Abstract
The CreBC (carbon source-responsive) two-component regulation system of Escherichia coli affects a number of functions, including intermediary carbon catabolism. The impacts of different creC mutations (a ΔcreC mutant and a mutant carrying the constitutive creC510 allele) on bacterial physiology were analyzed in glucose cultures under three oxygen availability conditions. Differences in the amounts of extracellular metabolites produced were observed in the null mutant compared to the wild-type strain and the mutant carrying creC510 and shown to be affected by oxygen availability. The ΔcreC strain secreted more formate, succinate, and acetate but less lactate under low aeration. These metabolic changes were associated with differences in AckA and LdhA activities, both of which were affected by CreC. Measurement of the NAD(P)H/NAD(P)+ ratios showed that the creC510 strain had a more reduced intracellular redox state, while the opposite was observed for the ΔcreC mutant, particularly under intermediate oxygen availability conditions, indicating that CreC affects redox balance. The null mutant formed more succinate than the wild-type strain under both low aeration and no aeration. Overexpression of the genes encoding phosphoenolpyruvate carboxylase from E. coli and a NADH-forming formate dehydrogenase from Candida boidinii in the ΔcreC mutant further increased the yield of succinate on glucose. Interestingly, the elimination of ackA and adhE did not significantly improve the production of succinate. The diverse metabolic effects of this regulator on the central biochemical network of E. coli make it a good candidate for metabolic-engineering manipulations to enhance the formation of bioproducts, such as succinate.
INTRODUCTION
The survival of an organism depends, at least in part, on its ability to sense and respond to changes in the environment. In bacteria, global regulators control the transcription of genes in response to specific external stimuli and metabolic signals, finely tuning different aspects of their physiology to overcome environmental challenges. In Escherichia coli, seven global regulators (ArcA, Crp, Fis, Fnr, Ihf, Lrp, and NarL) directly modulate the expression of about one-half of all genes (1). This facultative aerobe is able to adapt its metabolism to different oxygen availability conditions through the concerted actions of a network of regulators, including the global regulators ArcAB and Fnr (2–4). These regulators affect many metabolic pathways, allowing the cells to reach an adequate redox balance under any given conditions. There is a very close association between carbon and electron flows, and even small differences in oxygen availability have been observed to elicit profound effects on the distribution of carbon fluxes (5).
CreBC (for carbon source responsive) is a global sensing and regulation system affecting genes involved in a variety of functions, including enzymes of intermediary catabolism (6). Previous studies have shown that the creABCD operon is activated (i) during growth in minimal medium when glycolytic carbon sources are being fermented or (ii) during aerobic growth when low-molecular-weight fermentation products are used as carbon sources (6). While creB and creC encode the two-component system (i.e., a cytoplasmic response regulator and a membrane-associated sensor kinase, respectively), creA is a hypothetical open reading frame, and creD encodes an inner membrane protein of unknown function (7). CreC, originally designated PhoM, was first described as a phosphate donor for the PhoB protein, a response regulator that controls the expression of the pho regulon. The regulon includes genes involved in cytoplasmic inorganic phosphate homeostasis, such as phoA, encoding an alkaline phosphatase, and is controlled by PhoBR (8, 9). PhoR autophosphorylates when the concentration of inorganic phosphate falls below a critical threshold. In phoR-null mutants, activation of the pho regulon depends on CreC (10).
The genes that are known to be under the control of CreBC (i.e., the cre regulon) are as follows: (i) the ackA-pta operon, the products of which catalyze the conversion of acetyl-coenzyme A (CoA) into acetate and ATP; (ii) talA, which encodes one of the two transaldolases of the pentose phosphate pathway; (iii) radC, which encodes a RecG-like DNA recombination/repair function; (iv) malE, the first gene in the malEFG maltose transporter operon; (v) trgB, which encodes an ADP-ribose pyrophosphorylase; and (vi) three other genes (creD, cbrA, and cbrB), potentially related to resistance to colicin and other antimicrobials, that have not yet been assigned a specific function (6, 7, 11). A direct-repeat consensus DNA sequence, termed the cre tag (5′-TCACnnnnnnTTCAC-3′, where n represents any nucleotide), was defined based on analysis of the region upstream from the genes known to form the cre regulon and was observed to be required for the control of gene expression in vivo (7). Genome-wide expression profiling with DNA microarrays has revealed that CreBC also affects the expression of the following genes: cbrC, responsible for colicine E2 resistance; mokB, encoding an overlapping regulatory peptide that enables hokB expression; and the uncharacterized genes mppA, ynaI, yafU, and yafE (12).
The capability of global regulators to affect multiple metabolic pathways makes them useful tools for metabolic engineering, as they can be used to change the flow of carbon and reducing power simultaneously. This strategy has been used to manipulate bacterial metabolism to enhance the synthesis of different bioproducts, especially reduced metabolites (13). On the other hand, concern about the costs of energy used for aeration in bioprocesses has renewed attention to the regulation of aerobic and anaerobic bacterial metabolism as a means to achieve the sustainable synthesis of a variety of bioproducts under these conditions (14). Among the different regulatory systems of E. coli, ArcAB has attracted significant attention in the last few years, as manipulations in this sensor/regulator pair offer the possibility of directing carbon flow toward the synthesis of reduced bioproducts under low-aeration conditions (15–17). In our laboratory, we analyzed the effects of arcA mutants on the central carbon catabolism of E. coli using glucose and glycerol in microaerobiosis and anaerobiosis and observed that mutations in arcA resulted in significant increases in the synthesis of polyhydroxyalkanoates (18–21) and ethanol (22, 23). In these studies, the constitutive allele creC510 was observed to affect the intracellular redox state, enhancing carbon catabolism in an arcA genetic background so that part of the excess reducing power generated by the arcA mutants was consumed by the augmented amount of carbon intermediates due to creC510, further increasing the synthesis of reduced products (18).
The results obtained with the creC510 arcA double mutants suggested that the CreBC system could also be an interesting target for metabolic manipulations in E. coli and prompted us to further investigate its metabolic effects. The aim of this work was to evaluate the potential of CreC as a new tool for the design of bacterial strains suitable for the synthesis of different bioproducts. For this purpose, we characterized the effects of the regulator on central carbon metabolism, analyzing physiological traits, carbon flow, and redox balance, focusing on low oxygen availability conditions in the presence of excess carbon source. Succinate was selected as a model metabolite, and several metabolic manipulations were implemented in a ΔcreC mutant to evaluate the potential of this genetic background for the synthesis of the carboxylic acid.
MATERIALS AND METHODS
Bacterial strains, mutant construction, plasmids, and oligonucleotides.
All E. coli strains were derivatives of K1060, a K-12 strain, and are listed in Table 1, along with the plasmids used in this study. All null mutants reported in this work were constructed by allelic replacement (24). Briefly, a kanamycin resistance cassette (FLP recombination target [FRT]-kan-FRT) was amplified by PCR from template plasmid pKD4 with the corresponding primers (Table 1). The purified PCR product was electroporated into E. coli K1060 carrying pKD46 (a helper plasmid that expresses the λ-Red functions). Insertion of the FRT-kan-FRT cassette into the correct locus was confirmed by colony PCR of kanamycin-resistant recombinants. For the construction of mutant K1060C, bearing the constitutive creC510 allele, a prior step was needed. E. coli strain 58-161, F− Strr (creC510), carrying pKD46 was transformed with the FRT-kan-FRT cassette in order to add a selection marker near the creABCD operon in the intergenic region limited by the convergent open reading frames yjjX and trpR, located 2.5 kb upstream from creABCD. The resulting mutant, E. coli 58KF, was used as the donor strain in P1 transduction, in which K1060 was the recipient strain. Kanamycin-resistant clones were selected, and the presence of the creC510 allele was confirmed by PCR and DNA sequencing. The kan cassette was removed in all the strains by transforming them with the thermosensitive plasmid pCP20 (25), encoding the Saccharomyces cerevisiae FLP recombinase. The excision of kan, as well as gene deletion, was confirmed by PCR. Plasmid pCP20 was removed by two consecutives passages at 42°C. Inactivation of ackA in E. coli CEA1060 (ΔcreC ΔadhE ΔackA) was further corroborated by determining the amount of acetate present in the supernatants of cultures grown in M9 minimal medium supplemented with glucose at 30 g liter−1, as described below (the CEA1060 mutant produced 50% less acetate than its parental strain, CE1060).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in the study
| Strain, plasmid, or oligonucleotide | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. coli strains | ||
| K1060b | F− fadE62 lacI60 tyrT58(AS) fabB5 mel-1 | 69 |
| K1060C | Same as K1060, but creC510 by K1060 × P1(58-161, F− Strr) | This work |
| DB1060 | Same as K1060, but ΔcreB by allelic replacement | This work |
| DC1060 | Same as K1060, but ΔcreC by allelic replacement | This work |
| DBC1060 | Same as K1060, but ΔcreB ΔcreC by allelic replacement | This work |
| CE1060 | Same as DC1060, but ΔadhE by allelic replacement | This work |
| CEA1060 | Same as CE1060, but ΔackA by allelic replacement | This work |
| 58-161, F− Strrb | F− relA1 rpsL100(Strr) spoT1 metB1 creC510 | 70 |
| 58KF | Same as 58-161, F− Strr, but Kmr by insertion of kan in an intergenic region between yjjX and trpR | This work |
| Plasmids | ||
| pCP20 | Helper plasmid used for kan excision; Saccharomyces cerevisiae FLP λ cI857 λPR repA(Ts); Apr Cmr | 25 |
| pKD4 | Template plasmid carrying the FRT-kan-FRT cassette; Kmr | 24 |
| pKD46 | Helper plasmid expressing the λ-Red functions; Apr | 24 |
| pSBF2 | Plasmid pDHK30 (71) carrying FDH1 from Candida boidinii under the control of the lac promoter; Kmr | 43 |
| pEcPpc | Plasmid pTrc99A (72) carrying ppc from E. coli under the control of the lac promoter; Apr | 39 |
| Oligonucleotidesc (5′→3′) | ||
| cre/F | TAG GCC TGA TAA GAC GTG GCG CAT CAG GCA TCG TGC ACC GAA TGC CGG ATG TGT AGG CTG GAG CTG CTT C (K1060C construction) | This work |
| cre/R | GCC GCG TCT TAT CAT GCC TAC CAA ACA TAT TGA AAT TAC GGG TAT TTG TAC ATA TGA ATA TCC TCC TTA G (K1060C construction) | This work |
| creB-KF/F | TTA GCG CGG TTC CTG TCA TGC CGT GGC GGC AAT AAC AGA GGC GAT TTA TGG TGT AGG CTG GAG CTG CTT C (DB1060 and DBC1060 construction) | This work |
| creB-KF/R | GCC CAG CAA CAA CCG CAT GCC GAT ACG CAT TAC AGG CCC CTC AGG CTA TAC ATA TGA ATA TCC TCC TTA (DB1060 construction) | This work |
| creC-KF/F | GTC AAA GAA GTT AAA CCG GGC GTG CGA AGA GCA ACG GAG GGG ACG TTG ATC GTG TAG GCT GGA GCT GCT TC (DC1060 construction) | This work |
| creC-KF/R | GAC GTG TTC CTG ATC CAC TTC GGC GCT TAG CGT GAT GCA ACC GCT CTC GGG CAT ATG AAT ATC CTC CTT AG (DC1060 and DBC1060 construction) | This work |
| DW-ack/F | AAC TCA GCG GGA CAA CG (CEA1060 construction) | This work |
| DW-ack/R | GAA AGC AGA CCT TCA ACG (CEA1060 construction) | This work |
| DW-adh/F | AGA CGC GCT GAC AAT ACG (CE1060 and CEA1060 construction) | This work |
| DW-adh/R | GCC ACC AGA CGC ATA ACC (CE1060 and CEA1060 construction) | This work |
Ap, ampicillin; Cm, chloramphenicol; Km, kanamycin; Str, streptomycin.
