Abstract
Alginate lyases are important tools for oligosaccharide preparation, medical treatment, and energy bioconversion. Numerous alginate lyases have been elucidated. However, relatively little is known about their substrate degradation patterns and product-yielding properties, which is a limit to wider enzymatic applications and further enzyme improvements. Herein, we report the characterization and module truncation of Aly5, the first alginate lyase obtained from the polysaccharide-degrading bacterium Flammeovirga. Aly5 is a 566-amino-acid protein and belongs to a novel branch of the polysaccharide lyase 7 (PL7) superfamily. The protein rAly5 is an endolytic enzyme of alginate and associated oligosaccharides. It prefers guluronate (G) to mannuronate (M). Its smallest substrate is an unsaturated pentasaccharide, and its minimum product is an unsaturated disaccharide. The final alginate digests contain unsaturated oligosaccharides that generally range from disaccharides to heptasaccharides, with the tetrasaccharide fraction constituting the highest mass concentration. The disaccharide products are identified as ΔG units. While interestingly, the tri- and tetrasaccharide fractions each contain higher proportions of ΔG to ΔM ends, the larger final products contain only ΔM ends, which constitute a novel oligosaccharide-yielding property of guluronate lyases. The deletion of the noncatalytic region of Aly5 does not alter its M/G preference but significantly decreases the enzymatic activity and enzyme stability. Notably, the truncated protein accumulates large final oligosaccharide products but yields fewer small final products than Aly5, which are codetermined by its M/G preference to and size enlargement of degradable oligosaccharides. This study provides novel enzymatic properties and catalytic mechanisms of a guluronate lyase for potential uses and improvements.
INTRODUCTION
Alginate is a linear polysaccharide composed of alternating residues of β-d-mannuronic acid (M) and its C-5 epimer, α-l-guluronic acid (G) (1). The uronic acid residues are arranged into homopolyuronic blocks of M residues (M block), G residues (G block), or heteropolyuronic blocks (MG or GM blocks). Alginate has been identified as a cell wall component of seaweeds belonging to Phaeophyta, such as kelp and sargassum (2, 3). Due to its ability to form a strong gel after absorbing water, algal alginate has been widely used as a supporting material in food, medical, and industrial applications (4, 5). Alginates containing acetyl modifications at the O-2 or O-3 positions have been purified from the extracellular matrix of some bacteria, such as the pathogen Pseudomonas aeruginosa and Azotobacter soil bacteria (6, 7). Understanding how to prevent Pseudomonas pathogens from synthesizing and secreting extracellular high-molecular-weight alginates is important for clinical therapy protocols (8, 9).
Alginate lyase can break the 1 to 4 O linkages between the uronic acid residues of alginate via a β-elimination mechanism. The reaction forms C-4=C-5 double bonds within the sugar residue linked at the C-4 position, thereby yielding soluble oligosaccharides with unsaturated nonreducing (nr) ends, which is usually designated Δ to show the differences from the G and M residues (10–13). The enzymatic degradation of alginate can provide energies and carbon sources to alga-consuming animals or microbes, demonstrating the potential of alginate lyases in preparing oligosaccharides or converting alginates into biomasses (14, 15). Moreover, the enzymatic degradation of bacterial alginate using alginate lyases may assist antibiotics in killing Pseudomonas pathogens or may facilitate more efficient inhibition of bacterial growth (16–18). Hence, alginate lyases are potential enzyme tools for the clinical treatment of human cystic fibrosis syndrome.
During the last decade, nearly 100 alginate lyases have been discovered in ocean animals, bacteria, and bacterial phages (10, 11, 19–21). Most of the elucidated enzymes are endo-type lyases that internally and randomly degrade alginate, producing a series of unsaturated oligosaccharides with different degrees of polymerization (19, 22, 23). Only a few of these enzymes have been reported as exo-type enzymes that gradually cut the polysaccharide, yielding predominantly monosaccharides as products (24–29). Some alginate lyases show strict selectivity for M- or G-enriched blocks of alginate, whereas others demonstrate broader substrate preferences (10, 11, 19). However, relatively few data (e.g., nuclear magnetic resonance [NMR] spectral properties of final oligosaccharide products, sizes of degradable oligosaccharide substrates, and degradation patterns of oligosaccharides) have been reported in detail, which limits broader and extended uses of alginate lyases as efficient tools.
Alginate lyases are module-organized proteins that usually contain catalytic and noncatalytic regions (NCRs). Based on the sequence similarities of the catalytic modules, most alginate lyases have been assigned to the polysaccharide lyase families PL5, PL6, PL7, PL14, PL15, PL17, and PL18, although a few remain unclassified (19). Recent studies have revealed the conservative catalytic motifs and the three-dimensional structural characteristics of alginate lyases within the PL5, PL7, PL14, PL15, PL17, and PL18 families (19, 24–26). In most module-organized polysaccharide depolymerases, the catalytic modules are responsible for recognizing, binding, and degrading substrates and thus determine the enzymes' biochemical characteristics. In contrast, the NCR fragments cannot degrade substrates but may affect the enzymes' properties. In this regard, the effects of NCRs on alginate lyases are relatively unknown, except for a CBM13 module (30).
Bacteria within the Flammeovirga genus have been identified from costal seawater, deep-sea sediment, and ocean animal gut (31–34). Most of these Flammeovirga strains can efficiently digest various polysaccharides, such as agarose, alginate, and starch, indicating that the strains contain abundant enzyme resources related to carbohydrate metabolism. Thus far, three β-agarases and one α-amylase have been identified in three different Flammeovirga strains (35–38), although little is known about other types of polysaccharide depolymerases, such as alginate lyases. Due to the excellent polysaccharide-degrading capabilities of bacteria, the draft genome sequences of Flammeovirga sp. strain OC4 and the Flammeovirga pacifica strain WPAGA1 have recently been sequenced and annotated; however, only one putative alginate lyase gene has been identified individually (39, 40). Interestingly, the whole genome of Flammeovirga sp. strain MY04 encoded at least five putative alginate lyases. In this study, we present Aly5 as the first alginate lyase to be identified from the genus Flammeovirga. Furthermore, the effects of the NCR fragment on the enzymatic and alginate-degrading properties of Aly5 were observed through gene truncation, protein purification, and enzyme characterization.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Unless otherwise noted, Escherichia coli strains were cultured at 37°C in Luria-Bertani (LB) medium, supplemented when necessary with antibiotics: e.g., ampicillin (100 μg/ml) or kanamycin (50 μg/ml). Flammeovirga sp. strain MY04 (CGMCC no. 2777) was cultured at 30°C in a medium (pH 7.0) containing (wt/vol) 0.40% tryptone, 0.25% yeast extract, and 3.0% NaCl. Agar powder (1.5% [wt/vol]) was used to prepare the solid media.
Agarose, alginate (viscosity, ≥2,000 cP, 2% at 25°C), chondroitin, heparin, hyaluronan, xanthan, and xylan were purchased from Sigma-Aldrich. M- and G-enriched disaccharide, trisaccharide, tetrasaccharide, pentasaccharide, and hexasaccharide (>95% promised purities) were purchased from Qingdao BZ Oligo Biotech Co., Ltd. (Qingdao, China).
Sequence analyses of the genes and proteins.
Using the software BioEdit version 7.2.5 (41), the DNA sequences of the open reading frames (ORFs) were translated into the amino acid sequences of the corresponding proteins, and GC contents (G+C percentage) were calculated. For functional annotation of the predicted proteins, similarity searches of amino acid sequences were performed using the BLAST algorithm on the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov). Signal peptides and their types were identified using the SignalP 4.1 server and the LipoP 1.0 server (http://www.cbs.dtu.dk/services/), respectively. Molecular weights of the putative proteins were estimated using the peptide mass tool on the ExPASy server of the Swiss Institute of Bioinformatics (http://swissmodel.expasy.org/). Protein modules and domains were identified in the products using the Simple Modular Architecture Research Tool (https://en.wikipedia.org/wiki/Simple_Modular_Architecture_Research_Tool), the Pfam database (http://pfam.xfam.org), and the Carbohydrate-Active Enzyme (CAZy) database (http://www.cazy.org). Multiple sequence alignments and phylogenetic analyses were performed using MEGA version 5.05 (42).
Construction of expression vectors.
