Abstract
Shotgun metagenomic sequencing does not depend on gene-targeted primers or PCR amplification; thus, it is not affected by primer bias or chimeras. However, searching rRNA genes from large shotgun Illumina data sets is computationally expensive, and no approach exists for unsupervised community analysis of small-subunit (SSU) rRNA gene fragments retrieved from shotgun data. We present a pipeline, SSUsearch, to achieve the faster identification of short-subunit rRNA gene fragments and enabled unsupervised community analysis with shotgun data. It also includes classification and copy number correction, and the output can be used by traditional amplicon analysis platforms. Shotgun metagenome data using this pipeline yielded higher diversity estimates than amplicon data but retained the grouping of samples in ordination analyses. We applied this pipeline to soil samples with paired shotgun and amplicon data and confirmed bias against Verrucomicrobia in a commonly used V6-V8 primer set, as well as discovering likely bias against Actinobacteria and for Verrucomicrobia in a commonly used V4 primer set. This pipeline can utilize all variable regions in SSU rRNA and also can be applied to large-subunit (LSU) rRNA genes for confirmation of community structure. The pipeline can scale to handle large amounts of soil metagenomic data (5 Gb memory and 5 central processing unit hours to process 38 Gb [1 lane] of trimmed Illumina HiSeq2500 data) and is freely available at https://github.com/dib-lab/SSUsearch under a BSD license.
INTRODUCTION
Microbial phylogeny, identification, and evolution studies were revolutionized by the introduction of small-subunit (SSU) rRNA analysis 25 years ago (1), and with the advent of PCR and high-throughput sequencing, community structure studies now are commonplace (2–5). The growing sizes of SSU rRNA gene databases provide a rich ecological and phylogenetic context for SSU rRNA gene-based community structure surveys (6, 7). However, the accuracy of PCR-based amplicon approaches is reduced by primer bias and chimeras (8, 9).
Unlike gene-targeted amplicon sequencing, shotgun sequencing takes samples from the entire community by sequencing randomly sheared fragments of DNA (10, 11). Hence, while amplicon sequencing can provide far deeper coverage of SSU rRNA genes with the same amount of sequencing, shotgun sequencing may provide a more accurate characterization of microbial diversity, including functional diversity (12). In particular, shotgun sequencing may provide an improved means to detect divergent sequences not recovered by standard SSU rRNA gene primers, such as those of Verrucomicrobia, as well as eukaryotic members of the community (8, 12–14). Note that both approaches remain prone to sequencing error and bias from environmental DNA extraction (9).
The challenges for using shotgun DNA for rRNA analyses are in efficiently searching for these fragments in large sequence data sets and the subsequent analysis of the matching short reads. Several methods have been developed for SSU rRNA retrieval in large data sets (15–18), but speed improvements still are needed to match the growth in data size; moreover, none of them provide further community analysis using the identified rRNA gene sequences. In addition, traditional community analysis tools (6, 19, 20) are largely designed to handle sequences that are amplified by PCR primers. There are two primary types of community analyses: reference based (supervised) and operational taxonomic unit (OTU) based (unsupervised). The reference-based method assigns SSU rRNA gene sequences to bins based on the taxonomy of their closest reference sequences, while OTU-based methods assign overlapping gene sequences to bins based on de novo clustering with a specified similarity cutoff (e.g., 97%). The reference-based method can be applied easily to shotgun data once SSU rRNA gene fragments are retrieved (21) and several tools are available for this (22–26), but the OTU-based approach still remains challenging with shotgun data because reads are from randomly sheared fragments.
The main goal of this study is to enable unsupervised OTU-based analysis of large shotgun metagenomic data sets from soil. We improved speed and memory efficiency with a hidden Markov model (HMM)-based method, which already has been shown to be fast and accurate for SSU rRNA searches (16–18), using a well-curated and up-to-date training reference sequence collection from SILVA (7). Our unsupervised clustering method first was tested on a synthetic community with shotgun data of 100-bp reads. We next applied the method to soil data sets, where we assembled longer reads from the overlapping paired-end Illumina HiSeq reads and mapped those to 150-bp small hypervariable regions of SSU rRNA genes for de novo clustering and further diversity analysis. We retrieved and analyzed the large-subunit (LSU) ribosomal gene for confirmatory analysis. Finally, we went beyond traditional primer evaluation (in silico database search) by evaluating primer biases using the paired shotgun and amplicon data produced from the same DNA extract (27, 28).
MATERIALS AND METHODS
Soil samples, DNA extraction, and sequencing.
Two sets of soil samples were used. The first sample, which was used to develop the method, was a bulk (non-root-influenced) soil sample (SB1) taken in 2009 from between rows of switchgrass. The method then was applied to the second sample set taken in 2012, which consisted of seven replicate rhizosphere samples from both corn (C) and Miscanthus (M) plots. All samples were from the Great Lakes Bioenergy Research Center (GLBRC) Cropping System Comparison Site at the Kellogg Biological Station in southwest Michigan (http://data.sustainability.glbrc.org/pages/1.html). The rhizosphere samples were closely associated with the roots (<1 mm).
DNA extraction and SSU rRNA gene amplification methods were described previously (29). The SSU rRNA gene amplicons from the first sample were sequenced by the Joint Genome Institute (JGI) in their standard work flow, which used 454 GS FLX and Titanium platforms and a primer set (926F, AAACTYAAAKGAATTGACGG; 1392R, ACGGGCGGTGTGTRC) that targeted the V6-V8 variable region of bacteria, archaea, and eukaryotes. The second set also was sequenced at the JGI but at a later time, so the Illumina MiSeq platform and a primer set (515F, GTGCCAGCMGCCGCGGTAA; 806R, GGACTACHVGGGTWTCTAAT) that targeted the V4 variable region were used. Shotgun sequencing also was done by JGI using Illumina GAII platforms for the first set and HiSeq 2500 with 250-bp insert libraries and two 150-bp reads for the second set. We had about 8 Gb of data for the first set and about 300 Gb of data each for corn and Miscanthus for the second set.
Data preprocessing.
Data preprocessing is necessary for both shotgun and amplicon data due to sequencing errors. However, it is not included as part of the core pipeline, because users have their own preferences. We trimmed trailing bases with quality score 2, called the read segment quality control indicator (encoded by ASCII 66 “B” in Illumina GAII or ASCII 35 “#” in Illumina HiSeq shotgun data), and discarded reads shorter than 30 bp and with “N.” The reads then were quality trimmed with fastq-mcf (version 1.04.662) (http://code.google.com/p/ea-utils) with “-l 50 -q 30 -w 4 -x 10 -max-ns 0 -X.” The paired-end reads overlapping by more than 10 bp were assembled into one long read by FLASH (version 1.2.7) (30) with “-m 10 -M 120 -x 0.20 -r 140 -f 250 -s 25.” Roche 454 pyrotag amplicon data were processed using the RDP Pipeline PIPELINE INITIAL PROCESS and CHIMERA CHECK tools (6). Reads were sequenced from the reverse primer end to the forward primer end. Since the targeted region is about 467 bp (926F/1392R), most reads were not long enough to reach the forward primer. Thus, only the reverse primer product was used for quality trimming. The minimum length was set to 400 bp, and defaults were used for other parameters.
