Abstract
Mitochondrial oxidative and thermogenic functions in brown and beige adipose tissues modulate rates of energy expenditure. It is unclear, however, how beige or white adipose tissue contributes to brown fat thermogenic function or compensates for partial deficiencies in this tissue and protects against obesity. Here, we show that the transcription factor Yin Yang 1 (YY1) in brown adipose tissue activates the canonical thermogenic and uncoupling gene expression program. In contrast, YY1 represses a series of secreted proteins, including fibroblast growth factor 21 (FGF21), bone morphogenetic protein 8b (BMP8b), growth differentiation factor 15 (GDF15), angiopoietin-like 6 (Angptl6), neuromedin B, and nesfatin, linked to energy expenditure. Despite substantial decreases in mitochondrial thermogenic proteins in brown fat, mice lacking YY1 in this tissue are strongly protected against diet-induced obesity and exhibit increased energy expenditure and oxygen consumption in beige and white fat depots. The increased expression of secreted proteins correlates with elevation of energy expenditure and promotion of beige and white fat activation. These results indicate that YY1 in brown adipose tissue controls antagonistic gene expression programs associated with energy balance and maintenance of body weight.
INTRODUCTION
The incidence of obesity has become a global epidemic, leading to an increase in associated pathologies such as cardiovascular disease, type 2 diabetes, and cancer (1). Obesity is the result of a net imbalance between the energy intake and the energy expenditure (2). An important component of energy expenditure is adaptive thermogenesis caused by the heat dissipated in response to lower temperatures or diet and produced in the mitochondria of brown adipose tissue (BAT) and skeletal muscle. In BAT, this thermogenic process is largely dependent on inducible mitochondrial uncoupling respiration mediated by uncoupling protein 1 (UCP1) (3, 4). In addition to brown adipocytes, the presence of clusters of inducible thermogenic adipocytes within subcutaneous white fat depots, referred to as browning, has been proposed as an additional or compensatory site of energy dissipation and body weight control (5–7). Although these adipocytes, named “beige” (or “brite”), are derived from a different developmental lineage than BAT, they express UCP1 and similar thermogenic components (5, 8, 9). Moreover, under some conditions, white adipocytes activate thermogenic processes that contribute to energy expenditure (6, 10). In humans, the presence of these inducible thermogenic adipocytes opens the possibility to identify targets to therapeutically treat obesity. Toward this goal, it is necessary to identify the molecular and cellular mechanisms governing beige or white adipocyte thermogenesis that, despite recent advances (11–15), are currently not completely understood.
At the transcriptional level, activation of the canonical thermogenic program in BAT (often in response to adrenergic agonism) (16), involves inducible coactivation of peroxisome proliferator-activated receptor gamma (PPARγ) coactivator-1α (PGC-1α) with transcription factors such as PPARα and PPARγ, leading to Ucp1 expression, along with the concomitant expression of mitochondrial genes through estrogen-related receptor α (ERRα) and nuclear respiratory factors (NRFs) (17, 18). We recently identified the transcription factor Yin Yang 1 (YY1) as part of the PGC-1α transcriptional complex driving mitochondrial gene expression (19, 20). YY1 can act as a transcriptional activator or repressor, depending on its cofactor recruitment (21–24). In skeletal muscle, YY1 activates nucleus-encoded mitochondrial genes through PGC-1α binding; in contrast, YY1 represses insulin signaling genes via recruitment of Polycomb proteins and increased histone H3 trimethyl K27 (H3K27me3) (25). Loss of YY1 in skeletal muscle results in a global decrease of mitochondrial gene expression, decreasing oxidative function, and exercise intolerance (19). These studies suggest that the transcriptional activities of YY1 in BAT might be important to promote the canonical mitochondrial thermogenic pathway and impact energy balance and body weight.
Recently, systemic factors have been shown to regulate brown and beige adipose tissue thermogenesis, including fibroblast growth factor 21 (FGF21) (26), bone morphogenetic protein 4 (BMP4) (27), BMP7 (28), BMP8b (29), FGF19 (30), growth differentiation factor 5 (GDF5) (31), natriuretic peptides (32), prostaglandins (33, 34), vascular endothelial growth factor (VEGF) (35), β-aminoisobutyric acid (BAIBA) (36), meteorin-like (14), and irisin (37). These factors are involved in the control of energy expenditure and body weight through modulation of BAT or beige fat. Within this context, whether BAT-secreted factors can contribute to energy expenditure or to compensate for defective BAT thermogenesis through activation of other thermogenic tissues, including beige or white adipose tissue, is poorly understood.
Here, we report that loss of YY1 in BAT leads to a strong suppression of mitochondrial and thermogenic gene expression. Despite reduced BAT thermogenic function, YY1-deficient mice are protected against diet-induced obesity and have activated thermogenic beige and white adipose tissue. We show that YY1 has an antagonistic control of BAT genes, in that it activates genes linked to adaptive thermogenesis through activation of the canonical thermogenic pathway but suppresses a series of secreted proteins, including FGF21, BMP8b, and GDF15, which activate whole-body energy expenditure.
MATERIALS AND METHODS
Animal experiments.
All experiments and protocols were approved by the Institutional Animal Care and Use Committees of Dana Farber Cancer Institute or Beth Israel Deaconess Medical Center. YY1-Ucp1Cre and YY1-AdipoCre mice were generated by breeding animals harboring a floxed YY1 allele (38) with transgenic mice expressing Ucp1 Cre (39) or Adiponectin Cre recombinase, respectively (40). For wild-type mouse experiments, 8-week-old C57BL/6 mice were purchased from Taconic Farms. Mice were maintained on standard chow or 60% high-fat diet (HFD) (Research Diets) with 12-h cycles. For cold exposure experiments, mice were placed in 4°C or 30°C incubators at the indicated time points, and body temperature was measured with a rectal probe. For metabolic studies, energy expenditure was analyzed using a comprehensive lab animal monitoring system (Columbus Instruments). Mice were acclimated for 24 h before measurements were taken. Whole-mouse magnetic resonance imaging (MRI) measurements were performed in a CITI-scan instrument.
Gene expression and Western blot analysis.
Total RNA from cultured cells or tissues was purified using TRIzol (Invitrogen) for cDNA synthesis (ABI high-capacity kit). Relative mRNA expression was quantified by quantitative PCR (qPCR) using SYBR green dye (ABI) and specific primers (data not shown). For Western blotting, whole-cell lysates were prepared with radioimmunoprecipitation assay (RIPA) buffer, separated by SDS-PAGE, and transferred to Immobilon-P membranes (Millipore). For detection of circulating GDF15, 1 μl of plasma was used for SDS-PAGE. The following antibodies were used: anti-YY1 (Santa-Cruz), anti-UCP1 (Abcam), anti-NDUFA9 (Abcam), anti-succinate dehydrogenase (anti-SDHA) (Abcam), antiaconitase (Abcam), anti-GDF15 (Abcam), anti-MTCO1 (Abcam), anti-UQCRC2 (Abcam), pCREB (Cell Signaling), total CREB (Cell Signaling), and antitubulin (Millipore).
Coimmunoprecipitations.
A pool of interscapular brown adipose tissue from 5 mice was homogenized with a motorized pestle in 4 ml of nuclear isolation buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol [DTT], Complete protease inhibitors [Roche]) and incubated on ice for 10 min. Nuclei were pelleted by centrifugation at 3,200 × g for 5 min and washed in 4 ml of the same buffer before centrifugation at 3,200 × g for 5 min. Nucleus pellets were then resuspended in 600 μl of immunoprecipitation buffer (0.1% NP-40, 150 mM NaCl, 20 mM HEPES, 1 mM EDTA, Complete protease inhibitors [Roche]). Two hundred micrograms of protein was incubated with 3 μg of YY1 antibody (sc1703; Santa Cruz Biotechnology, Inc.) or rabbit IgG for controls overnight at 4°C with rotation. Immunocomplexes were precipitated with magnetic protein G beads (DynaBeads; Invitrogen) during a 1-h rotation at 4°C. Samples were then washed 5 times in immunoprecipitation buffer. Samples were finally boiled, and the supernatants were run on SDS-PAGE, including input samples for Western blot detection using PGC-1α antibody (Santa Cruz), YY1 antibody (Santa Cruz Biotechnology, Inc.), and lamin B1 (Abcam).
The brown fat cell line De2.3 was treated with dimethyl sulfoxide (DMSO) or 10 μM forskolin at 10 μM for 4 h. The nuclei were directly isolated by incubation of cell pellets with nuclear isolation buffer. The same protocol was used for brown fat tissue to coimmunoprecipitate YY1–PGC-1α complex.
