Abstract
Development of protection tools targeting Dickeya species is an important issue in the potato production. Here, we present the identification and the characterization of novel biocontrol agents. Successive screenings of 10,000 bacterial isolates led us to retain 58 strains that exhibited growth inhibition properties against several Dickeya sp. and/or Pectobacterium sp. pathogens. Most of them belonged to the Pseudomonas and Bacillus genera. In vitro assays revealed a fitness decrease of the tested Dickeya sp. and Pectobacterium sp. pathogens in the presence of the biocontrol agents. In addition, four independent greenhouse assays performed to evaluate the biocontrol bacteria effect on potato plants artificially contaminated with Dickeya dianthicola revealed that a mix of three biocontrol agents, namely, Pseudomonas putida PA14H7 and Pseudomonas fluorescens PA3G8 and PA4C2, repeatedly decreased the severity of blackleg symptoms as well as the transmission of D. dianthicola to the tuber progeny. This work highlights the use of a combination of biocontrol strains as a potential strategy to limit the soft rot and blackleg diseases caused by D. dianthicola on potato plants and tubers.
INTRODUCTION
The Pectobacterium and Dickeya pectinolytic bacteria are phytopathogens responsible for several macerating diseases on a wide range of crop and ornamental plants (1–3). The damage caused by these pathogens remains an important issue in many countries worldwide. In Europe, the pectinolytic pathogens on potato crops include Pectobacterium atrosepticum, Pectobacterium carotovorum subsp. carotovorum, Dickeya dianthicola, and Dickeya solani (4–8). Recently, new Pectobacterium taxa, i.e., P. wasabiae and P. carotovorum subsp. brasiliense, have been characterized (2, 9, 10). Pectobacterium sp. populations are qualified as endemic, as their presence has been observed for almost a century in Europe, while the presence of Dickeya spp. as a cause of the symptoms in potato fields was sporadic until the 2000s and the Dickeya sp. damages were probably exclusively due to the species D. dianthicola. In contrast, D. solani seems to have emerged in the 2000s (3).
Pectobacterium and Dickeya potato pathogens induce blackleg on stems and soft rot on tubers (11, 12). A polyphyletic pathogen population may be isolated from a single sample of symptomatic plant tissues (13). The pathogens penetrate in host plants through natural pores or wounds (i.e., the lenticels, the elongation zones of roots, or insect wounds) as well as mechanical wounds. Insects can act as vectors; surface water and aerosols may also contribute to the dissemination of the pathogens (12, 14–21). Once the pathogens have infected the plant, they may propagate throughout the whole plant and son tubers by the vascular vessels. Losses due to Pectobacterium and Dickeya, related to stem and tuber rot, are major causes for downgrading and rejection of potatoes during seed potato certification. The certification is based on visual inspections in fields or on lots, ranging in Europe from 0 to 4% blackleg tolerance (depending on the seed generation) and from 0 to 0.5% rotted tuber tolerance in storage (Directives 2014/20/UE and 2014/21/UE). Economic impacts differ from country to country; annual direct losses have been estimated to be up to €30 million in the Netherlands (3, 22).
While our knowledge of the taxonomy, epidemiology, and structural and functional genomics of the Pectobacterium sp. and Dickeya sp. pathogens is steadily increasing, the plant protection toolbox remains undeveloped. No fully resistant potato variety is available, although some of them appeared to be sensitive to the pathogens (23–25). A major preventive approach consists of planting certified seed potato tubers. Some other preventive and curative methods have been proposed, such as the use of (i) plant defense elicitors, (ii) natural “predators,” including bacteriophages and bacteria, (iii) bacterial competitors, and (iv) quorum-quenching bacteria (reviewed in references 14, 26, and 27). Different compounds (natural or synthetic), such as salicylic acid, 2-aminobenzoic acid, alpha-elicitins, and complex polysaccharides, have been described as elicitors of plant defense responses against the pectinolytic bacteria. Some natural elicitors are produced by biocontrol bacteria (28–34). So far, only a few bacteria and bacteriophages are described as predators of Dickeya and Pectobacterium (14, 35). Notably, Podoviridae and Myoviridae bacteriophages were tested in vitro, in planta, and in field assay (36–38). In addition, antibiosis activities directed at the pectinolytic bacteria have been reported in several bacterial competitors from the Pseudomonas, Bacillus, Serratia, Lactobacillus, and Lactococcus genera (39–48). These biocontrol bacteria produce a large variety of antimicrobial molecules, such as hydrogen peroxide, siderophores, antibiotics, and volatile compounds. More recently, some bacteria and low-molecular-weight compounds were showed to inactivate the quorum-sensing signals N-acylhomoserine lactones, which modulate the expression of the virulence factors in Pectobacterium and Dickeya potato pathogens (49–54). The quorum-quenching bacteria identified so far belong to the Delftia, Ochrobactrum, Bacillus, and Rhodococcus genera. The colonization of potato plant roots by the quorum-quenching Rhodococcus was shown to be enhanced using the growth-stimulating agents gamma-caprolactone and gamma-heptalactone (55–61).
In this study, we combined microbiological, molecular, and genomic tools to isolate and characterize novel biocontrol agents directed at members of the Dickeya and Pectobacterium species. We evaluated their protective effect against the D. dianthicola-induced tuber soft rot and blackleg diseases as well as against pathogen transmission to the tuber progeny over 3-year greenhouse assays. This work led us to propose a combination of biocontrol Pseudomonas strains to increase the robustness and reproducibility of the plant protection effect.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used are listed in Table 1 (50, 51, 62–64). All of them were routinely cultivated in TY medium (Bacto tryptone, 5 g/liter; yeast extract, 3 g/liter; agar, 15 g/liter) at 30°C for the Dickeya strains and at 25°C for the other bacteria. Tryptic soy agar (TSA) medium was used to isolate bacteria from environmental samples.
TABLE 1.
Bacterial strains
| Strain | Isolation yr | Isolation conditionsa | Source(s) (reference[s])b |
|---|---|---|---|
| Plant pathogens | |||
| Dickeya dianthicola RNS04.9 | 2004 | Blackleg symptoms | V. Hélias, FN3PT and RD3PT collection (49, 50) |
| Dickeya solani 3337 | 2008 | Blackleg symptoms | V. Hélias, FN3PT and RD3PT collection (49, 50, 61, 62) |
| Pectobacterium atrosepticum CFBP6276 | 1999 | Blackleg symptoms | CFBP collection (63) |
| Pectobacterium carotovorum subsp. carotovorum RNS98.1 | 1998 | Blackleg symptoms | V. Hélias, FN3PT and RD3PT collection (49, 50, 62) |
| Biocontrol agents | |||
| Bacillus simplex BA2H3 | 2011 | Potato field soil | This study (see references 65 and 66 for genome sequences) |
| Pseudomonas brassicacearum PA1G7 | 2011 | Potato field soil | |
| Pseudomonas brassicacearum PP1-210F | 2010 | Tuber peels | |
| Pseudomonas fluorescens PA3G8 | 2011 | Tuber peels | |
| Pseudomonas fluorescens PA4C2 | 2011 | Potato roots | |
| Pseudomonas putida PA14H7 | 2010 | Potato field soil |
All isolates were collected in France.
CFBP, French Collection of Plant-Pathogenic Bacteria (Angers, France); FN3PT and RD3PT, Fédération Nationale des Producteurs de Plants de Pomme de Terre and Recherche, Développement et Promotion du Plant de Pomme de Terre (France).
Isolation of bacterial strains from environmental samples.
In 2010 and 2011, environmental samples (rhizospheric soils, asymptomatic plant tissues, and blackleg- and soft rot-infected tissues) were collected from several potato fields by the French regional associations of seed potato growers, i.e., Bretagne-Plant (BP) (Hanvec), Comité Centre & Sud (CCS) (Laurière), and Comité Nord Plants de Pomme de Terre (CNPPT) (Achicourt). One gram (fresh soil or tissue) of each sample was mixed with 10 ml of isotonic saline solution (NaCl, 8 g/liter), and 100 μl of the appropriate dilutions was spread onto TSA plates and incubated from 24 h to 48 h at 25°C. Approximately 10,000 bacterial isolates were picked from the TSA plates, inoculated in liquid TY into 96-microwell plates, and incubated at 25°C for 48 h.
Screening for antagonistic activity against Dickeya and Pectobacterium pathogens.
Two successive screens were performed on agar plates. The first one was performed on the 10,000 isolates which were cultivated in 96-microwell plates. One drop (10 μl) of the bacterial cultures was spotted on square petri plates (120 by 120 mm) which were previously inoculated using 300 μl of P. atrosepticum CFBP6276 or D. dianthicola RNS04.9 cell suspension (3.105 CFU/ml). After incubation at 25°C for 24 h and 48 h, the bacterial growth inhibition zone was measured.
In the next screen, all bacterial isolates that exhibited growth inhibition were tested a second time against P. atrosepticum CFBP6276 and D. dianthicola RNS04.9 and against two other pathogens, i.e., P. carotovorum subsp. carotovorum RNS98.1 and D. solani 3337, according to a similar growth inhibition assay. The bacterial isolates that exhibited growth-inhibitory activity against the Pectobacterium sp. or Dickeya sp. strains were individually stored in 25% glycerol solution at −80°C.
Screening for bioprotection on potato tubers.
The isolates that exhibited growth-inhibitory activity against Pectobacterium sp. and Dickeya sp. in the agar plate assay were tested for bioprotection in a potato tuber maceration assay. Potato tubers were surface sterilized with a bleach solution (1.3% of active chlorine) and cut into slices (1.0 cm in thickness). A well (4-mm diameter and 2-mm depth) was dug into the tuber slice. Each slice (4 replicates per condition) was inoculated with 20 μl of a isotonic saline solution-washed culture of each tested pathogen strain (P. atrosepticum CFBP6276, P. carotovorum subsp. carotovorum RNS98.1, D. dianthicola RNS04.9, and D. solani 3337) which had been cultivated alone or in the presence of the tested biocontrol agents (1/1 ratio) in TY medium for 24 h. The infected slices were incubated at 25°C for 36 h. The measurement of the maceration zone diameters allowed the determination of 3 qualitative symptom categories: no maceration, medium maceration, or strong maceration of the tuber tissues. This screening allowed (i) the exclusion of bacterial isolates that provoked symptoms on potato tubers and (ii) the identification of bacterial isolates that showed sufficient growth inhibition activity to reduce disease symptoms on tubers. Procedures for inoculation of the pathogen and biocontrol agents in this tuber maceration assay differed from the methodology used in the greenhouse assays.
