Abstract
Clostridium sporogenes PA 3679 is a nonpathogenic, nontoxic model organism for proteolytic Clostridium botulinum used in the validation of conventional thermal food processes due to its ability to produce highly heat-resistant endospores. Because of its public safety importance, the uncertain taxonomic classification and genetic diversity of PA 3679 are concerns. Therefore, isolates of C. sporogenes PA 3679 were obtained from various sources and characterized using pulsed-field gel electrophoresis (PFGE) and whole-genome sequencing. The phylogenetic relatedness and genetic variability were assessed based on 16S rRNA gene sequencing and whole-genome single nucleotide polymorphism (SNP) analysis. All C. sporogenes PA 3679 isolates were categorized into two clades (clade I containing ATCC 7955 NCA3679 isolates 1961-2, 1990, and 2007 and clade II containing PA 3679 isolates NFL, UW, FDA, and Campbell and ATCC 7955 NCA3679 isolate 1961-4). The 16S maximum likelihood (ML) tree clustered both clades within proteolytic C. botulinum strains, with clade I forming a distinct cluster with other C. sporogenes non-PA 3679 strains. SNP analysis revealed that clade I isolates were more similar to the genomic reference PA 3679 (NCTC8594) genome (GenBank accession number AGAH00000000.1) than clade II isolates were. The genomic reference C. sporogenes PA 3679 (NCTC8594) genome and clade I C. sporogenes isolates were genetically distinct from those obtained from other sources (University of Wisconsin, National Food Laboratory, U.S. Food and Drug Administration, and Campbell's Soup Company). Thermal destruction studies revealed that clade I isolates were more sensitive to high temperature than clade II isolates were. Considering the widespread use of C. sporogenes PA 3679 and its genetic information in numerous studies, the accurate identification and genetic characterization of C. sporogenes PA 3679 are of critical importance.
INTRODUCTION
Food-borne botulism is a neuroparalytic disease that results from the ingestion of botulinum neurotoxin (BoNT) produced by clostridia, including Clostridium botulinum and rare strains of Clostridium baratii and Clostridium butyricum. C. botulinum is a heterogeneous species comprised of four distinct groups (groups I to IV) of strains that differ based on genetic and phenotypic properties (1). Group I (proteolytic) C. botulinum strains can also be uniquely differentiated from the other groups based on their ability to form highly heat-resistant endospores. Spores of this organism are ubiquitously found in the environment and are generally presumed to be a natural contaminant of foods that come in contact with the soil (2). The geographical distribution of C. botulinum varies by serotype. For example, in the United States, spores of C. botulinum strains producing type B toxin are typically found in the soils east of the Mississippi River, and type A strains are found in the western region of the United States. Serotype E strains are commonly found in the soil sediments of the Great Lakes regions and in Alaska (2, 3).
Spore germination and outgrowth of group I C. botulinum can result in media and foods that have a pH greater than 4.6 (4–6). As a result, commercial processing technologies are designed to deliver a process that eliminates the threat of toxin production by group I C. botulinum spores, the target pathogen, in shelf-stable low-acid canned food products (7). However, production of C. botulinum spore crops required to validate these processes is exceedingly challenging due to potentially low spore yields and variable resistance to high temperature between batches, thus highlighting the need for a nontoxigenic surrogate that produces spores of a consistent yield and that are highly heat resistant.
Clostridium sporogenes PA 3679 is a nonpathogenic, nontoxic, Gram-positive putrefactive anaerobe that produces spores that are more heat resistant than spores of group I C. botulinum (8). Therefore, PA 3679 was widely adopted and used as the nontoxigenic model organism for proteolytic group I C. botulinum for validating thermal processing technologies for low-acid canned foods. PA 3679 was first isolated in 1927 from spoiled canned corn by E. J. Cameron at the National Canner's Association (NCA) (9), and the origin and identity of PA 3679 have been thoroughly reviewed by Brown et al. (10). Although PA 3679 is classified as a strain of C. sporogenes, its true identity remains uncertain. On the basis of serological testing, McClung (8) argued that PA 3679 was not a nontoxic C. botulinum strain but an undescribed species. Gross et al. (11) reported a private communication from Cameron that although PA 3679 resembled C. sporogenes, it is likely not identical to that species. That study also showed that PA 3679 was serologically identical to two other putrefactive anaerobes, S2 and 9x, isolated from spoiled canned meats; however, all three strains were culturally and serologically distinct from C. sporogenes strain Spray (11). The nontoxigenic counterpart for group I C. botulinum is considered to be C. sporogenes (12, 13). Studies investigating the genetic relationship of C. sporogenes and C. botulinum that employed molecular techniques such as multiple-locus variable-number tandem-repeat analysis (MLVA) (14, 15), DNA microarrays and analysis of the flagellar glycosylation island (FGI) (16), DNA-rRNA hybridization (17), 16S rRNA gene sequencing (18, 19), and multilocus sequence typing (MLST) (20, 21) have been reported and included other strains of C. sporogenes, but not PA 3679. A DNA-DNA hybridization study by Lee and Riemann (22) revealed that PA 3679 was 100% homologous to C. botulinum strain 62A, yet in a similar study, PA 3679 shared only 83% homology to C. botulinum strain A190 and 68% homology with C. sporogenes strain J-53 (23). Most recently, in a study by Weigand et al. (24), whole-genome sequencing (WGS) was employed to show that strains of C. sporogenes and group I C. botulinum were distinctly grouped into two clades based on 2,016 core orthologues shared between these organisms. The PA 3679 strain included in that study was C. sporogenes PA 3679 (NCTC8594) that was sequenced by Bradbury et al. (25).
Since 1927, PA 3679 has been globally distributed and studied in numerous laboratories; it was also deposited into the American Type Culture Collection (ATCC) in 1941 or earlier (10) and is listed as ATCC 7955. Additionally, the draft genome sequence of Clostridium sporogenes PA 3679 (National Collection of Type Cultures [NCTC] 8594) has been completed and is available through GenBank (accession number AGAH00000000.1) (25). According to their analysis, the 16S rRNA gene sequence of NCTC8594 shared 99 to 100% nucleotide similarity with that of group I C. botulinum and C. sporogenes strains, and MLST using seven group I C. botulinum housekeeping genes showed that NCTC8594 clustered with C. botulinum type A strain A207 (25). In this study, isolates of PA 3679 were collected from five different sources, where they were maintained by various laboratory culturing techniques over the past 88 years. Pulsed-field gel electrophoresis (PFGE) and whole-genome sequencing were used to examine the genetic diversity of these isolates. A whole-genome single nucleotide polymorphism (SNP)-based approach to assess the relatedness among the isolates and with C. sporogenes PA 3679 (NCTC8594) and C. sporogenes ATCC 15579 (GenBank accession number NZ_ABKW00000000.2) has been performed.
MATERIALS AND METHODS
Bacterial strains.
Spore crops of C. sporogenes PA 3679 were obtained from five different sources, and the details are presented in Table 1. Three distinct freeze-dried spore crop lots of ATCC 7955 were received from the American Type Culture Collection (ATCC) and labeled for the year the crop was preserved (1961, 1990, and 2007). ATCC 15579 was also purchased from the ATCC and included in this study because it is a non-PA 3679 C. sporogenes strain and the nucleotide sequence is available in GenBank (accession number NZ_ABKW00000000.2). The freeze-dried vials received from ATCC were opened aseptically according to the instructions provided by ATCC, inoculated into sterile TPGY (Trypticase-peptone-glucose-yeast extract) broth and incubated anaerobically at 37°C. The Campbell isolate was received from the Campbell's Soup Company as a liquid culture containing ∼108 spores/ml, harvested in December 2003. The NFL isolate was received as a liquid spore culture from the National Food Laboratory (NFL). The UW isolate was received from the Eric A. Johnson laboratory at the University of Wisconsin—Madison as a 72-h TPGY broth culture revived from a freeze-dried sample from 13 June 1972 labeled “PA 3679-66.” The FDA isolate was obtained from the U.S. Food and Drug Administration (FDA) culture collection (Bedford Park, IL) as a spore crop prepared in 2006 and suspended in water. All cultures from the various sources were inoculated (1:100) into 10 ml of TPGY broth and incubated anaerobically at 37°C for 24 to 48 h. Each culture was subsequently streaked for isolation onto egg yolk agar (EYA) and incubated anaerobically at 37°C for 24 to 48 h to assess the purity of the cultures by examining colony morphology.
