Abstract
Snake venom protein from Deinagkistrodon acutus (DA protein), one of the major venomous species in Taiwan, causes hemorrhagic symptoms that can lead to death. Although horse-derived antivenin is a major treatment, relatively strong and detrimental side effects are seen occasionally. In our study, yolk immunoglobulin (IgY) was purified from eggs, and DA protein was recognized using Western blotting and an enzyme-linked immunosorbent assay (ELISA), similar to therapeutic horse antivenin. The ELISA also indicated that specific IgY antibodies were elicited after the fifth booster, plateaued, and lasted for at least 3 months. To generate monoclonal single-chain variable fragment (scFv) antibodies, we used phage display technology to construct two libraries with short or long linkers, containing 6.24 × 108 and 5.28 × 108 transformants, respectively. After four rounds of biopanning, the eluted phage titer increased, and the phage-based ELISA indicated that the specific clones were enriched. Nucleotide sequences of 30 individual clones expressing scFv were analyzed and classified into four groups that all specifically recognized the DA venom protein. Furthermore, based on mass spectrometry, the scFv-bound protein was deduced to be snake venom metalloproteinase proteins. Most importantly, both IgY and mixed scFv inhibited the lethal effect in mice injected with the minimum lethal dosage of the DA protein. We suggest that together, these antibodies could be applied to the development of diagnostic agents or treatments for snakebite envenomation in the future.
INTRODUCTION
Envenomation from venomous snakebites is a frequently discussed medical issue globally because of the frequent overlapping of human habitats with snake habitats, particularly in tropical and subtropical regions. Approximately 2.5 million people are bitten by venomous snakes, and more than 100,000 die every year (1). Snake venom elicits high mortality because of various complications, depending on the species, type, and injected quantity. In general, snake venom contains a mixture of proteins, polypeptides, and metal ions and has various functions, such as inducing paralysis and death and digesting prey (2). Currently, snake venom proteins have been divided into three major types: hemotoxins, resulting in hemorrhage; neurotoxins, affecting the nervous system; and myotoxins, affecting the muscular system. More than 40 terrestrial snake species exist in Taiwan, of which 15 are venomous (3). Among them, the bites of six venomous snakes, including Deinagkistrodon acutus (formerly Agkistrodon acutus, hundred-pace snake), which is a monotypic Viperiae, are the most common in a clinical context (4). The toxin protein from D. acutus (DA protein) consists of a complex of proteins with various biological activities, including phospholipase A2, metalloproteinases, peptidases, nucleotidases, nucleases, and phosphatases, that causes hemorrhagic symptoms, resulting in death (5–7). Among these proteins, snake venom metalloproteinase proteins (SVMPs) are considered to play a crucial role in the hemorrhagic activity, which includes the digestion of the basement membrane of vessels or elements of the extracellular matrix, such as collagen and fibronectin (8). In addition to SVMPs, numerous different components in crude venom with biological activity react synergistically or autonomously in their effects. This suggests that polyclonal antivenin immune therapy is an effective treatment against venomous snakebites.
At present, horse-derived hyperimmune antivenin is the major treatment against snakebites, including those of D. acutus, for inactivating these components. However, the cost for generating therapeutic horse sera is relatively high, and detrimental side effects, such as anaphylactic shock or serum sickness, are observed occasionally (9). To reduce the cost and side effects, chicken yolk immunoglobulin (IgY) from eggs could be an alternative to horse sera. IgY in eggs is derived from serum IgG molecules transferred to egg yolk and show a function equivalent to that of mammalian IgG (10). Using female chickens as immunization hosts for antibody production has numerous advantages, including the requirement of a smaller feeding space and small amount of antigens for immunization, as well as the obtainment of a relatively high titer that persists for a longer period (11, 12). In addition, the noninvasive collection of IgY isolation is cost-effective, as 2% to 10% are specific antibodies could be obtained from total IgY harvested (13, 14). In addition to these advantages, a chicken can provide more than 40 g IgY per year (15). Thus far, numerous studies on IgY used for immunotherapy in clinical and experimental treatments have been reported (16–20). However, polyclonal antibodies, including IgY, are prone to cross-reactions and are of inconsistent quality. Alternatively, specific diagnostic agents also help in rapidly confirming the type of snake that delivered the bite for selecting precise therapeutic agents. Among them, monoclonal antibodies against one particular epitope and with higher specificity have more latent capacity for being developed into diagnostic snake venom agents capable of rapid diagnosis.
To generate monoclonal antibodies, the phage display system is an effective in vitro method for constructing animal or human antibody libraries in terms of time and costs (21, 22). Among various antibody forms, the antigen-binding fragment or single-chain variable fragment (scFv) displayed on the phage is highly suitable and fast for selecting specific antibodies (23–25). Among the many animals that are adequate as animal models for producing antibodies against immunizing antigens, chickens are the most convenient and rapid host for constructing antibody libraries for selecting specific scFvs against various targets for therapy or diagnosis (24, 26, 27). Monoclonal scFv is a small protein with favorable tissue penetration, maintaining the variable regions of light and heavy chains that are joined with a flexible peptide linker, and it has a specific antigen-binding ability (28, 29). Although monoclonal antibodies are considered to have a lower efficacy against snake venom because of specificity for only one epitope, a combination of numerous monoclonal antibodies as therapy still has the potential to reduce symptoms, increase the survival rate, and prevent death (30). Monoclonal antibodies also are more promising for the development of specific diagnostic agents for the rapid diagnosis of snakebite envenomation.
In an attempt to develop a substitute to horse-derived antivenin to neutralize snake venom proteins and develop rapid diagnostic reagents, in this study, we sought to generate polyclonal and monoclonal antibodies with neutralizable efficacy from chickens, including polyclonal IgY from eggs and monoclonal scFv. These antibodies were isolated using phage display technology after immunizing female chickens with DA venom proteins. We not only analyzed the generated polyclonal IgY but also tested the protective efficacy of specific monoclonal scFv antibodies in mice treated with a lethal dose of DA venom protein. In addition, the scFv antibodies also were analyzed for specific binding activity with six snake venom proteins to examine potential application as rapid diagnostic reagents. We anticipate that the polyclonal IgY and monoclonal scFv antibodies could be applied to the development of diagnostic or therapeutic agents for snakebite envenomation for better prophylaxis in the future.
MATERIALS AND METHODS
Animals.
All animal experimental protocols in this study had been approved by the Institutional Animal Care and Use Committee of the Taipei Medical University before study initiation. The female White Leghorn (Gallus domesticus) chickens and ICR mice, 12 to 14 g in body weight, were purchased from the National Laboratory Animal Center, Taiwan, and maintained in the animal facility of the Taipei Medical University.
Preparation and analysis of the DA venom protein for chicken immunization.
DA venom protein was kindly provided by the vaccine center of the Centers for Disease Control (CDC), Ministry of Health and Welfare, and was dissolved in phosphate-buffered saline (PBS) to a final concentration of 10 mg/ml. The DA protein was analyzed using SDS-PAGE under reducing conditions and stained with Coomassie brilliant blue. Before immunization, the DA protein was attenuated using a solution of 0.125% (final concentration) glutaraldehyde (GA) (Sigma) in the dark for 1 h at room temperature (25°C). For immunizing female White Leghorn chicken by intramuscular injection, the first immunization dose was 100 μg of the DA protein in 500 μl PBS with an equal volume of Freund's complete adjuvant, and the subsequent immunization doses each were 80 μg of the DA protein in incomplete adjuvant, administered at intervals of 7 days. Eggs from preimmunization, each immunization, and each month after the seventh immunization were collected, and the polyclonal IgY antibodies were purified from the egg yolk using dextran sulfate and sodium sulfate as described previously (31, 32).
Construction of phage display scFv antibody libraries and biopanning.
