three experiments. (D) Quantitation of repeat addition processivity (RAP) reconstituted by mutant and WT telomerases expressed in HEK 293 cells as for panel A was performed by measuring the extension products with 10 or more telomeric repeats over total product. RAP of WT telomerase in the absence of coexpressed TPP1-POT1 was set at 100%. A P value of 0.001 to 0.01 is very significant (**), and a value of 0.0001 to 0.001 is extremely significant (***). Values are from three experiments.
These data suggest that interaction with TPP1 may be completely abolished by the valine-to-tyrosine amino acid substitution at position 791, while a less efficient TPP1-hTERT-L805A interaction can be partially compensated by the overexpression of TPP1-POT1, as reflected by an increase in the level of total DNA synthesis and RAP. The enhancement of DNA synthesis and processivity of hTERT-V763S by TPP1-POT1 suggests that alteration of the valine at position 763 may also partially affect interaction with TPP1.
Specific IFD residues regulate telomerase localization and recruitment to telomeres.
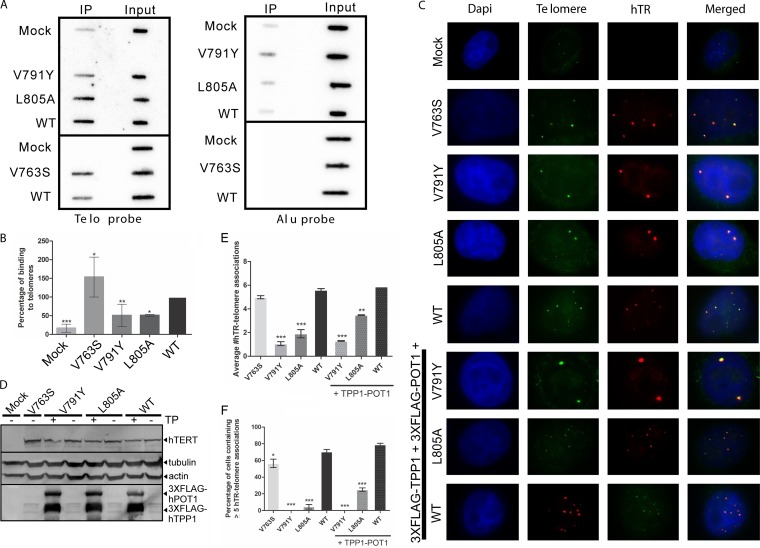
The binding of telomerase to the telomere is a crucial step for nucleotide addition to chromosome ends by telomerase. Thus, we first assessed the binding of the hTERT IFD variants and hTERT-WT to telomeres by chromatin immunoprecipitation (ChIP). We cotransfected HeLa cells with hTR and 3×FLAG-tagged hTERT (WT or variants), performed an IP against the FLAG tag, and isolated and analyzed the coprecipitated DNA. Telomeric DNA was detected using a radiolabeled (T2AG3)3 probe, and a probe against Alu repeats was used as a control (Fig. 3A). While hTERT-V763S displays an increased telomere binding affinity (153.39% ± 53.37%), both hTERT-V791Y (50.78% ± 29.98%) and hTERT-L805A (58.98% ± 2.51%) display significantly decreased binding to telomeres compared with hTERT-WT (set at 100%) (Fig. 3B). Our data suggest that the reduced DNA synthesis and RAP levels for the latter two variants may be due to impaired interaction with the telomere, resulting from potential defects in telomerase recruitment.
FIG 3.
Overexpression of TPP1-POT1 partially rescues the defective localization of hTERT-L805A, but not hTERT-V791Y to telomeres. (A) Representative ChIP experiment using M2 anti-FLAG antibody against 3×FLAG-tagged hTERT mutants or hTERT-WT coexpressed with hTR in HeLa cells (supertelomerase system). Immunoprecipitated chromatin was probed with oligonucleotide probes against telomeric DNA sequences [telo; (T2AG3)3] or Alu repeats. (B) Quantitation of binding of mutant telomerases and WT telomerase to telomeric DNA, as represented in panel A. Quantitation was performed by normalizing to input, and nonspecific binding to Alu sequences was taken into account for the calculations. Binding of hTERT-WT to telomeres was set at 100%. A P value of 0.01 to 0.05 is significant (*), a value of 0.001 to 0.01 is very significant (**), and a value of 0.0001 to 0.001 is extremely significant (***). Values are from three experiments. (C) Representative hTR-FISH experiment using mutant and WT telomerases expressed and reconstituted with coexpressed hTR in HeLa cells (supertelomerase system) with or without coexpressed TPP1-POT1. An untransfected sample (mock) was used as a negative control to assess nonspecific signals. DAPI was used for detection of the nucleus. An Oregon green-conjugated telomeric probe, (T2AG3)3, was used for the detection of telomeres. Three different Cy3-labeled hTR probes were used to detect telomerase localization. Yellow foci indicate colocalization of hTR (telomerase) at the telomeres. (D) Immunoblot of hTERT, 3×FLAG-hTPP1, and 3×FLAG-hPOT1 using 50 μg of total protein from the HeLa supertelomerase cells used for panel C to confirm equal protein expression. Blots were probed using antibodies against hTERT, FLAG (to detect hTPP1 and hPOT1), or the loading controls actin and tubulin. An untransfected sample (mock) was used as a negative control for nonspecific signals. (E) Quantitation of average number of hTR-telomere associations per cell, as represented in panel C. A P value of 0.001 to 0.01 is very significant (**), and a value of 0.0001 to 0.001 is extremely significant (***). Values are from two experiments, with ≥200 cells counted in each. (F) Quantitation of percentage of cells containing 5 or more hTR-telomere associations, as represented in panel C. A P value of 0.01 to 0.05 is significant (*), and a value of 0.0001 to 0.001 is extremely significant (***). Values are from two experiments, with ≥200 cells counted in each.