Histological analysis.
Freshly harvested tissues were fixed in 4% paraformaldehyde overnight. Paraffin embedding, sectioning, and hematoxylin and eosin staining were performed by the Dana Farber/Harvard Cancer Research Pathology core facility.
Fatty acid and glutamine oxidation.
Brown adipose tissue was Dounce homogenized in cold STE buffer (0.25 M sucrose, 10 mM Tris-HCl, 1 mM EDTA) and centrifuged at 420 × g for 10 min. The supernatant was added to 370 μl of a reaction mixture containing a final concentration of 100 mM sucrose, 10 mM Tris-HCl, 5 mM KH2PO4, 0.2 mM EDTA, 80 mM KCl, 1 mM MgCl2, 2 mM l-carnitine, 0.1 mM malate, 0.05 mM coenzyme A, 2 mM ATP, 1 mM DTT, and bovine serum albumin (BSA)-palmitate solution (0.7% BSA, 500 μM oleic acid, 0.4 μCi [14C]oleic acid or [14C]glutamine). Samples were incubated at 37°C for 1 h, and the reaction was stopped by adding 200 μl of 1 M perchloric acid. Next, 2-phenylethylamine-saturated Whatman paper was placed under the tube cap in order to trap radiolabeled CO2 during overnight incubation. Finally, the Whatman paper was placed in scintillation liquid, and radioactive counts were measured in a scintillation counter.
Oxygen consumption.
Direct ex vivo tissue respiration was performed using a Clark electrode (Strathkelvin Instruments). Freshly isolated tissue was minced in respiration buffer (1.5 mM pyruvate, 25 mM glucose, 2% BSA) and placed in electrode chambers. The O2 consumption rate was normalized to tissue weight.
For direct mitochondrial respiration from brown adipose tissue, mitochondria were isolated in STE buffer (0.25 M sucrose, 5 mM Tris, 2 mM EDTA) and centrifuged for 10 min at 8,500 × g. The pellet was resuspended in STE buffer and centrifuged again twice at 8,500 × g. Pelleted mitochondria were resuspended in 0.5 ml of STE buffer, and the total protein concentration was measured. Respirometry was performed using the XF24-3 platform from Seahorse Biosciences as previously described (19).
Primary adipocyte cell culture.
The stromal vascular fraction (SVF) of brown adipose tissue of 6-week-old mice was isolated by collagenase digestion followed by two alternative filtration steps (using 100 and 40 μM strainers) and centrifugations for 5 min at 500 × g. Then cells were plated and differentiated upon confluence with adipogenic cocktail (0.5 mM 3-isobutyl-1-methylxanthine [IBMX], 1 μM dexamethasone, 1 μM rosiglitazone, 0.02 μM insulin, 1 nM T3) for 48 h. The cells were maintained in 0.02 μM insulin and 1 nM T3 and harvested at days 6 to 8 postdifferentiation.
ChIP.
Brown adipose tissue was dissected, cut in small pieces and cross-linked in 1% formaldehyde–phosphate-buffered saline (PBS) for 10 min; the reaction was then quenched in 0.125 M glycine. BAT was then homogenized in isolation buffer (250 mM sucrose, 5 mM Tris, 2 mM EDTA) using a motorized pestle followed by chromatin isolation in buffer containing 50 mM HEPES, 140 mM NaCl, 1 mM EDTA, 1%Triton X-100, 0.1%, Na-deoxycholate, 0.1% SDS, and Complete protease inhibitors (Roche). Samples were sonicated in a Diagenode Bioruptor for 5 cycles of 5 min with a duty cycle of 30 s “on” and 30 s “off.” Samples were immunoprecipitated with specific antibodies to YY1 (Santa Cruz) and H3K27me3 (Abcam), and DNA was isolated for qPCR analysis as described previously (25).
Detection of plasmatic norepinephrine.
Thirty microliters of plasma was used to measure plasmatic norepinephrine by enzyme-linked immunosorbent assay (ELISA) (Rocky Mountain Diagnostics) according to the manufacturer's guidelines.
Arrays and gene set enrichment analysis.
Extracted RNA from BAT or inguinal subcutaneous white adipose tissue (IWAT) was used to perform gene arrays using a Mouse 430A 2.0 GeneChip at the Microarray Core Facility at the Dana Farber Cancer Institute. To generate the gene expression files (GCT), CEL files were used as input for the Gene Pattern (http://genepattern.broadinstitute.org/) module Expression File Creator using robust multiarray average (RMA) and quantile normalization. For gene set enrichment analysis (GSEA), GCT files were used as input using the GSEA 2.0 software (http://www.broadinstitute.org/gsea) using the default parameters (41).
Microarray data accession number.
The Gene Expression Omnibus accession number for the gene expression data reported is the super series no. GSE68443.
RESULTS
Genetic deletion of YY1 in adipose tissue leads to protection against diet-induced obesity.
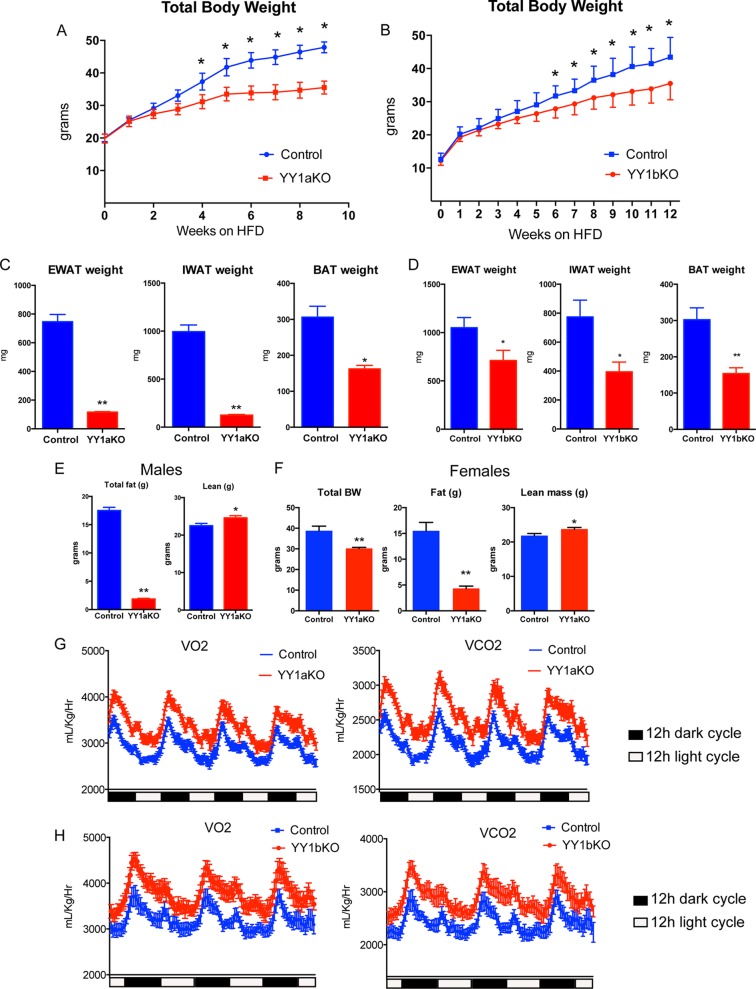
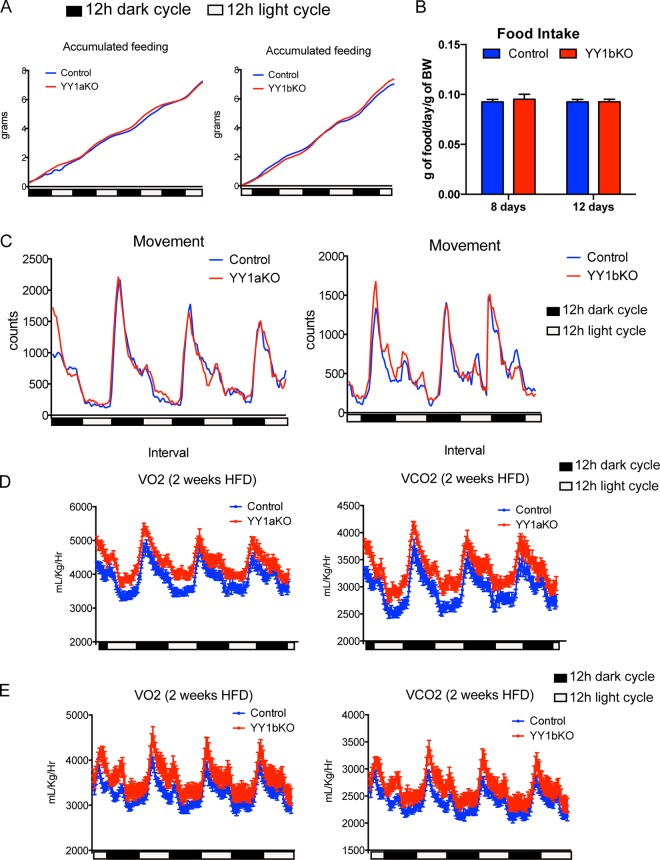
We have previously shown that YY1 loss of function in skeletal muscle of mice leads to a defective mitochondrial function that results in a decreased oxidative capacity and exercise intolerance (19). Adipose tissue largely contributes to energy balance by either storing excessive nutrient intake or by uncoupling mitochondrial respiration in specialized brown and beige adipocytes (3). In order to determine the YY1-dependent transcriptional networks of genes linked to energy balance in adipose tissue and how they might affect body weight, we generated adipose-specific YY1 KO mouse models using a Cre recombinase-based conditional strategy with Adiponectin Cre (AdipoCre) (40) or Ucp1 Cre (Ucp1Cre) (39) in YY1 floxed mice. This strategy allows the deletion of YY1 in all adipocytes, using AdipoCre (YY1aKO mice), or in brown adipocytes, using Ucp1Cre (YY1bKO mice). When both YY1aKO and YY1bKO mice were challenged with a high-fat diet, they exhibited decreased body weight compared to respective YY1 floxed littermate control mice (Fig. 1A and B). In addition, YY1 KO mice fat depots had lower weights (Fig. 1C and D) that were consistent with decreased adiposity assessed using MRI (Fig. 1E). Female mice also showed similar reduced body weight and decreased fat accumulation (Fig. 1F). To further analyze