Abstract
Cronobacter sakazakii is an important pathogen that causes high mortality in infants. Due to its occasional antibiotic resistance, a bacteriophage approach might be an alternative effective method for the control of this pathogen. To develop a novel biocontrol agent using bacteriophages, the C. sakazakii-infecting phage CR5 was newly isolated and characterized. Interestingly, this phage exhibited efficient and relatively durable host lysis activity. In addition, a specific gene knockout study and subsequent complementation experiment revealed that this phage infected the host strain using the bacterial flagella. The complete genome sequence analysis of phage CR5 showed that its genome contains 223,989 bp of DNA, including 231 predicted open reading frames (ORFs), and it has a G+C content of 50.06%. The annotated ORFs were classified into six functional groups (structure, packaging, host lysis, DNA manipulation, transcription, and additional functions); no gene was found to be related to virulence or toxin or lysogen formation, but >80% of the predicted ORFs are unknown. In addition, a phage proteomic analysis using SDS-PAGE and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) revealed that seven phage structural proteins are indeed present, supporting the ORF predictions. To verify the potential of this phage as a biocontrol agent against C. sakazakii, it was added to infant formula milk contaminated with a C. sakazakii clinical isolate or food isolate, revealing complete growth inhibition of the isolates by the addition of phage CR5 when the multiplicity of infection (MOI) was 105.
INTRODUCTION
Enterobacter sakazakii was first defined in 1980 (1) and reclassified into a new genus, Cronobacter, in 2007 based on its fluorescent amplified fragment length polymorphism (f-AFLP) fingerprints, ribopatterns, and 16S rRNA sequencing (2). Cronobacter sakazakii is a generally well-known pathogen in infant milk formula powders and causes bacteremia, meningitis, and necrotizing enterocolitis in neonates, with high fatality rates (3, 4). Due to this high risk to infants, C. sakazakii pathogenesis has attracted broad public attention. This pathogen was recently reported to infect the elderly in Taiwan, suggesting that there may be various routes of C. sakazakii infection (5). When C. sakazakii was defined, antibiotic susceptibility tests revealed that this species had occasional antibiotic resistance (1, 6), suggesting that the antibiotic resistance of C. sakazakii has resulted in limited antibiotic therapies for the control of this pathogen. Therefore, novel alternative biocontrol agents should be developed.
Bacteriophages are bacterial viruses that infect and lyse specific host bacteria for their replication and propagation (7). Due to their host specificity and lysis activity, phages have been considered alternative biocontrol agents for the control of pathogenic bacteria. Although this approach has been used as a therapeutic approach in the former Soviet Union and eastern Europe for several decades, the application of bacteriophages for the control of pathogens has been suggested and evaluated only in Western countries over the last decade (8). Because of the high risk of C. sakazakii infection and the emergence of antibiotic-resistant strains, a bacteriophage agent could be very useful for the control of C. sakazakii. Previous U.S. Food and Drug Administration (FDA) approval of bacteriophages for food applications, such as ListShield (Intralytix, Baltimore, MD, USA) and Listex P100 (Micros Food Safety, Wageningen, The Netherlands), supports this.
As of September 2015, 18 complete genome sequences of bacteriophages specific for C. sakazakii have been reported (9–19). These phage genome analyses have revealed that they have no virulence-related genes encoding virulence factors and toxins. A few attempts using C. sakazakii phages have been made in food applications to control C. sakazakii infection. Selected C. sakazakii phages have been tested in reconstituted infant formula, and the results show that they can efficiently prevent the bacterial growth of C. sakazakii in formula (20). In addition, 67 newly isolated C. sakazakii phages were tested, and some of them reduced C. sakazakii in pure broth culture up to 4 log (CFU/ml), highlighting their potential as biocontrol agents against C. sakazakii in foods (21). Furthermore, phage therapy using C. sakazakii phages showed high efficiency in alleviating Cronobacter-induced urinary tract infections in mice (22).
In this study, the C. sakazakii-infecting bacteriophage CR5 was isolated and purified, and its general features were experimentally characterized to evaluate its possibility as a novel efficient biocontrol agent. In addition, its host receptor was confirmed using a specific gene knockout of the host strain to elucidate its host infection mechanisms. Furthermore, its genome was completely sequenced, and its core structural proteins were analyzed to further understand its characteristics at the genomic and proteomic levels. In addition, the application of phage CR5 to infant formula milk was assessed, suggesting its potential as a novel biocontrol agent against C. sakazakii infection in foods.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. C. sakazakii ATCC 29544 was used for the isolation and propagation of bacteriophage CR5. All of the bacteria were grown at 37°C (except Enterobacter cloacae and Enterobacter agglomerans, which were grown at 30°C) for 12 h in tryptic soy broth (TSB) medium (Difco, Detroit, MI, USA).
TABLE 1.