Flammeovirga sp. strain MY04 cells were treated with lysozymes, and genomic DNA was extracted using SDS and proteinase K treatment (43). To express Aly5, the full-length Aly5 gene was amplified using the primers E30L-5f (5′-GGATCCTTCCTAACAGGAAGCATCATAGG-3′) and E30L-5r (5′-CTCGAGTGTGACTTACTTTTAGGTAAC-3′) and the high-fidelity PrimeSTARHS DNA polymerase (TaKaRa, Dalian, China). Primer pairs with restriction enzyme sites (underlined) for NdeI and XhoI were designed to generate a 6×His tag at the C terminus of the recombinant protein (rAly5). DNA amplification products were cloned into the expression vector pET-30a(+), and the recombinant plasmid (pE30-Aly5) was transformed into E. coli BL21(DE3) cells. The integrities of the nucleotide sequences of newly constructed plasmids were confirmed by DNA sequencing.
Heterologous expression and purification of rAly5.
E. coli BL21(DE3) cells harboring the plasmid pE30-Aly5 were initially cultured in LB broth. When the cell density reached an A600 value of 0.6 to 0.8, the broth was supplemented with isopropyl 1-thio-β-d-galactoside at a final concentration of 0.05 mM to initiate rAly5 expression. After continual cultivation for an additional 24 h at 16°C, cells were harvested by centrifugation at 6,000 × g for 10 min, washed twice with ice-cold buffer A (50 mM Tris, 150 mM NaCl [pH 8.0]), resuspended in buffer A, and disrupted by sonication (60 repetitions, 5 s) in an ice-water bath. After centrifugation at 15,000 × g for 30 min, the supernatant containing soluble proteins was loaded onto a Ni-nitrilotriacetic acid agarose (Ni-NTA) column (Novagen) that was preequilibrated with buffer A. Subsequently, the column was eluted with buffer A supplemented with increasing gradient concentrations of imidazole (0, 10, 50, and 250 mM). Fractionated protein samples were analyzed using SDS-PAGE. To obtain active alginate lyase, the purified 63-kDa proteins were diluted and dialyzed against buffer B (50 mM Tris, 50 mM NaCl, 5% glycerol [vol/vol] [pH 8.0]).
SDS-PAGE was performed using 13.2% (wt/vol) polyacrylamide gels according to the methods of Sambrook and Russell (43). Proteins were detected by staining each gel with Coomassie brilliant blue R-250. Protein concentrations were determined by the Folin-Lowry method using Folin Ciocalteu's phenol reagent (Sigma-Aldrich) and bovine serum albumin as a standard.
Enzyme activity assay.
To determine the substrate preferences of rAly5, various polysaccharides (e.g., agarose, chondroitin, heparin, hyaluronan, xanthan, xylan, and alginate) and M- and G-enriched oligosaccharides were individually dissolved in deionized water to prepare stock solutions (10 mg/ml). Each stock solution (30 μl) was mixed with 100 μl of 150 mM NaAc-HAc buffer (pH 6.0), 140 μl of water, and 30 μl of the appropriately diluted enzyme and then incubated at 37°C for 12 h. Enzyme-treated polysaccharide samples were heated in boiling water for 10 min and then ice cooled. After centrifugation at 15,000 × g for 15 min, the supernatant was collected and analyzed by measuring the absorbance at 235 nm. One unit was defined as the amount of enzymes required to increase the absorbance at 235 nm by 0.1 per min (10).
Biochemical characterization of rAly5.
To determine the optimal pH for rAly5 activity, enzymatic reactions were performed using buffers with different pH values, including 150 mM NaAc-HAc buffer (pH 4.0 to 6.5), 150 mM NaH2PO4-Na2HPO4 buffer (pH 6.0 to 8.0), and 150 mM Tris-HCl buffer (pH 7.5 to 10), each with a total volume of 300 μl. To determine the optimal temperature for rAly5 activity, enzyme reactions were performed in 150 mM NaAc-HAc buffer (pH 6.0) at temperatures ranging from 0 to 80°C for 90 min. The thermostability of rAly5 was evaluated by measuring the residual activity of the enzyme after incubation at various temperatures for 2 h. The effect of pH on rAly5 stability was determined by measuring the residual activity of rAly5 after incubation at 4°C at various pH values (4.0 to 10) for 2 h. The effects of metal ions and chelating agents on rAly5 activity were examined by determining rAly5 activities in the presence of 1 and 10 mM concentrations of various chemicals, respectively.
Analyses of the polysaccharide-degrading properties of rAly5.
The digestion pattern of alginate (1.0 mg/ml) by rAly5 (1.0 U/ml) at 40°C was traced over 72 h. Similar experiments were conducted using different final alginate concentrations, ranging from 1.0 to 10 mg/ml. Aliquots of the digests were removed for time course analysis. To determine the molar ratio of the individual oligosaccharide fractions in the products, samples (1.0 mg/ml) were analyzed by gel filtration on a Superdex peptide 10/300 GL column (GE Healthcare) and monitored at 235 nm using a UV detector. The mobile phase was 0.2 M NH4HCO3, and the flow rate was 0.4 ml/min. Online monitoring and data analysis were performed using the software LCsolution version 1.25.
To determine the oligosaccharide compositions of the final digests, 100 mg alginate (1.0 mg/ml) was digested using excess rAly5 (5 U/ml) at 40°C for 72 h. The reaction mixture was heated in boiling water for 10 min, subsequently cooled to 4°C, and centrifuged at 15,000 × g for 30 min. The supernatant was concentrated by rotary evaporation at 40°C. The resulting products were analyzed by gel filtration as described above, loaded onto a preequilibrated Superdex peptide 10/300G column (GE Healthcare), and eluted with 0.20 M NH4HCO3 at a flow rate of 0.4 ml/min. Size-defined unsaturated oligosaccharide fractions were collected and freeze-dried repeatedly to remove NH4HCO3 for further analysis. The molecular mass of each oligosaccharide fraction was determined by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Axima-CFR Plus; Shimadzu, Japan). For 1H-NMR spectroscopy, each purified oligosaccharide fraction (2 mg) was dissolved in 0.3 ml of D2O in 5-mm NMR tubes. The spectra were recorded on a JNM-ECP600 (JEOL, Japan) apparatus set at 600 MHz, using trimethylsilane (TMS) as the internal standard.
Analyses of the oligosaccharide-degrading properties of rAly5.
To determine the smallest substrate of rAly5, a total of 150 mg alginate was partially digested by rAly5 at 40°C. The resulting unsaturated oligosaccharides with different degrees of polymerization (DPs)—UDP2, UDP3, UDP4, UDP5, UDP6, and UDP7—were fractionated using gel filtration and a Superdex peptide 10/300 GL column. Each oligosaccharide fraction (∼20 μg) was digested with rAly5 (0.01 U) in a total volume of 20 μl at 40°C for 12 h, and the digest was analyzed by high-performance liquid chromatography (HPLC) using a Superdex peptide 10/300 GL column with monitoring at 235 nm.
To determine the enzymatic degradation pattern of rAly5, purified oligosaccharides of UDP5 (200 μg) were fluorescently labeled at their reducing ends using excess 2-aminobenzamide (2-AB) (Sigma), as described by Bigge et al (44). The labeled products (∼1 μg each) were purified by gel filtration HPLC and further degraded with rAly5 (5 U) in a total volume of 1 ml. The substrate and degradation products were subjected to the gel filtration assay described above, using a fluorescence detector and excitation and emission wavelengths of 330 and 420 nm, respectively.
Analyses of the effects of the noncatalytic module on enzymatic properties.
To identify the effects of the NCR on enzyme properties, the partial gene encoding the catalytic module (Leu337 to His566) of Aly5 was amplified using primers E30L5-T336Nf (5′-GGATCCGTGTTAGGGTTACAAAATTGGAAGC-3′) and E30L5-T336Nr (5′-CTCGAGTGTGACTTACTTTTAGGTAAC-3′). DNA products were gel recovered and cloned into plasmid pET-30a(+). The derived vector pE30-Aly5-T336N was transformed into BL21(DE3) cells to express the catalytic module of Aly5, rAly5-T336N (hereafter called the “T336N protein”). The soluble protein was expressed, purified, and characterized using a strategy identical to that described previously for rAly5.
RESULTS
Information regarding the Aly5 gene and protein sequences.
ORF 5101 in the genome of Flammeovirga sp. strain MY04 was predicted to be an alginate lyase gene (GenBank accession no. KT266807). The gene was 1,701 bp in length, with a GC content of 38%, and encoded a putative protein (Aly5) composed of 566 amino acid residues.
