Abstract
Human chorionic gonadotropin (hCG) is composed of a common α subunit and a placenta-specific β subunit. Importantly, hCG is highly expressed in the differentiated and multinucleated syncytiotrophoblast, which is formed via trophoblast cell fusion and stimulated by cyclic AMP (cAMP). Although the ubiquitous activating protein 2 (AP2) transcription factors TFAP2A and TFAP2C may regulate hCGβ expression, it remains unclear how cAMP stimulates placenta-specific hCGβ gene expression and trophoblastic differentiation. Here we demonstrated that the placental transcription factor glial cells missing 1 (GCM1) binds to a highly conserved promoter region in all six hCGβ paralogues by chromatin immunoprecipitation-on-chip (ChIP-chip) analyses. We further showed that cAMP stimulates GCM1 and the CBP coactivator to activate the hCGβ promoter through a GCM1-binding site (GBS1), which also constitutes a previously identified AP2 site. Given that TFAP2C may compete with GCM1 for GBS1, cAMP enhances the association between the hCGβ promoter and GCM1 but not TFAP2C. Indeed, the hCG-cAMP-protein kinase A (PKA) signaling pathway also stimulates Ser269 and Ser275 phosphorylation of GCM1, which recruits CBP to mediate GCM1 acetylation and stabilization. Consequently, hCG stimulates the expression of GCM1 target genes, including the fusogenic protein syncytin-1, to promote placental cell fusion. Our study reveals a positive feedback loop between GCM1 and hCG regulating placental hCGβ expression and cell differentiation.
INTRODUCTION
Successful pregnancy requires a variety of hormones, growth factors, and cytokines to regulate uterine decidualization, embryo implantation, and pregnancy maintenance. For instance, progesterone and estrogen steroid hormones from the ovary prepare the uterine endometrium for embryo implantation. Human chorionic gonadotropin (hCG) is a critical hormone for pregnancy maintenance in humans. hCG is a glycoprotein hormone comprising α and β subunits. The hCGα subunit is shared with other glycoprotein hormones, including thyroid-stimulating hormone, follicle-stimulating hormone, and luteinizing hormone (LH), whereas the hCGβ subunit is specifically expressed in placenta and is unique to hCG. While hCGα is encoded by a single gene on chromosome 6, hCGβ can be encoded by a gene cluster of six (hCGβ1, -2, -3, -5, -7, and -8) paralogues on chromosome 19 (1). Recent studies have suggested that the expression of hCGβ5 and -8 may account for the majority of hCGβ transcripts given that all hCGβ paralogues share high sequence homology in their promoter regions (2, 3).
hCG expression can be detected in early 6- to 8-cell embryos, and this may serve as an embryonic signal for the rendering of appropriate maternal physiology for pregnancy (1). After implantation, the serum hCG level increases dramatically as pregnancy proceeds, reaching its peak at 9 or 10 weeks of gestation, and then decreases and remains at ∼20% of the peak value until term (4). One of the crucial hCG functions is to stimulate progesterone synthesis from the corpus luteum in the early stage of gestation (5, 6). A G-protein-coupled receptor has been identified as the receptor for hCG and LH. When hCG binds to its receptor, the coupled G protein(s) activates adenylyl cyclase, leading to an increase in the concentration of intracellular cyclic AMP (cAMP) that activates protein kinase A (PKA) (5, 7, 8).
The expression profile of hCG during pregnancy is physiologically relevant to the rate of placental growth and the degree of syncytiotrophoblast differentiation. Human placenta is composed of villous tissues, of which the outer surface is a multinucleated syncytiotrophoblast (STB) layer overlying mononucleated cytotrophoblasts (CTBs) (9). Indeed, the latter may differentiate and undergo cell-cell fusion to form a multinucleated STB layer, which is a primary hCG producer and is responsible for gas and nutrient exchange between fetus and mother. It has been known that activation of the cAMP-PKA signaling pathway stimulates trophoblastic differentiation in terms of hCG expression and CTB cell fusion (10, 11). The observation that CTBs and the STB layer express hCG and its receptor suggests that hCG may impose an autocrine and/or a paracrine effect on trophoblastic differentiation through cAMP and PKA (12). Shi et al. (13) showed that inhibition of PKA by H89 blocks hCG-induced trophoblastic differentiation.
The mammalian glial cells missing (GCM) family of transcription factors contains two members, GCM1 and GCM2, which are essential for the development of placenta and parathyroid gland, respectively (14–16). Human GCM1 is primarily expressed in placenta, controls trophoblastic differentiation, and functions via the transcriptional regulation of genes encoding the syncytin-1 and -2 fusogenic proteins, placental growth factor, and the HtrA4 serine protease (17–20). Activation of cAMP signaling by the cAMP stimulant forskolin (FSK) stimulates GCM1 activity as well the expression of its target genes (21, 22). At the molecular level, cAMP activates PKA to phosphorylate Ser269 and Ser275 on GCM1, which recruits dual-specificity phosphatase 23 to dephosphorylate Ser322 (22, 23). Dephosphorylation of Ser322 prevents FBW2-mediated GCM1 ubiquitination and facilitates interactions between GCM1 and the CBP coactivator, resulting in GCM1 acetylation and stabilization by CBP (24, 25).
Because hCG is an essential pregnancy hormone, the mechanism of placenta-specific expression of hCGβ genes has been a subject of intensive studies. The promoter activity of hCGβ5 was stimulated by 8-bromo-cAMP in human JEG-3 placental cells but not hamster BHK fibroblast or INR1-G9 islet cells (26). It was later reported that the activating protein 2 (AP2) transcription factors TFAP2A and TFAP2C bind to GC-rich DNA sequences in the cAMP-responsive region the of hCGβ5 promoter (27–30). Subsequently, the Ets2 and Sp1 transcription factors were shown to activate the above-mentioned hCGβ5 promoter region (30–32). As TFAP2A, TFAP2C, Ets2, and Sp1 are expressed in various types of tissues and cells, it is difficult to reconcile their ubiquitous expression patterns with the cAMP-stimulated and placenta-specific expression pattern of hCGβ genes. In this study, we revisit this issue and unravel positive regulatory circuitry between GCM1 and hCG that is very likely responsible for the placenta-specific expression of hCGβ genes, the exponential rise of the hCG level, and fast trophoblast differentiation in early pregnancy.
MATERIALS AND METHODS
Plasmid constructs.
A DNA fragment encoding human GCM1 with an N-terminal triple-hemagglutinin (HA) tag or a C-terminal FLAG tag was subcloned into a pEF1 expression vector (Invitrogen, Carlsbad, CA) under the control of the EF1 promoter to generate pHA-GCM1 or pGCM1-FLAG. Plasmid pcDNA3-TFAP2C was kindly provided by Shoumo Bhattacharya (University of Oxford, United Kingdom) and used for construction of plasmid pTFAP2C-FLAG, which encodes human TFAP2C with a C-terminal FLAG tag in the pEF1 vector. Plasmid pCBP-HA was described previously (22). A DNA fragment encoding human luteinizing hormone/choriogonadotropin receptor (LH/CGR) with an HA tag after the signaling peptide was subcloned into pEF1 to generate pHA-LH/CGR. The pGL4hCGβ reporter plasmid was constructed by subcloning an hCGβ5 genomic fragment from nucleotides −1674 to +103 relative to the transcription start site into the pGL4 luciferase vector (Promega, Madison, WI). Three potential GCM1-binding sites (GBS1 to -3) in the pGL4hCGβ reporter plasmid were individually mutated to generate the M1, M2, and M3 reporter plasmids. A genomic fragment containing the 600 nucleotides upstream of the human GCM1 transcription start site was subcloned into pE1bLUC to generate the pE1bLUCGCM1-600 reporter plasmid. The p(GBS)4E1bLuc reporter plasmid, which contains four copies of the GBS derived from the syncytin-1 gene, was described previously (22).
Cell culture, transfection, and lentivirus transduction.
HepG2, 293T, BeWo, and JAR cells were obtained from the American Type Culture Collection (Manassas, VA). BeWo31 cells were described previously (25). HepG2, 293T, and JAR cells were maintained at 37°C in minimal essential alpha medium with 10% fetal bovine serum (FBS), 100 μg/ml streptomycin, and 100 U/ml penicillin. BeWo and BeWo31 cells were maintained at 37°C in F-12K medium supplemented with 15% FBS and the above-mentioned antibiotics. For transient expression, cells were transfected with expression plasmids by using the Lipofectamine 2000 reagent (Invitrogen). For luciferase reporter assays, cells were harvested and analyzed with a commercial kit (Promega). Specific luciferase activities were normalized by the protein concentration, which was measured by using a bicinchoninic acid (BCA) protein assay kit (Pierce, Rockford, IL).
Placental villous tissues were obtained from early or term placentas of healthy women undergoing elective dilatation and curettage or scheduled cesarean section with informed consent according to the study protocol (CT-100058) approved by the Institutional Review Board of Cathay General Hospital of Taiwan. The placental tissues were either freshly fixed for immunohistochemistry or subjected to explant culture in Iscove's modified Dulbecco's medium with 20% FBS, 2 mM l-glutamine, 100 μg/ml streptomycin, 100 U/ml penicillin, and 0.25 μg/ml amphotericin B. Primary cytotrophoblast cells isolated from elective termination of midtrimester (21 weeks of gestation) uncomplicated pregnancies were purchased from ScienCell Research Laboratories (Carlsbad, CA) and cultured according to the supplier's instructions.