TABLE 1.
Clostridium sporogenes isolates used in this study
| Strain | Spore crop production date | Source or reference |
|---|---|---|
| 1961a | 1961 | American Type Culture Collection (ATCC) |
| ATCC 7955 NCA3679 1961-2b | This study | |
| ATCC 7955 NCA3679 1961-4b | This study | |
| ATCC 7955 NCA3679 1990a | 1990 | American Type Culture Collection (ATCC) |
| ATCC 7955 NCA3679 2007a | 2007 | American Type Culture Collection (ATCC) |
| Campbell | 2003 | Campbell's Soup Company |
| UWc | 1972 | University of Wisconsin—Madison |
| FDA | 2006 | U.S. Food and Drug Administration |
| NFL | 2012 | National Food Laboratory |
| ATCC 15579 | American Type Culture Collection |
ATCC 7955 NCA3679.
1961-2 and 1961-4 were isolated from the 1961 spore crop of ATCC 7955 NCA3679.
The UW isolate was a freeze-dried spore preparation (obtained from the Eric A. Johnson laboratory) that was rehydrated and cultured in TPGY broth.
Spore crop production.
Spore crops were cultivated by inoculating 1 to 2 liters of TPGY broth (1:100) with an 18- to 24-h culture of each strain. The TPGY broth was incubated at 37°C anaerobically for up to 28 days. Spores were harvested by centrifuging the culture at 10,000 × g for 15 min at 4°C using a Sorvall Evolution RC (Thermo Scientific, Waltham, MA) centrifuge fitted with an SLA-3000 rotor. The spore pellet was washed four times with sterile deionized water. To eliminate vegetative cells, the spore samples were sonicated using a Vibracell VC-750 sonicator (Sonics and Materials, Inc., Newton, CT) at 50% amplitude for 20 min in an ice-water bath. The sonicated spore suspensions were centrifuged at 10,000 × g for 15 min at 4°C and washed six times in deionized water. The final spore pellet was resuspended in 10 ml of sterile deionized water. Spore crops were heat shocked at 80°C for 10 min and enumerated by pour plating using peptone-yeast extract-glucose-starch (PYGS) agar medium without glucose (26). The plates were incubated anaerobically at 37°C for 5 days, and then spore-forming units (SFU)/ml were counted.
Heat resistance studies.
The thermal resistance of selected spore crops was evaluated at various temperatures (94 to 121°C). Spore crops were diluted to approximately 105 to 106 spores/ml in phosphate buffer (0.067 M, pH 7.0), and 1-ml quantities of diluted spores were dispensed into duplicate sterile nuclear magnetic resonance (NMR) tubes (Kimble Chase, Portsmouth, NH). The NMR tubes were heat sealed and completely submerged in a calibrated, heated Fluke high-precision bath (model 7103; Fluke, Everett, WA) with silicone oil as the heat transfer fluid. The spores were processed at 94°C, 97°C, 100°C, 103°C, 105°C, 108°C, 111°C, 114°C, 117°C, and 121°C for 5 min. The tubes were immediately placed in ice water to stop the heat treatment. The spore suspensions were removed from the tubes using a sterile blunt needle syringe, and the contents were transferred to duplicate 10-ml TPGY broth tubes. The samples were incubated anaerobically at 37°C for 5 days and visually examined for growth as exhibited by turbidity.
Isolates ATCC 7955 NCA3679 1961-2 and 2007, as well as isolate FDA, were selected for additional thermal studies to determine the D-values (the time required to reduce a population by 90%) at the highest temperatures at which survivors were obtained for each particular isolate (i.e., D97°C [the D-value at 97°C, which was the highest temperature at which survivors were found] for isolate 1961-2; D100°C for isolate 2007; D117°C for isolate FDA). The spore crops of the C. sporogenes isolates 1961-2, 2007, and FDA were diluted to 106 to 107 spores/ml in phosphate buffer (0.067 M, pH 7.0) and placed into sterile NMR tubes (Kimble Chase). The NMR tubes containing spores of selected isolates were processed at 97°C ± 0.1°C for 2, 4, 6, 8, 10, 12, and 14 min (isolate 1961-2), 100°C ± 0.1°C for 2, 3, 4, 5, 6, 8, and 10 min (isolate 2007), or 117°C ± 0.1°C for 2, 4, 5, 6, and 8 min (isolate FDA). After each time interval, NMR tubes containing spores were immediately placed in ice water to halt further heating of the samples. The spore suspensions were removed from the tubes as described above, serially diluted in 0.1 M Butterfield's phosphate buffer (BPB) (pH 7.2), and pour plated using PYGS agar. The plates were incubated anaerobically for 5 to 7 days at 37°C prior to enumeration. Survivors were enumerated as log CFU per milliliter. The D-values of each spore crop isolate were calculated from the negative slope of the regression lines using the linear portions of the survivor curves. At least six replicates of each experiment were conducted.
PFGE.
Isolated colonies of each isolate were selected and inoculated into fresh 10 ml of TPGY broth and incubated anaerobically at 37°C until an optical density at 600 nm (OD600) of 0.6 was reached. Pulsed-field gel electrophoresis (PFGE) plug samples were prepared using 1.5-ml culture aliquots, and restriction digestion of the DNA samples embedded in agarose plugs was performed using restriction endonucleases XhoI (at 37°C) and SmaI (at 25°C) as previously described (26). The sample plugs were rinsed with 0.5× Tris-borate-EDTA (TBE) (45 mM Tris-borate, 1 mM EDTA [pH 8.0]), loaded onto a 1% agarose gel, and electrophoresed in 0.5× TBE containing thiourea (4.5 μg/ml) using a clamped homogenous electric field system (CHEF Mapper; Bio-Rad, Hercules, CA) with the following parameters: initial switch time of 0.5 s, final switch time of 40 s, voltage of 6 V/cm at 14°C for 22 h. The gels were stained with ethidium bromide (1 μg/ml) for 20 min, destained in ultrapure water for 1 h, and visualized using a GelDocXRS+ camera. The restriction banding patterns generated using XhoI and SmaI were analyzed using the BioNumerics software, version 6.5 (Applied Maths, Sint-Martens-Latem, Belgium), according to the standardized PulseNet protocol (27, 28). Similarity between the restriction banding patterns was determined using the Dice coefficient correlation. The dendrograms were constructed using the average of the composite data sets generated by SmaI and XhoI and the unweighted pair group method using average linkages (UPGMA) using the average UPGMA linkage clustering tool. The optimization value and position tolerance were 1.5%.
Genomic DNA extraction.