Calculation of the absolute fitness value in competition assays.
Growth inhibition efficiency was precisely quantified in a competitive assay. TY medium (5 ml) was inoculated with each of the rifampin-resistant derivatives of the pathogens P. atrosepticum CFBP6276, P. carotovorum subsp. carotovorum RNS98.1, D. dianthicola RNS04.9, and D. solani 3337 at a final concentration of 106 CFU/ml in the absence or presence of the biocontrol bacterial agents, which were added at a 1/1 ratio. Experiments were performed in triplicate. Bacterial enumerations on TY agar plates supplemented with rifampin (100 μg/ml) were performed at the inoculation time (Nt = 0) and at the final time after 24 h of incubation at 25°C (Nt = 24). The fitness value (W) of the pathogen was calculated according to the formula W = Nt = 24/Nt = 0. For each pathogen, the W values were compared using a Kruskal-Wallis test (α = 0.05), and then a pairwise comparison test was performed (α = 0.05) using the package Rcmdr (R software).
Genus identification of the antagonist strains by 16S rRNA sequencing.
The nucleotide sequences of the 16S rRNA gene rrs of the antagonist isolates were amplified using the universal bacterial 16S rRNA primers pA (5′-AGAGTTTGATCCTGGCTCAG) and pH (5′-AAGGAGGTGATCCAGCCGCA) (65). The amplicon was sequenced by GATC-Biotech Company (Germany), and the 16S rRNA sequences were analyzed online at the Ribosomal Database Project website (http://rdp.cme.msu.edu/) using the BLASTN program (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
MLSA of the biocontrol agents.
Draft genomes of the 6 characterized biocontrol agents Pseudomonas brassicacearum strains PP1-210F (AYJR00000000) and PA1G7 (JBON00000000), Pseudomonas fluorescens strains PA4C2 (AXDA00000000) and PA3G8 (JBOO00000000), Pseudomonas putida strain PA14H7 (JBOP00000000), and Bacillus simplex strain BA2H3 (AXBR00000000) were available (66, 67). A multilocus sequence analysis (MLSA) of the four concatenated housekeeping genes gyrB (2,418 bp), recA (1,053 bp), rpoB (4,074 bp), and 16S rRNA (1,536 bp) was performed using ClustalW for sequence alignment, MEGA V6 for phylogeny, and the neighbor-joining method for tree construction (68).
Greenhouse assays.
During 3 years (2012 to 2014), four independent plant protection assays were performed in two different greenhouses, one located at the INRA-FN3PT experimental site (UMT Inno-Plant, Le Rheu, Brittany, France) and the other at the SIPRE-CNPPT experimental site (Achicourt, Nord-Pas-de-Calais, France). The assays were named according to their location and year: B1 and B2 (2012 and 2013 in Brittany, respectively) and N1 and N2 (2013 and 2014 in Nord-Pas-de-Calais, respectively). All assays were conducted under similar conditions, i.e., long day (16 h) and day and night temperatures of 25°C and 20°C, respectively, using the same potato cultivar (i.e., Bintje) and class (Super Elite; size grade length, 28 to 35 cm) of Solanum tuberosum seed tubers (CNPPT, Achicourt). Each tuber was planted in a pot (16 cm in diameter) containing 1 kg of soil. In the Brittany (INRA-FN3PT) assays B1 and B2, we used a reconstituted, heated soil (neutral pH) that consisted of a mixture of brown peat (60%), sand (40%), and clay (40 kg/m3 of the total mass prepared), while in the Nord-Pas-de-Calais (SIPRE-CNPPT) assays N1 and N2, a field soil (2/3) mixed with sterilized sand (1/3) was used. For the N1 and N2 experiments, the soil was clay-silt (70% silt, 19% clay, and 11% sand) and of neutral pH (6.8). To prevent soil deficiencies, the fertilizer Hakaphos was added 20 days after the plant emergence, according to the recommendation of the solution provider (Compo Expert; 500 ml per tray of a diluted powder solution (1:5, wt/vol).
In each assay (B1, B2, N1, and N2), six conditions were compared: uninfected plants and plants infected with D. dianthicola RNS04.9 alone and in combination with each of the biocontrol P. fluorescens strains PA3G8 and PA4C2 and P. putida PA14H7 or with a mixture of them at a 1:1:1 ratio. For each condition, 20 plants were used. The pathogen and biocontrol agents were inoculated separately. Three weeks after tubers were planted (100% plant emergence), except for the uninfected plants, the pathogen D. dianthicola RNS04.9 was inoculated at 109 CFU/plant by watering the pots with 100 ml cell suspension adjusted to 107 CFU/ml. The biocontrol agents were introduced three times during the experiment: (i) before planting, tubers were immersed in a biocontrol cell suspension at 108 CFU/ml for 2 h and then air dried at room temperature for 2 h, and (ii) 2 weeks after planting and (iii) 4 days after pathogen infection, the biocontrol strains were inoculated at 1010 CFU/plant by watering with 100 ml cell suspension at 108 CFU/ml. The 1:1:1 mixture of the three biocontrol strains P. fluorescens PA3G8 and PA4C2 and P. putida PA14H7 contained each of the strains at 3 × 107 CFU/ml. The cell number (CFU/ml) of each inoculum was verified by counting on TY agar plates.
Survey and analysis of blackleg symptoms in greenhouse assays.
Twice a week, blackleg symptoms were noted by visual observation and analyzed as described by Andrivon et al. (69) and Rouffiange et al. (70). A disease progress curve (DPC) was plotted and used to measure the delay (Δt in days) between the emergence of the first symptoms in the presence versus absence of the biocontrol agents. In addition, the DPCs allowed the calculation of the areas under the DPCs (AUDPCs). AUDPC values were divided by experiment duration in days for the calculation of relative AUDPC values (AUDPCr values), and then the ratio between AUDPCr values in the presence versus absence of the biocontrol agents was used to compare the different conditions. We evaluated data homogeneity between B1 and B2 assays and between N1 and N2 assays using Levene's test (alpha, R), and then each parameter (Δt and AUDPCr ratio) was analyzed by analysis of variance (ANOVA) (α = 0.05) and a pairwise comparison test (α = 0.05) using the R software.
Detection of pathogenic populations in the tuber progeny in greenhouse assays.
At the end of the B2 and N2 greenhouse assays, all the asymptomatic tubers were collected from each asymptomatic plant. For each asymptomatic plant, stolon ends of the asymptomatic tubers were collected and pooled into 10 ml of phosphate buffer (2.7 g/liter of Na2HPO4 · 12H2O, 0.4 g/liter of NaH2PO4 · 2H2O, pH 7.2). After 2 h at room temperature with shaking at 200 rpm, 200 μl of the macerates was mixed with 1,800 μl of PEB medium (16, 63) and incubated at 26°C for 48 h to perform the enrichment step. Bacteria were centrifuged, and total DNA extraction was conducted using a Genomic DNA purification kit (Qiagen). The presence of the Dickeya pathogen was tested by PCR using the Dickeya-specific primers ADE1 (5′-GATCAGAAAGCCCGCAGCCAGAT) and ADE2 (5′-CTGTGGCCGATCAGGATGGTTTTGTCGTGC) as described by Nassar et al. (71). Data were analyzed using Fisher's statistical test (α = 0.05).
Monitoring of the pathogenic and biocontrol populations in greenhouse assays.
In the course of the B2 and N2 greenhouse assays, soil samples (n = 3) were collected before tubers were planted, before pathogen inoculation (ca. 23 days), after the last introduction of the biocontrol agents (ca. 37 days), and after the last symptom observation. One gram of the soil was weighed and placed in a 2-ml Eppendorf tube. Three hundred microliters of lysozyme (25 mg/ml) was added, and samples were incubated for 30 min at 37°C in a water bath. A few ceramic beads were added to the tubes, and the samples were homogenized for 5 min using a Bead Beater tool (Mixer Mill MM 400; Retschat) at maximal speed. DNA was extracted using a kit (Macherey-Nagel NucleoSpin soil kit) following the supplier's recommendations. Finally, the DNA was resuspended in 30 μl TE (pH 7.5) (Tris-HCl, 10 mM; EDTA, 1 mM). The quantity and quality of the extracted DNA were analyzed using a NanoDrop ND-1000 spectrophotometer and by migration in agarose gels (1%, wt/vol).