Host range of C. sakazakii phage CR5
| Bacterial straina | Plaque formationb | Source or reference |
|---|---|---|
| C. sakazakii type strains | ||
| ATCC 29544 | +++ | ATCC |
| ATCC 29544 ΔflgK | − | This study |
| ATCC 29544 ΔflgK pBAD18::flgK | +++++ | This study |
| ATCC 51329 | + | ATCC |
| BAA-894 | I | 44 |
| ST4 (ATCC 29004) | I | ATCC |
| C. sakazakii isolates (sample type) | ||
| 1-2 (dried fish) | +++++ | 45 |
| 3-2 (dried fish) | I | 45 |
| 4-1 (dried fish) | ++++ | 45 |
| 5-2 (dried fish) | ++ | 45 |
| 7-1 (dried fish) | I | 45 |
| 10-2 (dried fish) | I | 45 |
| 2-1 (dried powdered vegetables) | +++ | 45 |
| 16-2 (sunsik) | +++ | 45 |
| 20-2 (sunsik) | +++ | 45 |
| 27-1 (sunsik) | I | 45 |
| 29-1 (sunsik) | I | 45 |
| 31-3 (sunsik) | ++++ | 45 |
| Other Cronobacter type strains | ||
| C. malonaticus DSM 18702 | + | DSM |
| C. turicensis DSM 18703 | + | DSM |
| C. dublinensis DSM 18705 | I | DSM |
| C. dublinensis subsp. lactaridi DSM 18707 | + | DSM |
| Gram-negative bacteria | ||
| Klebsiella pneumoniae KCTC 2242 | − | KCTC |
| Klebsiella oxytoca KCTC 1686 | − | KCTC |
| K. oxytoca ATCC 43863 | − | ATCC |
| Enterobacter aerogenes ATCC 35028 | − | ATCC |
| E. cloacae ATCC 7256 | I | ATCC |
| E. agglomerans ATCC 27987 | I | ATCC |
| Salmonella enterica serovar Typhimurium ATCC 19586 | − | ATCC |
| Salmonella enterica serovar Enteritidis ATCC 13076 | − | ATCC |
| Escherichia coli DH5α | − | Invitrogen |
| Vibrio fischeri ATCC 700601 | − | ATCC |
| Pseudomonas aeruginosa ATCC 27853 | − | ATCC |
| Gram-positive bacteria | ||
| Listeria monocytogenes ATCC 19114 | − | ATCC |
| Staphylococcus aureus ATCC 29213 | − | ATCC |
| Bacillus cereus ATCC 14579 | − | ATCC |
| Enterococcus faecalis ATCC 29212 | − | ATCC |
ST4, sequence type 4.
+++++, EOP of 4 to 2; ++++, EOP of 2 to 1; +++, EOP of 1 to 0.5; ++, EOP of 0.5 to 0.2; +, EOP < 0.2; I, formation of inhibition zone; −, not susceptible to phage CR5.
Bacteriophage isolation and purification.
Environmental samples from farm soil in Suwon, South Korea, were used for the isolation of C. sakazakii-targeting bacteriophages. To isolate the bacteriophages, 25 g of each sample was homogenized with 225 ml of sodium chloride-magnesium sulfate (SM) buffer (100 mM NaCl, 10 mM MgSO4·7H2O, and 50 mM Tris-HCl [pH 7.5]). After homogenization, 25 ml of each homogenized sample was diluted twice with 25 ml of 2×-concentrated TSB, and the mixture was incubated with vigorous shaking at 37°C for 12 h. The collected samples were centrifuged at 6,000 × g for 10 min, and the supernatants were filtered using 0.22-μm-diameter-pore filters (Millipore, Billerica, MA, USA). Ten milliliters of each filtrate was mixed with 50 ml of TSB medium containing 109 CFU/ml of an overnight culture of C. sakazakii ATCC 29544 as the propagation strain. The mixture was then incubated at 37°C for 12 h with vigorous shaking. The culture was centrifuged at 6,000 × g for 10 min, and the supernatant containing the phages was filtered using a 0.22-μm-diameter-pore filter to remove the residual bacterial cells. Next, 10-fold serial dilutions of the supernatant were spotted on molten 0.4% TSB soft agar containing 109 CFU/ml of an overnight culture of C. sakazakii ATCC 29544. Individual plaques were picked, and the phages were resuspended with SM buffer. These plaque isolation and phage resuspension steps were repeated at least five times to isolate and purify the individual phages. When the optical density at 600 nm (OD600) of the culture of C. sakazakii ATCC 29544 reached 1.0, bacteria were infected with the purified phage CR5 at a multiplicity of infection (MOI) of 1 and incubated at 37°C for 4 h. For the purification of phage CR5 after propagation, the host cell debris was removed by subsequent centrifugation at 6,000 × g for 10 min, the supernatant was filtered with 0.22-μm-diameter-pore filters, and the phage particles were precipitated by treatment with polyethylene glycol (PEG) 6000 (Junsei, Tokyo, Japan). As a final step, cesium chloride (CsCl) density gradient ultracentrifugation (HIMAC CP100β; Hitachi, Tokyo, Japan) with different CsCl steps (step density = 1.3, 1.45, 1.5, and 1.7 g/ml) was performed at 78,500 × g and 4°C for 2 h. The band containing the viral particles was recovered by puncturing the centrifuge tube with a sterilized needle, followed by dialysis using standard dialysis buffer (10 mM NaCl, 10 mM MgSO4, and 1 M Tris-HCl [pH 8.0]). The purified phages were stored at 4°C for further experiments.
Transmission electron microscopy.
The morphology of the purified phage CR5 was observed using transmission electron microscope (TEM) analysis. This analysis was conducted in accordance with the procedure described by Kim and Ryu (23), with some modifications. Briefly, 10 μl of CR5 phage stock was loaded onto a copper grid and incubated for 1 min, and excess solution was removed. Next, the same amount of 2% uranyl acetate (pH 4.0) was applied and washed with ultrapure water. The stained grid was subjected to TEM analysis with JEM-2100F (JEOL, Tokyo, Japan) at 200 kV. The identification and classification of CR5 were conducted according to the guidelines of the International Committee on the Taxonomy of Viruses (ICTV) (24).
Host range analysis.
Five milliliters of molten 0.4% TSB agar containing 100 μl of each test bacterial culture was overlaid on 1.5% TSB agar plates. Ten microliters of each serially diluted phage CR5 suspension from 102 to 109 PFU/ml was spotted on the overlaid plates. The host lysis activity of the test bacteria by phage CR5 was determined by measuring the formation of plaques in the spots. The efficiency of plating (EOP) was calculated by a comparison of titers between the selected test bacteria and the propagation host strain C. sakazakii ATCC 29544.
Bacterial challenge test.
