Abstract
Background
The present meta-analysis aimed to summarize the inconsistent findings on the association of apolipoprotein M gene (ApoM) rs805296 polymorphism with the risk of coronary artery disease (CAD), and to obtain a more authentic result about this topic.
Material/Methods
A total of 7 available articles were identified through electronic databases – PubMed, EMBASE, and Chinese National Knowledge Infrastructure (CNKI) – and their useful data were carefully extracted. The relationship between ApoM rs805296 polymorphism and CAD risk was assessed by odds ratios (ORs) and corresponding 95% confidence intervals (95% CIs), which were calculated using the fixed- or random-effects model, according to the degree of heterogeneity. Hardy-Weinberg equilibrium test, sensitivity test, and publication bias examination were also performed in this meta-analysis.
Results
According to the pooled results, ApoM rs805296 polymorphism conferred an increased risk of CAD under all the genetic contrasts: CC versus TT, CC + TC versus TT, CC versus TT+TC, C versus T, and TC versus TT (OR=2.13, 95% CI=1.16–3.91; OR=1.80, 95% CI=1.50–2.17; OR=1.91, 95% CI=1.04–3.51; OR=1.72, 95% CI=1.45–2.04; OR=1.78, 95% CI=1.47–2.15).
Conclusions
ApoM rs805296 polymorphism may be a risk factor for developing CAD.
MeSH Keywords: Apolipoproteins; Coronary Artery Disease; Polymorphism, Genetic; Risk Factors
Background
Coronary artery disease (CAD), one of the most common cardiovascular diseases, ranks first among fatal diseases in adults around the world [1–3]. Several risk factors have been confirmed, such as smoking, hypertension, diabetes, high blood cholesterol, excessive alcohol drinking, depression, and lack of exercise [4,5]. As for the underlying mechanism of CAD, it is reported that cardiac atherosclerosis may have a significant influence on the occurrence and progression of the disease [6,7]. However, genetic and environmental risk factors have been widely researched in the etiology of CAD, and their remarkable effects on susceptibility to CAD have also been identified.
In recent years, accumulating evidence indicates that genetic polymorphisms may be implicated in individual susceptibility to CAD, including polymorphisms within genes of APLNR [8], interleukin-6 [9], CYP7A1 [10], and PAI-1 [11]. In addition, the apolipoprotein M gene (ApoM), located on human chromosome 6 p21.31, has been reported to be significantly related to the occurrence of CAD [12].
The ApoM gene codes a 22-kDa protein that belongs to the apolipoprotein superfamily in structure. ApoM protein was first identified and determined in a study on lipoprotein by Xu et al. in 1993 [13]. Human ApoM cDNA, with 734 base pairs, encodes a residue-long protein with 188 amino acids [14]. ApoM is reportedly related to lower high-density lipoprotein (HDL) cholesterol, triglyceride-rich lipoproteins, lipoproteins containing ApoB, and very low-density lipoprotein (VLDL). Only expressed in the kidneys and liver [15], ApoM has been confirmed to have great influence on the transportation of reverse cholesterol [16].
Previous studies have suggested that one of the polymorphisms in ApoM gene, rs805296, was related to the susceptibility to CAD [17–20]; however, the number of studies is relatively limited, and the results are divergent rather than conclusive due to various reasons. Therefore, we comprehensively summarized all the findings on the association of ApoM rs805296 polymorphism with risk of CAD so as to reach a more authentic conclusion by performing the present meta-analysis.
Material and Methods
Study source and search strategy
The electronic databases searched for all the usable studies were: PubMed, EMBASE, and Chinese National Knowledge Infrastructure (CNKI). The words and items for literature searching contained “Apolipoprotein M” or “ ApoM, “polymorphism” or “variant” or “mutation”, and “coronary artery disease” or “CAD” or “atherosclerosis”. In case of missing any adequate studies, we screened the articles in the reference lists of relevant studies by manual searching. All the studies were restricted to those in English or Chinese.
