Abstract
Hydrocarbons released during oil spills are persistent in marine sediments due to the absence of suitable electron acceptors below the oxic zone. Here, we investigated an alternative bioremediation strategy to remove toluene, a model monoaromatic hydrocarbon, using a bioanode. Bioelectrochemical reactors were inoculated with sediment collected from a hydrocarbon-contaminated marine site, and anodes were polarized at 0 mV and +300 mV (versus an Ag/AgCl [3 M KCl] reference electrode). The degradation of toluene was directly linked to current generation of up to 301 mA m−2 and 431 mA m−2 for the bioanodes polarized at 0 mV and +300 mV, respectively. Peak currents decreased over time even after periodic spiking with toluene. The monitoring of sulfate concentrations during bioelectrochemical experiments suggested that sulfur metabolism was involved in toluene degradation at bioanodes. 16S rRNA gene-based Illumina sequencing of the bulk anolyte and anode samples revealed enrichment with electrocatalytically active microorganisms, toluene degraders, and sulfate-reducing microorganisms. Quantitative PCR targeting the α-subunit of the dissimilatory sulfite reductase (encoded by dsrA) and the α-subunit of the benzylsuccinate synthase (encoded by bssA) confirmed these findings. In particular, members of the family Desulfobulbaceae were enriched concomitantly with current production and toluene degradation. Based on these observations, we propose two mechanisms for bioelectrochemical toluene degradation: (i) direct electron transfer to the anode and/or (ii) sulfide-mediated electron transfer.
INTRODUCTION
The large number of oil spills from offshore extraction platforms, ships, or more diffuse sources poses major concerns to aquatic ecosystems (1). Volatile organic compounds (VOCs), such as hexane, benzene, toluene, ethylbenzene, and xylenes (BTEX), are a large fraction of petroleum hydrocarbons, representing up to 15% of all hydrocarbon contaminants (2). These compounds can have a broad range of toxicity on a large number of organisms (3), affecting natural biota and ecosystems (4), fisheries (5), and human health (6). Although the most dramatic effects usually are observed when a large volume of oil is spilled, it also has been shown that small quantities of oil can severely impact marine ecosystems (7).
When an oil spill occurs, a variety of hydrocarbon removal strategies can be applied to minimize negative environmental impacts. Biological methods now are widely used due to their low cost, adaptability to local conditions, and potential for full contaminant mineralization (8). In confined environments, such as marine sediments and aquifers, the excess of carbon source due to the presence of contaminants leads to rapid depletion of both nutrients (9) and thermodynamically favorable terminal electron acceptors (10), which reduce the biodegradation rate of pollutants. The degradation of contaminants is stimulated by overcoming the limitations to the metabolism of naturally present microorganisms.
One of the most used bioremediation approaches is the stimulation of aerobic metabolism by adding oxygen to the environment (11). However, oxygen diffusion is limited, and oxygen losses can occur due to reaction with reduced species, such as Fe2+ or Mn2+, present in these environments (12, 13). Furthermore, oxygen naturally escapes from the contaminated area. Continuous amendment with electron acceptors therefore is required, which increases the overall process cost and field implications (14).
Electron acceptor limitations in oil-contaminated sediments can be overcome by using bioelectrochemical systems (BESs) (15, 16). Benthic BESs have been described already as a suitable technology to power oceanographic devices by harvesting energy from anaerobic sediments from the ocean floor (17, 18). A BES is a device in which electrogenic microorganisms oxidize an electron donor using an electrode as a sole electron acceptor (19). Recently, laboratory-scale studies demonstrated that BES-based technologies can be successfully applied to remove both aliphatic and aromatic hydrocarbons from oil-contaminated sediments (14, 20–22), groundwater (21), and refinery wastewater (23) with simultaneous current production. Very few studies have discussed the possibility of bioelectrochemically degrading toluene by either inoculating BES reactors with hydrocarbon contaminated sediment (14) or using pure cultures of, e.g., Geobacter metallireducens (14) and Pseudomonas putida (24). Linking electrocatalysis in complex systems to the microbial communities involved still is lacking, as is an in-depth knowledge of the microbial ecology and energy flows associated with hydrocarbon degradation using an anode. There are indications that iron-reducing (21), nitrate-reducing (16, 25), and aerobic bacteria (25) play a significant role in hydrocarbon degradation, as they have been found in biofilms formed on anodes. Earlier research also demonstrated that sulfide can serve as a shuttle to convey electrons from acetate to an anode (26). Considering the high concentration of sulfate in marine systems (∼28 mM), anaerobic hydrocarbon degradation linked to sulfate reduction may occur, leading to the formation of sulfur intermediates after partial oxidation at anodes. It has been speculated that sulfur metabolism can play a role in electron transfer between the electron donor and the anode during benzene degradation in a BES (21).
In this study, bioelectrochemical toluene degradation occurring at the marine sediment-water interface was assessed using BESs inoculated with marine sediment from a petroleum-contaminated site. The objectives of this study were to (i) investigate the biodegradability of toluene in BESs inoculated with the marine contaminated sediment, (ii) evaluate the effect of anode potential on toluene biodegradation, (iii) identify the most abundant microbial populations involved in bioelectrochemical toluene degradation process, and (iv) investigate the role of the sulfur cycle on toluene biodegradation in BESs.
MATERIALS AND METHODS
Bioelectrochemical reactor setup, operation, and experimental conditions.
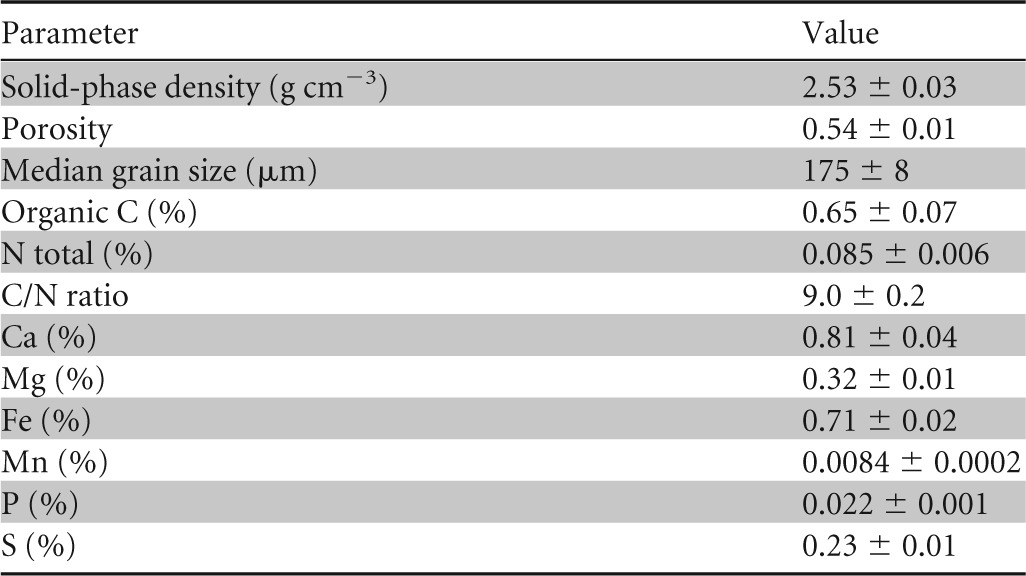
The bioelectrochemical experiments were carried out using custom-made five-neck glass BES reactors with 250 ml total volume (27). The anodic chamber was filled with 190 ml of artificial ocean water (Instant Ocean sea salt) as the growth medium to simulate marine environments (pH 6.4) and 10 ml of marine sediment as the microbial inoculum (Mokbaai, Netherlands) (Table 1) provided by the Royal Netherlands Institute for Sea Research. The marine sediment was collected from a hydrocarbon-contaminated site in which the presence of the so-called cable bacteria has been described previously (28). Prior to the electrochemical experiment setup, the anode chamber was flushed with N2:CO2 (90:10) for 30 min to ensure anoxic conditions. Toluene (23 μl; ∼40 mg liter−1) acted as the sole source of carbon. Following each depletion of toluene from the medium, toluene again was spiked into the anode chamber using a microsyringe. The cathodic chamber was filled with diluted artificial ocean water (1:4 dilution with distilled water; pH 8.2) in order to avoid the oxidation of the cathode material. Anodes were graphite plates (Mersen, Bay City, MI) with a geometric surface area of 8 cm2. A titanium wire (1 mm diameter; Advent Research Materials, Oxford, England) fixed in the top center of the graphite plate served as a current collector. The wire was covered with a heat-shrinkable polytetrafluorethylene (PTFE) tube (Sigma-Aldrich) in order to prevent corrosion due to the salinity of the medium. Cathodes were constructed using stainless steel mesh (4.5 by 5.0 cm) connected to titanium wire current collectors. The anodic chamber and the cathodic chamber were separated using a cation exchange membrane (Ultrex) soaked in 5% NaCl for at least 24 h before use. If not stated otherwise, all potential values refer to the Ag/AgCl (3 M KCl) reference electrode. All bioelectrochemical experiments were conducted at room temperature (22 ± 2°C) in duplicate or triplicate with the aid of a potentiostat (VSP; BioLogic, France). Anodes were poised at 0 mV (reactors R0a and R0b) and +300 mV (reactors R300a, R300b, and R300c), and the current profiles were recorded using chronoamperometry. Two abiotic controls (ocean water plus toluene, here referred to as Ab0 and Ab300) and one control without toluene (sediment plus ocean water, referred to as Sed300) also were set up. Further details of the experimental design are provided in Table S1 in the supplemental material. Current density profiles and concentrations of toluene, sulfate, and sulfide were monitored over time. The current density was calculated considering the geometric surface area of the electrodes. Cyclic voltammetry (CV) was conducted under substrate turnover (presence of toluene at ∼5 mg liter−1) and substrate non-turnover conditions (absence of toluene) (29) using a scan rate of 1 mV s−1 in order to detect redox-active moieties in the electrochemical process. At the end of the experiments, bioanodes and bulk anolyte samples were collected and stored at −20°C for the analysis of the microbial communities.