Building SSU and LSU rRNA gene models.
We quality trimmed SILVA (7) SSU and LSU Ref NR database (version 115) sequences by discarding all sequences with ambiguous DNA bases and converting U to T. We then clustered them at a 97% similarity cutoff using pick_otus.py (default with UCLUST) and pick_rep_set.py from QIIME (version 1.8.0) (20). We chose the longest representative in each OTU to be further clustered at an 80% similarity cutoff. We collected the longest sequence in each OTU, resulting in 4,027 representative sequences for the SSU rRNA gene and 1,295 for the LSU rRNA gene to obtain a phylogenetically diverse set of reference genes. We further grouped these sequences into two groups, one combining Bacteria and Archaea and the other containing only Eukaryota (see Discussion). Each group was used to make two HMMs (hidden Markov models), one with sequences from a previous step and the other with reverse complement, using hmmbuild in HMMER version 3.1 (31). Finally, the HMM files were concatenated into a single file for each gene. This step is not part of the pipeline, and the resulting HMMs were included in the database of this pipeline.
Identification of rRNA gene fragments from metagenomic data.
The analysis framework is shown in Fig. S1 in the supplemental material, as are the reasons for our choices of pipeline components. We searched Illumina shotgun metagenomic data with hmmsearch in HMMER version 3.1 (31) using the LSU and SSU HMMs. For testing the sensitivity of newly built models, we analyzed our tool with meta-rna (16) and metaxa (18). We used an E value of 10 for hmmsearch with the newly built HMMs. Meta-rna (rna_hmm3.py) (the package was not assigned a version number; the most recent version update, 21 October 2011, was used) was run with flags “-k euk,bac,arc -e 0.00001” (16), and metaxa (metaxa_x) version 2.0.2 was run with flags “–allow_single_domain 1e-5,0 -N 1 -E 1e-5.” A bulk soil (SB1), a rhizosphere soil (M1), and a synthetic community sample (12) were used as test data. We aligned the HMMER hits from the E value cutoff of 10 using the multiple-sequence aligner align.seqs in mothur (version 1.33.3) (32). For SSU rRNA gene fragments, 18,491 full-length SSU rRNA gene sequences (14,956 from Bacteria, 2,297 from Archaea, and 1,238 from Eukaryota) from the SILVA database (release 102) (7) were downloaded from the mothur website (http://www.mothur.org/wiki/Silva_reference_files) and used as the template with flags “threshold = 0.5” and “flip = t.” For LSU rRNA, multiple-sequence alignments (MSA) of representative sequences of the SILVA LSU Ref NR database, clustered at a 97% similarity cutoff, were used as the template with the same flags as those for SSU. Based on alignment information provided in the align.seqs output report file, those shotgun reads with more than 50% mapped to a reference gene were designated SSU rRNA or LSU rRNA gene fragments.
Testing the effect of target region size and variable region on clustering.
We used shotgun data of a synthetic community comprised of 64 species, which were sequenced by the paired-end 100-bp method on an Illumina HiSeq 2000 (12). For testing the effect of target region size on clustering, we picked V4 with starting position 577 in Escherichia coli. Sizes from 50 to 180 bp with a 10-bp increment were chosen. The minimum read length was set to the target region size minus 5 bp if the region size was less than 100 bp, and it was set to 95 bp when the region size was greater than 100 bp. We used the pre.cluster command in mothur (19) with 1 edit distance to collapse reads with errors and their original reads. De novo clustering then was achieved by RDP McClust with an algorithm for unweighted pair group method using average linkages (UPGMA) and a minimum read overlapping length of 25 bp (33). We chose McClust as the clustering tool due to its speed and memory efficiency (33, 34). We chose V2, V3, V4, V5, V6, and V8, starting at positions 127, 427, 577, 787, 987, and 1227 in E. coli, respectively (35), to test the hypervariable region effect on clustering results. Target region sizes of 80 bp and 120 bp and a distance cutoff of 5% were chosen. Further, the analyses described above also were applied to 16S rRNA genes from the 64 species comprising the community to obtain the true OTU numbers.
Community structure comparison based on OTUs from de novo clustering.
For the clustering analysis of shotgun and amplicon data, 150 bases corresponding to the V8 region (E. coli positions 1227 to 1377) were aligned. Reads shorter than 100 bp were removed from the alignment and the remainder clustered using McClust with a minimum overlap of 25 bp and the UPGMA method (6). The clustering result was converted to the mothur format, and community structure comparisons were done using make.shared with a label of 0.05, dist.shared(calc = thetayc), and the pcoa command. E. coli positions 127 to 277, 577 to 727, and 997 to 1147 were chosen for V2, V4, and V6, respectively, for comparisons of different regions (35). Procrustes analysis as implemented in QIIME (20) was used to transform V2, V6, and V8 PCoA (principal-coordinate analysis) results and to minimize the distances between corresponding points in V4. The bulk soil sample (SB1) was sequenced in six lanes from one Illumina plate using DNA from the same extraction. We used these as technical replicates for testing the reproducibility of de novo OTU-based analysis on shotgun data. Since sequencing depth is critical for reproducibility testing, we pooled these into two samples of three lanes each, with the first three lanes as SB1_123 and the remaining three lanes as SB1_456.
Comparison of OTU-based microbial community structures inferred from shotgun and amplicon SSU rRNA gene sequences.
The abundance of each OTU in shotgun data and amplicon data (V6-V8 for SB1 and V4 for M1) from the same DNA extraction were compared to check the consistency between the two sequencing approaches. Pearson's correlation coefficient and linear regressions were used to evaluate the correlation between the two types of data and between technical replicates. All of the abundances of each OTU were increased by a pseudocount of one to allow display on a log scale (avoiding zeros).
Comparison of taxonomy-based microbial community structures inferred from shotgun and amplicon SSU rRNA gene sequences.
The SSU rRNA fragments from shotgun data and amplicon data were classified using RDP Classifier (21). The reference SSU rRNA genes from RDP and SILVA are provided on the mothur website and were used as training sets, with a bootstrap confidence cutoff for classification of 50%. Representative sequences of SILVA LSU Ref NR clustered at a 97% similarity cutoff were used as a training set with taxonomy information built from the sequence file for the LSU rRNA gene. The bacterial taxonomy profiles from shotgun data and amplicon data were compared at the phylum level.
Copy number correction.
We used the SSU rRNA gene copy number database in CopyRighter (36), which provides the copy number for each taxon in the Greengenes database (37). In the taxonomic summary, the abundance of each taxon is weighted by the inverse of its SSU rRNA gene copy number. Similarly, in OTU-based analysis, the abundance of each OTU is weighted by the inverse of SSU rRNA gene copy number of its consensus taxon. Unclassified sequences are weighted by the inverse of the average copy number of all taxa in the data set.