C. sakazakii ATCC 29544 was inoculated into TSB broth medium and grown at 37°C for 12 h with agitation (220 rpm), and then 1% of this culture was subinoculated into 50 ml of fresh TSB broth and incubated at 37°C with agitation. To confirm the bacterial lytic activity of the phage, CR5 (MOI, 1.0) was added to an exponentially growing C. sakazakii ATCC 29544 culture at an OD600 of 1.0, and the culture OD at 600 nm was monitored at 1-h intervals. A culture of C. sakazakii ATCC 29544 without the phage was used as a control. All of the tests were conducted in triplicate.
Mutant construction and complementation for identification of host receptor.
The mutants of C. sakazakii ATCC 29544 used for the identification of host receptors were constructed using the one-step gene inactivation method previously described by Kim et al. (25). For construction of the mutant, five host receptor-associated genes, including rfaC for the O antigen of lipopolysaccharide (LPS), lamB or ompC for outer membrane protein, fhuA for ferric ion uptake transporter, and flgK for flagella, were selected, and their specific primer sets were designed (Table 2). The kanamycin resistance cassette for integration into each selected gene in the host chromosome was PCR amplified from pKD13 using one of the specific gene-targeting primer sets, which contains identical small DNA sequences upstream of the start codon and downstream of the stop codon of each selected gene. The PCR product was then electroporated into C. sakazakii ATCC 29544 containing pKD46 with an integrase gene for integration of the PCR product into the host chromosome via homologous recombination. After integration of the PCR product, the mutant of each gene was selected using TSB agar plates containing 50 μg/ml kanamycin sulfate (Sigma, St. Louis, MO). The kanamycin resistance gene was removed from the selected mutant using the pCP20 plasmid (26). For complementation of the flgK mutant, the host flgK gene was PCR amplified using the flgK-comple-F forward primer (HindIII) (5′-GTG TAC TAA TAA GCT TCG TTC GGT TCC CTG-3′) and flgK-comple-R reverse primer (NheI) (5′-TAA GCG CTA GCG ATA ATT ATC GTC AGG ACC-3′). After purification of the PCR product, it was cloned into the pBAD18 expression vector. After the transformation of pBAD18-flgK into the C. sakazakii flgK mutant and selection with 50 μg/ml ampicillin, arabinose (0.2% final concentration) was used as an induction reagent for complementation of the flgK gene in the mutant.
TABLE 2.
Primers used for construction of C. sakazakii mutants
| Gene target | Primer name | Oligonucleotide sequence (5′–3′)a |
|---|---|---|
| rfaC | rfaC-red-F | AAC GGA TGT TTC CCC GCA AAG CCA GGG ACG CAG TTG TTC AAA AAC GGT AGC GGC GTG TAG GCT GGA GCT GCT TCG |
| rfaC-red-R | TGC CCG CGT TCT GGA GAC GCT CAA CGA ACT GCT GCT GAA CGA GGA AGC CTG ACG GAT TCC GGG GAT CCG TCG ACC | |
| lamB | lamB-red-F | GAG ATA GAA TGA TGA TAA CTC TGC GTA AAC TCC CTC TGG CTG TGG CCG TGA TGG CTG TAG GCT GGA GCT GCT TCG |
| lamB-red-R | TAC CAC CAG ATT TCC ATC TGG GCA CCG AAG GTC CAC TCA TCA TTG TCG CCA CGG CAT TCC GGG GAT CCG TCG ACC | |
| ompC | ompC-red-F | TCG GAC AAT GGA TTT GCC CGC TAG TTC CCT GAA TTA GTG AGC AGT GGC AAT AAT ATG TAG GCT GGA GCT GCT TCG |
| ompC-red-R | GGA GCC CGC AGG CTC CTT TTG CAC ATC AGG TCG GGG ATT AGA ACT GGT AAA CCA GAT TCC GGG GAT CCG TCG ACC | |
| fhuA | fhuA-red-F | TCA AAC AGG TTA TTG ACG TTT AAG GCG ACA GAC GAG CCC GGC AGG CCT AAA CGC GTG TAG GCT GGA GCT GCT TCG |
| fhuA-red-R | TAG CAT GGC GCG TTC CAC TCA CAC TCA GAT CAA TAC CAG GAT TTG CAG ACT GGC GAT TCC GGG GAT CCG TCG ACC | |
| flgK | flgK-red-F | CGC ATG TTC TGC TGA TAC ATC ATT TGT GTA CTA ATA CGC ATC GTT CGG TTC CCT GTG TAG GCT GGA GCT GCT TCG |
| flgK-red-R | GCG CTG CCG ATA ATT ATC GTC AGG ACC CGC ATA TGA ATG TTC AAA AGG AAC CTC CAT TCC GGG GAT CCG TCG ACC |
Sequences of the priming sites in pKD13 are underlined.
Bacteriophage DNA isolation and purification.
Before isolation of the phage genomic DNA, the phage particles were treated with DNase I and RNase A at 37°C for 30 min to remove any bacterial DNA and RNA, respectively. The overall isolation and purification of the phage genomic DNA of CR5 were conducted according to Wilcox et al. (27) and Sambrook and Russell (28).
Bacteriophage genome sequencing and bioinformatics analysis.
The phage genomic DNA was sheared using a HydroShear DNA machine (Digilab, Holliston, MA, USA) and then sequenced using the Genome Sequencer FLX (GS-FLX) instrument (Roche, Mannheim, Germany) at Macrogen, Inc. (Seoul, South Korea). After pyrosequencing, the filtered sequence reads were assembled with Newbler version 2.3 (Roche). All of the open reading frames (ORFs) were predicted with the bacterial genetic code parameter using the Glimmer version 3.02 (29), GeneMarkS (30), and FgenesB software programs (Softberry, Inc., Mount Kisco, NY, USA), and their ribosomal binding sites were confirmed by RBSfinder (J. Craig Venter Institute, Rockville, MD, USA). The annotation and functional analysis of the predicted ORFs were conducted using the BLASTP (31) and InterProScan databases (32). Virulence factor analysis was performed using the Virulence Factor Database (http://www.mgc.ac.cn/VFs/). The complete genome sequence and ORF annotations were handled using Artemis version 14 (33).