Strain obtained through the E. coli Genetic Stock Center, Yale University, New Haven, CT.
The use of each oligonucleotide in the construction of mutant strains is indicated in parentheses.
Growth media and culture conditions.
The medium used for shaken-flask experiments was M9 minimal medium containing (per liter of deionized H2O) 6.0 g of Na2HPO4, 3.0 g of KH2PO4, 0.5 g of NaCl, 1.0 g of NH4Cl, 0.4 g of MgSO4, 0.01 g of CaCl2, and 0.06 g of ammonium iron(III) citrate. MgSO4, CaCl2, and ammonium iron(III) citrate were added to the medium after autoclaving and cooling. Glucose was used at 30 g liter−1 as the sole carbon source in all experiments. The aeration level was adjusted by a combination of rotational agitation and the relationship between the volume of culture medium (Vm) and the volume of the flask (Vf) as follows: for high aeration, 250 rpm and a Vm/Vf ratio of 1/10; for low aeration, 125 rpm and a Vm/Vf ratio of 1/2. For high aeration, 250-ml Erlenmeyer flasks were used, while 10- or 50-ml cylindrical bottles (penicillin bottles) with cotton plugs were used for low aeration. Nonaerated cultures were implemented in sealed tubes filled with culture medium (10 ml) and agitated at 4 rpm to keep the cells in suspension. Working cultures were inoculated from overnight precultures (started from single colonies grown overnight on LB agar) in the same culture medium and under the same growth conditions used in the experiment (i.e., with the same Vm/Vf ratio). Nonaerated precultures were inoculated with a low-aerated prepreculture to ensure adaptation of the cells to this condition. Isopropyl-β-d-1-thiogalactopyranoside (IPTG) (0.1 or 1 mM), NaHCO3 (100 mM), and antibiotics (kanamycin and ampicillin at 50 and 100 μg ml−1, respectively) were added whenever needed.
Analytical determinations.
The cell dry weight (CDW) was determined in cell pellets of 10-ml culture samples that were centrifuged for 10 min at 4°C and 10,000 × g. The supernatant was separately stored for metabolite analysis. The cells were washed twice with the same volume of 150 mM NaCl and finally dried in an oven at 65°C until a constant weight was reached. The supernatant was filtered through a 0.22-μm-pore-size syringe filter (Chrom Tech Inc., Apple Valley, MN) and stored at 4°C for high-pressure liquid chromatography analysis (in a LC-20AT Prominance [Shimadzu Corp., Kyoto, Japan] chromatograph equipped with an Aminex column [HPX-87-H; catalog no. 125-0140; Bio-Rad Laboratories Inc., Hercules, CA] operated at 50°C). A UV detector (SPD-20AV; Shimadzu Corp.) set to 215 nm was used for the quantification of organic acids. The mobile phase consisted of 5 mM H2SO4 run at a flow rate of 0.6 ml min−1. Peaks were identified by their characteristic retention times against a set of standards of known organic acids (Sigma-Aldrich Co., St. Louis, MO). The ethanol concentration was measured by using an enzymatic kit based on alcohol dehydrogenase (Sigma-Aldrich Co.). Glucose was measured in supernatants by the glucose oxidase-peroxidase method utilizing a commercial kit (Wiener Laboratorios SAIC, Rosario, Argentina).
The NADH/NAD+ and NADPH/NADP+ ratios were obtained from the content of each nucleotide, quantified in the pellet fractions of 1-ml culture samples. The samples were transferred to precooled plastic tubes, and the metabolic activity was quenched by immersion of the tubes in liquid N2. Thawed samples were treated with 300 μl of either 0.2 M HCl (NAD[P]H extraction) or 0.2 M NaOH (NAD[P]+ extraction). Acid/alkaline extraction was carried out at 50°C for 10 min, and samples were rapidly placed on ice to cool them to 0°C afterwards. Suspensions were neutralized by dropwise addition of 1 M HCl or NaOH, and cellular debris was removed by centrifuging at 14,000 × g for 5 min. The supernatants were then transferred to new tubes and immediately used for cofactor measurements. Nucleotide determination was performed as described by Bernofsky and Swan (26), using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide as the final electron acceptor, as modified by Nikel et al. (22, 27, 28). The dinucleotide content was normalized to the CDW as indicated by Nikel and Chavarría (29). All reagents were purchased from Sigma-Aldrich Co.
In vitro enzyme activity measurements.
The acetate kinase (AckA) assay is based on the formation of acetyl-hydroxamate (30, 31). The assay mixture (1-ml final volume) consisted of 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 10 mM ATP, 800 mM CH3CO2K, and 700 mM freshly neutralized NH2OH. Working solutions of neutralized NH2OH were freshly prepared by dropwise addition of 3.5 M KOH to an equal volume of 4.0 M NH2OH·HCl. The cell extract was added to the reaction mixture and incubated for 5 min at room temperature, after which 1 ml of 10% (wt/vol) trichloroacetic acid was added, immediately followed by 1 ml of freshly prepared 1.25% (wt/vol) FeCl3 in 1 N HCl. After another 5-min incubation period at room temperature, the mixture was centrifuged at maximal velocity for 1 min, and the absorbance of the supernatant was read at 540 nm. For d-lactate dehydrogenase (LdhA), the activity was assayed by measuring the pyruvate-dependent reduction of NADH (32). The assay mixture consisted of 50 mM sodium phosphate buffer (pH 7.5), 25 mM pyruvate, and 7.5 mM NADH. The cell extract was added to the reaction mixture to initiate the assay, and the rate of change in the absorption at 340 nm was recorded at 30°C using a microtiter plate reader. Calibration curves were performed using lithium-potassium acetyl-phosphate and NADH for AckA and LdhA, respectively. One unit of AckA or LdhA activity was defined as the quantity of enzyme that catalyzes the formation of 1 μmol product in 1 min at 30°C. All reagents were purchased from Sigma-Aldrich Co.
Statistical analysis.
The reported experiments were independently repeated at least twice (as indicated in the corresponding figure legend or table), and the mean value of the corresponding parameter ± standard deviation is presented. The statistical significance between multiple comparisons was obtained by a two-tailed Student's t test. Data were considered statistically significant when P was <0.05.
RESULTS
CreC affects the growth and the fermentation profile of E. coli under low oxygen availability conditions.
Previous reports suggested that CreC-dependent regulation is affected by aeration. To analyze this effect further, an E. coli strain carrying the wild-type creC (K1060), a creC deletion derivative (DC1060), and another strain carrying the constitutive creC510 allele (K1060C) (Table 1) were grown at three levels of aeration (high, low, and no aeration). The growth and production of different metabolites were determined to characterize the metabolic responses of each strain to oxygen availability in M9 minimal medium cultures supplemented with 30 g liter−1 glucose.