Strain-specific primers were developed for monitoring the biocontrol Pseudomonas strains PA3G8, PA4C2, and PA14H7 in the extracted soil DNA by quantitative PCR (qPCR). Using CLC Genomics Workbench 5.5, Illumina reads (from 9 to 12 millions of reads) of each of the Pseudomonas strains were mapped against the genomes of the other strains found in the NCBI database (length fraction, 0.5; similarity, 0.8). The unmapped reads were collected and assembled into contigs which represented the strain-specific regions. Several pairs of primers were designed using Primer3web version 4.0.0, and their specificity was tested by PCR on bacterial strains. All strain-specific primers were designed to amplify a DNA fragment of ca. 200 bp in length. Finally, promising candidates were evaluated for qPCR using a LightCycler 480 system (Roche Life Science) and total DNAs extracted from soils which were inoculated with known concentrations of the targeted bacteria. Primers that generated a nonspecific or a weak amplification signal were not retained. We finally selected primers 33aF (5′-TTCGGAGCTTTGCCAGGAAT) and 33aR (5′-TGCCTCATCGAAACACTGCT) for the detection of P. fluorescens PA4C2, 1bF (5′-ACCAAGACAGCCCTACTCCT) and 1bR (5′-CCGACTCTTGCACCGTGATA) for P. fluorescens PA3G8, and 1cF (5′-CTGAAAGCCCACTCATGTCG) and 1cR (5′-CAACACAGGGAAACACAGCA) for P. putida PA14H7. In addition, using the genome sequences of D. dianthicola RNS04.9 (72), D. dianthicola NCPPB 3534 (PRJNA202755), and GBBP 2039 (PRJNA202756) as well as those of D. solani 3337 (62) and P. atrosepticum CFBP6276 (64), we also identified the strain-specific primers of Ddi5bF (5′-GGGTCAGGAGATGTTTTCGC) and Ddi5bR (5′-TGGGACTCATGTATACCGGC) for the pathogen D. dianthicola RNS04.9. The qPCR detection thresholds were 610 DNA copies/g of dry soil for D. dianthicola RNS04.9 and 700, 820, and 390 DNA copies/g of dry soil for the Pseudomonas strains PA3G8, PA14H7, and PA4C2, respectively.
The qPCR sample mix was prepared as follows. First, 8.5 μl of master mix (Roche) was vortexed with 3 μl of the specific forward primer (0.2 μM) and 3 μl of the specific reverse primer (0.2 μM). Half a microliter of extracted DNA from one gram of the soil (Macherey-Nagel NucleoSpin soil kit) was added to the qPCR mix. qPCR was initiated with a denaturation step (10 min at 95°C), followed by 40 cycles of 15 s at 95°C, 15 s at 60°C, and 20 s at 72°C. A melting step was next performed for 15 s at 95°C and 30 s at 60°C, and finally a fluorescence read step was performed continuously from 60 s to 95°C to assess the dissociation characteristics of double-stranded DNA during heating. This last step determined the melting temperature (Tm). Analysis of the data was performed with qPCR-480 LightCycler software (Roche).
The raw results of qPCR were expressed in grams per microliter and corresponded to the DNA concentration of the amplified product, denoted [PPCR]. Initially, it was necessary to convert [PPCR] in number of DNA copies per microliter of reaction medium (denoted [DNAc]), using the formula [DNAc] = (Na × [PPCR])/(MMADN × TPCR), where Na is the Avogadro number (6.02 × 1023), MMADN is the average molecular mass of a DNA base pair (equal to 660 g/mol), and TPCR is the corresponding genome size in base pair units. Genome sizes were 4,721,506 bp, 6,210,847 bp, 6,391,599 bp, and 5,878,513 bp for D. dianthicola RNS04.9 and Pseudomonas strains PA4C2, PA3G8, and PA14H7, respectively (as assessed using the RAST annotation database tool). Finally, a second conversion had to be done. [DNAc] was converted into number of DNA copies per gram of dry soil (denoted [DNAsoil]) according to the formula [DNAsoil] = [DNAc] × (Vef/Wsoil), where Vef is the final volume after DNA extraction, i.e., 30 μl (see above), and Wsoil is the weight of dry soil deduced for the experiment. To achieve a dry soil weight, the fresh soil weighed initially was dried at 65°C for 48 h, and soil moisture was calculated. Wsoil is expressed as grams of dry soil.
RESULTS
Identification of bacteria inhibiting growth of Dickeya and Pectobacterium spp.
Environmental samples (rhizospheric soil and healthy or symptomatic plant tissues) were collected in the course of 11 samplings performed during 3 different campaigns which were conducted by the three regional seed potato organizations (BP, CNPPT, and CCS). Each sample was spread onto TSA medium and contained about 3× 108 CFU/g of fresh plant tissues or rhizospheric soil. Over 10,000 environmental isolates were screened for antagonistic activity directed at P. atrosepticum CFBP6276 and D. dianthicola RNS04.9. Among them, 241 isolates (2.4% of the screened isolates) exhibited the capacity to inhibit the growth of one or both of the tested pathogens.
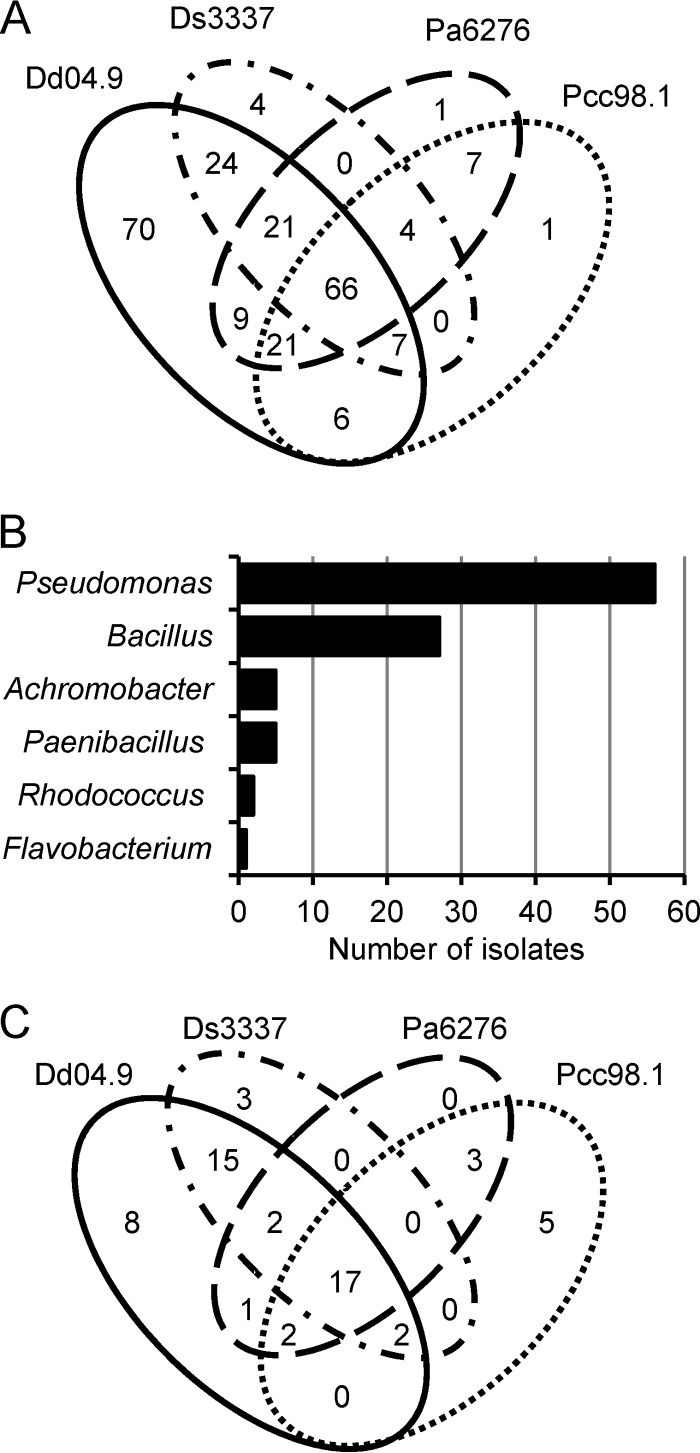
In a second round of tests, the antagonistic activity of these 241 isolates was confirmed using P. atrosepticum CFBP6276, D. dianthicola RNS04.9, P. carotovorum subsp. carotovorum RNS98.1, and D. solani 3337. Most (93%) of the 241 growth-inhibiting isolates were active against D. dianthicola RNS04.9, whereas 46 to 54% of them were able to inhibit at least one of the three other pathogens (Fig. 1A). Remarkably, 66 isolates (27%) exhibited growth-inhibitory properties against all four pathogens.
FIG 1.
Identification of Dickeya- and Pectobacterium-antagonistic strains. (A) Distribution (Venn diagram) of the 241 isolates according to their capacity for growth inhibition against D. dianthicola RNS04.9 (Dd04.9), D. solani 3337 (Ds3337), P. atrosepticum CFBP6276 (Pa6276), and P. carotovorum subsp. carotovorum RNS98.1 (Pcc98.1). (B) Distribution of the retained 96 bacterial isolates according to their genus identification. (C) Distribution (Venn diagram) of the retained 58 isolates according to their capacity for limiting symptoms induced by each pathogen in potato tuber assays. The successive screening and selection to keep 241, then 96, and finally 58 isolates are described in the text.
From the 241-isolate collection, a subset of 96 bacterial isolates was retained according to their efficiency (inhibition halo diameter of ≥2.5 cm) and ability to inhibit at least two phytopathogens. All of these 96 isolates were identified by 16S rRNA sequencing. Most of them belonged to the Pseudomonas (59%) and Bacillus (28%) genera (Fig. 1B).
Identification of bacterial isolates reducing soft rot symptoms on potato tubers.
Each of the 96 16S rRNA-identified isolates was inoculated alone and in the presence of each of the four pathogens D. dianthicola RNS04.9, D. solani 3337, P. atrosepticum CFBP6276, and P. carotovorum subsp. carotovorum RNS98.1 on tuber slices. When inoculated alone, 25% of the 96 tested strains induced an alteration of the plant tissues. Furthermore, after 1 day of cocultivation with the pathogens, 15% of the 96 tested strains were not able to reduce the pathogen-induced soft rot symptoms. All the tuber-altering strains and those unable to reduce tuber symptoms after cocultivation with the pathogens were not retained for further experiments.
In contrast with the above, 58 isolates (60% of the 96 tested strains) reduced the soft rot symptoms caused by at least one of the four phytopathogens. The antagonistic range of these 58 isolates is shown in Fig. 1C. Among the 17 isolates which were able to inhibit all pathogens in this assay, 6 bacterial strains that exhibited prominent protection were retained. P. fluorescens strains PA3G8 and PA4C2 and P. putida strain PA14H7 were especially efficient against Dickeya-induced symptoms, while B. simplex BA2H3 and P. brassicacearum strains PA1G7 and PP1-210F were especially efficient against Pectobacterium-induced symptoms.