The antibody library was constructed as described in detail previously (33, 34). In brief, chicken spleens were harvested after the last immunization and homogenized in 5 ml of TRIzol solution (Invitrogen) to extract RNA according to the manufacturer's protocol, and 20 μg of the total RNA was used to synthesize first-strand cDNA by using HiScript I reverse transcriptase (Bionovas, Canada). The variable regions of immunoglobulin light and heavy chains (VL and VH) were amplified using chicken-specific primers, and the chains were linked with a short (scFv-S) or long (scFv-L) linker to form the full-length scFv antibodies by performing a second round of PCR. The scFv fragments were digested with SfiI (New England Biolabs) and cloned into the pComb3X vector. Recombinant phagemids were transformed into ER2738 Escherichia coli through electroporation (MicroPulser from Bio-Rad), and the bacteria then were infected with the VCS-M13 helper phage. After overnight culture, the phages in the supernatant were precipitated using 4% polyethylene glycol 8000 and 3% NaCl. After centrifugation, the phages were resuspended in PBS containing 1% bovine serum albumin (BSA) and 20% glycerol for storage at −20°C.
Biopanning of antibody libraries against the DA antigen was performed using 96-well microplates. In brief, wells were coated with the DA protein (0.5 μg/well) overnight at 4°C and then blocked with 3% BSA in PBS for 1 h at 37°C. The recombinant phage display scFv antibody libraries were added at 1011 to 1012 PFU, and the plates were incubated for 2 h at 37°C. After washing to remove unbound phages by PBST (PBS containing 0.05% Tween 20), bound phages were eluted using 0.1 M glycine-HCl (pH 2.2) and neutralized with 2 M Tris-base buffer. Small amounts of eluted phages after each round of biopanning were used to calculate the eluted phage titers. The remaining eluted phages were used to infect E. coli ER2738 to amplify the phages, and the amplified phages were collected as described previously. Four rounds of biopanning were performed; amplified phages from each round were used to carry out the phage-based enzyme-linked immunosorbent assay (ELISA).
Protein expression and purification of scFv antibodies.
After biopanning, purified total phagemid DNA from E. coli was transformed into TOP10F′ E. coli. Randomly selected clones that had been cultured overnight were diluted 1:100 in super broth containing 1 mM MgCl2 and ampicillin (50 μg/ml) and were incubated for 8 h. After adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and induction overnight, the bacteria were harvested through centrifugation, resuspended in histidine (His)-binding buffer (20 mM sodium phosphate, 0.5 M NaCl, 20 mM imidazole, pH 7.4), and lysed by sonication for SDS-PAGE and Western blot analysis. The scFv antibodies were purified using Ni2+ Sepharose (GE Healthcare Bio-Sciences AB, Sweden) according to the manufacturer's instructions. The purified scFv antibodies were further concentrated, their buffer was replaced with PBS by using Amicon Ultra-4 Centrifugal Filter Devices (Merck Millipore, Germany), and they were used for binding assays in Western blotting, ELISA, and neutralization assay in vivo.
Western blotting.
DA protein was immobilized on polyvinylidene fluoride (PVDF) membranes (Amersham Biosciences, United Kingdom) after SDS-PAGE. The PVDF membranes were blocked with 5% skim milk in PBS for 1 h at 25°C. On the positive-control blot, therapeutic horse antivenin from the CDC at a 1:1,000 dilution was added, and the blot was incubated for 1 h at 25°C and washed three times with PBST. Bound antibodies were detected by adding horseradish peroxidase (HRP)-conjugated goat anti-horse IgG (Jackson ImmunoResearch). After three washes, the membranes were developed using the diaminobenzidine (DAB) substrate until the desired intensity was reached. For the IgY binding assay, purified IgY at a 1:1,000 dilution was used and detected using HRP-conjugated donkey anti-chicken IgY (Jackson ImmunoResearch). For determining the scFv binding activity, purified scFvs (1 μg/ml) were added to six snake venom proteins, including DA, BM (Bungarus multicinctus), TS (Trimeresurus stejnegeri), TM (Trimersurus mucrosquamatus), NNA (Naja naja atra), and DRF (Daboia russellii formosensis), from the CDC that were immobilized on PVDF membranes. After incubation, the goat anti-chicken light-chain IgG (Bethyl) and HRP-conjugated donkey anti-goat IgG (Jackson ImmunoResearch) were used. Steps including blocking, washing, incubation, and color development were performed under the same conditions as those described previously.
ELISA and competitive ELISA.
To analyze the IgY binding activity, ELISA wells were coated with the DA protein (0.5 μg/well) overnight at 4°C and blocked with 5% skim milk in PBS for 1 h at 37°C. The preimmunization- or seventh-immunization-induced IgY was 2-fold serially diluted from 500× to 256,000×, added to the ELISA wells, and incubated for 1 h at 37°C. After removing unbound IgY, the wells were washed with PBST six times, the HRP-conjugated donkey anti-chicken IgY was added, and the wells incubated for another 1 h. After washing to remove unbound antibodies, the wells were developed by adding 3,3′,5,5′-tetramethylbenzidine (TMB), and the reaction was stopped by adding HCl. On phage-based ELISA, amplified phages from each round of biopanning were added at 1011 to 1012 PFU as binders for 1 h at 37°C after coating the wells with DA protein and blocking. After washing, the HRP-conjugated mouse anti-M13 antibody (Amersham Biosciences) was added for detection. For the competitive ELISA, the DA protein was 2-fold serially diluted, from 400 μg/ml to 0.39 μg/ml, and mixed with the scFv antibody (0.05 μg/ml) at a 1:1 ratio. After incubation for 1 h at 25°C, the mixed agents were added to the plates, which had immobilized DA protein and were blocked with 5% skim milk in PBS, and the plates were incubated for another 1 h at 37°C. After washing, goat anti-chicken light-chain antibodies and HRP-conjugated donkey anti-goat IgG were added. For the specific binding assay, purified scFv (1 μg/ml) was incubated with six snake venom proteins (DA, BM, TS, TM, NNA, and DRF) immobilized on ELISA plates. The subsequent procedure was the same as that described previously for ELISA for detecting scFv antibodies. Steps including blocking, washing, incubation, and color development were performed under the same conditions as those described above. All ELISA analyses were performed in duplicate.
Sequence analysis.
The nucleotide sequences of light and heavy chains from randomly selected clones were determined with an autosequencer machine (ABI 3730 XL) by using ompseq (5′-AAGACAGCTATCGCGATTGCAGTG-3′) and HRML-F (5′-GGTGGTTCCTCTAGATCTTCC-3′) primers. The sequence results were translated into amino acid sequences and aligned with the chicken immunoglobulin germ line gene by using the BioEdit program.
Mass spectrometric analysis.
DA protein was analyzed using SDS-PAGE under reducing or nonreducing conditions and stained with Coomassie brilliant blue. The bands, which were recognized by monoclonal scFv antibodies by Western blotting, were cut from the SDS-PAGE gel to perform the mass spectrometry LTQ Orbitrap XL MS (Thermo Fisher) analysis. The database used was Swiss-Prot 2011 (533,049 sequences; 189,064,225 residues).
Neutralization assay of chicken antibodies against DA snake venom protein in vivo.
The ICR mice (12 to 14 g) were distributed into groups of nine each. The minimum lethal dose (MLD) was tested according to the method described by the CDC, Taiwan. Various amounts of DA venom (66.5 μg, 99.75 μg [1.5 times the 66.5-μg amount], and 133 μg [2.0 times the 66.5-μg amount]) were dissolved in 200 μl PBS, incubated at 37°C for 1 h, and injected intraperitoneally. PBS (200 μl) alone was used as a negative control. The 99.75-μg amount of DA caused the death of all mice. Therefore, 99.75 μg DA venom was combined with 4 mg of preimmunization or immunization-induced polyclonal IgY antibodies or with 4 mg of therapeutic horse antivenin, mixed with scFv at a low dose of 1 mg or a high dose of 4 mg in 200 μl PBS, and incubated at 37°C for 1 h. Subsequently, the mixed agents were injected intraperitoneally into mice. The mice were monitored for survival at 1-h intervals for 36 h.
RESULTS
Characterization of DA venom protein and chicken polyclonal IgY.