For stable gene knockdown, BeWo or JAR cells were infected with lentiviruses harboring lentiviral pLKO.1 short hairpin RNA (shRNA) expression plasmids (provided by the National RNAi Core Facility of Taiwan) containing a scramble sequence (5′-CCTAAGGTTAAGTCGCCCTCG-3′) or target sequences for GCM1 (5′-CCTCAGCAGAACTCACTAAAT-3′), TFAP2A (5′-GCTGAATTTCTCAACCGACAA-3′), TFAP2C (5′-CCTCAGCTCTACGTCTAAATA-3′), LH/CGR (5′-CCACTCTCTCACAAGTCTATA-3′), and hCGβ (5′-CCGTGGCTCTCAGCTGTCAAT-3′). Infected cells were subjected to antibiotic selection using 1 μg/ml of puromycin; puromycin-resistant clones were pooled for studies. Rescue of hCGβ expression in GCM1 knockdown BeWo cells was conducted with a recombinant lentivirus harboring a pCDH (SBI, Mountain View, CA) expression cassette for RNA interference (RNAi)-resistant HA-GCM1 (33).
ChIP-chip analysis.
Chromatin immunoprecipitation-on-chip (ChIP-chip) analysis in HA-GCM1-expressing BeWo31 cells was performed as described previously (20). In brief, cells were treated with or without 50 μM FSK for 24 h before being subjected to assays using normal mouse IgG or HA antibody (Ab). The immunopurified genomic fragments were amplified by ligation-mediated PCR, and the resulting amplicons were fragmented and labeled for hybridization with human promoter 1.0R array chips (Affymetrix), followed by data analysis using the Partek software package (Partek, St. Louis, MO). Furthermore, RefSeq mapping data were collected from the UCSC Genome Browser's RefFlat table, and the closest RefSeq 5′ end for each ChIP region was identified.
Immunohistochemistry.
For cellular localization of GCM1, TFAP2C, and hCGβ in vivo, early and term human placental biopsy specimens were fixed in 4% paraformaldehyde, dehydrated, embedded in paraffin, and sectioned at 5 μm. The sections were deparaffinized, rehydrated, and subjected to immunostaining by incubation with normal rabbit/mouse serum, rabbit anti-hCGβ Ab (Dako, Glostrup, Denmark), mouse anti-TFAP2C Ab (Santa Cruz Biotechnology, Santa Cruz, CA), and rabbit anti-GCM1 Ab, respectively. The sections were then incubated sequentially with biotinylated secondary Ab and horseradish peroxidase (HRP)-conjugated streptavidin (Jackson ImmunoResearch, West Grove, PA). Antigenic detection was performed by using an SK-4100 3,3′-diaminobenzidine (DAB) peroxidase substrate kit (Vector Laboratories, Burlingame, CA), and the sections were further counterstained with hematoxylin.
Regulation of hCGβ expression.
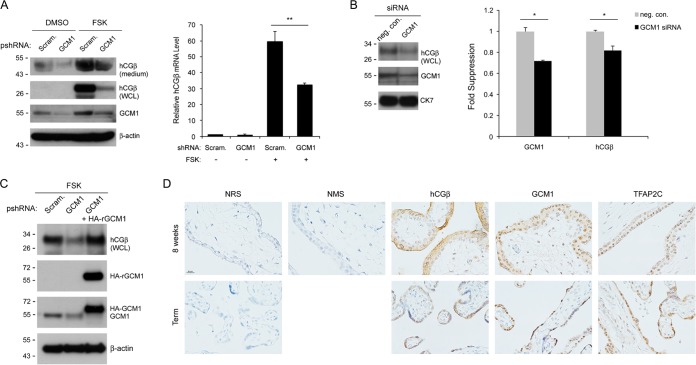
Placental explants were incubated with a small interfering RNA (siRNA) duplex targeting GCM1 (5′-UUCAGAAUCAAAGUCGUCAGGUUCC-3′ and 5′-GGAACCUGACGACUUUGAUUCUGAA-3′) or Silencer Select negative-control siRNA1 (Invitrogen) and then subjected to immunoblotting with hCGβ, GCM1, and cytokeratin 7 (CK7; Millipore, Billerica, MA) Abs or quantitative real-time PCR (qRT-PCR) analyses of GCM1 and hCGβ transcripts. BeWo cells were treated with or without 50 μM FSK for different periods of time to compare the effects of cAMP on hCGβ, GCM1, and TFAP2 gene expression. BeWo cells were also treated with FSK with or without 20 μM H89 for 24 h to assess the effect of H89 on FSK-stimulated GCM1 and hCGβ gene expression. BeWo cells stably expressing scramble, GCM1, TFAP2A, and TFAP2C shRNAs were generated and treated with or without FSK, followed by immunoblot analysis of the hCGβ protein level. Densitometric analysis of immunoblot band intensities was performed by using the ImageJ program (http://imagej.nih.gov/ij/).
On the other hand, TFAP2C knockdown or GCM1 knockdown BeWo cells were transduced with lentiviruses harboring the pCDH-HA-GCM1 or pCDH-TFAP2C-FLAG expression cassette in the presence of FSK to examine whether GCM1 or TFAP2C complements the shortage of hCGβ expression due to TFAP2C or GCM1 knockdown. Regulation of GCM1 promoter activity by TFAP2C was studied in HepG2 cells transfected with pTFAP2C-FLAG and the pE1bLUCGCM1-600 reporter plasmid. Regulation of hCGβ promoter activity by GCM1 or TFAP2C was studied by using HepG2 cells transfected with pGL4hCGβ or its mutant (M1 to M3) reporter plasmid plus different combinations of pHA-GCM1, pTFAP2C-FLAG, and pCBP-HA in the presence or absence of FSK.
Because of low endogenous GCM1 activity, JAR cells were used for gain-of-function experiments for GCM1-mediated hCGβ gene expression. JAR cells were transfected with the indicated amounts of pHA-GCM1 in the presence or absence of 20 μM H89, followed by immunoblotting and qRT-PCR analyses of hCGβ protein and transcript levels. JAR cells stably expressing HA-GCM1 or GCM1 shRNA were also generated with lentiviruses harboring the pCDH-HA-GCM1 or pLKO.1-GCM1 expression cassette and subjected to immunoblotting with hCGβ Ab.
EMSA and ChIP analysis.
The direct interaction between GBS1 and GCM1 or TFAP2C was studied by an electrophoretic mobility shift assay (EMSA). In brief, recombinant GCM1-FLAG and TFAP2C-FLAG proteins were purified from 293T cells transfected with pGCM1-FLAG and pTFAP2C-FLAG, respectively. A radiolabeled oligonucleotide probe harboring GBS1 (5′-GCACCTCCTGCGGGCCTATT-3′) was incubated with GCM1-FLAG or TFAP2C-FLAG alone or TFAP2C plus increasing amounts of GCM1-FLAG. In a separate experiment, nuclear fractions of BeWo cells treated with or without FSK were prepared and used for EMSAs in the presence of GCM1 or TFAP2C Ab. The assay conditions were described previously (17), and the reaction mixtures were analyzed by electrophoresis on 5% nondenaturing polyacrylamide gels. The band intensities were quantified by using the ImageJ program.
Associations of GCM1 or TFAP2C with the syncytin-1 and hCGβ promoters in BeWo or BeWo31 cells were measured by ChIP analysis with HA, GCM1, and TFAP2C Abs. The primer sequences used for ChIP analysis of GBS1 in the hCGβ gene are 5′-GCGGGGAAGGGACTAAG-3′ and 5′-CCAACGAGGGATTCAGC-3′, and those used for ChIP analysis of the proximal GBS in the syncytin-1 gene are 5′-CTCTCTGGAGAGTGAATTACTGAGTC-3′ and 5′-CCTGTCTCTCAGTTGCAAGATAATTGC-3′.
Immunofluorescence.
For colocalization of hCGβ and GCM1, JAR cells were transfected with the pHA-GCM1 expression plasmid. At 48 h posttransfection, cells were fixed, sequentially stained with mouse anti-hCGβ Ab (Santa Cruz Biotechnology) and Cy2-labeled anti-mouse IgG (Jackson ImmunoResearch), and then stained with tetramethylrhodamine isothiocyanate (TRITC)-labeled anti-HA Ab (Sigma, St. Louis, MO). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Immunofluorescence was examined under a Leica TCS SP5 confocal microscope (Leica, Wetzlar, Germany).
Immunoassays of cAMP and hCGβ.
Because commercial hCG preparations from urine of pregnant women may be contaminated with a growth factor(s) such as epidermal growth factor (EGF) (34), three different commercial hCG proteins, no. 1 (catalogue number GWB-791987; GenWay, San Diego, CA), no. 2 (catalogue number GC10; Sigma), and no. 3 (catalogue number C0434; Sigma), were screened for tyrosine phosphorylation of EGF receptor (EGFR) in 293T cells, and no EGFR activation was detected (data not shown). Unless otherwise indicated, the no. 1 commercial hCG protein was used for the most part in this study. The levels of intracellular cAMP induced by hCG were measured by using a cAMP direct immunoassay kit (BioVision, Mountain View, CA) according to the manufacturer's instructions. For quantitation of secretory hCGβ, the culture media were collected and secretory hCBβ was measured with an Access Total hCGβ assay kit (Beckman Coulter, Miami, FL).
GCM1 phosphorylation and acetylation.
To study the effect of hCG on GCM1 modification, 293T cells were transfected with pGCM1-FLAG and pHA-LH/CGR for 24 h, followed by treatment with the above-described commercial hCG proteins at 50 or 100 IU/ml. After 24 h, cells were harvested for measurement of cAMP levels or subjected to coimmunoprecipitation with a phosphor-specific Ab to Ser269 and Ser275 (p-Ser269275-GCM1) and FLAG Ab. In a separate experiment, cells were subjected to coimmunoprecipitation with anti-acetylated lysine (Ac-K) Ab (Cell Signaling, Beverly, MA) and FLAG Ab for analysis of GCM1 acetylation. The effect of hCG on GCM1 phosphorylation and acetylation was also investigated by using BeWo31 cells and primary cytotrophoblast cells. In brief, cells were treated with the indicated concentrations of hCG, followed by coimmunoprecipitation with different combinations of p-Ser269275-GCM1, Ac-K, HA, and GCM1 Abs. The p-Ser269275-GCM1 Ab raised in guinea pigs was described previously (23). In a separate experiment, BeWo31 cells were transfected with p(GBS)4E1bLuc in the presence or absence of hCG to study the effect of hCG on GCM1 transcriptional activity.