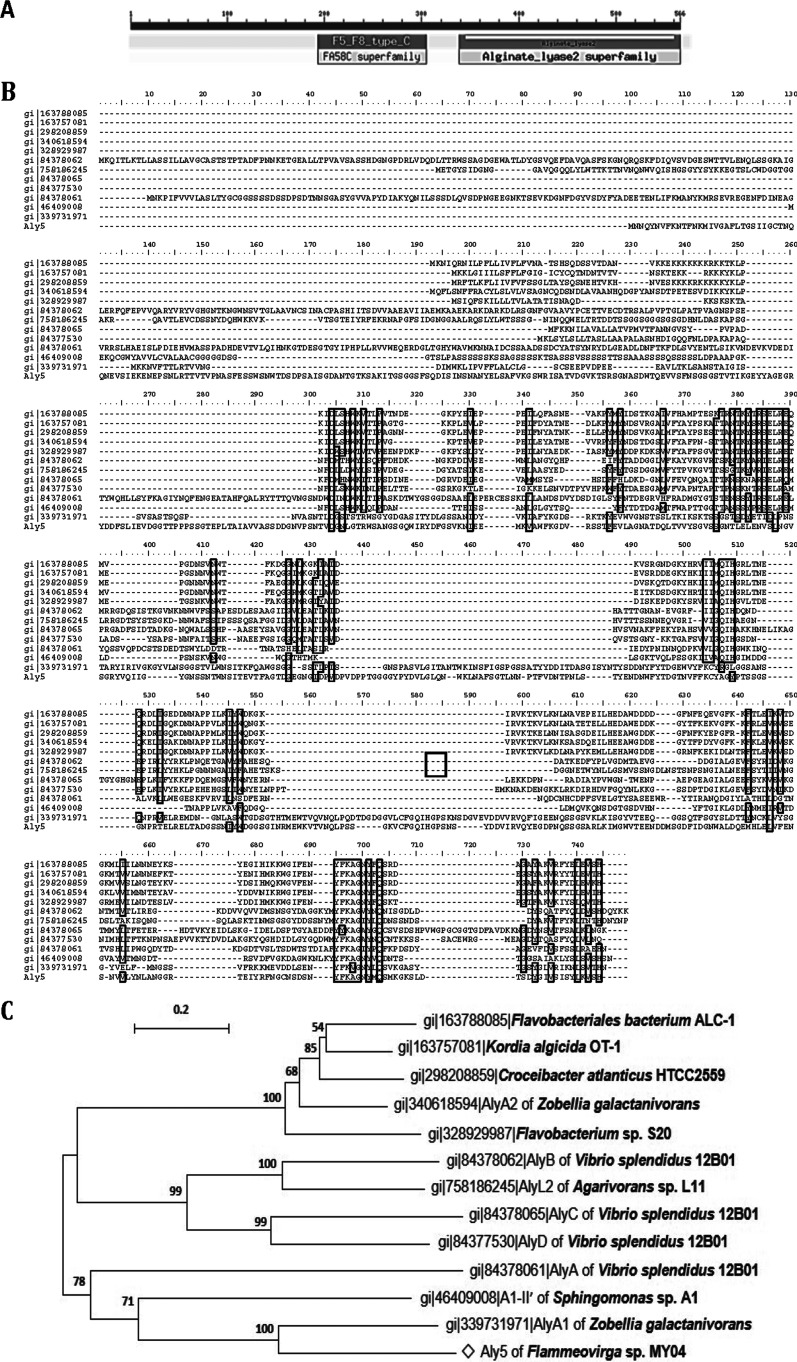
The molecular mass of the Aly5 protein was ∼61 kDa. The isoelectric point (pI) was 4.08. A BLASTp search showed that among elucidated alginate lyases, the Aly5 protein demonstrated a sequence identity higher than 30% with only the AlyA1 protein (gi no. 339731971) of Zobellia galactanivorans (41%) (25, 45). SignalP 4.1 and LipoP 1.0 analyses indicated that the type II signal peptide of Aly5 contained 27 amino acid residues (Met1 to Gly27). Analyses using the Carbohydrate-Active Enzyme database and the Simple Modular Architecture Research Tool indicated that the Aly5 protein contained an NCR fragment (Cys28 to V336), including an F5/8-type C module (Ile188 to Val305), for which the function was unknown, and a putative catalytic module, Alg2 (Leu337 to His566) (Fig. 1A). Protein sequence alignment showed that the Alg2 module contained one catalytic motif (Gln441-Ile442-His443 [QIH]) that was conserved in elucidated PL7 alginate lyases (Fig. 1B). Further phylogenic analysis suggested that Aly5 is a novel alginate lyase from the genus Flammeovirga and belongs to a PL7 subclass that remains to be defined (Fig. 1C).
FIG 1.
Sequence properties of the alginate lyase Aly5 from Flammeovirga sp. strain MY04. (A) Module organization of the alginate lyase Aly5. The F5/8-type C module is a predicted discoidin noncatalytic domain (Ile188 to Val305). The alginate_lyase2 module (Alg2) is a putative catalytic domain (Leu337 to His566). The full-length protein and the Alg2 module were expressed to yield the recombinant protein rAly5 and its truncated version, rAly5-T336N, respectively. (B) Protein sequence alignment of Aly5 and elucidated PL-7 alginate lyases. Amino acid residues with homologies of ≥75% are shaded in a black frame and with a gray background. (C) Phylogenic analysis of alginate lyases based on protein sequence alignments and substrate preferences. The tree was created using the neighbor-joining method and MEGA version 5.05 software. The numbers on the branches indicate the bootstrap confidence values from 1,000 replicates. The bar is equal to the distance corresponding to 1 amino acid substitution per 10 amino acids.
Heterologous expression of Aly5 in E. coli.
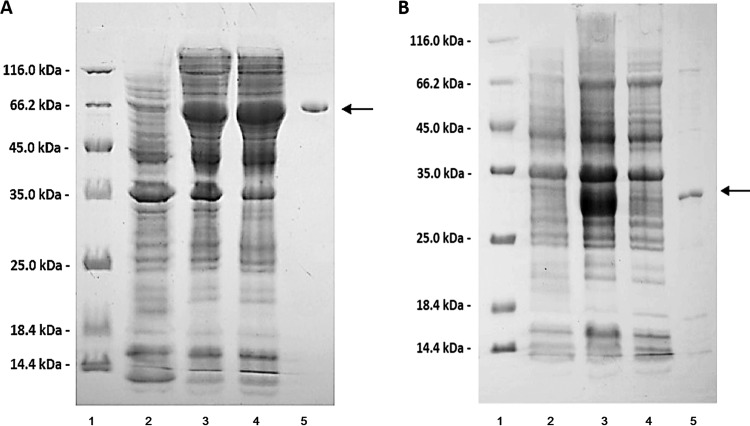
The full-length Aly5 gene was amplified directly from the genomic DNA of Flammeovirga sp. strain MY04. The 1.7-kb PCR products were recovered and cloned into the pET-30a(+) vector, downstream of a T7 promoter. A His6 tag was successively added to the C terminus of the protein product (rAly5) in the expression vector (pE30-Aly5). SDS-PAGE analysis indicated that BL21(DE3) cells harboring pE30-Aly5 plasmids could yield soluble recombinant proteins (∼800 mg/liter) that exhibit the appropriate molecular mass (i.e., 62 kDa) (Fig. 2A). By sonication and centrifugation, crude enzymes were extracted from the E. coli cell cultures harboring pE30-Aly5. The soluble protein fraction of Aly5 could be eluted from the Ni-NTA column using imidazole at concentrations greater than 50 mM. The rAly5 protein was further purified by gel filtration chromatography. SDS-PAGE analysis suggested that the purified soluble protein of rAly5 had a purity greater than 99%, a recovery of ∼60%, and an initial concentration of ∼6 mg/ml (Fig. 2A).
FIG 2.
Purification of recombinant Aly5 (A) and the truncated protein rAly5-T336N (B) from E. coli using Ni2+ chelation affinity chromatography. The enzyme purity following each fractionation step was assessed by SDS-PAGE using 13.2% polyacrylamide gels, followed by staining with Coomassie brilliant blue. Lane 1 corresponds to the unstained protein molecular weight marker SM0431 (Thermo), lane 2 corresponds to the induced cell lysate of the E. coli strain containing the control plasmid pET-30a(+), lane 3 corresponds to the induced cell lysate of the E. coli strain containing the recombinant plasmid pE30-Aly5 (A) or pE30-Aly5-T336N (B), lane 4 corresponds to the supernatant fluid of the induced cell lysate, and lane 5 corresponds to the rAly5 (A) or rAly5-T336N (B) purified protein obtained from the supernatant.
Enzymatic characteristics of rAly5.
The recombinant protein rAly5 did not digest agarose, chondroitin, heparin, hyaluronan, xanthan, or xylan but could effectively digest alginate to produce oligosaccharides, exhibiting strong absorbances at 235 nm, suggesting that Aly5 is an alginate lyase (Fig. 3A).