We also investigated the ability of the hTERT IFD variant complexes to localize to the telomeres by assessing the localization of hTR to the telomere using fluorescence in situ hybridization (FISH) experiments. Localization of hTR to the telomere, which is dependent on the presence of hTERT at the telomere (23), has been used extensively as a marker of telomerase localization or recruitment to the telomere (5, 8, 11, 14, 29, 34). Using HeLa cells overexpressing hTERT (mutant or WT) and hTR, we performed FISH using an Oregon green fluorescently labeled telomere sequence-specific probe and three different Cy3 fluorescently labeled hTR-specific probes to assess the localization of telomerase to the telomeres (Fig. 3C). Equal protein expression was verified by Western analysis (Fig. 3D). Quantitation of the average number of hTR-telomere associations per cell revealed that HeLa cells expressing hTERT-V791Y had significantly fewer hTR-telomere associations (1.07 ± 0.17) than cells expressing hTERT-WT (5.60 ± 0.11), which is comparable to the previously reported number of hTR-telomere associations for hTERT-V791Y (1.86 ± 0.18) and hTERT-WT (7.14 ± 0.74) (5) (Fig. 3E). HeLa cells expressing hTERT-L805A also displayed significantly fewer hTR-telomere associations (1.91 ± 0.35) than the WT enzyme, whereas colocalization of hTR and telomeres in cells expressing the hTERT-V763S enzyme (4.99 ± 0.13) was similar and not statistically different from the number of colocalizations in hTERT-WT-expressing cells (Fig. 3E). hTR was observed at five or more telomeres in 70.50% ± 2.83% of HeLa cells coexpressing hTERT-WT and in 56.50% ± 4.95% of hTERT-V763S-expressing cells (Fig. 3F). The percentage of cells containing hTR associations with five or more telomeres decreased to 0.00% ± 0.00% in cells coexpressing hTERT-V791Y and hTR and to 4.50% ± 2.83% in cells coexpressing hTERT-L805A and hTR (Fig. 3F). These results, and those obtained with ChIP, indicate that IFD domain V791Y and L805A mutant telomerase enzymes are defective in recruitment to telomeres, possibly due to disrupted interaction with TPP1.
Overexpression of TPP1-POT1 partially rescues the defective localization of hTERT-L805A but not hTERT-V791Y to telomeres.
Based on the known role of TPP1 in recruitment of telomerase to telomeres (8, 9) and our results showing stimulation of telomerase catalytic activity of the IFD domain hTERT-L805A mutant enzyme by TPP1-POT1, we addressed the question of whether overexpression of TPP1-POT1 or of TPP1 or POT1 individually could stimulate recruitment of hTERT-L805A and hTERT-V791Y to the telomeres in HeLa cells (Fig. 3C). Overexpression of either TPP1 or POT1 individually with telomerase (WT or variants) had no significant effect on telomere localization compared to the overexpression of telomerase alone (data not shown). Colocalization of hTR to telomeres in HeLa cells coexpressing hTR and WT hTERT was similar in the absence and presence of overexpressed TPP1-POT1 (average of 5.60 ± 0.11 versus 5.88 ± 0.00 colocalizations/cell, respectively) (Fig. 3E). hTR was observed at five or more telomeres in 70.50% ± 2.83% of HeLa cells coexpressing WT telomerase and in 78.99% ± 1.56% of hTERT-WT-expressing cells also coexpressing TPP1 and POT1 (Fig. 3F). Overexpression of TPP1 and POT1 significantly increased the number of hTR-telomere localizations in cells expressing hTERT-L805A (average of 3.48 ± 0.03 versus 1.91 ± 0.35 colocalizations/cell in the absence of TPP1 and POT1) but not to levels observed in cells expressing hTERT-WT (Fig. 3E). The percentage of cells expressing hTERT-L805A containing five or more hTR-telomere associations increased when TPP1-POT1 were also coexpressed (25.00% ± 2.12%) versus 4.50% ± 2.83% (Fig. 3F). However, the low average number of hTR-telomere colocalizations in cells expressing hTERT-V791Y could not be rescued by overexpression of TPP1-POT1 (1.29% ± 0.03% versus 1.07% ± 0.17%) (Fig. 3E). The overexpression of TPP1-POT1 in cells expressing hTERT-V791Y did not increase the percentage of cells containing five or more hTR-telomere colocalizations (Fig. 3F). These results indicate that the hTERT-V791Y variant is defective in recruitment to the telomeres, most probably due to its inability to interact with TPP1. This finding, together with the absence of complete recovery of hTERT-L805A localization to telomeres upon overexpression of TPP1-POT1, suggests that residues of the IFD domain can mediate telomerase recruitment to telomeres in a TPP1-dependent manner. The enhancement of DNA synthesis and processivity of hTERT-V763S by TPP1-POT1 is consistent with a role of TPP1-POT1 in regulating activity/processivity (15, 17, 31–33, 35).
Limited-life-span cells expressing IFD mutant enzymes display growth defects accompanied by an increase in apoptosis, signal-free ends, and DNA damage at telomeres.