the energy balance of YY1 KO mice, we subjected these mice to metabolic analysis using a comprehensive laboratory analysis system (CLAMS). The higher O2 consumption and CO2 production observed in both types of YY1 KO mice revealed increased total energy expenditure compared to that in control mice (Fig. 1G and H). Importantly, the increased oxygen consumption of YY1 mutant mice occurred without differences in food intake measured by CLAMS over 3 days or by monitoring the weight of consumed food for 8 and 12 days (Fig. 2A and B). In addition, movement behaviors were similar in control and YY1 mutant mice (Fig. 2C). The changes in energy expenditure were already observed as soon as 2 weeks after initiation of high-fat feeding when body weights between control and mutant mice were still comparable (Fig. 2D and E). Collectively, these results show that loss of YY1 in adipose tissue protects from diet-induced obesity due to increased energy expenditure.
FIG 1.
YY1 deficiency in adipose tissue protects against diet-induced obesity through increased energy expenditure. (A and B) Total body weight curves of (A) YY1aKO and (B) YY1bKO mice versus littermate controls when chronically fed a high-fat diet. (C and D) Decreased fat content measured by adipose tissue depot weights from (C) YY1aKO and (D) YY1bKO mice. (E and F) Total body weight (BW), adiposity, or lean mass measured by MRI citi-scan in (E) male and (F) female mice chronically fed a high-fat diet. (G and H) Increased energy expenditure of (G) YY1aKO and (H) YY1bKO mice versus control mice as indicated by O2 consumption (VO2) and CO2 production (VCO2) analyzed by CLAMS. Data show representative results from two independent experiments for body weight measurements (n = 8 to 10). Data are represented as means ± standard errors of the means (SEM). *, P < 0.05, and **, P < 0.01, by t test.
FIG 2.
YY1 deficiency in adipose tissue leads to increased energy expenditure without changes in food intake or physical activity. (A and B) Accumulated food intake measured by CLAMS analysis (A) and by weighing food pellets (B) for individually caged mice at the indicated time points. (C) Total spatial mouse movement in 3 dimensions analyzed by CLAMS. (D and E) Total O2 consumption and CO2 production in (D) YY1aKO and (E) YY1bKO mice versus control mice fed a high-fat diet for 2 weeks. Data are represented as means ± SEM. *, P < 0.05, and **, P < 0.01, by t test (n = 8).
Loss of YY1 leads to defective mitochondrial/thermogenic gene expression and function of brown adipose tissue.
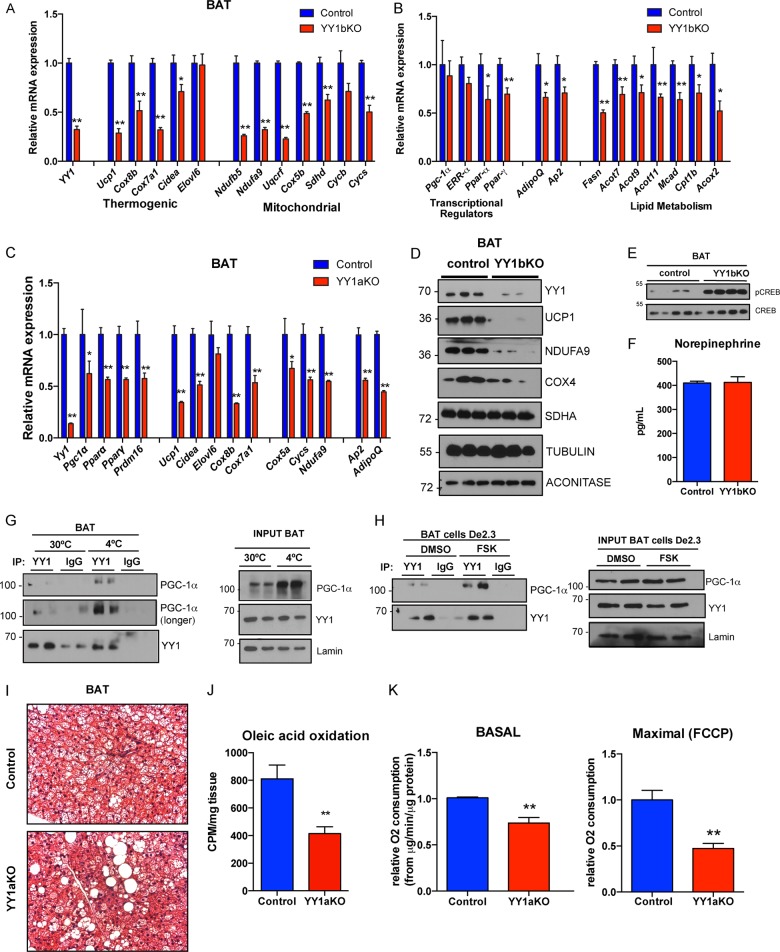
To determine the molecular mechanisms that could explain the increased energy expenditure in adipose-specific YY1 KO mice, we analyzed YY1-dependent gene expression programs in brown adipose tissue. Consistent with our previous findings in skeletal muscle that YY1 activates mitochondrial genes (19), YY1aKO and YY1bKO mutant mice exhibited downregulation of several mitochondrial genes encoding oxidative phosphorylation (OXPHOS) proteins, including Ndufa9, Cox5b, Cycs (Fig. 3A, B, and C), canonical thermogenic genes, including Ucp1, Cox8b, Cox7a1, and Cidea (Fig. 3B, C, and D), and the fatty acid oxidation genes Cpt1b, Acox2, and Mcad (Fig. 3B and C) in BAT. In addition, expression of key transcriptional regulators of brown fat thermogenic activation and function, including the Ppar-α and Ppar-γ genes, was also decreased (Fig. 3C). Interestingly, BAT of YY1 mutant mice exhibited augmented phosphorylation of CREB, a downstream target of adrenergic signaling involved in UCP1 and mitochondrial gene expression (42) (Fig. 3E), independent of changes in plasma norepinephrine (Fig. 3F) suggesting activation of a transcriptional mechanism to compensate for defective thermogenic function. Next, to support the mechanistic regulatory control of YY1 in the physiological thermogenic process, we tested whether endogenous YY1 and PGC-1α interacted in brown adipocytes. In agreement with our previous results that YY1 interacts with PGC-1α in different cell types (19, 20), Fig. 3G shows by immunoprecipitation analysis that both endogenous proteins form part of a complex, and importantly, this interaction is increased during cold exposure. Similar results were obtained using the brown fat cell line De2.3 (43): treatment with forskolin strongly induced the capacity of YY1 to immunoprecipitate PGC-1α at early time points when the amount of PGC-1α remained unchanged (Fig. 3H). These results suggest that during activation of the thermogenic process, adrenergic agonism causes YY1 to recruit PGC-1α, driving activation of mitochondrial and thermogenic genes. Consistent with these molecular results, histological analysis of BAT revealed increased lipid content from YY1aKO and YY1bKO mutant mice compared to controls, suggesting a decreased mitochondrial oxidative function (Fig. 3I; data not shown). Functionally, we measured fatty acid oxidation rates by quantifying the release of radioactive 14CO2 in brown fat homogenates incubated with 14C-radiolabeled oleic acid. Interestingly, YY1aKO and YY1bKO mice showed reduced fatty oxidation rates in BAT compared to control mice (Fig. 3J; data not shown). In addition, BAT mitochondria isolated from YY1 mutant mice had reduced oxygen consumption in both basal and maximal respiration (Fig. 3K). These results show that loss of YY1 in brown adipocytes compromises the mitochondrial thermogenic capacity and the fatty acid oxidation in BAT cells, and this is consistent with the physical interaction between YY1 and PGC-1α. Because YY1aKO and YY1bKO mice are protected against diet-induced obesity by increasing energy expenditure, these results indicate that other tissues, including white fat depots, compensate for the defective BAT thermogenic capacity and may account for the leaner phenotype.