TABLE 1.
Chemical characterization of sea sediment used as the microbial inoculum
Chemical analyses.
Samples for toluene analysis were acidified (pH 2) and stored at 4°C until analysis (EPA method 5035A) (30). The analysis was carried out by a gas chromatograph-flame ionization detector GC-FID (Agilent 6890N) equipped with a headspace autosampler (Agilent 7697A) (EPA method 5021A) (31). The toluene removal rate was estimated by the first-order kinetic constant calculated during the second batch cycle, when the current reached the highest peak, to minimize the effects due to toluene adsorption onto the graphite electrode. Rates of toluene removal were reported as the mass of toluene removed per working volume of reactor per day (and as the mass of toluene removed per mass of sediment per day).
In order to monitor sulfate consumption, samples were filtered and stored at 4°C until the analysis using an ion chromatograph (Metroohm 761 Compact IC). Sulfide was measured immediately after sampling with the Nanocolor sulfide standard test (Macherey-Nagel, Düren, Germany) according to the manufacturer's instructions.
Microscopy analysis.
Scanning electron microscopy-energy dispersive spectrometry (SEM-EDS) (JEOL JSM-7600F) and fluorescence in situ hybridization (FISH) (Nikon A1R) analyses were performed on bioanode samples at the end of the experiments. For SEM-EDS, the bioanode was fixed in 2% paraformaldehyde and 1.5% glutaraldehyde for 2 h at 4°C, subsequently washed with phosphate-buffered saline (PBS) (130 mM NaCl, 10 mM phosphate buffer, pH 7.2), and fixed with 1% OsO4 in PBS for 90 min. The treated bioanode then was washed twice with distilled water and 25% ethanol and dehydrated in a series of ethanol solutions (25%, 40%, 50%, 60%, 70%, 80%, 90%, and 100%) for 5 to 10 min each. Finally, the bioanode was critical point dried using a gradient series of ethanol-hexamethyldisilazane (HMDS) solutions (25%, 50%, 75%, and 3 rounds at 100%) for 10 min each, dried overnight, and covered with a thin layer of Au. Samples for FISH were fixed with 4% paraformaldehyde for 4 h, washed in PBS, and stored in PBS-ethanol (50%) at −20°C until the analysis. FISH was performed using a mixture of Cy3-labeled SRB385 and SRB385Db (32) probes targeting the 16S rRNA of deltaproteobacterial sulfate-reducing bacteria (SRB). Probe coverages were estimated by RDP ProbeMatch tools using the RDP database (33). Coverage data for targeted SRB Deltaproteobacteria are reported in Table S2 in the supplemental material.
qPCR assays for the quantification of toluene-degrading and sulfate-reducing bacteria.
The microbial biofilm was aseptically removed from the anodes, and total bacterial DNA was extracted using the FastDNA spin-for-soil kit (MP Biomedicals, Solon, OH). DNA also was extracted from the bulk anolyte collected from the reactors. Quantitative PCR (qPCR) assays targeting genes encoding 16S rRNA, the α-subunit of the dissimilatory sulfite reductase (dsrA), and the α-subunit of the benzylsuccinate synthase (bssA) were carried out in 10-μl reaction mixtures consisting of 3 μl template DNA, 500 nM each primer, and 7 μl of SsoAdvanced universal SYBR green supermix (Bio-Rad, United Kingdom) on a CFX96 Touch qPCR machine (Bio-Rad, United Kingdom). Details of primer sequences and annealing temperatures are given in Table 2 (34–37). Amplification conditions were the following: initial denaturation at 95°C for 5 min, followed by 40 cycles consisting of 95°C for 30 s, annealing for 30 s, and 72°C for 40 s, after which the data were acquired. All DNA samples were tested for amplification inhibition by sample dilution. Sample, standards, and no-template control (NTC) reactions were carried out in triplicate. The specificity of amplification was verified via the generation of melting curves (65 to 95°C with a 1°C hold for 5 s). Gene abundances were estimated following the one-point calibration method (OPC) in order to account for possible discrepancies between PCR efficiency of the standards and the DNA extracted from the anodes and the bulk anolyte (38). From every qPCR assay, raw fluorescence data (non-baseline corrected) from individual PCRs were imported into LinRegPCR (2014.x) for optimal baseline subtraction and estimation of threshold cycle number (Cq) and efficiency (E) (39). OPC was performed by defining one standard containing a known number of target genes (N0 standard, equivalent to 103 to 105 genes). The template concentration of a sample, N0 sample, was estimated from standards showing similar Cq values using the equation
(38). Estandard and Esample were calculated from the arithmetic mean of triplicate reactions. N0 sample values were converted to gene number per nanogram of extracted DNA to allow direct comparison between anode and bulk samples.
TABLE 2.
Primers sets used for the qPCR assays
| Target gene and primer | Annealing tempa (°C) | Primer sequence (5′–3′) | Size (bp) | Reference |
|---|---|---|---|---|
| 16S rRNA | ||||
| 1055F | 58 | ATGGCTGTCGTCAGCT | 351 | 34 |
| 1392R | ACGGGCGGTGTGTAC | |||
| dsrA | ||||
| DSR67F | 58 | SCACTGGAARCACGG | 943 | 35 |
| Del1075R | GYTCVCGGTTCTTDC | 36 | ||
| bssA | ||||
| 7768F | 53 | CAAYGATTTAACCRACGCCAT | 795 | 37 |
| 8543R | TCGTCRTTGCCCCAYTTNGG |
Primer annealing temperature was optimized for this work.
Standards were prepared using plasmids (pCR 2.1-TOPO) containing sequence-verified dsrA, bssA, and 16S rRNA genes. Using the vector-specific primers pUC-F (5′-GTT TTC CCA GTC ACG AC-3′) and pUC-R (5′-CAG GAA ACA GCT ATG AC-3′) (Invitrogen, Paisley, United Kingdom), standards were obtained by amplification following an initial denaturation at 95°C for 10 min, 35 cycles consisting of 94°C for 1 min, 57°C for 1 min, and 72°C for 1.5 min, and then final elongation at 72°C for 10 min. Amplicon size was verified by agarose gel electrophoresis, purified with a GeneJET PCR purification kit (Life Technologies, United Kingdom), and quantified with a Quant-iT PicoGreen double-stranded DNA assay kit using a fluorescence-based microplate reader (Infinite 200 PRO; Tecan, United Kingdom). Standard gene abundances were calculated from the DNA concentration, amplicon length, and mean mass of a base pair (1.1 × 10−21 g). The linear range of quantification and the detection limits were determined by analyzing triplicate dilution series of the standards (107 to 101 target molecules per 1 μl).
Amplification of 16S rRNA genes, sequencing, and sequence analyses.
The V5-V6 hypervariable regions of the 16S rRNA gene were PCR amplified and sequenced by MiSeq Illumina (Illumina, Inc., San Diego, CA) using a 250-bp-by-2-paired-end protocol. The multiplexed libraries were prepared using a dual PCR amplification protocol as previously described (40). Briefly, the first PCR was performed in three 75-μl volume reaction mixes with GoTaq Green master mix (Promega Corporation, Madison, WI) and 1 μM each primer. 783F and 1046R primers were used (41, 42). The second PCR was performed in three 50-μl volume reaction mixes by using 23 μl of the purified amplicons (Wizard SV gel and PCR cleanup system; Promega Corporation, Madison, WI) from the first step as the template and 0.2 μM each primer. Primer sequences contained the standard Nextera indexes (Illumina, Inc., San Diego, CA). DNA quantity after the amplification was measured using a Qubit (Life Technologies, Carlsbad, CA). DNA sequencing was carried out at Parco Tecnologico Padano (Lodi, Italy). Reads from sequencing were demultiplexed according to the indexes. The Uparse pipeline was used for the following elaborations (43). Forward and reverse reads were merged with perfect overlapping and quality filtered with default parameters. Singleton sequences (i.e., sequences appearing only one time in the whole data set) were removed both from the whole data set and from each sample data set. Operational taxonomic units (OTUs) were defined for the whole data set, clustering the sequences at 97% sequence identity and defining a representative sequence for each cluster. A subset of 6,000 random sequences was chosen from each sample, and the abundance of each OTU was estimated by mapping the sequences of each sample against the representative sequence of each OTU at 97% sequence identity. Classification of the sequences representative of each OTU at different taxonomic ranks was done using the RDP classifier (33).