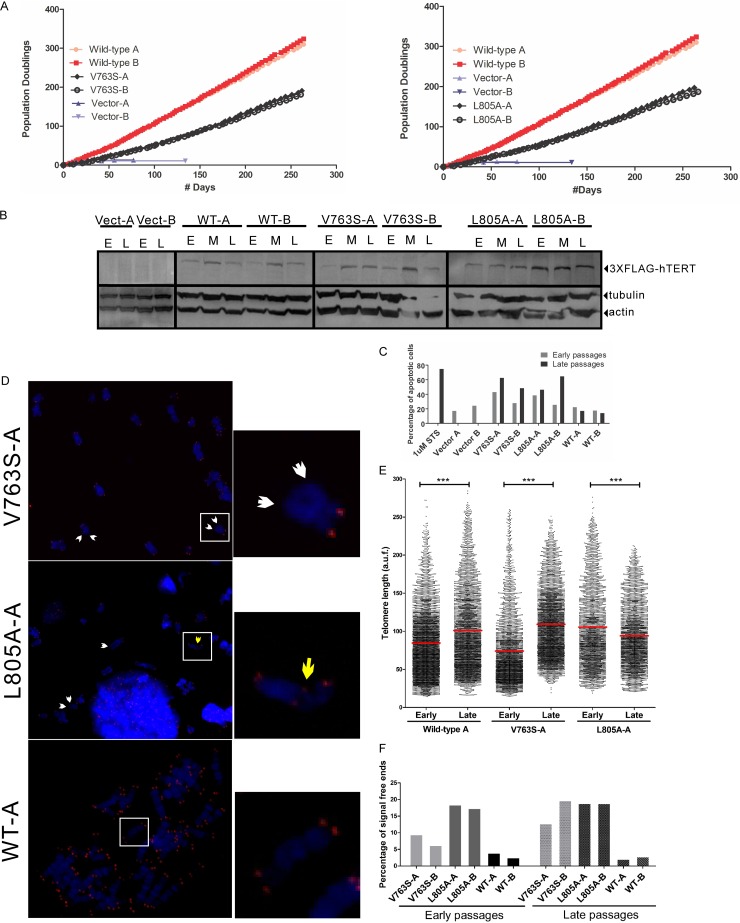
We previously reported that the IFD domain variant hTERT-V791Y is unable to immortalize limited-life-span HA5 cells, resulting in high levels of apoptosis accompanied by an increase in the frequency of short telomeres (5). To determine if other IFD residues regulate cell survival and short telomere maintenance, we retrovirally infected HA5 cells with hTERT-V763S, hTERT-L805A, or hTERT-WT. HA5 cells are life span-limited hTERT-negative human embryonic kidney cells transformed with simian virus 40 (SV40) and thus are senescence defective but can undergo apoptosis resulting from the inability to maintain telomere length in the absence of a functional telomerase enzyme (5, 6, 36). Following infection, we selected two clones expressing each hTERT variant or hTERT-WT and followed their growth in culture for ∼300 days (Fig. 4A). The expression of hTERT, assessed by Western blotting, was comparable between WT- and variant-expressing cells and remained relatively constant at early (20 days postinfection), middle (100 days postinfection) and late (200 days postinfection) passages (Fig. 4B). When the number of population doublings was plotted against number of days (Fig. 4A), we observed a significant growth defect for both hTERT-V763S- and hTERT-L805A-expressing cells compared to hTERT-WT-expressing cells.
FIG 4.
hTERT-V763S- and hTERT-L805A-expressing cells display growth defects, an increase in signal free ends and apoptosis. (A) Growth curves of HA5 cells expressing hTERT-V763S, hTERT-L805A, or hTERT-WT. Growth of two clones for each hTERT variant or hTERT-WT (clones A and B) was monitored for >250 days. Two clones of cells infected with empty vector control (clones A and B) did not survive past 140 days. For comparison purposes, data for hTERT-WT-expressing clones and hTERT vector-containing clones are shown in both graphs. (B) Representative immunoblot of hTERT using 50 μg of total protein from HA5 cells expressing hTERT variants or hTERT-WT and assessed for growth, as reported in panel A, to confirm similar protein expression at early (E), middle (M), and late (L) passages. Blots were probed using antibodies against FLAG (3×FLAG-hTERT) or the loading controls actin and tubulin. Results are representative of three experiments. (C) FACS analysis was performed at early (20 days postinfection) and late passages (200 days postinfection) of HA5 cells containing vector or expressing hTERT mutant enzymes or hTERT-WT (clones A and B for each) to assess the percentage of apoptotic cells. hTERT-WT-expressing HA5 cells treated with staurosporine (STS) were used as a positive control for cell death. (D) Representative metaphase spreads were followed by Q-FISH analysis of HA5 cells expressing hTERT mutant enzymes or hTERT-WT at early and late passages to assess the levels of signal free ends (short telomeres). Individual chromosomes were stained with DAPI, and the telomeric DNA was probed with a Cy3-labeled PNA telomeric probe. White arrows indicate signal free ends (short telomeres). The yellow arrow indicates telomeric fusion. Magnification, ×63. (E) Quantitation of telomere length in HA5 cells expressing hTERT mutant enzymes or hTERT-WT (clone A from each), expressed as arbitrary units of fluorescence (a.u.f.). The red line indicates the mean telomere length. A P value of 0.0001 to 0.001 is extremely significant (***). The total number of telomeres analyzed ranged from 3,500 to 4,000. (F) Quantitation of percentage of signal free ends at early and late passages. Two clones for each hTERT variant and hTERT-WT were analyzed. At least 1,000 telomeres were counted per clone for each passage.