C. sporogenes isolates were inoculated (1/100) into 10 ml of TPGY broth and incubated anaerobically overnight at 37°C. From the 18-h cultures, 3.0 ml was centrifuged using an Eppendorf microcentrifuge 5415R (Eppendorf, Hauppauge, NY) at 10,000 × g for 10 min to pellet the bacteria. The cell pellet was washed twice with TES (50 mM Tris-HCl, 25 mM EDTA [pH 8.0], 6.7% sucrose) buffer and stored at −80°C prior to DNA extraction using the MagAttract high-molecular-weight DNA kit for Gram-positive bacteria (Qiagen, Valencia, CA) according to the manufacturer's instructions, with some modifications. The cell pellets were thawed in a 37°C water bath and resuspended in 300 μl of buffer P1 (50 mM Tris, 10 mM EDTA [pH 8.0]). Lysozyme (100 mg/ml) was prepared in water, and 40 μl of the lysozyme solution was added to the cell suspension. The cells were incubated at 37°C for 2 h using a ThermoMixer C (Eppendorf) set at 900 rpm. After cell lysis, 40 μl of proteinase K, as supplied, was added and incubated at 56°C and 900 rpm for 1 h. RNase A (2 mg/ml) was added to the cell suspension, which was subsequently incubated at 25°C for 2 min. Next, 300 μl of the kit buffer AL was added, followed by 30 μl of the MagAttract suspension G and 560 μl of buffer MB. The samples were mixed by pulse vortexing and incubated at 25°C at 1,400 rpm for 3 min. The remaining steps of the DNA extraction were followed according to the manufacturer's instructions. The DNA was eluted in 50 μl of ultrapure water. The concentration and quality of each DNA sample were evaluated using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE) and the Qubit dsDNA BR assay kit (Life Technologies, Carlsbad, CA) using a Qubit 2.0 fluorometer (Life Technologies). The DNA was stored at 4°C prior to further analysis.
Whole-genome sequencing (WGS) and phylogenetic analysis.
Paired-end libraries were prepared using the Nextera XT DNA sample preparation kit version 2 (Illumina, San Diego, CA). Tagmentation of the input DNA (1 ng) and PCR amplification to add indexing primers were performed according to the manufacturer's instructions. The library insert size (range, 300 to 1,000 bp) was analyzed on an Agilent 2100 bioanalyzer using the Agilent high-sensitivity DNA chip (Agilent, Santa Clara, CA). The “Library Normalization” step was omitted, and instead the individual libraries were diluted to 4 nM and pooled into a single tube. The pooled amplicon libraries (5 μl) were denatured into single strands by adding an equal volume of freshly prepared 0.2 N NaOH. The DNA sample was mixed briefly by vortexing, centrifuged at 280 × g for 1 min using an Eppendorf centrifuge 5430R (Eppendorf), and incubated at 25°C for 5 min. Meanwhile, the PhiX DNA control library was prepared by mixing 2 μl of PhiX control DNA with 3 μl of 10 mM Tris-HCl (pH 8.5) and 0.1% Tween 20. The PhiX control DNA library was denatured by adding 5 μl of 0.2 N NaOH, briefly vortexed, centrifuged at 280 × g for 1 min, and incubated at 25°C for 5 min. The PhiX control DNA library was diluted to 12.5 pM using cold HT1 buffer, and the sample DNA library was diluted to 15 pM prior to loading onto the MiSeq reagent cartridge. Sequencing by synthesis of the paired-end 250-bp reads was conducted on a MiSeq benchtop sequencing platform (Illumina).
Reads were analyzed using FastQC 0.11.2 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and NexteraXT transposon sequences were found to be present in less than 0.14% of the reads across all data sets. Low-quality bases were filtered out with sickle 1.290 (a sliding-window, adaptive, quality-based trimming tool for FastQ files developed by N. A. Joshi and J. N. Fass and available at https://github.com/najoshi/sickle) using the quality options -q 33 and -t Sanger. Reads were then trimmed to the minimum length of 150 nucleotides (nt) with sickle using the option -l 150. Both trimmed and raw reads were each assembled using two de novo assemblers: Spades 3.1 (29) and Ray 2.3.1 (30). For the Ray assemblies, kmer size was determined using Kmergenie 1.6663 (31). Assemblies were compared using Quast 2.3 (32). For each genome, the best assembly was selected based on the number of contigs, N50, largest contig size, and total assembly size (see Table S1 in the supplemental material). Additional assembly statistics were calculated with the assemblathon_stats.pl script (33). Draft assemblies excluded contigs below 5 kbp. Draft annotation statistics were generated using Prokka 1.10 (34), and tRNA quantities were amended using tRNAscan-SE 1.3.1 (35).
For final genome assemblies, the minimum contig size was set at 1 kbp. Assemblies were reannotated with Prokka 1.10, and tRNAs were verified and amended using tRNAscan-SE 1.3.1. Coding sequence (CDS) annotations were checked using InterProScan 5.3-46.0 (36) and manually corrected for improper product names. Consecutive product duplicates were verified with BLAST (37) protein homology searches against the NCBI nonredundant database to check for potential pseudogenes, which were then verified by read mapping in Geneious 7.1 (38). Genuine pseudogenes were then reannotated as such manually.
None of the eight isolates had a complete 16S rRNA gene sequence in its assembly. To generate these sequences, the contigs from each isolate's assembly were mapped to a reference 16S rRNA gene (Paenibacillus sp. strain 32O-W) using Geneious 7.1. A subset of these contigs which together spanned the full 16S rRNA genes were selected and de novo assembled using Geneious 7.1 (minimum overlap of 25 bp). The original sequencing reads were then mapped to this assembly to verify complete coverage, and a consensus sequence was then generated in Geneious 7.1 and appended to the final genome assembly. 16S rRNA gene sequences for each isolate were concatenated with related sequences retrieved from GenBank (as of 5 September 2014), the data set was aligned with MUSCLE 3.8.31 (39), and the resulting alignment was manually inspected for sequences presenting a wrong reverse complement orientation with SeaView 4.5.2 (40). Incorrectly oriented sequences were removed and reverse complemented with the FASTX-Toolkit 0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit), reintroduced in the proper orientation, and then realigned with MUSCLE. Ambiguous regions in the alignment were removed with BMGE 1-1 (41) using the default parameters. A best maximum likelihood (ML) tree was computed with PhyML 3.0 release (released 26 March 2015) (42) under the general time reversible (GTR) model of nucleotide substitution (four gamma categories) using a total of 100 bootstraps.
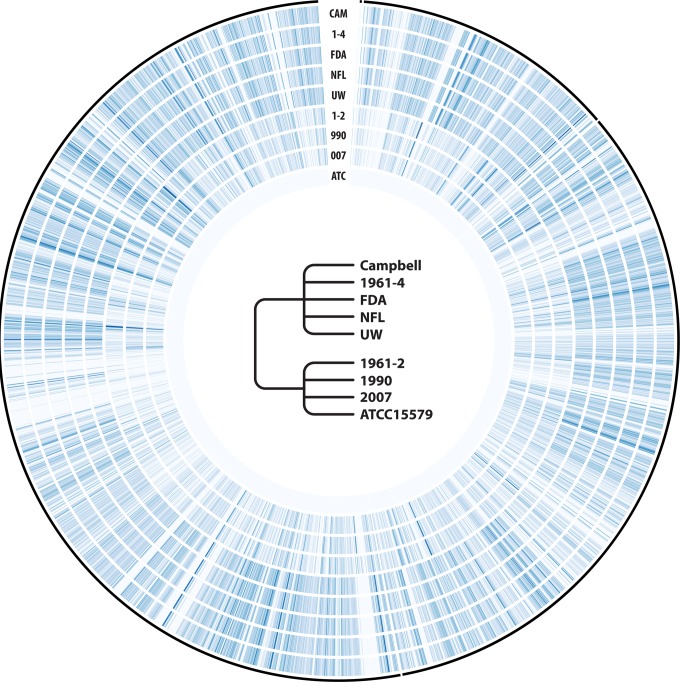
Reads from each data set were mapped on the ATCC 15579 genomic reference with BWA-SW 0.7.9a (43) and converted from SAM (sequence alignment) files to BAM (binary form of SAM) files with SAMtools 0.1.19 (44). SNPs were inferred with VarScan 2.3.7 (45) and converted to binary files (0 for absence; 1 for presence) with custom Perl scripts. SNP sliding windows were inferred with a step and window size of 1,000 bp using scripts from Pombert et al. (46) and displayed using Circos (47) (see Fig. 3).
FIG 3.