A principal component analysis (PCA) based on Hellinger-transformed family relative abundance data and post hoc comparison of mean abundances (Tukey-Kramer test) at family and OTU levels (95% confidence level) were performed using STAMP (44). The post hoc tests were performed by applying the Benjamini-Hochberg false discovery rate (FDR) correction (45).
Nucleotide sequence accession number.
The sequences were deposited in the European Nucleotide Archive under accession number PRJEB7900.
RESULTS
Toluene degradation and current production.
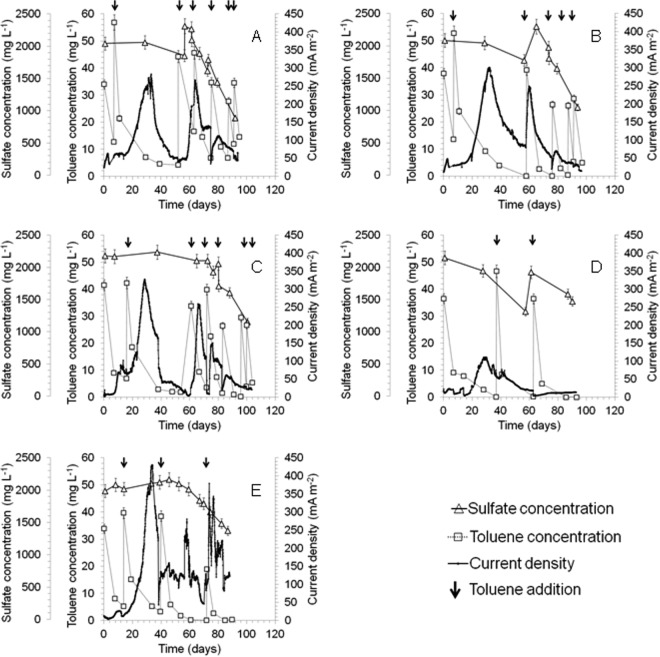
Current production in reactors R0a and R0b with the anodes polarized at 0 mV was initiated after a short lag phase (Fig. 1A and B), albeit to a low level, and likely was associated with residual organic carbon in the sediment. After 7 days of the experiment, these reactors were spiked with toluene. A maximum current density of 283 mA m−2 (day 33) and 301 mA m−2 (day 32) was reached in reactors R0a and R0b, respectively, after the second cycle of toluene addition. Following toluene depletion, the current decreased and toluene was spiked again to observe a reproducible current production (third batch cycle). The current density increased immediately after spiking to 258 ± 12 mA m−2 in the third cycle and decreased again in a reproducible fashion after toluene depletion in these reactors. After further toluene additions, the current density did not return to the same extent, while after 90 days of operation the current density decreased to about 50 mA m−2 in both reactors. This decrease in the electrochemical performance was correlated with the onset of sulfate reduction at this stage of the experiment. Whereas the sulfate concentration decreased only slightly (257 ± 52 mg liter−1) during the first 58 days of reactor operation, it dropped sharply when the current started to decrease on day 64 (R0a) and on day 61 (R0b) (Fig. 1).
FIG 1.
Current density, toluene concentration, and sulfate concentration profiles in bioanodes polarized at 0 mV (versus an Ag/AgCl [3 M KCl] reference electrode) (R0a [A] and R0b [B]) and at +300 mV (R300a [C], R300b [D], R300c [E]). Toluene degradation was concomitant with current production regardless of the anode polarization, and sulfate removal after around 60 to 80 days coincided with toluene removal, suggesting a link between SRB and hydrocarbon removal.
Toluene measurements indicated that the concentration of toluene in solution decreased sharply after addition, without a lag phase, which likely relates to initial adsorption to the graphite anode. This assumption was confirmed by the results of the abiotic controls where toluene concentration dropped immediately after addition, without current production (see Fig. S1 in the supplemental material). These results also confirm that toluene degradation is coupled with current production during the first three cycles. Toluene removal in the subsequent cycles most likely was linked to microbial sulfate reduction. The toluene degradation rate was calculated during the second batch cycle at the highest current production at ∼1 mg liter−1 day−1 (∼16 mg kgsediment−1 day−1) both for R0a and R0b reactors.
The anodes polarized at +300 mV in the duplicate reactors R300a and R300b exhibited discrepancies in terms of current production (Fig. 1C and D). In both of these reactors, a common lag phase of about 10 days was observed before the current production started. After the lag phase, the current density increased to 100 ± 14 mA m−2. When the reactors both were spiked with toluene, the current in R300a increased up to 328 mA m−2 (day 29), but no further increase was observed in R300b. When toluene was consumed in R300a the current dropped, but it recovered up to 260 mA m−2 after toluene addition in the third cycle. Further toluene additions did not result in any current production. The decreasing trend in the bioelectrochemical performance in both R300a and R300b also could be attributed to microbial sulfate reduction as also observed for the two replicates at 0 mV (R0a and R0b). To investigate which of the two profiles (R300a or R300b) was representative, a third replicate (R300c) (Fig. 1E) was set up. Similar to R300a and R300b, an initial lag phase of approximately 1 week was observed in the R300c reactor before the current density increased to 27 mA m−2. When the current decreased, toluene was spiked in the reactor and the current increased up to 431 mA m−2 (day 34). The current dropped when toluene was depleted and increased after a fresh toluene spike. Poor stability of current output was observed after the toluene spiking. Any other fluctuation of the current density (see Fig. 1E) was attributed to stirrer malfunction and sulfate reduction. Initial toluene removal without current production could be related to toluene adsorption onto the graphite electrode, as observed for all replicates at +300 mV and on the basis of data from abiotic controls (see Fig. S1 in the supplemental material). The toluene degradation rate, calculated for the second cycle of toluene addition, was ∼1 mg liter−1 day−1 (∼16 mg kgsediment−1 day−1), which was exactly the same as that for the bioanodes polarized at 0 mV.
To further elucidate whether toluene was used for current production and to determine the contribution of the inherent content of sediment to current production, a control reactor without toluene addition (Sed300) was set up, and the anode was polarized at +300 mV (see Fig. S1 in the supplemental material). The current density remained under 50 mA m−2 during 1 month of operation, indicating that the presence of toluene was necessary for current production.
Cyclic voltammograms were recorded on reactor R300c under substrate turnover conditions (at maximum current density, day 35, and during the period of decreased current, day 38) and non-turnover (no substrate addition) conditions (day 38). A catalytic wave was observed between +170 to +340 mV, and an oxidation peak was detected at +200 mV (Fig. 2), indicating the presence of a redox-active moiety with an oxidation potential just below the anode potential. No reduction current was observed in the backward scan, indicating a nonreversible oxidation reaction at the anode.
FIG 2.
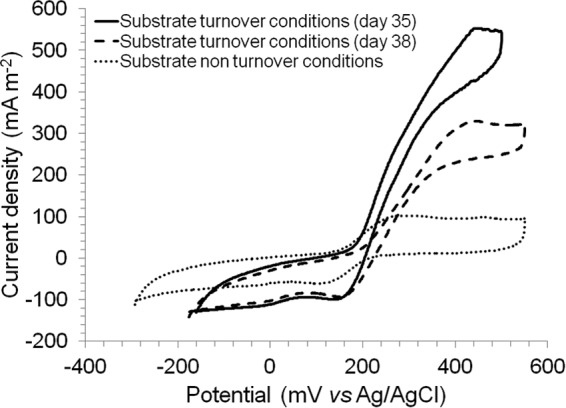
Cyclic voltammograms recorded with reactor R300c at +300 mV (versus an Ag/AgCl [3 M KCl] reference electrode) during substrate turnover and non-turnover conditions. The scan rate was 1 mV s−1. A catalytic wave (between +170 and +340 mV) and an oxidation peak (+200 mV) were observed.
Microscopy analysis.
FISH analysis performed using SRB385 and SRB385Db probes on the anode of R300c revealed the presence of a biofilm containing SRB (Fig. 3). SRB were detected in all of the observed fields, confirming their important role in the process of toluene removal from the reactors. SEM-EDS analysis of samples collected from the same anode to assess the extent of the electrochemical oxidation of sulfide (produced from sulfate) and reduction to elemental sulfur on the anode (46) and to quantify the amount of sulfur on the electrode surface revealed sulfur deposition, ranging from 1.1% to 2.0% (see Fig. S2A in the supplemental material), and FeS2 particles on the anode surface (see Fig. S2B).
FIG 3.
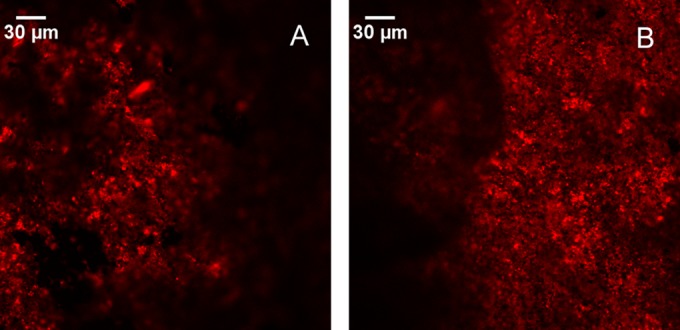
FISH images of the anode surface of reactor R300c at +300 mV (versus an Ag/AgCl [3 M KCl] reference electrode). Red fluorescence indicated the presence of sulfate-reducing bacteria. The hybridization was performed using a mixture of Cy3-labeled SRB385 and SRB385Db probes targeting the 16S rRNA of sulfate-reducing bacteria.