Regulation of GCM1 target gene expression by hCG.
The regulation of GCM1 and expression of its target genes by hCG signaling was investigated by different approaches. First, JAR cells expressing scramble or LH/CGR shRNA were transfected with the empty vector pEF1 or pHA-GCM1 for 48 h, followed by immunoblotting with hCGβ, HA, GCM1, and LH/CGR Abs. Second, BeWo cells treated with 4 μg/ml normal IgG or LH/CGR Ab for 72 h and BeWo cells stably expressing hCGβ shRNA were subjected to immunoblotting with GCM1, hCGβ, syncytin-1, and HtrA4 Abs. Third, BeWo cells were mock treated or treated with 100 IU/ml hCG or hCG plus 5 μM H89 for 4 or 7 days, followed by immunoblotting and qRT-PCR analyses of GCM1 and the indicated GCM1 target genes. The LH/CGR Ab was raised in rabbits against a synthetic oligopeptide corresponding to amino acids 127 to 143 (LifeSpan Biosciences, Seattle, WA).
qRT-PCR.
Cells were lysed by using a RealTime Ready cell lysis kit (Roche Applied Science, Indianapolis, IN) and reverse transcribed, followed by analysis of the indicated transcripts with LightCycler 480 real-time PCR instrument II (Roche Applied Science). The sequences of the primer sets for PCR analysis are as follows: 5′-CTGACAAGGCTTTTTTCTTCACA-3′ and 5′-CCAGACGGGACAGGTTT-3′ for endogenous GCM1, 5′-CCTCTCATCCTCATCAGCAAT-3′ and 5′-TCGTCGTCCTTGTAATCTGGT-3′ for HA-GCM1, 5′-CTGAGTCTCTGAGGTCACTT-3′ and 5′-TGATAGGATGCTGGGGT-3′ for hCGβ, 5′-CATTCCGCTCCTGATGT-3′ and 5′-AGTGGACTCTGAGGTGAC-3′ for hCGα, 5′-GTTGGACAAGATTGGGTTGA-3′ and 5′-TTACACAGTTGCTGGGC-3′ for TFAP2C, 5′-GAAGGCCCTTCATAACCAATGA-3′ and 5′-GATATTTGGCTAAGGAGGTGATGTC-3′ for syncytin-1, 5′-GTCAGCACCAAACAGCG-3′ and 5′-GGAGATTCCATCAGTCACCC-3′ for HtrA4, and 5′-AACTCCATCATGAAGTGTGACG-3′ and 5′-GATCCACATCTGCTGGAAGG-3′ for β-actin.
Cell-cell fusion assay.
Stimulation of cell-cell fusion by hCG was examined in scramble or GCM1 shRNA-expressing BeWo cells treated with or without 100 IU/ml hCG for 96 h, followed by immunofluorescence staining with E-cadherin Ab (BD Biosciences, San Jose, CA) and Alexa Fluor 546-conjugated goat anti-mouse IgG (Invitrogen) and nuclear staining with DAPI. Images were captured by using a confocal microscope as described above. Three microscopic fields per sample were randomly selected for examination in each of three independent experiments. Quantification of cell fusion was calculated as a fusion index of (N − S)/T, where N is the number of nuclei in the syncytia, S is the number of syncytia, and T is the total number of nuclei counted.
Statistical analysis.
Statistical analysis of the data was performed by using Student's t test or analysis of multiple variance (ANOVA) with a Bonferroni post hoc test (see Fig. 4B and D and 5C and D). We also used the Spearman rank correlation for testing trends in the data shown in Fig. 7C. A P value of <0.05 was considered statistically significant. A P value of >0.05 was considered to be of no statistical significance.
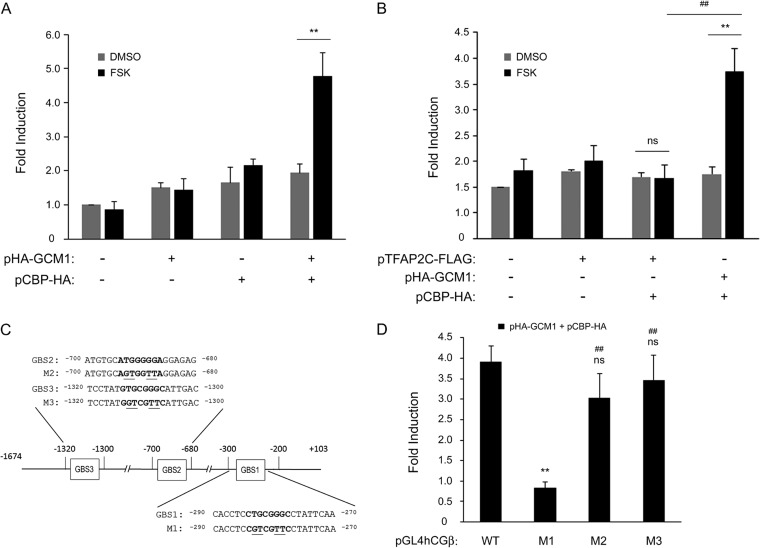
FIG 4.
Regulation of hCGβ promoter activity by GCM1. (A) GCM1 and CBP collaborate to regulate hCGβ promoter activity in response to cAMP stimulation. HepG2 cells were transfected with pGL4hCGβ, pHA-GCM1, and pCBP-HA for 24 h. Cells were mock treated (dimethyl sulfoxide [DMSO]) or treated with 50 μM FSK for an additional 24 h before being harvested for luciferase assays. Mean values and standard deviations obtained from four independent experiments are presented. (B) TFAP2C is not involved in cAMP-induced hCGβ promoter activation. Different combinations of pGL4hCGβ, pHA-GCM1, pTFAP2C-FLAG, and pCBP-HA were transfected into HepG2 cells for hCGβ promoter activity analysis as described above for panel A. ** and ##, P < 0.01. (C) Schematic representation of potential GCM1-binding sites in the hCGβ promoter. Sequences of the potential GBSs are listed in boldface type. Note that the nucleotide residues essential for GCM1 binding in each GBS were changed by site-directed mutagenesis and are underlined. (D) Identification of a GCM1-responsive element in the hCGβ promoter. HepG2 cells were transfected with pHA-GCM1 and pCBP-HA plus wild-type or mutant pGL4hCGβ (M1 to M3) for 24 h. Cells were mock treated or treated with 50 μM FSK for an additional 24 h before being harvested for luciferase assays. Mean values and standard deviations obtained from three independent experiments are presented. **, P < 0.01 (compared with the wild type); ns, not significant (compared with the wild type); ##, P < 0.01 (compared with M1).
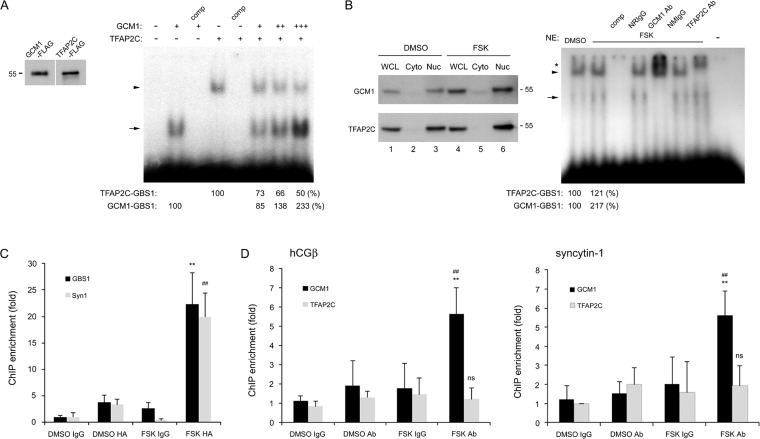
FIG 5.
Association of the hCGβ promoter with GCM1 and TFAP2C. (A and B) GCM1 competes with TFAP2C for GBS1 in the hCGβ promoter. (A) Approximately 1 ng of the radiolabeled GBS1 oligonucleotide probe was incubated with 0.3 μg of recombinant TFAP2C-FLAG alone or with increasing amounts of recombinant GCM1-FLAG (0.5, 1, and 2 μg) for EMSAs. As a control, unlabeled GBS1 (comp) was added at a 100-fold excess for competition. The arrowhead and arrow indicate the TFAP2C-GBS1 complex and the GCM1-GBS1 complex, respectively. The numbers at the bottom indicate the protein-DNA complex band intensities. Recombinant TFAP2C-FLAG and GCM1-FLAG proteins were analyzed by immunoblotting with anti-FLAG Ab (left). (B) Nuclear (Nuc) and cytosolic (Cyto) fractions were prepared from BeWo cells treated with dimethyl sulfoxide (DMSO) or FSK and subjected to immunoblotting with GCM1 or TFAP2C Ab. Approximately 5 μg of the nuclear fraction from mock-treated or FSK-treated BeWo cells was incubated with the radiolabeled GBS1 oligonucleotide probe in the presence of normal rabbit IgG (NRIgG), normal mouse IgG (NMIgG), TFAP2C, or GCM1 Ab for EMSAs. The arrowhead and arrow indicate the TFAP2C-GBS1 complex and the GCM1-GBS1 complex, respectively. The asterisk indicates the supershifted TFAP2C-GBS1 or GCM1-GBS1 complex. The numbers at the bottom indicate the protein-DNA complex band intensities. NE, nuclear extract. (C and D) cAMP stimulation enhances the occupancy of the hCGβ promoter by GCM1. (C) BeWo31 cells treated with dimethyl sulfoxide or FSK were subjected to ChIP analysis using normal mouse IgG or HA Ab. The immunopurified genomic fragments were analyzed by qRT-PCR with primer sets specific for hCGβ GBS1 and the proximal GBS in the syncytin-1 (Syn1) promoter. Mean values and standard deviations obtained from three independent experiments are presented. ** and ##, P < 0.01 (compared with dimethyl sulfoxide-HA Ab treatment for GBS1 and Syn1, respectively). (D) BeWo cells treated with dimethyl sulfoxide or FSK were subjected to ChIP analysis using normal rabbit IgG, GCM1, or TFAP2C Ab. The immunopurified genomic fragments were quantitated as described above for panel C. Mean values and standard deviations obtained from three independent experiments are presented. **, P < 0.01 (compared with dimethyl sulfoxide-GCM1 or TFAP2C Ab treatment); ns, not significant (compared with dimethyl sulfoxide-GCM1 or TFAP2C Ab treatment); ##, P < 0.01 (compared with FSK and TFAP2C Ab treatment).