Implementation, reproducibility, and sequence data.
The SSUsearch pipeline can be found at https://github.com/dib-lab/SSUsearch as a tutorial with ipython notebooks (38). Scripts for reproducing the figures in this paper can be found at https://github.com/dib-lab/2014-ssu-search/blob/master/analysis-in-paper.Makefile. The synthetic community data for testing can be downloaded from the NCBI Sequence Read Archive (SRA) under accession number SRR606249.
Accession numbers.
The amplicon data for C1 to C7 and M1 to M7 have been deposited in the JGI genome portal under project identifier (ID) 1025756 with library ID M2094 and M2113, respectively, and SB1 was deposited in the NCBI SRA under accession number SRX902929. The shotgun data for the same three data sets were deposited in the JGI portal (C1 to C7 are under project ID 1023764, 1023767, 1023770, 1023773, 1023776, 1023779, and 1023782, and M1 to M7 are under project ID 1023785, 1023788, 1023791, 1023794, 1023797, 1023800, and 1018623; SB1 is under project ID 402775).
RESULTS
We developed an optimized pipeline that readily analyzes large data sets (see Fig. S1 in the supplemental material). The pipeline has two major steps: SSU rRNA gene fragment search and unsupervised OTU analysis. HMMER-based methods search with HMMs and scale with increasing sizes of SSU rRNA gene databases (6), so we chose them for the first search step. We used meta-rna (16) but could not run it on large data sets due to its poor memory management. Therefore, we simplified and optimized the approach used by meta-rna. Since the search step is still the computational bottleneck (see Fig. S1), our interest here was to make an improvement on search speed and memory efficiency while retaining accuracy. Our implementation is about 4 times faster and 100 times more memory efficient than meta-rna and is 10 times faster and 15 times more memory efficient than metaxa (18) (Table 1). The speed improvement is realized from two modifications. (i) We reduce the number of HMMs to search with by merging Bacteria and Archaea models. SSU rRNA genes are highly conserved; thus, the merged model still has high sensitivity. Even more so, we can increase the sensitivity by using a more relaxed E value cutoff, since false positives are tolerable in this initial search step. (ii) We use reverse-complement HMMs rather than reverse complementing the reads, because the latter scales poorly with large data sets. The newly built HMMs for Bacteria and Archaea together and HMMs for Eukaryota cover most hits found by separate HMMs of the three domains (Bacteria, Archaea, and Eukaryota) in meta-rna in all three test data sets, the two soil data sets in this study, and one synthetic community (Table 1). Our method identified 15,566 (0.03%) of 44,787,632 quality-trimmed and paired-end merged sequences as SSU rRNA gene fragments in the bulk soil sample (SB1) and 112,402 (0.04%) of 274,060,925 reads in the first Miscanthus replicate (M1).
TABLE 1.
Comparison of search results from SSUsearch, metaxa, and meta-rnaa
| Search toolb | Mock | SB1 | M1 |
|---|---|---|---|
| SSUsearch (no. of hits) | 6,432 | 2,789 | 2,612 |
| Meta-rna (no. of hits) | 6,455 | 2,781 | 2,600 |
| Metaxa (no. of hits) | 5,322 | 2,649 | 2,444 |
| SSUsearch∩meta-rna (no. of hits) | 6,300 | 2,759 | 2,576 |
| SSUsearch∩metaxa (no. of hits) | 5,286 | 2,642 | 2,442 |
| metarna∩metaxa (no. of hits) | 5,304 | 2,649 | 2,436 |
| SSUsearch∩meta-rna∩metaxa (no. of hits) | 5,268 | 2,642 | 2,435 |
| SSUsearch CPU time (min) | 1.6 | 4 | 4.8 |
| Meta-rna CPU time (min) | 8.6 | 16.5 | 16.9 |
| Metaxa CPU time (min) | 17.5 | 47.2 | 34.8 |
| SSUsearch memory (Mb) | 35 | 33 | 30 |
| Meta-rna memory (Mb) | 3,406 | 4,005 | 4,234 |
| Metaxa memory (Mb) | 452 | 456 | 572 |
Subsets of 5 million reads from mock (metagenome of a synthetic community of 48 Bacteria and 16 Archaea samples), SB1 (bulk soil metagenome), and M1 (Miscanthus rhizosphere metagenome) were used as testing data.
The symbol ∩ indicates overlap.
Unsupervised OTU analysis with shotgun data is not available in any current pipeline, so we developed a method for OTU clustering around a small region where all reads overlap. To show the validity of our unsupervised method, we did tests on effects of target region sizes and different variable regions with shotgun data from a synthetic community. We found all region sizes from 50 bp to 160 bp in V4 had an OTU number that approached the species number at a distance cutoff of 4% or 5% (see Fig. S2 in the supplemental material) when testing target region size effect on OTU number. We also did a similar test with only full-length SSU rRNA genes from 64 species to make sure the OTU number is close to the species number and confirmed that the OTU number was close to the species number (at a range of 50 to 60) when a cutoff of 0 to 0.06 was chosen (see Fig. S3). When the target region size was larger than 170 bp, the clustering tool (McClust) (33) did not cluster because the percentage of nonoverlapping reads exceeded its threshold. On the basis of the results described above, we chose 80 bp or 120 bp as the target region size and a 5% distance cutoff for testing different hypervariable regions. The number of OTUs created in all variable regions is close to the real number of species in the synthetic community except when using V3.
Reproducibility between technical replicates is important and is a basic feature of a sequencing method (39, 40). We evaluated it by comparing the correlation of OTU abundance between technical replicates from the bulk soil sample and found high correlation between them (Pearson's correlation coefficient of 0.997). The consistency was better for the more abundant OTUs (see Fig. S4A in the supplemental material). Log transformation could reduce the effect of high-abundance OTUs on the overall correlation. Even with log-transformed abundance, the two replicates have a Pearson's correlation coefficient of 0.91 and linear regression (R2) of 0.88 when OTUs with less than 25 total counts were discarded (see Fig. S4B). The choice of a cutoff of 25 was chosen to compare data to those from another study on amplicon reproducibility (41) and as mentioned in Discussion.
We also compared OTU-based microbial community structures inferred from shotgun and amplicon SSU rRNA gene sequences. OTU abundances in shotgun and amplicon data, however, do not correlate as well (Pearson's correlation coefficient of 0.87 for bulk soil sample SB1 [see Fig. S4C in the supplemental material] and 0.58 for Miscanthus rhizosphere sample M1 [see Fig. S4D]), showing that the amplicon and shotgun methods do not provide the same information. The classification of OTUs with total abundance higher than 10 and with a ratio between two data types higher than 5-fold shows bias against Verrucomicrobia in the bulk soil sample (SB1_PT) amplified by the V6-V8 primer, while it shows a favorable bias for Verrucomicrobia in the rhizosphere sample (M1_PT) amplified by the V4 primer. The classification showed a bias against Actinobacteria in M1_PT amplified by the V4 primer (see Fig. S5). These results were consistent with the taxonomy-based comparison of the two data types (see below) that suggested primer bias in amplicon data.