Fitness decrease of the phytopathogens in coculture assay.
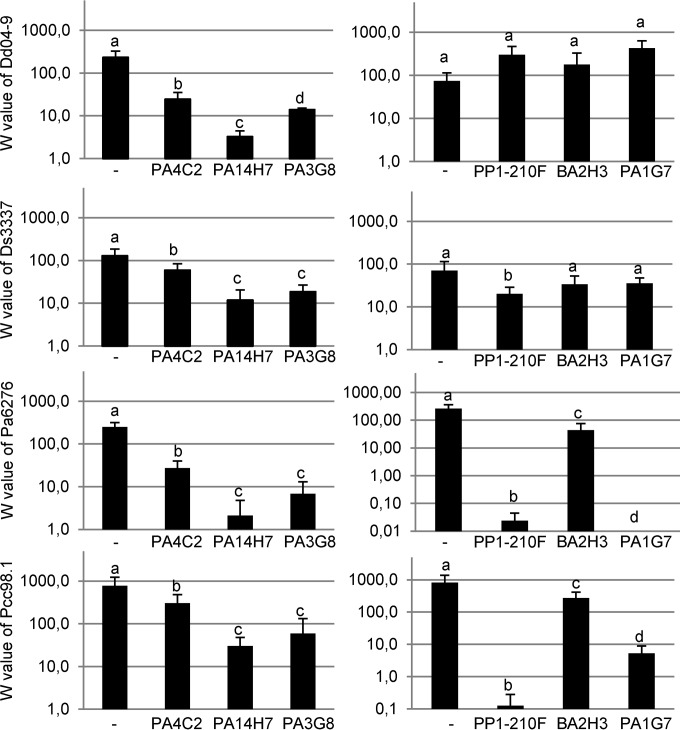
Liquid cultures were used to calculate the absolute fitness values for the four Dickeya and Pectobacterium phytopathogens in the absence or presence of each of the six bacterial isolates P. brassicacearum PP1-210F and PA1G7, P. fluorescens PA4C2 and PA3G8, P. putida PA14H7, and B. simplex BA2H3 (Fig. 2). The antagonistic bacteria P. fluorescens PA4C2 and especially P. fluorescens PA3G8 and P. putida PA14H7 strongly reduced the fitness of all the tested Dickeya and Pectobacterium pathogens in liquid coculture assays. In contrast, the antagonistic bacteria B. simplex BA2H3 and P. brassicacearum PA1G7 and PP1-210F affected the fitness of the Pectobacterium pathogens but exerted a weak or no effect on the Dickeya pathogens. Furthermore, we tested for the presence of antibacterial activity in the culture supernatants of these 6 strains. Each of the 100-fold-concentrated supernatants inhibited Dickeya and Pectobacterium strains as judged from a halo assay performed on TY agar plates.
FIG 2.
Pathogen fitness in the presence and absence of the growth-inhibiting strains The means of absolute fitness (W) of the pathogens (from the top, Dd04.9, Ds3337, Pa6276, and Pcc98.1) were calculated in the absence (−) or presence of the bacterial strains P. fluorescens PA4C2 and PA3G8 and P. putida PA14H7 (left panels) and P. brassicacearum PP1-210F and PA1G7 and B. simplex BA2H3 (right panels). Significantly different W values (by Kruskal-Wallis test then pairwise comparison test [α = 0.05]) are indicated by different letters.
MLSA characterization of the biocontrol agents.
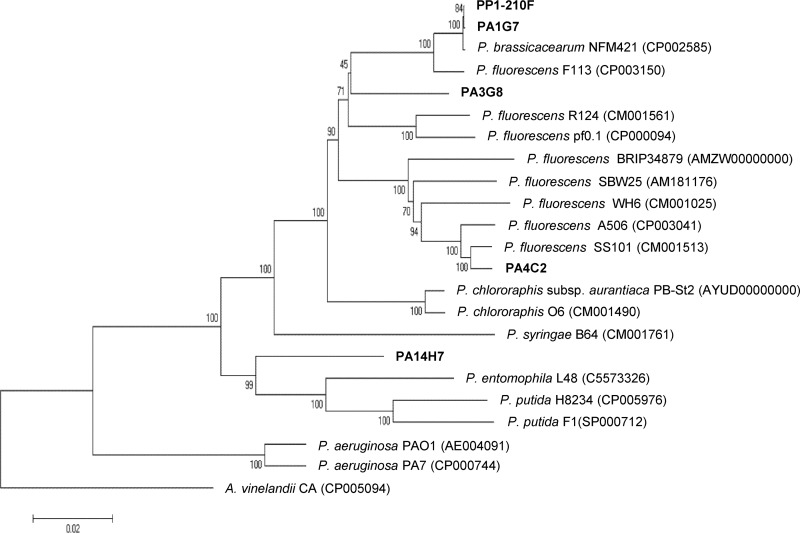
Draft genomes of the Pseudomonas strains PA3G8, PA14H7, PA4C2, PA1G7, and PP1-210F and Bacillus strain BA2H3 (66, 67) were used for MLSA characterization with four housekeeping genes, i.e., gyrB, recA, rpoB, and the 16S rRNA gene. The isolate BA2H3 is proposed to belong the B. simplex species, while PA14H7 belongs to P. putida, PA4C2 and PA3G8 to P. fluorescens, and PP1-210F and PA1G7 to P. brassicacearum (Fig. 3).
FIG 3.
MLSA-based relationship tree of the Pseudomonas strains. Concatenated gyrB, recA, rpoB, and 16S rRNA genes were used to construct a relationship tree of the Pseudomonas antagonistic strains (P. fluorescens PA3G8 and PA4C2, P. putida PA14H7, and P. brassicacearum PA1G7 and PP1-210F) and several well-identified strains of the Pseudomonas genus. An Azotobacter strain concatenated sequence was used to root the phylogenetic tree. The sequences were aligned with ClustalW, and then a neighbor-joining tree was created. The accession numbers of the NCBI sequences are indicated in parentheses.
Reduction of blackleg symptoms by the biocontrol agents P. fluorescens PA3G8 and PA4C2 and P. putida PA14H7.
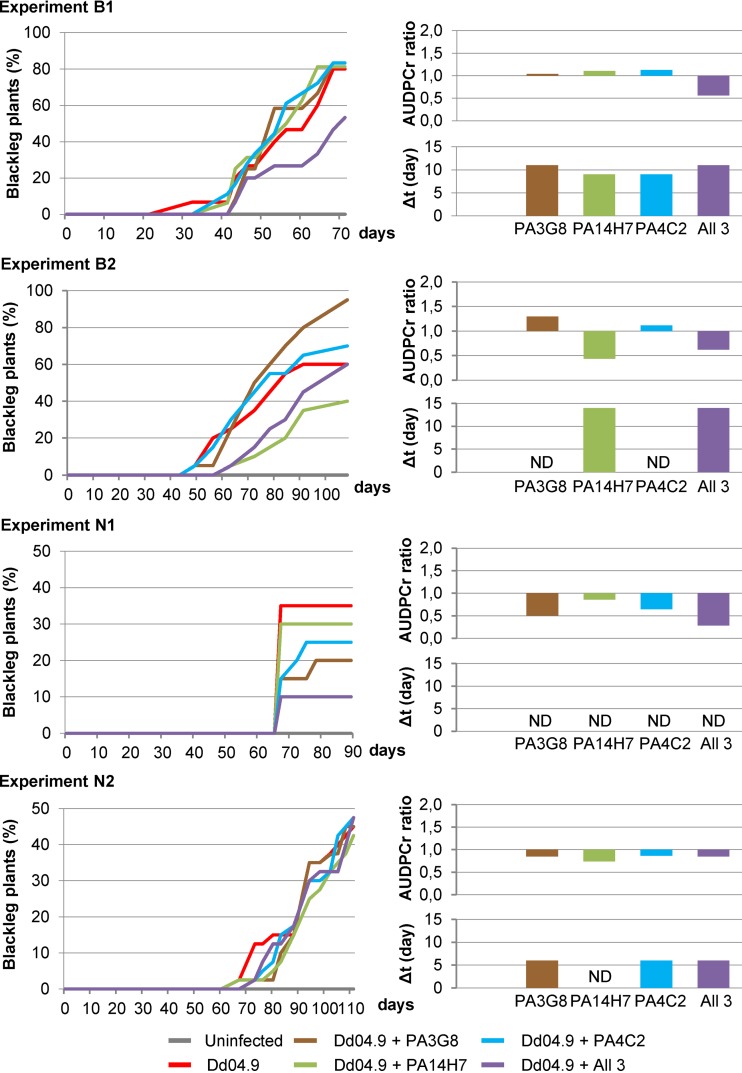
Over 3 years (2012 to 2014), four independent experiments (B1, B2, N1, and N2) were conducted in two greenhouses located in Brittany and Nord-Pas-de-Calais (Fig. 4). In all these experiments, no symptoms occurred under the control condition, i.e., when the plants were not infected by the pathogen (gray lines in the DPCs in Fig. 4). In contrast, D. dianthicola RNS04.9 induced symptoms in up to 80% of the plants in experiments B1 and B2 and in up to 45% in experiments N1 and N2 (red lines in the DPCs in Fig. 4). It should be noted that a reconstituted soil was used in the Brittany assays, while a native field soil mixed with sand was used in the Nord-Pas-de-Calais assays. In the presence of the biocontrol strains, variations in DPC were analyzed using the Δt and AUDPCr ratio.
FIG 4.
Blackleg symptom dynamics in greenhouse assays. Two independent assays (B1 and B2) were conducted in Brittany and two others (N1 and N2) in Nord-Pas-de-Calais. In the four assays, a set of 20 plants was used under each of the 6 compared conditions: uninfected plants and plants infected with D. dianthicola RNS04.9 alone (Dd04.9) and in combination with each of the biocontrol P. fluorescens strains PA3G8 and PA4C2 and P. putida strain PA14H7 and a mixture of them (All 3). Left panels, disease progression curves (DPCs); right panels, Δt values (in days) and AUDPCr ratios.