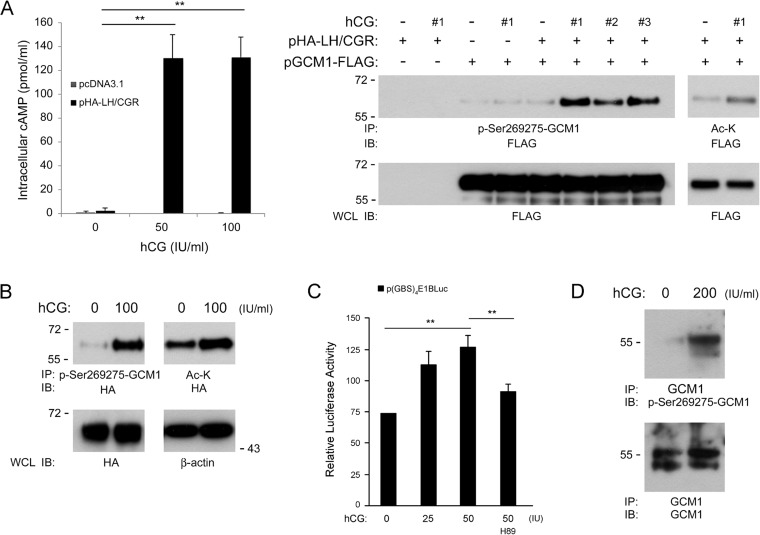
FIG 7.
Regulation of GCM1 posttranslational modification and activity by hCG. (A) Stimulation of Ser269 and Ser275 phosphorylation in GCM1 by hCG. (Left) 293T cells were transfected with the empty vector or pHA-LH/CGR for 24 h. Cells were then treated without or with 50 or 100 IU/ml of hCG for an additional 24 h before they were harvested for cAMP assays. Mean values and standard deviations obtained from three independent experiments are presented. **, P < 0.01. (Right) On the other hand, 293T cells were transfected with pGCM1-FLAG and pHA-LH/CGR and treated without or with 100 IU/ml of hCG from different commercial suppliers (#1 to #3). Cells were subjected to immunoprecipitation (IP) with p-Ser269275-GCM1 or Ac-K Ab followed by immunoblotting (IB) with FLAG Ab. (B) Stimulation of GCM1 phosphorylation and acetylation in placental cells by hCG. BeWo31 cells were mock treated or treated with the indicated amounts of hCG for 24 h before being harvested for immunoprecipitation analysis with p-Ser269275-GCM1 or Ac-K Ab followed by immunoblotting with HA Ab. (C) Stimulation of GCM1 transcriptional activity by hCG. BeWo31 cells were transfected with the GCM1 reporter plasmid p(GBS)4E1bLuc for 24 h. Cells were then treated with hCG or hCG plus 5 μM H89 for an additional 24 h before being harvested for luciferase assays. Mean values and standard deviations obtained from three independent experiments are presented. There was a significant correlation (by the Spearman rank correlation test) between GCM1-mediated transcriptional activation and hCG treatment. **, P < 0.01. (D) hCG stimulates Ser269 and Ser275 phosphorylation of GCM1 in primary trophoblast cells. Human primary trophoblast cells were mock treated or treated with 200 IU/ml hCG and then harvested for immunoprecipitation with GCM1 Ab, followed by immunoblotting with p-Ser269275-GCM1 Ab.
RESULTS
Expression of GCM1 and hCGβ in placenta.
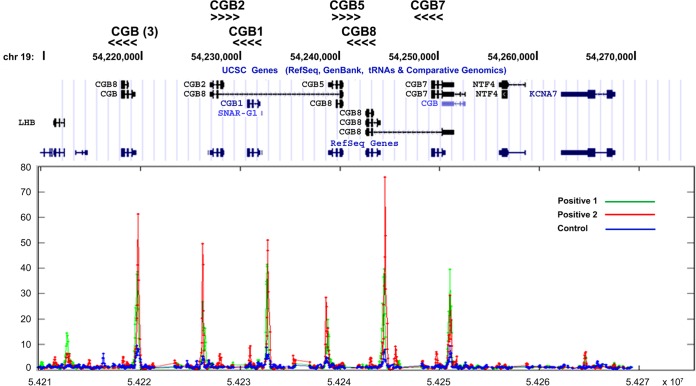
Human GCM1 is a novel placental transcription factor, and its target genes are very likely responsible for the unique phenotypes in placenta. The trophoblast-like BeWo cell line has been widely used as an in vitro model for human trophoblasts. Accordingly, we performed ChIP-chip experiments with FSK-treated BeWo31 cells, which are BeWo cells stably expressing HA-tagged GCM1, and identified hCGβ as a candidate GCM1 target gene. As shown in Fig. 1, specific hybridization signals were detected in all members of the hCGβ gene cluster but not in flanking genes such as KCNA7 (potassium voltage-gated channel), NTF4 (neurotrophin 4), and LHB (luteinizing hormone β subunit). Given that the intensity of hybridization signals and the gene orientation are different among hCGβ paralogues, HA-GCM1 binding was localized to a similar position in the proximal promoter regions of all hCGβ paralogues, including the two pseudogenes hCGβ1 and -2 (CGB1 and -2 in Fig. 1). This observation is most likely due to a high degree of sequence homology in the proximal promoter regions of all hCGβ paralogues. To support the ChIP-chip observations, we further conducted ChIP analyses using normal rabbit IgG or GCM1 Ab in BeWo cells treated with FSK. The genomic fragments immunopurified by GCM1 Ab were subjected to TA cloning for sequencing. Indeed, we detected an association of GCM1 with all members of the hCGβ gene cluster (see Fig. S1 in the supplemental material).
FIG 1.
hCGβ is a target gene of GCM1. BeWo31 cells stably expressing HA-GCM1 were treated with 50 μM FSK for 24 h and then subjected to ChIP-chip analysis using anti-HA Ab (positive) or normal mouse IgG (control). The immunopurified genomic fragments were amplified and labeled for hybridization to chips of the human promoter 1.0R array. Note that significant hybridization signals were detected in the proximal promoter region of all six paralogues of the hCGβ gene cluster but not flanking genes such as KCNA7, NTF4, and LHB. The orientations of hCGβ paralogues are marked by arrowheads.
Because cAMP signaling is crucial for the activation of hCGβ expression, we tested whether GCM1 is involved in cAMP-induced hCGβ gene expression. BeWo cells stably expressing scramble or GCM1 shRNA were mock treated or treated with FSK, followed by immunoblotting and qRT-PCR analyses of hCGβ protein and transcript levels. The hCGβ protein levels in both culture medium and whole-cell lysates were upregulated by FSK in scramble control BeWo cells (Fig. 2A, left). Correspondingly, FSK treatment also increased the hCGβ transcript level in scramble control BeWo cells (Fig. 2A, right). In contrast, the stimulatory effect of FSK on hCGβ expression was significantly compromised in GCM1 knockdown BeWo cells at both the protein and transcript levels (Fig. 2A). Moreover, treatment of term placental villous explants with GCM1 siRNA also decreased hCGβ expression (Fig. 2B). We further transduced GCM1 knockdown BeWo cells with RNAi-resistant HA-GCM1-expressing lentiviruses and were able to reinstate the stimulation of hCGβ expression by FSK (Fig. 2C). Finally, immunohistochemistry indicated that both GCM1 and hCGβ are highly expressed in the syncytiotrophoblast layer of placentas at 8 weeks of gestation and full term (Fig. 2D). Taken together, these results suggested that GCM1 is required for the regulation of hCGβ expression by cAMP in placenta.
FIG 2.
Regulation of hCGβ expression by GCM1. (A) GCM1 knockdown suppresses hCGβ expression in placental cells. BeWo cells stably expressing scramble or GCM1 shRNA were treated with dimethyl sulfoxide (DMSO) or 50 μM FSK for 24 h. Culture supernatants (medium) and cells (WCL [whole-cell lysate]) were harvested for immunoblotting with GCM1, hCGβ, and β-actin Abs. Note that secreted and glycosylated hCGβ has a higher molecular mass. In a separate experiment, cells were harvested for qRT-PCR analysis of hCGβ mRNA. (B) GCM1 knockdown suppresses hCGβ expression in placental explants. Human full-term placental explants were incubated with negative-control (neg. con.) or GCM1 siRNA, followed by immunoblotting with the indicated Abs or qRT-PCR analyses of GCM1 and hCGβ transcripts. (C) Complementation of hCGβ expression in GCM1 knockdown BeWo cells. Lentiviruses harboring an RNAi-resistant HA-GCM1 (HA-rGCM1) expression cassette were transduced into GCM1-knockdown BeWo cells treated with 50 μM FSK. Cells were harvested for immunoblotting with the indicated Abs. (D) Immunohistochemistry of hCGβ, GCM1, and TFAP2C in human placentas. First-trimester (8 weeks) and full-term placental sections were incubated with hCGβ, GCM1, and TFAP2C Abs, respectively. As a control, sections were incubated with normal rabbit serum (NRS) or normal mouse serum (NMS). Bar, 20 μm. Mean values and standard deviations obtained from three independent experiments are presented in panels A and B. *, P < 0.05; **, P < 0.01.