FIG 3.
Loss of YY1 in brown adipose tissue leads to a mitochondrial and thermogenic defect. (A to C) Expression of mitochondrial and thermogenic genes (A) and transcriptional regulator and lipid metabolism genes (B) of BAT from YY1bKO and YY1aKO (C) mice versus control mice fed a high-fat diet. (D and E) Western blot analysis (D) of mitochondrial proteins and CREB signaling (E) of BAT from control and YY1bKO mice. (F) Plasmatic norepinephrine levels measured by ELISA in YY1bKO versus control mice fed a high-fat diet for 2 weeks (n = 10). (G and H) Endogenous coimmunoprecipitation of YY1 or the IgG control with specific antibodies followed by pulldown with protein G magnetic beads from brown adipose tissue of mice housed at thermoneutrality (30°C) or cold (4°C) for 3 h (G) and from brown adipose cells of the De2.3 line (H) treated with DMSO or 10 μM forskolin (FSK) and detection of PGC-1α by Western blot analysis. (I) Histological analysis of BAT cross sections stained with hematoxylin and eosin (H&E). (J) Fatty acid oxidation rates by [14C]oleic acid oxidation from BAT extracts of YY1aKO versus control mice. (K) Oxygen consumption of isolated mitochondria of BAT from YY1aKO versus control mice measured by Seahorse Bioscience technology. FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone. Data are represented as means ± SEM. *, P < 0.05, and **, P < 0.01, by t test (n = 8 to 10).
YY1-Ucp1 KO mice have a compensatory browning of subcutaneous white adipose tissue.
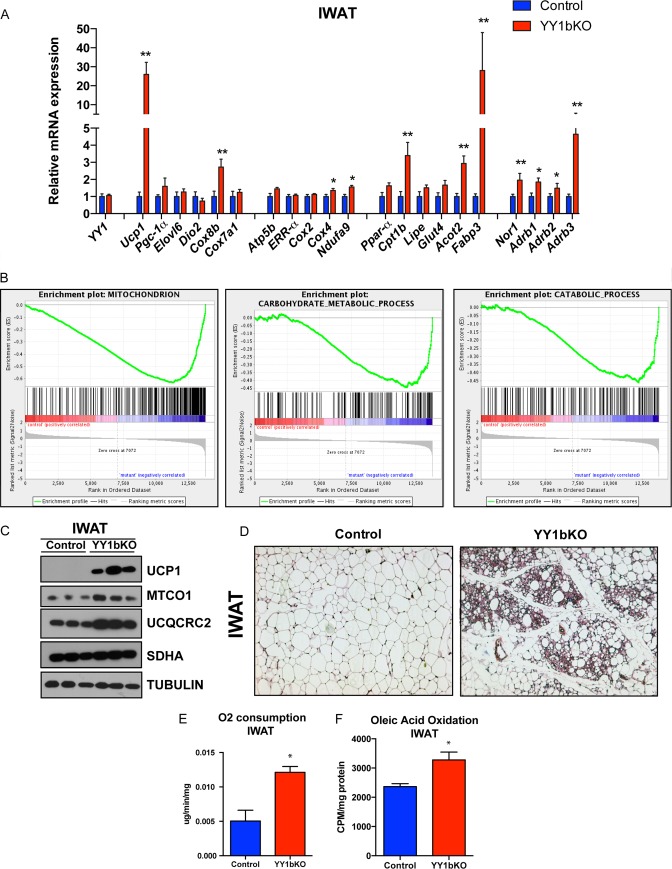
Defective brown fat function often leads to obesity (44, 45); however, despite the mitochondrial and thermogenic deficiencies of YY1aKO and YY1bKO mice, they are partially resistant to diet-induced weight gain (Fig. 1A and B). In order to determine what tissues and mechanisms contribute to this energetic compensation, we first measured gene expression in different tissues linked to energy expenditure. Gene expression analysis of liver and skeletal muscle from control and YY1aKO and YY1bKO mice did not reveal significant differences in fatty acid oxidation, oxygen consumption, or metabolic gene expression (data not shown). Browning of inguinal subcutaneous white adipose tissue (IWAT) relies on the emergence of thermogenic beige cells, a process that is induced by adrenergic stimulation (46, 47). Notably, when YY1bKO mice were challenged with a high-fat diet, the expression of Ucp1 in IWAT was elevated 25-fold compared to control mice (Fig. 4A). Additional thermogenic markers such as Cox8b and fatty acid oxidation gene Cpt1b were also increased (Fig. 4A). This gene expression pattern linked to mitochondrial oxidative and thermogenic pathways was also confirmed using gene expression analysis. Gene set enrichment analysis (GSEA) showed that pathways involved in mitochondrial biology and catabolic processes such as breakdown of lipids and carbohydrates were highly enriched (Fig. 4B). UCP1 was also strongly increased at the protein level along with mitochondrial proteins COX4 and UQCRC2 (Fig. 4C). Moreover, histological analysis of IWAT from mutant mice revealed the typical clustering of beige cells with appearance of multilocular lipid droplet cells (Fig. 4D). The oxygen consumption of IWAT measured in tissue extracts using the Clark electrode and fatty acid oxidation rates measured by radiolabeled [14C]oleic acid were increased in mutant mice (Fig. 4E and F). Remarkably, when YY1aKO and YY1bKO mice were exposed to cold, they did not show signs of dramatic initial temperature drop (data not shown) as it occurs in Ucp1 KO mice (48), and they survived for as long as 2 weeks after cold exposure (data not shown). Collectively, these results suggest that browning of IWAT in YY1 mutant mice compensates for BAT defective thermogenic function through elevation of energy expenditure in response to a high-fat diet that leads to protection against obesity.
FIG 4.
Browning of subcutaneous white adipose tissue in YY1 KO mice fed a high-fat diet. (A) Gene expression of IWAT from YY1bKO versus control mice fed a high-fat diet. (B) Gene Set Expression Analysis (GSEA) of IWAT from YY1bKO versus control mice indicating increased catabolic pathways. (C) Western blot analysis of mitochondrial proteins in IWAT from YY1bKO versus control mice on a high-fat diet. (D) H&E staining of IWAT cross sections from YY1bKO versus control mice fed a high-fat diet. (E) Ex vivo oxygen consumption of IWAT homogenates from YY1bKO versus control mice measured by Clark electrode. (F) Fatty acid oxidation rates measured by released 14CO2 from IWAT homogenates incubated with 14C-radiolabeled oleic acid in YY1bKO and control mice. Data are represented as means ± SEM. *, P < 0.05, and **, P < 0.01, by t test (n = 6 to 10).
Adipose YY1 KO mice activate white fat nutrient uptake and energy consumption pathways.