Selection standards
All the publications meeting the following requirements were included into our meta-analysis: (1) possessing the case and control subjects at the same time; (2) studying the correlation between ApoM rs805296 polymorphism and CAD risk; (3) with sufficient data describing genotype and allele frequencies of the polymorphism in case and control groups; (4) human studies; and (5) the genotype distribution in control group conforming to Hardy-Weinberg equilibrium. Those studies with case-only design, inadequate information, or duplicating other articles were excluded. For publications with similar datasets, the one with the largest amount of information was included.
Data extraction
An identical form for data extraction was designed in advance, and the whole process was completed by 2 authors separately. Information to be extracted from each included article incorporated the following aspects: year of publication, name of first author, country of origin, ethnicity, genotyping method, sample sizes of cases and controls, genotypic and allelic distribution in case and control groups, and P value for Hardy-Weinberg equilibrium in control group.
Statistical analyses
STATA software (V.12.0) was used in all statistical analyses. Since all the publications would have control groups with genotypic and allelic frequencies consistent with Hardy-Weinberg equilibrium according to the selection criteria, we checked the compliance degree of Hardy-Weinberg equilibrium using the chi-square test for those not stating relevant data. The strength of relationship between ApoM rs805296 polymorphism and CAD risk was evaluated with odds ratio (OR) and its corresponding 95% confidence interval (95% CI) under 5 genetic models (CC versus TT, CC + TC versus TT, CC versus TT+TC, C versus T and TC versus TT). The absence or presence of statistically significant inter-study heterogeneity, tested by the χ2-test-based Q statistic, determined the use of fixed-effects model (Mantel-Haenszel method) or random-effects model (DerSimonian and Laird method). In sensitivity analysis, each single study was deleted in turn to observe the alterations of the overall results. Through Begg’s funnel plots and Egger’s test, we detected if there existed significant publication bias across eligible studies. For all the statistical tests, the significance level was set at P<0.05.
Results
Publication characteristics
Figure 1 displays the detailed process of study selection and reasons for study exclusion. Initially, 81 records were identified through the computer search, and 36 remained after excluding 8 studies not on humans and 37 apparently irrelevant ones. Through the subsequent exclusion for reviews and letters (7), without full texts (3), duplicates (5), not about ApoM rs805296 polymorphism (8) and without original data (6), we eventually included 7 studies in the quantitative synthesis [18,21–26]. The primary features of eligible studies are presented in Table 1.
Figure 1.
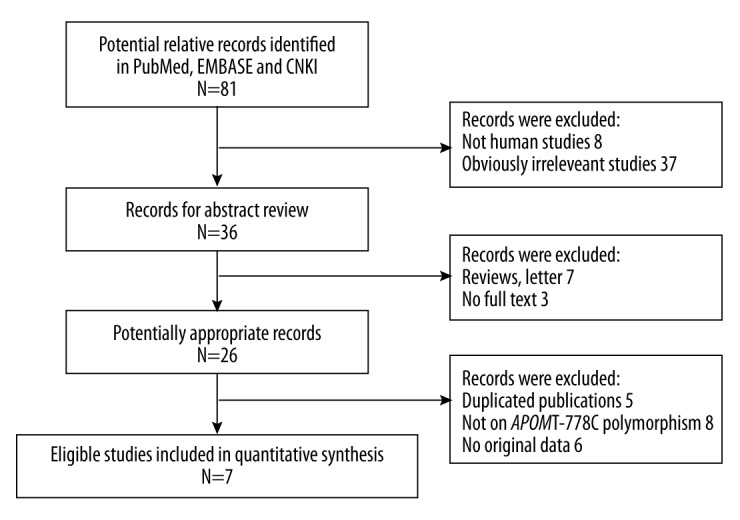
Flow diagram of study selection.
Table 1.