Proteomic analysis of the phage structural proteins.
To analyze the total protein profiles of phage CR5 using SDS-PAGE, a purified phage stock (1011 PFU/ml) was suspended in loading buffer (0.05 M Tris-HCl [pH 8.0], 1.6% SDS, 25% glycerol, 5% 2-mercaptoethanol, 0.003% bromophenol blue, final concentrations). Next, this sample was treated in boiling water for 5 min, and the denatured phage proteins were subsequently separated using a 12% SDS-polyacrylamide gel. After SDS-PAGE, gels containing eight major visible bands were excised, destained with 50% acetonitrile containing 10 mM NH4HCO3, and dehydrated with 100% acetonitrile. The dried gels were rehydrated with 25 mM NH4HCO3 and digested with 400 mM trypsin overnight at 37°C. The digested peptides were extracted, dried, and redissolved in equilibration buffer (5% acetonitrile and 0.5% acetic acid). ZipTip C18 pipette tips (Millipore, Billerica, MA, USA) were used for desalting and cleanup of the samples. The matrix-assisted laser desorption ionization (MALDI) matrix was α-cyano-4-hydroxycinnamic acid (CHCA) in a mixture of 50% acetonitrile and 0.1% trifluoroacetic acid. Mass spectrometry was performed using the AB Sciex TOF/TOF 5800 system (Framingham, MA, USA) in the positive reflector mode at the Korea Basic Science Institute (KBSI) (Seoul, South Korea). The scan parameters in the MS mode were a mass range of 800 to 4,000 Da and a total number of laser shots of 400. After the MS scan, fragmentation of the selected precursors was performed at 1-kV collision energy with air. The metastable suppressor mode was selected. The output peptide sequences were searched against a database containing all protein sequences of phage CR5. The protein identification was processed with the ProteinPilot 4.0 software using the Paragon algorithm (AB Sciex). Next, the statistical cutoff values were used at a peptide confidence of 95%.
Food application.
Reconstituted infant formula milk was prepared according to the manufacturer's specifications. Approximately 33.6 g of premium goat infant formula stage 1 powder (Ildong Foodis, Seoul, South Korea) was resuspended in 240 ml of sterilized water, according to the manufacturer's instructions. The host strain (C. sakazakii ATCC 29544 as a clinical isolate, 31-3 as a food isolate, and their mixture at 102 CFU/ml final concentration) and phage CR5 at different MOI values (104 to 105) were added to 20 ml of the prepared infant formula milk. Bacterial cultures without the phage were used as controls. The mixture was incubated with shaking (200 rpm) at 37°C for up to 10 h. The number of viable cells was counted at 2-h intervals using the standard serial dilution and viable cell counting method. All of the tests were conducted in triplicate.
Nucleotide sequence accession number.
The complete genome sequence of C. sakazakii-infecting phage CR5 is available in the GenBank database under the accession no. JX094500.
RESULTS
Isolation of bacteriophage CR5.
Phage CR5 was isolated and purified from a soil sample from a cow farm in Suwon, South Korea, having lytic activity against C. sakazakii ATCC 29544. The TEM analysis of phage CR5 revealed that it belongs to the Myoviridae family (Fig. 1). The diameter of the isomeric head and the length of the tail were approximately 98 nm and 200 nm, respectively.
FIG 1.

TEM image of phage CR5 belonging to the family Myoviridae. The black triangles and the white triangle indicate the contracted and noncontracted tails, respectively. The phages were negatively stained with 2% (wt/vol) uranyl acetate and observed using TEM JEM-2100 (JEOL, Tokyo, Japan) at 200 kV.
Host range analysis and bacterial challenge test.
The host range analysis of phage CR5 with C. sakazakii type strains, C. sakazakii isolates from the ingredients of infant foods, and other Cronobacter species type strains revealed that this phage can infect or inhibit the growth of C. sakazakii and a few other Cronobacter species type strains, indicating its high specificity for Cronobacter infections (Table 1). Interestingly, while Cronobacter and Enterobacter have a close taxonomic relationship, E. cloacae ATCC 7256 and E. agglomerans ATCC 27987 showed very weak turbid growth inhibition zones, indicating that phage CR5 does not infect them (Table 1). The host lysis activity of phage CR5 was determined using the bacterial challenge method. As soon as phage CR5 (MOI, 1) was added to the culture of C. sakazakii ATCC 29544 at an OD600 of 0.5, the host strain was efficiently lysed, and the host growth inhibition activity was sustained, even after 10 h (Fig. 2), indicating that the duration of host growth inhibition activity of phage CR5 is longer than that of other C. sakazakii phages that have been reported (20).
FIG 2.
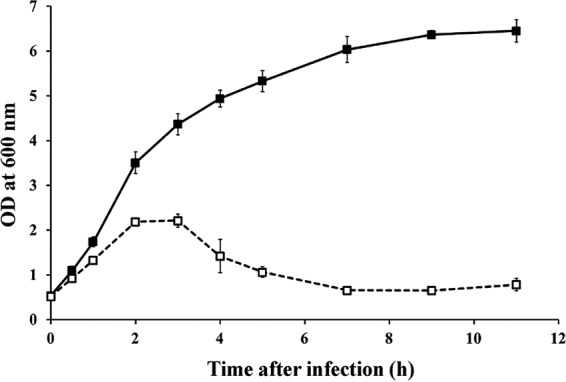
Bacterial challenge test of phage CR5 with C. sakazakii ATCC 29544 (MOI, 1). The closed squares indicate non-phage-treated C. sakazakii ATCC 29544, and the open squares indicate phage CR5-treated C. sakazakii ATCC 29544. The error bars indicate standard deviations from the results of triplicate experiments.