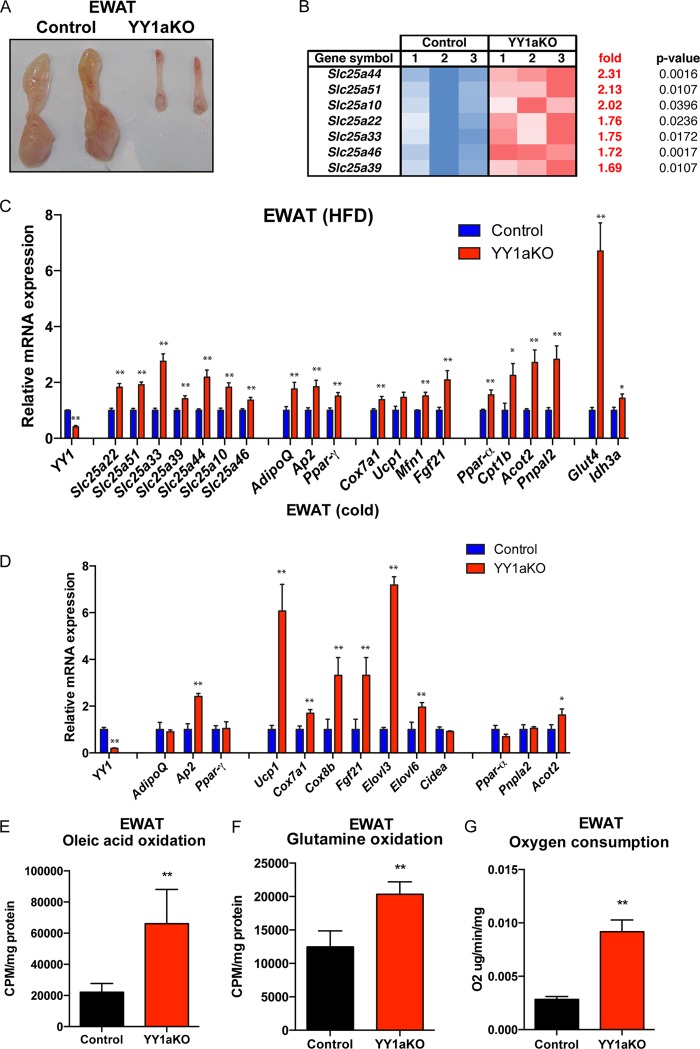
In addition to subcutaneous white adipose browning, visceral white fat can switch the energy storage to an energy dissipation program that contributes to reduce body weight (6, 10). Based on the fact that epididymal white adipose tissue (EWAT) was decreased in YY1 KO mice (Fig. 5A and Fig. 1C and D), we performed GSEA from visceral adipose tissue from wild-type and YYaKO mice. Interestingly, expression of several members of the SLC25 family of nuclear-encoded mitochondrial carriers was increased in EWAT from YY1aKO mice (Fig. 5B). The nucleus-encoded SLC25 family of genes are involved in a multiple import of solutes into the mitochondria. Interestingly, amino acid (Slc25a22 and Slc25a44), nucleotide (Slc25a33), and carboxylate (Slc25a10) import carriers were elevated in EWAT of YY1aKO mice fed a high-fat diet, as shown by qPCR in Fig. 5C. In addition, major changes in mRNA transcripts linked to nutrient intake (Glut4 and Fabp3) and mitochondrial energy utilization (Ppar-α, Acot2, Cpt1b, and Elovl3) were increased in YY1aKO mice in either the high-fat-diet-fed or cold-exposed groups (Fig. 5C and D).
FIG 5.
Increased nutrient uptake and energy consumption pathways in visceral white adipose tissue of YY1aKO mice. (A) Images of dissected visceral white adipose tissue from control and YY1aKO mice. (B) Gene expression array heat map showing the identification of genes of the Slc25 family increased in EWAT of YY1aKO mice versus control mice. (C and D) Gene expression of EWAT from YY1aKO mice versus control mice fed a high-fat diet for 3 months (C) or exposed to cold for 4 days (D). (E and F) Oleic acid oxidation (E) and glutamine oxidation (F) rates measured by released 14CO2 of EWAT tissue homogenates from YY1aKO mice versus control mice fed a high-fat diet for 2 weeks. (G) Ex vivo oxygen consumption of EWAT homogenates from YY1aKO versus control mice measured by Clark electrode. Data are represented as means ± SEM. *, P < 0.05, and **, P < 0.01, by t test (n = 5 to 8).
To determine if these elevated levels of gene expression translated to functional energetics, fatty acid or glutamine substrate oxidation was measured in white adipose tissue homogenates. Oleic acid and glutamine oxidation rates measured by released of radiolabeled 14CO2 were elevated in EWAT of YY1aKO mice (Fig. 5E and F). Consistent with the increased complete substrate oxidation, ex vivo oxygen consumption using epididymal white fat showed increased respiration in YY1aKO mice (Fig. 5G). Together, these results indicate that YY1 deficiency in adipose tissue caused an increase in gene expression programs linked to energy catabolism and expenditure in visceral fat. These specific energetic changes in visceral adipose tissue might also contribute to the lower body weight of adipose YY1-deficient mice.
Secreted factors linked to energy expenditure are increased in brown fat of YY1-Ucp1 KO mice.
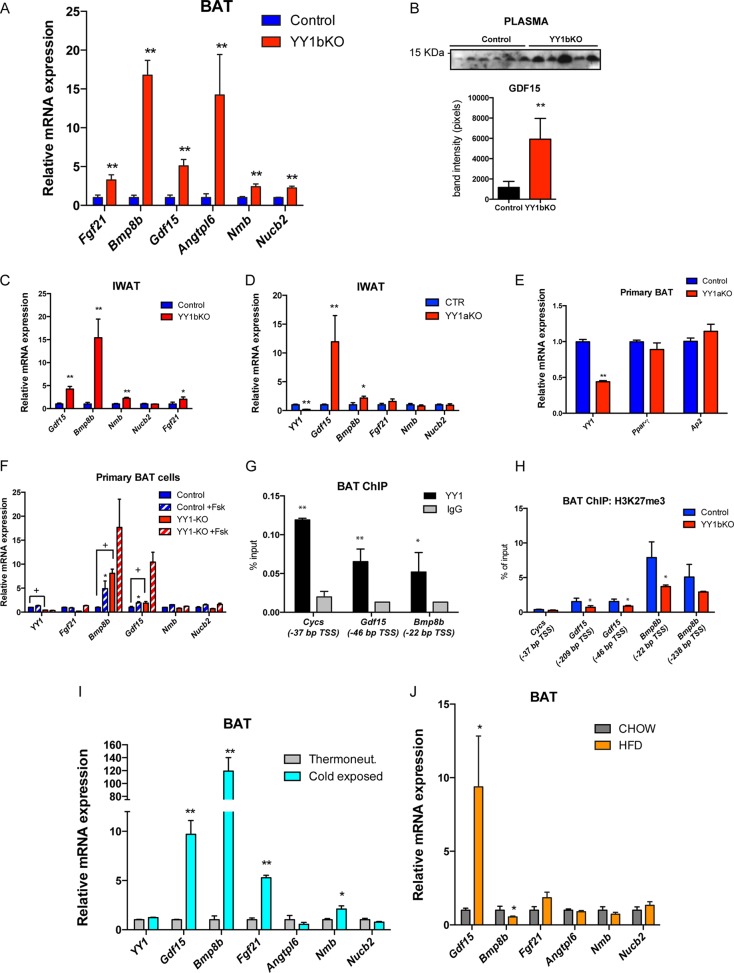
The decreased thermogenic capacity of BAT and the increased thermogenic activity of beige and white fat in YY1 mutant mice suggest that there may be alternative compensatory mechanisms driven by BAT, but independent of BAT thermogenesis, that protect against obesity. Based on the data that mitochondrial oxidation and thermogenic pathways were activated in white adipose tissue but defective in BAT, we hypothesized that BAT-secreted proteins might account for increased energy expenditure in YY1 mutant mice. To test this, whole-genome gene expression profiling comparing BAT from control and YY1bKO mice was analyzed to identify upregulated genes encoding secreted proteins. We used the Signal P4.1 algorithm (49) to detect peptide signals (indicative of secreted proteins) on protein sequences of upregulated genes in BAT YY1 KO mice. Among the 167 genes that were upregulated more than 2-fold, 24 genes were found to contain a signal peptide. At the top of this list, there were 6 secreted proteins that have been linked to energy expenditure, including FGF21 (26), BMP8b (29), GDF15 (50, 51), angiopoietin-like 6 (ANGPTL6) (52), neuromedin B (NMB) (53), and nesfatin (NUCB2) (54), and verified by qPCR (Fig. 6A). Plasmatic protein levels of GDF15 were also increased (Fig. 6B). In addition, the expression of Gdf15, Bmp8b, Nmb, and Fgf21 was also increased in IWAT of YY1bKO (Fig. 6C) and Gdf15 and Bmp8b in IWAT of YY1aKO (Fig. 6D), suggesting that these factors could mediate an enhanced local action within IWAT. To assess whether YY1 directly suppresses the expression of the genes encoding these secreted proteins in a cell-autonomous manner, we isolated preadipocytes from interscapular brown fat of wild-type and YY1bKO mice and differentiated them in vitro. Brown adipocytes from both genotypes exhibited the same degree of differentiation as assessed by intracellular lipid accumulation (data not shown) and markers such as aP2 and PPARγ (Fig. 6E). Among the top six brown fat genes encoding increased expression of secreted proteins in YY1bKO mice, the expression of the BMP8b and GDF15 genes was increased in YY1-deficient brown adipocytes, while the expression of the other four genes remained unchanged (Fig. 6F). These results suggest that within the list of elevated transcripts encoding secreted proteins in YY1 KO brown fat, the BMP8b and GDF15 transcripts were regulated by YY1 in a cell-autonomous manner; however, the FGF21, neuromedin B, nesfatin, and Angptl6 genes were regulated through the actions of other tissues. To confirm that BMP8b and GDF15 were direct YY1 targets, we searched for the presence of YY1 DNA binding sites (ATGGCG) (20) within the proximal promoter (1.5 kb upstream of the transcription start site, TSS) using the Alibaba 2.1 algorithm (TRANSFAC). The first two strong hits were Gdf15 and Bmp8b as they harbor strong binding sites at bp −46 and −22, respectively (data not shown). Chromatin immunoprecipitation analysis performed with BAT showed that YY1 was recruited to the proximal TSS regions of Gdf15 and Bmp8b (Fig. 6G), further supporting a direct transcriptional repression of these genes. Our previous studies showed that YY1 recruits Polycomb proteins, including the histone methyltransferase EZH2 that forms H3K27me3 marks in insulin signaling promoter genes in skeletal muscle (25). Thus, we analyzed H3K27me3 epigenetic marks by ChIP on Bmp8b and Gdf15 promoters comparing wild-type and YY1 mutant mice. Figure 6H shows that Bmp8b and Gdf15 promoters from BAT of YY1 mutant mice have decreased H3K27me3 marks, indicating that YY1 functions as a repressor of Bmp8b and Gdf15 genes.