Single nucleotide polymorphism (SNP) profiling of the sequenced Clostridium sporogenes strains against the ATCC 15579 reference genome (GenBank accession number NZ_ABKW00000000.2). The four C. sporogenes contigs in the ATCC 15579 reference accessions are represented by the outer thin black lines. SNP densities (SNPs per kilobase pair) in the inner concentric heatmaps are color coded according to a bluish gradient (lightest for 0, darkest for 200; the maximum observed was 194 SNPs/kbp). All regions are drawn to scale. The cladogram in the center illustrates the relationships inferred between the various strains from our 16S rRNA gene maximum likelihood tree.
Nucleotide sequence accession number.
The sequences of all eight PA 3679 genomes were submitted to DBBJ/EMBL/GenBank under the BioProject accession number PRJNA278445.
RESULTS
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed on the ATCC 7955 NCA3679 1961 spore crop prior to streaking for isolation to ascertain the purity of the culture. Both PFGE and culturing methods indicated that this crop contained at least two different isolates, which were designated 1961-2 and 1961-4. Isolate 1961-2 produced large, pale, irregularly shaped colonies on EYA, whereas isolate 1961-4 was characterized by small, white, irregularly shaped colonies with an iridescent sheen indicating lipase activity, a common phenotypic trait for C. sporogenes. These isolates also displayed distinct restriction banding patterns (Fig. 1). PFGE analysis of the C. sporogenes PA 3679 isolates revealed three clusters based on the restriction banding patterns generated for each isolate (Fig. 1). All of the ATCC 7955 NCA3679 isolates except isolate 1961-4 formed one cluster. Within this cluster, isolate 1961-2 showed 80% similarity to the 1961 spore crop and only 77% similarity to the 1990 and 2007 spore crops, which shared an identical pulsotype. The restriction banding pattern of isolate 1961-4 shared 95% similarity to the second cluster comprised of isolates Campbell, FDA, NFL, and UW, which shared the same pulsotype. C. sporogenes ATCC 15579 was used as a genomic reference to understand the similarity between PA 3679 and another strain of C. sporogenes. Strain ATCC 15579 was only 57% similar to the other PA 3679 isolates examined and formed its own cluster.
FIG 1.
Dendrogram of nine C. sporogenes PA 3679 isolates and one C. sporogenes ATCC 15579 strain. Similarity between the restriction banding patterns was determined using the Dice coefficient correlation. The dendrogram was constructed using the average of the composite data sets generated by SmaI and XhoI and the unweighted pair group method using average linkages (UPGMA) using the average UPGMA linkage clustering tool. The optimization value and position tolerance were 1.5%.
Whole-genome sequencing.
The genomes of C. sporogenes ATCC 7955 NCA3679 isolates 1961-2, 1961-4, 1990, and 2007, PA 3679 isolates FDA, UW, NFL, and Campbell, and strain ATCC 15579 were sequenced, and the genome assembly and annotation statistics are presented in Tables 2 and 3. The average genome size of clade I organisms was 4.15 Mb, and the %GC content was 27.85. Clade II isolates shared a slightly smaller average genome size of ∼3.98 Mb and a %GC content of 28.
TABLE 2.
Genome assembly statistics of sequenced Clostridium sporogenes isolatesa
| Strain | Est. size (bp) | No. of reads (bp) | No. of bases (bp) | Est. coverage | Max. N50 (bp) | Max. length (bp) | Total size (bp) | No. of contigs of the following size: |
% genome | % GC content | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| >1 kbp | >5 kbpb | >100 kbp | ||||||||||
| PA 3679c | 4,173,562 | - | - | ∼200 | - | - | - | 181 | 131 | 4 | - | 27.83 |
| ATCC 15579c | 4,102,125 | - | - | - | - | - | - | 4 | 3 | 3 | - | 28.02 |
| ATCC 15579d | 4,102,125 | 1,538,278 | 384,569,500 | 94 | 146,994 | 366,529 | 4,060,754 | 58 | 45 | 15 | 98.99 | 27.85 |
| 1961-2 | 4,173,562 | 3,298,626 | 824,656,500 | 198 | 213,496 | 811,671 | 4,095,436 | 42 | 27 | 14 | 98.13 | 27.88 |
| 1990 | 4,173,562 | 3,927,744 | 981,936,000 | 235 | 187,341 | 545,953 | 4,167,596 | 59 | 49 | 11 | 99.86 | 27.85 |
| 2007 | 4,173,562 | 4,641,772 | 1,160,443,000 | 278 | 321,923 | 1,217,917 | 4,174,960 | 33 | 21 | 12 | 100.03 | 27.83 |
| 1961-4 | 4,173,562 | 3,388,606 | 847,151,500 | 203 | 115,686 | 306,290 | 3,975,480 | 88 | 46 | 16 | 95.25 | 28.01 |
| Campbell | 4,173,562 | 7,846,204 | 1,961,551,000 | 470 | 159,201 | 424,558 | 3,991,459 | 60 | 34 | 19 | 95.64 | 28.01 |
| FDA | 4,173,562 | 4,780,078 | 1,195,019,500 | 286 | 151,701 | 306,291 | 4,023,031 | 87 | 38 | 18 | 96.39 | 28.01 |
| NFL | 4,173,562 | 2,954,430 | 738,607,500 | 177 | 136,601 | 306,614 | 3,958,149 | 77 | 49 | 18 | 94.84 | 28.00 |
| UW | 4,173,562 | 19,211,592 | 4,802,898,000 | 1,151 | 150,182 | 306,290 | 3,930,977 | 48 | 36 | 19 | 94.19 | 27.99 |
Est., estimated; Max. maximum. Hyphens in the table indicate that the data were not available in GenBank.
Contigs smaller than 5 kbp are potentially spurious.
NCBI reference assemblies NZ_AGAH00000000.1 (PA 3679) and NZ_ABKW00000000.2 (ATCC 15579).
This study.
TABLE 3.
Annotation statistics (draft annotations using >5-kbp contigs) of sequenced Clostridium sporogenes strains
| Strain | No. of proteins | No. of rRNAs | No. of tRNAs | No. of other RNAs | No. of genes | No. of repeat regions (CRISPR)a | No. of signaling peptides |
|---|---|---|---|---|---|---|---|
| PA 3679b | 4,045 | 2 | 43 | 0 | 4,090 | - | - |
| ATCC 15579b | 3,767 | 23 | 77 | 6 | 3,873 | - | - |
| ATCC 15579c | 3,657 | 4 | 61 | 0 | 3,722 | 2 | 175 |
| 1961-2 | 3,768 | 5 | 59 | 0 | 3,832 | 2 | 174 |
| 1990 | 3,870 | 5 | 55 | 0 | 3,930 | 2 | 175 |
| 2007 | 3,866 | 5 | 59 | 0 | 3,930 | 2 | 174 |
| 1961-4 | 3,634 | 4 | 76 | 0 | 3,714 | 5 | 150 |
| Campbell | 3,703 | 4 | 58 | 0 | 3,765 | 5 | 154 |
| FDA | 3,693 | 4 | 77 | 0 | 3,774 | 5 | 153 |
| NFL | 3,646 | 6 | 78 | 0 | 3,730 | 5 | 151 |
| UW | 3,636 | 4 | 59 | 0 | 3,699 | 5 | 152 |
CRISPR, clustered regularly interspaced short palindromic repeat. Hyphens indicate that the data were not available in GenBank.
NCBI reference assemblies NZ_AGAH00000000.1 (PA 3679) and NZ_ABKW00000000.2 (ATCC 15579).
This study.