GCM1 and TFAP2C are required for cAMP-stimulated hCGβ expression.
We then compared the functional roles of TFAP2A, TFAP2C, and GCM1 in the regulation of hCGβ expression. We reasoned that the expression level of a transcription factor might be correlated with that of hCGβ should the factor be involved in the induction of hCGβ expression by cAMP stimulation. As shown in Fig. 3A, immunoblotting of BeWo cells treated with FSK for different periods of time revealed that hCGβ upregulation is correlated with GCM1 but not with either TFAP2A or TFAP2C. Notably, elevated GCM1 protein and transcript levels were detected 4 h after FSK treatment, which occurred prior to the rise of hCGβ protein levels (i.e., 8 h after FSK treatment) (Fig. 3A, left and middle). The upregulation of GCM1 and hCGβ protein and transcript levels by FSK treatment lasted for 24 h, the longest treatment time in the present study (Fig. 3A, left and middle). Of note, the PKA inhibitor H89 was able to suppress FSK-stimulated GCM1 and hCGβ expression (Fig. 3A, right).
FIG 3.
GCM1 is required for cAMP-induced hCGβ expression. (A) Correlated expression of hCGβ and GCM1 in response to cAMP stimulation. BeWo cells were treated with or without 50 μM FSK for the indicated periods of time. Cells were then harvested for immunoblotting with GCM1, hCGβ, TFAP2A, TFAP2C, and β-actin Abs or for qRT-PCR analysis of GCM1 and hCGβ mRNAs. The numbers at the bottom indicate the protein band intensities. In a separate experiment, BeWo cells were treated with FSK plus 20 μM H89 for 24 h for immunoblotting with GCM1, hCGβ, and β-actin Abs. Mean values and standard deviations obtained from three independent experiments are presented. ** and ##, P < 0.01 (compared with dimethyl sulfoxide [DMSO] treatment for GCM1 and hCGβ mRNA, respectively). (B) GCM1 and TFAP2C are involved in the stimulation of hCGβ expression by cAMP. BeWo cells stably expressing scramble, GCM1, TFAP2A, or TFAP2C shRNA were treated with 50 μM FSK for 24 h. Cells were then harvested for immunoblotting with the indicated Abs. (C) TFAP2C knockdown suppresses GCM1 expression. Scramble control and TFAP2C knockdown BeWo cells were mock treated or treated with 50 μM FSK for 24 h and then harvested for qRT-PCR analysis of GCM1 mRNA. Mean values and standard deviations obtained from three independent experiments are presented. *, P < 0.05; **, P < 0.01. (D) GCM1 is downstream of TFAP2C in stimulation of hCGβ expression by cAMP. TFAP2C knockdown or GCM1 knockdown BeWo cells were transduced with lentiviruses harboring an HA-GCM1 or TFAP2C-FLAG expression cassette. At 72 h postransduction, cells were treated with 50 μM FSK for an additional 24 h. Cells were then harvested for immunoblotting using hCGβ, GCM1, TFAP2C, HA, FLAG, and β-actin Abs. (E) TFAP2C regulates GCM1 promoter activity. HepG2 cells were transfected with 0.1 μg of pE1bLUCGCM1-600 and different amounts of pTFAP2C-FLAG. In a separate experiment, HepG2 cells were transfected with 0.2 μg of TFAP2C-FLAG plus 0.1 μg of pE1bLUCGCM1-600 or TFAP2C site-deleted (mutated) pE1bLUCGCM1-600. At 48 h posttransfection, cells were harvested for luciferase assays. Mean values and standard deviations obtained from three independent experiments are presented. *, P < 0.05; **, P < 0.01.
As a complementary approach, loss-of-function experiments were conducted in BeWo cells stably expressing TFAP2A, TFAP2C, or GCM1 shRNA to compare its role in regulation of hCGβ expression in response to cAMP stimulation. As expected, GCM1 knockdown impeded FSK-upregulated hCGβ expression (Fig. 3B). Interestingly, knockdown of TFAP2C but not of TFAP2A also diminished the stimulation of GCM1 and hCGβ expression by FSK (Fig. 3B). The diminishment of GCM1 expression by TFAP2C knockdown was further confirmed at the transcriptional level by qRT-PCR analysis (Fig. 3C). We transduced TFAP2C knockdown BeWo cells with HA-GCM1-expressing lentiviruses and were able to restore FSK-upregulated hCGβ expression (Fig. 3D, left). Moreover, transduction of GCM1 knockdown BeWo cells with TFAP2C-FLAG-expressing lentiviruses failed to reverse hCGβ expression in the presence of FSK (Fig. 3D, right). These observations suggested that GCM1 might be downstream of TFAP2C in the control of hCGβ expression. To test this hypothesis, we further tested whether TFAP2C regulates GCM1 promoter activity by transient-expression assays. HepG2 cells, which lack endogenous TFAP2C activity (27, 35), were transfected with pTFAP2C-FLAG and pE1bLUCGCM1-600. As shown in Fig. 3E (left), the luciferase activity directed by the 0.6-kb promoter region of the GCM1 gene in pE1bLUCGCM1-600 was stimulated by TFAP2C-FLAG. A functional TFAP2C site (nucleotides −361 to −351 relative to the transcriptional start site) was identified in the GCM1 promoter, as the deletion of this site significantly reduced TFAP2C-driven transactivation (Fig. 3E, right). In addition, the expression pattern of TFAP2C was similar to that of GCM1 in trophoblasts of first-trimester and full-term placentas by immunohistochemistry (Fig. 2D). Collectively, these results suggested that TFAP2C is upstream of GCM1 and may indirectly modulate hCGβ gene expression through GCM1.
Regulation of hCGβ gene expression by GCM1.
We then studied how GCM1 regulates hCGβ at the transcriptional level. To this end, we examined whether GCM1 regulates hCGβ promoter activity in HepG2 cells, which do not express endogenous GCM1 either. Our previous studies indicated that cAMP signaling stimulates GCM1 activity by enhancing the interaction between GCM1 and the transcriptional coactivator CBP (22). Therefore, HepG2 cells were transfected with pGL4hCGβ, pHA-GCM1, and pCBP-HA in the presence or absence of FSK. While the luciferase activity directed by pGL4hCGβ was marginally affected by GCM1 or CBP alone or both GCM1 and CBP in the absence of FSK, it was significantly stimulated by both GCM1 and CBP in the presence of FSK (Fig. 4A). As a control, we failed to detect any significant effect of both TFAP2C and CBP on hCGβ promoter activity under FSK treatment (Fig. 4B). We inspected the hCGβ genomic sequence and identified three potential GCM1-binding sites (GBS1 to -3) in its promoter region from nucleotides −1674 to +103 upstream of the transcription start site (Fig. 4C). Each of the three potential GBSs was individually mutated in the pGL4hCGβ reporter plasmid to generate the M1, M2, and M3 reporter constructs (Fig. 4C). By transient-expression assays, we observed that GBS1 mutation dramatically diminishes the synergistic effect of GCM1 and CBP on hCGβ promoter activity under FSK treatment (Fig. 4D). These results suggest that GCM1 and CBP are very likely to upregulate hCGβ expression via GBS1 in response to cAMP stimulation.
The above-mentioned GBS1 element (CTGCGGGC) is a GCM1-responsive element crucial for hCGβ activation; it also overlaps a previously reported TFAP2-binding site (AP2 [CCTGCGGGC]) (27–30). To characterize the potential interplay between GCM1 and TFAP2C at GBS1, an EMSA was performed by using recombinant GCM1-FLAG and TFAP2C-FLAG and an oligonucleotide probe harboring the GBS1 and AP2 sequences. As shown in Fig. 5A, both GCM1 and TFAP2C were able to bind the GBS1 probe. Interestingly, under a constant amount of TFAP2C, the formation of the TFAP2C-GBS1 complex was decreased by increasing amounts of GCM1 with a concomitant increase in GCM1-GBS1 complex formation (a 2-fold increase, as determined by densitometry) (Fig. 5A). We also conducted EMSAs using nuclear extracts from mock- and FSK-treated BeWo cells. As expected, the amount of nuclear GCM1 but not TFAP2C was stimulated by FSK treatment (Fig. 5B, left, lanes 3 and 6). Correspondingly, the formation of the GCM1-GBS1 complex was increased 2-fold with nuclear extracts of FSK-treated BeWo cells (Fig. 5B, right). The associations between GCM1 and the hCGβ promoter were further compared in mock- and FSK-treated BeWo31 cells by ChIP analysis. Using the syncytin-1 promoter as a positive control, we observed that FSK treatment stimulates the occupancy of GBS1 in the hCGβ promoter and the proximal GBS in the syncytin-1 promoter by HA-GCM1 (Fig. 5C). We further compared the efficiencies of binding of GCM1 and TFAP2C to the hCGβ and syncytin-1 promoters in BeWo cells treated with or without FSK. Indeed, FSK treatment enhanced the association between the hCGβ promoter and GCM1 but not TFAP2C (Fig. 5D, left). As expected, the association between the syncytin-1 promoter and GCM1 was also enhanced by FSK (Fig. 5D, right). Therefore, given that GBS1 is a composite GBS/AP2 site, cAMP enhances GCM1 binding to the hCGβ promoter to upregulate hCGβ gene expression.
Regulation of GCM1 activity by hCG signaling.