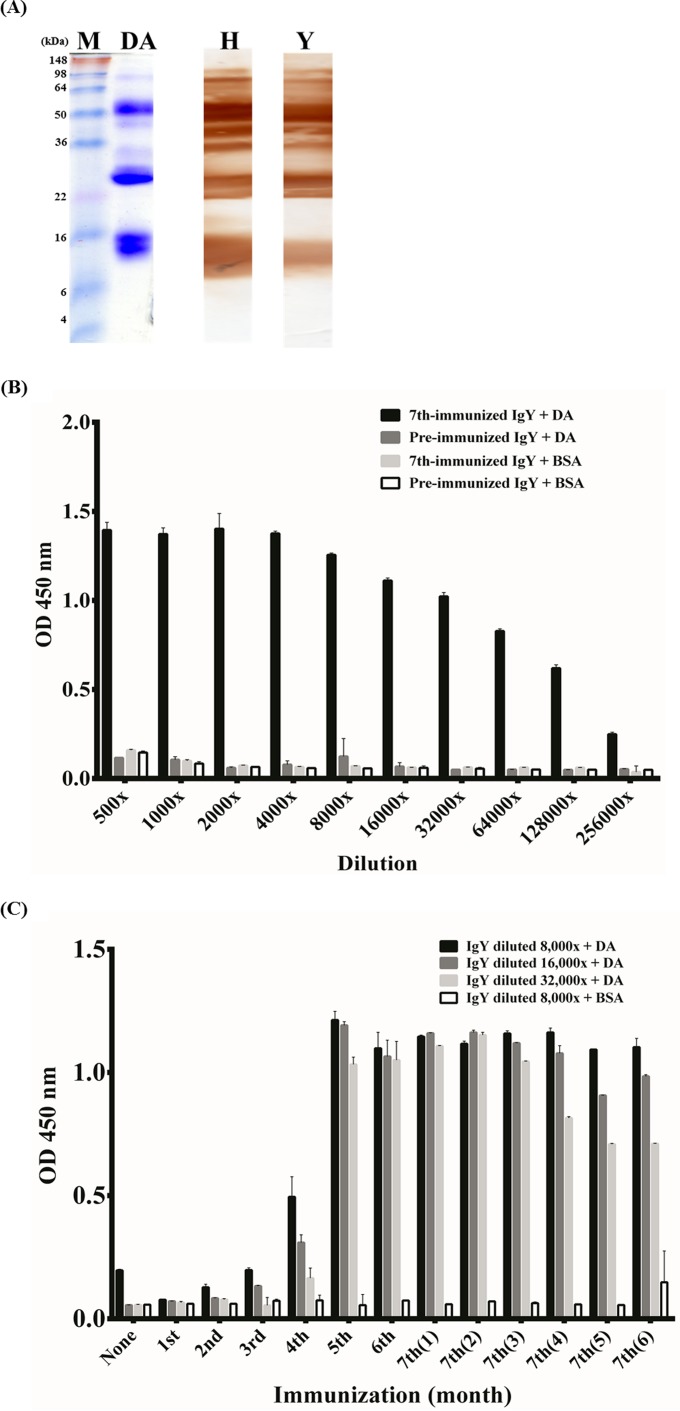
DA snake venom was kindly provided by the CDC and used to immunize chickens. After electrophoresis under reducing conditions and staining the SDS-PAGE gel with Coomassie brilliant blue, the DA protein was observed to contain a complex of proteins (Fig. 1A). The PAGE showed that four of the proteins, two with molecular masses of approximately 50 and 25 kDa and two with molecular masses of approximately 15 kDa, and were detected at high concentrations. After analyzing purified IgY from immunized chicken eggs (data not shown), we used IgY antibodies as the primary antibody to detect the DA protein (Fig. 1A). According to these data, this anti-DA IgY (lane Y) could recognize the DA protein and showed binding patterns similar to those of the therapeutic horse antivenin (lane H) by Western blotting. The ELISA analysis also showed that anti-DA IgY had a strong binding ability (with optical densities [ODs] exceeding 0.8) up to a 64,000× dilution, an intermediate level (ODs between 0.4 and 0.8) of binding ability at 128,000× dilution, and a weak reaction (ODs between 0.2 and 0.4) at 256,000× dilution; however, no signal was obtained with BSA (Fig. 1B). In contrast, no reactions were observed between the purified preimmunized IgY against the DA venom or BSA protein (ODs below 0.2). To understand the immune response in chicken, ELISA was performed to analyze the purified IgY from preimmunization, each immunization, and each month after the seventh booster (Fig. 1C). The ELISA data indicated that specific IgY antibodies were elicited after the fifth booster, and the titers reached a plateau. This suggested that a strong humoral antibody response was elicited in chicken. It also showed that the immune response lasted for at least 3 months after the seventh booster and started decreasing in the fourth month.
FIG 1.
Binding activity of chicken polyclonal IgY against DA venom proteins by using Western blotting and ELISA. (A) After separating proteins using SDS-PAGE under a reducing condition, DA protein was stained with Coomassie brilliant blue dye (lane DA). After being transferred onto the PVDF membrane, the DA proteins were detected using horse antivenin as the positive control (lane H) or using polyclonal IgY from the seventh immunization (lane Y). (B) The ELISA plates were coated with DA or BSA proteins. Purified IgY antibodies were 2-fold serially diluted and added to the wells to test their binding specificity. (C) The antibody response in chicken was monitored for 6 months after the seventh immunization. The IgY from the seventh immunization was diluted 8,000×, 16,000×, or 32,000× and incubated with DA. The 8,000×-diluted IgY was incubated with BSA as a control. ELISA data were represented as the means from duplicate experiments.
Construction of the scFv antibody library and biopanning of the recombinant phage display scFv antibody library.
To construct the antibody library, chicken was sacrificed after final immunization, and total RNA was extracted from the enlarged spleen to synthesize first-strand cDNA. The VH (approximately 400 bp) with a short or long linker or VL (approximately 350 bp) was successfully amplified using specific primers and joined to form full-length scFv gene fragments with short (scFv-S) or long (scFv-L) linkers (approximately 750 bp) by using an overlap extension PCR (data not shown). Thus, two libraries, scFv-S and scFv-L, containing 6.24 × 108 and 5.28 × 108 transformants, respectively, were constructed and infected by M13 helper phage to form phage display scFv antibody libraries.
To select high-affinity DA-specific scFv antibodies, the constructed phage libraries were subjected to four rounds of biopanning. Eluted phage titers were determined by infecting E. coli after each round of biopanning. Compared with the first eluted titer containing 104 CFU, the second eluted titer decreased to approximately 103 CFU, and the third eluted titer returned to approximately 104 CFU. The fourth eluted titer increased to 105 CFU. To confirm the biopanning results, a phage-based ELISA was performed. All amplified phages were used as detecting binders for recognizing the DA protein, and the signals of the phage-based ELISA revealed that the specific phage clones were amplified after the second round of biopanning (Fig. 2). Both eluted phage titers and the phage-based ELISA data demonstrated that high-affinity specific clones were successfully enriched through the biopanning procedure.
FIG 2.
Specific binding assay of total amplified phages after each round of biopanning. Amplified phages, containing 1011 to 1012 PFU, were used as detection binders against the DA proteins or BSA on phage-based ELISA. Purified IgY was used as a positive control. ELISA data are represented as the means from duplicate experiments.
Expression and sequence analysis of scFv antibodies.
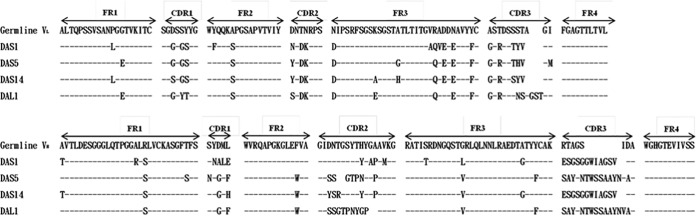
After biopanning and transforming the total DNA into E. coli TOP10F′, 20 clones were randomly selected from the short and long linker libraries for analyzing protein expression, which was confirmed using Western blotting. All of these clones expressed scFv antibodies of various sizes and patterns (data not shown). Fifteen clones expressing scFv antibodies randomly selected from the short or long linker library were analyzed to predict the amino acid sequence. BLAST alignment analyses were performed for each VL and VH sequence with their respective chicken germ line sequences. They were classified into three short linker groups, groups 1 (60%), 2 (33.33%), and 3 (6.67%), and one dominantly expressed long linker group (Table 1). The first clones from each short linker group were named DAS1, DAS5, and DAS14, and that from the long linker group was named DAL1. The amino acid sequences in framework regions (FRs) of VL and VH were rarely variable compared with those of the chicken germ line (Fig. 3). Compared with the FRs, complementarity-determining regions (CDRs) of VL and VH showed high variability. In the VL region, their CDRs showed about 46% variation in DAS1 and DAS14 and 50% variation in DAS5 and DAL1. On the other hand, the VH region analysis showed more than 70% variation (DAS1, 73%; DAS5, 83%; DAS14, 70%; and DAL1, 90%). The lengths in CDR3 of VL and VH also differed. The respective VL CDR1, CDR2, and CDR3 coding sequences of the four clones were similar, except that DAL1 had three amino acids more in CDR3 than the other clones. These clones showed similar VH CDR1 coding sequences, except DAS1, which showed more variability. The clones also showed high variability in VH CDR2 and CDR3 compared with that of the chicken germ line. In VH CDR2, DAS1 and DAS14 had the same lengths of these regions as those of the chicken germ line, whereas DAS5 and DAL1 had two fewer amino acids at different positions. Otherwise, DAS1 and DAS14 had the same coding sequences in VH CDR3. In addition, DAS5 and DAL1 differed in only one amino acid at position 14 in VH CDR3. Because CDR3 had almost the same sequence and more variability, the activity of VH CDR3 against the DA protein was assumed to be crucial. These observations suggest that the sequences of these four selected scFv monoclonal antibodies all were generated from the antigen-induced immune response and were not directly selected from naive IgM-like sequences.