Clear differences were observed in growth and in metabolite secretion, mainly between E. coli DC1060 and the other two strains, both of which showed similar behavior under the three culture conditions (Table 2). Biomass formation levels at 24 h were similar under high and low aeration for all strains, but surprisingly, strain DC1060 grew twice as much as the other strains with no aeration (see Fig. S1 in the supplemental material). Metabolite distributions in highly aerated cultures were similar for all strains, with acetate as the main secreted product. Under low aeration, E. coli K1060 and K1060C showed similar trends, while strain DC1060 secreted more formate (2-fold), succinate (13-fold), and acetate (1.5-fold) than the other strains but approximately half the lactate (P < 0.05). Differences in succinate production in nonaerated cultures were even more marked, as the null mutant produced 36% more of the metabolite than under low aeration, while the other strains (K1060 and K1060C) had very low accumulation levels (Table 2). Succinate formation in nonaerated cultures of the ΔcreC strain was around 50-fold higher than in cultures of E. coli K1060 grown under these conditions (9.4 versus 0.2 mmol gCDW−1). Interestingly, and in contrast to what was observed under low aeration, in 24-h nonaerated cultures, the null mutant accumulated more lactate (+60%) and less acetate (−60%) than the other two strains, while all the strains accumulated similar amounts of formate (Table 2). While all the strains produced larger amounts of ethanol when no aeration was supplied than under the other growth conditions, K1060C was the strain that accumulated the highest levels, followed by wild-type E. coli K1060, and strain DC1060 had the lowest ethanol formation.
TABLE 2.
Growth and fermentation profiles of the strains under study under different conditions of oxygen availabilitya
| Aeration condition | E. coli strainb | Biomass (gCDW liter−1) | Yield of fermentation metabolites on biomass (mmol gCDW−1) |
||||
|---|---|---|---|---|---|---|---|
| Succinate | Lactate | Formate | Acetate | Ethanol | |||
| High | K1060 | 1.29 ± 0.02 | 0.4 ± 0.1 | 1.0 ± 0.1 | 0 | 14 ± 2 | 9.0 ± 1.0 |
| K1060C | 1.25 ± 0.01 | 1.0 ± 1.0 | 1.0 ± 0.1 | 0 | 12 ± 1 | 9.0 ± 0.1 | |
| DC1060 | 1.25 ± 0.01 | 0.3 ± 0.1 | 1.0 ± 0.1 | 0 | 11 ± 1 | 6.0 ± 0.1 | |
| Low | K1060 | 0.38 ± 0.02 | 0.5 ± 0.2 | 46 ± 8 | 17 ± 3 | 89 ± 13 | 18 ± 5 |
| K1060C | 0.39 ± 0.02 | 0.4 ± 0.1 | 48 ± 7 | 22 ± 3 | 91 ± 9 | 17 ± 2 | |
| DC1060 | 0.36 ± 0.01 | 6.9 ± 0.3 | 22 ± 2 | 44 ± 3 | 127 ± 10 | 19 ± 3 | |
| DB1060 | 0.39 ± 0.01 | 6.3 ± 0.2 | 19 ± 1 | 48 ± 1 | 120 ± 4 | ND | |
| DBC1060 | 0.36 ± 0.01 | 6.7 ± 0.3 | 20 ± 1 | 50 ± 5 | 125 ± 9 | ND | |
| None | K1060 | 0.14 ± 0.01 | 0.2 ± 0.1 | 27 ± 8 | 61 ± 2 | 118 ± 4 | 68 ± 10 |
| K1060C | 0.11 ± 0.04 | 0.1 ± 0.1 | 19 ± 8 | 41 ± 11 | 150 ± 7 | 120 ± 23 | |
| DC1060 | 0.26 ± 0.01 | 9.4 ± 0.9 | 45 ± 5 | 58 ± 5 | 46 ± 9 | 29 ± 12 | |
| DB1060 | 0.23 ± 0.02 | 11.0 ± 1.0 | 46 ± 4 | 56 ± 4 | 53 ± 10 | ND | |
| DBC1060 | 0.24 ± 0.02 | 11.6 ± 0.2 | 51 ± 6 | 61 ± 5 | 55 ± 3 | ND | |
Cells were grown in M9 minimal medium supplemented with 30 g liter glucose−1 for 24 h. ND, not determined. The results represent the mean values ± standard deviations of three determinations from at least four independent cultures. CDW, cell dry weight.
The E. coli strains used were K1060 (wild type), K1060C (creC510; constitutive creC allele), DC1060 (ΔcreC), DB1060 (ΔcreB), and DBC160 (ΔcreB ΔcreC).
The effect of CreC on the fermentation profile and enzyme activities is mediated by its partner response regulator, CreB.
As CreC is the sensor protein of a two-component signal transduction system (the partner response regulator is encoded by creB), the possibility that the changes observed could be due to cross talk with other regulators was considered. To investigate this possibility, the fermentation profile of a ΔcreB derivative of strain K1060 (E. coli DB1060) was analyzed under low aeration and no aeration, conditions under which the most significant differences between E. coli DC1060 and the parental strain had been observed. The fermentation profile was also analyzed in a ΔcreB ΔcreC double mutant (E. coli DBC1060).
While under low aeration the final biomass was the same for all the strains, 24-h nonaerated cultures of the three null mutants produced 2-fold more biomass than the wild-type strain under this condition (Table 2). When comparing the specific production of organic acids under both low aeration and no aeration, all the deletion mutant strains produced comparable amounts of all the metabolites, with similar differences compared to the wild-type strain. The same metabolic profile was obtained in strains in which either component or both were inactivated, clearly indicating that the effects observed were due to the regulation exerted by the CreBC two-component system.
CreC influences the NADH/NAD+ ratio.
When no aeration was supplied, ethanol levels were lower in cultures of the ΔcreC strain, while the creC510 mutant accumulated the largest amounts. These results suggested that CreC affects the redox state of the cells, as the formation of this metabolite is associated with high availability of reducing equivalents. To obtain an accurate measurement of the intracellular redox state of each strain, the levels of the cofactors NAD(P)H and NAD(P)+ were quantified in vitro, and their ratios were determined.
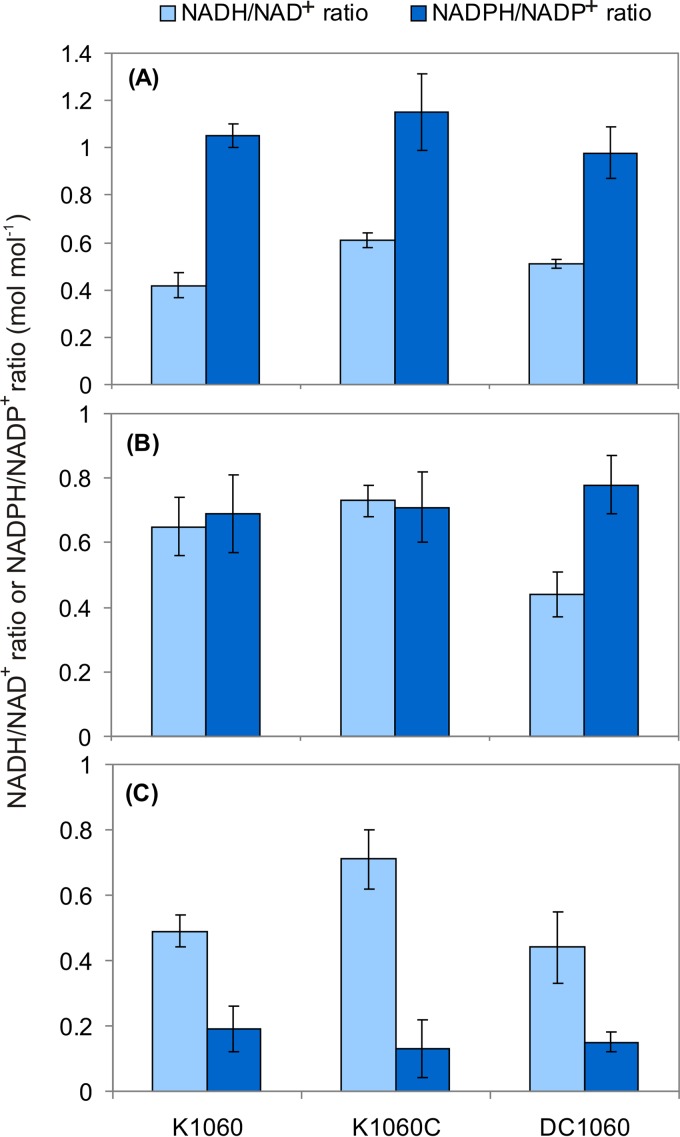
While under high aeration the NADPH/NADP+ ratios were higher than the NADH/NAD+ ratios for all strains, the opposite was observed in nonaerated cultures. The ratios of the two types of cofactors were similar under low aeration, except for the null mutant, in which this relationship was approximately the same under high and low aeration. Interestingly, the NADPH/NADP+ ratios were observed to vary over a much wider range under the conditions tested, while variations in the NADH/NAD+ ratio were more discrete. However, when the values obtained for each of the strains were compared, no significant variations were detected among the NADPH/NADP+ ratios, while differences in the NADH/NAD+ ratios were observed under the three aeration conditions. In all cases, the highest NADH/NAD+ ratio was observed for the mutant carrying the constitutive allele creC510 (E. coli K1060C), and under low aeration, the deletion mutant (E. coli DC1060) had the lowest redox ratio values (Fig. 1). Under high aeration, a condition under which CreC is believed to be inactive, the NADH/NAD+ ratios differed slightly, and the highest value was observed for E. coli K1060C, which has the constitutively active regulator. Under low aeration, where CreC is expected to be active, the differences were more conspicuous (Fig. 1). Under this condition, E. coli DC1060 showed clear differences from both the wild-type strain and the creC510 mutant, with a NADH/NAD+ ratio 47% lower than the value for E. coli K1060 and 66% lower than that of E. coli K1060C. These results indicate that, under oxygen limitation, CreC affects the availability of redox cofactors, promoting a more reduced intracellular environment. When no aeration was supplied, the wild-type strain and the null mutant behaved similarly, but strain K1060C showed a significantly higher NADH/NAD+ ratio than the other two E. coli strains (P < 0.05).
FIG 1.