Main information extracted from eligible studies in the meta-analysis.
| Author/year | Country | Ethnicity | Control source | Genotyping method | Case | Control | P (HWE) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | TT | TC | CC | T | C | Number | TT | TC | CC | T | C | ||||||
| Jiao/2007 | China | Asian | Hospital-based | PCR-RFLP | 118 | 86 | 29 | 3 | 201 | 35 | 225 | 194 | 31 | 0 | 419 | 31 | 0.267 |
| Huang/2009 | China | Asian | Hospital-based | PCR-RFLP | 220 | 145 | 66 | 9 | 356 | 84 | 195 | 150 | 41 | 4 | 341 | 49 | 0.548 |
| Wang/2009 | China | Asian | Hospital-based | PCR-RFLP | 45 | 29 | 15 | 1 | 73 | 17 | 60 | 51 | 9 | 0 | 111 | 9 | 0.530 |
| Zheng/2009 | China | Asian | Hospital-based | Real-time PCR | 126 | 99 | 25 | 2 | 223 | 29 | 118 | 100 | 18 | 0 | 218 | 18 | 0.370 |
| Ma/2011 | China | Asian | Hospital-based | Allele-specific PCR | 46 | 28 | 17 | 1 | 73 | 19 | 56 | 47 | 9 | 0 | 103 | 9 | 0.513 |
| Zhang/2012 | China | Asian | Hospital-based | PCR-RFLP | 675 | 530 | 135 | 10 | 1195 | 155 | 636 | 556 | 74 | 6 | 1186 | 86 | 0.052 |
| Zheng/2014 | China | Asian | Hospital-based | Direct sequencing | 206 | 165 | 39 | 2 | 369 | 43 | 209 | 170 | 35 | 4 | 375 | 43 | 0.180 |
PCR – polymerase chain reaction; PCR-RFLP – PCR-restriction fragment length polymorphism; P (HWE) – P value for Hardy-Weinberg equilibrium test.
Study results
Table 2 shows the ORs with 95% CIs and P values for heterogeneity test under all the genetic models. Overall, the ApoM rs805296 polymorphism elevated the CAD risk in all the genetic contrasts (CC versus TT: OR=2.13, 95% CI=1.16–3.91; CC + TC versus TT: OR=1.80, 95% CI=1.50–2.17; CC versus TT+TC: OR=1.91, 95% CI=1.04–3.51; C versus T: OR=1.72, 95% CI=1.45–2.04; TC versus TT: OR=1.78, 95% CI=1.47–2.15). Figure 2 describes the forest plot for the association of ApoM rs805296 polymorphism with CAD risk under the CC versus TT model.
Table 2.
Meta-analysis results about APOM rs805296 polymorphism and CAD risk.
| APOM rs805296 | CC versus TT | CC + TC versus TT | CC versus TT+TC | C versus T | TC versus TT | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | Ph | OR | 95% CI | Ph | OR | 95% CI | Ph | OR | 95% CI | Ph | OR | 95% CI | Ph | |
| Fixed-effects model value | 2.13 | 1.16–3.91 | 0.491 | 1.80 | 1.50–2.17 | 0.204 | 1.91 | 1.04–3.51 | 0.557 | 1.72 | 1.45–2.04 | 0.126 | 1.78 | 1.47–2.15 | 0.361 |
Ph – P value of heterogeneity test.
Figure 2.
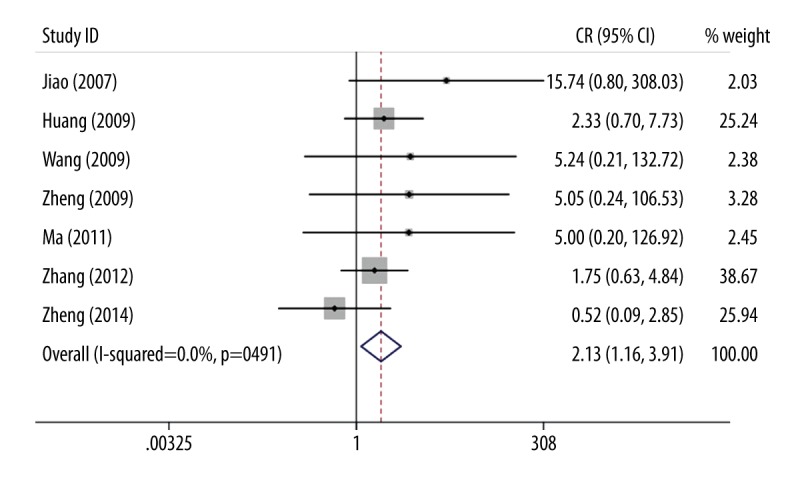
Risk of CAD and APOM rs805296 polymorphism under the CC versus TT contrast.