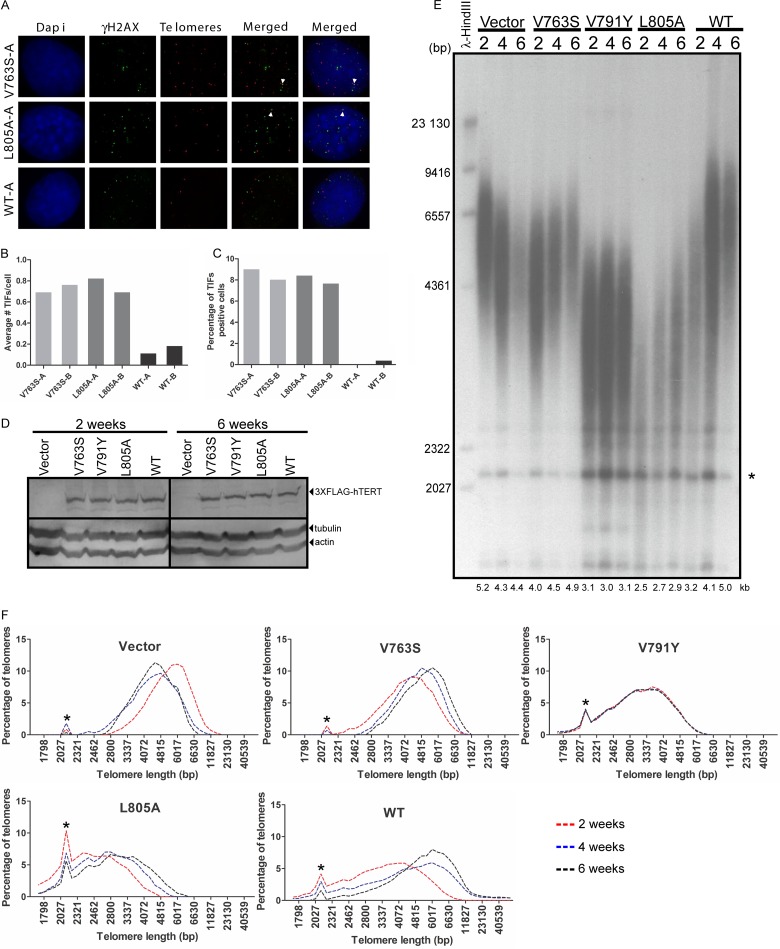
To determine if the decrease in cell growth rate was due to an increase in apoptosis, we performed fluorescence-activated cell sorting (FACS) analysis (Fig. 4C). hTERT-V763S- and hTERT-L805A-expressing cells displayed high levels of apoptosis compared to hTERT-WT-expressing cells that were particularly evident at 200 days postinfection. Based on the defects of hTERT-L805A and hTERT-V791Y in binding to telomeres (ChIP) and associating with telomeres (hTR-FISH) and the increase in short telomeres previously observed in hTERT-V791Y-expressing cells (5), we predicted that an increase in cellular apoptosis possibly resulted from the inability of IFD domain hTERT mutant enzymes to be recruited to short telomeres. We prepared metaphase spreads of hTERT-expressing cells at 20 and 200 days postinfection, followed by quantitative FISH (Q-FISH) analysis using a PNA-telomeric probe and costaining with DAPI for chromosome visualization (Fig. 4D). Stable expression of hTERT-WT and hTERT-V763S led to an increase in the mean telomere length of late-passage cells compared to early-passage cells (84.65 to 101.10 arbitrary units of fluorescence [AUF] and 74.08 to 109.20 AUF, respectively) while hTERT-L805A-expressing cells failed to elongate telomeres, resulting in a decrease from 105.5 AUF at early passage to 94.11 AUF at late passage (Fig. 4E). We also quantified the percentage of chromosome ends that lack a telomeric signal, or signal-free ends (SFEs), as a measure of short telomeres and found that persistently higher levels of SFEs could be detected in cells expressing hTERT-V763S or hTERT-L805A than in the hTERT-WT-expressing cells (Fig. 4F). At late passages (∼200 days postinfection), hTERT-V763S clone A and hTERT-L805A clone A displayed frequencies of 12.77% and 18.84% SFEs, respectively, compared to hTERT-WT-expressing cells, which had only 2.03% SFEs (Fig. 4E). Levels of SFEs were higher in hTERT-L805A-expressing cells than in hTERT-V763S-expressing cells, consistent with lower binding of hTERT-L805A to telomeres than of hTERT-V763S, which we observed by ChIP and hTR-FISH. Importantly, at late passages, hTERT-L805A-expressing cells also displayed observable telomeric fusion events (Fig. 4D, middle), which likely resulted from the inability of the shortened telomere to perform the end-capping function.
When telomeres shorten sufficiently to affect T-loop formation, telomere uncapping occurs, and telomeres are recognized as double-strand breaks by the DNA damage repair machinery. This leads to the formation of telomere dysfunction-induced foci (TIF). Since we observed high levels of SFEs in hTERT variant-expressing cells, we measured TIF levels in HA5 cells expressing these variants at 200 days postinfection using combined immunofluorescence and FISH (Fig. 5A). An antibody against γ-H2AX foci in combination with a Cy3-labeled telomeric PNA probe was used to assess colocalization of DNA damage at the telomeres. Quantitation of the average number of TIF per cell and percentage of TIF-positive cells (≥3 TIF/cell) indicates that both parameters were elevated in hTERT-V763S (clone A, 9.09% TIF-positive cells and 0.70 TIF/cell; clone B, 8.18% TIF-positive cells and 0.77 TIF/cell) and hTERT-L805A (clone A, 8.49 TIF-positive cells and 0.83 TIF/cell; clone B, 7.72% TIF-positive cells and 0.70 TIF/cell) compared with the hTERT-WT expressing cells (clone A, 0.00% TIF-positive cells and 0.12 TIF/cell; clone B, 0.45% TIF-positive cells and 0.19 TIF/cell) (Fig. 5B and C). These data provide evidence that the residues at position 763 and 805 are critical in regulating short-telomere maintenance and cellular survival.