Determination of the NADH/NAD+ and NADPH/NADP+ ratios in E. coli K1060 (wild-type strain), K1060C (carrying the constitutive creC510 allele), and DC1060 (ΔcreC). Cells were grown in M9 minimal medium containing 30 g liter−1 glucose under high aeration (A), low aeration (B), and no aeration (C). Cells were harvested at mid-exponential phase. The results represent the averages ± standard deviations of duplicate measurements of at least two independent cultures.
AckA and LdhA activities are subjected to CreC regulation.
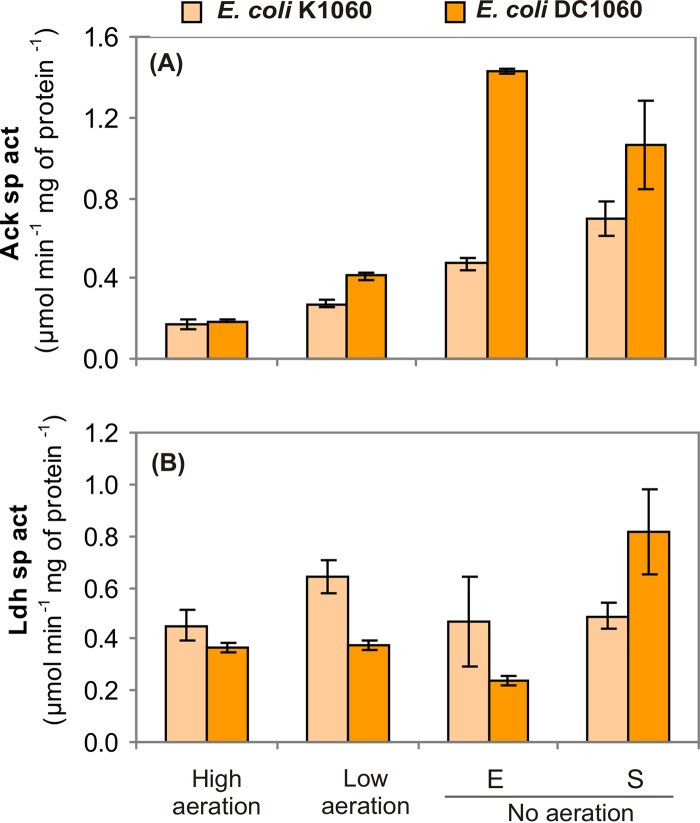
The metabolic profiles of the strains showed differences in metabolite distribution affecting several organic acids, mainly succinate, formate, lactate, and acetate. CreC has been reported to activate the pta-ackA operon (7), so the increase in acetate observed in the absence of CreC led us to investigate whether the variations in acetate synthesis could be attributed to differences in AckA (acetate kinase) activity. On the other hand, variations in the amount of lactate could be attributed to metabolic regulation or to differences in LdhA (lactate dehydrogenase) activity. However, LdhA is not considered to be regulated by CreBC (at least at the transcriptional level), since the ldhA promoter region does not present a cre tag. In an attempt to elucidate these interrogants, strains K1060 and DC1060 were grown under the three aeration conditions mentioned above, and the specific activities of AckA and LdhA were determined in vitro during exponential growth (Fig. 2).
FIG 2.
Acetate kinase (A) and lactate dehydrogenase (B) in vitro specific activities (sp act) of cells grown in M9 minimal medium containing 30 g liter−1 glucose under high aeration, low aeration, and no aeration. The samples were harvested at mid-exponential phase, except under anaerobic conditions, where the activity was measured both in the exponential (E) and early stationary (S) phases (see Fig. S1 in the supplemental material for detailed information on sampling times). The results represent the averages ± standard deviations of duplicate measurements of at least two independent cultures.
Under high oxygen availability, no significant differences were observed in the specific AckA or LdhA activities between the two strains, while under low aeration, the enzyme activities reflected the metabolic profile shown in Table 2. That is, the ΔcreC strain had higher AckA activity and lower LdhA activity than the wild-type strain, in agreement with the higher acetate and lower lactate production under these conditions. The specific activities of AckA and LdhA in the ΔcreB mutants were also analyzed under low aeration to compare them with the corresponding metabolic profiles, and as expected, the two strains behaved identically (data not shown). From these experiments, it can be concluded that, either directly or indirectly, CreC affects the activities of both AckA and LdhA.
In nonaerated cultures, however, there was no clear correspondence between enzyme activity and metabolite levels. Although E. coli DC1060 produced more lactate and less acetate than the parental strain in 24-h cultures, the LdhA and AckA activities (measured during the exponential phase) in the nonaerated cultures were approximately 50% lower and 200% higher, respectively (P < 0.05). The discrepancy observed in nonaerated cultures led us to make additional measurements in order to consider possible variations due to culture age: (i) metabolite levels were determined in the exponential-phase cultures used for the initial enzyme determinations, and (ii) enzyme activities were also measured at the onset of the stationary phase of growth. Similar amounts of acetate were detected in the supernatants of exponential-phase nonaerated cultures of both strains (15.6 mM in K1060 versus 12.6 mM in DC1060), but K1060 produced more lactate than DC1060 (0.7 mM versus undetectable levels, respectively) under these conditions, in accordance with the differences observed in LdhA activity.
It must be noted that, in the absence of aeration, E. coli DC1060 produced more biomass and grew faster than the wild-type strain, making it difficult to choose the best moment along the growth curve for comparisons. For this reason, the cultures used for enzyme determinations were harvested in what could be considered exponential and early stationary phase in each case (the sampling times are shown in Fig. S1 in the supplemental material).
Although in early-stationary-phase nonaerated cultures the AckA activity did not match the observed trend in acetate formation, differences between the two parameters were more moderate than in the exponential phase (Fig. 2A). In contrast, the results obtained for LdhA in the early stationary phase of nonaerated cultures were quite different from those obtained in the exponential phase. The early-stationary-phase cultures of strain DC1060 displayed higher LdhA activity than the wild-type K1060 (Fig. 2B) and produced more lactate (6.3 mM in DC1060 versus undetectable levels in K1060), in accordance with the larger amounts of lactate measured in 24-h nonaerated cultures (Table 2).
To further investigate this, we measured pH in the cultures, as this parameter is known to affect LdhA activity (33). Under high and low aeration, the pH was observed to decrease throughout the exponential phase for all the strains and to remain stable upon entry into the stationary phase. No differences in pH evolution were observed among the strains except in the unaerated cultures. Under this condition, the wild type (K1060) grew very little, and the pH decreased only slightly, while the null mutant (DC1060) grew more and had a much greater drop in pH (see Fig. S1 in the supplemental material).
Given that (i) the ΔcreC mutant exhibited lower LdhA activity and lactate levels than the wild-type strain under all conditions except in stationary-phase nonaerated cultures and (ii) appreciable differences in pH were observed only under this condition, it is apparent that the low pH could be at least partially responsible for the increase in LdhA activity observed in DC1060, which in turn correlates with higher lactate levels.
Manipulation of CreC as a genetic tool for the improvement of succinate formation.
One of the most interesting properties of the creC-null mutant was the augmented production of succinate, since the organic acid is a biotechnologically interesting compound (34). Many strategies have been employed to optimize the production of the metabolite (35), such as overexpressing carboxylating enzymes (36–39) and supplying carbon dioxide by the addition of sodium bicarbonate to the culture medium (39–41). Since the creC mutant produced larger amounts of succinate than the parental strain, additional strategies were tested to increase the titer of the metabolite in E. coli DC1060. Two plasmids were introduced into the strains: plasmid pEcPpc, carrying the carboxylating enzyme phosphoenolpyruvate carboxylase (PpcEc) from E. coli (42), and plasmid pSBF2, carrying a NAD+-dependent formate dehydrogenase from the methylotrophic yeast Candida boidinii (FDH1Cb) (43–45). In both plasmids, the corresponding genes are under the control of the lac promoter, and their expression can be induced by IPTG. Taking into account that the ΔcreC mutant produces large amounts of formate under low aeration, expression of FDH1Cb could help convert the excess formate to CO2 and NADH, further increasing the supply of precursors necessary for succinate synthesis. This surplus of CO2, together with that provided by NaHCO3 (added to the culture medium), could be funneled to the pyruvate-oxaloacetate node by means of the overexpressed PpcEc, which fixes carbon dioxide to pyruvate, forming oxaloacetate, which, in turn, could be converted to succinate (Fig. 3).
FIG 3.
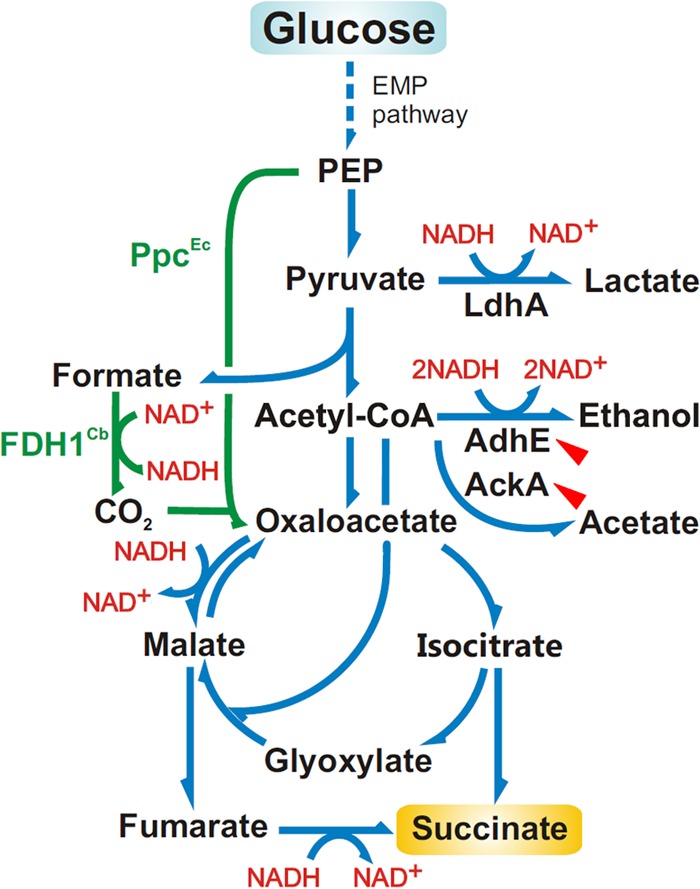
Diagram of the main metabolic pathways leading to succinate formation from glucose in E. coli. The genes encoding PpcEc (phosphoenolpyruvate carboxylase) from E. coli and FDH1Cb (NADH-forming formate dehydrogenase) from C. boidinii were overexpresed in plasmids pEcPpc and pSBF2, respectively (the corresponding reactions are highlighted in green). The genes encoding AdhE (alcohol dehydrogenase) and AckA (acetate kinase) were knocked out in an attempt to enhance succinate accumulation (indicated by the red arrowheads). Note that several reactions within the biochemical network were grouped for the sake of simplicity. EMP pathway, Embden-Meyerhof-Parnas pathway; PEP, phosphoenolpyruvate; Acetyl-CoA, acetyl coenzyme A.