Heterogeneity examination
As shown in Table 2, P values for heterogeneity under all the genetic models were larger than 0.05 (P=0.491 for CC versus TT model; P=0.204 for CC + TC versus TT model; P=0.557 for CC versus TT+TC model; P=0.126 for C versus T model; P=0.361 for TC versus TT model). Therefore, there was no marked heterogeneity and the fixed-effects model was used for pooling the results.
Publication bias test
Begg’s funnel plots and Egger’s test were used to detected possible publication bias among the included studies from the visual and statistical perspective, respectively, and neither the shapes of funnel plots (Figure 3) nor the statistical data of Egger’s test (P=0.260) provided evidence for the presence of obvious publication bias.
Figure 3.
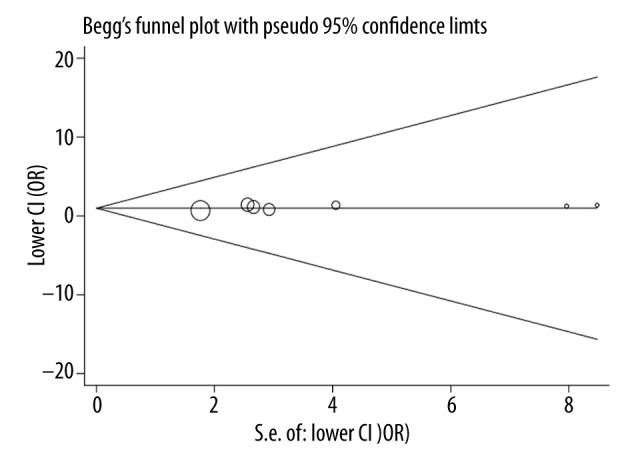
Funnel plot for publication bias examination.
Sensitivity analysis
In the process of sensitivity analysis, every individual study was omitted in sequence, and the changed results were observed correspondingly. No radical alteration occurred in the pooled results, suggesting that no single study substantially affected the results, and our meta-analysis outcomes were statistically robust.
Discussion
CAD is a complex multi-genetic disease caused by synergistic effects of genetic and environmental risk factors [27,28]. Hereditary epidemiological studies have suggested that genetic mutations may elevate individual risk of developing CAD [29–31]. Initially separated and cloned from chylomicrons [13], ApoM in plasma mainly exists in HDL particles, and very little is in triglyceride-rich lipoprotein (TGRLP) and low-density lipoprotein (LDL), suggesting ApoM may be associated with lipid transportation and metabolism [15]. Richter et al. found an important role of ApoM in the formation of HDL, and confirmed its protective effects against atherosclerosis [16,32]. In the study by Xu et al., the correlation between ApoM and indexes of lipid indicated that ApoM levels in plasma had a positive relation with factors against the progression of atherosclerosis, such as ApoA I and HDLC, and was negatively related with factors promoting atherosclerosis development, such as triglyceride, total cholesterol, and lipoprotein (a), and that elevated levels of ApoM could prevent and slow the progression of atherosclerosis [33].
The human ApoM gene is located in a region adjacent to that of major histocompatibility complex (MHC) in which multiple genes are related to immune response; therefore, the ApoM gene is likely to participate in the regulation of immune defense [34]. Among a number of polymorphisms within the ApoM gene, the rs805296 variant in the proximal promoter region has been verified to have a link with plasma cholesterol, and may increase individual susceptibility to CAD [35].