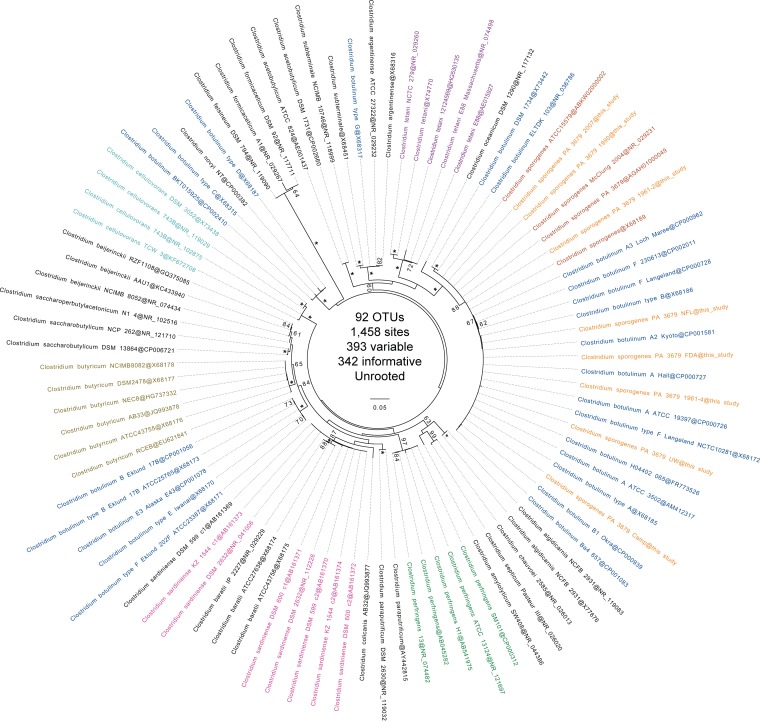
A maximum likelihood (ML) tree of 16S rRNA gene sequences of Clostridium (Fig. 2) was constructed, and all C. sporogenes isolates analyzed in this study were included among group I (proteolytic) Clostridium botulinum strains. However, isolates 1961-2, 2007, and 1990 were identified as C. sporogenes based on 16S rRNA gene sequencing because they were located within a tight cluster comprised of other C. sporogenes strains including McClung 2004 (NR_029231), ATCC 15579, and C. sporogenes PA 3679 NCTC8594 (25). The 16S sequences NR_029231 and X68189 were both derived from strain ATCC 3584 L. S. McClung 2004 (18), which was deposited into ATCC by I. C. Hall (http://www.atcc.org/products/all/3584.aspx#history). Isolates FDA, UW, NFL, Campbell, and 1961-4 were not included within this group but instead were clustered with other group I (proteolytic) C. botulinum strains.
FIG 2.
Maximum likelihood (ML) tree of Clostridium 16S rRNA gene sequences. The best ML tree computed with PhyML under the general time reversible (GTR) model of nucleotide substitution (four gamma categories) with 100 bootstraps. Bootstrap values greater than 60 are shown at their respective nodes, while nodes present in all bootstraps are indicated with an asterisk. Species/strain names are indicated before the @ symbol, and accession numbers are indicated after the @ symbol. The Clostridium sporogenes strains from this study are colored light orange. The previous Clostridium sporogenes, Clostridium botulinum, Clostridium perfringens, Clostridium sardiniense, Clostridium butyricum, Clostridium cellulovorans, and Clostridium tetani species are colored dark orange, navy, green, pink, gold, cyan, and purple, respectively. OTUs, operational taxonomic units.
Single nucleotide polymorphism (SNP) profiling of the sequenced C. sporogenes isolates against the ATCC 15579 reference genome (GenBank accession number NZ_ABKW00000000.2) is illustrated in Fig. 3. The total numbers of SNPs, SNPs/kilobase, and indels inferred from VarScan 2.3 between each isolate and the genomic references ATCC 15579 and PA 3679 (NCTC8594) (GenBank accession number AGAH00000000.1) are presented in Table 4. Similar to the results obtained by PFGE analysis, the isolates were categorized into two clades. Clade I included isolates ATCC 15579, 1961-2, 1990, and 2007. The ATCC 15579 sequenced in this study contained only 85 SNPs and 31 indels compared with the reference genome sequence for this strain. These SNPs are likely due to assembly or sequencing errors. Similarly, when the reads were mapped to their own assembly for any particular isolate in either clade I or II, an average of 100 to 200 SNPs was typically found, except for isolate 1990, which had 421 SNPs. Isolates 1961-2, 1990, and 2007 were found to have a higher number of SNPs compared to ATCC 15579 than when these isolates were compared to PA 3679 (NCTC8594) (Table 4). The isolates in clade II included isolates 1961-4, Campbell, FDA, NFL, and UW and contained an average of 189,260 SNPs compared to ATCC 15579 and an average of 202,292 SNPs compared to PA 3679 (NCTC8594) (Table 4). Unlike the pairwise comparison of isolates in clade I, strains within clade II showed very low numbers of SNPs and indels, suggesting that they are highly similar to each other. Comparison of the FDA and NFL isolates showed the highest number of SNPs at 198 (0.05 SNPs/kb) but only 4 indels.
TABLE 4.
Comparison of total number of SNPs, SNPs/kilobase, and indels between C. sporogenes isolates within clades I and II and the genomic reference strains ATCC 15579 and PA 3679 (NCTC8594)
| Clade and isolate/strain | Total no. of SNPs, SNPs/kb, and indels in the following pairwise comparisona: |
|||||||
|---|---|---|---|---|---|---|---|---|
| 1961-2 | 1990 | 2007 | 1961-4 | Campbell | FDA | NFL | UW | |
| Clade I | ||||||||
| 1961-2 | 230 | 6,815 | 7,099 | |||||
| 0.058 | 1.703 | 1.775 | ||||||
| 6 | 124 | 139 | ||||||
| 1990 | 7,382 | 421 | 562 | |||||
| 1.846 | 0.105 | 0.141 | ||||||
| 138 | 11 | 17 | ||||||
| 2007 | 7,085 | 202 | 223 | |||||
| 1.771 | 0.051 | 0.056 | ||||||
| 131 | 5 | 5 | ||||||
| Reference strains | ||||||||
| PA 3679b | 27,899 | 25,298 | 22,746 | 201,870 | 199,573 | 203,654 | 199,528 | 206,839 |
| 6.975 | 6.325 | 5.687 | 50.468 | 49.893 | 50.914 | 49.882 | 51.710 | |
| 615 | 500 | 363 | 2,219 | 2,103 | 2,304 | 2,155 | 2,529 | |
| ATCC 15579c | 143,110 | 139,899 | 124,943 | 188,803 | 187,081 | 190,471 | 186,800 | 193,146 |
| 35.778 | 34.975 | 31.236 | 47.201 | 46.770 | 47.618 | 46.700 | 48.287 | |
| 2,112 | 19,008 | 1,259 | 2,268 | 2,127 | 2,370 | 2,152 | 2,578 | |
| Clade II | ||||||||
| 1961-4 | 144 | 117 | 155 | 128 | 120 | |||
| 0.036 | 0.029 | 0.039 | 0.032 | 0.030 | ||||
| 2 | 5 | 3 | 3 | 3 | ||||
| Campbell | 136 | 103 | 141 | 117 | 117 | |||
| 0.034 | 0.026 | 0.035 | 0.029 | 0.029 | ||||
| 4 | 3 | 4 | 5 | 6 | ||||
| FDA | 168 | 136 | 157 | 150 | 138 | |||
| 0.042 | 0.034 | 0.039 | 0.038 | 0.035 | ||||
| 3 | 3 | 2 | 3 | 3 | ||||
| NFL | 187 | 172 | 198 | 137 | 176 | |||
| 0.047 | 0.043 | 0.050 | 0.034 | 0.044 | ||||
| 5 | 8 | 4 | 4 | 6 | ||||
| UW | 123 | 98 | 127 | 100 | 94 | |||
| 0.031 | 0.025 | 0.032 | 0.025 | 0.024 | ||||
| 4 | 7 | 6 | 4 | 5 | ||||
For each pairwise comparison, the first line shows the total number of SNPs, the second line shows the number of SNPs per kilobase, and the third line shows the number of indels. The number of SNPs per kilobase is based on 4,000,000-bp genome size estimate.
NCTC8594; GenBank accession number AGAH00000000.1 (25).