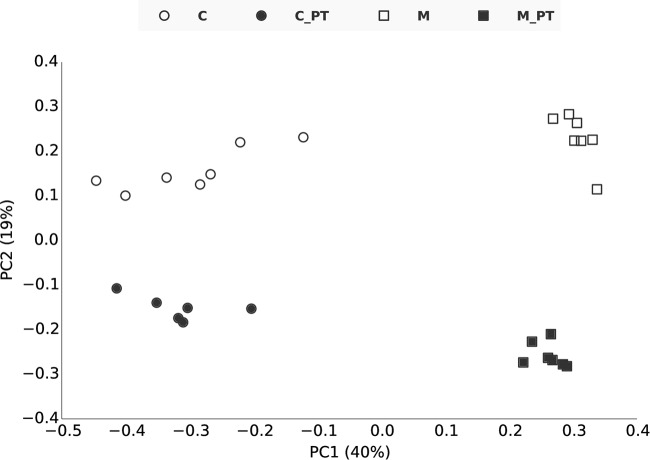
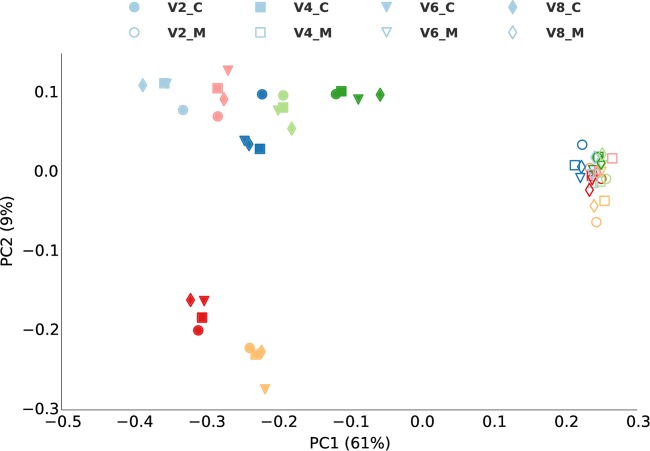
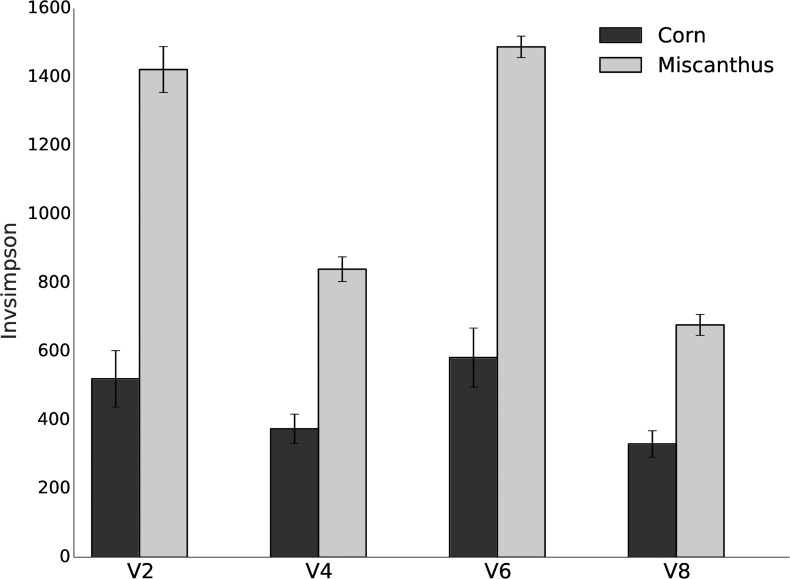
We applied ordination analysis to OTU tables from an unsupervised analysis of corn and Miscanthus rhizosphere samples. OTUs from shotgun and amplicon data both showed separation of rhizosphere communities of corn and Miscanthus (Fig. 1, horizontal dimension) as well as a significant difference between the two data types (Fig. 1, vertical dimension), confirming the difference between shotgun and amplicon data. Significant separation (P < 0.001 by analysis of molecular variance [AMOVA] test in mothur) of corn and Miscanthus samples also was observed when V2, V4, V6, and V8 shotgun data were used for clustering (Fig. 2), but the sample groupings were the same for all variable regions. Figures 1 and 2 showed that the dispersion among the seven corn replicates was much higher than that for the Miscanthus replicates. Miscanthus samples had higher alpha diversity than corn samples, as shown for each of the V2, V4, V6, and V8 regions, although there were variations among these regions (Fig. 3).
FIG 1.
Principal-coordinate analysis (PCoA) of amplicon- and shotgun-derived data from seven field replicates. There are significant differences between amplicon (filled)- and shotgun (unfilled)-derived data (along the y axis) and between corn (circle) and Miscanthus (square) rhizosphere samples (along the x axis) (P < 0.001 by AMOVA [analysis of molecular variance]). PCoA was applied to the OTU table that resulted from clustering with shotgun data and amplicon data using 150 bp of the V4 region. Labels with suffix “_PT” indicate amplicon data, and others indicate shotgun data.
FIG 2.
Comparison of ordination analysis with different variable regions. PCoA of OTUs from different SSU rRNA variable regions (V2, V4, V6, and V8) was applied on corn (“_C”) and Miscanthus (“_M”) rhizosphere samples. Different colors indicate the seven replicates. OTU tables from the clustering of shotgun data using 150 bp of V2, V4, V6, and V8 regions were used for PCoA, and Procrustes analysis in QIIME was used to transform the PCoA results from different regions and plot them in the same figure.
FIG 3.
Alpha diversity comparisons between corn and Miscanthus samples using V2, V4, V6, and V8 regions. All variable regions showed Miscanthus samples are more diverse than corn samples, even though there was variation of diversity among rRNA gene regions. Alpha diversity was calculated with inverse Simpson (Invsimpson) using OTUs resulting from clustering with different variable regions.