In the Brittany experiments B1 and B2, Levene's test indicated that AUDPCr ratios were not homogeneous in experiments conducted with P. putida strain PA14H7. Levene's test also revealed that the Δt values were also not homogeneous in experiments conducted with P. fluorescens strains PA3G8 and PA4C2. However, the values (<1) of the AUDPCr ratio indicated a slower kinetics of blackleg symptom appearance in the presence of a mixture of the 3 Pseudomonas biocontrol agents PA3G8, PA14H7, and PA4C2, although when inoculated alone, these three strains did not affect the blackleg symptom kinetics. In addition, the Δt values were consistently higher when P. putida PA14H7 was introduced alone and when all the three agents were simultaneously introduced.
In the Nord-Pas-de-Calais experiments N1 and N2, Levene's test indicated that AUDPCr ratios were homogeneous under all compared conditions. Moreover, all the AUDPCr ratios (<1) revealed a decrease in blackleg symptom kinetics. However, Levene's test indicated that none of the Δt values were homogeneous between the two experiments N1 and N2. The Δt values increased in experiment N2 only in experiments conducted with P. fluorescens strains PA3G8 and PA4C2 and the mixture of all three antagonistic strains, P. fluorescens PA3G8 and PA4C2 and P. putida PA14H7.
Limitation of dissemination of the pathogen D. dianthicola to tuber progeny.
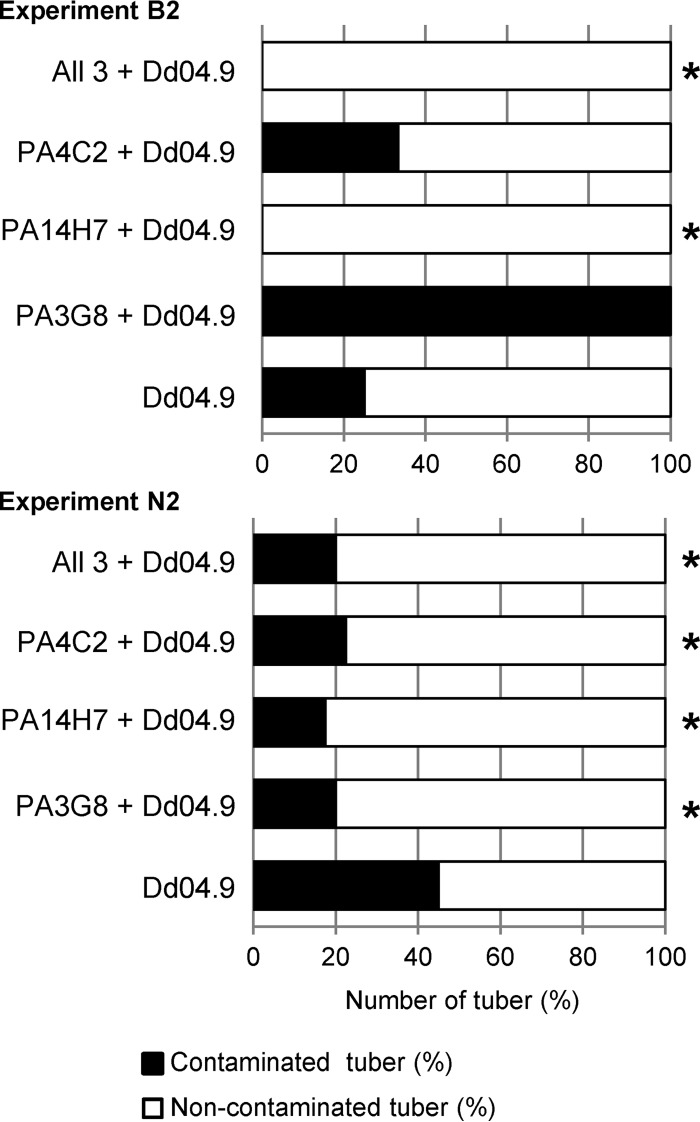
At the end of the B2 and N2 experiments, asymptomatic tubers of the asymptomatic plants were collected and screened for the presence of the pathogen D. dianthicola RNS04.9 by PCR (Fig. 5). In experiment N2, all the combinations of the biocontrol strains led to reduced pathogen propagation in the tuber progeny. In addition, when considering both experiments B2 and N2, a consistent reduction of pathogen propagation was observed when P. putida PA14H7 and the mixture of all three antagonistic strains were introduced. The results of treatment with P. fluorescens PA3G8 in experiment B2 did not significantly differ from those for the control condition due to a large difference of the number of analyzed symptomatic plants (1 versus 10, respectively), which affected the power of the Fisher test.
FIG 5.
Pathogen propagation in potato tubers. In experiments B2 in Brittany (upper graph) and N2 in Nord-Pas-de-Calais (lower graph), PCR detection of D. dianthicola RNS04.9 (Dd04.9) was performed on asymptomatic tubers harvested from asymptomatic plants which were cultivated in the presence and absence of the biocontrol strains (P. fluorescens PA3G8 and PA4C2, P. putida PA14H7, and all the three). Statistically significant decreases (Fisher's test; α = 0.05) of Dd04.9 presence are indicated by asterisks.
Dynamics of the pathogenic and biocontrol agents in greenhouse assays.
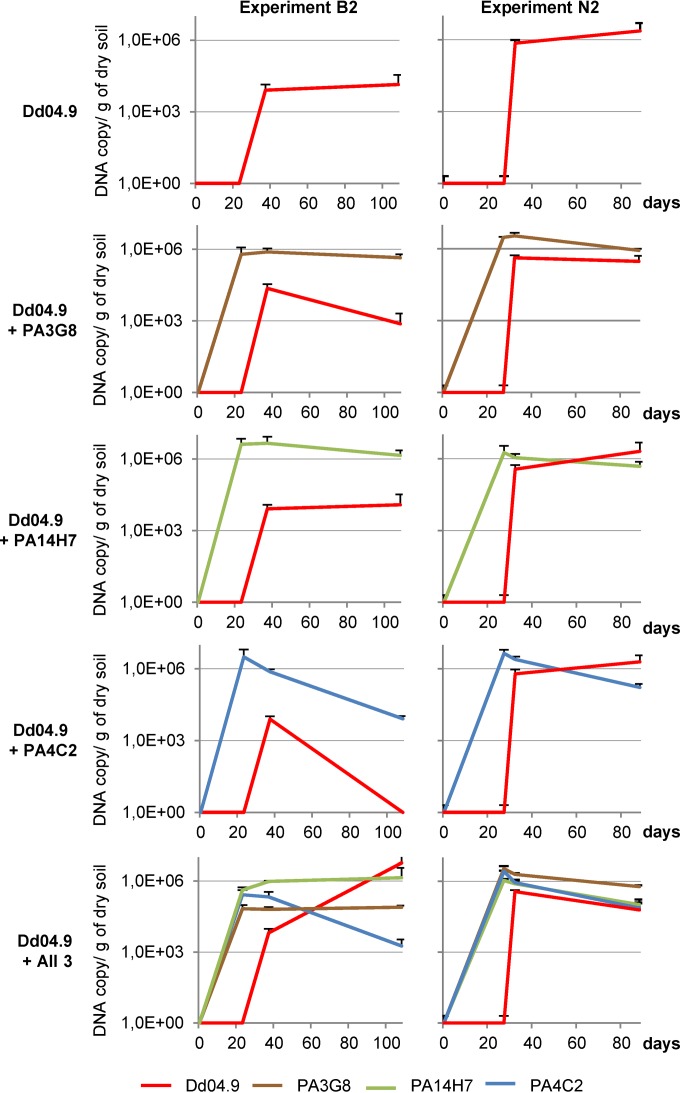
In the B2 and N2 experiments, antagonistic and pathogenic populations in the rhizosphere were quantified using strain-specific qPCR primers that targeted D. dianthicola RNS04.9 and each of the biocontrol strains. While the cell concentration of P. fluorescens strain PA4C2 (blue lines in Fig. 6) decreased by 1 and 2 orders of magnitude after its introduction, the other biocontrol strains P. fluorescens PA3G8 and P. putida PA14H7 were detected at a level which ranged between 105 and 106 DNA copies/g of dry soil (Fig. 6). Under all the tested conditions, the D. dianthicola RNS04.9 population (red lines in Fig. 6) was maintained at a level ranging between 104 and 106 DNA copies/g of dry soil. The presence of the biocontrol strains did not induce a consistent decrease of the pathogen concentration in the rhizosphere.
FIG 6.
Dynamics of the pathogenic and biocontrol populations. In experiments B2 (left graphs) and N2 (right graphs), the population level in soil was quantified by qPCR when D. dianthicola RNS04.9 was inoculated alone (D.d04-9), and in combination with each of the biocontrol P. fluorescens strains PA3G8 and PA4C2 and P. putida strain PA14H7 and a mixture of all of them (All 3).
DISCUSSION
The development of biocontrol tools aimed at D. dianthicola remains an important issue in plant protection, as it is the dominant Dickeya species recovered from plants with blackleg and soft rot symptoms in temperate climates in Europe (73, 74). Moreover, most of the published biocontrol assays were directed at the control of the P. atrosepticum, P. carotovorum subsp. carotovorum, and, to a lesser extent, Dickeya sp. pathogens (14, 28, 34, 36–38, 42, 45, 55, 75, 76). Here, we report the identification and characterization of Pseudomonas isolates which control the virulence of D. dianthicola on potato plants and tubers.