Identification of the host receptor.
As mentioned previously, phage CR5 infects C. sakazakii ATCC 29544 and lyses the host cells (Fig. 3A). To determine the host receptor of phage CR5, five well-known host receptor genes, which encode the O antigen of lipopolysaccharides (LPS), LamB (maltose transporter), OmpC (outer membrane protein), FhuA (ferric ion transporter), and FlgK of flagella (flagellar hook-filament junction protein), were specifically deleted to find the specific C. sakazakii ATCC 29544 host receptor of phage CR5 (data not shown), and only the ΔflgK mutant showed resistance to phage CR5 (Fig. 3B), suggesting that the specific host receptor of phage CR5 is the flagellum of C. sakazakii ATCC 29544. To confirm this host receptor, a subsequent complementation experiment using the expression of the flgK gene was conducted. The flgK gene was cloned into the pBAD18 expression vector and expressed in the ΔflgK mutant, and the results showed the recovery of the sensitivity to phage CR5, thereby substantiating the finding that the host flagellum is the host receptor of phage CR5 (Fig. 3C). Flagellum formation or motility has been known to be associated with the virulence of C. sakazakii (34), and most phage-resistant bacteria using flagella as a receptor tend to lose their motility (35), suggesting that C. sakazakii that is resistant to the flagella targeting CR5 is expected to be avirulent. Therefore, CR5 could be a good candidate for the control of pathogenic C. sakazakii.
FIG 3.
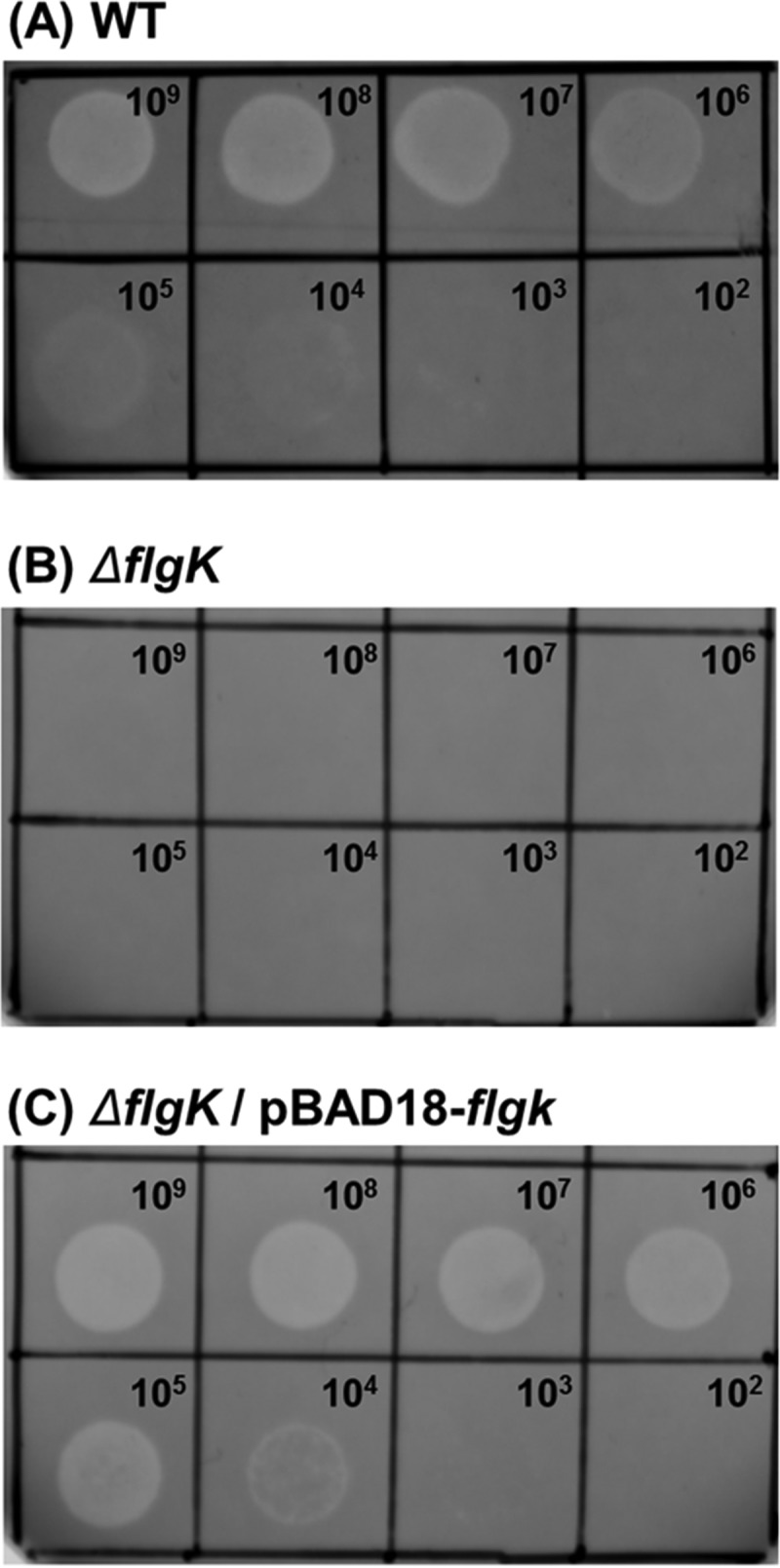
Identification of the C. sakazakii host receptor by deletion and complementation of the flgK gene. (A) Wild-type (WT) C. sakazakii ATCC 29544. (B) C. sakazakii ATCC 29544 ΔflgK mutant. (C) C. sakazakii ATCC 29544 ΔflgK mutant containing pBAD18::flgK for complementation. Tenfold serially diluted phage CR5 samples were dotted from 102 to 109 PFU/ml in each chamber.
Genome sequence analysis.