In this present study, we referred to previous studies and analyzed the association between ApoM rs805296 polymorphism and CAD risk. Our results indicate that ApoM rs805296 polymorphism under all the comparisons could elevate the risk of CAD, suggesting this polymorphism might act as a promoter for CAD onset. Several case-control studies have investigated the significance of ApoM rs805296 in CAD risk in Chinese populations, and obtained useful findings. Using the method of polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), Huang et al. carried out a screening for ApoM rs805296 in 220 CAD cases and 195 normal controls, and observed the frequency of C allele in case and control groups was 19.1% and 12.6% respectively, and this difference was statistically significant (P=0.011), which proved the polymorphism rs805296 might be a susceptible factor for CAD [21]. Zhang et al. performed a large study recruiting 675 patients with acute coronary syndrome (ACS) and 636 healthy control subjects, and found that the frequencies of both C allele and CC genotype of ApoM rs805296 polymorphism in the case group were significantly higher than those in the control group (P<0.01). Subsequently, after the adjustment of susceptibility factors for CAD, the C allele was found to be an independent risk factor for the occurrence of ACS [25]. In addition, some other studies also obtained results similar to those mentioned above [18,22,23,26]. In contrast, Zheng et al. found no statistically significant difference in distribution of 3 genotypes of ApoM rs805296 polymorphism, including TT, TC, and CC, between case and control groups, and concluded that rs805296 might not be correlated with the development of CAD [24]. Conducted in a Chinese population, the study of Zheng et al. obtained results that are in contrast with our present study and the other case-control studies listed above, which might be attributed to differences in number of samples, methods of genotyping, correction factors, and other risk elements.
Absence of heterogeneity and publications bias is the biggest strength of this meta-analysis. However, as in previous studies and meta-analyses, our meta-analysis also has some weaknesses that should be clearly presented. Because all the prior studies on the association of ApoM rs805296 polymorphism with CAD risk only focused on Chinese populations, our meta-analysis solely discussed this association among Chinese people, which might not be representative in other ethnic groups. In addition, the limited number of included studies and the relatively small sample size might lessen the statistical power of our results. Another important aspect that should be stated is that some potential risk factors such as family history, smoking status, body mass index (BMI) and other environmental influences [36] were not incorporated into the discussion of the present study due to limited original information of included studies.
Conclusions
In conclusion, our meta-analysis results revealed a significant correlation of ApoM rs805296 polymorphism with CAD risk, and showed rs805296 polymorphism might confer increased risk of CAD in the Chinese population. The association between ApoM rs805296 and onset risk of CAD needs to be further verified by studies containing combined effects of genetic and environmental factors and larger sample size in multiple ethnicities.
Footnotes
Source of support: Departmental sources
References
- 1.McGovern PG, Pankow JS, Shahar E, et al. Recent trends in acute coronary heart disease – mortality, morbidity, medical care, and risk factors. The Minnesota Heart Survey Investigators. N Engl J Med. 1996;334:884–90. doi: 10.1056/NEJM199604043341403. [DOI] [PubMed] [Google Scholar]
- 2.GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117–71. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Padmanabhan S, Hastie C, Prabhakaran D, Dominczak AF. Genomic approaches to coronary artery disease. Indian J Med Res. 2010;132:567–78. [PMC free article] [PubMed] [Google Scholar]