TABLE 1.
Classification of anti-DA scFv clones according to the identity of heavy- and light-chain variable regions
| Group | Variable regions in: |
|||||
|---|---|---|---|---|---|---|
| Short linker |
Long linker |
|||||
| Light chain | Heavy chain | Proportion (%) of all regions | Light chain | Heavy chain | Proportion (%) of all regions | |
| 1 | 1, 2, 3, 4, 6, 7, 8, 12, 13 | 1, 2, 3, 4, 6, 7, 8, 12, 13 | 60 | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15 | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15 | 100 |
| 2 | 5, 9, 10, 11, 15 | 5, 9, 10, 11, 15 | 33.33 | |||
| 3 | 14 | 14 | 6.67 | |||
FIG 3.
Sequence alignment of variable regions of immunoglobulin light- and heavy-chain domains of anti-DA scFv antibodies. Nucleotide sequences of 15 scFv-S or 15 scFv-L clones randomly selected from the libraries after four rounds of biopanning were analyzed. The amino acid sequences predicted from DNA were aligned with those of the chicken germ line. Classification of the clones based on light- and heavy-chain variable regions are summarized in Table 1 and named according to the first clone of each group. Sequence gaps were introduced to maximize the alignment by blank space. Dashes indicate identical sequences. FR and CDR boundaries are indicated above the germ line sequences.
Purification of scFv antibodies and their binding assay by using competitive ELISA.
We used Ni2+ Sepharose to purify the expressed scFv antibodies, which were fused with His tags. These four scFvs, DAS1, DAS5, DAS14, and DAL1, were successfully purified and showed different patterns by Western blot assay (Fig. 4A). For the binding activity assay, a competitive ELISA was carried out. The four scFv antibodies, with NNAS1 (scFv against NNA venom protein) as a negative control, were premixed with the free form of the DA protein and added to the wells. The four scFv antibodies showed different binding activities with DA protein based on different rates of absorbance reduction with increasing amounts of the free form of the DA protein (Fig. 4B). The negative control, NNAS1, showed no difference. By using the absence of DA free-form protein as a standard, the four scFv antibodies showed different binding affinities with 50% inhibitory effects as indicated by the absorbance values. DAS1 and DAS14 showed 72% and 83% inhibitory effects, respectively, under a concentration of 200 μg/ml of DA free-form protein. DAL1 showed the strongest binding affinity, with 55% inhibitory effects under a concentration of only 1.56 μg/ml of DA free-form protein, whereas the affinity of DAS5 resulted in only 46% inhibitory effects even at a concentration of 200 μg/ml DA free-form protein.
FIG 4.
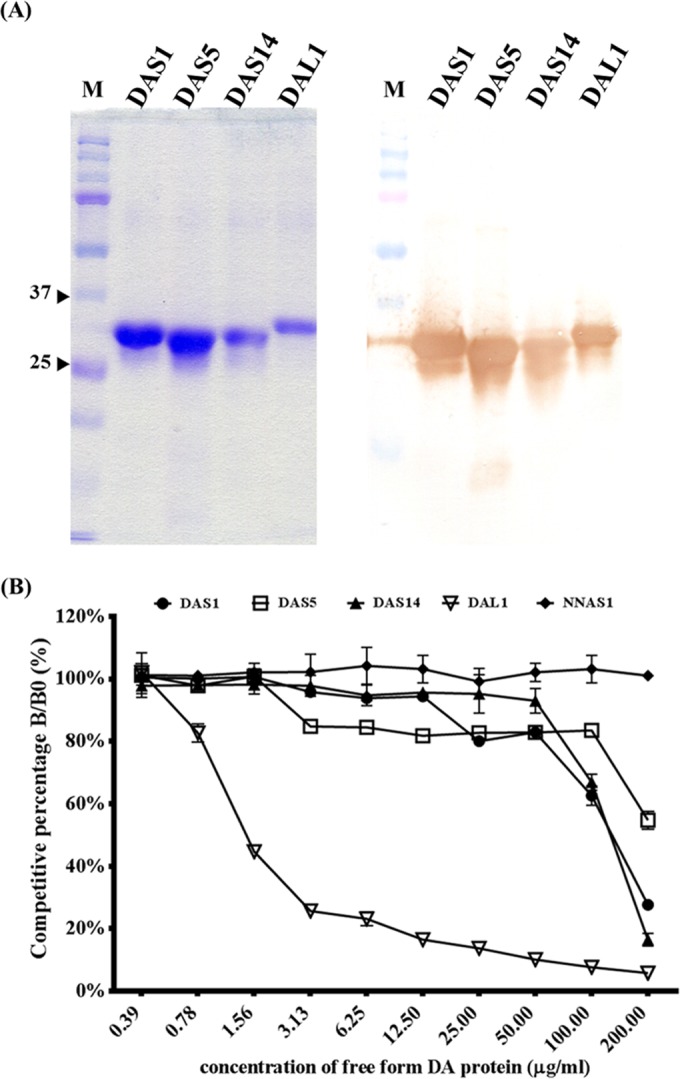
Protein expression and purification of anti-DA scFv antibodies and binding activity assay using competitive ELISA. (A, left) After expression in E. coli, the four His-fused scFv antibodies (lanes DAS1, DAS5, DAS14, and DAL1) were purified using Ni2+ Sepharose and analyzed by Coomassie brilliant blue-stained SDS-PAGE. Molecular masses (in kDa) are shown on the left. (A, right) Their identities were further confirmed by probing with goat anti-chicken light antibody by Western blotting. (B) These four scFv antibodies, along with NNAS1 (scFv against NNA venom protein) as the negative control, were premixed with different concentrations of DA proteins and added to the wells containing immobilized DA protein. B and B0 indicate the amounts of bound scFv in the presence and absence of DA protein, respectively. ELISA data are represented as the means from duplicate experiments.
Specific binding assay of anti-DA scFv antibodies with six snake venom proteins.
For the specific binding assay, venom proteins of six major venomous snake species in Taiwan were kindly provided by the CDC. These four monoclonal scFv antibodies were used as detecting antibodies for ELISA and Western blot assay. On ELISA, four scFv monoclonal antibodies strongly and specifically (ODs above 0.8) recognized the DA venom protein but not the other venom proteins (ODs below 0.2) (Fig. 5A). Even the TS and TM snake venom proteins, which also cause hemorrhagic symptoms resulting in death, could not be recognized by the scFv antibodies. Similarly, on Western blot assay, all antibodies demonstrated a high affinity and specificity to a protein of approximately 50 kDa in DA protein but not to the other snake venom proteins (Fig. 5B). Taking these findings together, these four monoclonal antibodies likely recognized the same protein in the DA snake venom protein. The results suggested that these four selected scFv antibodies against the DA protein had specific binding activities.
FIG 5.