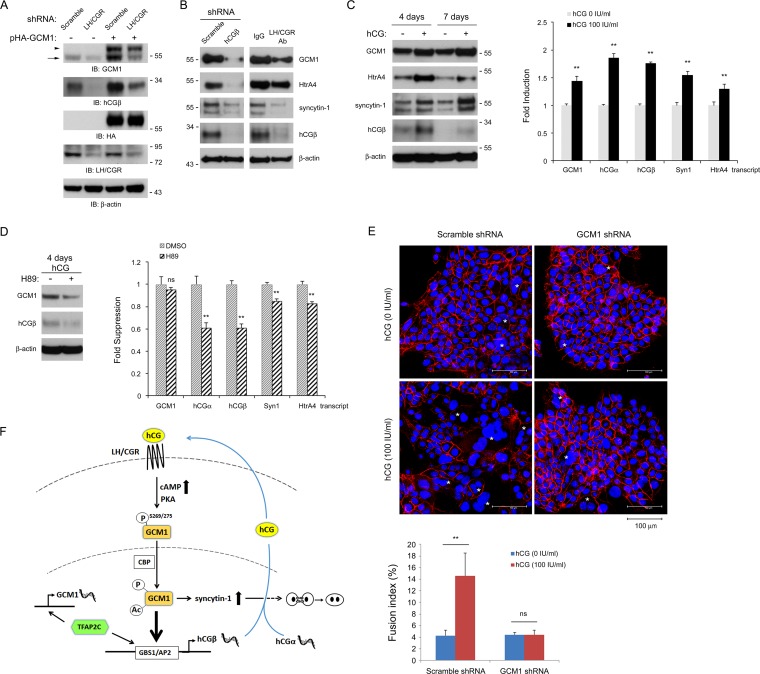
The regulation of hCGβ expression by GCM1 was further investigated by gain-of-function experiments in trophoblast-like JAR cells, which exhibit lower endogenous GCM1 activity. Increasing amounts of pHA-GCM1 were transfected into JAR cells, and a positive correlation between HA-GCM1 and hCGβ expression was observed in a dose-dependent manner (Fig. 6A, left). Unexpectedly, the level of endogenous GCM1 protein (Fig. 6A, left, bottom band) was also elevated in HA-GCM1-expressing JAR cells in a dose-dependent manner, given that its transcript level was not significantly altered (Fig. 6C). This lower band was unlikely derived from internal translation or N-terminal truncation of HA-GCM1, as it was not detected in HepG2 cells expressing HA-GCM1 (Fig. 6A, right). The positive effect of HA-GCM1 on hCGβ and endogenous GCM1 expression was further blocked by H89 (Fig. 6A, middle). Moreover, immunofluorescence microscopy showed that hCGβ expression was upregulated in JAR cells expressing HA-GCM1 (Fig. 6B), and qRT-PCR revealed correlated expression of HA-GCM1 and hCGβ mRNA (Fig. 6C). Stable HA-GCM1-expressing and GCM1 knockdown JAR cells were generated, respectively. By immunoblotting or enzyme-linked immunosorbent assays (ELISAs), we confirmed that the level of hCGβ secretion was increased by HA-GCM1 overexpression and decreased by GCM1 knockdown in JAR cells (Fig. 6D, right). In line with data from the transient-expression experiments, the endogenous GCM1 protein level was increased in HA-GCM1-expressing JAR cells (Fig. 6D, left).
FIG 6.
Autoregulation of GCM1. (A) Expression of endogenous hCGβ and GCM1 proteins is elevated by exogenous HA-GCM1. JAR or HepG2 cells were transfected with the indicated amounts of pHA-GCM1. At 48 h posttransfection, cells were harvested for immunoblotting with hCGβ, GCM1, HA, and β-actin Abs. In a separate experiment, JAR cells transfected with pHA-GCM1 were treated with 20 μM H89 for 24 h before harvest for immunoblot analysis. (B) JAR cells transfected with pHA-GCM1 were subjected to immunofluorescence staining with HA (a) and hCGβ (b) Abs. Nuclei were stained with DAPI (c) (blue). (d) Merge image of panels a, b, and c. Bars, 20 μm. (C) Expression of endogenous hCGβ but not GCM1 transcript is elevated by exogenous HA-GCM1. Transfected JAR cells, as described above for panel A, were harvested for RNA purification and qRT-PCR analyses of HA-GCM1, GCM1, hCGβ, and hCGα transcripts. (D) Correlation of GCM1 and hCGβ expression. JAR cells stably expressing scramble or GCM1 shRNA and JAR cells stably expressing the empty pCDH vector or HA-GCM1 were established. Cells were harvested for immunoblotting with hCGβ, GCM1, HA, and β-actin Abs. In addition, the hCGβ levels in culture supernatants were measured by an ELISA (right). Mean values and standard deviations obtained from three independent experiments are presented in panels C and D. **, P < 0.01.
We speculated that the induction of hCGβ expression by HA-GCM1 results in elevated hCG production, which may impose a paracrine and/or autocrine effect on the endogenous GCM1 protein level. To study whether hCG regulates GCM1 expression and activity, we first tested whether GCM1 modification is regulated by hCG signaling. To this end, 293T cells were transfected with pGCM1-FLAG and pHA-LH/CGR, followed by treatment with or without hCG. As expected, the intracellular cAMP level was induced by hCG in pHA-LH/CGR-transfected but not mock-transfected 293T cells (Fig. 7A, left). Our previous studies indicated that cAMP activates PKA to phosphorylate GCM1 at Ser269 and Ser275 (23). Indeed, enhanced Ser269 and Ser275 phosphorylation in GCM1-FLAG was detected in 293T cells coexpressing GCM1-FLAG and HA-LH/CGR that were treated with three different commercial hCG preparations (Fig. 7A, right). Phosphorylation of the Ser269 and Ser275 residues promotes GCM1 acetylation and stabilization by CBP (23). Correspondingly, the acetylation of GCM1-FLAG was also stimulated by hCG in 293T cells coexpressing GCM1-FLAG and HA-LH/CGR (Fig. 7A, right). We further demonstrated that hCG treatment stimulates HA-GCM1 phosphorylation and acetylation in BeWo31 cells (Fig. 7B). Moreover, the transcriptional activity of GCM1 on the p(GBS)4E1bLuc reporter plasmid was stimulated by hCG, which was counteracted by the PKA inhibitor H89 (Fig. 7C). Finally, we demonstrated that hCG facilitates Ser269 and Ser275 phosphorylation of GCM1 in primary trophoblast cells prepared from human placentas (Fig. 7D). Our results support that GCM1 activity is upregulated by hCG through GCM1 phosphorylation and acetylation.
Regulation of GCM1 target gene expression and placental cell differentiation by hCG.
We then investigated the role of hCG signaling in the control of GCM1 and GCM1 target gene expression. Stable LH/CGR knockdown JAR cells were transfected with or without pHA-GCM1, followed by immunoblot analysis with GCM1 Ab. While HA-GCM1 expression in scramble control JAR cells increased the endogenous GCM1 protein level, this positive effect was compromised in LH/CGR knockdown cells (Fig. 8A). Correspondingly, hCGβ expression was decreased and barely stimulated by HA-GCM1 in LH/CGR knockdown cells (Fig. 8A). We further tested the effects of the hCG-LH/CGR signaling pathway on GCM1 target gene expression in BeWo cells. Knockdown of hCGβ decreased the levels of the GCM1 protein and its target genes, including HtrA4 and syncytin-1, in BeWo cells (Fig. 8B, left). Likewise, the protein levels of GCM1, hCGβ, HtrA4, and syncytin-1 were decreased in BeWo cells treated with Ab against the external N-terminal domain of LH/CGR (Fig. 8B, right). Furthermore, the protein and transcript levels of GCM1 and its target genes were measured in BeWo cells under hCG treatment for 4 or 7 days. As shown in Fig. 8C, the protein and transcript levels of GCM1, hCGβ, HtrA4, and syncytin-1 were upregulated by hCG. The PKA inhibitor H89 counteracted the hCG-induced upregulation of GCM1 and hCGβ proteins (Fig. 8D, left). Interestingly, H89 treatment significantly decreased the levels of hCGα, hCGβ, syncytin-1, and HtrA4 transcripts but not GCM1 transcripts in BeWo cells treated with hCG for 4 days (Fig. 8D, right). Therefore, H89 might block PKA-mediated phosphorylation and stabilization of GCM1.
FIG 8.
Regulation of GCM1 target gene expression and placental cell fusion by hCG. (A) Autoregulation of GCM1 is linked to the hCG-LH/CGR signaling pathway. JAR cells stably expressing scramble or LH/CGR shRNA were transfected with or without pHA-GCM1 for 48 h, followed by immunoblotting with the indicated Abs. Note that endogenous GCM1 (arrow) was not induced in LH/CGR knockdown JAR cells in the presence of HA-GCM1 (arrowhead). (B and C) hCG stimulates GCM1 and GCM1 target gene expression. (B) Scramble control or hCGβ knockdown BeWo cells were subjected to immunoblotting with the indicated Abs. On the other hand, BeWo cells were incubated with 4 μg/ml normal IgG or LH/CGR Ab for 72 h and then harvested for immunoblotting with the indicated Abs. Note that the endogenous GCM1 level decreases when hCGβ is knocked down or in the presence of LH/CGR Ab. (C) BeWo cells were mock treated or treated with 100 IU/ml hCG for 4 or 7 days. Cells were then harvested for immunoblotting with the indicated Abs. In separate experiments, BeWo cells treated with 100 IU/ml hCG for 4 days were subjected to qRT-PCR analyses of GCM1, hCGα, hCGβ, syncytin-1 (Syn1), and HtrA4 transcripts. (D) BeWo cells were treated with 100 IU/ml hCG plus 5 μM H89 for 4 days. Cells were then harvested for immunoblotting and qRT-PCR analyses as described above for panel C. Mean values and standard deviations obtained from three independent experiments are presented in panels C and D. **, P < 0.01 (compared with mock or dimethyl sulfoxide [DMSO] treatment). (E) Regulation of placental cell fusion by hCG. BeWo cells expressing scramble or GCM1 shRNA were treated with or without 100 IU/ml hCG for 4 days and then subjected to immunofluorescence staining with E-cadherin Ab. Fusion events and syncytium formation (denoted by asterisks) were examined by fluorescence microscopy. Quantification of the level of cell fusion is described in Materials and Methods. Mean values and standard deviations obtained from three independent experiments are presented. **, P < 0.01. (F) A positive feedback loop governs GCM1 activity, hCGβ expression, and placental cell differentiation. The binding of hCG to LH/CGR elevates the cAMP level to activate PKA, which phosphorylates GCM1 on Ser269 and Ser275. These phosphorylation events facilitate the interaction between GCM1 and CBP, which acetylates, stabilizes, and collaborates with GCM1 to activate hCGβ expression through GBS1. TFAP2C may mediate GCM1 expression and compete with GCM1 for GBS1 as well because it also harbors an AP2-binding site. Nevertheless, the activity of GCM1, but not that of TFAP2C, is stimulated by the cAMP-PKA signaling pathway to support the placenta-specific expression of hCGβ genes. Expression of the hCGα gene is also stimulated by hCG, and together with hCGβ expression, more hCG may be released to further activate the expression of GCM1 and its target genes such as syncytin-1 to promote placental cell fusion and differentiation.