GenBank accession number NZ_ABKW00000000.2.
Heat resistance studies.
The thermal resistance of isolates 1961-2, 2007, 1961-4, FDA, NFL, UW, and Campbell was assessed at temperatures ranging from 94°C to 121°C, and the growth of survivors in TPGY broth was recorded (Table 5). Isolates representing clade I, isolates 1961-2 and 2007, were more sensitive to high temperature than the other isolates because no survivors were detected after 5 min at 100°C (isolate 1961-2) and 105°C (isolate 2007) (Table 5). Isolates of clade II were more resistant to high temperature, as isolates Campbell, FDA, and NFL survived heat treatment at 121°C, which is typical of the thermal resistance of C. sporogenes PA 3679. Isolates UW and 1961-4 were slightly less heat resistant, because survivors were obtained after heat treatment for 5 min at 117°C but not at 121°C.
TABLE 5.
Growth of Clostridium sporogenes PA 3679 spores after heat treatment at different temperatures for 5 min
| Temp (°C) | Growtha of indicated isolate (initial concnb) |
||||||
|---|---|---|---|---|---|---|---|
| 1961-2 (7.65 × 105) | 2007 (1.01 × 106) | 1961-4 (6.65 × 105) | FDA (1.64 × 106) | NFL (7.65 × 106) | UW (7.25 × 106) | Campbell (3.95 × 106) | |
| 94 | +/+ | +/+ | NT | NT | NT | NT | NT |
| 97 | +/+ | +/+ | NT | NT | NT | NT | NT |
| 100 | −/− | +/+ | NT | NT | NT | NT | NT |
| 103 | −/− | +/+ | NT | NT | NT | NT | NT |
| 105 | −/− | −/− | +/+ | +/+ | +/+ | NT | +/+ |
| 108 | −/− | −/− | +/+ | +/+ | +/+ | +/+ | +/+ |
| 111 | NT | −/− | +/+ | +/+ | +/+ | +/+ | +/+ |
| 114 | NT | −/− | +/+ | +/+ | +/+ | +/+ | +/+ |
| 117 | NT | NT | +/− | +/+ | +/+ | +/+ | +/+ |
| 121 | NT | NT | −/− | +/− | +/+ | −/− | +/+ |
The growth of isolates was assessed by turbidity of TPGY broth tubes in duplicate. The results are shown as follows: +/+, growth was observed in both TPGY broth tubes; −/−, growth was not observed in either TPGY broth tube; +/−, growth was observed in only one TPGY broth tube; NT, not tested.
The initial concentration is shown in parentheses after the isolate. Non-heat-treated controls were heat shocked at 80°C for 10 min and are reported as spore-forming units per milliliter.
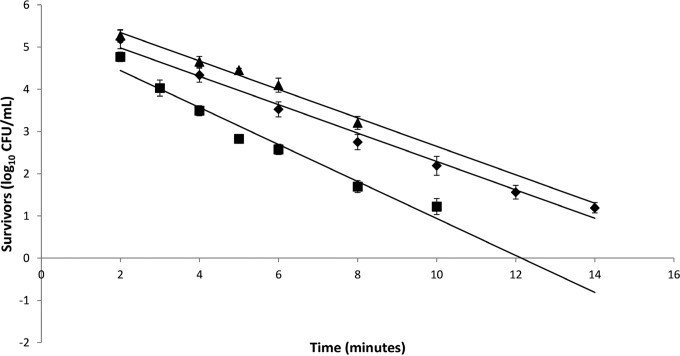
Additional thermal studies of isolates 1961-2, 2007, and FDA were conducted at 97°C, 100°C, and 117°C, respectively, for the calculation of D-values (the time required to reduce a population by 90%) as a measure of heat resistance of these isolates representative of the two genetic clades identified via PFGE and whole-genome sequencing (Fig. 4). Initially, thermal studies of isolate 2007 were conducted at 103°C; however, enumeration of spores after 4 min was consistently below our level of detection using the plate count method. Therefore, thermal studies on this isolate were carried out at 100°C. Isolate 1961-2 had a D97°C of 2.97 min, whereas isolate 2007 was found to have a D100°C of 2.28 min. Isolate FDA was much more heat resistant than the other two strains, with a D117°C of 2.97 min.
FIG 4.
Survival curve of C. sporogenes strains ATCC 7955 NCA3679 1961-2 (◆) and 2007 (■) and PA 3679 FDA (▲). D-values were calculated from the negative reciprocal of the slopes of the regression lines. Data for the survival curves were generated by heat treatment at 97°C (isolate 1961-2), 100°C (isolate 2007), and 117°C (isolate FDA).
DISCUSSION
The strict USDA/CDC regulations required for working with C. botulinum and its toxins and the extreme biohazardous risk of this organism obstruct the routine use of this organism to validate thermal processing technologies for shelf-stable low-acid canned foods in industry and research facilities despite it being the primary health hazard for these products. C. sporogenes PA 3679 has been adopted as a suitable surrogate for group I (proteolytic) C. botulinum because it is phenotypically and genetically similar to C. botulinum, it is nontoxic, and its spores are more resistant to high temperature.
Over the last 8 decades, C. sporogenes PA 3679 has been distributed to and shared with a multitude of laboratories worldwide and subcultured routinely using various techniques and growth media. It is likely that the genetic makeup of this organism has drifted from the original isolate discovered in 1927. Furthermore, other putrefactive anaerobes isolated from spoiled canned foods have been identified and potentially used synonymously with PA 3679 (10, 11). PFGE and SNP analyses of the C. sporogenes isolates examined in this study categorized the strains into two clades. On the basis of the data presented in this study, we conclude that clade II isolates (isolates FDA, Campbell, NFL, UW, and 1961-4) are most likely to be the classical C. sporogenes PA 3679 strain described by Townsend et al. (9). Upon initial examination of the PA 3679 isolates using PFGE, it was evident that the ATCC 7955 1961 spore crop originally contained PA 3679 (as isolate 1961-4) but that it was contaminated with at least one other organism (isolate 1961-2). Isolate 1961-2 was found to be the most similar, yet it was not identical to other clade I isolates (isolates 1990 and 2007) based on both PFGE and SNP analyses. Strains 1990, 2007, and 1961-2 are likely strains of C. sporogenes, since they clustered with other C. sporogenes strains based on 16S rRNA gene sequencing (Fig. 2). SNP comparisons between isolates 1990 and 2007, which number in the hundreds (Table 4), suggest that these isolates may be clonal, since a similar number of SNPs is observed for a particular strain when the reads are mapped against its own assembly. Approximately 7,000 SNPs were found between isolate 1961-2 and either isolate 1990 or isolate 2007, suggesting that this isolate is of similar origin but has since diverged slightly. Interestingly, clade I isolates are more similar to the genomic reference PA 3679 (NCTC8594) than clade II isolates, since an almost 10-fold increase in SNPs was observed between clade II isolates and NCTC8594. However, the levels of divergence observed between clade I isolates and NCTC8594 suggest that they have been drifting apart for multiple generations. Although clade I isolates were more genetically similar to NCTC8594, the thermal screening data generated in this study showed that they are sensitive to high temperature and thus, by definition, should not be classified as PA 3679.
In a study compiling 911 D-values of proteolytic C. botulinum and C. sporogenes PA 3679 collected from 38 studies, PA 3679 was found to have an estimated D121°C of 1.28 min and a z-value (the temperature increase needed to achieve a 10-fold decrease in D-values) of 11.1°C (48). Comparatively, the mean D121°C and z-value for C. botulinum were found to be 0.19 min and 11.3°C, respectively (48). Isolates 1961-2 and 2007 were significantly sensitive to high temperatures and had a D97°C value and a D100°C value of 2.97 min and 2.28 min, respectively. Production of spores that are resistant to high temperatures is the defining trait for C. sporogenes PA 3679 and an essential requirement for this strain to be used as a model organism for group I C. botulinum to validate thermal processing technologies. The thermal resistance of C. sporogenes PA 3679 (NCTC8594) was not determined in this study, and thermal resistance data for this particular strain were not available in the literature. However, a study by Bull et al. (49) examined the resistance of NCTC8594 spores to combined high temperature and pressure in various food matrices and compared it with that of spores of C. botulinum strains 7273B, 62-A, 213B, and NCTC2916 under the same conditions. They found that NCTC8594 was more sensitive to combined high temperature and pressure than the majority of C. botulinum spores tested and concluded that NCTC8594 may not be a suitable surrogate for C. botulinum, at least for high-pressure thermal process validation studies (49).