Quantification of anaerobic toluene-degrading and sulfate-reducing bacteria.
The α-subunit of the dissimilatory sulfite reductase (dsrA) and the α-subunit of the benzylsuccinate synthase (bssA) are genetic markers commonly used for the identification and quantification of sulfate reducers and anaerobic toluene degraders, respectively, that use the fumarate addition pathway for toluene degradation (37, 47). In order to assess the extent of the enrichment of anaerobic toluene-degrading and sulfate-reducing bacteria, the quantification of 16S rRNA, dsrA, and bssA genes was carried out on DNA samples extracted from the anode and bulk anolyte of reactors polarized at 0 mV and +300 mV. A linear response (R2 > 0.98) from 101 to 107 genes μl−1 was observed for every qPCR assay. The detection limit (i.e., the lowest standard concentration that is significantly different from that of the NTCs) was 10 genes. 16S rRNA and bssA gene assays did not show discrepancies in the efficiency of amplification of standards and samples (E16S rRNA = 1.9 and EbssA = 1.6, respectively, for both standards and samples). In contrast, the efficiency of amplification of dsrA standards was significantly lower than that for the samples (Estandard = 1.49 and Esample = 1.61). In order to account for this discrepancy, which otherwise would lead to the overestimation of dsrA gene abundance (48), the gene abundances were corrected by following the one-point calibration method (OPC). The bioelectrochemical treatment led to a significant enrichment of dsrA and bssA genes (Fig. 4; also see Table S3 in the supplemental material). All bulk and anode samples retrieved from the reactors showed from 3 to 6 orders of magnitude more dsrA gene copies than the sediment used as the microbial inoculum. bssA gene abundances also increased after treatment, accounting for 2- to 5-orders-of-magnitude-higher values than those for the inoculum. In most of the samples, dsrA and bssA values were of the same order of magnitude. The highest dsrA and bssA gene abundances were observed in samples retrieved from the reactors in which anodes had been polarized at +300 mV (R300). For example, R300c-bulk and R300b-anode contained four orders of magnitude or more 16S rRNA, dsrA, and bssA genes than the sediment used as the inoculum (Fig. 4; see Table S3). However, reactors at +300 mV (R300a to R300c) also showed greater variability between replicates regarding catabolic gene content and the extent of the bacterial colonization of the anodes. The biofilms formed on the anodes of R300a and R300b during the incubation showed 16S rRNA, dsrA, and bssA gene abundances as high as those of the bulk sample. In contrast, R300c showed a stronger SRB enrichment in the bulk than in the anode sample, and it was the only reactor showing this behavior (see Table S3). Both reactors with anodes polarized at 0 mV produced similar results. Both anodes (R0a and R0b) showed similar 16S rRNA, dsrA, and bssA gene abundances that were two orders of magnitude higher than those found in the bulk anolyte (2 × 106 ± 0.4 × 106 and 4 × 104 ± 0 × 104 16S rRNA copies per ng of DNA, respectively [means ± standard errors]). A strong correlation between dsrA and bssA gene abundances was observed (R2 = 0.785). The correlation was stronger (R2 = 0.997) when only the microbial communities in the bulk anolyte were considered (see Fig. S3B). The slope was between 0.72 and 0.77, suggesting that around 80% of the SRB also carry bssA and therefore could be involved in toluene degradation (see Fig. S3A). Although this is a compelling observation, it is not possible to state definitively that the same organisms harbor both bssA and dsrA; however, making the reasonable assumption that both dsrA and bssA genes are present as single copies on the genomes of the organisms detected (43), this is a valid interpretation of the data.
FIG 4.
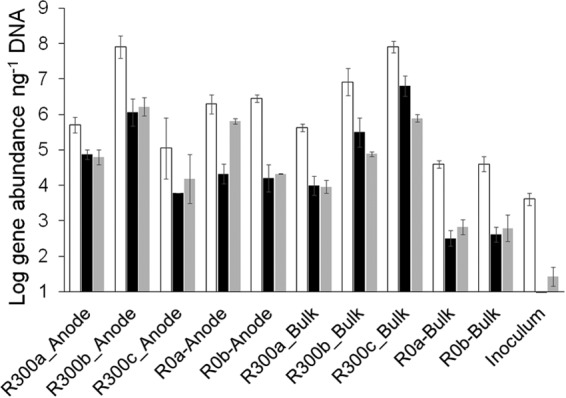
Quantification of toluene-degrading bacteria and SRB in the anode and bulk anolyte of reactors polarized at 0 mV and +300 mV (versus an Ag/AgCl [3 M KCl] reference electrode). White, black, and gray bars represent 16S rRNA, dsrA, and bssA gene abundances, respectively. Bars are averages from triplicate measurements, and standard deviations are shown. Sulfate reducers and toluene degraders were enriched during the bioelectrochemical process.
Structure and taxonomic composition of the microbial communities.
PCA based on Hellinger-transformed data for the relative abundance of taxa at the family level showed two main groups within the samples (Fig. 5). Microbial communities from the bulk anolyte of all reactors and from the start-up inoculum formed a cluster in the lower part of the graph. The microbial communities grown on the anodes during the experiments were positioned in the upper part of the graph, with the only exception being the microbial community on the anode of the reactor R300b, which was close to the communities in the bulk anolyte and in the inoculum. PCA was further performed at the OTU level and the previous observation was confirmed (see Fig. S4 in the supplemental material).
FIG 5.
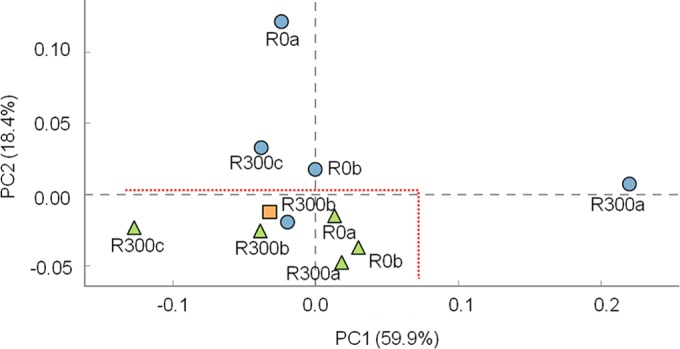
Principal component analysis on Hellinger-transformed data of family relative abundance calculated for microbial communities enriched at the anodes, in the bulk anolyte of reactors polarized at 0 mV and +300 mV (versus an Ag/AgCl [3 M KCl] reference electrode), and in the start-up sediment used as the inoculum. (Blue circles denote samples collected from the anodes, orange square denotes microbial inoculum, and green triangles denote samples collected from the bulk anolyte in the reactors at the end of the treatment.) The microbial communities enriched on the anodes and in the bulk anolyte formed separate groups.
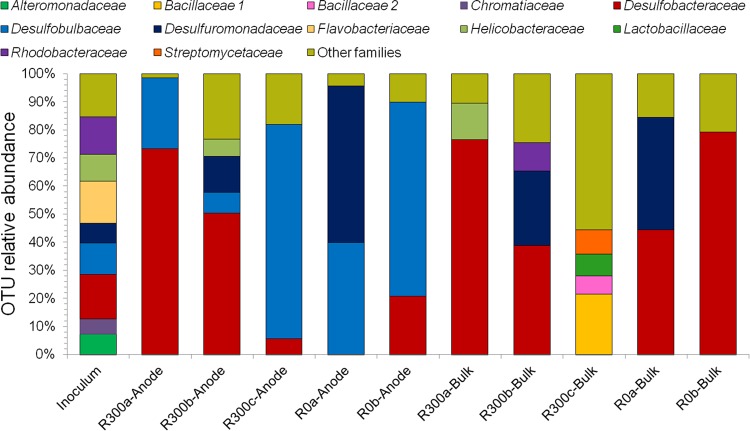
The microbial community in the sediment used as an inoculum had higher diversity than the communities selected during toluene biodegradation (Fig. 6; also see Table S4 in the supplemental material). The microbial communities in reactors were enriched with respect to SRB. The Shannon index calculated on the OTU relative abundance decreased from 4.6 in the inoculum to 3.4 ± 1.0 in the reactors after the process. Desulfobulbaceae were abundant on the anodes of the reactors R0a and R0b. The enrichment of Desulfuromonadaceae was observed in R0a both on the anode and in the bulk samples. The family Desulfobacteraceae was selected in the bulk samples of R0a and R0b and, to a lesser extent, on the anode of R0b. Similarly, bioanodes polarized at +300 mV were dominated by sulfate reducers: the Desulfobacteraceae were the most abundant microorganisms on the anodes of R300a and R300b, while the family Desulfobulbaceae dominated the anode of R300c and, to a lesser extent, were present on the anode of R300a. In the bulk anolyte, Desulfobacteraceae were the most highly represented microorganisms in the samples collected from R300a and R300b, whereas the most abundant microorganisms in the bulk sample collected from R300c belonged to the family Bacillaceae.
FIG 6.