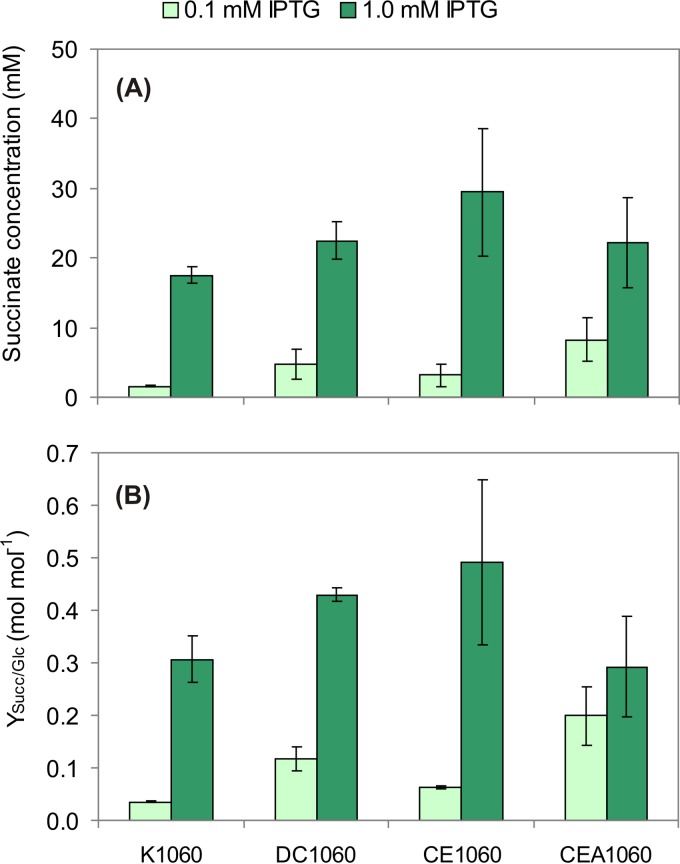
To test this hypothesis, strains K1060 and DC1060, cotransformed with plasmids pSBF2 and pEcPpc, were grown for 48 h under low aeration with the addition of 100 mM NaHCO3. Two different concentrations of IPTG (0.1 mM and 1.0 mM) were used to get a better estimation of the relative weights of the conversions catalyzed by PpcEc and FDH1Cb in succinate production. With the lowest concentration of IPTG, the creC mutant produced 2.7-fold more succinate than the parental strain (Fig. 4A). The yield of succinate on glucose followed this trend, being 0.04 and 0.12 mol mol−1 for K1060 and DC1060, respectively (Fig. 4B). When IPTG was supplied at 1.0 mM, succinate production was triggered, with marked increases in both titer and yield in both strains. The succinate yield of the ΔcreC mutant strain overexpressing ppcEc and FDH1Cb was around 40% higher than that of the wild type (K1060/pSBF2/pEcPpc) under the same culture conditions.
FIG 4.
Profile of succinate formation in the E. coli strains under study. Cells were grown in M9 minimal medium containing 30 g liter−1 glucose and 100 mM NaHCO3 under low aeration for 48 h. The E. coli strains tested were K1060 (wild-type strain), DC1060 (ΔcreC), CE1060 (ΔcreC ΔadhE), and CEA1060 (ΔcreC ΔadhE ΔackA). All the bacteria were transformed with plasmids pSBF2 (carrying FDH1Cb, encoding a NADH-forming formate dehydrogenase from C. boidinii) and pEcPpc (carrying ppcEc, encoding the endogenous phosphoenolpyruvate carboxylase from E. coli). The expression of the genes in these plasmids was induced by addition of IPTG at two concentrations (0.1 mM and 1 mM). Succinate was assayed in culture supernatants, and the results are reported as the final concentration (A) and yield of succinate on glucose (YSucc/Glc) (B). The results represent the averages ± standard deviations of duplicate measurements of at least two independent cultures.
The production of other metabolites in the strains carrying the plasmids (see Fig. S2 in the supplemental material) showed some modifications compared to the strains without plasmids (Table 2). For example, formate synthesis was lower in the strains containing plasmids than in the plasmidless counterparts, in accordance with the results expected from FDH1Cb overexpression.
In order to eliminate side products and further increase reducing power availability to stimulate succinate production, additional modifications in the genetic background of strain DC1060 were tested. The ethanol pathway was deleted to increase NADH availability, and ackA was also eliminated to save carbon atoms in the form of acetyl-CoA, a substrate for succinate formation via the glyoxylate pathway (46). The double mutant CE1060 (ΔcreC ΔadhE) and the triple mutant CEA1060 (ΔcreC ΔadhE ΔackA) were cotransformed with plasmids pEcPpc and pSBF2, and succinate was measured in cultures of these strains grown under the same conditions described above. In contrast to what was expected, the strains did not present much larger amounts or yields of succinate than E. coli DC1060/pSBF2/pEcPpc (Fig. 4). In all cases, a sharp increase in succinate production was observed with larger amounts of IPTG. These results indicate that overexpression of ppcEc and FDH1Cb had a marked effect on succinate production, while the mutations in adhE and ackA did not.
Acetate, formate, lactate, and pyruvate were measured in the cultures of the different strains (see Fig. S2 in the supplemental material). Acetate formation in the ackA mutant was reduced compared to the parental strain but not completely abolished, probably due to the activity of alternative pathways for acetate formation (e.g., PoxB). In all cases, higher expression of the heterologous enzymes (1.0 mM IPTG) tended to reduce the differences in the metabolic profiles among the mutants and caused a decrease in the amounts of formate compared to the low induction level (0.1 mM IPTG).
DISCUSSION
While analyzing different arcA mutants, a constitutive creC allele was discovered to be responsible for the peculiar phenotypic traits of one of the mutants that grew better than the others (18). In that study, the constitutive creC510 allele was observed to ameliorate some of the phenotypic characteristics of ΔarcA mutants, and the effect was attributed to increased substrate consumption. Further work analyzed the carbon fluxes of strains harboring arcA and creB mutations in microaerobic glucose-limited chemostat cultures (47), revealing that these regulators share the control of carbon catabolism under these conditions. These results raised questions concerning the contribution of the CreBC system to central carbon metabolism under different oxygen availability conditions when the carbon source does not limit bacterial growth. This question was of special interest, since it was reported that CreC does not respond only to the composition of the medium (i.e., whether it is rich or mineral medium), as was initially believed (7), but its activation is also dependent on the aeration level and the carbon source (6).
To further analyze the metabolic effects of CreBC, three different aeration levels (high, low, and no aeration) were tested to determine the behavior of creC mutants considering different aspects of cell metabolism, such as the metabolite profile, enzyme activities, and redox state. These experiments showed that CreC has a clear effect on carbon distribution that varies under different oxygen availability conditions. CreC was observed to affect the accumulation of formate, lactate, acetate, ethanol, and succinate. These effects were shown to be mediated by CreB, ruling out the possibility that they could be due to interaction of CreC with other noncognate response regulators. This observation is not trivial, since CreC was originally associated with the pho regulon (48), interacting with the response regulator PhoB. This affirmation, however, does not exclude the possibility that other regulators could affect the expression of genes in common.
Differences in the distributions of metabolites under the aeration conditions analyzed in this work were also associated with the intracellular redox state. In previous studies that analyzed ΔarcA creC510 double mutants, the constitutive creC510 allele was observed to contribute to a high ethanol/acetate ratio (18), while the opposite was observed in creB mutants (47). These differences were also observed in this study. For example, ethanol accumulation, which is normally correlated with a reduced intracellular state (i.e., an increased NADH/NAD+ ratio), was lower in the Δcre mutants and higher in the mutant carrying the constitutive creC510 allele when aeration was suppressed compared to the parental strain.
The effects of CreC on the redox state of the cells were further analyzed by measuring the ratios between reduced and oxidized cofactors. Under all oxygen conditions, the strain carrying creC510 had a higher NADH/NAD+ ratio than the wild-type strain and the ΔcreC mutant, showing that CreC contributes to a more reduced intracellular redox state. While the null mutant had significant (P < 0.05) differences in NADH/NAD+ ratios from both the parental strain and the creC510 mutant under low aeration, this was not observed under high aeration or no aeration, indicating that the effect of CreC on redox potential was stronger under intermediate oxygen availability conditions.