FIG 6.
Identification of brown fat-secreted factors, including the products of the Gdf15 and Bmp8b genes, elevated in brown adipose tissue of YY1 mutant mice. (A) Gene expression validation of identified secreted proteins by qPCR (n = 8). (B) Detection of plasma GDF15 by Western blotting and quantification of band intensities from YY1bKO versus control mice fed a high-fat diet (2 weeks). (C and D) Gene expression of IWAT from YY1bKO (C) and YY1aKO (D) mice fed a high-fat diet for 2 weeks (n = 6 to 8). (E) Gene expression of primary BAT cells from YY1aKO mice showing differentiation markers. (F) Gene expression of primary BAT adipocytes from control and YY1aKO mice treated with DMSO or 10 μM forskolin (n = 3). (G) YY1 ChIP in BAT from wild-type mice showing the relative binding versus IgG in Gdf15 and Bmp8b promoters. Cytochrome c (Cycs) was used as a positive control. The genomic position of YY1 binding is indicated as relative distance from the transcription start site (TSS) in base pairs. (H) ChIP of the repressive H3K27me3 mark in BAT from YY1bKO versus control mice (n = 3) in Gdf15 and Bmp8b promoters indicating the relative position of the TSS. Cytochrome c was used as the negative control. (I) BAT gene expression of wild-type mice exposed to cold or thermoneutrality for 4 days (n = 5). (J) BAT gene expression of wild-type mice fed on chow or a high-fat diet (HFD) (n = 6). Data are represented as means ± SEM. In panels A to E and G to H, * indicates P < 0.05 and ** indicates P < 0.01 (t test). Analysis of variance (ANOVA) with Bonferroni multiple comparisons was used in panel F (*, P < 0.05; +, P < 0.05).
Finally, to strengthen the previous roles of Bmp8b and GDF15 in energy balance, we investigated their regulatory gene expression in BAT from mice exposed to cold and fed with high-fat diet. Notably, Gdf15 and Bmp8b were induced in cold-exposed mice, but only Gdf15 was upregulated upon high-fat-diet feeding (Fig. 6I and J). Experiments using primary brown fat cells showed that GDF15 is induced by forskolin, indicating that the cyclic AMP (cAMP) pathway that is activated by both cold exposure and a short-term high-fat diet controls GDF15 expression in BAT (Fig. 6I and J).
Taken in aggregate, these results show that among the genes with elevated expression, those encoding secreted proteins in BAT from YY1 mutant mice, Bmpb8 and Gdf15, are direct repressive targets of YY1 maintaining H3K27me3 in their promoters. These data suggest that the repression function of YY1 on Bmp8b and Gdf15 promoters might be attenuated during an elevation in energy demand. In addition, YY1 deficiency in BAT caused an increase in gene expression of secreted proteins, including expression of the FGF21, Neuromedin B, Nesfatin, and Angptl6 genes; however, these genes are not direct YY1 targets and are controlled through the action of other tissues.
DISCUSSION
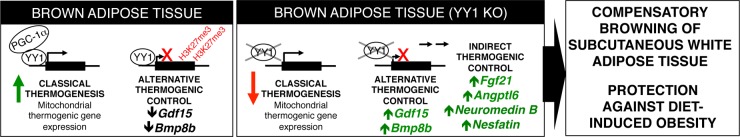
Increasing energy expenditure by inducing mitochondrial biogenesis and thermogenesis of brown/beige or white adipose tissue has become a promising strategy to treat obesity (55). Within this context, it is not completely understood how BAT communicates with beige and white adipose tissue to contribute to energy expenditure or compensate partial deficiencies of BAT thermogenic function. In these studies, we have found a compensatory thermogenic mechanism associated with secreted BAT factors that protects against diet-induced obesity. The transcription factor YY1 plays an antagonistic function in the control of the thermogenic gene expression in BAT. Using the activator function, YY1 controls the canonical thermogenic and mitochondrial gene programs; in contrast, YY1 uses its repressor function to suppress a series of genes encoding secreted proteins that are linked to energy expenditure. Despite a relevant defective BAT thermogenic function, mice lacking YY1 in adipose tissue are protected against diet-induced obesity and show increased thermogenic activity in beige and white adipose tissue. We propose a model by which YY1 controls a set of BAT thermogenic factors that promote beige adipose tissue recruitment, leading to an increase in energy expenditure and protection against obesity (Fig. 7).
FIG 7.
Model. YY1 recruits PGC-1α and induces classical thermogenesis by activating mitochondrial and canonical thermogenic genes in brown fat. However, YY1 plays a direct suppressor role in the control of alternative thermogenic genes, including Bmp8b and Gdf15, and has indirect control of the genes Fgf21, Angptl6, Neuromedin B, and Nesfatin linked to energy expenditure. With excessive caloric intake and loss of YY1, these factors are secreted from BAT, increase thermogenesis in beige and white fat cells, and protect against diet-induced obesity.
Consistent with our previous findings that YY1 controls a transcriptional program of mitochondrial function in skeletal muscle (19, 20), here we show that in addition to mitochondrial OXPHOS genes, YY1 also increases expression of the canonical thermogenic program in BAT, including strong expression of the specific marker UCP1. Mechanistically, YY1 binds to the promoter of these genes and recruits the transcriptional coactivator PGC-1α, suggesting that the elevation of PGC-1α levels under energy-demanding conditions mediated by adrenergic signaling, such as cold, provides a transcriptional activation by YY1 of thermogenic and mitochondrial genes. Whether additional proteins involved in the BAT bioenergetic program, including PRDM16, contribute to this YY1 activation function is unknown. Conversely, YY1 can also function as a repressor of transcription (21). In previous studies, we identified a series of Polycomb proteins of the PRC1 and PRC2 complexes, including Pc2 and Ezh2 methyltransferase, that mediated repression of several genes in the insulin/insulin-like growth factor (IGF) signaling in skeletal muscle (25). Here, we show that in BAT, YY1 represses expression of several secreted proteins that are associated with energy expenditure. Specifically, YY1 was bound to the promoters of Bmp8b and Gdf15 maintaining the repressive epigenetic mark H3K27 trimethylation that is produced through EZH2. At the mechanistic level, how YY1 can activate or repress genes in brown fat could be explained through different recruitment of coactivators or corepressors. However, an important question to address in the future is how these antagonistic transcriptional activities of YY1 are regulated under conditions where energy expenditure is required, such as cold or a high-fat diet.