Two successive screenings allowed us to retain 58 isolates (mainly Pseudomonas), which represent 0.6% of the 10,000 tested isolates. This percentage is in line with the reported percentage (0.1 to 1%) of antibiotic-producing Pseudomonas spp. in natural environments (77). A first in vitro screening allowed the identification of 241 isolates (out of 10,000) which inhibited the growth of Dickeya sp. and/or Pectobacterium sp. pathogens. Among them, the 96 most efficient strains were subjected to a second screening which permitted us to discard the strains provoking damage on potato tubers and those inefficiently reducing symptoms on tubers after a 24-h cocultivation with the pathogens. Finally, among this collection of 58 isolates, only five Pseudomonas isolates (PA3G8, PA14H7, PA4C2, PA1G7, and PP1-210F) and one Bacillus strain (BA2H3) were retained because of their efficient in vitro activity against Pectobacterium and Dickeya strains. The culture supernatants of all these strains inhibited the growth of the pathogens in an in vitro halo assay. In contrast, when the fitness values were calculated in the cocultivation experiment, three isolates, i.e., B. simplex BA2H3 and P. brassicacearum PA1G7 and PP1-210F, preferentially antagonized the growth of the Pectobacterium strains but did not exhibit any effect against Dickeya growth. Moreover, the three others Pseudomonas strains, PA3G8, PA14H7, and PA4C2, antagonized the Dickeya and Pectobacterium strains. These observations suggested that these 6 strains could express different kinds of antibacterial activities targeting Dickeya and/or Pectobacterium pathogens. We recently published Illumina draft genome sequences of these 6 strains, which permitted the assembly of 30 to 90 contigs per genome (66, 67). RAST annotation allowed the identification of potential functions that could account for antibiosis activity of these strains. All the five Pseudomonas genomes exhibit genes encoding biosynthesis of the biofilm adhesin as well as the antibiotics bacteriocin and colicin V. P. fluorescens PA3G8 and P. putida PA14H7 could also produce the antibiotic or bacterial cytostatic compound paerucumarin, while B. simplex BA2H3 could produce bacitracin. Five of the strains contain siderophore operons, coding for achromobactin in P. fluorescens PA3G8 and P. brassicacearum PA1G7, achromobactin, enterobactin, and aerobactin in P. putida PA14H7, achromobactin and pyoverdine in P. brassicacearum PP1-210F, and anthrachelin in B. simplex BA2H3. Another relevant trait revealed by the analysis of the genomes is the resistance to antibiotics and toxic compounds. All these strains potentially could be resistant to fluoroquinolones and fosfomycin, and they could produce beta-lactamase. The Pseudomonas strains could also inhibit lysozyme activity and be resistant to streptothricin, and the Bacillus strain BA2H3 could be resistant to vancomycin. The involvement of these putative functions in biocontrol efficiency remains, however, to be investigated.
The biocontrol effects of P. putida PA14H7 and P. fluorescens PA4C2 and PA3G8 were investigated using in vitro and in planta assays: a growth inhibition assay in pure culture (fitness value calculation), a soft rot assay on potato tuber, blackleg development in a greenhouse experiment, and transmission to tuber progeny and population dynamics in the course of four independent greenhouse assays. In the greenhouse assays, a repeated reduction of the blackleg symptoms and of transmission to tuber progeny was observed when the three strains P. putida PA14H7 and P. fluorescens PA4C2 and PA3G8 were mixed. This observation suggested that they acted together to control the D. dianthicola-induced disease and pathogen transmission. Protection was also observed with P. putida PA14H7 alone in some but not all of the experiments. In our greenhouse experiments, the biocontrol agents were not able to compete with the pathogen in the rhizosphere, even though a decrease of the blackleg symptoms and transmission to daughter tubers were observed. This may be explained by some environmental conditions which may have limited the anti-Dickeya activities in the rhizosphere compartments (such as repression of a putative antibiosis, spatial compartmentation, nutrients under nonlimiting conditions, etc.). This observation could also suggest that plant protection is dependent of a tight contact between the plant and biocontrol agents. As a consequence, the biocontrol agents would directly act by preventing the penetration of the pathogens in the plant tissues. An alternative, but nonexclusive, hypothesis would be that the biocontrol agents would exert a plant defense-stimulating effect which limits the pathogen-induced symptoms.
Further work will aim at investigating potato protection in field assays, using either single-strain inoculation or a combination of the identified biocontrol agents. These biocontrol agents could be also evaluated in combination with the quorum-quenching Rhodococcus erythropolis (55–57) or with other proposed strategies using plant defense stimulation, predators, or competitors (28–34, 39-48). In regard to the practicality of using such findings, this strategy used only 3 treatments by the biocontrol agents. This frequency is relatively low compared to that for some other treatments current used in potato fields. Moreover, the high inoculum density which was used in this study may be easily obtained with Pseudomonas strains (>1010 CFU/ml in standard culture of Enterobacteriaceae). Some Pseudomonas strains are already commercialized as biocontrol agents.
To our knowledge, few experiments have been conducted in a greenhouse for the evaluation of putative biocontrol agents directed at D. solani (30, 45, 76). Most of the other assays that targeted the pectinolytic enterobacteria were performed on potato tubers only or on other plant hosts for the greenhouse assessments (78). This work is a first report of a significant reduction of D. dianthicola-induced blackleg disease symptoms and transmission of this pathogen to the tuber progeny. Previously, combinations between one chemical (chitosan or an antibiotic compound) and one biological agent (Pseudomonas, Trichoderma virdi, or Bacillus) were tested against P. carotovorum subsp. carotovorum during tuber storage (40, 48, 79). However, to our knowledge, no combination of bacterial biocontrol agents has yet been reported to protect potato against both blackleg and soft rot diseases. Clearly, our results demonstrated that a mix of several Pseudomonas agents increased the reproducibility and efficiency of the biocontrol. The combination of several biocontrol agents would reinforce reproducibility of the plant protection but also would potentially increase the range of the target pathogens.
ACKNOWLEDGMENTS
We thank Yvan Rahbe (INSA-Lyon), Brice Dupuis (Agroscope-Changins), and Sylvie Marhadour (RD3PT-FN3PT) for advice on the statistical analyses. Y.R.D.E. particularly thanks Yves Le Hingrat and Yves Dessaux for their review and correction of the manuscript. We are grateful to Sébastien Vast (CNPPPT), Plilippe Laty (CCS), and Philippe Dolo (BP) for providing the environmental samples from which antagonists were obtained.
REFERENCES
- 1.Pérombelon MCM. 1992. Potato blackleg: epidemiology, host-pathogen interaction and control. Netherlands J Plant Pathol 98:135–146. doi: 10.1007/BF01974480. [DOI] [Google Scholar]
- 2.Ma B, Hibbing ME, Kim H-S, Reedy RM, Yedidia I, Breuer J, Breuer J, Glasner JD, Perna NT, Kelman A, Charkowski AO. 2007. Host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya.. Phytopathology 97:1150–1163. doi: 10.1094/PHYTO-97-9-1150. [DOI] [PubMed] [Google Scholar]
- 3.Toth IK, van der Wolf JM, Saddler G, Lojkowska E, Hélias V, Pirhonen M, Tsror Lahkim L, Elphinstone JG. 2011. Dickeya species: an emerging problem for potato production in Europe. Plant Pathol 60:385–399. doi: 10.1111/j.1365-3059.2011.02427.x. [DOI] [Google Scholar]
- 4.Hauben L, Moore ER, Vauterin L, Steenackers M, Mergaert J, Verdonck L, Swings J. 1998. Phylogenetic position of phytopathogens within the Enterobacteriaceae.. Syst Appl Microbiol 21:384–397. doi: 10.1016/S0723-2020(98)80048-9. [DOI] [PubMed] [Google Scholar]
- 5.Gardan L. 2003. Elevation of three subspecies of Pectobacterium carotovorum to species level: Pectobacterium atrosepticum sp. nov., Pectobacterium betavasculorum sp. nov. and Pectobacterium wasabiae sp. nov. Int J Syst Evol Microbiol 53:381–391. doi: 10.1099/ijs.0.02423-0. [DOI] [PubMed] [Google Scholar]
- 6.Samson R. 2005. Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int J Syst Evol Microbiol 55:1415–1427. doi: 10.1099/ijs.0.02791-0. [DOI] [PubMed] [Google Scholar]
- 7.van der Wolf JM, De Boer HS. 2007. Potato biology and biotechnology, p 595–617. Elsevier, New York, NY. [Google Scholar]
- 8.van der Wolf JM, Nijhuis EH, Kowalewska MJ, Saddler GS, Parkinson N, Elphinstone JG, Pritchard L, Toth IK, Lojkowska E, Potrykus M, Waleron M, de Vos P, Cleenwerck I, Pirhonen M, Garlant L, Helias V, Pothier JF, Pfluger V, Duffy B, Tsror L, Manulis S. 2014. Dickeya solani sp. nov., a pectinolytic plant-pathogenic bacterium isolated from potato (Solanum tuberosum). Int J Syst Evol Microbiol 64:768–774. doi: 10.1099/ijs.0.052944-0. [DOI] [PubMed] [Google Scholar]