Some of the factors that contribute to the overall resistance of bacterial spores include the type, pH, and mineral content of sporulation medium used (50–53), heating menstruum (54), and sporulation temperature (55, 56). It is also well established that the resistance of spore crops of a particular strain to high temperature and combined high temperature and pressure can vary from batch to batch as well as being strain dependent (9, 26, 49, 57, 58). Based on the results of this study, a reexamination and reevaluation of the thermal destruction data reported in the literature based on the “isolate” of PA 3679 used in a particular study are warranted.
The second isolate purified from the ATCC 7955 1961 spore crop, isolate 1961-4, shared the most similarity with other clade II isolates, including isolates NFL, UW, Campbell, and FDA. The spores produced by clade II isolates showed a higher degree of heat resistance based on the thermal studies performed, and it is likely that clade II isolates more accurately represent E. J. Cameron's PA 3679 from 1927 (9). C. sporogenes PA 3679 was deposited into the ATCC around 1941 (9) and used in various laboratories as a model organism for group I (proteolytic) C. botulinum. It is possible that the 1961 crop contained E. J. Cameron's PA 3679 strain, but that the crop was contaminated and the strain was outcompeted over time. The later 1990 and 2007 spore crops of ATCC 7955 may have been distributed to various laboratories, and data published using subcultures of those spore crops may not adequately represent PA 3679.
Weigand et al. (24) identified a subset of C. sporogenes clade-specific genes. We are currently in the process of screening clade II C. sporogenes PA 3679 isolates for C. sporogenes clade-specific genes identified by Weigand et al. (24). A thorough genomic comparison between clade II isolates of C. sporogenes PA 3679 and group I C. botulinum strains to highlight the gene content differences between these organisms, especially genes that may be involved in increased spore heat resistance, is also in progress. Understanding the genetic mechanisms of spore heat resistance is paramount to the design and development of adequate countermeasures to ensure the production of wholesome and safe, minimally processed foods.
Supplementary Material
Funding Statement
Y.W. was supported by an appointment to the Research Participation Program at the Center for Food Safety and Applied Nutrition administered by the Oak Ridge Institute for Science and Education via an interagency agreement between the U.S. Department of Energy and the FDA. The sponsor had no role in the study design, data collection and analysis, the decision to publish, or the preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02616-15.
REFERENCES
- 1.Collins MD, East AK. 1998. Phylogeny and taxonomy of the food-borne pathogen Clostridium botulinum and its neurotoxins. J Appl Microbiol 84:5–17. doi: 10.1046/j.1365-2672.1997.00313.x. [DOI] [PubMed] [Google Scholar]
- 2.Dodds KL, Hauschild AH. 1989. Distribution of Clostridium botulinum in the environment and its significance in relation to botulism, p 472 In Recent advances in microbial ecology. Proceedings of the 5th International Symposium on Microbial Ecology. International Society for Microbial Ecology, Kyoto, Japan. [Google Scholar]
- 3.Meyer KF, Dubovsky BJ. 1922. The distribution of the spores of B. botulinus in the United States. IV. J Infect Dis 31:559–594. doi: 10.1093/infdis/31.6.559. [DOI] [Google Scholar]
- 4.Townsend CT, Yee L, Mercer WA. 1954. Inhibition of the growth of Clostridium botulinum by acidification. Food Res 19:536–542. doi: 10.1111/j.1365-2621.1954.tb17486.x. [DOI] [Google Scholar]
- 5.Ingram M, Robinson HM. 1951. A discussion of the literature on botulism in relation to acid foods. Proc Soc Appl Bacteriol 14:73–84. doi: 10.1111/j.1365-2672.1951.tb01995.x. [DOI] [Google Scholar]
- 6.Tanaka N. 1982. Toxin production by Clostridium botulinum in media at pH lower than 4.6. J Food Prot 45:679. [DOI] [PubMed] [Google Scholar]
- 7.Stumbo CR, Purohit KS, Ramakrishna TV. 1975. Thermal process lethality guide for low-acid foods in metal containers. J Food Sci 40:1316–1323. doi: 10.1111/j.1365-2621.1975.tb01080.x. [DOI] [Google Scholar]
- 8.McClung LS. 1937. Studies on anaerobic bacteria. X. Heat stable and heat labile antigens in the botulinum and related groups of spore-bearing anaerobes. J Infect Dis 60:128–134. [Google Scholar]
- 9.Townsend CT, Esty JR, Baselt FC. 1938. Heat-resistance studies on spores of putrefactive anaerobes in relation to determination of safe processes for canned foods. J Food Sci 3:323–346. doi: 10.1111/j.1365-2621.1938.tb17065.x. [DOI] [Google Scholar]
- 10.Brown JL, Tran-Dinh N, Chapman B. 2012. Clostridium sporogenes PA 3679 and its uses in the derivation of thermal processing schedules for low-acid shelf stable foods and as a research model for proteolytic Clostridium botulinum.. J Food Prot 75:779–792. doi: 10.4315/0362-028X.JFP-11-391. [DOI] [PubMed] [Google Scholar]
- 11.Gross CE, Vinton C, Stumbo CR. 1946. Bacteriological studies relating to thermal processing of canned meats. V. Characteristics of putrefactive anaerobe used in thermal resistance studies. Food Res 11:405–410. [DOI] [PubMed] [Google Scholar]
- 12.Sebaihia M, Peck MW, Minton NP, Thomson NR, Holden MTG, Mitchell WJ, Carter AT, Bentley SD, Mason DR, Crossman L, Paul CJ, Ivens A, Wells-Bennik MHJ, Davis IJ, Cerdeno-Tarraga AM, Churcher C, Guail MA, Chillingworth T, Feltwell T, Fraser A, Goodhead I, Hance Z, Jagels K, Larke N, Maddison M, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Sanders M, Simmonds M, White B, Whithead S, Parkhill J. 2007. Genome sequence of proteolytic (group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res 17:1082–1092. doi: 10.1101/gr.6282807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peck MW, Stringer SC, Carter AT. 2011. Clostridium botulinum in the post-genomic era. Food Microbiol 28:183–191. [DOI] [PubMed] [Google Scholar]
- 14.Macdonald TE, Helma CA, Ticknor LO, Jackson PJ, Okinaka RT, Smith LA, Smith TJ, Hill KK. 2008. Differentiation of Clostridium botulinum serotype A strains by multiple-locus variable-number tandem-repeat analysis. Appl Environ Microbiol 74:875–882. doi: 10.1128/AEM.01539-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fillo S, Giordani F, Anniballi F, Gorge O, Ramisse V, Vergnaud G, Riehm JM, Scholz HC, Splettstoesser WD, Kieboom J, Olsen J-S, Fenicia L, Lista F. 2011. Clostridium botulinum group I strain genotyping by 15-locus multilocus variable-number tandem-repeat analysis. J Clin Microbiol 49:4252–4263. doi: 10.1128/JCM.05396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter AT, Paul CJ, Mason DR, Twine SM, Alston MJ, Logan SM, Austin JW, Peck MW. 2009. Independent evolution of neurotoxin and flagellar genetic loci in proteolytic Clostridium botulinum.. BMC Genomics 10:115. doi: 10.1186/1471-2164-10-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson JL, Francis BS. 1975. Taxonomy of the clostridia: ribosomal ribonucleic acid homologies among the species. J Gen Microbiol 88:229–244. doi: 10.1099/00221287-88-2-229. [DOI] [PubMed] [Google Scholar]