The determination of reduced and oxidized cofactors under the three aeration conditions tested revealed additional interesting data. While under high aeration the NADPH/NADP+ ratios were higher than the NADH/NAD+ ratios for all the strains, the opposite was observed in unaerated cultures. This observation could have two different implications. The first is that the degrees of oxidation of the two cofactors under high aeration and no aeration reflect differences in cofactor usage. NADP(H) is considered the preferred cofactor in anabolism and stress resistance mechanisms, whereas NAD(H) is mainly related to catabolism and fermentation processes (49). Therefore, the lower NADPH/NADP+ ratio compared to the NADH/NAD+ ratio in nonaerated cultures is possibly related to a decrease in the NADPH content, which in turn is reflected in the low biomass formation under these conditions. Another possible explanation could be that cells regulate NADH/NAD+ ratios more tightly than NADPH/NADP+ ratios, as the latter were observed to vary over a much wider range with respect to oxygen availability. In this scenario, the ratio between reduced and oxidized NAD(H) is maintained with modest variations under different oxygen availability conditions, while the pool of NADP(H) is mostly reduced in highly aerated cultures and mostly oxidized in nonaerated ones.
In this work, no significant variations were detected when the NADPH/NADP+ ratios obtained for the different strains were compared under each condition, while differences in the NADH/NAD+ ratios were observed under all aeration conditions. The oxidation state of NAD(H) has previously been reported to vary greatly in different genetic backgrounds (49–52), such as in redox regulatory mutants (53–55), so the differences in NADH/NAD+ ratios between the strains carrying the cre variants support the hypothesis that CreC affects the intracellular redox state. Additionally, the results obtained in this work could reflect the fact that the content of NADP+ responds more strongly to variations in oxygen availability than to different genetic or metabolic backgrounds.
At least 120 proteins have been shown to change their expression in response to a shift from aerobic to anaerobic conditions (2). The expression of over 30 operons (more than 70 genes) is under the control of the Fnr regulator (56), and at least 40 operons are regulated by the ArcAB two-component regulatory system (57, 58), including 16 genes that encode proteins playing roles in carbon metabolism (16, 59). The ArcAB and Fnr global regulation systems are major controlling factors of gene expression, and in most cases, they operate coordinately to fine-tune catabolism in response to oxygen availability (60). The metabolic changes observed in this work suggest that CreBC might also contribute to the regulation of the intracellular redox state, although it is likely that its role does not involve direct sensing of the redox state of the cells, but rather, as has been suggested in previous work, a metabolic signal, such as a carbon catabolism intermediary (6).
The CreBC system was first reported to respond to growth in minimal medium (7) and was later shown to be active when cells are grown in gluconeogenic C sources or fermenting glucose (6). The results presented in this study, in which all experiments were performed using glucose as the C source, clearly indicate that the regulation mediated by CreBC is affected by oxygen availability. In a mineral culture medium with excess glucose, high aeration did not offer a propitious environment for CreC activation, as shown by similar metabolic profiles (characterized by high acetate formation) and enzyme activities among the three strains. Mutations in CreC gave more conspicuous phenotypes when low or no aeration was supplied. This result is in accordance with the observations by Cariss et al. (6), which indicated that CreC was activated when cells were cultured in sealed flasks, limiting the oxygen supply. However, there was not a clear trend across the different aeration levels. The in vitro enzyme activity levels, as well as the formation of some metabolites, were not affected in the same manner by the absence of CreC at different aeration levels. Except for succinate, which had higher yields in the absence of CreC under both low and no aeration, the relative values of other organic acids differed under the two conditions. For example, under low aeration, the null mutant accumulated more formate and acetate but less lactate than the parental strain, while in the absence of aeration the opposite relationship was observed for acetate and lactate, with similar amounts of formate.
In an attempt to further characterize this effect, the activities of AckA and LdhA were determined, and both were observed to be affected by CreC. While ackA was previously shown to be under transcriptional regulation exerted by CreBC (7), ldhA was not thought to be affected by this regulatory system. The gene has also been observed to be negatively regulated by Mlc (which affects the expression of pts genes) (61) and CsrA (a regulator of carbohydrate metabolism that influences glycogen biosynthesis, gluconeogenesis, glycolysis, and glycogen degradation) (62, 63) and positively regulated by ArcAB and CsrB (which antagonizes CsrA) (64). The differences in lactate production, together with those observed in LdhA activity, suggest that CreC could also have a regulatory effect on ldhA, in spite of the lack of a consensus cre tag in the region upstream from the gene. Further experiments need to be performed to assess whether these differences are directly mediated by CreBC regulation at the transcriptional level. For all cultures grown under low aeration, a correlation between enzyme activities and metabolite levels was observed, as higher acetate and lower lactate values in the ΔcreC mutant than in the parental strain corresponded to higher AckA and lower LdhA activities, but this correlation was not observed in nonaerated cultures.
Although enzymatic studies are typically performed in exponential cultures, enzyme activity was also determined in early-stationary-phase nonaerated cultures to investigate whether there was a correlation between in vitro enzyme activities in this growth phase and metabolite levels (measured in 24-h cultures). When analyzing this point of the growth phase, lactate production in strains K1060 and DC1060 correlated with LdhA activity, as higher enzymatic activity was observed for E. coli DC1060, which accumulated more lactate in nonaerated exponential cultures. The conditions under which ldhA is normally expressed are anaerobiosis and acidic pH (33). Given that DC1060 grew more than K1060 and that cultures of DC1060 had a marked reduction in pH, it is possible that in the stationary phase the effects of the lower pH contributed to the higher LdhA activity observed. This, in turn, could result in higher lactate accumulation. In the exponential phase of nonaerated cultures, which had only slight differences in pH, both LdhA activity and lactate formation were lower for the null mutant, as observed in the cultures grown with high and low aeration. The only condition under which the ΔcreC mutant had higher LdhA activity and produced more lactate than the parental strain was when these cultures had much lower pH, so it is possible that this parameter affected LdhA activity, leading to increased lactate levels. Under all other conditions, where no differences in culture pH were observed between the strains, the ΔcreC mutant produced less lactate and had lower LdhA activity, so it can be proposed that CreC has a positive regulatory effect on ldhA, the nature of which remains to be elucidated.
Concerning acetate, while the DC1060 mutant produced larger amounts of the metabolite than the wild-type strain under low aeration, but not when aeration was suppressed, the in vitro AckA activity was higher for the mutant strain under both conditions, indicating that there was not a direct correlation between AckA activity and acetate accumulation. This suggests there must be additional factors that affected acetate levels in the cultures, which could be due to postranscriptional, allosteric, or even metabolic regulation. The possibility that other enzymes might contribute to the effect observed, such as pyruvate oxidase (PoxB), which has been reported to be the main pathway for acetate production in the stationary phase (30), although the enzyme is supposedly repressed under anaerobiosis (65), cannot be ruled out.
In previous studies, it was seen that ackA is transcriptionally activated by CreC (7) and that in the absence of CreB there is a reduced acetate flux when the cells are grown in continuous cultures under carbon-limited conditions and restricted oxygen supply (47). However, differences in aeration conditions have been shown to have dramatic effects on CreC regulation (6). In our work, performed in the presence of an excess carbon source, the creC-null mutants displayed higher AckA activities than the wild-type strain, while the absence of CreC had different effects on acetate accumulation depending on the aeration level (low or no aeration). Furthermore, the relative weights of the effects of Cre on other physiological parameters analyzed in this study also depended greatly on oxygen availability. While the strongest effects on redox levels were observed under low aeration, a notable influence was seen over growth in nonaerated cultures, and differences in metabolite distribution compared to the other strains also varied appreciably under the conditions analyzed. These results suggest that the metabolic effects of the cre variants are highly susceptible to changes in culture conditions, including oxygen availability.
The ΔcreC mutation proved useful to increase the production of succinate. This compound is an intermediary metabolite in the tricarboxylic acid cycle and, under anaerobic conditions, can be produced via the reductive arm of the cycle and the glyoxylate shunt, although the latter route contributes less than the former. As NADH is required for its formation, different strategies have been used to increase cofactor availability and to improve succinate production (66). It has been proposed that the manipulation of the CreBC system (as well as ArcAB) could provide a relevant tool for the modulation of central metabolism and reducing power availability aimed at biotechnological purposes (16), such as succinate production. Although the positive effect of the creC deletion on succinate formation may not seem obvious from the point of view of the redox balance, the increase in succinate production in E. coli DC1060 compared to the parental strain was observed under all aeration conditions, and also when plasmids overexpressing decarboxylating enzymes were added to both strains. The complementary strategy of enhancing CO2 fixation (through PpcEc) and increasing the pool of NADH (through FDH1Cb) at the expense of excess formate worked satisfactorily. Additional mutations in structural genes did not produce significant increases in the titer of succinate, even though they affected the secretion of other acids. In general, the differences in succinate production between the mutants became less relevant when other modifications were introduced, such as the expression of FDH1Cb and PpcEc, indicating that the relative weights of the mutations affecting the different metabolic steps were lower than those of the overexpression of the two heterologous genes. However, the creC mutants continued to accumulate more succinate than the strain carrying the wild-type allele, even when the enzymes were overproduced in all the strains, evidencing the role of the creC mutation in boosting succinate synthesis. A hitherto unknown regulation exerted by CreBC on the enzymes involved in the synthesis and/or consumption of succinate in E. coli, as observed in this work for LdhA, cannot be ruled out. The relatively simple strategy of modifying the global regulation exerted by CreBC offers an appealing alternative to the traditional gene-by-gene metabolic-engineering strategies for the production of succinate (40, 46, 67, 68).