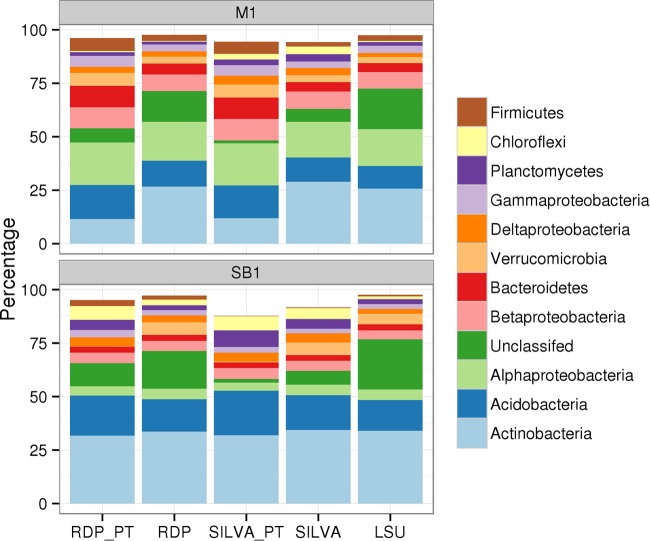
We compared the taxonomy-based microbial community structures inferred from shotgun data with those from amplicon SSU rRNA gene sequences (12,163 amplicons for SB1 and 60,148 amplicons for M1), and we confirmed known primer biases and revealed a new bias. Before comparing two data types, we looked at the taxonomy profile of shotgun data using different variable regions. Shotgun data mapped to different variable regions show similar taxonomies at the bacterial phylum level (Pearson's correlation coefficient of >0.96), except that V6 has more unclassified sequences (see Fig. S6 in the supplemental material). Since different variable regions may provide different levels of taxonomic precision for certain groups (42), taxonomy information from all regions may better represent the taxonomy profile. Thus, we used all SSU rRNA gene fragments for taxonomy comparison with amplicon data. Both shotgun and amplicon data show Actinobacteria, Proteobacteria, and Acidobacteria as the three most abundant phyla, as is expected for soil (43). Since shotgun data are more accurate at estimating community structure than other methods, we accepted the shotgun data as the reference (9, 12). The 926F/1392R (V6-V8) primer set is biased against Verrucomicrobia (0.3% in amplicon data versus 5.8% in shotgun data by RDP database) in bulk soil sample SB1 (Fig. 4), and the 515F/806R (V4) primer set is biased against Actinobacteria (11.6% in amplicon data versus 26.6% in shotgun data) and in favor of Verrucomicrobia (5.9% in amplicon data versus 3.2% in shotgun data) in rhizosphere sample M1 (Fig. 4).
FIG 4.
Taxonomy profiles of bacterial phyla of shotgun fragments from SSU and LSU rRNA genes and of amplicon reads. Classifications were done using both RDP and SILVA reference databases. The suffix “_PT” indicates amplicon data. The lower graph depicts bulk soil sample (SB1); the amplicon data used the V6-V8 primer set (515F/806R) and showed fewer Verrucomicrobia detected using both databases. The upper graph depicts the rhizosphere sample (M1), and its amplicon data used the V4 primer set (515F/806R) and showed fewer Actinobacteria and more Verrucomicrobia using both databases.
To take advantage of the fact that shotgun data are untargeted, we retrieved and classified the LSU rRNA genes, which are cotranscribed with SSU rRNA genes. Their taxonomy profile was similar to that of SSU rRNA genes (Pearson's correlation coefficient of 0.87 for SB1 and 0.91 for M1), except that more reads (19.6%) remain unclassified (Fig. 4). This is expected because of the much lower number of reference LSU rRNA genes in the SILVA database. The two genes show consistent community profiles at the bacterial phylum level, and they also confirm the known primer bias against Verrucomicrobia in the 926F/1392R (V6-V8) primer set and that against Actinobacteria in the 515F/806R (V4) primer set (Fig. 4). Further, both the LSU and SSU HMMs show the ability to identify members of the Eukaryota and give about the same taxonomy profile at the domain level (see Fig. S7 in the supplemental material). It is worth noting that both LSU and SSU shotgun data show Archaea to be twice as numerous (2% versus 1%) in the bulk soil as in the rhizosphere and that Eukaryota were much more numerous (6% versus 1%) in the Miscanthus rhizosphere soil than in the bulk soil (see Fig. S7). A higher fungal percentage (2.54% versus 0.39%), fungus/bacterium ratio (0.027 versus 0.004), and arbuscular mycorrhizal fungus (AMF) percentage in fungi (0.18% versus 0) were found in the rhizosphere sample (M1) than in the bulk soil sample (SB1) (see Table S1 in the supplemental material). Copy correction was applied to two soil samples (SB1 and M1). Both samples showed that Firmicutes and Bacteroidetes had the highest fold change after copy number correction (see Fig. S8). Despite copy number correction, the clustering of our soil samples did not change (see Fig. S9) compared to that without copy number correction (Fig. 1), probably because of the low proportion of taxa with large rrn number corrections.
DISCUSSION
We present, characterize, and validate an efficient method for retrieving and analyzing SSU rRNA gene fragments from shotgun metagenomic sequences. The pipeline enables unsupervised diversity analysis with copy number correction on multiple variable regions, has the scalability to handle large soil metagenomes, is expandable to other phylogenetic marker genes, and is publicly available on GitHub.
We apply a two-step approach for retrieving SSU rRNA gene fragments, a loose HMM filtering step followed by a more stringent step that screens by identity to the best-match reference. The first step leverages HMMER (31); thus, it should have better scalability than existing shotgun analysis pipelines that use BLAST-like tools such as MG-RAST (44). MG-RAST annotates shotgun reads by BLAT search against rRNA databases, and the taxonomy of reads is inferred from the best hit or least common ancestor of several top hits (24, 45, 46). BLAT or BLAST-like tools are not scalable for large data sets and must be run in parallel on large computer clusters, because BLAST-like tools typically do pairwise comparisons of reads against large and growing rRNA databases, such as RDP, SILVA, and Greengenes (6, 7, 37), while HMM-based methods compare reads to only a fixed number of models (commonly one for each domain) and are more scalable (44). Moreover, these pipelines lack unsupervised community analysis. An HMM-based search has been used before, as it is fast and sensitive for rrn retrieval (16–18, 22), and current existing implementations, such as meta-rna, RNASelector, and metaxa, all are wrappers around HMMER (31). We chose meta-rna and metaxa for comparison, because RNASelector can run only in a graphic interface that is not suitable for large data sets.
The second step evaluates hmmsearch results based on identity to their best-match reference and also prepares the alignment of SSU RNA gene fragments for clustering. Since there is no clear sequence identity threshold for SSU rRNA genes, the choice of identity cutoff is arbitrary (the default is 50%). This is also a common practice for amplicon analysis platforms, where reads with low identity to reference sequences are discarded prior to clustering (19, 20). For consistency in comparison, sequences in our amplicon data sets with less than 50% identity to reference sequences also are discarded. An alignment of the SSU rRNA gene fragments is essential for the later unsupervised analyses. Compared to methods that use only the 16S rRNA gene in E. coli as an alignment template (47), our method takes advantage of the rich phylogenetic diversity of SSU rRNA genes provided by the SILVA database. Increasing the number of reference sequences can improve the quality of alignment but also linearly increases the memory required (32, 48).