Consequently, we tested whether the hCG-GCM1 axis regulates trophoblastic differentiation in terms of BeWo cell fusion. As shown in Fig. 8E, immunofluorescence staining of cell surface E-cadherin for distinguishing syncytia from unfused mononucleated cells revealed that hCG significantly stimulated cell-cell fusion in scramble shRNA-expressing BeWo cells (fusion index of 14.6% for treated cells versus 4.3% for untreated cells). In contrast, cell fusion events were not affected by hCG in GCM1 knockdown BeWo cells (Fig. 8E). Taken together, the hCG-LH/CGR signaling pathway is required for autoregulation of GCM1 and hCG in placental cell differentiation.
DISCUSSION
The hCGβ subunit distinguishes hCG from the other glycoprotein hormones and is encoded by a gene cluster specifically expressed in placental trophoblasts. Notably, rising hCG levels are correlated with the rapid proliferation and differentiation of syncytiotrophoblasts in the early gestation stage. As hCG is essential for the maintenance of pregnancy, elucidation of the regulatory mechanism of hCG expression in placenta is of crucial clinical importance. It has long been known that cAMP signaling induces placental syncytiotrophoblast differentiation and hCG expression. Previous studies indicated that AP2-binding sites in the promoters of the hCGα and hCGβ genes are involved in the regulation of their expression under cAMP stimulation (27, 29). Interestingly, some of these AP2-binding sites have also been reported to be trophoblast-specific elements (TSEs). Steger et al. (36) reported a 56-kDa protein in JEG-3 placental cells that binds TSEs in both the hCGα and hCGβ genes, and later on, the same group further identified TFAP2C as the 56-kDa nuclear protein by affinity chromatography with the hCGα TSE (29). On the other hand, Knofler et al. (28) reported that TFAP2A is involved in the regulation of hCGβ promoter activity. Given that TFAP2A and TFAP2C are abundant in placental cells, evidence supporting a stimulating effect of cAMP on TFAP2A or TFAP2C expression at the protein and transcript levels is scant or incoherent. In addition, the fact that TFAP2A and TFAP2C are ubiquitously expressed in human tissues further complicates the issue of how hCGβ gene expression is specifically regulated in placenta.
We revisited this issue and reasoned that the mechanism underlying hCGβ gene expression and trophoblastic differentiation should comply with the following aspects: first, the transcription factor specifically drives hCGβ gene expression in placenta; second, the activity of the transcription factor is stimulated by cAMP; and third, target genes of the transcription factor are associated with placental cell differentiation.
In the present study, we provided multiple lines of evidence showing that GCM1 is responsible for cAMP-stimulated hCGβ gene expression in placenta. We demonstrated that cAMP induces temporal changes in hCGβ expression that are coordinated with GCM1, but not TFAP2A or TFAP2C, expression. This notion is substantiated by loss-of-function (i.e., RNA interference) experiments showing that GCM1 knockdown impairs the stimulation of hCGβ expression by cAMP and by ChIP-chip experiments showing that GCM1 recognizes a highly conserved promoter region in all hCGβ paralogues. We further attributed a key GCM1-responsive element, GBS1, in the proximal promoters of all hCGβ paralogues for GCM1-mediated hCGβ promoter activation in response to cAMP stimulation. Indeed, GBS1 harbors an AP2 site and was also previously defined as the hCGβ TSE. Given that TFAP2C may compete with GCM1 for binding to GBS1 (or the TSE), we demonstrated that the efficiency of binding of GCM1 to GBS1 is enhanced under cAMP stimulation in EMSAs and ChIP analyses (Fig. 5B to D). This notion can be attributed to the fact that GCM1 expression and stability are upregulated by the cAMP-PKA signaling pathway. Because GCM1 is specifically expressed in trophoblasts, we believe that GCM1, but not TFAP2C or TFAP2A, is the bona fide placental factor collaborating with CBP to modulate hCGβ gene expression under cAMP stimulation.
Unexpectedly, the stimulation of GCM1 and hCGβ expression by cAMP in BeWo placental cells was impeded when TFAP2C was knocked down. Because GCM1 overexpression counteracts the observed negative effect of TFAP2C knockdown and TFAP2C regulates GCM1 promoter activity, it is very likely that TFAP2C maintains the basal expression level of GCM1 in placenta and thereby indirectly regulates hCGβ gene expression through GCM1. Gene-targeting studies have reported that TFAP2C may be located upstream of Cdx2 and Eomes and is essential for the development of extraembryonic tissues (37–39). Therefore, TFAP2C may regulate placental development via genetic programs executed by key placental transcription factors, including GCM1, Cdx2, and Eomes. TFAP2A has also been implicated in the regulation of hCGβ gene expression. The facts that TFAP2A knockdown fails to affect cAMP-induced hCGβ expression in the present study and that TFAP2A knockout mice exhibit defective phenotypes irrelevant to placental development (40) indicate that TFAP2 is unlikely to modulate hCGβ gene expression in trophoblasts.
This study also demonstrated that hCG signaling may enhance GCM1 stability and activity by promoting GCM1 phosphorylation and acetylation. Specifically, gain-of-function experiments revealed that hCGβ transcript and protein expression levels are elevated in JAR cells expressing exogenous HA-GCM1. Interestingly, the endogenous GCM1 protein level was also increased in HA-GCM1-expressing JAR cells, given that the endogenous GCM1 transcript level was not changed. We speculated that HA-GCM1 expression imposes a posttranslational effect on endogenous GCM1 expression through the hCG signaling pathway because the observed positive effect of HA-GCM1 is impaired by LH/CGR knockdown. Moreover, hCGβ knockdown or treatment with LH/CGR Ab in BeWo cells decreases the expression of GCM1 and its target genes, further supporting that hCG regulates GCM1 activity. Indeed, this notion was corroborated by evidence that the activation of LH/CGR by hCG elevates intracellular cAMP levels to stimulate PKA-mediated Ser269 and Ser275 phosphorylation of GCM1 in BeWo31 cells, primary trophoblast cells, and 293T cells coexpressing LH/CGR and GCM1. Phosphorylation of Ser269 and Ser275 enhances the interaction of GCM1 and CBP to promote GCM1 acetylation and stabilization and transactivation of GCM1 target genes (Fig. 8F).
The rapid proliferation and differentiation of trophoblastic villi coincide with rising hCG levels during the first trimester of gestation. In this scenario, hCG activates the cAMP-PKA signaling pathway via its G-protein-coupled transmembrane receptor LH/CGR to upregulate GCM1 expression and activity in placental trophoblasts. On the other hand, hCG also stimulates hCGα gene expression (Fig. 8C). Together with the transactivation of the hCGβ gene by GCM1, more hCG is produced and released to further stimulate the expression of GCM1 and its target genes. Consequently, the level of the syncytin-1 fusogenic protein is elevated to mediate trophoblast fusion to meet physiological requirements in the early stage of placental development (Fig. 8F). Therefore, our study reveals a positive feedback loop between GCM1 and hCG that regulates placental development and functions.
Supplementary Material
ACKNOWLEDGMENT
We thank Hui-Ju Tsai at the Division of Biostatistics and Bioinformatics, National Health Research Institutes, for assistance in statistical analysis.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00655-15.