- 4.Mehta PK, Wei J, Wenger NK. Ischemic heart disease in women: a focus on risk factors. Trends Cardiovasc Med. 2015;25:140–51. doi: 10.1016/j.tcm.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charlson FJ, Moran AE, Freedman G, et al. The contribution of major depression to the global burden of ischemic heart disease: a comparative risk assessment. BMC Med. 2013;11:250. doi: 10.1186/1741-7015-11-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saikku P, Leinonen M, Tenkanen L, et al. Chronic Chlamydia pneumoniae infection as a risk factor for coronary heart disease in the Helsinki Heart Study. Ann Intern Med. 1992;116:273–78. doi: 10.7326/0003-4819-116-4-273. [DOI] [PubMed] [Google Scholar]
- 7.Simons M, Bonow RO, Chronos NA, et al. Clinical trials in coronary angiogenesis: issues, problems, consensus: An expert panel summary. Circulation. 2000;102:E73–86. doi: 10.1161/01.cir.102.11.e73. [DOI] [PubMed] [Google Scholar]
- 8.Wang P, Xu C, Wang C, et al. Association of SNP Rs9943582 in APLNR with left ventricle systolic dysfunction in patients with coronary artery disease in a Chinese Han GeneID population. PLoS One. 2015;10:e0125926. doi: 10.1371/journal.pone.0125926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K, Dong PS, Zhang HF, et al. Role of interleukin-6 gene polymorphisms in the risk of coronary artery disease. Genet Mol Res. 2015;14:3177–83. doi: 10.4238/2015.April.10.29. [DOI] [PubMed] [Google Scholar]
- 10.Iwanicki T, Balcerzyk A, Niemiec P, et al. CYP7A1 gene polymorphism located in the 5′ upstream region modifies the risk of coronary artery disease. Dis Markers. 2015;2015:185969. doi: 10.1155/2015/185969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang Z, Jiang W, Ouyang M, Yang K. PAI-1 4G/5G polymorphism and coronary artery disease risk: a meta-analysis. Int J Clin Exp Med. 2015;8:2097–107. [PMC free article] [PubMed] [Google Scholar]
- 12.Visvikis-Siest S, Marteau JB. Genetic variants predisposing to cardiovascular disease. Curr Opin Lipidol. 2006;17:139–51. doi: 10.1097/01.mol.0000217895.67444.de. [DOI] [PubMed] [Google Scholar]
- 13.Xu N, Dahlback B. A novel human apolipoprotein (apoM) J Biol Chem. 1999;274:31286–90. doi: 10.1074/jbc.274.44.31286. [DOI] [PubMed] [Google Scholar]
- 14.Xie T, Rowen L, Aguado B, et al. Analysis of the gene-dense major histocompatibility complex class III region and its comparison to mouse. Genome Res. 2003;13:2621–36. doi: 10.1101/gr.1736803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo G, Zhang X, Nilsson-Ehle P, Xu N. Apolipoprotein M. Lipids Health Dis. 2004;3:21. doi: 10.1186/1476-511X-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfrum C, Poy MN, Stoffel M. Apolipoprotein M is required for prebeta-HDL formation and cholesterol efflux to HDL and protects against atherosclerosis. Nat Med. 2005;11:418–22. doi: 10.1038/nm1211. [DOI] [PubMed] [Google Scholar]
- 17.Niu N, Zhu X, Liu Y, et al. Single nucleotide polymorphisms in the proximal promoter region of apolipoprotein M gene (apoM) confer the susceptibility to development of type 2 diabetes in Han Chinese. Diabetes Metab Res Rev. 2007;23:21–25. doi: 10.1002/dmrr.641. [DOI] [PubMed] [Google Scholar]
- 18.Jiao GQ, Yuan ZX, Xue YS, et al. A prospective evaluation of apolipoprotein M gene T-778C polymorphism in relation to coronary artery disease in Han Chinese. Clin Biochem. 2007;40:1108–12. doi: 10.1016/j.clinbiochem.2007.04.023. [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Niu N, Brismar K, et al. Apolipoprotein M promoter polymorphisms alter promoter activity and confer the susceptibility to the development of type 1 diabetes. Clin Biochem. 2009;42:17–21. doi: 10.1016/j.clinbiochem.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Zhao D, He Z, Qin X, et al. Association of apolipoprotein M gene polymorphisms with ischemic stroke in a Han Chinese population. J Mol Neurosci. 2011;43:370–75. doi: 10.1007/s12031-010-9453-7. [DOI] [PubMed] [Google Scholar]