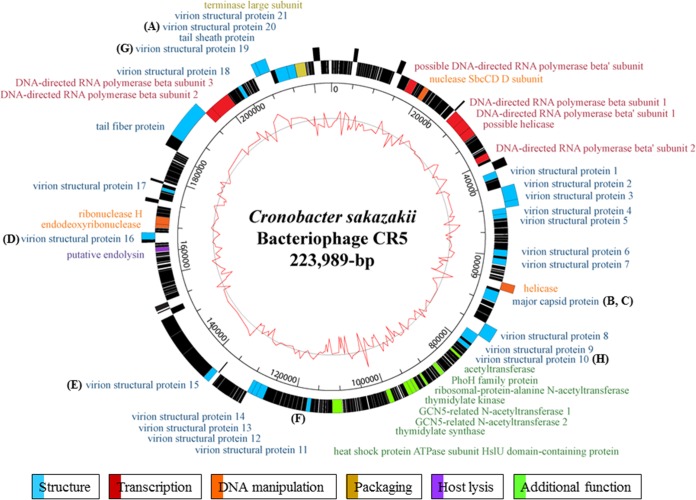
The genome of bacteriophage CR5 contains 223,989 bp of DNA, with a G+C content of 50.06%, 231 predicted ORFs, and no tRNA genes (Fig. 4). The average gene length is 912 bp, and the gene coding percentage is 94.1%. The functional ORFs were classified into six groups: structure (major capsid protein, tail fiber protein, tail sheath protein, and many virion structural proteins), packaging (terminase large subunit), host lysis (endolysin), DNA manipulation (nuclease SbcCD subunit D, helicases, endodeoxyribonuclease, and RNase H), transcription (RNA polymerase β- and β′-subunits), and additional functions (N-acetyltransferases, PhoH-like protein, thymidylate synthase, thymidylate kinase, and heat shock protein). However, >80% of the products of predicted ORFs in this genome (185 of the 231 ORFs) remain hypothetical proteins, likely due to insufficient annotation data of C. sakazakii bacteriophage genomes in the genome databases.
FIG 4.
Genome map of phage CR5. The inner circle with the red line indicates the G+C content. The outer circle indicates the predicted ORFs by strand. The functional categories and annotation of the ORFs are indicated by specific colors, according to the legend. Black ORFs indicate hypothetical proteins. Letters A to H indicate the major structural proteins. The scale units are base pairs.
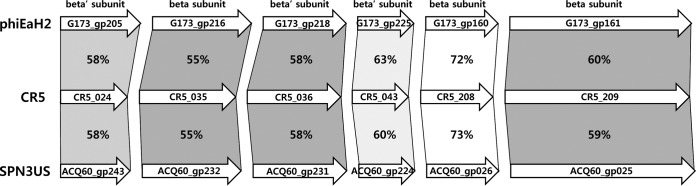
Because the flagella were identified in this study as the host receptors of phage CR5, it is likely that the tail fiber protein targets these host flagella for phage infection. This genome has only one gene encoding a tail fiber protein, and it is likely associated with the host specificity of phage CR5. Although the host lysis activity of phage CR5 is strong and consistent, this genome has only one host lysis-related gene encoding endolysin, without any genes encoding holin and Rz/Rz1, which is different from other Gram-negative bacterium-infecting phages. The thymidylate synthase and thymidylate kinase of the phage may help the folate metabolism of the host as an additional function for the host strain. Interestingly, the phage genome has six phage RNA polymerase β/β′-subunits, and they did not share protein domains with each other. Each predicted phage RNA polymerase of CR5 showed high amino acid sequence identities (55 to 73%) with previously reported RNA polymerase β/β′-subunits of other two phages, Salmonella phage SPN3US and Erwinia phage phiEaH2 (36, 37), suggesting that those phages may share a phage gene transcription mechanism (Fig. 5). However, the comparative sequence analysis of RNA polymerase β/β′-subunits between phage CR5 and the host strain C. sakazakii ATCC 29544 revealed that the amino acid sequences of RNA polymerase β-subunits are quite different (data not shown), suggesting that they may have different gene transcription mechanisms. Furthermore, the presence of multiple copies of RNA polymerase β/β′-subunits in phage CR5 suggests that phage gene transcription may be dominant, rather than host gene transcription with a single copy of the host RNA polymerase β/β′-subunit predominating (36). However, the functions of the phage RNA polymerase β/β′-subunits remain to be elucidated.
FIG 5.
Comparative sequence analysis of RNA polymerase β/β′-subunits in the CR5 phage with those of Erwinia phage phiEaH2 and Salmonella phage SPN3US. The protein sequence identities are indicated between the arrows. The locus tag of each gene is indicated in the middle of each arrow.
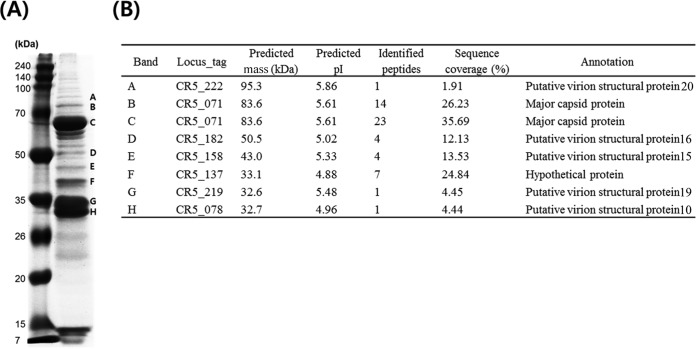
Proteomic analysis of the phage structural proteins.