FIG 3.
Polysaccharide degradation patterns of rAly5 and the truncated enzyme rAly5-T336N. (A) Time course treatment of alginate (1.0 mg/ml) using rAly5 (1.0 U/ml) at 40°C. (B) Final main products of alginate (∼20 μg) degraded by rAly5 and rAlgL-T336N (40°C, 1.0 U/ml). The oligosaccharide products were gel filtered through a Superdex peptide 10/300 GL column and monitored using a wavelength of 235 nm. The degree of polymerization of each oligosaccharide is indicated on the peak (e.g., UDP2 represents unsaturated disaccharide). The molar ratio of each oligosaccharide was calculated based on peak areas.
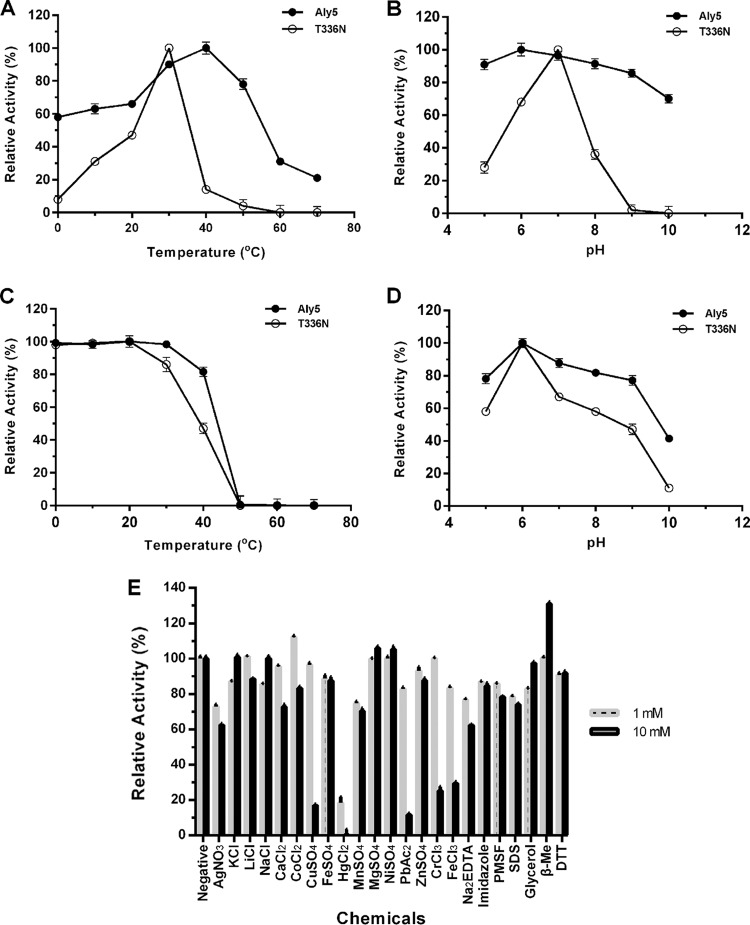
The full-length enzyme rAly5 demonstrated its highest activity at 40°C when alginate was used as the substrate (Fig. 4A). The rAly5 enzyme retained more than 80% of its residual activity after incubation for 2 h at temperatures ranging from 0 to 40°C (Fig. 4B). The optimal pH, determined at 40°C in 50 mM NaAc-HAc buffer, was 6.0 (Fig. 4C). The enzyme retained more than 80% of its residual activity after incubation for 2 h in pH environments ranging from pH 5 to 8 (Fig. 4D).
FIG 4.
Biochemical characteristics of rAly5 and its truncated enzyme rAly5-T336N. (A) Effects of temperature on enzyme activities. (B) Enzyme thermostabilities. The enzymes were preincubated at pH 6 for 2 h at different temperatures. (C) Effects of pH on enzyme activities. (D) Stabilities of enzymes as a function of pH. The enzymes were preincubated for 2 h at 0°C under different pH conditions. The residual activities were determined under optimal temperature conditions. (E) Effects of various compounds on the enzyme activity of rAly5.
The alginate lyase activities of rAly5 were strongly inhibited by 10 mM Ag+, Cu2+, Hg2+, Pb2+, Cr3+, or Fe3+, whereas Hg2+ produced strong inhibition at much lower concentrations (i.e., 1.0 mM) (Fig. 4E). In contrast, rAly5 activities were increased to 116% by 1.0 mM Co2+ and to 135% by a 10 mM concentration of the reducing agent β-mercaptoethanol. Notably, the enzyme activity of rAly5 was only weakly affected by changing NaCl concentrations from 0 to 1 M (data not shown).
Under optimal conditions (40°C in 50 mM NaAc-HAc [pH 6.0]), the specific activity of rAly5 with regard to alginate was measured as described in Materials and Methods and was ∼620 U/mg of protein.
Alginate degradation pattern and oligosaccharide-yielding properties of rAly5.
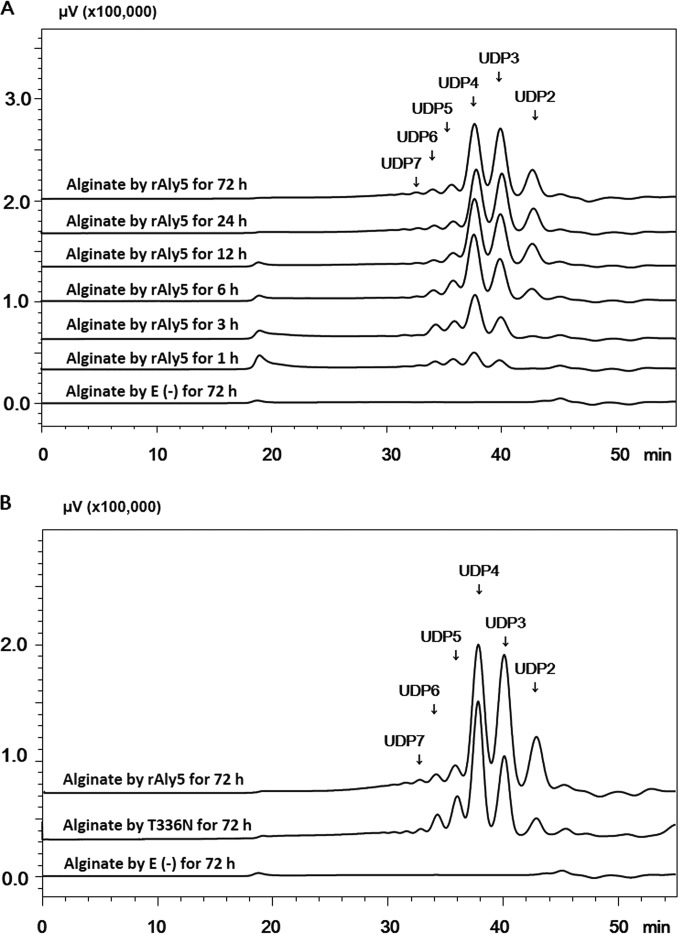
To determine the polysaccharide-degrading pattern of rAly5, the digestion of alginate (1 mg/ml) by rAly5 (1 U/ml) was monitored at 40°C. The reaction time varied from 0 to 1, 3, 6, 12, or 72 h. The digests (∼20 μg) were loaded onto a gel filtration column (Superdex peptide 10/300 GL) and monitored via absorbance at 235 nm. When the alginate polysaccharide was digested, rAly5 initially produced high-molecular-weight oligosaccharides and then converted them into smaller unsaturated oligomers (Fig. 3A). These results indicated that the Aly5 protein is an endo-type alginate lyase when degrading polysaccharides.
To identify the final oligosaccharide products, the alginate was digested using excess rAly5 (5 U/ml) at 40°C for 72 h. The resulting unsaturated oligosaccharide products were analyzed and purified through gel filtration. Further MS analyses indicated that the molecular masses of the oligosaccharide fractions in the final main products were 352, 528, 704, 880, 1,056, and 1,232 Da. The results indicate that the final alginate digests obtained using rAly5 are composed mainly of unsaturated disaccharide (UDP2), trisaccharide (UDP3), tetrasaccharide (UDP4), pentasaccharide (UDP5), hexasaccharide (UDP6), and heptasaccharide (UDP7) fractions. Based on peak areas, the molar ratio of these final products (UDP2/UDP3/UDP4/UDP5/UDP6/UDP7) was determined to be 1:2.2:2.3:0.42:0.28:0.17 (Fig. 3B). These results indicated that the UDP4 fraction was the final product having the highest mass concentration (∼40% [wt/wt]).