In conclusion, this work shows that CreC affects both carbon catabolism and the intracellular redox state. The creC mutants were shown to exhibit different behaviors depending on the degree of oxygen limitation when grown in a mineral medium, with diverse effects on growth, metabolite secretion, and redox balance. These results also reveal that, like other global regulators, Cre influences many different aspects of bacterial physiology, although more research is needed to elucidate the mechanisms by which CreC exerts its regulation. The diverse metabolic effects of this regulator over the central biochemical network of E. coli make it a good candidate for genetic manipulations to improve the formation of compounds of commercial interest, such as succinate.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to G. N. Bennett (Rice University, Houston, TX) and to P. Kim (Catholic University of Korea, Bucheon, Gyunggi, South Korea) for the kind gifts of plasmids pSBF2 and pEcPpc, respectively.
M.J.P. is a career investigator from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina).
We declare no conflict of interest.
Funding Statement
CONICET funded Manuel S. Godoy through a postdoctoral fellowship.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02984-15.
REFERENCES
- 1.Martínez-Antonio A, Collado-Vides J. 2003. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr Opin Microbiol 6:482–489. doi: 10.1016/j.mib.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Bettenbrock K, Bai H, Ederer M, Green J, Hellingwerf KJ, Holcombe M, Kunz S, Rolfe MD, Sanguinetti G, Sawodny O, Sharma P, Steinsiek S, Poole RK. 2014. Towards a systems level understanding of the oxygen response of Escherichia coli.. Adv Microb Physiol 64:65–114. doi: 10.1016/B978-0-12-800143-1.00002-6. [DOI] [PubMed] [Google Scholar]
- 3.Rolfe MD, Ter Beek A, Graham AI, Trotter EW, Asif HMS, Sanguinetti G, Teixeira de Mattos MJ, Poole RK, Green J. 2011. Transcript profiling and inference of Escherichia coli K-12 ArcA activity across the range of physiologically relevant oxygen concentrations. J Biol Chem 286:10147–10154. doi: 10.1074/jbc.M110.211144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J, Shalel-Levanon S, Bennett GN, San KY. 2006. Effect of the global redox sensing/regulation networks on Escherichia coli and metabolic flux distribution based on C-13 labeling experiments. Metab Eng 8:619–627. doi: 10.1016/j.ymben.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Nikel PI, de Almeida A, Giordano AM, Pettinari MJ. 2010. Redox driven metabolic tuning: carbon source and aeration affect synthesis of poly(3-hydroxybutyrate) in Escherichia coli.. Bioeng Bugs 1:291–295. doi: 10.4161/bbug.1.4.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cariss SJ, Tayler AE, Avison MB. 2008. Defining the growth conditions and promoter-proximal DNA sequences required for activation of gene expression by CreBC in Escherichia coli.. J Bacteriol 190:3930–3939. doi: 10.1128/JB.00108-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avison MB, Horton RE, Walsh TR, Bennett PM. 2001. Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. J Biol Chem 276:26955–26961. doi: 10.1074/jbc.M011186200. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh YJ, Wanner BL. 2010. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol 13:198–203. doi: 10.1016/j.mib.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wanner BL. 1996. Signal transduction in the control of phosphate-regulated genes of Escherichia coli.. Kidney Int 49:964–967. doi: 10.1038/ki.1996.136. [DOI] [PubMed] [Google Scholar]
- 10.Wanner BL, Wilmes MR, Young DC. 1988. Control of bacterial alkaline phosphatase synthesis and variation in an Escherichia coli K-12 phoR mutant by adenyl cyclase, the cyclic AMP receptor protein, and the phoM operon. J Bacteriol 170:1092–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou L, Lei XH, Bochner BR, Wanner BL. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J Bacteriol 185:4956–4972. doi: 10.1128/JB.185.16.4956-4972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cariss SJ, Constantinidou C, Patel MD, Takebayashi Y, Hobman JL, Penn CW, Avison MB. 2010. YieJ (CbrC) mediates CreBC-dependent colicin E2 tolerance in Escherichia coli.. J Bacteriol 192:3329–3336. doi: 10.1128/JB.01352-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bidart GN, Ruiz JA, de Almeida A, Méndez BS, Nikel PI. 2012. Manipulation of the anoxic metabolism in Escherichia coli by ArcB deletion variants in the ArcBA two-component system. Appl Environ Microbiol 78:8784–8794. doi: 10.1128/AEM.02558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlson R, Wlaschin A, Srienc F. 2005. Kinetic studies and biochemical pathway analysis of anaerobic poly-(R)-3-hydroxybutyric acid synthesis in Escherichia coli.. Appl Environ Microbiol 71:713–720. doi: 10.1128/AEM.71.2.713-720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pettinari MJ, Nikel PI, Ruiz JA, Méndez BS. 2008. ArcA redox mutants as a source of reduced bioproducts. J Mol Microbiol Biotechnol 15:41–47. doi: 10.1159/000111991. [DOI] [PubMed] [Google Scholar]
- 16.Ruiz JA, de Almeida A, Godoy MS, Mezzina MP, Bidart GN, Méndez BS, Pettinari MJ, Nikel PI. 2012. Escherichia coli redox mutants as microbial cell factories for the synthesis of reduced biochemicals. Comput Struct Biotechnol J 3:e201210019. doi: 10.5936/csbj.201210019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz JA, Fernández RO, Nikel PI, Méndez BS, Pettinari MJ. 2006. dye (arc) mutants: insights into an unexplained phenotype and its suppression by the synthesis of poly(3-hydroxybutyrate) in Escherichia coli recombinants. FEMS Microbiol Lett 258:55–60. doi: 10.1111/j.1574-6968.2006.00196.x. [DOI] [PubMed] [Google Scholar]
- 18.Nikel PI, de Almeida A, Pettinari MJ, Méndez BS. 2008. The legacy of HfrH: mutations in the two-component system CreBC are responsible for the unusual phenotype of an Escherichia coli arcA mutant. J Bacteriol 190:3404–3407. doi: 10.1128/JB.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikel PI, Pettinari MJ, Galvagno MA, Méndez BS. 2006. Poly(3-hydroxybutyrate) synthesis by recombinant Escherichia coli arcA mutants in microaerobiosis. Appl Environ Microbiol 72:2614–2620. doi: 10.1128/AEM.72.4.2614-2620.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikel PI, Pettinari MJ, Galvagno MA, Méndez BS. 2008. Poly(3-hydroxybutyrate) synthesis from glycerol by a recombinant Escherichia coli arcA mutant in fed-batch microaerobic cultures. Appl Microbiol Biotechnol 77:1337–1343. doi: 10.1007/s00253-007-1255-7. [DOI] [PubMed] [Google Scholar]
- 21.López NI, Pettinari MJ, Nikel PI, Méndez BS. 2015. Polyhydroxyalkanoates: much more than biodegradable plastics. Adv Appl Microbiol 93:73–106. doi: 10.1016/bs.aambs.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Nikel PI, Pettinari MJ, Galvagno MA, Méndez BS. 2010. Metabolic selective pressure stabilizes plasmids carrying biosynthetic genes for reduced biochemicals in Escherichia coli redox mutants. Appl Microbiol Biotechnol 88:563–573. doi: 10.1007/s00253-010-2774-1. [DOI] [PubMed] [Google Scholar]
- 23.Nikel PI, Ramírez MC, Pettinari MJ, Méndez BS, Galvagno MA. 2010. Ethanol synthesis from glycerol by Escherichia coli redox mutants expressing adhE from Leuconostoc mesenteroides.. J Appl Microbiol 109:492–504. doi: 10.1111/j.1365-2672.2010.04668.x. [DOI] [PubMed] [Google Scholar]
- 24.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 26.Bernofsky C, Swan M. 1973. An improved cycling assay for nicotinamide adenine dinucleotide. Anal Biochem 53:452–458. doi: 10.1016/0003-2697(73)90094-8. [DOI] [PubMed] [Google Scholar]
- 27.Nikel PI, Pérez-Pantoja D, de Lorenzo V. 2013. Why are chlorinated pollutants so difficult to degrade aerobically? Redox stress limits 1,3-dichloroprop-1-ene metabolism by Pseudomonas pavonaceae. Philos Trans R Soc Lond B Biol Sci 368:20120377. doi: 10.1098/rstb.2012.0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikel PI, Chavarría M, Fuhrer T, Sauer U, de Lorenzo V. 2015. Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and pentose phosphate pathways. J Biol Chem 290:25920–25932. doi: 10.1074/jbc.M115.687749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikel PI, Chavarría M. 2015. Quantitative physiology approaches to understand and optimize reducing power availability in environmental bacteria. In McGenity TJ. (ed), Hydrocarbon and lipid microbiology protocols, in press Humana Press, New York, NY. doi: 10.1007/8623_2015_84. [DOI] [Google Scholar]
- 30.Dittrich CR, Bennett GN, San KY. 2005. Characterization of the acetate-producing pathways in Escherichia coli.. Biotechnol Prog 21:1062–1067. [DOI] [PubMed] [Google Scholar]
- 31.Lipmann F, Tuttle LC. 1945. The detection of activated carboxyl groups with hydroxylamine as interceptor. J Biol Chem 161:415. [PubMed] [Google Scholar]
- 32.Tarmy EM, Kaplan NO. 1968. Chemical characterization of D-lactate dehydrogenase from Escherichia coli B. J Biol Chem 243:2579–2586. [PubMed] [Google Scholar]
- 33.Jiang GR, Nikolova S, Clark DP. 2001. Regulation of the ldhA gene, encoding the fermentative lactate dehydrogenase of Escherichia coli.. Microbiology 147:2437–2446. doi: 10.1099/00221287-147-9-2437. [DOI] [PubMed] [Google Scholar]