Taxonomic composition at the family level of the microbial communities enriched at the anodes, in the bulk anolyte of reactors polarized at 0 mV and +300 mV (versus an Ag/AgCl [3 M KCl] reference electrode), and in the sediment used as the inoculum. The families with a relative abundance of 5% (or higher) are reported. Sulfate reducers and sulfur reducers were enriched during the bioelectrochemical treatment.
A post hoc test (Tukey-Kramer test; 95% confidence) was performed in order to detect families associated with specific conditions. The conditions applied during bioelectrochemical toluene degradation led to the selection on the anodic biofilms of bacteria belonging to the family Desulfobulbaceae, which had significantly higher relative abundance in anode communities (43.6% ± 29.1%) than in communities from non-anode samples (1.6% ± 1.0%) (see Fig. S5 in the supplemental material). The only anodic sample that showed a poor enrichment of Desulfobulbaceae was collected from the reactor R300b, which likely explains the low current production even after repeated toluene additions in this reactor. Therefore, the presence of Desulfobulbaceae may be linked to the current production coupled with the biodegradation of toluene. When the Tukey-Kramer test was repeated, excluding the samples collected from R300b, a stronger difference between anodic and bulk communities was observed (see Fig. S6). This enforced the hypothesis that the family Desulfobulbaceae has a role in current production.
The sequences of the most abundant OTUs (see Fig. S7 in the supplemental material) were used to determine the best match to sequences contained in the RDP database (see Data Set S1 in the supplemental material). The results showed that the microorganisms enriched in the reactors were phylogenetically related to the sulfate reducers Desulfosarcina ovata, Desulfococcus multivorans, and Desulfobacterium catecholicum and included sulfate-reducing hydrocarbon degraders (Desulfotignum toluenicum and Desulfobacula toluolica) (49–52). Furthermore, the Tukey-Kramer test showed that OTUs phylogenetically related to Desulfobulbus rhabdoformis and Desulfobulbus propionicus were more abundant on the anodes polarized at 0 mV (up to 28% of reads), while OTUs phylogenetically related to Desulfotignum balticum, Desulfotignum toluenicum, and Desulfobacula toluolica were more abundant in the bulk anolyte (up to 46% of reads) (see Fig. S8).
DISCUSSION
Current production and toluene degradation.
Toluene degradation in marine environments was studied in batch reactors by applying different anodic potentials (0 mV and +300 mV). Regardless of the applied potential, toluene degradation coupled with current production was observed. So far, only one study has examined the effect of electrode potential on hydrocarbon degradation. Firman and colleagues assessed toluene degradation by inoculating BESs with a pure culture of Pseudomonas putida F1 and applied anode potentials in the range of +75 to 500 mV. Current production between 11 mA m−2 and 94 mA m−2 was directly correlated with the potential applied, but no information about toluene degradation rates at the different potentials was reported (24). Studies with more degradable substrates (i.e., acetate) showed a discrepancy in the reported results. In some cases a positive correlation between anodic potential and current production was observed (53), while other studies have shown the opposite trend (53, 54).
The maximum current outputs of 301 mA m−2 (0 mV) and 431 mA m−2 (+300 mV) obtained in this study were higher than the current densities obtained in other studies with toluene as the carbon source. Lin and colleagues studied bioelectrochemical toluene degradation with neutral red and ferricyanide as redox mediators (55). The power generation was increased when the mediators were added, and the maximum voltage obtained was 110.4 mV. The voltage harvested in that case, corresponding to a current density of less than 2 mA m−2, was significantly lower than the values obtained in our experiments. In this case, however, the anode was not poised, which could have affected current production (55). A higher current density of about 150 mA m−2 has been reported when a pure culture of Geobacter metallireducens was used with a graphite electrode polarized at +300 mV as the sole electron acceptor (14). Very low current densities have been reported in the case of other hydrocarbons. In a recent study, a sediment microbial fuel cell was used to stimulate total petroleum hydrocarbon degradation, and a peak current of 86 mA m−2 was reached by using a biochar anode (25). However, the high current densities obtained in this study could be attributed to the lower sediment content, to salinity, and to relatively high sulfate concentrations in the medium that reduced the internal resistance.
The toluene removal rate was calculated to be ∼1 mg liter−1 day−1 (∼16 mg kgsediment−1 day−1) for both anode potentials tested in this study. This value was slightly higher than the degradation rates observed in studies with other mixed cultures able to oxidize toluene by using an anode as the sole electron acceptor, either with or without redox mediators (14, 55). The higher rate found in our experiments could be linked to the adsorption of toluene on graphite electrodes. The adsorption of hydrocarbons often occurs when graphite electrodes are used; however, it has been demonstrated that toluene degradation is not restricted to adsorption onto the electrodes (14). To minimize adsorption effects, the degradation rate was calculated based on the results of the second batch of toluene addition.
The cyclic voltammograms obtained under substrate turnover conditions and under substrate non-turnover conditions with reactor R300c (+300 mV) showed a nonreversible reaction. The midpoint potential of the redox-active moiety in both turnover and non-turnover conditions was located at around +200 mV. This result suggests the involvement of this moiety in the electron exchange process. The microbial community in this reactor was dominated by members of the families Bacillaceae (bulk) and Desulfobulbaceae (anode). Bacillus subtilis (family Bacillaceae) has been reported to produce an undetermined electron shuttle with an oxidation peak at +40 to +60 mV (56), a potential lower than that observed in our study. To our knowledge, the nature and the redox potential of the external electron transfer mediators in the family Desulfobulbaceae have not been described yet in the literature. The nature of the redox-active moiety detected in R300c in this study also is undetermined.
With the exception of reactor R300b, current production was attributed to toluene degradation during the first three batch cycles. After the third batch cycle, the current density decreased and toluene removal most probably was coupled to sulfate reduction (although the sulfide concentration was below the detection limit). This shift in the microbial metabolism in a sulfate-rich medium was previously described during bioelectrochemical benzene and sulfide removal, but the current production did not decrease over time (21). Sulfate was abundantly present (2 g liter−1) in the bulk anolyte in our experiments, providing no substrate limitations to sulfate-reducing microorganisms. The small anode surface (8 cm2 geometric area) compared to the total working volume (200 ml) favors such microorganisms that are not in direct contact with the electrode. Thereafter, anode biofilm is enriched with sulfate reducers that outcompete electroactive microorganisms, limiting toluene diffusion to the anodic surface. A hypothetical pathway of this limitation can be summarized as follows: when the biofilm grows and becomes thicker, the cells on the outer surface of the biofilm are the ones to degrade toluene; thus, the thickness of the biofilm body limits electron transfer, resulting in a decrease in current production. Electron transfer throughout a biofilm has been shown to be facilitated when Geobacter is present (57); however, Geobacter was not enriched in our reactors. Previous studies showed that when sulfate is present in the growth medium, bacteria can oxidize organic substrates by sulfate reduction and concomitant sulfide production (58). Sulfide can be electrochemically oxidized to elemental sulfur on the anodic surface (46, 59, 60). In this work, sulfide was oxidized to sulfur and deposited on the anodic surface, as confirmed by both SEM results (see Fig. S2 in the supplemental material) and microbial community findings. 16S rRNA gene sequencing revealed that the family Desulfuromonadaceae was enriched during treatment. Members of this family have been found to reduce elemental sulfur to sulfide (61), which can be reoxidized to elemental sulfur on the anodic surface, enhancing current production as long as there is active sulfur cycling (26). On the basis of the amount of sulfate reduced in the reactors and assuming the complete reduction of sulfate to sulfide followed by the complete oxidation of sulfide to elemental sulfur on the electrode, between 60 and 120 mg of sulfur should have been produced in the reactors at +300 mV (R300a, R300b, and R300c) and between 80 and 110 mg in the reactors at 0 mV (R0a and R0b).
Microbial communities in toluene-degrading BES.
Sulfur metabolism was found to be linked with toluene degraders in the BES reactors. The reduction of sulfate to sulfide most probably was occurring both on the electrode surface and in the bulk of the reactor, whereas subsequent oxidation of sulfide to elemental sulfur most probably was happening on the electrode surface (bio)electrochemically. The presence of SRB in the anodic biofilm was demonstrated by FISH and confirmed by 16S rRNA gene sequencing and qPCR using dsrA-specific primers. SRB belonging to the families Desulfobulbaceae and Desulfobacteraceae were found in the microbial inoculum and were further enriched in BESs. Members of these families have been described previously as key players during hydrocarbon degradation under sulfate-reducing conditions (62, 63). The same families (Desulfobulbaceae and Desulfobacteraceae) have been reported previously to be enriched on anodes during bioelectrochemical benzene degradation in sulfide-rich groundwater (21). Organisms related to Desulfobulbus propionicus were able to produce current in sulfide-rich marine sediment (64, 65). Similarly, in our work, organisms from the family Desulfobulbaceae were found at significantly higher relative abundances in the microbial communities of the anodic biofilms than in those of the bulk anolyte. Filamentous Desulfobulbaceae, the so-called cable bacteria, have been reported recently to perform electron transfer over centimeter-long distances in marine sediments, delivering the electrons obtained by sulfide oxidation (64, 65). Here, we observe that cable bacteria (Desulfobulbaceae) might be involved in toluene degradation via sulfur metabolism, although these bacteria generally are thought to oxidize sulfide in sediments rather than organic matter (64). The role of Desulfobulbaceae in current production is supported by the fact that one of the bioanodes (reactor R300b) did not show current production over the last month of operation despite repeated toluene spiking. Microbial analysis showed the lowest abundance of Desulfobulbaceae in this reactor (R300b). On the basis of these observations, it is possible that bacteria related to the families Desulfobulbaceae and Desulfobacteraceae were involved in hydrocarbon degradation and/or in sulfide/sulfur oxidation with simultaneous electron transfer to the anode.