FIG 5.
hTERT-V763S- and hTERT-L805A-expressing HA5 cells display higher levels of DNA damage at telomeres compared to hTERT-WT-expressing HA5 cells. (A) Representative combined FISH-immunofluorescence (IF) experiment for the detection of DNA damage at the telomeres in HA5 cells expressing hTERT mutant enzymes or hTERT-WT at ∼200 days postinfection (late passage). DAPI stains the nucleus. FITC-conjugated donkey anti-mouse antibody against anti-γH2AX antibody detects γH2AX (IF). A Cy3-labeled PNA probe against the telomeres was used for FISH. Yellow foci represent colocalization of γH2AX at the telomeres, which represents telomere dysfunction-induced foci (TIF). (B) Quantitation of average number of TIF per cell. At least 200 cells were counted per clone. Two clones for each hTERT variant and hTERT-WT were analyzed. (C) Quantitation of percentage of TIF positive cells. Cells containing 3 or more TIF were counted as positive. At least 200 cells were counted per clone. Two clones for each hTERT variant and hTERT-WT were analyzed. (D) Immunoblot of hTERT using 100 μg of total protein from HeLa cells expressing hTERT-variants, hTERT-WT and vector control, assessed for telomere elongation, to confirm similar protein expression at 2 and 6 weeks postinfection. Blots were probed using antibodies against hTERT or the loading controls actin and tubulin. (E) Representative TRF analysis of genomic DNA digested with HinfI/RsaI to evaluate telomere length of HeLa cells at 2, 4, and 6 weeks postinfection. Average telomere length at each time point is shown below the gel. λ-HindIII marker sizes are shown on the left of the gel. The asterisk represents signal due to internal telomeric sequences. (F) Plot of telomere length versus percentage of telomeres to assess telomere length distribution from the TRF gel shown in panel E. The asterisk represents signal due to internal telomeric sequences.
The hTERT-V763S mutant enzyme does not appear to be defective in enzyme processivity, binding to telomeres (ChIP), or localization to telomeres (hTR-FISH). Interestingly, HA5 cells expressing hTERT-V763S displayed significantly higher levels of short telomeres than hTERT-WT-expressing cells. Therefore, we speculated that the hTERT-V763S enzyme might be defective in elongating short telomeres but not in maintenance of average telomere length. To determine the effects of the mutant telomerases on telomere maintenance, we stably expressed the mutant and WT hTERTs in HeLa cells, which have longer telomeres than HA5 cells. We confirmed equal hTERT expression in the different clones by Western analysis (Fig. 5D) and measured the effect of mutant and WT telomerase expression on telomere length by telomeric restriction fragment length (TRF) analysis (Fig. 5E). The telomere length of hTERT-WT-expressing cells progressively increased over 6 weeks, consistent with a previous report (19). Similarly, the hTERT-V763S-expressing cells displayed a progressive increase (from 4.0 to 4.9 kb) in telomere length, suggesting that this mutant enzyme is not defective in telomere elongation per se (Fig. 5E and F). In contrast, cells expressing hTERT-L805A displayed only a slight increase in telomere length (from 2.5 to 2.9 kb) (Fig. 5F), and neither the empty vector control nor hTERT-V791Y induced telomere elongation (Fig. 5E and F). These results strongly suggest that hTERT-V763S is capable of extending telomeres with a longer starting length, such as in HeLa cells (compared to HA5 cells). The growth defects observed in hTERT-V763S-expressing HA5 cells might be due to the inability of this mutant to elongate/maintain telomeres with short initial lengths. Altogether, our data support an important role for the IFD in the maintenance of short telomeres.
DISCUSSION
Telomerase biogenesis, trafficking, and recruitment to telomeres are prerequisites for the enzyme's ability to act at telomeres in a processive manner and immortalize cells. In this study, we sought to determine the role of the unique human telomerase IFD in the regulation of telomerase-specific functions in vitro as well as in vivo and demonstrated that this region of hTERT is involved in mediating telomerase activity, processivity, and recruitment to telomeres in a TPP1-dependent manner.
A recent study identified specific basic residues within the hTERT TEN domain that directly interact with the acidic TEL patch of TPP1 (19). Although an interaction interface between the hTERT IFD residues and the TEL-patch of TPP1 OB fold is improbable due to the nonpolar nature of the valine and leucine residues (V763, V791, and L805), the hTERT IFD could possibly interact indirectly with TPP1. Several studies suggest that interaction with TPP1 for recruitment and telomerase catalytic activation may involve regions other than the TEN domain (14, 15, 33). Furthermore, mutations in the hTERT C-terminal extension prevent association of telomerase with the TPP1-OB fold (14), while the interaction of telomerase with TPP1 is not abolished upon TEN domain deletion (15), suggesting that other hTERT regions, perhaps the IFD, may be important to work synergistically with the TEN domain for maximum catalytic activation and recruitment of telomerase. Consistent with these reports, in coimmunoprecipitation experiments using overexpressed TPP1 and telomerase as well as immunofluorescence/FISH of endogenous TPP1 with overexpressed telomerase, we found no difference in the binding to TPP1 (immunoprecipitation) or colocalization with endogenous TPP1 (immunofluorescence/FISH) when comparing the mutant or WT telomerases (data not shown).
Based on the Tribolium castaneum crystal structure (37), we speculate that the substitution of these amino acids may indirectly affect the interaction with the incoming DNA substrate, resulting in altered telomerase catalytic functions. Although the beetle TERT lacks the TEN domain and a large region of the IFD motif (37), the antiparallel α-helices 13 and 14, constituting the IFD motif, are implicated in the structural coordination of α-helices 10 and 15, which are involved in mediating interactions with the DNA substrate and RNA template, respectively (37). Immunofluorescence experiments show that the variant hTERTs can localize to the Cajal bodies, suggesting that hTERT-V763s, hTERT-V791Y, and hTERT-L805A are proficient in telomerase assembly (Fig. 1E). In agreement with an earlier study on the S. cerevisiae TERT IFD that demonstrated the importance of this motif as a determining factor for primer recognition and binding (4), our findings suggest that the hTERT IFD can potentially be involved in mediating the binding to the telomeric substrate. Indeed, our in vitro data demonstrate that mutations in the IFD region lead to defective DNA synthesis and altered levels of processivity. Furthermore, ChIP experiments show that the ability of hTERT-L805A and hTERT-V791Y to localize and bind telomeres is compromised.