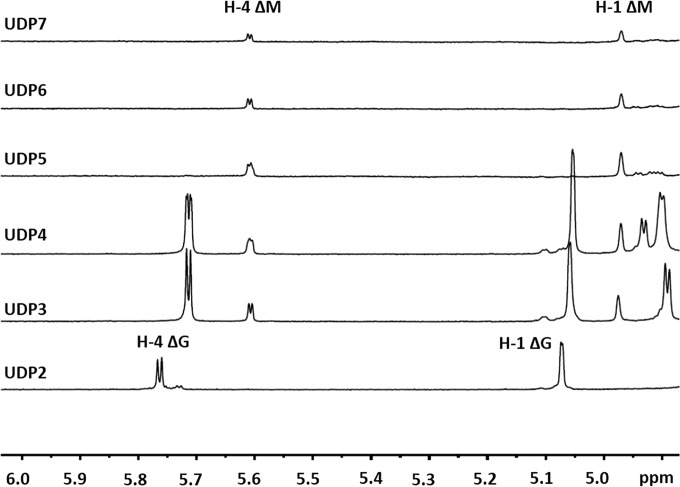
Through gel filtration, the final alginate digests were purified into size-defined oligosaccharide fractions and further analyzed by 1H-NMR spectroscopy. Interestingly, based on the presence of strong H-4ΔG signals at 5.76 ppm, the UDP2 fraction was identified as the ΔG disaccharide (25), suggesting that Aly5 mainly yields ΔG as a disaccharide product when degrading alginate. The UDP3 and UDP4 fractions showed high proportions of ΔG in their nr ends: e.g., the ratios of ΔG to ΔM units were individually determined to be ∼2.7:1 and 2.5:1, respectively, by integrating the H-4ΔG (δ 5.72 ppm) and H-4ΔM (δ 5.62 ppm) signals (Fig. 5) (25). Moreover, the two signals at 3.47 and 3.54 ppm in the UDP3 fraction indicate the existence of H-2 of G at the reducing end, neighboring G and M, respectively. Thus, the ΔGG and ΔMG units were determined to have a molar ratio of ∼2.7:1, indicating that each contributed to a mass concentration of 72.9% and 27.1% (wt/wt), respectively. In contrast, large oligosaccharide fractions (e.g., UDP5, UDP6, and UDP7) contained ΔM rather than ΔG units in their nr ends (Fig. 5). Therefore, although Aly5 yielded large products containing ΔM ends, Aly5 preferred to produce small oligosaccharides in which the nr ends primarily contained ΔG units.
FIG 5.
1H-NMR analyses (600 MHz, 28°C) of the final oligosaccharide products of alginate digested by rAly5. The oligosaccharide products were gel filtered using a Superdex peptide 10/300 GL column and were monitored using a wavelength of 235 nm. Fractions with the same molecular mass (retention time) were collected for NMR analyses. The H-4Δ signals at 5.70 or 5.75 ppm indicate that the residue neighboring the unsaturated residue is a G, meaning that ΔG constitutes the first two sugar residues at the nr ends. The H-4Δ signal at 5.60 ppm indicated that ΔM constitutes the first two residues at the nr ends. As shown for the UDP3 and UDP4 fractions, the intensity of the H-4ΔG signal is much higher than that of the H-4ΔM signal.
Degradation pattern for unsaturated oligosaccharides degraded by rAly5.
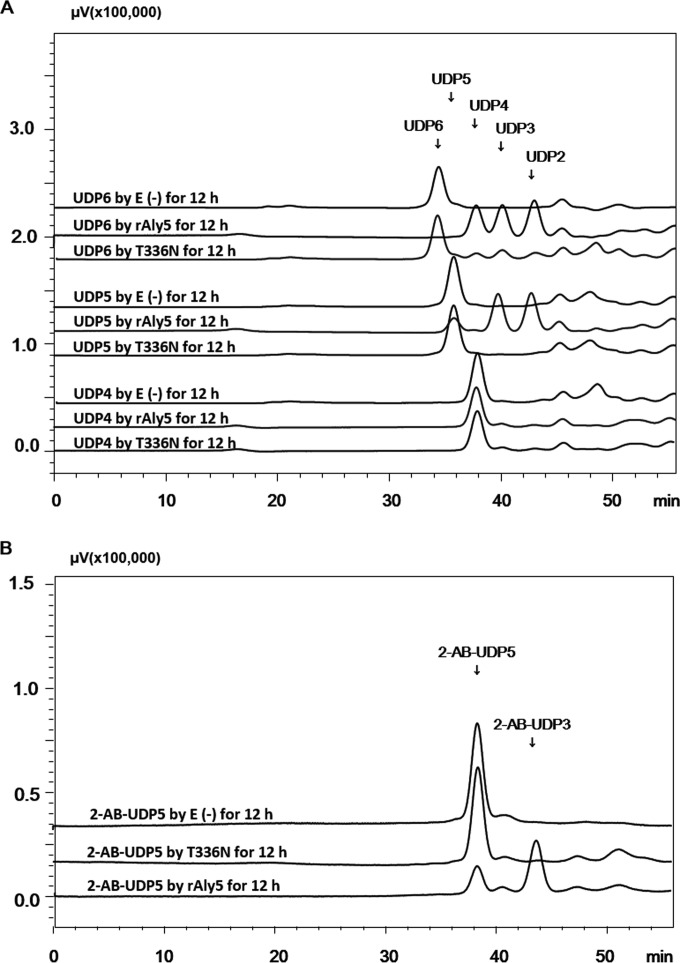
To investigate associated oligosaccharide degradation properties, alginate was initially partially digested by rAly5. Subsequently, the intermediate unsaturated oligosaccharide fractions were purified through gel filtration using a Superdex peptide 10/300 GL column and individually characterized using MS analysis. After further enzymatic reactions with rAly5, the final digest of each size-defined oligosaccharide fraction was analyzed via gel filtration HPLC. The results showed that rAly5 could partially degrade the UDP5 fraction and could completely degrade larger unsaturated oligosaccharide fractions (e.g., UDP6), but that rAly5 could not degrade small oligosaccharides, including the UDP2, UDP3, and UDP4 fractions (Fig. 6A). When cleaving the UDP5 fraction, rAly5 produced UDP2 and UDP3 fractions with equal molar proportions, whereas rAly5 yielded two UDP3 molecules or two oligosaccharides, UDP2 and UDP4, when digesting UDP6 oligosaccharides (Fig. 6A).
FIG 6.
Oligosaccharide degradation patterns of rAly5 and its truncated protein T336N. (A) Degradation of unsaturated oligosaccharides (∼20 μg) by rAly5 (A) at 40°C for 12 h. E (−), without the enzyme; E (+), with the enzyme at 0.1 U/ml. HPLC analyses were performed using a Superdex peptide 10/300 GL column monitored at a wavelength of 235 nm. (B) Degradation of 2-AB-labeled unsaturated pentosaccharides (2-AB-UDP5) by rAly5 and rAly5-T336N. Fluorescent HPLC analyses were performed using a Superdex peptide 10/300 GL column with an excitation wavelength of 330 nm and a monitoring wavelength of 420 nm.
To further elucidate the oligosaccharide degradation pattern of rAly5, each intermediate unsaturated oligosaccharide fraction was labeled using excess 2-aminobenzamide (2-AB), after which the products were purified via gel filtration and digested with rAly5. Fluorescent HPLC analyses indicated that rAly5 did not degrade 2-AB-labeled UDP2, UDP3, or UDP4 but that rAly5 partially digested 2-AB-labeled UDP5 into UDP2 and 2-AB-labeled DUP3 (Fig. 6B).
Therefore, the oligosaccharide fraction of alginate-derived UDP5 was the smallest substrate of rAly5, and the UDP2 fraction was the minimal product yielded from the nr ends of 2-AB-labeled substrates.
Substrate preference of rAly5.