Microorganisms of the family Desulfuromonadaceae were enriched mainly in R0a and, to a lesser extent, in R300b. Desulfuromonas acetoxidans has been reported to use both elemental sulfur and the anode of a BES as terminal electron acceptors (66). The abundance of this family suggests that metabolism related to elemental sulfur reduction also plays an important role during toluene removal in our reactors. However, the role of sulfur cycle on hydrocarbon degradation in BESs has not been completely elucidated.
Members of the family Bacillaceae (phylum Firmicutes) were relatively abundant in the bulk anolyte of reactor R300c. Their ability to degrade aromatic hydrocarbons in aerobic conditions was described previously (67). Members of the phylum Firmicutes also were selected during the electrochemical degradation of petroleum hydrocarbons in soil (25). Another recent study reported the presence of Bacillus species in an anodic biofilm during cis-dichloroethene oxidation; however, in this work the microbial degradation was triggered by the presence of oxygen produced due to high anodic potential (+1,300 mV) (68). Another study showed that Bacillus subtilis was able to use an electrode as the electron acceptor by excreting external mediators (56).
Interestingly, iron-reducing microorganisms, which usually dominate the anodic communities in BES fed with more readily degradable substrates, such as acetate, were scarce (69, 70).
To strengthen the role of sulfur metabolism in hydrocarbon degradation, other SRBs, closely related to Desulfobulbus rhabdoformis and Desulfobulbus propionicus, were selected at the anodes and were significantly more abundant at the anode of the reactors operated at 0 mV. Similarly, D. rhabdoformis was isolated from a water-oil separation system (71), and the presence of this genus has been widely described in petroleum reservoirs (72, 73). D. propionicus also has been shown to grow using an electrode as the sole electron acceptor (74).
Mechanisms of toluene degradation and final conclusions.
A maximum current density of 283 to 431 mA m−2 was recorded, and a toluene degradation rate of ∼1 mg liter−1 day−1 (∼16 mg kgsediment−1 day−1) was observed regardless of the tested potential (0 and +300 mV). The most abundant microorganisms enriched both in the anodic and in the bulk communities were SRB, some of which were phylogenetically related to known anaerobic hydrocarbon degraders. This is logical if the environmental conditions are taken into account; oxygen is quickly depleted when an oil spill occurs due to the prominent presence of organic matter (75), while under anaerobic conditions sulfate reduction typically is enhanced (75). Toluene degradation rates under sulfate-reducing conditions can vary by orders of magnitude. For example, microcosm experiments assessing toluene removal in soils showed a degradation rate of ∼1 μg kg−1 day−1 (76), whereas the toluene removal rate in microcosms inoculated with contaminated soil and amended with sulfate as the electron acceptor was between 1 and 4 mg liter−1 day−1 (77). The latter rates are similar to the values obtained in our study (∼1 mg liter−1 day−1). Our results confirmed that sulfur species may be involved in the production of current during the bioelectrocatalytic degradation of toluene. However, based on our findings, it cannot be ruled out that BES-based toluene degradation contributed to toluene removal from the sediment, as the anode might have served only as a sulfide scavenger. Rakoczy and colleagues suggested that bioelectrochemical benzene degradation in sulfate-rich environments could occur in the bulk anolyte rather than on the anodic surface. The authors hypothesized that the electron transfer was mediated via electron shuttling related to the sulfur cycle (21), which corroborates earlier findings of Dutta and colleagues in which acetate consumption in BES was accelerated in the presence of sulfide (26). A similar mechanism was described during ferric iron reduction, where ferrihydrite was reduced to ferrous iron by sulfide produced during microbial thiosulfate reduction (78, 79). The microbial community characterization showed that different groups of microorganisms were enriched in the reactors, suggesting that complex electron transfer pathways occurred during toluene removal (see Fig. S9 in the supplemental material). Elucidating the complex microbial interactions that occurred during bioelectrochemical toluene removal was not an explicit aim of the present work; however, the main mechanisms can be hypothesized. Both biotic and abiotic processes occurred on and in the proximity of the anode. The cyclic voltammograms in our study indicate that direct bioelectrocatalytic activity exists. Assuming that only sulfur-mediated electron transfer occurred, we estimate that between 700 and 1,100 mg liter−1 of sulfate should have been consumed. This value is much higher than the sulfate reduction experimentally observed, suggesting that direct electron transfer or sulfur cycling was the prevalent mechanism during the first weeks of reactor operation.
The enrichment of SRB suggests their involvement in toluene degradation coupled to sulfide production. Under these conditions, sulfide can be electrochemically oxidized to elemental sulfur on the anodic surface, which can further serve as the mediator (46). The abundance of Desulfobulbaceae, however, indicates that sulfide oxidation to elemental sulfur on the anodic surface is a biologically mediated process. Furthermore, members of the Desulfuromonadaceae identified in the BES may have a role in the reduction of sulfur to sulfide, leading to internal sulfur cycling. Nevertheless, further studies are needed to completely elucidate the electron transfer mechanisms involved in toluene degradation in the BES.
BES have been demonstrated to be a suitable technology to power oceanographic devices by harvesting energy from the ocean floor (18, 19). The results presented in this paper reinforce earlier studies that suggest the potential of buried anodes to stimulate hydrocarbon degradation in anoxic marine sediments (15). The anode thus can serve both as a direct electron acceptor for microbial respiration and as a scavenger to reduce the concentration of toxic sulfide. Further studies should be focused on designing BES configurations to achieve the best hydrocarbon removal performance. Furthermore, nutrients could represent a limiting factor to hydrocarbon degradation in marine environments (9), and future works should explore the effect of inorganic nutrient limitation on bioelectrochemical hydrocarbon removal in order to better establish the advantages and the drawbacks of this technology.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in the framework of the Kill-Spill project under the FP7 program of the European Commission and the OILSPORE project funded by the UK NERC (NE/J024325/1).
We thank Filip Meysman of the Royal Netherlands Institute for Sea Research for providing the microbial inoculum. M.D. gratefully thanks the Science Doctoral School of the University of Milano–Bicocca for funding his research stay at Ghent University.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02250-15.