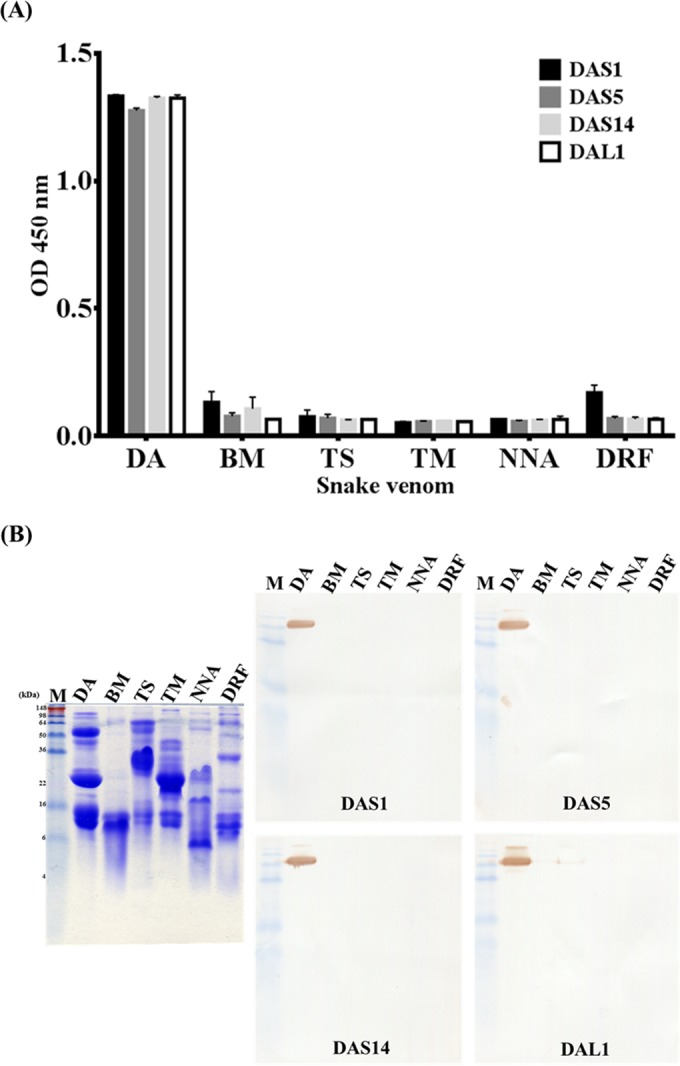
Specific binding assay of anti-DA scFv antibodies with six snake venom proteins by Western blotting and ELISA. Snake venom proteins from six species (Deinagkistrodon acutus [DA], Bungarus multicinctus [BM], Trimeresurus stejnegeri [TS], Trimersurus mucrosquamatus [TM], Naja naja atra [NNA], and Daboia russellii formosensis [DRF]) were immobilized on ELISA plates (A) or PVDF membranes (B). The four purified scFv antibodies containing DAS1, DAS5, DAS14, and DAL1 were used as primary antibodies at 1 μg/ml. ELISA data are represented as the means from duplicate experiments.
Mass spectrometry.
To predict which protein was recognized by the scFv antibodies, we analyzed the DA protein using SDS-PAGE under reducing or nonreducing conditions (arrows mark bands from the SDS-PAGE gel according to the Western blot results used to perform mass spectrometry) (Fig. 6). The matched amino acid sequences are shown in boldface, and the percentages were the following: m1, 24%; m2, 37%; m3, 24% (see Fig. S1 in the supplemental material). All three results indicated that the scFv antibodies recognize the zinc metalloproteinase-disintegrin acutolysin, also called SVMP, of the DA snake venom protein. The details of amino acids matched with SVMP are provided in the supplemental material.
FIG 6.
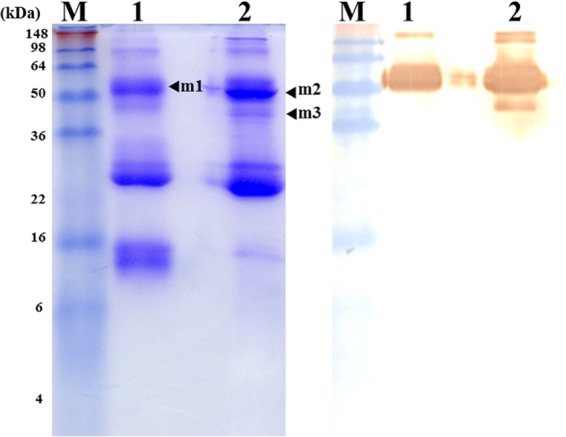
Mass spectrometric verification of the DA venom protein recognized using scFv antibodies. The major protein bands, including m1, m2, and m3 (left), recognized using anti-DA scFv antibodies by Western blotting (right) under reducing (lane 1) or nonreducing (lane 2) conditions were severed from the SDS-PAGE gel (left) and used to perform a mass spectrometric analysis. According to alignment with the Swiss-Prot 2011 database, the matched amino acid sequences (shown in Fig. S1) may belong to the zinc metalloproteinase-disintegrin acutolysin.
Neutralization assay of antibodies against DA proteins in vivo.
The average MLD of DA protein which had been determined by the CDC was 66.5 μg. Thus, to confirm the MLD, we tested multiple doses containing 66.5 μg, 99.75 μg, and 133 μg of DA protein by intraperitoneal injection; PBS alone was used as a control (Fig. 7A). The 66.5-μg dose of DA protein resulted in the death of two mice within 1 h and that of five mice within 2 h, whereas two mice survived. The 99.75-μg and 133-μg doses of DA protein resulted in the death of all mice within 1 h. The control PBS-treated mice all survived. Thus, the 99.75-μg dose of DA protein was used in the ensuing study. To further test whether chickens are suitable animal models for producing neutralizing antibodies against snake venom, polyclonal IgY antibodies purified from the eggs before immunization or after the seventh immunization, or therapeutic horse antivenin, was mixed with the DA snake venom protein, incubated for 1 h at 37°C, and then injected intraperitoneally (Fig. 7B). Not only the therapeutic horse antivenin but also the immunization-induced IgY protected the mice from the lethal DA snake venom protein treatment. In contrast, the preimmunized IgY had no protective effect. To test the effect of monoclonal antibodies, 4 mg or 1 mg of the mixture of the four monoclonal scFv antibodies then was incubated with the DA protein for 1 h at 37°C and then injected intraperitoneally (Fig. 7C). The results showed that the 4-mg dose of the mixed scFv antibodies inhibited the lethal effect in mice injected with the DA venom protein, except that only two mice died within 5 and 8 h, respectively. Even the 1-mg dose of the scFv antibodies showed a partial protective effect based on the result that one of the mice had its life prolonged for 1 h, two of them had their lives prolonged for 3 h, two of them had their lives prolonged for 4 h, two of them had their lives prolonged for 10 h, and one of them survived venom treatment. The in vivo data indicated that the mixture of the four scFv antibodies could increase the survival rate of mice treated with the lethal dose of the DA venom protein.
FIG 7.