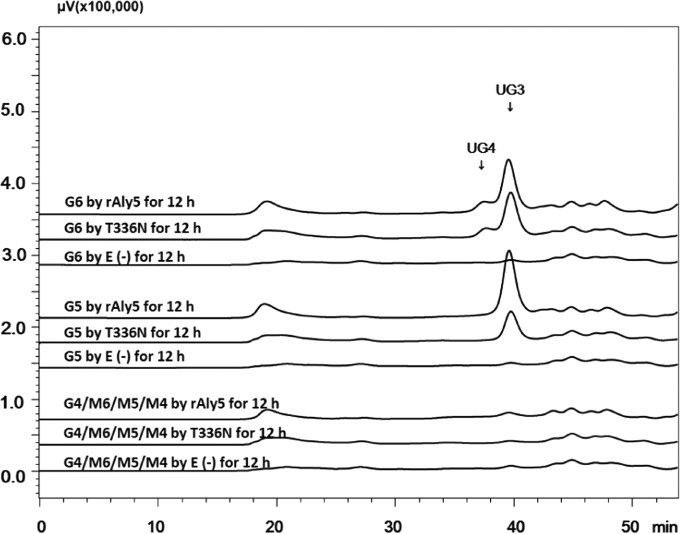
To determine the selectivity of Aly5 for guluronate or mannuronate, size-defined saturated oligosaccharides were used as testing substrates using a procedure that was similar to that described for unsaturated oligosaccharides. As shown in Fig. 7, rAly5 could digest G-enriched saturated oligosaccharides that were larger than tetrasaccharides in size. When degrading G5, rAly5 produced the saturated disaccharide G2 and the unsaturated trisaccharide UG3, whereas digesting G6, rAly5 yielded G2 and the unsaturated tetrasaccharide UG4 or G3 and UG3 as the final products. These results indicated that Aly5 cleaved G2, the disaccharide product, from the nonreducing end of the saturated oligosaccharide substrate. Moreover, the enzyme could not degrade mannuronate oligosaccharides, such as the tetrasaccharide M4, the pentasaccharide M5, and the hexasaccharide M6, regardless of their degrees of polymerization (Fig. 7). These results indicate that Aly5 is an alginate lyase with G rather than M preference.
FIG 7.
Substrate preferences of rAly5 and its truncated T336N protein. Degradation of saturated oligosaccharides (∼ 20 μg) by enzymes at their optimal temperatures for 12 h. E (−), without the enzyme (control group). HPLC analyses were performed using a Superdex peptide 10/300 GL column monitored at a wavelength of 235 nm. G6, hexasaccharide of guluronate; G5, pentasaccharide of guluronate; M5, pentasaccharide of mannuronate.
Effects of the noncatalytic module of the enzyme.
To investigate possible effects of the NCR module of the enzyme, in particular the substrate degradation properties of Aly5, we amplified the encoding gene of the catalytic module (Leu337 to His566) and cloned the 0.7-kb DNA fragment into a pET-30a(+) plasmid. The truncated T336N protein, named rAly5-T336N, was expressed in E. coli BL21(DE3) cells and purified using procedures similar to the methods described for rAly5.
SDS-PAGE analyses showed that under similar conditions, the catalytic-module-encoding gene of Aly5 produced soluble proteins at concentrations of <10 mg/liter, much lower than the concentrations produced by the full-length gene (Fig. 2B). The truncated T336N protein could degrade alginate into unsaturated oligosaccharides (Fig. 3B), demonstrating maximal activity at 30°C, which is 10°C lower than the temperature for rAly5 (Fig. 4A). The T336N protein retained its thermostability at temperatures ranging from 0 to 30°C, a range more narrow than that of rAly5 (Fig. 4B). The truncated enzyme demonstrated highest activity at pH 7.0, differing from rAly5 (pH 6) (Fig. 4C). T336N also retained stability within a narrower pH range (from 5.5 to 6.5) (Fig. 4D). The optimized enzyme activity of T336N toward alginate was determined to be ∼264 U/mg of protein, much lower than that of the wild-type enzyme. These results suggest that the NCR fragment of Aly5 is essential to maintain structure coordination, thereby affecting enzyme stabilities and biochemical characteristics.
Comparison of the substrate degradation patterns of rAly5 and T336N.
Similar to rAly5, the truncated protein T336N can degrade alginate to produce UDP2, DUP3, UP4, UDP5, UDP6, and UDP7 fractions as final products with a molar ratio of 1:3.7:6.1:1.8:1:0.35, which differs significantly from the molar ratio produced by the full-length enzyme (Fig. 3B). For T336N, the proportions of the UDP2 and UDP3 fractions are significantly decreased, whereas the proportions of larger oligosaccharides (e.g., UDP4, UDP5, UDP6, and UDP7) are increased. To confirm these results, we performed the catalytic reaction under various conditions, including increasing the amount of T336N protein by 5 times, changing the temperature to 30 or 40°C, and doubling the reaction time, but none of these conditions altered the oligosaccharide products or their composition ratios. Thus, compared with the wild-type enzyme, the capacity of the T336N enzyme to produce small oligosaccharides was weakened due to the deletion of the NCR domain.
To investigate the oligosaccharide-degrading properties of the T336N protein, size-defined intermediate oligosaccharide products of alginate were used as testing substrates. As shown in Fig. 6A, the T336N enzyme cannot degrade the UDP2, UDP3, and UDP4 fractions, as is the case with rAly5. However, unlike rAly5, which can partially digest both UDP5 and 2-AB-labeled UDP5, the truncated T336N enzyme could not degrade either (Fig. 6A and B), although it could weakly cleave UDP6 to produce UDP2, UDP3, and UDP4 fractions (Fig. 6A), suggesting that the UDP6 fraction rather than the UDP5 fraction is the minimal substrate for the T336N protein. Moreover, the truncated T336N enzyme could completely degrade the saturated guluronate oligosaccharide G6, producing G3 and UG3 or G2 and UG4 fractions (Fig. 7). The T336N enzyme could also partially digest G5 to produce saturated disaccharides (G2) and unsaturated trisaccharides (UG3), thereby exhibiting a pattern similar to that of rAly5 but digesting a lower proportion (Fig. 7). Moreover, the T336N enzyme did not digest any of the mannuronate oligosaccharides tested (Fig. 7). Therefore, the catalytic module of Aly5 was the key element that determined its M/G preference and substrate-degrading pattern, whereas the NCR fragment was essential for enzyme binding and degradation of small substrates.
DISCUSSION
Like most reported Flammeovirga bacteria, the MY04 strain was efficient with regard to enzymatic degradation and bacterial utilization of multiple polysaccharides, including alginate (34). Therefore, Flammeovirga sp. strain MY04 constituted a source of abundant carbohydrate-active enzymes. We have constructed a fosmid DNA library of MY04 and have found an agarase gene, named agaG4, clustered with several cellulase- and mannase-encoding genes (36). However, the efficient alginate degradation system of MY04 has not yet been elucidated, nor has that of any other Flammeovirga strain. In the present study, an alginate lyase gene (the Aly5 gene), was identified in the genome of a Flammeovirga strain and was expressed in E. coli cells to produce proteins for enzyme characterization.
The full-length protein of Aly5 is module-organized and contains an N-terminal NCR fragment and a C-terminal catalytic module in its sequence (Fig. 1A). The catalytic module contained a Gln441-Ile442-His443 function motif that was highly conserved among alginate lyases belonging to various PL families (11, 19). However, the full-length protein exhibited ≤30% sequence identities with most previously elucidated alginate lyases, except for AlyA1 of Z. galactanivorans, with which Aly5 exhibited an identity of 41%. The AlyA1 protein was classified as a member of the PL7 superfamily (25, 45). However, the catalytic module of Aly5 exhibited lower homologies to those of other PL7 alginate lyases, including the type member, A1-II′ from Sphingomonas sp. strain A1 (gi no. 46409008) (46), and the double-catalytic-module-containing enzyme, AlyA, from Vibrio splendidus 12B01 (gi no. 84378061) (Fig. 1B) (22). Furthermore, due to their extraordinary sequence properties, Aly5 of Flammeovirga sp. strain MY04, AlyA1 of Z. galactanivorans, A1-II′ of Sphingomonas sp. strain A1, and AlyA of Vibrio splendidus 12B01 (gi no. 84378061) were clustered as a novel endolytic branch within the PL7 superfamily (Fig. 1C). Notably, both AlyA and A1-II′ could degrade M- as well as G-enriched blocks (22, 46), while AlyA1 was G specific (25).
The present study demonstrated the recombinant protein of Aly5 to be an endo-type alginate lyase when degrading the associated polysaccharide (Fig. 3A) or oligosaccharides (Fig. 6A), with a G rather than M preference (Fig. 7). The enzyme could completely degrade alginate into a series of unsaturated oligosaccharides with molecular sizes ranging from di- to heptasaccharides, in which the UDP4 fraction exhibited the highest mass concentration (Fig. 3B). Interestingly, based on 1H-NMR spectral analyses, we have identified two prominent characteristics of the final alginate digests obtained via Aly5, and both of these characteristics are in accordance with Aly5's G preference. First, the enzyme primarily produced ΔG as the disaccharide product during the alginate digestion (Fig. 5), thereby exhibiting a pattern similar to that of AlyA1, a PL7 guluronate lyase from Z. galactanivorans (25). Second, in the small final oligosaccharide products, the ΔG proportions are much higher than the proportions of ΔM units, whereas only ΔM ends are found in large products (Fig. 5). To the best of our knowledge, this second property is a novel oligosaccharide-producing property of guluronate lyase.