To confirm the expression and identification of the major phage structural proteins, a proteomic analysis using SDS-PAGE and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) was performed. The comparative analysis of these partial peptide sequences with the genome sequence analysis results allowed for the identification of these proteins (Fig. 6). Interestingly, eight protein bands were identified: two as a putative major capsid protein (CR5_071, bands B and C), five as putative virion structural proteins (CR5_078, band H; CR5_158, band E; CR5_182, band D; CR5_219, band G; and CR5_222, band A), and one as a hypothetical protein (CR5_137, band F). Interestingly, one major band (band C, ∼68 kDa) and one minor band (band B, ∼83 kDa) were detected and identified as a putative major capsid protein using protein sequence analysis (Fig. 6). The protein sequence analysis predicted the protein size of the putative major capsid protein to be 83.6 kDa (Fig. 6B), indicating that the minor band may be the procapsid protein before maturation. Previously, it was reported that the cleavage of the procapsid protein by a proteolysis enzyme is essential for maturation of the major capsid protein (38). Therefore, overall proteomic experiments of phage CR5 may be required to extend our understanding of proteomic characteristics and the maturation process of these structural phage proteins.
FIG 6.
Proteomic analysis of the structural proteins of phage CR5 using SDS-PAGE (A) and MALDI-TOF MS (B). In the SDS-PAGE, eight major bands were picked and labeled A to H. In a comparison with the genome annotation information of phage CR5, these major bands were identified with molecular weights and partial peptide sequences using MALDI-TOF MS. In addition, letters A to H are also added to the CR5 genome map to indicate the locations of major structural proteins (see Fig. 4).
Food application.
To verify the potential of using phage CR5 as a novel biocontrol agent against C. sakazakii in food samples, one or two C. sakazakii strains, ATCC 29544 and/or food isolate 31-3, were added to an infant formula milk sample containing phage CR5 at an MOI of 104 or 105, and the viable cell numbers of C. sakazakii in the sample were monitored. After phage CR5 at an MOI of 104 was added to the sample containing the clinical isolate ATCC 29544, the bacterial strain was lysed in 2 h but recovered. However, after addition at an MOI of 105, it was also lysed in 2 h and never recovered up to 10 h, suggesting that the addition of phage CR5 at an MOI of at least 105 is required to completely control the C. sakazakii ATCC 29544 strain in the sample (Fig. 7A). Furthermore, after phage CR5 at an MOI of 104 or 105 was added to the sample containing the food isolate 31-3, the bacterial strain was lysed and never recovered, even though the lysis times of CR5 at an MOI of 104 and 105 were 6 h and 2 h, respectively (Fig. 7B).
FIG 7.
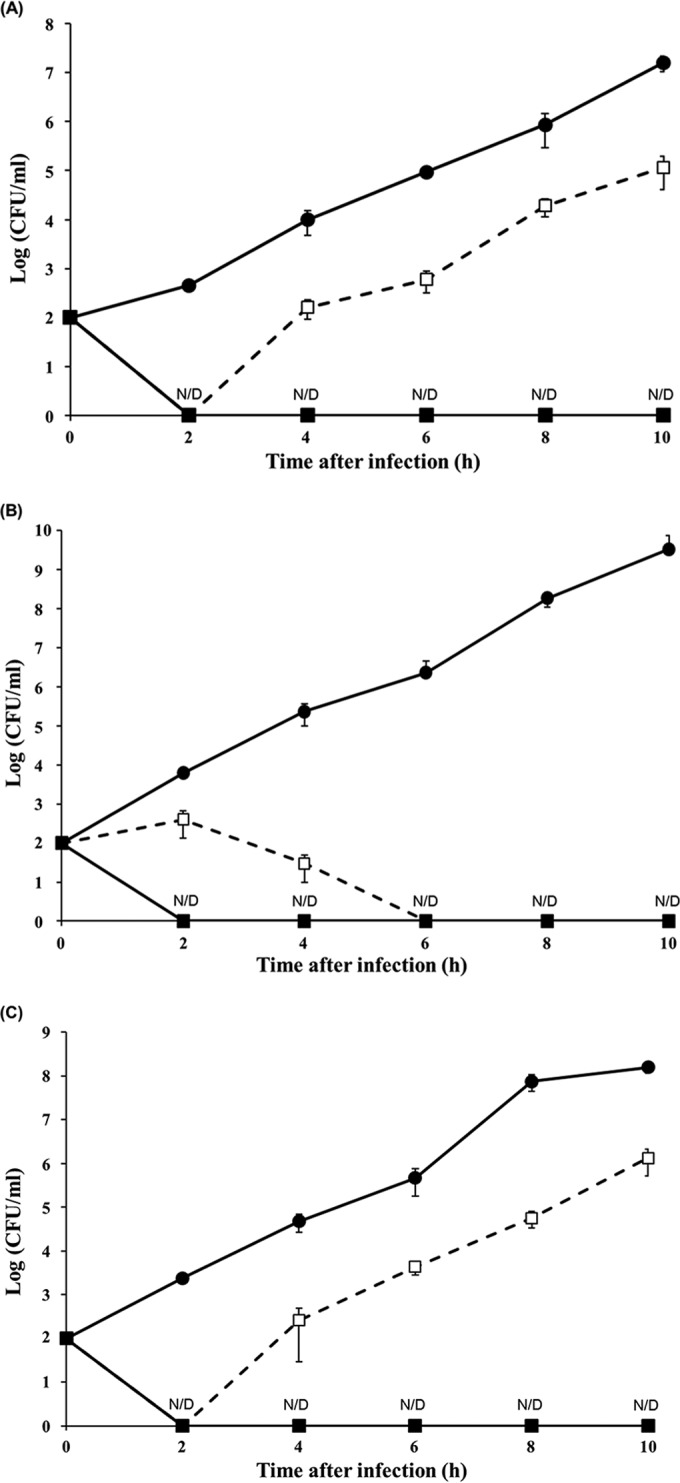
Food application of phage CR5 against C. sakazakii ATCC 29544 (A), food isolate 31-3 (B), and their mixture (C) in infant formula milk. The closed circles indicate the non-phage-treated samples used as negative controls. The open squares with dashed lines indicate phage CR5-treated samples with an MOI of 104, and the closed squares indicate phage CR5-treated samples with an MOI of 105. N/D, not detected.