Our previous work showed that the hTERT-V791Y fails to immortalize HA5 cells (5). Despite similar defects of the hTERT-V791Y and hTERT-L805A in reconstituting telomerase activity and processivity and in localization to the telomere, hTERT-L805A is able to immortalize HA5 cells. These results are consistent with the stimulation of hTERT-L805A's enzymatic activity, processivity, and recruitment to telomeres, but not of hTERT-V791Y, by the overexpression of TPP1-POT1. These data support the hypothesis that the IFD is involved in interacting with the telomeric substrate and more specifically, in a manner that is dependent on TPP1. Importantly, in spite of the significantly lower levels of telomerase activity, processivity, and decreased recruitment of hTERT-L805A to telomeres, this mutant enzyme was nevertheless able to immortalize HA5 cells, possibly relying on a distributive mode of telomere maintenance rather than a processive one when telomeres are short (38). The growth defects observed in hTERT-L805A-expressing limited-life-span cells can be explained by defects in the maintenance of short telomeres, represented by elevated levels of SFEs, which subsequently lead to telomere uncapping, TIF formation, and increased levels of apoptosis. Furthermore, enhancement of hTERT-L805A catalytic functions and partial rescue in telomere association mediated by overexpression of TPP1-POT1 suggests that the mutated amino acid may potentially induce a subtle conformational change that renders the interaction of hTERT-L805A with TPP1 suboptimal, but the presence of excess TPP1 can compensate for a less efficient hTERT-L805A-TPP1 interaction. Alternatively, overexpression of TPP1-POT1 may offset a less efficient interaction of the mutant enzyme with other proteins involved in telomerase recruitment, such as HOT1 (13) or TIN2 (10).
The glycine residue at position 100 of the hTERT TEN domain is important for stimulation of telomerase processivity in vitro by TPP1-POT1, as its substitution by a valine residue abolishes such enhancement (33). Furthermore, hTERT-G100V fails to localize to telomeres, accumulating instead in Cajal bodies (14), and is unable to immortalize HA5 cells (33). These data led to the speculation that this mutant enzyme was defective in recruitment due to impaired interaction with the TPP1 OB fold. The hTERT-V791Y variant displays the same phenotypes as hTERT-G100V and is the second hTERT variant reported to be insensitive to stimulation by TPP1-POT1, suggesting that the V791 residue is important for mediating binding to telomeres through a direct or indirect interaction with TPP1.
It has previously been reported that telomerase activity and processivity can be regulated independently (27). This phenomenon was also observed with the V763S mutant enzyme, which displays lower levels of DNA synthesis but higher levels of RAP than the WT enzyme. Additionally, although the rate of repeat addition processivity regulation was previously shown to be independent of the levels of processivity in hTERT motif 3 mutant enzymes (27), we observed a higher rate of RAP for the hTERT-V763S variant, which could have potentially contributed to the higher level of processivity. Since no defect in localization or binding to the telomeres was observed with the hTERT-V763S mutant enzyme, and since the overexpression of TPP1-POT1 was able to enhance telomerase activity and processivity of this mutant enzyme, we speculate that the hTERT-V763S variant is capable of interacting with TPP1 to allow telomerase recruitment to telomeres.
The higher levels of RAP and lower activity displayed by hTERT-V763S suggest that interactions of hTERT-V763S with other telomerase-associated proteins might be affected, such as that with the negative regulator of telomerase activity, hPif1 (39). A putative binding site for the mouse Pif1 was mapped to residues 536 to 928 of hTERT, which encompasses the IFD region (amino acids 721 to 822) (40). More recently, hPif1 was reported to preferentially dislodge telomerase from long telomeres in order to recycle the telomerase to shorter telomeres for elongation (41). In accordance with this model, if hTERT-V763S interaction with hPif1 is altered, the mutant enzyme might continue to elongate the same telomere at the expense of other short telomeres. This also suggests that this variant might be less able to interact and elongate telomeres that are short, consistent with the observed increase in short telomeres, or SFEs, in HA5 cells expressing hTERT-V763S. Moreover, the higher level of processivity displayed by this enzyme might be due to an enhanced ability of the hTERT-V763S to interact with TPP1-POT1.
Despite the high levels of processivity exhibited by hTERT-V763S in vitro and the ability of hTERT-V763S to be recruited to telomeres, HA5 cells expressing hTERT-V763S exhibited growth defects in comparison with hTERT-WT-expressing HA5 cells. Growth defects were likely due to increased apoptosis caused by uncapped telomeres and a failure to elongate short telomeres, evident by the higher levels of SFEs and TIF displayed by these cells than by the hTERT-WT-expressing cells. In human cells, telomerase was previously proposed to favorably elongate short telomeres (42–45). Our data support the idea that hTERT-V763S cannot be recruited to the short telomeres, possibly due to a preferred binding to longer substrates or its inability to be displaced from the elongated telomeres by hPif1 and, thus, failure to be recycled to shorter ones.
Consistent with the ChIP data, which showed efficient binding of hTERT-V763S to the telomeres in HeLa cells, this mutant enzyme was also able to induce telomere elongation when stably expressed in HeLa cells. This strongly supports the hypothesis that in HA5 cells, which display shorter telomeres than HeLa cells, hTERT-V763S is unable to bind to short telomeres for telomere maintenance, suggesting an important implication of the IFD in the maintenance of short telomeres.
In summary, our data propose an important role for the human telomerase IFD in the regulation of telomerase-specific functions, recruitment to telomeres, and cell survival. We demonstrate for the first time that the IFD can mediate telomerase activity, processivity, and telomerase recruitment to telomeres in a TPP1-dependent manner. Additional studies of other associated proteins as potential candidates that can interact with the hTERT IFD, such as the recruitment protein HOT1 or the telomerase trafficking protein TCAB1, and hPif1 will allow a better definition of the exact function of the IFD in telomerase regulation, telomere maintenance, and cellular survival.