- 34.McKinlay JB, Vieille C, Zeikus JG. 2007. Prospects for a bio-based succinate industry. Appl Microbiol Biotechnol 76:727–740. doi: 10.1007/s00253-007-1057-y. [DOI] [PubMed] [Google Scholar]
- 35.Thakker C, Martínez I, San KY, Bennett GN. 2012. Succinate production in Escherichia coli.. Biotechnol J 7:213–224. doi: 10.1002/biot.201100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao YP, Liao JC. 1993. Alteration of growth yield by overexpression of phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxykinase in Escherichia coli.. Appl Environ Microbiol 59:4261–4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gokarn RR, Eiteman MA, Altman E. 2000. Metabolic analysis of Escherichia coli in the presence and absence of the carboxylating enzymes phosphoenolpyruvate carboxylase and pyruvate carboxylase. Appl Environ Microbiol 66:1844–1850. doi: 10.1128/AEM.66.5.1844-1850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D, Li Q, Mao Y, Xing J, Su Z. 2010. High-level succinic acid production and yield by lactose-induced expression of phosphoenolpyruvate carboxylase in ptsG mutant Escherichia coli.. Appl Microbiol Biotechnol 87:2025–2035. doi: 10.1007/s00253-010-2689-x. [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Li Z, Xie J, Ye Q. 2009. Production of succinate by a pflB ldhA double mutant of Escherichia coli overexpressing malate dehydrogenase. Bioprocess Biosyst Eng 32:737–745. doi: 10.1007/s00449-009-0298-9. [DOI] [PubMed] [Google Scholar]
- 40.Jantama K, Haupt MJ, Svoronos SA, Zhang X, Moore JC, Shanmugam KT, Ingram LO. 2008. Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol Bioeng 99:1140–1153. doi: 10.1002/bit.21694. [DOI] [PubMed] [Google Scholar]
- 41.Samuelov NS, Lamed R, Lowe S, Zeikus JG. 1991. Influence of CO2-HCO3− levels and pH on growth, succinate production, and enzyme activities of Anaerobiospirillum succiniciproducens.. Appl Environ Microbiol 57:3013–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon YD, Kwon OH, Lee HS, Kim P. 2007. The effect of NADP-dependent malic enzyme expression and anaerobic C4 metabolism in Escherichia coli compared with other anaplerotic enzymes. J Appl Microbiol 103:2340–2345. doi: 10.1111/j.1365-2672.2007.03485.x. [DOI] [PubMed] [Google Scholar]
- 43.Berríos-Rivera SJ, Bennett GN, San KY. 2002. The effect of increasing NADH availability on the redistribution of metabolic fluxes in Escherichia coli chemostat cultures. Metab Eng 4:230–237. doi: 10.1006/mben.2002.0228. [DOI] [PubMed] [Google Scholar]
- 44.Sakai Y, Murdanoto AP, Konishi T, Iwamatsu A, Kato N. 1997. Regulation of the formate dehydrogenase gene, FDH1, in the methylotrophic yeast Candida boidinii and growth characteristics of an FDH1-disrupted strain on methanol, methylamine, and choline. J Bacteriol 179:4480–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balzer GJ, Thakker C, Bennett GN, San KY. 2013. Metabolic engineering of E. coli to minimize byproduct formate and improving succinate productivity through increasing NADH availability by heterologous expression of NAD+-dependent formate dehydrogenase. Metab Eng 20:1–8. doi: 10.1016/j.ymben.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 46.Sánchez AM, Bennett GN, San KY. 2005. Novel pathway engineering design of the anaerobic central metabolic pathway in Escherichia coli to increase succinate yield and productivity. Metab Eng 7:229–239. doi: 10.1016/j.ymben.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Nikel PI, Zhu J, San KY, Méndez BS, Bennett GN. 2009. Metabolic flux analysis of Escherichia coli creB and arcA mutants reveals shared control of carbon catabolism under microaerobic growth conditions. J Bacteriol 191:5538–5548. doi: 10.1128/JB.00174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amemura M, Makino K, Shinagawa H, Nakata A. 1990. Cross talk to the phosphate regulon of Escherichia coli by PhoM protein: PhoM is a histidine protein kinase and catalyzes phosphorylation of PhoB and PhoM-open reading frame 2. J Bacteriol 172:6300–6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holm AK, Blank LM, Oldiges M, Schmid A, Solem C, Jensen PR, Vemuri GN. 2010. Metabolic and transcriptional response to cofactor perturbations in Escherichia coli.. J Biol Chem 285:17498–17506. doi: 10.1074/jbc.M109.095570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y, Wang L, Yang F, Lin X, Zhang S, Zhao ZK. 2011. Determining the extremes of the cellular NAD(H) level by using an Escherichia coli NAD+-auxotrophic mutant. Appl Environ Microbiol 77:6133–6140. doi: 10.1128/AEM.00630-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vemuri GN, Altman E, Sangurdekar DP, Khodursky AB, Eiteman MA. 2006. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl Environ Microbiol 72:3653–3661. doi: 10.1128/AEM.72.5.3653-3661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bühler B, Park JB, Blank LM, Schmid A. 2008. NADH availability limits asymmetric biocatalytic epoxidation in a growing recombinant Escherichia coli strain. Appl Environ Microbiol 74:1436–1446. doi: 10.1128/AEM.02234-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nikel PI, Pettinari MJ, Ramírez MC, Galvagno MA, Méndez BS. 2008. Escherichia coli arcA mutants: metabolic profile characterization of microaerobic cultures using glycerol as a carbon source. J Mol Microbiol Biotechnol 15:48–54. doi: 10.1159/000111992. [DOI] [PubMed] [Google Scholar]
- 54.Shalel-Levanon S, San KY, Bennett GN. 2005. Effect of oxygen on the Escherichia coli ArcA and FNR regulation systems and metabolic responses. Biotechnol Bioeng 89:556–564. doi: 10.1002/bit.20381. [DOI] [PubMed] [Google Scholar]
- 55.Alexeeva S, Hellingwerf KJ, Teixeira de Mattos MJ. 2003. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J Bacteriol 185:204–209. doi: 10.1128/JB.185.1.204-209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unden G, Achebach S, Holighaus G, Tran HQ, Wackwitz B, Zeuner Y. 2002. Control of FNR function of Escherichia coli by O2 and reducing conditions. J Mol Microbiol Biotechnol 4:263–268. [PubMed] [Google Scholar]
- 57.Liu X, De Wulf P. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J Biol Chem 279:12588–12597. doi: 10.1074/jbc.M313454200. [DOI] [PubMed] [Google Scholar]
- 58.Salmon KA, Hung SP, Steffen NR, Krupp R, Baldi P, Hatfield GW, Gunsalus RP. 2005. Global gene expression profiling in Escherichia coli K-12: effects of oxygen availability and ArcA. J Biol Chem 280:15084–15096. doi: 10.1074/jbc.M414030200. [DOI] [PubMed] [Google Scholar]
- 59.Perrenoud A, Sauer U. 2005. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli.. J Bacteriol 187:3171–3179. doi: 10.1128/JB.187.9.3171-3179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Green J, Paget MS. 2004. Bacterial redox sensors. Nat Rev Microbiol 2:954–966. doi: 10.1038/nrmicro1022. [DOI] [PubMed] [Google Scholar]
- 61.Plumbridge J. 2002. Regulation of gene expression in the PTS in Escherichia coli: the role and interactions of Mlc. Curr Opin Microbiol 5:187–193. doi: 10.1016/S1369-5274(02)00296-5. [DOI] [PubMed] [Google Scholar]
- 62.Sabnis NA, Yang H, Romeo T. 1995. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J Biol Chem 270:29096–29104. doi: 10.1074/jbc.270.49.29096. [DOI] [PubMed] [Google Scholar]
- 63.Yang H, Liu MY, Romeo T. 1996. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J Bacteriol 178:1012–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baker CS, Morozov I, Suzuki K, Romeo T, Babitzke P. 2002. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli.. Mol Microbiol 44:1599–1610. doi: 10.1046/j.1365-2958.2002.02982.x. [DOI] [PubMed] [Google Scholar]
- 65.Chang YY, Wang AY, Cronan JE. 1994. Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS (katF) gene. Mol Microbiol 11:1019–1028. doi: 10.1111/j.1365-2958.1994.tb00380.x. [DOI] [PubMed] [Google Scholar]
- 66.Berríos-Rivera SJ, Sánchez AM, Bennett GN, San KY. 2004. Effect of different levels of NADH availability on metabolite distribution in Escherichia coli fermentation in minimal and complex media. Appl Microbiol Biotechnol 65:426–432. doi: 10.1007/s00253-004-1609-3. [DOI] [PubMed] [Google Scholar]
- 67.Kim TY, Park JM, Kim HU, Cho KM, Lee SY. 2015. Design of homo-organic acid producing strains using multi-objective optimization. Metab Eng 28:63–73. doi: 10.1016/j.ymben.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 68.Sánchez AM, Bennett GN, San KY. 2006. Batch culture characterization and metabolic flux analysis of succinate-producing Escherichia coli strains. Metab Eng 8:209–226. doi: 10.1016/j.ymben.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 69.Overath P, Schairer HU, Stoffel W. 1970. Correlation of in vivo and in vitro phase transitions of membrane lipids in Escherichia coli.. Proc Natl Acad Sci U S A 67:606–612. doi: 10.1073/pnas.67.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hayes W. 1953. Observations on a transmissible agent determining sexual differentiation in Bacterium coli. J Gen Microbiol 8:72–88. doi: 10.1099/00221287-8-1-72. [DOI] [PubMed] [Google Scholar]
- 71.Phillips GJ, Park SK, Huber D. 2000. High copy number plasmids compatible with commonly used cloning vectors. Biotechniques 28:400–402. [DOI] [PubMed] [Google Scholar]
- 72.Amann E, Ochs B, Abel KJ. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli.. Gene 69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.