BAT is a central thermogenic tissue that accounts for a large fraction of energy expenditure associated with thermoregulation or body weight maintenance (16). However, compensatory mechanisms have been reported through adrenergic activation of beige adipose tissue (7). In addition, there are conditions under which increased beige or white adipose tissue-dependent energy expenditure occurs without substantial thermogenic contribution of BAT (6, 11, 14, 56). Adipose-specific YY1 KO mice provide a genetic model that illustrates how BAT defective thermogenic function can be compensated for through activation of beige and white fat and that this compensation is sufficient to protect against diet-induced obesity. Interestingly, BAT YY1 deficiency exhibits increases in a number of secreted proteins, including FGF21, BMP8b, GDF15, neuromedin B, nesfatin, and ANGPTL6, that are linked to energy expenditure (26, 29, 51, 53, 54). As discussed above, BMP8b and GDF15 are direct repressive YY1 targets; however, the rest of genes encoding secreted proteins appear to be indirect targets and to be targeted through non-cell-autonomous regulation. It is unclear what other tissues and mechanisms operate in the adipose YY1-deficient mice and how BAT YY1 causes repression of these genes. The precise contribution of each of these secreted proteins to the lean phenotype of adipose-specific YY1 KO mice is unclear. The nature of this endocrine regulation could act through the central nervous system or by targeting peripheral organs, as is the case with BMP8b (29). In contrast, FGF21 could also play a direct role in adipocytes through induction of UCP1 (26). In addition, increases in circulating levels of GDF15 (51) and ANGPTL6 (52) lead to global increased energy expenditure. Finally, although neuromedin B and nesfatin have an anorexic effect (53, 54), the food intake of YY1 KO mice is not affected, suggesting that other mechanisms such as the increased energy expenditure or action of the other secreted proteins compensate for the food intake. Therefore, it is likely that the synergistic action of these secreted factors is necessary for the protection of diet-induced obesity.
In summary, we have uncovered an antagonistic function of the transcription factor YY1 in BAT through activation of thermogenic genes and by suppression of genes encoding secreted factors associated with energy expenditure. Targeting components of this pathway may be therapeutically useful to treat obesity or associated metabolic diseases.
ACKNOWLEDGMENTS
We thank members of the Puigserver laboratory for advice and fruitful discussions and the BIDMC Animal Research Facility technicians for mouse care. We thank Linus T. Tsai and Evan D. Rosen for providing us the Ucp1-Cre mouse line.
These studies were supported by NIH/NIDDK RO1DK081418 (P.P.) and by postdoctoral fellowships: EMBO Long-Term (F.V.), NIH-1F32DK105679-01 (M.S.S.), and DFG, German Research Foundation HA 7246/1-1 (M.H.).
F.V. designed and performed all of the experiments and wrote the manuscript. M.S.S assisted in mouse organ collection and revised the manuscript. M.H. contributed to the experiments performed during the review process. S.M.B. revised the manuscript and provided conceptual advice and reagents. D.M. assisted with mouse genotyping and tissue oxygen consumption experiments. J.B. revised the manuscript and provided conceptual advice and reagents. P.P. conceived the study, provided conceptual advice, supervised the project, and wrote and revised the manuscript.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Haslam DW, James WP. 2005. Obesity. Lancet 366:1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 2.Lowell BB, Spiegelman BM. 2000. Towards a molecular understanding of adaptive thermogenesis. Nature 404:652–660. [DOI] [PubMed] [Google Scholar]
- 3.Kajimura S, Saito M. 2013. A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annu Rev Physiol 76:225–249. doi: 10.1146/annurev-physiol-021113-170252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicholls DG, Bernson VS, Heaton GM. 1978. The identification of the component in the inner membrane of brown adipose tissue mitochondria responsible for regulating energy dissipation. Exper Suppl (Basel) 32:89–93. doi: 10.1007/978-3-0348-5559-4_9. [DOI] [PubMed] [Google Scholar]
- 5.Wu J, Boström P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, Huang K, Tu H, van Marken Lichtenbelt WD, Hoeks J, Enerbäck S, Schrauwen P, Spiegelman BM. 2012. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiefer FW, Vernochet C, O'Brien P, Spoerl S, Brown JD, Nallamshetty S, Zeyda M, Stulnig TM, Cohen DE, Kahn CR, Plutzky J. 2012. Retinaldehyde dehydrogenase 1 regulates a thermogenic program in white adipose tissue. Nat Med 18:918–925. doi: 10.1038/nm.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz TJ, Huang P, Huang TL, Xue R, McDougall LE, Townsend KL, Cypess AM, Mishina Y, Gussoni E, Tseng YH. 2013. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature 495:379–383. doi: 10.1038/nature11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scimè A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. 2008. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishibashi J, Seale P. 2010. Medicine. Beige can be slimming. Science 328:1113–1114. doi: 10.1126/science.1190816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polak P, Cybulski N, Feige JN, Auwerx J, Rüegg MA, Hall MN. 2008. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab 8:399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Lee MW, Odegaard JI, Mukundan L, Qiu Y, Molofsky AB, Nussbaum JC, Yun K, Locksley RM, Chawla A. 2014. Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell 160:74–87. doi: 10.1016/j.cell.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiu Y, Nguyen KD, Odegaard JI, Cui X, Tian X, Locksley RM, Palmiter RD, Chawla A. 2014. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell 157:1292–1308. doi: 10.1016/j.cell.2014.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, Lo JC, Zeng X, Ye L, Khandekar MJ, Wu J, Gunawardana SC, Banks AS, Camporez JP, Jurczak MJ, Kajimura S, Piston DW, Mathis D, Cinti S, Shulman GI, Seale P, Spiegelman BM. 2014. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell 156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao RR, Long JZ, White JP, Svensson KJ, Lou J, Lokurkar I, Jedrychowski MP, Ruas JL, Wrann CD, Lo JC, Camera DM, Lachey J, Gygi S, Seehra J, Hawley JA, Spiegelman BM. 2014. Meteorin-like is a hormone that regulates immune-adipose interactions to increase beige fat thermogenesis. Cell 157:1279–1291. doi: 10.1016/j.cell.2014.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, Castellot JJ, Rosen ED, Spiegelman BM. 2014. A smooth muscle-like origin for beige adipocytes. Cell Metab 19:810–820. doi: 10.1016/j.cmet.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosen ED, Spiegelman BM. 2014. What we talk about when we talk about fat. Cell 156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92:829–839. doi: 10.1016/S0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 18.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 19.Blättler SM, Verdeguer F, Liesa M, Cunningham JT, Vogel RO, Chim H, Liu H, Romanino K, Shirihai OS, Vazquez F, Rüegg MA, Shi Y, Puigserver P. 2012. Defective mitochondrial morphology and bioenergetic function in mice lacking the transcription factor Yin Yang 1 in skeletal muscle. Mol Cell Biol 32:3333–3346. doi: 10.1128/MCB.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. 2007. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 21.Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. 2003. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J 22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bushmeyer S, Park K, Atchison ML. 1995. Characterization of functional domains within the multifunctional transcription factor, YY1. J Biol Chem 270:30213–30220. doi: 10.1074/jbc.270.50.30213. [DOI] [PubMed] [Google Scholar]