ACKNOWLEDGMENTS
We thank Gerardo Ferbeyre (Université de Montréal, Montréal, Canada) for the plasmid construct pMSCV-puromycin, Thomas R. Cech (University of Colorado Boulder, Boulder, CO) for the pVAN3XFLAG-hTPP1, pVAN3XFLAG-hPOT1 and pVAN3XFLAG-hTERT constructs, and Joachim Lingner (Swiss Institute for Experimental Cancer Research, Lausanne, Switzerland) for the pcDNA6/myc-His C-hTERT and pBluescript II SK(+)-hTR plasmids. We thank Silvia Bacchetti (Istittuto Regina Elena, Rome, Italy) for the HA5 cells. We are grateful to Peter Lansdorp (British Columbia Cancer Center, Vancouver, Canada) for providing us with the TFL-Telo, version 2.0, software. We thank Michael Terns (The University of Georgia, Athens, GA) for providing us with the anticoilin antibody. We thank Stephane Richard (McGill University, Montreal, Canada) for providing the Axio Imager M1 microscope. We are thankful to Paul Maddox (University of North Carolina, Chapel Hill, NC) for providing help with microscopy analysis and Christian Young for assisting us with FACS analysis.
C.A. was supported by a Chercheur National of the Fonds de Recherche du Québec-Santé (FRQS). Support to T.W.C. was provided by a McGill University Faculty of Medicine Internal Studentship, a Canadian Institute for Health Research McGill Integrated Cancer Research Training Program Studentship, and an FRQS studentship, and support to Y.D. was provided by a Canadian Institute for Health Research Canada graduate scholarship.
ADDENDUM
While the manuscript was under review, a cryo-electron microscopy structure of Tetrahymena telomerase subunits was published (46) in which the authors suggest that p50 is a structural and functional paralog of TPP1. Furthermore, the cryo-electron microscopy density attributed to the IFD apparently independently contacts both p50 and the TERT TEN domain. Based on the Tetrahymena telomerase structure, the authors propose that the human TERT IFD may interact with the TPP1 OB fold and hTERT TEN domain, in support of our findings that the human TERT IFD regulates telomerase and telomere function in a TPP1-dependent manner.
REFERENCES
- 1.Shay JW, Reddel RR, Wright WE. 2012. Cancer and telomeres—an ALTernative to telomerase. Science 336:1388–1390. doi: 10.1126/science.1222394. [DOI] [PubMed] [Google Scholar]
- 2.D'Souza Y, Lauzon C, Chu TW, Autexier C. 2013. Regulation of telomere length and homeostasis by telomerase enzyme processivity. J Cell Sci 126:676–687. doi: 10.1242/jcs.119297. [DOI] [PubMed] [Google Scholar]
- 3.Schuller AP, Harkisheimer MJ, Skordalakes E. 2011. In vitro reconstitution of the active T. castaneum telomerase. J Vis Exp 2011(53):e2799. doi: 10.3791/2799: e2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lue NF, Lin YC, Mian IS. 2003. A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol Cell Biol 23:8440–8449. doi: 10.1128/MCB.23.23.8440-8449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Souza Y, Chu TW, Autexier C. 2013. A translocation-defective telomerase with low levels of activity and processivity stabilizes short telomeres and confers immortalization. Mol Biol Cell 24:1469–1479. doi: 10.1091/mbc.E12-12-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armbruster BN, Banik SS, Guo C, Smith AC, Counter CM. 2001. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol Cell Biol 21:7775–7786. doi: 10.1128/MCB.21.22.7775-7786.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armbruster BN, Etheridge KT, Broccoli D, Counter CM. 2003. Putative telomere-recruiting domain in the catalytic subunit of human telomerase. Mol Cell Biol 23:3237–3246. doi: 10.1128/MCB.23.9.3237-3246.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP. 2010. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol 30:2971–2982. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. 2007. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 10.Frank AK, Tran DC, Qu RW, Stohr BA, Segal DJ, Xu L. 2015. The shelterin TIN2 subunit mediates recruitment of telomerase to telomeres. PLoS Genet 11:e1005410. doi: 10.1371/journal.pgen.1005410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stern JL, Zyner KG, Pickett HA, Cohen SB, Bryan TM. 2012. Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol Cell Biol 32:2384–2395. doi: 10.1128/MCB.00379-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung DH, Kung HF, Huang JJ, Shaw PC. 2012. PinX1 is involved in telomerase recruitment and regulates telomerase function by mediating its localization. FEBS Lett 586:3166–3171. doi: 10.1016/j.febslet.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 13.Kappei D, Butter F, Benda C, Scheibe M, Draskovic I, Stevense M, Novo CL, Basquin C, Araki M, Araki K, Krastev DB, Kittler R, Jessberger R, Londono-Vallejo JA, Mann M, Buchholz F. 2013. HOT1 is a mammalian direct telomere repeat-binding protein contributing to telomerase recruitment. EMBO J 32:1681–1701. doi: 10.1038/emboj.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong FL, Batista LF, Freund A, Pech MF, Venteicher AS, Artandi SE. 2012. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150:481–494. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nandakumar J, Bell CF, Weidenfeld I, Zaug AJ, Leinwand LA, Cech TR. 2012. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492:285–289. doi: 10.1038/nature11648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, Lei M. 2011. Human telomere POT1-TPP1 complex and its role in telomerase activity regulation. Methods Mol Biol 735:173–187. doi: 10.1007/978-1-61779-092-8_17. [DOI] [PubMed] [Google Scholar]