- 9.Duarte V, De Boer SH, Ward LJ, Oliveira AMR. 2004. Characterization of atypical Erwinia carotovora strains causing blackleg of potato in Brazil. J Appl Microbiol 96:535–545. doi: 10.1111/j.1365-2672.2004.02173.x. [DOI] [PubMed] [Google Scholar]
- 10.Pitman AR, Harrow SA, Visnovsky SB. 2010. Genetic characterisation of Pectobacterium wasabiae causing soft rot disease of potato in New Zealand. Eur J Plant Pathol 126:423–435. doi: 10.1007/s10658-009-9551-y. [DOI] [Google Scholar]
- 11.Perombelon MC, Kelman A. 1980. Ecology of the soft rot erwinias. Annu Rev Phytopathol 18:361–387. doi: 10.1146/annurev.py.18.090180.002045. [DOI] [Google Scholar]
- 12.Pérombelon MCM. 2002. Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathol 51:1–12. doi: 10.1046/j.0032-0862.2001.Shorttitle.doc.x. [DOI] [Google Scholar]
- 13.Perombelon MCM. 1972. The extent and survival of contamination of potato stocks in Scotland by Erwinia carotovora var. carotovora and E. carotovora var. atroseptica.. Ann Appl Biol 71:111–117. doi: 10.1111/j.1744-7348.1972.tb02945.x. [DOI] [Google Scholar]
- 14.Czajkowski R, Pérombelon MCM, van Veen JA, van der Wolf JM. 2011. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: a review: control of Dickeya and Pectobacterium species in potato. Plant Pathol 60:999–1013. doi: 10.1111/j.1365-3059.2011.02470.x. [DOI] [Google Scholar]
- 15.Czajkowski R, de Boer WJ, van Veen JA, van der Wolf JM. 2010. Downward vascular translocation of a green fluorescent protein-tagged strain of Dickeya sp. (biovar 3) from stem and leaf inoculation sites on potato. Phytopathology 100:1128–1137. doi: 10.1094/PHYTO-03-10-0093. [DOI] [PubMed] [Google Scholar]
- 16.Czajkowski R, de Boer WJ, Velvis H, van der Wolf JM. 2010. Systemic colonization of potato plants by a soilborne, green fluorescent protein-tagged strain of Dickeya sp. biovar 3. Phytopathology 100:134–142. doi: 10.1094/PHYTO-100-2-0134. [DOI] [PubMed] [Google Scholar]
- 17.Pérombelon MCM, Fox RA, Lowe R. 1979. Dispersion of Erwinia carotovora in aerosols produced by the pulverization of potato haulm prior to harvest. J Phytopathol 94:249–260. doi: 10.1111/j.1439-0434.1979.tb01557.x. [DOI] [Google Scholar]
- 18.Muniz CA, Jaillard D, Lemaitre B, Boccard F. 2007. Erwinia carotovora Evf antagonizes the elimination of bacteria in the gut of Drosophila larvae. Cell Microbiol 9:106–119. doi: 10.1111/j.1462-5822.2006.00771.x. [DOI] [PubMed] [Google Scholar]
- 19.Nykyri J, Fang X, Dorati F, Bakr R, Pasanen M, Niemi O, Palva ET, Jackson RW, Pirhonen M. 2014. Evidence that nematodes may vector the soft rot-causing enterobacterial phytopathogens. Plant Pathol 63:747–757. doi: 10.1111/ppa.12159. [DOI] [Google Scholar]
- 20.Moh AA, Massart S, Lahlali R, Jijakli MH, Lepoivre P. 2011. Predictive modelling of the combined effect of temperature and water activity on the in vitro growth of Erwinia spp. infecting potato tubers in Belgium. Biotechnol Agron Soc Environ 15:379–386. [Google Scholar]
- 21.Reverchon S, Nasser W. 2013. Dickeya ecology, environment sensing and regulation of virulence programme: Dickeya dadantii pathogenicity. Environ Microbiol Rep 5:622–636. doi: 10.1111/1758-2229.12073. [DOI] [PubMed] [Google Scholar]
- 22.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 6:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapwood DH, Read PJ, Spokes J. 1984. Methods for assessing the susceptibility of potato tubers of different cultivars to rotting by Erwinia carotovora subspecies atroseptica and carotovora.. Plant Pathol 33:13–20. doi: 10.1111/j.1365-3059.1984.tb00581.x. [DOI] [Google Scholar]
- 24.Reeves AF, Olanya OM, Hunter JH, Wells JM. 1999. Evaluation of potato varieties and selections for resistance to bacterial soft rot. Am J Potato Res 76:183–189. doi: 10.1007/BF02854220. [DOI] [Google Scholar]
- 25.Gill ED, Schaerer S, Dupuis B. 2014. Factors impacting blackleg development caused by Dickeya spp. in the field. Eur J Plant Pathol 140:317–327. doi: 10.1007/s10658-014-0465-y. [DOI] [Google Scholar]
- 26.Haas D, Défago G. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 27.Diallo S, Crépin A, Barbey C, Orange N, Burini J-F, Latour X. 2011. Mechanisms and recent advances in biological control mediated through the potato rhizosphere: biological control mediated through the potato rhizosphere. FEMS Microbiol Ecol 75:351–364. doi: 10.1111/j.1574-6941.2010.01023.x. [DOI] [PubMed] [Google Scholar]
- 28.Song G, Ryu S, Kim Y, Lee J, Choi J, Ryu C-M. 2013. Elicitation of induced resistance against Pectobacterium carotovorum and Pseudomonas syringae by specific individual compounds derived from native Korean plant species. Molecules 18:12877–12895. doi: 10.3390/molecules181012877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palva TK. 1994. Salicylic acid induced resistance to Erwinia carotovora subsp. carotovora in tobacco. Mol Plant Microbe Interact 7:356–363. doi: 10.1094/MPMI-7-0356. [DOI] [Google Scholar]
- 30.Czajkowski R, van der Wolf JM, Krolicka A, Ozymko Z, Narajczyk M, Kaczynska N, Lojkowska E. 2015. Salicylic acid can reduce infection symptoms caused by Dickeya solani in tissue culture grown potato (Solanum tuberosum L.) plants. Eur J Plant Pathol 141:545–558. doi: 10.1007/s10658-014-0561-z. [DOI] [Google Scholar]
- 31.Saubeau G, Gaillard F, Legentil L, Nugier-Chauvin C, Ferrières V, Andrivon D, Val F. 2014. Identification of three elicitins and a galactan-based complex polysaccharide from a concentrated culture filtrate of Phytophthora infestans efficient against Pectobacterium atrosepticum.. Molecules 19:15374–15390. doi: 10.3390/molecules191015374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luzzatto T, Golan A, Yishay M, Bilkis I, Ben-Ari J, Yedidia I. 2007. Priming of antimicrobial phenolics during induced resistance response towards Pectobacterium carotovorum in the ornamental monocot calla lily. J Agric Food Chem 55:10315–10322. doi: 10.1021/jf072037+. [DOI] [PubMed] [Google Scholar]
- 33.Kloepper JW, Ryu C-M, Zhang S. 2004. Induced systemic resistance and promotion of plant growth by Bacillus spp. Phytopathology 94:1259–1266. doi: 10.1094/PHYTO.2004.94.11.1259. [DOI] [PubMed] [Google Scholar]
- 34.Yang SY, Park MR, Kim IS, Kim YC, Yang JW, Ryu C-M. 2011. 2-Aminobenzoic acid of Bacillus sp. BS107 as an ISR determinant against Pectobacterium carotovorum subsp. carotovotrum SCC1 in tobacco. Eur J Plant Pathol 129:371–378. doi: 10.1007/s10658-010-9687-9. [DOI] [Google Scholar]
- 35.Rendulic S. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 36.Lim J-A. 2013. Biocontrol of Pectobacterium carotovorum subsp. carotovorum using bacteriophage PP1. J Microbiol Biotechnol 23:1147–1153. doi: 10.4014/jmb.1304.04001. [DOI] [PubMed] [Google Scholar]
- 37.Czajkowski R, Ozymko Z, Lojkowska E. 2014. Isolation and characterization of novel soilborne lytic bacteriophages infecting Dickeya spp. biovar 3 (“D. solani”). Plant Pathol 63:758–772. doi: 10.1111/ppa.12157. [DOI] [Google Scholar]
- 38.Adriaenssens EM, Van Vaerenbergh J, Vandenheuvel D, Dunon V, Ceyssens P-J, De Proft M, Kropinski AM, Noben J-P, Maes M, Lavigne R. 2012. T4-related bacteriophage LIMEstone isolates for the control of soft rot on potato caused by “Dickeya solani.” PLoS One 7:e33227. doi: 10.1371/journal.pone.0033227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trias R, Bañeras L, Montesinos E, Badosa E. 2010. Lactic acid bacteria from fresh fruit and vegetables as biocontrol agents of phytopathogenic bacteria and fungi. Int Microbiol 11:231–236. [DOI] [PubMed] [Google Scholar]
- 40.Rahman MM, Ali ME, Khan AA, Akanda AM, Uddin MK, Hashim U, Abd Hamid SB. 2012. Isolation, characterization, and identification of biological control agent for potato soft rot in Bangladesh. Sci World J 2012:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shrestha A, Kim EC, Lim CK, Cho S, Hur JH, Park DH. 2009. Biological control of soft rot on Chinese cabbage using beneficial bacterial agents in greenhouse and field. Korean J Pesticide Sci 13:325–331. [Google Scholar]
- 42.Kloepper JW. 1983. Effect of seed piece inoculation with plant growth-promoting rhizobacteria on populations of Erwinia carotovora on potato roots and in daughter tubers. Phytopathology 73:217. doi: 10.1094/Phyto-73-217. [DOI] [Google Scholar]
- 43.Cronin D, Moënne-Loccoz Y, Fenton A, Dunne C, Dowling DN, O'Gara F. 2006. Ecological interaction of a biocontrol Pseudomonas fluorescens strain producing 2,4-diacetylphloroglucinol with the soft rot potato pathogen Erwinia carotovora subsp. atroseptica.. FEMS Microbiol Ecol 23:95–106. doi: 10.1111/j.1574-6941.1997.tb00394.x. [DOI] [Google Scholar]
- 44.Sharga BM, Lyon GD. 1998. Bacillus subtilis BS 107 as an antagonist of potato blackleg and soft rot bacteria. Can J Microbiol 44:777–783. [PubMed] [Google Scholar]
- 45.Czajkowski R, de Boer WJ, van Veen JA, van der Wolf JM. 2012. Characterization of bacterial isolates from rotting potato tuber tissue showing antagonism to Dickeya sp. biovar 3 in vitro and in planta. Plant Pathol 61:169–182. doi: 10.1111/j.1365-3059.2011.02486.x. [DOI] [Google Scholar]