- 21.Huang J, Zhang S, Huang YZ, et al. A study on the associativity of apolipoprotein M gene promoter T-778C polymorphism with coronary heart disease among Han People of Guizhou. Journal of Guiyang Medical College. 2009;34:297–99. [Google Scholar]
- 22.Ma LP, Li B, Wang X. The study on the interrelation between polymorphism of apolipoprotein M and in the ratio of apoB/apoA1 patients with coronary heart disease. China Modern Medicine. 2011;18:42–43. [Google Scholar]
- 23.Wang X, Li B. Correlation between apoM genotype and lipoprotein (a) in patients with coronary heart disease. Chinese Journal of Integrative Medicine on Cardio-/Cerebrov Ascular Diasese. 2009;7:654–56. [Google Scholar]
- 24.Zheng L, Luo G, Zhang X, et al. Determination of single-nucleotide polymorphism in the proximal promoter region of apolipoprotein M gene in coronary artery diseases. Int J Gen Med. 2009;2:177–82. doi: 10.2147/ijgm.s4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang XL, Liu TF, Cai WZ, et al. Polymorphisms in the ApoM gene confer the susceptibility to acute coronary syndrome. Shandong Medicine. 2012;52:1–3. [Google Scholar]
- 26.Zheng L, Luo G, Zhang J, et al. Decreased activities of apolipoprotein m promoter are associated with the susceptibility to coronary artery diseases. Int J Med Sci. 2014;11:365–72. doi: 10.7150/ijms.7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marenberg ME, Risch N, Berkman LF, et al. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–46. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 28.Nora JJ, Lortscher RH, Spangler RD, et al. Genetic – epidemiologic study of early-onset ischemic heart disease. Circulation. 1980;61:503–8. doi: 10.1161/01.cir.61.3.503. [DOI] [PubMed] [Google Scholar]
- 29.Hirashiki A, Yamada Y, Murase Y, et al. Association of gene polymorphisms with coronary artery disease in low- or high-risk subjects defined by conventional risk factors. J Am Coll Cardiol. 2003;42:1429–37. doi: 10.1016/s0735-1097(03)01062-3. [DOI] [PubMed] [Google Scholar]
- 30.Yokota M, Ichihara S, Lin TL, et al. Association of a T29–>C polymorphism of the transforming growth factor-beta1 gene with genetic susceptibility to myocardial infarction in Japanese. Circulation. 2000;101:2783–87. doi: 10.1161/01.cir.101.24.2783. [DOI] [PubMed] [Google Scholar]
- 31.Baroni MG, Berni A, Romeo S, et al. Genetic study of common variants at the Apo E, Apo AI, Apo CIII, Apo B, lipoprotein lipase (LPL) and hepatic lipase (LIPC) genes and coronary artery disease (CAD): variation in LIPC gene associates with clinical outcomes in patients with established CAD. BMC Med Genet. 2003;4:8. doi: 10.1186/1471-2350-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richter S, Shih DQ, Pearson ER, et al. Regulation of apolipoprotein M gene expression by MODY3 gene hepatocyte nuclear factor-1alpha: haploinsufficiency is associated with reduced serum apolipoprotein M levels. Diabetes. 2003;52:2989–95. doi: 10.2337/diabetes.52.12.2989. [DOI] [PubMed] [Google Scholar]
- 33.Xu N, Nilsson-Ehle P, Ahren B. Correlation of apolipoprotein M with leptin and cholesterol in normal and obese subjects. J Nutr Biochem. 2004;15:579–82. doi: 10.1016/j.jnutbio.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Xu N, Zhang XY, Dong X, et al. Effects of platelet-activating factor, tumor necrosis factor, and interleukin-1alpha on the expression of apolipoprotein M in HepG2 cells. Biochem Biophys Res Commun. 2002;292:944–50. doi: 10.1006/bbrc.2002.6755. [DOI] [PubMed] [Google Scholar]
- 35.Iwamoto T, Fukuda S, Shimizu S, Takasaki M. Long-term effects of lipoprotein(a) on carotid atherosclerosis in elderly Japanese. J Gerontol A Biol Sci Med Sci. 2004;59:62–67. doi: 10.1093/gerona/59.1.m62. [DOI] [PubMed] [Google Scholar]
- 36.Allen RA, Lee EM, Roberts DH, et al. Polymorphisms in the TNF-alpha and TNF-receptor genes in patients with coronary artery disease. Eur J Clin Invest. 2001;31:843–51. doi: 10.1046/j.1365-2362.2001.00907.x. [DOI] [PubMed] [Google Scholar]