This (second) novel property is determined by both the substrate preference and the substrate degradation pattern of Aly5. We found that Aly5 could degrade only oligosaccharides that were ≥UDP5 in size: in other words, Aly5 cleaved disaccharide units (e.g., ΔG and G2) from the nr ends of pentasaccharide substrates (e.g., UDP5, G5, and 2-AB-UDP5) or degraded hexasaccharide fractions (e.g., G6 and UDP6) to produce disaccharide (e.g., ΔG and G2), trisaccharide (e.g., G3 and UDP3), and tetrasaccharide (e.g., UDP4) fractions (Fig. 6A and B and Fig. 7). However, rAly5 could not digest oligosaccharides that were <UDP5 in size, nor could rAly5 degrade any size-defined mannuronate oligosaccharides to produce smaller products (Fig. 6A and 7). That is, if large intermediate oligosaccharide products (e.g., UDP5, UDP6, and UDP7) contain ΔG or G-enriched units in their nr ends, the G-specific lyase Aly5 will further degrade these oligomers into smaller ones; in contrast, if the oligosaccharide fractions contain ΔM or M-enriched units in their nr ends or their molecular sizes are smaller than UDP5, enzymatic degradation by rAly5 will be inhibited. Therefore, given the final oligosaccharide products yielded by rAly5, small fractions are markedly different from large ones, which are nondegradable by Aly5, at their nr ends. Despite the inability of rAly5 to digest mannuronate oligosaccharides, the above hypothesis is further supported by the observation that rAly5 cannot further degrade any of the ΔM-containing final oligosaccharide products, regardless of their sizes.
Recent studies have revealed that NCRs within polysaccharide depolymerases are important elements that are inactive during substrate degradation but that affect enzymatic characteristics and oligosaccharide productivities (30, 47). Among PL7 alginate lyases, the effects of most NCRs remain unclear, with the exception of the CBM13 module of AlyL2 of Agarivorans sp. strain L11 (30), although crystal structures and biochemical properties of many catalytic domains have been elucidated (25, 48). Deletion of the CBM13 domain from AlyL2 changed its substrate preferences, and the presence of this module increased the disaccharide proportion of its products. This research provided a new strategy for improving alginate lyase properties through module truncation. However, the corresponding mechanisms are still unclear. In the present study, the deletion of the N-terminal NCR fragment from rAly5 alters many of its biochemical characteristics and its enzyme activity, but its M/G preferences remain the same (Fig. 7). Notably, the deletion of the NCR from rAly5 significantly decreases the yields of small final products, such as the UDP2, UDP3, and UDP4 fractions, but significantly increases the yields of large oligosaccharides, such as the UDP5, UDP6, and UDP7 fractions (Fig. 3B).
As previously discussed, the enzymatic degradation of large oligosaccharide fractions by rAly5 produces small final products in the forms of the UDP2, UDP3, and UDP4 fractions (Fig. 6). Accordingly, the abnormal accumulation of large oligosaccharide fractions in the final alginate digests produced by T336N reduces the yields of small products. Two possible causes may be responsible for the accumulations of large products. First, as previously discussed, the accumulated large oligosaccharide fractions are M enriched, as demonstrated by the observation that rAly5 could not further degrade any of these large final oligosaccharide products produced by T336N, despite their molecular sizes (negative results [data not shown]). Second, through oligosaccharide-degrading tests, we found that the deletion of the NCR fragment from rAly5 leads to less degradation of G5 (Fig. 7) and no digestion of the UDP5 (Fig. 6A) and 2-AB-UDP5 fractions (Fig. 6B) by the truncated protein. These results suggest that due to the NCR truncation, the smallest substrate of the T336N enzyme has become larger than that of rAly5 (UDP5). This hypothesis is supported by the observation that the T336N enzyme could successfully digest G6, a larger saturated oligosaccharide (Fig. 7). Therefore, the results of the present study demonstrate that the presence of the NCR fragment of Aly5 is essential for the catalytic module to bind and degrade the smallest oligosaccharide substrates (e.g., UDP5 fractions) and that the presence or absence of the NCR fragment therefore affects substrate degradation patterns and product yields.
ACKNOWLEDGMENTS
This work was financially supported by the National High Technology Research and Development Program of China (grant no. 2012AA021504), the Major State Basic Research Development Program of China (grant no. 2012CB822102), the National Natural Science Foundation of China (grant no. 31300664 and 31570071), the Open Research Fund Program of Shandong Provincial Key Laboratory of Glycoscience and Glycotechnology (Ocean University of China) (grant no. KLGG [OUC] 201301), the Specialized Research Fund for the Doctoral Program of Higher Education of China (grant no. 20130131120079), and the Specialized Research Fund for Postdoctoral Innovation of Shandong Province of China (grant no. 201303104).
We declare that we have no competing interests.
W.H. designed the study under the guidance of Y.L. and F.L. F.L. and W.H. drafted and corrected the manuscript. W.H., J.G., Y.C., and H.L. carried out the experiments. All authors approved the final manuscript.
REFERENCES
- 1.Gacesa P. 1998. Alginates. Carbohydr Polym 8:161–182. [Google Scholar]
- 2.Moe ST, Draget K, Skjåk-Bræk G, Smidsrød O. 1995. Alginates, p 245–286. In Stephen AM. (ed), Food polysaccharides and their applications. Marcel Dekker, New York, NY. [Google Scholar]
- 3.Haug A, Larsen B, Smidsrød O. 1974. Uronic acid sequence in alginate from different sources. Carbohydr Res 32:217–225. doi: 10.1016/S0008-6215(00)82100-X. [DOI] [Google Scholar]
- 4.Kloareg B, Quatrano RS. 1988. Structure of the cell walls of marine algae and ecophysiological functions of the matrix polysaccharides. Oceanogr Mar Biol Annu Rev 26:259–315. [Google Scholar]
- 5.Onsøyen E. 1996. Commercial applications of alginates. Carbohydr Eur 14:26–31. [Google Scholar]
- 6.Rehm BHA, Valla S. 1997. Bacterial alginates: biosynthesis and applications. Appl Microbiol Biotechnol 48:281–288. doi: 10.1007/s002530051051. [DOI] [PubMed] [Google Scholar]
- 7.Rehm BHA. 2010. Bacterial polymers: biosynthesis, modifications and application. Nat Rev Microbiol 8:578–592. doi: 10.1038/nrmicro2354. [DOI] [PubMed] [Google Scholar]
- 8.Wilson M. 2001. Bacterial biofilms and human disease. Sci Prog 84:235–254. doi: 10.3184/003685001783238998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 10.Wong TY, Preston LA, Schiller NL. 2000. Alginate lyase: review of major sources and enzyme characteristics, structure-function analysis, biological roles, and application. Annu Rev Microbiol 54:289–340. doi: 10.1146/annurev.micro.54.1.289. [DOI] [PubMed] [Google Scholar]
- 11.Kim HS, Lee CG, Lee EY. 2011. Alginate lyase: structure, property, and application. Biotechnol Bioprocess Eng 16:843–851. doi: 10.1007/s12257-011-0352-8. [DOI] [Google Scholar]
- 12.Svanem BIG, Stran WI, Erstevåg H, Skjåk-Bræk G, Hartmann M, Barberon T, Valla S. 2001. The catalytic activities of the biofunctional Azotobacter vinelandii mannuronan C-5-epimerase and alginate lyase AlgE7 probably originate from the same active site in the enzyme. J Biol Chem 276:31542–31550. doi: 10.1074/jbc.M102562200. [DOI] [PubMed] [Google Scholar]
- 13.Haug A, Larsen B, Smidsrød O. 1967. Studies on the sequence of uronic acid residues in alginic acid. Acta Chem Scand 21:691–704. doi: 10.3891/acta.chem.scand.21-0691. [DOI] [Google Scholar]
- 14.Wargacki AJ, Leonard E, Win MN, Regitsky DD, Santos CNS, Kim PB, Cooper SR, Raisner R, Herman A. 2012. An engineered microbial platform for direct biofuel production from brown macroalgae. Science 335:308–313. doi: 10.1126/science.1214547. [DOI] [PubMed] [Google Scholar]