Our unsupervised analysis with shotgun data is a novel and important part of the pipeline. Our tests show that regions as small as 50 bp can be applied to clustering. Thus, short reads around 50 bp can be applied to this method as long as there are sufficient numbers of reads aligned to the target region (sequencing depth is a limiting factor; see below). This is consistent with pilot studies from 454 amplicon sequencing (5, 49). When sequencing depth is limited, there is flexibility to control the number of reads to include for clustering by adjusting the target region size and read length cutoff within certain limits (see Fig. S2B in the supplemental material). The caveat of using very short or very large target regions is that the overlapping portion of reads will decrease; thus, there will be a decrease in the accuracy of clustering and the impact of sequencing error will increase. For example, an error in a 50-bp read can cause 2% distance; accordingly, we need to set a larger distance cutoff for clustering. In addition, we can obtain longer sequences from overlapping paired ends, as shown in our soil data (see Fig. S10). Thus, reads from Illumina shotgun data (ranging from 75 to 250 bp) can be used for unsupervised analysis. Note that the flexibility on the choice of variable region for analysis is another advantage of shotgun data (see Fig. S2C).
We also found good reproducibility of OTU abundance between technical replicates, which is critical for the validity of our method (see Fig. S4A in the supplemental material). Generally, OTU-based analysis provides higher resolution than taxonomy-based diversity analysis for community comparison, largely due to the databases lacking reference sequences from uncultured microbes (50). The high correlation of OTU abundance in two technical replicates shows the reproducibility of the analysis of shotgun data, which is comparable to the reproducibility of amplicon data shown in another study in terms of Pearson's correlation coefficient and linear regression (R2) (41). Further, comparison of OTU abundances in shotgun data and amplicon data sequenced from the same DNA extraction also show that many OTUs have inconsistent abundances between the two types of data (see Fig. S4C and D), which agrees with the differences seen in the taxonomy-based comparison (Fig. 4).
Community comparison by ordination methods such as PCoA and NMDS (nonmetric multidimentional scaling) is one of the most common analyses in microbial ecology. To the best of our knowledge, the methods used in two previous studies (47, 51) are the only existing tools that are designed to deal with the clustering of SSU rRNA gene fragments from Illumina shotgun data. The method used in the first study could result in poor alignment by using only the 16S rRNA gene in E. coli as the alignment template. The method used in the second study (PhylOTU) was applied to larger shotgun sequences from Sanger sequencing. It determines the OTU clustering of SSU rRNA gene fragments aligned over the whole gene length, which can be problematic, because fragments aligned to different regions do not overlap and the clustering results are not reliable, even though the reference sequences included in the clustering process can improve the results. Since our tests show that a hypervariable region as small as 50 bp can be used for unsupervised analyses (see Fig. S2A and C in the supplemental material), we made sure all of the sequences overlapped by picking one small region (150 bp), and all sequences included in clustering have lengths longer than 100 bp in our clustering method. In addition, longer reads obtained by assembling overlapping paired-end reads (see Fig. S10) can make use of more overlap among reads and are more suitable for clustering. As read lengths increase with the improvement of sequencing technology, larger regions can be chosen and the clustering results will be even more reliable. Also, shotgun data provide the flexibility to choose any variable region (Fig. 2 and 3; also see Fig. S2C and S6), and the consistency of results from different variable regions provides more confidence in the biological conclusions as well as the method itself.
Primer bias is a major limitation of amplicon methods, and it was apparent in our comparisons of community profiles from amplicon versus shotgun data (Fig. 4). Commonly, it is difficult to tell if a bias is caused by primer or DNA extraction. The paired amplicon and shotgun data from the same DNA extract provide us a new opportunity to evaluate this issue, since the difference is only from the sequencing step. We used two main SSU rRNA gene databases, RDP and SILVA, to make sure the taxonomy distribution was not biased by the choice of reference databases, and we further confirmed by taxonomy the distribution of the LSU rRNA gene. The bias against Verrucomicrobia with V6-V8 primers is consistent with other studies showing that the abundance of Verrucomicrobia in soil samples is underestimated due to primer bias (8). Meanwhile, the bias toward Verrucomicrobia with V4 primers agrees with studies showing that the V4 primer set has better coverage of Verrucomicrobia (8). Furthermore, the V4 primer set shows bias against Actinobacteria. The V4 primer set has been reported to cover 92.4% of Actinobacteria in reference databases, the lowest level of coverage among nine common bacterial phyla (8). Bias against Actinobacteria also has been reported in a study on a synthetic community where no primer mismatch was found with members from Actinobacteria (12), and another study on environmental samples using Sanger sequencing (24F/1492R) (52) suggested that the in silico evaluation of primers is not sufficient and that factors other than primer mismatch are causing the bias, for example, competition between primers and melting temperature (53). Thus, primer bias detection by comparing paired amplicon and shotgun data is superior to methods that only search primers in the reference databases.
Another advantage of our method over amplicon approaches is that we can identify the SSU rRNA gene from Bacteria, Archaea, and Eukaryota. Fungi are of special interest in microbial ecology due their critical roles in ecosystems (54, 55). The fungus/bacterium ratio is an important indicator of C/N ratio and soil health (56, 57). Our shotgun metagenome (DNA based) shows a fungus/bacterium ratio of 0.004 in bulk soil (SB1) and 0.027 in rhizosphere soil (M1) (see Table S1 in the supplemental material), while studies using phospholipid fatty acid analysis (PLFA) at the same sampling site (KBS) typically show ratios of 1 to 1.3 (58). The difference can be explained by the higher biomass of fungi relative to that of DNA, since some fungal hyphae may not be filled with nuclei. In addition, we also found higher percentages of AMF of rhizosphere soil (M1) than of bulk soil (SB1), which is consistent with their symbiotic relationship with grass roots.
We also show that copy number correction can be achieved in our pipeline. Gene copy number is another source of bias that limits one's ability to accurately profile microbial communities. There are up to 15 SSU rRNA gene copies in some bacteria and up to 5 in archaea (59). This pipeline utilizes the SSU rRNA copy database in CopyRighter (36). As expected, Firmicutes in soil samples have the highest fold change (see Fig. S8 in the supplemental material). However, due to their low proportion in these soils, the impact of copy number correction on the overall community profile was minor (see Fig. S9). Copy number correction, however, is still an unresolved issue, because SSU rRNA gene copy number data for most species and/or OTUs are lacking and copy number can be incorrect even for species with complete genome sequences because of the misassembly of these repeated regions.
Sequencing depth is another important factor in considering this method for diversity analysis. The percentage of SSU rRNA gene fragments in shotgun data varies depending on the SSU rRNA gene copy number and genome size of each member. In our bulk soil sample (SB1) and the Miscanthus rhizosphere soil sample (M1), we classified about 0.03% and 0.04%, respectively, of the total shotgun data as SSU rRNA. In an ideal situation we want to obtain enough SSU rRNA gene fragments to see saturation of the rarefaction curve in OTU-based analysis, which is difficult for soil samples because of their high diversity and the presence of sequencing error. However, studies have shown that near-saturation sequencing of SSU rRNA amplicons is not necessary for beta-diversity analysis (4, 60). Thus, the empirical fold coverage of 3,000, based on the whole length (about 1,500 bp) of the SSU rRNA gene, is suggested for surface soil samples, which require 11.2 Gb [i.e., 1,500 bp × 3,000 bp ÷ 0.04%] of shotgun data, assuming the SSU rRNA gene comprises about 0.04% of total data.