- 23.Wilkinson FH, Park K, Atchison ML. 2006. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci U S A 103:19296–19301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas MJ, Seto E. 1999. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236:197–208. doi: 10.1016/S0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 25.Blättler SM, Cunningham JT, Verdeguer F, Chim H, Haas W, Liu H, Romanino K, Rüegg MA, Gygi SP, Shi Y, Puigserver P. 2012. Yin Yang 1 deficiency in skeletal muscle protects against rapamycin-induced diabetic-like symptoms through activation of insulin/IGF signaling. Cell Metab 15:505–517. doi: 10.1016/j.cmet.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher FM, Kleiner S, Douris N, Fox EC, Mepani RJ, Verdeguer F, Wu J, Kharitonenkov A, Flier JS, Maratos-Flier E, Spiegelman BM. 2012. FGF21 regulates PGC-1α and browning of white adipose tissues in adaptive thermogenesis. Genes Dev 26:271–281. doi: 10.1101/gad.177857.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian SW, Tang Y, Li X, Liu Y, Zhang YY, Huang HY, Xue RD, Yu HY, Guo L, Gao HD, Liu Y, Sun X, Li YM, Jia WP, Tang QQ. 2013. BMP4-mediated brown fat-like changes in white adipose tissue alter glucose and energy homeostasis. Proc Natl Acad Sci U S A 110:E798–E807. doi: 10.1073/pnas.1215236110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, Ahrens MJ, Dudley AT, Norris AW, Kulkarni RN, Kahn CR. 2008. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 454:1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whittle AJ, Carobbio S, Martins L, Slawik M, Hondares E, Vázquez MJ, Morgan D, Csikasz RI, Gallego R, Rodriguez-Cuenca S, Dale M, Virtue S, Villarroya F, Cannon B, Rahmouni K, López M, Vidal-Puig A. 2012. BMP8B increases brown adipose tissue thermogenesis through both central and peripheral actions. Cell 149:871–885. doi: 10.1016/j.cell.2012.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu L, John LM, Adams SH, Yu XX, Tomlinson E, Renz M, Williams PM, Soriano R, Corpuz R, Moffat B, Vandlen R, Simmons L, Foster J, Stephan JP, Tsai SP, Stewart TA. 2004. Fibroblast growth factor 19 increases metabolic rate and reverses dietary and leptin-deficient diabetes. Endocrinology 145:2594–2603. doi: 10.1210/en.2003-1671. [DOI] [PubMed] [Google Scholar]
- 31.Hinoi E, Nakamura Y, Takada S, Fujita H, Iezaki T, Hashizume S, Takahashi S, Odaka Y, Watanabe T, Yoneda Y. 2013. Growth differentiation factor-5 promotes brown adipogenesis in systemic energy expenditure. Diabetes 63:162–175. doi: 10.2337/db13-0808. [DOI] [PubMed] [Google Scholar]
- 32.Bordicchia M, Liu D, Amri EZ, Ailhaud G, Dessì-Fulgheri P, Zhang C, Takahashi N, Sarzani R, Collins S. 2012. Cardiac natriuretic peptides act via p38 MAPK to induce the brown fat thermogenic program in mouse and human adipocytes. J Clin Invest 122:1022–1036. doi: 10.1172/JCI59701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madsen L, Pedersen LM, Lillefosse HH, Fjaere E, Bronstad I, Hao Q, Petersen RK, Hallenborg P, Ma T, De Matteis R, Araujo P, Mercader J, Bonet ML, Hansen JB, Cannon B, Nedergaard J, Wang J, Cinti S, Voshol P, Døskeland SO, Kristiansen K. 2010. UCP1 induction during recruitment of brown adipocytes in white adipose tissue is dependent on cyclooxygenase activity. PLoS One 5:e11391. doi: 10.1371/journal.pone.0011391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vegiopoulos A, Müller-Decker K, Strzoda D, Schmitt I, Chichelnitskiy E, Ostertag A, Berriel Diaz M, Rozman J, Hrabe de Angelis M, Nüsing RM, Meyer CW, Wahli W, Klingenspor M, Herzig S. 2010. Cyclooxygenase-2 controls energy homeostasis in mice by de novo recruitment of brown adipocytes. Science 328:1158–1161. doi: 10.1126/science.1186034. [DOI] [PubMed] [Google Scholar]
- 35.Sun K, Kusminski CM, Luby-Phelps K, Spurgin SB, An YA, Wang QA, Holland WL, Scherer PE. 2014. Brown adipose tissue derived VEGF-A modulates cold tolerance and energy expenditure. Mol Metab 3:474–483. doi: 10.1016/j.molmet.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts LD, Boström P, O'Sullivan JF, Schinzel RT, Lewis GD, Dejam A, Lee YK, Palma MJ, Calhoun S, Georgiadi A, Chen MH, Ramachandran VS, Larson MG, Bouchard C, Rankinen T, Souza AL, Clish CB, Wang TJ, Estall JL, Soukas AA, Cowan CA, Spiegelman BM, Gerszten RE. 2014. β-Aminoisobutyric acid induces browning of white fat and hepatic β-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metab 19:96–108. doi: 10.1016/j.cmet.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boström P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Boström EA, Choi JH, Long JZ, Kajimura S, Zingaretti MC, Vind BF, Tu H, Cinti S, Højlund K, Gygi SP, Spiegelman BM. 2012. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Affar EB, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, Shi Y. 2006. Essential dosage-dependent functions of the transcription factor Yin Yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol 26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong X, Banks A, Liu T, Kazak L, Rao RR, Cohen P, Wang X, Yu S, Lo JC, Tseng YH, Cypess AM, Xue R, Kleiner S, Kang S, Spiegelman BM, Rosen ED. 2014. IRF4 is a key thermogenic transcriptional partner of PGC-1α. Cell 158:69–83. doi: 10.1016/j.cell.2014.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, Klein U, Maratos-Flier E, Rosen ED. 2011. Transcriptional control of adipose lipid handling by IRF4. Cell Metab 13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thonberg H, Fredriksson JM, Nedergaard J, Cannon B. 2002. A novel pathway for adrenergic stimulation of cAMP-response-element-binding protein (CREB) phosphorylation: mediation via alpha1-adrenoceptors and protein kinase C activation. Biochem J 364:73–79. doi: 10.1042/bj3640073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan D, Fujimoto M, Lopes A, Wang YX. 2009. Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell 137:73–86. doi: 10.1016/j.cell.2009.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lowell BB, S-Susulic V, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS. 1992. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature 366:740–742. [DOI] [PubMed] [Google Scholar]
- 45.Feldmann HM, Golozoubova V, Cannon B, Nedergaard J. 2009. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab 9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 46.Cousin B, Cinti S, Morroni M, Raimbault S, Ricquier D, Pénicaud L, Casteilla L. 1992. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J Cell Sci 103:931–942. [DOI] [PubMed] [Google Scholar]
- 47.Young P, Arch JR, Ashwell M. 1984. Brown adipose tissue in the parametrial fat pad of the mouse. FEBS Lett 167:10–14. doi: 10.1016/0014-5793(84)80822-4. [DOI] [PubMed] [Google Scholar]
- 48.Enerbäck S, Jacobsson A, Simpson EM, Guerra C, Yamashita H, Harper ME, Kozak LP. 1997. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 387:90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- 49.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 50.Tsai VW, Macia L, Johnen H, Kuffner T, Manadhar R, Jørgensen SB, Lee-Ng KK, Zhang HP, Wu L, Marquis CP, Jiang L, Husaini Y, Lin S, Herzog H, Brown DA, Sainsbury A, Breit SN. 2013. TGF-b superfamily cytokine MIC-1/GDF15 is a physiological appetite and body weight regulator. PLoS One 8:e55174. doi: 10.1371/journal.pone.0055174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnen H, Lin S, Kuffner T, Brown DA, Tsai VW, Bauskin AR, Wu L, Pankhurst G, Jiang L, Junankar S, Hunter M, Fairlie WD, Lee NJ, Enriquez RF, Baldock PA, Corey E, Apple FS, Murakami MM, Lin EJ, Wang C, During MJ, Sainsbury A, Herzog H, Breit SN. 2007. Tumor-induced anorexia and weight loss are mediated by the TGF-beta superfamily cytokine MIC-1. Nat Med 13:1333–1340. doi: 10.1038/nm1677. [DOI] [PubMed] [Google Scholar]
- 52.Oike Y, Akao M, Yasunaga K, Yamauchi T, Morisada T, Ito Y, Urano T, Kimura Y, Kubota Y, Maekawa H, Miyamoto T, Miyata K, Matsumoto S, Sakai J, Nakagata N, Takeya M, Koseki H, Ogawa Y, Kadowaki T, Suda T. 2005. Angiopoietin-related growth factor antagonizes obesity and insulin resistance. Nat Med 11:400–408. doi: 10.1038/nm1214. [DOI] [PubMed] [Google Scholar]
- 53.Ohki-Hamazaki H, Watase K, Yamamoto K, Ogura H, Yamano M, Yamada K, Maeno H, Imaki J, Kikuyama S, Wada E, Wada K. 1997. Mice lacking bombesin receptor subtype-3 develop metabolic defects and obesity. Nature 390:165–169. doi: 10.1038/36568. [DOI] [PubMed] [Google Scholar]
- 54.Oh-I S, Shimizu H, Satoh T, Okada S, Adachi S, Inoue K, Eguchi H, Yamamoto M, Imaki T, Hashimoto K, Tsuchiya T, Monden T, Horiguchi K, Yamada M, Mori M. 2006. Identification of nesfatin-1 as a satiety molecule in the hypothalamus. Nature 443:709–712. doi: 10.1038/nature05162. [DOI] [PubMed] [Google Scholar]
- 55.Tseng YH, Cypess AM, Kahn CR. 2010. Cellular bioenergetics as a target for obesity therapy. Nat Rev Drug Discov 9:465–482. doi: 10.1038/nrd3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. 2011. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest 121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]