REFERENCES
- 1.Sammarco PW, Kolian SR, Warby RAF, Bouldin JL, Subra WA, Porter SA. 2013. Distribution and concentrations of petroleum hydrocarbons associated with the BP/Deepwater Horizon oil spill, Gulf of Mexico. Mar Pollut Bull 73:129–143. doi: 10.1016/j.marpolbul.2013.05.029. [DOI] [PubMed] [Google Scholar]
- 2.Nelson-Smith A. 1972. Oil pollution and marine ecology. Elek Science, London, United Kingdom. [Google Scholar]
- 3.U.S. EPA 2011. Screening-level hazard characterization. U.S. EPA, Washington, DC. [Google Scholar]
- 4.Moreno R, Jover L, Diez C, Sardà F, Sanpera C. 2013. Ten years after the prestige oil spill: seabird trophic ecology as indicator of long-term effects on the coastal marine ecosystem. PLoS One 8:e77360. doi: 10.1371/journal.pone.0077360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sumaila RU, Cisneros-Montemayor AM, Dyck A, Huang L, Cheung W, Jacquet J, Kleisner K, Lam V, McCrea-Strub A, Swartz W, Watson R, Zeller D, Pauly D. 2012. Impact of the Deepwater Horizon well blowout on the economics of US Gulf fisheries. Can J Fish Aquat Sci 69:499–510. doi: 10.1139/f2011-171. [DOI] [Google Scholar]
- 6.Laffon B, Aguilera F, Ríos-Vázquez J, García-Lestón J, Fuchs D, Valdiglesias V, Pásaro E. 2013. Endocrine and immunological parameters in individuals involved in Prestige spill cleanup tasks seven years after the exposure. Environ Int 59:103–111. doi: 10.1016/j.envint.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 7.Wikelski M, Wong V, Chevalier B, Rattenborg N, Snell HL. 2002. Marine iguanas die from trace oil pollution. Nature 417:607–608. doi: 10.1038/417607a. [DOI] [PubMed] [Google Scholar]
- 8.Atlas RM. 1995. Petroleum biodegradation and oil spill bioremediation. Mar Pollut Bull 31:178–182. doi: 10.1016/0025-326X(95)00113-2. [DOI] [Google Scholar]
- 9.Head IM, Jones DM, Röling WFM. 2006. Marine microorganisms make a meal of oil. Nat Rev Microbiol 4:173–182. doi: 10.1038/nrmicro1348. [DOI] [PubMed] [Google Scholar]
- 10.Yang S-Z, Jin H-J, Wei Z, He R-X, Ji Y-J, Li X-M, Yu S-P. 2009. Bioremediation of oil spills in cold environments: a review. Pedosphere 19:371–381. doi: 10.1016/S1002-0160(09)60128-4. [DOI] [Google Scholar]
- 11.Lu L, Yazdi H, Jin S, Zuo Y, Fallgren PH, Ren ZJ. 2014. Enhanced bioremediation of hydrocarbon-contaminated soil using pilot-scale bioelectrochemical systems. J Hazard Mater 274:8–15. doi: 10.1016/j.jhazmat.2014.03.060. [DOI] [PubMed] [Google Scholar]
- 12.Broden RC, Goin RT, Kao C-M. 1997. Control of BTEX migration using a biologically enhanced permeable barrier. Ground Water Monit Remediat 17:70–80. [Google Scholar]
- 13.Tuxen N, Reitzel LA, Albrechtsen H-J, Bjerg PL. 2006. Oxygen-enhanced biodegradation of phenoxy acids in ground water at contaminated sites. Ground Water 44:256–265. doi: 10.1111/j.1745-6584.2005.00104.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhang T, Gannon SM, Nevin KP, Franks AE, Lovley DR. 2010. Stimulating the anaerobic degradation of aromatic hydrocarbons in contaminated sediments by providing an electrode as the electron acceptor. Environ Microbiol 12:1011–1020. doi: 10.1111/j.1462-2920.2009.02145.x. [DOI] [PubMed] [Google Scholar]
- 15.Lovley DR, Nevin KP. 2011. A shift in the current: new applications and concepts for microbe-electrode electron exchange. Curr Opin Biotechnol 22:441–448. doi: 10.1016/j.copbio.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Morris JM, Jin S, Crimi B, Pruden A. 2009. Microbial fuel cell in enhancing anaerobic biodegradation of diesel. Chem Eng J 146:161–167. doi: 10.1016/j.cej.2008.05.028. [DOI] [Google Scholar]
- 17.Nielsen ME, Reimers CE, White HK, Sharma S, Girguis PR. 2008. Sustainable energy from deep ocean cold seeps. Energy Environ Sci 1:584–593. doi: 10.1039/b811899j. [DOI] [Google Scholar]
- 18.Nielsen ME, Wu DM, Girguis PR, Reimers CE. 2009. Influence of substrate on electron transfer mechanisms in chambered benthic microbial fuel cells. Environ Sci Technol 43:8671–8677. doi: 10.1021/es9013773. [DOI] [PubMed] [Google Scholar]
- 19.Logan BE, Hamelers B, Rozendal R, Schröder U, Keller J, Freguia S, Aelterman P, Verstraete W, Rabaey K. 2006. Microbial fuel cells: methodology and technology. Environ Sci Technol 40:5181–5192. doi: 10.1021/es0605016. [DOI] [PubMed] [Google Scholar]
- 20.Morris JM, Jin S. 2012. Enhanced biodegradation of hydrocarbon-contaminated sediments using microbial fuel cells. J Hazard Mater 213-214:474–477. [DOI] [PubMed] [Google Scholar]
- 21.Rakoczy J, Feisthauer S, Wasmund K, Bombach P, Neu TR, Vogt C, Richnow HH. 2013. Benzene and sulfide removal from groundwater treated in a microbial fuel cell. Biotechnol Bioeng 110:3104–3113. doi: 10.1002/bit.24979. [DOI] [PubMed] [Google Scholar]
- 22.Holmes DE, Bond DR, O'Neil RA, Reimers CE, Tender LR, Lovley DR. 2004. Microbial communities associated with electrodes harvesting electricity from a variety of aquatic sediments. Microb Ecol 48:178–190. doi: 10.1007/s00248-003-0004-4. [DOI] [PubMed] [Google Scholar]
- 23.Ren L, Siegert M, Ivanov I, Pisciotta JM, Logan BE. 2013. Treatability studies on different refinery wastewater samples using high-throughput microbial electrolysis cells (MECs). Bioresour Technol 136:322–328. doi: 10.1016/j.biortech.2013.03.060. [DOI] [PubMed] [Google Scholar]
- 24.Friman H, Schechter A, Nitzan Y, Cahan R. 2012. Effect of external voltage on Pseudomonas putida F1 in a bio electrochemical cell using toluene as sole carbon and energy source. Microbiology 158:414–423. doi: 10.1099/mic.0.053298-0. [DOI] [PubMed] [Google Scholar]
- 25.Lu L, Huggins T, Jin S, Zuo Y, Ren ZJ. 2014. Microbial metabolism and community structure in response to bioelectrochemically enhanced remediation of petroleum hydrocarbon-contaminated soil. Environ Sci Technol 48:4021–4029. doi: 10.1021/es4057906. [DOI] [PubMed] [Google Scholar]
- 26.Dutta PK, Keller J, Yuan Z, Rozendal RA, Rabaey K. 2009. Role of sulfur during acetate oxidation in biological anodes. Environ Sci Technol 43:3839–3845. doi: 10.1021/es803682k. [DOI] [PubMed] [Google Scholar]
- 27.Patil SA, Harnisch F, Koch C, Hübschmann T, Fetzer I, Carmona-Martínez AA, Müller S, Schröder U. 2011. Electroactive mixed culture derived biofilms in microbial bioelectrochemical systems: the role of pH on biofilm formation, performance and composition. Bioresour Technol 102:9683–9690. doi: 10.1016/j.biortech.2011.07.087. [DOI] [PubMed] [Google Scholar]
- 28.Meysman FJR, Risgaard-Petersen N, Malkin SY, Nielsen LP. 2015. The geochemical fingerprint of microbial long-distance electron transport in the seafloor. Geochim Cosmochim Acta 152:122–142. doi: 10.1016/j.gca.2014.12.014. [DOI] [Google Scholar]
- 29.Patil SA, Hägerhäll C, Gorton L. 2012. Electron transfer mechanisms between microorganisms and electrodes in bioelectrochemical systems. Bioanal Rev 4:159–192. doi: 10.1007/s12566-012-0033-x. [DOI] [Google Scholar]
- 30.U.S. EPA 2002. Method 5035A, closed-system purge-and-trap and extraction for volatile organics in soil and waste samples. U.S. EPA, Washington, DC. [Google Scholar]
- 31.U.S. EPA 2003. Method 5021A, volatile organic compounds in various sample matrices using equilibrium headspace analysis. U.S. EPA, Washington, DC. [Google Scholar]
- 32.Loy A, Maixner F, Wagner M, Horn M. 2007. probeBase–an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res 35:D800–D804. doi: 10.1093/nar/gkl856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferris MJ, Muyzer G, Ward D. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol 62:340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki Y, Kelly SD, Kemner KM, Banfield JF. 2005. Direct microbial reduction and subsequent preservation of uranium in natural near-surface sediment. Appl Environ Microbiol 71:1790–1797. doi: 10.1128/AEM.71.4.1790-1797.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gittel A, Sørensen KB, Skovhus TL, Ingvorsen K, Schramm A. 2009. Prokaryotic community structure and sulfate reducer activity in water from high-temperature oil reservoirs with and without nitrate treatment. Appl Environ Microbiol 75:7086–7096. doi: 10.1128/AEM.01123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Von Netzer F, Pilloni G, Kleindienst S, Krüger M, Knittel K, Gründger F, Lueders T. 2013. Enhanced gene detection assays for fumarate-adding enzymes allow uncovering of anaerobic hydrocarbon degraders in terrestrial and marine systems. Appl Environ Microbiol 79:543–552. doi: 10.1128/AEM.02362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brankatschk R, Bodenhausen N, Zeyer J, Bürgmann H. 2012. Simple absolute quantification method correcting for quantitative PCR efficiency variations for microbial community samples. Appl Environ Microbiol 78:4481–4489. doi: 10.1128/AEM.07878-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruijter JM, Ramakers C, Hoogaars WMH, Karlen Y, Bakker O, van den Hoff MJB, Moorman AFM. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daghio M, Tatangelo V, Franzetti A, Gandolfi I, Papacchini M, Careghini A, Sezenna E, Saponaro S, Bestetti G. 2015. Hydrocarbon degrading microbial communities in bench scale aerobic biobarriers for gasoline contaminated groundwater treatment. Chemosphere 130:34–39. doi: 10.1016/j.chemosphere.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 41.Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA, Sogin ML. 2007. Microbial population structures in the deep marine biosphere. Science 318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Qian PY. 2009. Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One 4:e7401. doi: 10.1371/journal.pone.0007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 44.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. 2014. Genome analysis STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatic Adv 2014:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300. [Google Scholar]