- 18.Hutson RA, Thompson DE, Lawson PA, Schocken-Itturino RP, Böttger EC, Collins MD. 1993. Genetic interrelationships of proteolytic Clostridium botulinum types A, B and F and other members of the Clostridium botulinum complex as revealed by small-subunit rRNA gene sequences. Antonie Van Leeuwenhoek 64:273–283. [DOI] [PubMed] [Google Scholar]
- 19.Hill KK, Smith TJ, Helma CH, Ticknor LO, Foley BT, Svensson RT, Brown JL, Johnson EA, Smith LA, Okinaka RT, Jackson PJ, Marks JD. 2007. Genetic diversity among botulinum neurotoxin-producing clostridial strains. J Bacteriol 189:818–832. doi: 10.1128/JB.01180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobson MJ, Lin G, Whittam TS, Johnson EA. 2008. Phylogenetic analysis of Clostridium botulinum type A by multi-locus sequence typing. Microbiology 154:2408–2415. doi: 10.1099/mic.0.2008/016915-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen JS, Scholz H, Fillo S, Ramisse V, Lista F, Tromborg AK, Aarskaug T, Thrane I, Blatny JM. 2014. Analysis of the genetic distribution among members of Clostridium botulinum group I using a novel multilocus sequence typing (MLST) assay. J Microbiol Methods 96:84–91. doi: 10.1016/j.mimet.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Lee WH, Riemann H. 1970. The genetic relatedness of proteolytic Clostridium botulinum strains. J Gen Microbiol 64:85–90. doi: 10.1099/00221287-64-1-85. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura S, Okado I, Nakashio S, Nishida S. 1977. Clostridium sporogenes isolates and their relationship to C. botulinum based on deoxyribonucleic acid reassociation. J Gen Microbiol 100:395–401. doi: 10.1099/00221287-100-2-395. [DOI] [PubMed] [Google Scholar]
- 24.Weigand MR, Pena-Gonzalez A, Shirey TB, Broeker RG, Ishaq MK, Konstantinidis KT, Raphael BH. 2015. Implications of genome-based discrimination between Clostridium botulinum group I and Clostridium sporogenes strains for bacterial taxonomy. Appl Environ Microbiol 81:5420–5429. doi: 10.1128/AEM.01159-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradbury M, Greenfield P, Midgley D, Li D, Tran-Dinh N, Vriesekoop F, Brown J. 2012. Draft genome sequence of Clostridium sporogenes PA 3679, the common nontoxigenic surrogate for proteolytic Clostridium botulinum.. J Bacteriol 194:1631–1632. doi: 10.1128/JB.06765-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy NR, Marshall KM, Morrissey TM, Loeza V, Patazca E, Skinner GE, Krishnamurthy K, Larkin JW. 2013. Combined high pressure and thermal processing on inactivation of type A and proteolytic type B spores of Clostridium botulinum.. J Food Prot 76:1384–1392. doi: 10.4315/0362-028X.JFP-12-538. [DOI] [PubMed] [Google Scholar]
- 27.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 28.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahni N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boisvert S, Raymond F, Godzaridis E, Laviolette F, Corbeil J. 2012. Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol 13:R122. doi: 10.1186/gb-2012-13-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chikhi R, Medvedev P. 2014. Informed and automated k-mer size selection for genome assembly. Bioinformatics 30:31–37. doi: 10.1093/bioinformatics/btt310. [DOI] [PubMed] [Google Scholar]
- 32.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradnam KR, Fass JN, Alexandrov A, Baranay P, Bechner M, Birol I, Boisvert S, Chapman JA, Chapuis G, Chikhi R, Chitsaz H, Chou W-C, Corbeil J, Fabbro CD, Docking TR, Durbind R, Earl D, Emrich S, Fedotov P, Fonseca NA, Ganapathy G, Gibbs RA, Gnerre S, Godzaridis E, Goldstein S, Haimel M, Hall G, Haussler D, Hiatt JB, Ho IY, Howard J, Hunt M, Jackman SD, Jaffe DB, Jarvis ED, Jiang H, Kazakov S, Kersey PJ, Kitzman JO, Knight JR, Koren S, Lam T-W, Lavenier D, Laviolette F, Li Y, Li Z, Liu B, Liu Y, Luo R, MacCallum I, MacManes MD, et al. 2013. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. GigaScience 2:10. doi: 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 35.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong S-Y, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Mentjies P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 41.Criscuolo A, Gribaldo S. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol 10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller C, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pombert J-F, Xu J, Smith DR, Heiman D, Young S, Cuomo CA, Weiss LM, Keeling PJ. 2013. Complete genome sequences from three genetically distinct strains reveal high intraspecies genetic diversity in the microsporidian Encephalitozoon cuniculi.. Eukaryot Cell 12:503–511. doi: 10.1128/EC.00312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diao MM, Andre S, Membre J-M. 2014. Meta-analysis of D-values of proteolytic Clostridium botulinum and its surrogate strain Clostridium sporogenes PA 3679. Int J Food Microbiol 174:23–30. doi: 10.1016/j.ijfoodmicro.2013.12.029. [DOI] [PubMed] [Google Scholar]
- 49.Bull MK, Olivier SA, van Diepenbeek RJ, Kormelink F, Chapman B. 2009. Synergistic inactivation of spores of proteolytic Clostridium botulinum strains by high pressure and heat is strain and product dependent. Appl Environ Microbiol 75:434–445. doi: 10.1128/AEM.01426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reddy NR, Tetzloff RC, Skinner GE. 2010. Effect of media, additives, and incubation conditions on the recovery of high pressure and heat-injured Clostridium botulinum spores. Food Microbiol 27:613–617. doi: 10.1016/j.fm.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 51.Kihm DJ, Hutton MT, Hanlin JH, Johnson EA. 1990. Influence of transition metals added during sporulation on heat resistance of Clostridium botulinum 113B spores. Appl Environ Microbiol 56:681–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alderton G, Ito KA, Chen JK. 1976. Chemical manipulation of the heat resistance of Clostridium botulinum spores. Appl Environ Microbiol 31:492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mah J-H, Kang D-H, Tang J. 2008. Effects of minerals on sporulation and heat resistance of Clostridium sporogenes.. Int J Food Microbiol 128:385–389. doi: 10.1016/j.ijfoodmicro.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 54.Cameron MS, Leonard SJ, Barrett EL. 1980. Effect of moderately acidic pH on heat resistance of Clostridium sporogenes spores in phosphate buffer and in buffered pea puree. Appl Environ Microbiol 39:943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marshall KM, Nowaczyk L, Morrissey TR, Loeza V, Halik LA, Skinner GE, Reddy NR, Fleischman GJ, Larkin JW. 2015. Effect of sporulation temperature on the resistance of Clostridium botulinum type A spores to thermal and high pressure processing. J Food Prot 78:146–150. doi: 10.4315/0362-028X.JFP-14-186. [DOI] [PubMed] [Google Scholar]
- 56.Peck MW, Evans RI, Fairbairn DA, Hartley MG, Russell NJ. 1995. Effect of sporulation temperature on some properties of spores of non-proteolytic Clostridium botulinum.. Int J Food Microbiol 28:289–297. doi: 10.1016/0168-1605(95)00064-X. [DOI] [PubMed] [Google Scholar]
- 57.Esty JR, Meyer KF. 1922. The heat resistance of the spores of B. botulinum and allied anaerobes. XI. J Infect Dis 31:650–664. doi: 10.1093/infdis/31.6.650. [DOI] [Google Scholar]
- 58.Odlaug E, Pflug IJ. 1977. Thermal destruction of Clostridium botulinum spores suspended in tomato juice in aluminum thermal death time tubes. Appl Environ Microbiol 34:23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.