In this study, the LSU rRNA gene was used mainly as confirmation for SSU rRNA gene-based diversity analysis (Fig. 4; also see Fig. S7 in the supplemental material). However, the LSU rRNA gene offers additional stretches of variable and characteristic sequence regions due to its longer sequence length and yields better phylogenetic resolution (61). For this reason and because there are more available references for fungi, the LSU rRNA gene is used more commonly for fungal community studies (62–65). Currently, the use of the LSU rRNA gene is limited by reference sequences and available universal primer sets (61), but its increased resolution should not be overlooked, since a too-limited resolution of the SSU rRNA gene is a barrier in many ecological studies (54). In the future, other single-copy genes with phylogenetic references and finer resolution, such as rplB, gyrB, and recA, also could be recovered from metagenomic sequences and used for community structure analysis by a similar pipeline (66).
Conclusions.
We developed a fast and efficient pipeline that enables unsupervised diversity analysis with Illumina shotgun data. The pipeline has the scalability to analyze large data sets (5 central processing unit hours for 38 Gb data, with 4.8 Gb peak memory) and can be run on most desktops with more than 5 GB of memory. Since SSU rRNA-based community analysis is an important method in microbial ecology, this method can save projects with existing shotgun sequence data from the additional cost of SSU rRNA amplicon sequencing. Moreover, shotgun sequencing is not as affected by primer bias and chimeras as amplicon sequencing; thus, it can improve the measurement of microbial community structure. As read length and sequencing depth increase, longer and more SSU rRNA gene fragments can be recovered. Thus, clustering and diversity analysis by this pipeline will become even more reliable.
Supplementary Material
ACKNOWLEDGMENTS
We thank Aaron Garoutte, Bangzhou Zhang, Chao Xue, Eliane Gomes, and Caio Rachid for collecting and extracting DNA from soil samples, members of the Ribosomal Database Project (RDP) for discussions and suggestions, and the Institute for Cyber-Enabled Research (iCER) and High Performance Computing Center (HPCC) at Michigan State University for technical support.
J.G. performed the analyses under the supervision of C.T.B., J.M.T., and J.R.C., who also helped with analysis approaches and writing of the paper. J.G. and Q.Z. implemented the pipeline.
Funding Statement
This work was funded in part by the U.S. Department of Energy (DOE) Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494) and by DOE Office of Science grants BER DE-FG02-99ER62848 and DE-SC0004601.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02772-15.
REFERENCES
- 1.Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A 82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Streit WR, Schmitz RA. 2004. Metagenomics–the key to the uncultured microbes. Curr Opin Microbiol 7:492–498. doi: 10.1016/j.mib.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc Natl Acad Sci U S A 103:12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, Knight R, Fierer N. 2011. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem 43:1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Petrosino JF, Knight R, Birren BW. 2011. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 11.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD, Wang J, Antolin M, Artiguenave F, Blottiere H, Borruel N, Bruls T, Casellas F, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Forte M, Friss C, van de Guchte M, Guedon E, Haimet F, Jamet A, Juste C, Kaci G, Kleerebezem M, Knol J, Kristensen M, Layec S, Le Roux K, Leclerc M, Maguin E, Minardi RM, Oozeer R, Rescigno M, Sanchez N, Tims S, Torrejon T, Varela E, de Vos W, Winogradsky Y, Zoetendal E. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shakya M, Quince C, Campbell JH, Yang ZK, Schadt CW, Podar M. 2013. Comparative metagenomic and rRNA microbial diversity characterization using archaeal and bacterial synthetic communities. Environ Microbiol 15:1882–1899. doi: 10.1111/1462-2920.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. 2008. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol 74:2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmieder R, Lim YW, Edwards R. 2012. Identification and removal of ribosomal RNA sequences from metatranscriptomes. Bioinformatics 28:433–435. doi: 10.1093/bioinformatics/btr669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang Y, Gilna P, Li W. 2009. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 25:1338–1340. doi: 10.1093/bioinformatics/btp161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, Yi H, Chun J. 2011. rRNASelector: a computer program for selecting ribosomal RNA encoding sequences from metagenomic and metatranscriptomic shotgun libraries. J Microbiol 49:689–691. doi: 10.1007/s12275-011-1213-z. [DOI] [PubMed] [Google Scholar]
- 18.Bengtsson J, Eriksson KM, Hartmann M, Wang Z, Shenoy BD, Grelet GA, Abarenkov K, Petri A, Rosenblad MA, Nilsson RH. 2011. Metaxa: a software tool for automated detection and discrimination among ribosomal small subunit (12S/16S/18S) sequences of archaea, bacteria, eukaryotes, mitochondria, and chloroplasts in metagenomes and environmental sequencing datasets. Antonie Van Leeuwenhoek 100:471–475. doi: 10.1007/s10482-011-9598-6. [DOI] [PubMed] [Google Scholar]
- 19.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuczynski J, Stombaugh J, Walters WA, Gonzalez A, Caporaso JG, Knight R. 2012. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol Chapter 1:Unit 1E.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah N, Tang H, Doak TG, Ye Y. 2011. Comparing bacterial communities inferred from 16S rRNA gene sequencing and shotgun metagenomics. Pac Symp Biocomput 2011:165–176. [DOI] [PubMed] [Google Scholar]
- 23.Darling AE, Jospin G, Lowe E, Matsen FA, Bik HM, Eisen JA. 2014. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ 2:e243. doi: 10.7717/peerj.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huson DH, Auch AF, Qi J, Schuster SC. 2007. MEGAN analysis of metagenomic data. Genome Res 17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Logares R, Sunagawa S, Salazar G, Cornejo-Castillo FM, Ferrera I, Sarmento H, Hingamp P, Ogata H, de Vargas C, Lima-Mendez G, Raes J, Poulain J, Jaillon O, Wincker P, Kandels-Lewis S, Karsenti E, Bork P, Acinas SG. 2014. Metagenomic 16S rDNA Illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environ Microbiol 16:2659–2671. doi: 10.1111/1462-2920.12250. [DOI] [PubMed] [Google Scholar]
- 27.Mao DP, Zhou Q, Chen CY, Quan ZX. 2012. Coverage evaluation of universal bacterial primers using the metagenomic datasets. BMC Microbiol 12:66. doi: 10.1186/1471-2180-12-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.da Jesus E C, Susilawati E, Smith S, Wang Q, Chai B, Farris R, Rodrigues J, Thelen K, Tiedje J. 2010. Bacterial communities in the rhizosphere of biofuel crops grown on marginal lands as evaluated by 16S rRNA gene pyrosequences. BioEnergy Res 3:20–27. doi: 10.1007/s12155-009-9073-7. [DOI] [Google Scholar]
- 30.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eddy SR. 2009. A new generation of homology search tools based on probabilistic inference. Genome Infect 23:205–211. [PubMed] [Google Scholar]