- 46.Dutta PK, Rabaey K, Yuan Z, Keller J. 2008. Spontaneous electrochemical removal of aqueous sulfide. Water Res 42:4965–4975. doi: 10.1016/j.watres.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 47.Müller AL, Kjeldsen KU, Rattei T, Pester M, Loy A. 2014. Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. ISME J 9:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Callbeck CM, Sherry A, Hubert CRJ, Gray ND, Voordouw G, Head IM. 2013. Improving PCR efficiency for accurate quantification of 16S rRNA genes. J Microbiol Methods 93:148–152. doi: 10.1016/j.mimet.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Ommedal H, Torsvik T. 2007. Desulfotignum toluenicum sp. nov., a novel toluene-degrading, sulphate-reducing bacterium isolated from an oil-reservoir model column. Int J Syst Evol Microbiol 57:2865–2869. doi: 10.1099/ijs.0.65067-0. [DOI] [PubMed] [Google Scholar]
- 50.Rabus R, Nordhaus R, Ludwig W, Widdell F. 1993. Complete oxidation of toluene under strictly anoxic conditions by a new sulfate-reducing bacterium. Appl Environ Microbiol 59:1444–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Higashioka Y, Kojima H, Fukui M. 2011. Temperature-dependent differences in community structure of bacteria involved in degradation of petroleum hydrocarbons under sulfate-reducing conditions. J Appl Microbiol 110:314–322. doi: 10.1111/j.1365-2672.2010.04886.x. [DOI] [PubMed] [Google Scholar]
- 52.Harms G, Zengler K, Rabus R, Aeckersberg F, Minz D, Rossello-Mora R, Widdel F. 1999. Anaerobic oxidation of o-xylene, m-xylene, and homologous alkylbenzenes by new types of sulfate-reducing bacteria. Appl Environ Microbiol 65:999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner RC, Call DF, Logan BE. 2010. Optimal set anode potentials vary in bioelectrochemical systems. Environ Sci Technol 44:6036–6041. doi: 10.1021/es101013e. [DOI] [PubMed] [Google Scholar]
- 54.Aelterman P, Freguia S, Keller J, Verstraete W, Rabaey K. 2008. The anode potential regulates bacterial activity in microbial fuel cells. Appl Microbiol Biotechnol 78:409–418. doi: 10.1007/s00253-007-1327-8. [DOI] [PubMed] [Google Scholar]
- 55.Lin C-W, Wu C-H, Chiu Y-H, Tsai S-L. 2014. Effects of different mediators on electricity generation and microbial structure of a toluene powered microbial fuel cell. Fuel 125:30–35. doi: 10.1016/j.fuel.2014.02.018. [DOI] [Google Scholar]
- 56.Nimje VR, Chen C-Y, Chen C-C, Jean J-S, Reddy AS, Fan C-W, Pan K-Y, Liu H-T, Chen J-L. 2009. Stable and high energy generation by a strain of Bacillus subtilis in a microbial fuel cell. J Power Sources 190:258–263. doi: 10.1016/j.jpowsour.2009.01.019. [DOI] [Google Scholar]
- 57.Reguera G, Nevin KP, Nicoll JS, Covalla SF, Woodard TL, Lovley DR. 2006. Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl Environ Microbiol 72:7345–7348. doi: 10.1128/AEM.01444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jørgensen BB. 1977. The sulfur cycle of a coastal marine sediment (Limfjorden, Denmark). Limnol Oceanogr 22:814–832. doi: 10.4319/lo.1977.22.5.0814. [DOI] [Google Scholar]
- 59.Rabaey K, Van de Soempel K, Maignien L, Boon N, Aelterman P, Clauwaert P, De Schamphelaire L, Pham HT, Vermeulen J, Verhaege M, Lens P, Verstraete W. 2006. Microbial fuel cells for sulfide removal. Environ Sci Technol 40:5218–5224. doi: 10.1021/es060382u. [DOI] [PubMed] [Google Scholar]
- 60.Ryckelynck N, Stecher HA, Reimers CE. 2005. Understanding the anodic mechanism of a seafloor fuel cell: interactions between geochemistry and microbial activity. Biogeochemistry 76:113–139. doi: 10.1007/s10533-005-2671-3. [DOI] [Google Scholar]
- 61.Pfennig N, Biebl H. 1976. Desulfuromonas acetoxidans gen. nov. and sp. nov., a new anaerobic, sulfur-reducing, acetate-oxidizing bacterium. Arch Microbiol 110:3–12. doi: 10.1007/BF00416962. [DOI] [PubMed] [Google Scholar]
- 62.Cupples AM. 2011. The use of nucleic acid based stable isotope probing to identify the microorganisms responsible for anaerobic benzene and toluene biodegradation. J Microbiol Methods 85:83–91. doi: 10.1016/j.mimet.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 63.Kuppardt A, Kleinsteuber S, Vogt C, Lüders T, Harms H, Chatzinotas A. 2014. Phylogenetic and functional diversity within toluene-degrading, sulphate-reducing consortia enriched from a contaminated aquifer. Microb Ecol 68:222–234. doi: 10.1007/s00248-014-0403-8. [DOI] [PubMed] [Google Scholar]
- 64.Pfeffer C, Larsen S, Song J, Dong M, Besenbacher F, Meyer RL, Kjeldsen KU, Schreiber L, Gorby YA, El-Naggar MY, Leung KM, Schramm A, Risgaard-Petersen N, Nielsen LP. 2012. Filamentous bacteria transport electrons over centimetre distances. Nature 491:218–221. doi: 10.1038/nature11586. [DOI] [PubMed] [Google Scholar]
- 65.Schauer R, Risgaard-Petersen N, Kjeldsen KU, Tataru Bjerg JJ, Jørgensen BB, Schramm A, Nielsen LP. 2014. Succession of cable bacteria and electric currents in marine sediment. ISME J 8:1314–1322. doi: 10.1038/ismej.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bond DR, Holmes DE, Tender LM, Lovley DR. 2002. Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295:483–485. doi: 10.1126/science.1066771. [DOI] [PubMed] [Google Scholar]
- 67.Ling J, Zhang G, Sun H, Fan Y, Ju J, Zhang C. 2011. Isolation and characterization of a novel pyrene-degrading Bacillus vallismortis strain JY3A. Sci Total Environ 409:1994–2000. doi: 10.1016/j.scitotenv.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 68.Aulenta F, Verdini R, Zeppilli M, Zanaroli G, Fava F, Rossetti S, Majone M. 2013. Electrochemical stimulation of microbial cis-dichloroethene (cis-DCE) oxidation by an ethene-assimilating culture. N Biotechnol 30:749–755. doi: 10.1016/j.nbt.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 69.Kiely PD, Regan JM, Logan BE. 2011. The electric picnic: synergistic requirements for exoelectrogenic microbial communities. Curr Opin Biotechnol 22:378–385. doi: 10.1016/j.copbio.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 70.Daghio M, Gandolfi I, Bestetti G, Franzetti A, Guerrini E, Cristiani P. 2015. Anodic and cathodic microbial communities in single chamber microbial fuel cells. N Biotechnol 32:79–84. doi: 10.1016/j.nbt.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 71.Lien T, Madsen M, Steen IH, Gjerdevik K. 1998. Desulfobulbus rhabdoformis sp. nov., a sulfate reducer from a water-oil separation system. Int J Syst Bacteriol 48:469–474. doi: 10.1099/00207713-48-2-469. [DOI] [PubMed] [Google Scholar]
- 72.Guan J, Xia L-P, Wang L-Y, Liu J-F, Gu J-D, Mu B-Z. 2013. Diversity and distribution of sulfate-reducing bacteria in four petroleum reservoirs detected by using 16S rRNA and dsrAB genes. Int Biodeterior Biodegradation 76:58–66. doi: 10.1016/j.ibiod.2012.06.021. [DOI] [Google Scholar]
- 73.Wang L-Y, Ke W-J, Sun X-B, Liu J-F, Gu J-D, Mu B-Z. 2014. Comparison of bacterial community in aqueous and oil phases of water-flooded petroleum reservoirs using pyrosequencing and clone library approaches. Appl Microbiol Biotechnol 98:4209–4221. doi: 10.1007/s00253-013-5472-y. [DOI] [PubMed] [Google Scholar]
- 74.Holmes DE, Bond DR, Lovley DR. 2004. Electron transfer by Desulfobulbus propionicus to Fe(III) and graphite electrodes. Appl Environ Microbiol 70:1234–1237. doi: 10.1128/AEM.70.2.1234-1237.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Natter M, Keevan J, Wang Y, Keimowitz AR, Okeke BC, Son A, Lee M-K. 2012. Level and degradation of Deepwater Horizon spilled oil in coastal marsh sediments and pore-water. Environ Sci Technol 46:5744–5755. doi: 10.1021/es300058w. [DOI] [PubMed] [Google Scholar]
- 76.Noh S-L, Choi J-M, An Y-J, Park S-S, Cho K-S. 2003. Anaerobic biodegradation of toluene coupled to sulfate reduction in oil-contaminated soils: optimum environmental conditions for field applications. J Environ Sci Health A Tox Hazard Subst Environ Eng 38:1087–1097. doi: 10.1081/ESE-120019866. [DOI] [PubMed] [Google Scholar]
- 77.Dou J, Liu X, Hu Z, Deng D. 2008. Anaerobic BTEX biodegradation linked to nitrate and sulfate reduction. J Hazard Mater 151:720–729. doi: 10.1016/j.jhazmat.2007.06.043. [DOI] [PubMed] [Google Scholar]
- 78.Beller HR, Grbić-Galić D, Reinhard M. 1992. Microbial degradation of toluene under sulfate-reducing conditions and the influence of iron on the process. Appl Environ Microbiol 58:786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Straub KL, Schink B. 2004. Ferrihydrite-dependent growth of Sulfurospirillum deleyianum through electron transfer via sulfur cycling. Appl Environ Microbiol 70:5744–5749. doi: 10.1128/AEM.70.10.5744-5749.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.