REFERENCES
- 1.Jameson JL, Hollenberg AN. 1993. Regulation of chorionic gonadotropin gene expression. Endocr Rev 14:203–221. doi: 10.1210/edrv-14-2-203. [DOI] [PubMed] [Google Scholar]
- 2.Hollenberg AN, Pestell RG, Albanese C, Boers ME, Jameson JL. 1994. Multiple promoter elements in the human chorionic gonadotropin beta subunit genes distinguish their expression from the luteinizing hormone beta gene. Mol Cell Endocrinol 106:111–119. doi: 10.1016/0303-7207(94)90192-9. [DOI] [PubMed] [Google Scholar]
- 3.Rull K, Laan M. 2005. Expression of beta-subunit of HCG genes during normal and failed pregnancy. Hum Reprod 20:3360–3368. doi: 10.1093/humrep/dei261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole LA. 2009. New discoveries on the biology and detection of human chorionic gonadotropin. Reprod Biol Endocrinol 7:8. doi: 10.1186/1477-7827-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malassine A, Cronier L. 2002. Hormones and human trophoblast differentiation: a review. Endocrine 19:3–11. doi: 10.1385/ENDO:19:1:3. [DOI] [PubMed] [Google Scholar]
- 6.Keay SD, Vatish M, Karteris E, Hillhouse EW, Randeva HS. 2004. The role of hCG in reproductive medicine. BJOG 111:1218–1228. doi: 10.1111/j.1471-0528.2004.00412.x. [DOI] [PubMed] [Google Scholar]
- 7.Cronier L, Bastide B, Herve JC, Deleze J, Malassine A. 1994. Gap junctional communication during human trophoblast differentiation: influence of human chorionic gonadotropin. Endocrinology 135:402–408. doi: 10.1210/endo.135.1.8013377. [DOI] [PubMed] [Google Scholar]
- 8.Weedon-Fekjaer MS, Tasken K. 2012. Review: spatiotemporal dynamics of hCG/cAMP signaling and regulation of placental function. Placenta 33(Suppl):S87–S91. doi: 10.1016/j.placenta.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Benirschke K, Kaufmann P. 2001. Early development of the human placenta, p 42–49. In Benirschke K, Kaufmann P (ed), Pathology of the human placenta, 4th ed Springer-Verlag, New York, NY. [Google Scholar]
- 10.Keryer G, Alsat E, Tasken K, Evain-Brion D. 1998. Cyclic AMP-dependent protein kinases and human trophoblast cell differentiation in vitro. J Cell Sci 111:995–1004. [DOI] [PubMed] [Google Scholar]
- 11.Wice B, Menton D, Geuze H, Schwartz AL. 1990. Modulators of cyclic AMP metabolism induce syncytiotrophoblast formation in vitro. Exp Cell Res 186:306–316. doi: 10.1016/0014-4827(90)90310-7. [DOI] [PubMed] [Google Scholar]
- 12.Pidoux G, Gerbaud P, Tsatsaris V, Marpeau O, Ferreira F, Meduri G, Guibourdenche J, Badet J, Evain-Brion D, Frendo JL. 2007. Biochemical characterization and modulation of LH/CG-receptor during human trophoblast differentiation. J Cell Physiol 212:26–35. doi: 10.1002/jcp.20995. [DOI] [PubMed] [Google Scholar]
- 13.Shi QJ, Lei ZM, Rao CV, Lin J. 1993. Novel role of human chorionic gonadotropin in differentiation of human cytotrophoblasts. Endocrinology 132:1387–1395. doi: 10.1210/endo.132.3.7679981. [DOI] [PubMed] [Google Scholar]
- 14.Anson-Cartwright L, Dawson K, Holmyard D, Fisher SJ, Lazzarini RA, Cross JC. 2000. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat Genet 25:311–314. doi: 10.1038/77076. [DOI] [PubMed] [Google Scholar]
- 15.Schreiber J, Riethmacher-Sonnenberg E, Riethmacher D, Tuerk EE, Enderich J, Bosl MR, Wegner M. 2000. Placental failure in mice lacking the mammalian homolog of glial cells missing, GCMa. Mol Cell Biol 20:2466–2474. doi: 10.1128/MCB.20.7.2466-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunther T, Chen ZF, Kim J, Priemel M, Rueger JM, Amling M, Moseley JM, Martin TJ, Anderson DJ, Karsenty G. 2000. Genetic ablation of parathyroid glands reveals another source of parathyroid hormone. Nature 406:199–203. doi: 10.1038/35018111. [DOI] [PubMed] [Google Scholar]
- 17.Yu C, Shen K, Lin M, Chen P, Lin C, Chang GD, Chen H. 2002. GCMa regulates the syncytin-mediated trophoblastic fusion. J Biol Chem 277:50062–50068. doi: 10.1074/jbc.M209316200. [DOI] [PubMed] [Google Scholar]
- 18.Liang CY, Wang LJ, Chen CP, Chen LF, Chen YH, Chen H. 2010. GCM1 regulation of the expression of syncytin 2 and its cognate receptor MFSD2A in human placenta. Biol Reprod 83:387–395. doi: 10.1095/biolreprod.110.083915. [DOI] [PubMed] [Google Scholar]
- 19.Chang M, Mukherjea D, Gobble RM, Groesch KA, Torry RJ, Torry DS. 2008. Glial cell missing 1 regulates placental growth factor (PGF) gene transcription in human trophoblast. Biol Reprod 78:841–851. doi: 10.1095/biolreprod.107.065599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang LJ, Cheong ML, Lee YS, Lee MT, Chen H. 2012. High-temperature requirement protein A4 (HtrA4) suppresses the fusogenic activity of syncytin-1 and promotes trophoblast invasion. Mol Cell Biol 32:3707–3717. doi: 10.1128/MCB.00223-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knerr I, Schubert SW, Wich C, Amann K, Aigner T, Vogler T, Jung R, Dotsch J, Rascher W, Hashemolhosseini S. 2005. Stimulation of GCMa and syncytin via cAMP mediated PKA signaling in human trophoblastic cells under normoxic and hypoxic conditions. FEBS Lett 579:3991–3998. doi: 10.1016/j.febslet.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 22.Chang CW, Chuang HC, Yu C, Yao TP, Chen H. 2005. Stimulation of GCMa transcriptional activity by cyclic AMP/protein kinase A signaling is attributed to CBP-mediated acetylation of GCMa. Mol Cell Biol 25:8401–8414. doi: 10.1128/MCB.25.19.8401-8414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin FY, Chang CW, Cheong ML, Chen HC, Lee DY, Chang GD, Chen H. 2011. Dual-specificity phosphatase 23 mediates GCM1 dephosphorylation and activation. Nucleic Acids Res 39:848–861. doi: 10.1093/nar/gkq838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang CS, Yu C, Chuang HC, Chang CW, Chang GD, Yao TP, Chen H. 2005. FBW2 targets GCMa to the ubiquitin-proteasome degradation system. J Biol Chem 280:10083–10090. doi: 10.1074/jbc.M413986200. [DOI] [PubMed] [Google Scholar]
- 25.Chiang MH, Liang FY, Chen CP, Chang CW, Cheong ML, Wang LJ, Liang CY, Lin FY, Chou CC, Chen H. 2009. Mechanism of hypoxia-induced GCM1 degradation: implications for the pathogenesis of preeclampsia. J Biol Chem 284:17411–17419. doi: 10.1074/jbc.M109.016170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jameson JL, Lindell CM. 1988. Isolation and characterization of the human chorionic gonadotropin beta subunit (CG beta) gene cluster: regulation of transcriptionally active CG beta gene by cyclic AMP. Mol Cell Biol 8:5100–5107. doi: 10.1128/MCB.8.12.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson W, Albanese C, Handwerger S, Williams T, Pestell RG, Jameson JL. 1997. Regulation of the human chorionic gonadotropin alpha- and beta-subunit promoters by AP-2. J Biol Chem 272:15405–15412. doi: 10.1074/jbc.272.24.15405. [DOI] [PubMed] [Google Scholar]
- 28.Knofler M, Saleh L, Bauer S, Galos B, Rotheneder H, Husslein P, Helmer H. 2004. Transcriptional regulation of the human chorionic gonadotropin beta gene during villous trophoblast differentiation. Endocrinology 145:1685–1694. doi: 10.1210/en.2003-0954. [DOI] [PubMed] [Google Scholar]
- 29.LiCalsi C, Christophe S, Steger DJ, Buescher M, Fischer W, Mellon PL. 2000. AP-2 family members regulate basal and cAMP-induced expression of human chorionic gonadotropin. Nucleic Acids Res 28:1036–1043. doi: 10.1093/nar/28.4.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson BD, Cheng YH, Langland RA, Handwerger S. 2001. Differential expression of AP-2gamma and AP-2alpha during human trophoblast differentiation. Life Sci 69:2157–2165. doi: 10.1016/S0024-3205(01)01299-1. [DOI] [PubMed] [Google Scholar]
- 31.Johnson W, Jameson JL. 2000. Role of Ets2 in cyclic AMP regulation of the human chorionic gonadotropin beta promoter. Mol Cell Endocrinol 165:17–24. doi: 10.1016/S0303-7207(00)00269-0. [DOI] [PubMed] [Google Scholar]
- 32.Johnson W, Jameson JL. 1999. AP-2 (activating protein 2) and Sp1 (selective promoter factor 1) regulatory elements play distinct roles in the control of basal activity and cyclic adenosine 3′,5′-monophosphate responsiveness of the human chorionic gonadotropin-beta promoter. Mol Endocrinol 13:1963–1975. doi: 10.1210/mend.13.11.0386. [DOI] [PubMed] [Google Scholar]
- 33.Chang CW, Chang GD, Chen H. 2011. A novel cyclic AMP/Epac1/CaMKI signaling cascade promotes GCM1 desumoylation and placental cell fusion. Mol Cell Biol 31:3820–3831. doi: 10.1128/MCB.05582-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saleh L, Prast J, Haslinger P, Husslein P, Helmer H, Knofler M. 2007. Effects of different human chorionic gonadotrophin preparations on trophoblast differentiation. Placenta 28:199–203. doi: 10.1016/j.placenta.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Williams T, Admon A, Luscher B, Tjian R. 1988. Cloning and expression of AP-2, a cell-type-specific transcription factor that activates inducible enhancer elements. Genes Dev 2:1557–1569. doi: 10.1101/gad.2.12a.1557. [DOI] [PubMed] [Google Scholar]
- 36.Steger DJ, Buscher M, Hecht JH, Mellon PL. 1993. Coordinate control of the alpha- and beta-subunit genes of human chorionic gonadotropin by trophoblast-specific element-binding protein. Mol Endocrinol 7:1579–1588. doi: 10.1210/mend.7.12.7511787. [DOI] [PubMed] [Google Scholar]
- 37.Auman HJ, Nottoli T, Lakiza O, Winger Q, Donaldson S, Williams T. 2002. Transcription factor AP-2gamma is essential in the extra-embryonic lineages for early postimplantation development. Development 129:2733–2747. [DOI] [PubMed] [Google Scholar]
- 38.Werling U, Schorle H. 2002. Transcription factor gene AP-2 gamma essential for early murine development. Mol Cell Biol 22:3149–3156. doi: 10.1128/MCB.22.9.3149-3156.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuckenberg P, Buhl S, Woynecki T, van Furden B, Tolkunova E, Seiffe F, Moser M, Tomilin A, Winterhager E, Schorle H. 2010. The transcription factor TCFAP2C/AP-2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol Cell Biol 30:3310–3320. doi: 10.1128/MCB.01215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. 1996. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature 381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.