- 15.Martin M, Portetelle D, Michel G, Vandenbol M. 2014. Microorganisms living on macroalgae: diversity, interactions, and biotechnological applications. Appl Microbiol Biotechnol 98:2917–2935. doi: 10.1007/s00253-014-5557-2. [DOI] [PubMed] [Google Scholar]
- 16.Alkawash MA, Soothil JS, Schiller NL. 2006. Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. APMIS 114:131–138. doi: 10.1111/j.1600-0463.2006.apm_356.x. [DOI] [PubMed] [Google Scholar]
- 17.Bjarnsholt T, Ciofu O, Molin S, Givskov M, Høiby N. 2013. Applying insights from biofilm biology to drug development—can a new approach be developed? Nat Rev Drug Discov 12:791–808. doi: 10.1038/nrd4000. [DOI] [PubMed] [Google Scholar]
- 18.Maiorana A, Bugli F, Papi M, Torelli R, Ciasca G, Maulucci G, Palmieri V, Cacaci M, Sterbini FP, Posteraro B, Sanguinetti M, Spirito MD. 2015. Effect of alginate lyase on biofilm-grown Helicobacter pylori probed by atomic force microscopy. Int J Polym Sci 2015:989516. [Google Scholar]
- 19.Zhu BW, Yin H. 2015. Alginate lyase: review of major sources and classification, properties, structure—function analysis and application. Bioengineered 6:125–131. doi: 10.1080/21655979.2015.1030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan JL, Mao JX, Xie JP. 2014. Bacteriophage polysaccharide depolymerases and biomedical applications. Biodrugs 28:265–274. doi: 10.1007/s40259-013-0081-y. [DOI] [PubMed] [Google Scholar]
- 21.Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B. 2015. Bacteriophages and phage-derived proteins—application approaches. Curr Med Chem 22:1757–1773. doi: 10.2174/0929867322666150209152851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Badur AH, Jagtap SS, Yalamanchili G, Lee JK, Zhao HM, Rao CV. 2015. Alginate lyase from alginate-degrading Vibrio splendidus 12B01 are endolytic. Appl Environ Microbiol 81:1865–1873. doi: 10.1128/AEM.03460-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farrell EK, Tipton PA. 2012. Functional characterization of AlgL, an alginate lyase from Pseudomonas aeruginosa.. Biochemistry 51:10259–10266. doi: 10.1021/bi301425r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochiai A, Yamasaki M, Mikami B, Hashimoto W, Murata K. 2010. Crystal structure of exotype alginate lyase Atu3015 from Agrobacterium tumefaciens.. J Biol Chem 285:24519–24528. doi: 10.1074/jbc.M110.125450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas F, Lundqvist LCE, Jam M, Jeudy A, Barbeyron T, Sandstöm C, Michel G, Czjzek M. 2013. Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J Biol Chem 288:23021–23037. doi: 10.1074/jbc.M113.467217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park D, Jagtap S, Nair SK. 2014. Structure of a PL17 family alginate lyase demonstrates functional similarities among exotype depolymerases. J Biol Chem 289:8645–8655. doi: 10.1074/jbc.M113.531111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HT, Chung JH, Wang D, Lee J, Woo HC, Choi IG, Kim KH. 2012. Depolymerization of alginate into a monomeric sugar acid using Alg17c, an exo-oligoalginate lyase cloned from Saccharophagus degradans 2-40. Appl Microbiol Biotechnol 93:2233–2239. doi: 10.1007/s00253-012-3882-x. [DOI] [PubMed] [Google Scholar]
- 28.Ryu M, Lee EY. 2011. Saccharification of alginate by using exolytic oligoalginate lyase from marine bacterium Sphingomonas sp. MJ-3. J Ind Eng Chem 17:853–858. doi: 10.1016/j.jiec.2011.08.001. [DOI] [Google Scholar]
- 29.Li SY, Wang LN, Han F, Gong QH, Yu WG. 2015. Cloning and characterization of the first polysaccharide lyase family 6 oligoalginate lyase from marine Shewanella sp. Kz7. J Biochem doi: 10.1093/jb/mvv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li SY, Yang XM, Bao MM, Wu Y, Yu WG, Han F. 2015. Family 13 carbohydrate-binding module of alginate lyase from Agarivorans sp. L11 enhances its catalytic efficiency and thermostability, and alters its substrate preference and product distribution. FEMS Microbiol Lett 362:fnv054. [DOI] [PubMed] [Google Scholar]
- 31.Hosoya S, Yokota A. 2007. Flammeovirga kamogawensis sp. nov., isolated from coastal seawater in Japan. Int J Syst Evol Microbiol 57:1327–1330. doi: 10.1099/ijs.0.64977-0. [DOI] [PubMed] [Google Scholar]
- 32.Xu H, Fu XY, Yang N, Ding ZX, Lai QL, Zeng RY. 2012. Flammeovirga pacific sp. nov., isolated from deep-sea sediment. Int J Syst Evol Microbiol 62:937–941. doi: 10.1099/ijs.0.030676-0. [DOI] [PubMed] [Google Scholar]
- 33.Zhao J, Shi B, Jiang QR, Ke CH. 2012. Changes in gut-associated flora and bacterial digestive enzymes during the development stages of abalone (Haliotis diversicolor). Aquaculture 338–341:147–153. doi: 10.1016/j.aquaculture.2012.01.016. [DOI] [Google Scholar]
- 34.Han WJ, Gu JY, Li JG, Yan QJ, Wu ZH, Gu QQ, Li YZ. 2012. A polysaccharide-degrading marine bacterium Flammeovirga sp. MY04 and its extracellular agarase system. J Ocean Univ China 11:375–382. doi: 10.1007/s11802-012-1929-3. [DOI] [Google Scholar]
- 35.Yang JI, Chen LC, Shih YY, Hsieh CY, Chen CY, Chen WM, Chen CC. 2011. Cloning and characterization of a β-agarase from Flammeovirga yaeyamensis strain YT. J Biosci Bioeng 112:225–232. doi: 10.1016/j.jbiosc.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 36.Han WJ, Gu JY, Liu HH, Li FC, Wu ZH, Li YZ. 2013. An extra peptide within the catalytic module of a β-agarase affects the agarose degradation pattern. J Biol Chem 288:9519–9531. doi: 10.1074/jbc.M112.412247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou YP, Chen XL, Chan ZH, Zeng RY. 2015. Expression and characterization of a thermostable and pH-stable β-agarase encoded by a new gene from Flammeovirga pacifica WPAGA1. Process Biochem 50:1068–1075. doi: 10.1016/j.procbio.2015.04.005. [DOI] [Google Scholar]
- 38.Zhou GL, Jin M, Cai YP, Zeng RY. 2015. Characterization of a thermostable and alkali-stable α-amylase from deep-sea bacterium Flammeovirga pacifica.. Int J Biol Macromol 80:676–682. doi: 10.1016/j.ijbiomac.2015.07.042. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Yi ZW, Cai YP, Zeng RY. 2015. Draft genome sequence of algal polysaccharide degradation bacterium, Flammeovirga sp. OC4. Mar Genomics 21:21–22. doi: 10.1016/j.margen.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Chan ZH, Wang RP, Liu SL, Zhao CG, Yang SP, Zeng RY. 2015. Draft genome sequence of an agar-degrading marine bacterium Flammeovirga pacifica WPAGA1. Mar Genomics 20:23–24. doi: 10.1016/j.margen.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 42.Kumar S, Tamura K, Nei M. 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput Appl Biosci 10:189–191. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed, p A8.40–8.47 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 44.Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. 1995. Nonselective and efficient fluorescent labelling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem 230:229–238. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- 45.Thomas F, Barbeyron T, Tonon T, Génicot S, Czjzek M, Michel G. 2012. Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteria to their dependent transfers to marine Proteobacteria and human gut Bacteroides.. Environ Microbiol 14:2379–2394. doi: 10.1111/j.1462-2920.2012.02751.x. [DOI] [PubMed] [Google Scholar]
- 46.Miyake O, Ochiai A, Hashimoto W, Murata K. 2004. Origin and diversity of alginate lyases of families PL-5 and -7 in Sphingomonas sp. strain A1. J Bacteriol 186:2891–2896. doi: 10.1128/JB.186.9.2891-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han WJ, Gu JY, Liu HH, Li FC, Wu ZH, Li YZ. 2015. Deletion of a non-catalytic region increases the enzymatic activity of a β-agarase from Flammeovirga sp. MY04. J Ocean Univ China 14:841–848. doi: 10.1007/s11802-015-2800-0. [DOI] [Google Scholar]
- 48.Ogura K, Yamasaki M, Mikami B, Hashimoto W, Murata K. 2008. Substrate recognition by family 7 alginate lyase from Sphingomonas sp. A1. J Mol Biol 380:373–385. doi: 10.1016/j.jmb.2008.05.008. [DOI] [PubMed] [Google Scholar]