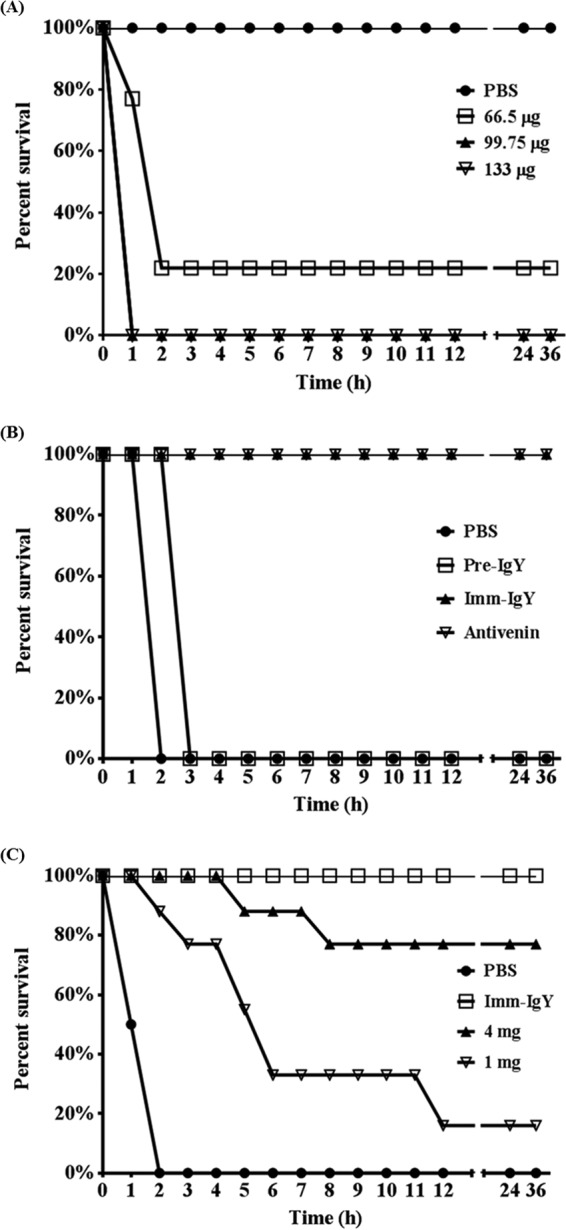
Neutralization assay of anti-DA antibodies against the DA venom proteins in vivo. (A) Groups of nine ICR mice were challenged with various doses of DA venom protein, including 66.5, 99.75, and 133 μg, by intraperitoneal injection to determine the MLD. (B) Preimmunized purified polyclonal IgY (Pre-IgY) antibodies and IgY after the seventh immunization (Imm-IgY), along with horse antivenin as a control, in an amount of 4 mg were mixed with the DA venom proteins, incubated for 1 h at 37°C, and used to challenge mice. (C) The mixture of the four purified scFv antibodies at a dose of 4 mg or 1 mg was mixed with the DA venom proteins, incubated for 1 h at 37°C, and used to challenge mice. Immunization-induced IgY was used as the control. All mice were monitored for survival at 1-h intervals for 36 h.
DISCUSSION
Taiwan has a tropical-subtropical climate with conditions suitable for the proliferation of many flora and fauna, including snakes (35). Poisoning from venomous snakebites is a global public health issue because of the resulting high mortality rate. At present, the main treatment for the bites of venomous snakes is purified and refined horse serum. However, producing antivenin from horses is complex in process and high in cost, and it may have detrimental side effects (9). Thus, developing an alternative therapy is imperative. The objective of this study was to present alternative treatments for venomous snakebites using polyclonal and monoclonal antibodies, which also would allow rapid diagnosis to help in therapy.
In the generation of antibodies, whether the process of antibody purification from animals is cost-effective or not is considered important (36). In our study, the collection of antibodies from chickens as the animal model was easier than collecting them from horses. As recommended by WHO procedures (www.who.int/bloodproducts/snakeantivenoms), in order to collect blood from horses by the venipuncture of the external jugular vein, areas with appropriate restraining devices, precautions for preventing infection, and a reinfusion of erythrocytes within 24 h after collecting and separating plasma from the whole blood are required. Therefore, using horses to generate antivenin is troublesome, and chickens have emerged as a superior option. In contrast to horses, the process of purifying IgY from chicken eggs, which are collected daily, is noninvasive, simple, and fast. In addition, IgY does not react with rheumatoid and the complement system that will cause injury to the body (37, 38). Thus, chickens are superior to other animals as an animal model to generate antibodies. In addition, snake venom proteins are difficult to collect because of numerous regionally adopted restrictions on venom collection. Thus, an effective technique is required to produce therapeutic antibodies for neutralizing snake venom protein. Before immunization, we used GA, which not only can attenuate the venom protein to prevent animal mortality but also is a useful adjuvant for increasing antigenicity to elicit an immune response more effectively to attenuate venom protein (3). In our study, we used about 800 μg/chicken venom protein for seven immunizations in 2 months. However, the procedure published by the WHO for immunizing horses requires about 15 to 35 mg/horse for 4 or 5 immunizations in 2 months. In the in vitro results, IgY showed binding patterns similar to those of horse antivenin containing the neutralizing antibodies (Fig. 1A). In addition, stronger immune responses were elicited in chicken, although the immune response only lasted for 3 months (Fig. 1B and C). In our other studies, the immune response in chickens could last at least 6 months. This may have been due to the weak immune response in this chicken or weak antigenicity of the DA proteins. Most importantly, immunization-induced IgY antibodies protected mice from the MLD of DA venom protein as effectively as the therapeutic horse antivenin (Fig. 7B). However, further investigation of the comparisons of the minimum required neutralization doses between IgY and horse therapeutic antivenin were not possible because of the difficulties faced in collecting venom proteins. Despite this restriction, we observed that chickens required a very small amount of venom protein for generating neutralizing polyclonal IgY antibodies against DA snake envenomation.
Many monoclonal antibodies, including scFvs, have been generated against a broad range of tumor-associated antigens or snake venom proteins by using phage display technology (30, 39). At minimal expense or time, recombinant antibodies fit the criteria more readily than traditional hybridoma, and the chicken scFv system is one of the simplest to exploit (33). We constructed two libraries from immunized chicken by PCR using 7- or 18-amino-acid peptide linker systems containing 6.24 × 108 and 5.28 × 108 transformants, respectively. In contrast, in order to generate effective naive antibody libraries, the size of library required to produce highly specific antibodies by using hyperimmune animals as the source of immunoglobulin cDNA was reduced (40). In this study, constructed antibody libraries proved highly effective, with specific panning responses observed on phage-based ELISA after four rounds of panning selection (Fig. 2) and yielding four antibodies against DA protein (Fig. 4). This illustrated that immunized libraries are more effective and rapid for selecting antibodies from the population, as most naive libraries carry out 4 to 6 rounds or more of biopanning to yield specific clones (41). Otherwise, biopanning and confirming the scFv binding activity took only 2 to 3 weeks in our study, which was faster than the hybridoma technology, which took 1 to 2 months (42). This indicates that the phage display system is fast and effective in selecting specific antibodies from antibody libraries with different combinations of variable regions of light chains and heavy chains. The amino acid sequences of the selected scFv antibodies, as predicted by comparison to the chicken germ line sequences, showed that CDRs had higher mutation rates than FRs; the mutation rates were higher than those of the chicken germ line, particularly in VH CDR3 (Fig. 3) (43). These high mutation rates demonstrated that these scFvs were derived from the DA antigen-induced immune response and were not directly selected from naive IgM sequences.
In contrast to polyclonal IgY antibodies, which were not specific to their corresponding snake venom (data not shown), the monoclonal antibodies, which are more specific, are necessary for developing rapid diagnostic reagents. In our study, both the ELISA and Western blot assay indicated that the four scFv antibodies recognized the DA protein specifically (Fig. 5). Although TS and TM also result in the same hemorrhagic symptoms as those of DA protein, the four scFv antibodies did not bind to them. In addition, the scFv antibodies were expressed in E. coli systems, making them cheaper, faster, and easier to produce and also allowing the preservation of the clones; the purified recombinant DNA needs to be preserved instead of monoclonal antibodies from hybridoma cells. It was suggested that diagnostic agents for the rapid diagnosis of snake venom from wound exudates resulting from venomous snakebites can be developed (44, 45). Otherwise, antibody affinity is crucial in diagnosis and therapy. By combining the results of the competitive ELISA and mass spectrometry, we could infer the dissociation constant (Kd) values of these four scFv antibodies based on the Klotz plot method (46). A study involving a cDNA library construction of the venom gland and its analysis indicated that metalloproteinase constituted approximately 30% of all assembled cDNAs in Bitis gabonica, which also belongs to the Viperidae family (47). Using ImageJ software, we deduced that the metalloproteinase constituted approximately 35% of the DA protein. Thus, based on the competitive ELISA results (Fig. 4B), we speculated that the amounts of DAS1, DAS5, DAS14, and DAL1 required for 50% inhibitory effects of the metalloproteinase were 47.45, 84.76, 45.30, and 0.69 μg/ml, respectively. Thus, the calculated Kd values of the four scFv antibodies, DAS1, DAS5, DAS14, and DAL1, were 7.08 × 10−7, 1.27 × 10−6, 6.76 × 10−7, and 1.03 × 10−8 M, respectively. However, the pure metalloproteinase and a more accurate method for determining Kd values are required to verify the binding affinity.