- 46.Wood EM, Miles TD, Wharton PS. 2013. The use of natural plant volatile compounds for the control of the potato postharvest diseases, black dot, silver scurf and soft rot. Biol Control 64:152–159. doi: 10.1016/j.biocontrol.2012.10.014. [DOI] [Google Scholar]
- 47.Helander IM, Nurmiaho-Lassila E-L, Ahvenainen R, Rhoades J, Roller S. 2001. Chitosan disrupts the barrier properties of the outer membrane of Gram-negative bacteria. Int J Food Microbiol 71:235–244. doi: 10.1016/S0168-1605(01)00609-2. [DOI] [PubMed] [Google Scholar]
- 48.Makhlouf AH, Abdeen R. 2014. Investigation on the effect of chemical and biological control of bacterial soft root disease of potato in storage. J Biol Agric Healthcare 4:31–44. [Google Scholar]
- 49.Faure D, Dessaux Y. 2007. Quorum sensing as a target for developing control strategies for the plant pathogen Pectobacterium.. Eur J Plant Pathol 119:353–365. doi: 10.1007/s10658-007-9149-1. [DOI] [Google Scholar]
- 50.Crépin A, Beury-Cirou A, Barbey C, Farmer C, Hélias V, Burini J-F, Faure D, Latour X. 2012. N-Acyl homoserine lactones in diverse Pectobacterium and Dickeya plant pathogens: diversity, abundance, and involvement in virulence. Sensors 12:3484–3497. doi: 10.3390/s120303484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Crépin A, Barbey C, Beury-Cirou A, Hélias V, Taupin L, Reverchon S, Nasser W, Faure D, Dufour A, Orange N, Feuilloley M, Heurlier K, Burini J-F, Latour X. 2012. Quorum sensing signaling molecules produced by reference and emerging soft-rot bacteria (Dickeya and Pectobacterium spp.). PLoS One 7:e35176. doi: 10.1371/journal.pone.0035176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uroz S. 2003. Novel bacteria degrading N-acylhomoserine lactones and their use as quenchers of quorum-sensing-regulated functions of plant-pathogenic bacteria. Microbiology 149:1981–1989. doi: 10.1099/mic.0.26375-0. [DOI] [PubMed] [Google Scholar]
- 53.Dong Y-H, Zhang X-F, Xu J-L, Zhang L-H. 2004. Insecticidal Bacillus thuringiensis silences Erwinia carotovora virulence by a new form of microbial antagonism, signal interference. Appl Environ Microbiol 70:954–960. doi: 10.1128/AEM.70.2.954-960.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jafra S, Przysowa J, Czajkowski R, Michta A, Garbeva P, Van der Wolf JM. 2006. Detection and characterization of bacteria from the potato rhizosphere degrading N-acyl-homoserine lactone. Can J Microbiol 52:1006–1015. doi: 10.1139/w06-062. [DOI] [PubMed] [Google Scholar]
- 55.Cirou A, Diallo S, Kurt C, Latour X, Faure D. 2007. Growth promotion of quorum-quenching bacteria in the rhizosphere of Solanum tuberosum.. Environ Microbiol 9:1511–1522. doi: 10.1111/j.1462-2920.2007.01270.x. [DOI] [PubMed] [Google Scholar]
- 56.Cirou A, Raffoux A, Diallo S, Latour X, Dessaux Y, Faure D. 2011. Gamma-caprolactone stimulates growth of quorum-quenching Rhodococcus populations in a large-scale hydroponic system for culturing Solanum tuberosum.. Res Microbiol 162:945–950. doi: 10.1016/j.resmic.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 57.Cirou A, Mondy S, An S, Charrier A, Sarrazin A, Thoison O, DuBow M, Faure D. 2012. Efficient biostimulation of native and introduced quorum-quenching Rhodococcus erythropolis populations is revealed by a combination of analytical chemistry, microbiology, and pyrosequencing. Appl Environ Microbiol 78:481–492. doi: 10.1128/AEM.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palmer AG, Streng E, Blackwell HE. 2011. Attenuation of virulence in pathogenic bacteria using synthetic quorum-sensing modulators under native conditions on plant hosts. ACS Chem Biol 6:1348–1356. doi: 10.1021/cb200298g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmer AG, Streng E, Jewell KA, Blackwell HE. 2011. Quorum sensing in bacterial species that use degenerate autoinducers can be tuned by using structurally identical non-native ligands. Chembiochem 12:138–147. doi: 10.1002/cbic.201000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manefield M, Welch M, Givskov M, Salmond GPC, Kjelleberg S. 2001. Halogenated furanones from the red alga, Delisea pulchra, inhibit carbapenem antibiotic synthesis and exoenzyme virulence factor production in the phytopathogen Erwinia carotovora.. FEMS Microbiol Lett 205:131–138. doi: 10.1111/j.1574-6968.2001.tb10936.x. [DOI] [PubMed] [Google Scholar]
- 61.Des Essarts YR, Sabbah M, Comte A, Soule're L, Queneau Y, Dessaux Y, Hélias V, Faure D. 2013. N,N′-Alkylated imidazolium-derivatives act as quorum-sensing inhibitors targeting the Pectobacterium atrosepticum-induced symptoms on potato tubers. Int J Mol Sci 14:19976–19986. doi: 10.3390/ijms141019976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khayi S, Mondy S, Beury-Cirou A, Moumni M, Helias V, Faure D. 2014. Genome sequence of the emerging plant pathogen Dickeya solani strain RNS 08.23.3.1A. Genome Announc 2:e01270-13. doi: 10.1128/genomeA.01270-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hélias V, Hamon P, Huchet E, Wolf JVD, Andrivon D. 2012. Two new effective semiselective crystal violet pectate media for isolation of Pectobacterium and Dickeya: isolating pectolytic bacteria on CVP. Plant Pathol 61:339–345. doi: 10.1111/j.1365-3059.2011.02508.x. [DOI] [Google Scholar]
- 64.Kwasiborski A, Mondy S, Beury-Cirou A, Faure D. 2013. Genome sequence of the Pectobacterium atrosepticum strain CFBP6276, causing blackleg and soft rot diseases on potato plants and tubers. Genome Announc 1:e00374-13. doi: 10.1128/genomeA.00374-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17:7843–7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khayi S, Raoul des Essarts Y, Mondy S, Moumni M, Hélias V, Beury-Cirou A, Faure D. 2015. Draft genome sequences of the three Pectobacterium-antagonistic bacteria Pseudomonas brassicacearum PP1-210F and PA1G7 and Bacillus simplex BA2H3. Genome Announc 3:e01497-14. doi: 10.1128/genomeA.01497-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cigna J, Raoul des Essarts Y, Mondy S, Hélias V, Beury-Cirou A, Faure D. 2015. Draft genome sequences of Pseudomonas fluorescens strains PA4C2 and PA3G8 and Pseudomonas putida PA14H7, three biocontrol bacteria against Dickeya phytopathogens. Genome Announc 3:e01503-14. doi: 10.1128/genomeA.01503-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andrivon D, Pellé R, Ellissèche D. 2006. Assessing resistance types and levels to epidemic diseases from the analysis of disease progress curves: principles and application to potato late blight. Am J Potato Res 83:455–461. doi: 10.1007/BF02883506. [DOI] [Google Scholar]
- 70.Rouffiange J, Gerardin D, Kellenberger I, Schaerer S, Dupuis B. 2013. Sensibilité de la pomme de terre aux pourritures de tiges provoquées par Dickeya spp. Rech Agron Suisse 4:424–431. [Google Scholar]
- 71.Nassar A, Darrasse A, Lemattre M, Kotoujansky A, Dervin C, Vedel R, Bertheau Y. 1996. Characterization of Erwinia chrysanthemi by pectinolytic isozyme polymorphism and restriction fragment length polymorphism analysis of PCR-amplified fragments of pel genes. Appl Environ Microbiol 62:2228–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.des Essarts YR, Mondy S, Hélias V, Faure D. 2015. Genome sequence of the potato plant pathogen Dickeya dianthicola strain RNS04.9. Genome Announc 3:e00581-15. doi: 10.1128/genomeA.00581-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lumb VM, Perombelon MCM, Zutra D. 1986. Studies of a wilt disease of the potato plant in Israel caused by Erwinia chrysanthemi.. Plant Pathol 35:196–202. doi: 10.1111/j.1365-3059.1986.tb02004.x. [DOI] [Google Scholar]
- 74.Laurila J, Hannukkala A, Nykyri J, Pasanen M, Hélias V, Garlant L, Pirhonen M. 2010. Symptoms and yield reduction caused by Dickeya spp. strains isolated from potato and river water in Finland. Eur J Plant Pathol 126:249–262. doi: 10.1007/s10658-009-9537-9. [DOI] [Google Scholar]
- 75.Kastelein P, Schepel EG, Mulder A, Turkensteen LJ, Van Vuurde JWL. 1999. Preliminary selection of antagonists of Erwinia carotovora subsp. atroseptica (Van Hall) Dye for application during green crop lifting of seed potato tubers. Potato Res 42:161–171. doi: 10.1007/BF02358406. [DOI] [Google Scholar]
- 76.Czajkowski R, de Boer WJ, van Veen JA, van der Wolf JM. 2012. Studies on the interaction between the biocontrol agent, Serratia plymuthica A30, and blackleg-causing Dickeya sp. (biovar 3) in potato (Solanum tuberosum). Plant Pathol 61:677–688. doi: 10.1111/j.1365-3059.2011.02565.x. [DOI] [Google Scholar]
- 77.Raaijmakers JM, Vlami M, De Souza JT. 2002. Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek 81:537–547. doi: 10.1023/A:1020501420831. [DOI] [PubMed] [Google Scholar]
- 78.Faquihi H, Mhand RA, Ennaji M, Benbouaza A, Achbani E. 2012. Aureobasidium pullulans (De Bary) G. Arnaud, a biological control against soft rot disease in potato caused by Pectobacterium carotovorum.. Int J Sci Res 3:1779–1786. [Google Scholar]
- 79.Abd-El-Khair H, Karima HEH. 2007. Application of some bactericides and bioagents for controlling the soft rot disease in potato. Res J Agric Biol Sci 3:463–473. [Google Scholar]