- 32.Schloss PD. 2009. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS One 4:e8230. doi: 10.1371/journal.pone.0008230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loewenstein Y, Portugaly E, Fromer M, Linial M. 2008. Efficient algorithms for accurate hierarchical clustering of huge datasets: tackling the entire protein space. Bioinformatics 24:i41–i49. doi: 10.1093/bioinformatics/btn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neefs JM, Van de Peer Y, De Rijk P, Chapelle S, De Wachter R. 1993. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res 21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angly FE, Dennis PG, Skarshewski A, Vanwonterghem I, Hugenholtz P, Tyson GW. 2014. CopyRighter: a rapid tool for improving the accuracy of microbial community profiles through lineage-specific gene copy number correction. Microbiome 2:11. doi: 10.1186/2049-2618-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pérez F, Granger BE. 2007. IPython: a system for interactive scientific computing. Comput Sci Eng 9:21–29. doi: 10.1109/MCSE.2007.53. [DOI] [Google Scholar]
- 39.Zhou J, Wu L, Deng Y, Zhi X, Jiang YH, Tu Q, Xie J, Van Nostrand JD, He Z, Yang Y. 2011. Reproducibility and quantitation of amplicon sequencing-based detection. ISME J 5:1303–1313. doi: 10.1038/ismej.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou J, He Z, Yang Y, Deng Y, Tringe SG, Alvarez-Cohen L. 2015. High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. mBio 6:e02288-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL. 2012. Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo F, Ju F, Cai L, Zhang T. 2013. Taxonomic precision of different hypervariable regions of 16S rRNA gene and annotation methods for functional bacterial groups in biological wastewater treatment. PLoS One 8:e76185. doi: 10.1371/journal.pone.0076185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janssen PH. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol 72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunagawa S, Mende DR, Zeller G, Izquierdo-Carrasco F, Berger SA, Kultima JR, Coelho LP, Arumugam M, Tap J, Nielsen HB, Rasmussen S, Brunak S, Pedersen O, Guarner F, de Vos WM, Wang J, Li J, Dore J, Ehrlich SD, Stamatakis A, Bork P. 2013. Metagenomic species profiling using universal phylogenetic marker genes. Nat Methods 10:1196–1199. doi: 10.1038/nmeth.2693. [DOI] [PubMed] [Google Scholar]
- 45.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kent WJ. 2002. BLAT–the BLAST-like alignment tool. Genome Res 12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo C, Rodriguez-R LM, Johnston ER, Wu L, Cheng L, Xue K, Tu Q, Deng Y, He Z, Shi JZ, Yuan MM, Sherry RA, Li D, Luo Y, Schuur EA, Chain P, Tiedje JM, Zhou J, Konstantinidis KT. 2014. Soil microbial community responses to a decade of warming as revealed by comparative metagenomics. Appl Environ Microbiol 80:1777–1786. doi: 10.1128/AEM.03712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. 2007. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res 35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schloss PD, Westcott SL. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol 77:3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharpton TJ, Riesenfeld SJ, Kembel SW, Ladau J, O'Dwyer JP, Green JL, Eisen JA, Pollard KS. 2011. PhylOTU: a high-throughput procedure quantifies microbial community diversity and resolves novel taxa from metagenomic data. PLoS Comput Biol 7:e1001061. doi: 10.1371/journal.pcbi.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farris MH, Olson JB. 2007. Detection of Actinobacteria cultivated from environmental samples reveals bias in universal primers. Lett Appl Microbiol 45:376–381. doi: 10.1111/j.1472-765X.2007.02198.x. [DOI] [PubMed] [Google Scholar]
- 53.Parada A, Needham DM, Fuhrman JA. 14 August 2015. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time-series and global field samples. Environ Microbiol doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 54.Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R, Koljalg U, Pennanen T, Rosendahl S, Stenlid J, Kauserud H. 2013. Fungal community analysis by high-throughput sequencing of amplified markers–a user's guide. New Phytol 199:288–299. doi: 10.1111/nph.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Porras-Alfaro A, Liu KL, Kuske CR, Xie G. 2014. From genus to phylum: large-subunit and internal transcribed spacer rRNA operon regions show similar classification accuracies influenced by database composition. Appl Environ Microbiol 80:829–840. doi: 10.1128/AEM.02894-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Vries FT, Hoffland E, van Eekeren N, Brussaard L, Bloem J. 2006. Fungal/bacterial ratios in grasslands with contrasting nitrogen management. Soil Biol Biochem 38:2092–2103. doi: 10.1016/j.soilbio.2006.01.008. [DOI] [Google Scholar]
- 57.Waring BG, Averill C, Hawkes CV. 2013. Differences in fungal and bacterial physiology alter soil carbon and nitrogen cycling: insights from meta-analysis and theoretical models. Ecol Lett 16:887–894. doi: 10.1111/ele.12125. [DOI] [PubMed] [Google Scholar]
- 58.da C Jesus E, Liang C, Quensen JF, Susilawati E, Jackson RD, Balser TC, Tiedje JM. 28 June 2015. Influence of corn, switchgrass, and prairie cropping systems on soil microbial communities in the upper Midwest of the United States. GCB Bioenergy doi: 10.1111/gcbb.12289. [DOI] [Google Scholar]
- 59.Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J Bacteriol 186:2629–2635. doi: 10.1128/JB.186.9.2629-2635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuczynski J, Liu Z, Lozupone C, McDonald D, Fierer N, Knight R. 2010. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Methods 7:813–819. doi: 10.1038/nmeth.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hunt DE, Klepac-Ceraj V, Acinas SG, Gautier C, Bertilsson S, Polz MF. 2006. Evaluation of 23S rRNA PCR primers for use in phylogenetic studies of bacterial diversity. Appl Environ Microbiol 72:2221–2225. doi: 10.1128/AEM.72.3.2221-2225.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mummey DL, Rillig MC. 2007. Evaluation of LSU rRNA-gene PCR primers for analysis of arbuscular mycorrhizal fungal communities via terminal restriction fragment length polymorphism analysis. J Microbiol Methods 70:200–204. doi: 10.1016/j.mimet.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 63.Liu KL, Porras-Alfaro A, Kuske CR, Eichorst SA, Xie G. 2012. Accurate, rapid taxonomic classification of fungal large-subunit rRNA genes. Appl Environ Microbiol 78:1523–1533. doi: 10.1128/AEM.06826-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Porter TM, Golding GB. 2012. Factors that affect large subunit ribosomal DNA amplicon sequencing studies of fungal communities: classification method, primer choice, and error. PLoS One 7:e35749. doi: 10.1371/journal.pone.0035749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Begerow D, Nilsson H, Unterseher M, Maier W. 2010. Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl Microbiol Biotechnol 87:99–108. doi: 10.1007/s00253-010-2585-4. [DOI] [PubMed] [Google Scholar]
- 66.Roux S, Enault F, Bronner G, Debroas D. 2011. Comparison of 16S rRNA and protein-coding genes as molecular markers for assessing microbial diversity (Bacteria and Archaea) in ecosystems. FEMS Microbiol Ecol 78:617–628. doi: 10.1111/j.1574-6941.2011.01190.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.