The mass spectrometry data suggested that the scFv-bound protein was zinc metalloproteinase-disintegrin acutolysin (approximately 67 kDa) of the DA venom protein (Fig. 6). SVMP are abundant in venom of snakes of the family Viperidae and are classified into P-I, P-II, and P-III classes according to their domain organization (48, 49). They are considered one of the major primary hemorrhagic toxic factors that interfere with the hemostatic system of the prey and increase blood loss. The P-III class showed more hemorrhagic and diverse biological activities than P-I and P-II (8, 49–51). Hemorrhagic P-III SVMPs, which comprise the metalloprotease domain and disintegrin-like (Dis) domain, followed by a cysteine-rich (CR) domain, gather in capillary blood and vessels by binding to the basement membrane through the Dis and CR domains to disrupt the microvascular system very effectively (52). In addition, the noncatalytic Dis and CR domains containing substrate binding sites were considered to confer greater hemorrhagic activity on P-III SVMPs (53). In our study, the mixture of the four scFv antibodies revealed the inhibition of the lethal effect of the DA venom protein in in vivo neutralization assays (Fig. 7C). Above all, we suggest that the anti-DA scFv antibodies bind to the Dis or CR domain to inhibit their hemorrhagic activities to protect mice. However, because of insufficient DA protein, further investigation of how these four scFv antibodies work in vivo was hard to implement. Further investigations also are required to determine the active binding sites of scFv antibodies that play a role in their inhibitory effects on the DA protein.
In summary, we used chickens as the animal model because it is cheaper, more convenient, and faster to generate polyclonal IgY antibodies, which are effective in neutralizing the DA venom. We constructed two scFv libraries from immunized chicken and successfully isolated four monoclonal antibodies by using phage display technology. The four scFv antibodies which recognized the SVMP based on the mass spectrometry results not only recognized the DA protein specifically but also inhibited the lethal effect of the DA venom. This indicates that chickens are more cost-effective than horses in generating antivenin. In addition, using phage display technology to generate monoclonal antibodies is more efficient in terms of cost and time. Taking the findings of this study together, we anticipate the application of polyclonal and monoclonal antibodies to the development of diagnostic agents or treatments for snakebite envenomation.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Center for Research, Diagnostics and Vaccine Development, Centers for Disease Control, Ministry of Health and Welfare, Taiwan, for providing the snake venom proteins.
We have no financial or commercial conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02608-15.
REFERENCES
- 1.Sajevic T, Leonardi A, Krizaj I. 2011. Haemostatically active proteins in snake venoms. Toxicon 57:627–645. doi: 10.1016/j.toxicon.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Aird SD. 2002. Ophidian envenomation strategies and the role of purines. Toxicon 40:335–393. doi: 10.1016/S0041-0101(01)00232-X. [DOI] [PubMed] [Google Scholar]
- 3.Ming-Yi L, Ruey-Jen H. 1997. Toxoids and antivenoms of venomous snakes in Taiwan. J Toxicol Toxin Rev 16:163–175. doi: 10.3109/15569549709016453. [DOI] [Google Scholar]
- 4.Xu X, Zhang L, Luo Z, Shen D, Wu H, Peng L, Song J, Zhang Y. 2010. Metal ions binding to NAD-glycohydrolase from the venom of Agkistrodon acutus: regulation of multicatalytic activity. Metallomics 2:480–489. doi: 10.1039/c0mt00017e. [DOI] [PubMed] [Google Scholar]
- 5.Ouyang C, Teng CM, Huang TF. 1990. Characterization of snake venom principles affecting blood coagulation and platelet aggregation. Adv Exp Med Biol 281:151–163. doi: 10.1007/978-1-4615-3806-6_15. [DOI] [PubMed] [Google Scholar]
- 6.Tu AT. 1996. Overview of snake venom chemistry. Adv Exp Med Biol 391:37–62. doi: 10.1007/978-1-4613-0361-9_3. [DOI] [PubMed] [Google Scholar]
- 7.Ouyang C, Teng CM, Huang TF. 1982. Characterization of the purified principles of Formosan snake venoms which affect blood coagulation and platelet aggregation. Taiwan Yi Xue Hui Za Zhi 81:781–790. [PubMed] [Google Scholar]
- 8.Markland FS Jr, Swenson S. 2013. Snake venom metalloproteinases. Toxicon 62:3–18. doi: 10.1016/j.toxicon.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Gold BS, Dart RC, Barish RA. 2002. Bites of venomous snakes. N Engl J Med 347:347–356. doi: 10.1056/NEJMra013477. [DOI] [PubMed] [Google Scholar]
- 10.Warr GW, Magor KE, Higgins DA. 1995. IgY: clues to the origins of modern antibodies. Immunol Today 16:392–398. doi: 10.1016/0167-5699(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 11.Gassmann M, Thommes P, Weiser T, Hubscher U. 1990. Efficient production of chicken egg yolk antibodies against a conserved mammalian protein. FASEB J 4:2528–2532. [DOI] [PubMed] [Google Scholar]
- 12.Hatta H, Tsuda K, Akachi S, Kim M, Yamamoto T. 1993. Productivity and some properties of egg yolk antibody (IgY) against human rotavirus compared with rabbit IgG. Biosci Biotechnol Biochem 57:450–454. doi: 10.1271/bbb.57.450. [DOI] [PubMed] [Google Scholar]
- 13.Hatta H, Kim M, Yamamoto T. 1990. A novel isolation method for hen egg yolk antibody, “IgY.” Agric Biol Chem 54:2531–2535. [PubMed] [Google Scholar]
- 14.Schade R, Burger W, Schoneberg T, Schniering A, Schwarzkopf C, Hlinak A, Kobilke H. 1994. Avian egg yolk antibodies. The egg laying capacity of hens following immunisation with antigens of different kind and origin and the efficiency of egg yolk antibodies in comparison to mammalian antibodies. ALTEX 11:75–84. [PubMed] [Google Scholar]
- 15.Mine Y, Kovacs-Nolan J. 2002. Chicken egg yolk antibodies as therapeutics in enteric infectious disease: a review. J Med Food 5:159–169. doi: 10.1089/10966200260398198. [DOI] [PubMed] [Google Scholar]
- 16.Kruger C, Pearson SK, Kodama Y, Vacca Smith A, Bowen WH, Hammarstrom L. 2004. The effects of egg-derived antibodies to glucosyltransferases on dental caries in rats. Caries Res 38:9–14. doi: 10.1159/000073914. [DOI] [PubMed] [Google Scholar]
- 17.Kovacs-Nolan J, Mine Y. 2012. Egg yolk antibodies for passive immunity. Annu Rev Food Sci Technol 3:163–182. doi: 10.1146/annurev-food-022811-101137. [DOI] [PubMed] [Google Scholar]
- 18.Nilsson E, Kollberg H, Johannesson M, Wejaker PE, Carlander D, Larsson A. 2007. More than 10 years' continuous oral treatment with specific immunoglobulin Y for the prevention of Pseudomonas aeruginosa infections: a case report. J Med Food 10:375–378. doi: 10.1089/jmf.2006.214. [DOI] [PubMed] [Google Scholar]
- 19.Nilsson E, Larsson A, Olesen HV, Wejaker PE, Kollberg H. 2008. Good effect of IgY against Pseudomonas aeruginosa infections in cystic fibrosis patients. Pediatr Pulmonol 43:892–899. doi: 10.1002/ppul.20875. [DOI] [PubMed] [Google Scholar]
- 20.Yokoyama K, Sugano N, Shimada T, Shofiqur RA, el Ibrahim SM, Isoda R, Umeda K, Sa NV, Kodama Y, Ito K. 2007. Effects of egg yolk antibody against Porphyromonas gingivalis gingipains in periodontitis patients. J Oral Sci 49:201–206. doi: 10.2334/josnusd.49.201. [DOI] [PubMed] [Google Scholar]
- 21.Smith GP. 1985. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 22.Barbas CF III, Kang AS, Lerner RA, Benkovic SJ. 1991. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci U S A 88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chi XS, Landt Y, Crimmins DL, Dieckgraefe BK, Ladenson JH. 2002. Isolation and characterization of rabbit single chain antibodies to human Reg Ialpha protein. J Immunol Methods 266:197–207. doi: 10.1016/S0022-1759(02)00117-5. [DOI] [PubMed] [Google Scholar]