- 17.Dalby AB, Hofr C, Cech TR. 2015. Contributions of the TEL-patch amino acid cluster on TPP1 to telomeric DNA synthesis by human telomerase. J Mol Biol 427:1291–1303. doi: 10.1016/j.jmb.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sexton AN, Youmans DT, Collins K. 2012. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J Biol Chem 287:34455–34464. doi: 10.1074/jbc.M112.394767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt JC, Dalby AB, Cech TR. 2014. Identification of human TERT elements necessary for telomerase recruitment to telomeres. eLife 2014(3):e03563. doi: 10.7554/eLife.03563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaug AJ, Crary SM, Fioravanti MJ, Campbell K, Cech TR. 2013. Many disease-associated variants of hTERT retain high telomerase enzymatic activity. Nucleic Acids Res 41:8969–8978. doi: 10.1093/nar/gkt653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Fong KW, Tang M, Han X, Gong Z, Ma W, Hebert M, Songyang Z, Chen J. 2014. Fam118B, a newly identified component of Cajal bodies, is required for Cajal body formation, snRNP biogenesis and cell viability. J Cell Sci 127:2029–2039. doi: 10.1242/jcs.143453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abreu E, Terns RM, Terns MP. 2011. Visualization of human telomerase localization by fluorescence microscopy techniques. Methods Mol Biol 735:125–137. doi: 10.1007/978-1-61779-092-8_12. [DOI] [PubMed] [Google Scholar]
- 23.Tomlinson RL, Abreu EB, Ziegler T, Ly H, Counter CM, Terns RM, Terns MP. 2008. Telomerase reverse transcriptase is required for the localization of telomerase RNA to cajal bodies and telomeres in human cancer cells. Mol Biol Cell 19:3793–3800. doi: 10.1091/mbc.E08-02-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loayza D, De Lange T. 2003. POT1 as a terminal transducer of TRF1 telomere length control. Nature 423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 25.Ourliac-Garnier I, Poulet A, Charif R, Amiard S, Magdinier F, Rezai K, Gilson E, Giraud-Panis MJ, Bombard S. 2010. Platination of telomeric DNA by cisplatin disrupts recognition by TRF2 and TRF1. J Biol Inorg Chem 15:641–654. doi: 10.1007/s00775-010-0631-4. [DOI] [PubMed] [Google Scholar]
- 26.Cohn M, Blackburn EH. 1995. Telomerase in yeast. Science 269:396–400. doi: 10.1126/science.7618104. [DOI] [PubMed] [Google Scholar]
- 27.Xie M, Podlevsky JD, Qi X, Bley CJ, Chen JJ. 2010. A novel motif in telomerase reverse transcriptase regulates telomere repeat addition rate and processivity. Nucleic Acids Res 38:1982–1996. doi: 10.1093/nar/gkp1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cristofari G, Lingner J. 2006. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J 25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. 2006. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell 17:955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jady BE, Richard P, Bertrand E, Kiss T. 2006. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell 17:944–954. doi: 10.1091/mbc.E05-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latrick CM, Cech TR. 2010. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J 29:924–933. doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. 2007. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 33.Zaug AJ, Podell ER, Nandakumar J, Cech TR. 2010. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev 24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J. 2007. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell 27:882–889. doi: 10.1016/j.molcel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 35.Sexton AN, Regalado SG, Lai CS, Cost GJ, O'Neil CM, Urnov FD, Gregory PD, Jaenisch R, Collins K, Hockemeyer D. 2014. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Genes Dev 28:1885–1899. doi: 10.1101/gad.246819.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moriarty TJ, Marie-Egyptienne DT, Autexier C. 2005. Regulation of 5′ template usage and incorporation of noncognate nucleotides by human telomerase. RNA 11:1448–1460. doi: 10.1261/rna.2910105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillis AJ, Schuller AP, Skordalakes E. 2008. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 455:633–637. doi: 10.1038/nature07283. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, Abreu E, Kim J, Stadler G, Eskiocak U, Terns MP, Terns RM, Shay JW, Wright WE. 2011. Processive and distributive extension of human telomeres by telomerase under homeostatic and nonequilibrium conditions. Mol Cell 42:297–307. doi: 10.1016/j.molcel.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang DH, Zhou B, Huang Y, Xu LX, Zhou JQ. 2006. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res 34:1393–1404. doi: 10.1093/nar/gkl029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snow BE, Mateyak M, Paderova J, Wakeham A, Iorio C, Zakian V, Squire J, Harrington L. 2007. Murine pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol Cell Biol 27:1017–1026. doi: 10.1128/MCB.01866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li JR, Yu TY, Chien IC, Lu CY, Lin JJ, Li HW. 2014. Pif1 regulates telomere length by preferentially removing telomerase from long telomere ends. Nucleic Acids Res 42:8527–8536. doi: 10.1093/nar/gku541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Britt-Compton B, Capper R, Rowson J, Baird DM. 2009. Short telomeres are preferentially elongated by telomerase in human cells. FEBS Lett 583:3076–3080. doi: 10.1016/j.febslet.2009.08.029. [DOI] [PubMed] [Google Scholar]
- 43.Ouellette MM, Liao M, Herbert BS, Johnson M, Holt SE, Liss HS, Shay JW, Wright WE. 2000. Subsenescent telomere lengths in fibroblasts immortalized by limiting amounts of telomerase. J Biol Chem 275:10072–10076. doi: 10.1074/jbc.275.14.10072. [DOI] [PubMed] [Google Scholar]
- 44.Hemann MT, Strong MA, Hao L-Y, Greider CW. 2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107:67–77. doi: 10.1016/S0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 45.Samper E, Flores JM, Blasco M. 2001. Restoration of telomerase activity rescues chromosomal instability and premature aging in Terc−/− mice with short telomeres. EMBO Rep 2:800–807. doi: 10.1093/embo-reports/kve174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang J, Chan H, Cash DD, Miracco EJ, Ogorzalek Loo RR, Upton HE, Cascio D, O'Brien Johnson R, Collins K, Loo JA, Zhou ZH, Feigon J. 15 October 2015. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science doi: 10.1126/science.aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]