- 24.Park KJ, Park DW, Kim CH, Han BK, Park TS, Han JY, Lillehoj HS, Kim JK. 2005. Development and characterization of a recombinant chicken single-chain Fv antibody detecting Eimeria acervulina sporozoite antigen. Biotechnol Lett 27:289–295. doi: 10.1007/s10529-005-0682-8. [DOI] [PubMed] [Google Scholar]
- 25.Pavoni E, Flego M, Dupuis ML, Barca S, Petronzelli F, Anastasi AM, D'Alessio V, Pelliccia A, Vaccaro P, Monteriu G, Ascione A, De Santis R, Felici F, Cianfriglia M, Minenkova O. 2006. Selection, affinity maturation, and characterization of a human scFv antibody against CEA protein. BMC Cancer 6:41. doi: 10.1186/1471-2407-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fehrsen J, van Wyngaardt W, Mashau C, Potgieter AC, Chaudhary VK, Gupta A, Jordaan FA, du Plessis DH. 2005. Serogroup-reactive and type-specific detection of bluetongue virus antibodies using chicken scFvs in inhibition ELISAs. J Virol Methods 129:31–39. doi: 10.1016/j.jviromet.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Finlay WJ, Shaw I, Reilly JP, Kane M. 2006. Generation of high-affinity chicken single-chain Fv antibody fragments for measurement of the Pseudonitzschia pungens toxin domoic acid. Appl Environ Microbiol 72:3343–3349. doi: 10.1128/AEM.72.5.3343-3349.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huston JS, Levinson D, Mudgett-Hunter M, Tai MS, Novotny J, Margolies MN, Ridge RJ, Bruccoleri RE, Haber E, Crea R. 1988. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci U S A 85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holliger P, Hudson PJ. 2005. Engineered antibody fragments and the rise of single domains. Nat Biotechnol 23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 30.Kulkeaw K, Sakolvaree Y, Srimanote P, Tongtawe P, Maneewatch S, Sookrung N, Tungtrongchitr A, Tapchaisri P, Kurazono H, Chaicumpa W. 2009. Human monoclonal ScFv neutralize lethal Thai cobra, Naja kaouthia, neurotoxin. J Proteomics 72:270–282. doi: 10.1016/j.jprot.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Akita EM, Nakai S. 1993. Comparison of four purification methods for the production of immunoglobulins from eggs laid by hens immunized with an enterotoxigenic E. coli strain. J Immunol Methods 160:207–214. doi: 10.1016/0022-1759(93)90179-B. [DOI] [PubMed] [Google Scholar]
- 32.Akita EM, Nakai S. 1993. Production and purification of Fab′ fragments from chicken egg yolk immunoglobulin Y (IgY). J Immunol Methods 162:155–164. doi: 10.1016/0022-1759(93)90380-P. [DOI] [PubMed] [Google Scholar]
- 33.Andris-Widhopf J, Rader C, Steinberger P, Fuller R, Barbas CF III. 2000. Methods for the generation of chicken monoclonal antibody fragments by phage display. J Immunol Methods 242:159–181. doi: 10.1016/S0022-1759(00)00221-0. [DOI] [PubMed] [Google Scholar]
- 34.Yamanaka HI, Inoue T, Ikeda-Tanaka O. 1996. Chicken monoclonal antibody isolated by a phage display system. J Immunol 157:1156–1162. [PubMed] [Google Scholar]
- 35.Meier J, Stocker KF. 1995. Biology and distribution of venomous snakes of medical importance and the composition of snake venoms, p 367–412. In White J, Meier J (ed), Handbook of clinical toxicology of animal venoms and poisons. CRC Press, Boca Raton, FL. [Google Scholar]
- 36.Dias da Silva W, Tambourgi DV. 2010. IgY: a promising antibody for use in immunodiagnostic and in immunotherapy. Vet Immunol Immunopathol 135:173–180. doi: 10.1016/j.vetimm.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davalos-Pantoja L, Ortega-Vinuesa JL, Bastos-Gonzalez D, Hidalgo-Alvarez R. 2000. A comparative study between the adsorption of IgY and IgG on latex particles. J Biomater Sci Polym Ed 11:657–673. doi: 10.1163/156856200743931. [DOI] [PubMed] [Google Scholar]
- 38.Carlander D, Larsson A. 2001. Avian antibodies can eliminate interference due to complement activation in ELISA. Ups J Med Sci 106:189–195. doi: 10.3109/2000-1967-145. [DOI] [PubMed] [Google Scholar]
- 39.Stewart CS, MacKenzie CR, Hall JC. 2007. Isolation, characterization and pentamerization of alpha-cobrotoxin specific single-domain antibodies from a naive phage display library: preliminary findings for antivenom development. Toxicon 49:699–709. doi: 10.1016/j.toxicon.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 40.Maynard J, Georgiou G. 2000. Antibody engineering. Annu Rev Biomed Eng 2:339–376. doi: 10.1146/annurev.bioeng.2.1.339. [DOI] [PubMed] [Google Scholar]
- 41.Gao C, Mao S, Kaufmann G, Wirsching P, Lerner RA, Janda KD. 2002. A method for the generation of combinatorial antibody libraries using pIX phage display. Proc Natl Acad Sci U S A 99:12612–12616. doi: 10.1073/pnas.192467999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pandey S. 2010. Hybridoma technology for production of monoclonal antibodies. Hybridoma 1:017. [Google Scholar]
- 43.Xu JL, Davis MM. 2000. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 13:37–45. doi: 10.1016/S1074-7613(00)00006-6. [DOI] [PubMed] [Google Scholar]
- 44.Rucavado A, Escalante T, Shannon JD, Ayala-Castro CN, Villalta M, Gutierrez JM, Fox JW. 2012. Efficacy of IgG and F(ab′)2 antivenoms to neutralize snake venom-induced local tissue damage as assessed by the proteomic analysis of wound exudate. J Proteome Res 11:292–305. doi: 10.1021/pr200847q. [DOI] [PubMed] [Google Scholar]
- 45.Rucavado A, Escalante T, Shannon J, Gutierrez JM, Fox JW. 2011. Proteomics of wound exudate in snake venom-induced pathology: search for biomarkers to assess tissue damage and therapeutic success. J Proteome Res 10:1987–2005. doi: 10.1021/pr101208f. [DOI] [PubMed] [Google Scholar]
- 46.Friguet B, Chaffotte AF, Djavadi-Ohaniance L, Goldberg ME. 1985. Measurements of the true affinity constant in solution of antigen-antibody complexes by enzyme-linked immunosorbent assay. J Immunol Methods 77:305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- 47.Francischetti IM, My-Pham V, Harrison J, Garfield MK, Ribeiro JM. 2004. Bitis gabonica (Gaboon viper) snake venom gland: toward a catalog for the full-length transcripts (cDNA) and proteins. Gene 337:55–69. doi: 10.1016/j.gene.2004.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fox JW, Serrano SM. 2005. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon 45:969–985. doi: 10.1016/j.toxicon.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 49.Takeda S, Takeya H, Iwanaga S. 2012. Snake venom metalloproteinases: structure, function and relevance to the mammalian ADAM/ADAMTS family proteins. Biochim Biophys Acta 1824:164–176. doi: 10.1016/j.bbapap.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 50.Gutierrez JM, Rucavado A, Escalante T, Diaz C. 2005. Hemorrhage induced by snake venom metalloproteinases: biochemical and biophysical mechanisms involved in microvessel damage. Toxicon 45:997–1011. doi: 10.1016/j.toxicon.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 51.Calvete JJ, Marcinkiewicz C, Monleon D, Esteve V, Celda B, Juarez P, Sanz L. 2005. Snake venom disintegrins: evolution of structure and function. Toxicon 45:1063–1074. doi: 10.1016/j.toxicon.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 52.Baldo C, Jamora C, Yamanouye N, Zorn TM, Moura-da-Silva AM. 2010. Mechanisms of vascular damage by hemorrhagic snake venom metalloproteinases: tissue distribution and in situ hydrolysis. PLoS Negl Trop Dis 4:e727. doi: 10.1371/journal.pntd.0000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Escalante T, Rucavado A, Fox JW, Gutierrez JM. 2011. Key events in microvascular damage induced by snake venom hemorrhagic metalloproteinases. J Proteomics 74:1781–1794. doi: 10.1016/j.jprot.2011.03.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.