In addition, a mixed culture of the clinical strain and food isolate was tested with phage CR5.
After phage CR5 at an MOI of 104 was added to the mixed culture, the growth curve of the host strains was very similar to that of a clinical strain, probably due to the presence of the clinical strain in the mixed culture. The host strains were lysed in 2 h but regrew (Fig. 7C). However, when phage CR5 at an MOI of 105 was added, the bacterial strains in the mixed culture were lysed in 2 h and never regrew up to 10 h (Fig. 7C), which is very similar to results with both the clinical strain and food isolate. Therefore, the mixed culture has characteristics of both strains for the control of C. sakazakii using phage CR5. These results suggest that the MOI of CR5 for complete control of both clinical and food isolates is ≥105, which is much lower than that of previously reported C. sakazakii phages (107 to 109) (20). Therefore, the high host lysis activity of phage CR5 against both clinical and food isolates of C. sakazakii in infant formula milk implies that it is a possible candidate for the development of a novel biocontrol agent or natural food preservative against C. sakazakii.
DISCUSSION
C. sakazakii is a fatal food-borne pathogen causing high mortality rates and is generally found in infant milk formula powders (3). Therefore, C. sakazakii urgently needs to be controlled in foods. However, a biocontrol agent or a natural food preservative to control this pathogen is not yet available, because antibiotic usage is not allowed in foods. The bacteriophage approach has been revisited to control various pathogens because of its bacterial host specificity, bactericidal activity, and human safety (39).
In this study, we isolated and characterized a novel C. sakazakii-infecting phage, CR5, with host lysis activity. In addition, phage CR5 maintained growth inhibition activity against C. sakazakii ATCC 29544 for up to >10 h (Fig. 2). The host growth inhibition activity of phage CR5 is longer than that of other C. sakazakii phages that have been reported (20). The duration of this host growth inhibition activity of phage CR5 in the bacterial challenge assay suggests that phage CR5 may be a candidate for a biocontrol agent against C. sakazakii. In addition, the host receptor of this C. sakazakii phage was identified through specific gene knockout studies and subsequent complementation experiments to understand the host-phage interaction for phage infection. To date, one receptor identification experiment has been conducted, and it confirmed that only one C. sakazakii phage, CR3, infects the host strains using flagella as the host receptors (13).
Interestingly, the genome study of phage CR5 revealed a few new molecular characteristics. A comparative genome analysis of CR5 with 17 other C. sakazakii phages showed extremely low sequence identities, suggesting that they may not share genomic characteristics (data not shown). The absence of endolysin-supporting proteins (e.g., holin and Rz/Rz1 proteins) in this genome suggests that the single endolysin may lyse the host cells with full function. To make sure there is no holin gene in the CR5 genome, the GenBank annotation data of all available 17 C. sakazakii genomes were collected and checked to see if there is any holin gene in those genomes. Only two genomes (those of phages ENT47670 and vB_CsaM_GAP161) have holin genes, implying that the presence of a holin gene may not be common in C. sakazakii phage genomes. A DNA sequence alignment analysis of these holin genes with the CR5 complete genome sequence showed that there is no DNA sequence match in the genome, indicating that there may be no holin genes in the CR5 genome. A subsequent protein domain analysis of these two holin proteins using the InterProScan program (32) showed that they have conserved protein domains (PF16083, LydA phage holin, holin superfamily III for holin protein from the phage ENT47670; PF11031, bacteriophage T holin for holin protein from the phage vB_CsaM_GAP161). Additional InterProScan analysis of all 231 ORFs of the CR5 phage genome revealed that there is no gene containing these conserved protein domains, suggesting that there is no holin gene in the CR5 genome. However, it is still possible that holin or holin-like genes in C. sakazakii phage genomes will be detected when more C. sakazakii phage genome annotation results are available in the sequence databases. Interestingly, additional SignalP analysis of the endolysin proteins in four C. sakazakii phage genomes without holin genes (CR5, ESP2949-1, phiES15, and vB_CskP_GAP227) revealed that they have signal peptides in the N terminus for secretion without holin, supporting this (data not shown). In addition, the phage CR5 genome has six copies of RNA polymerase β/β′-subunits. The presence of multiple copies of RNA polymerase β/β′-subunits in the genome and the lack of similarity of RNA polymerase β/β′-subunits between the phage and host genomes suggest that the phage has different gene transcription mechanisms, and phage gene transcription may be dominant rather than gene transcription of the host. To further understand the functions of multiple copies of RNA polymerase β/β′-subunits in the genome of phage CR5, three different Pseudomonas phage genomes, including those of 201phi2-1, phiKZ, and phiPA3 (40–42), were analyzed and compared. Pfam protein domain analysis of genes encoding RNA polymerase β/β′-subunits in two phages, 201phi2-1 and phiPA3, revealed that a gene encoding the RNA polymerase β′-subunit in each phage genome (gp275 of 201phi2-1 and ORF54 of phiPA3) has a highly conserved protein sequence motif, Rbp1_domain_2 (NADFDGD), associated with Mg2+ binding (40, 42), while no Rbp1_domain_2 motif was detected in the phage phiKZ. Three aspartic acid (D) residues in the motif (in bold) have been suggested to be necessary for holding Mg2+ in the active center of the RNA polymerase β′-subunit. These Mg2+ ions in the active center were reported to attract the negatively charged phosphate of nucleoside triphosphates (NTPs) and to allow it to interact with the 3′-OH end of the RNA transcript for RNA elongation (43). Interestingly, the gene encoding the RNA polymerase β′-subunit (CR5_024 of CR5) also contains this Rbp1_domain_2 motif in its protein sequence, suggesting that this RNA polymerase β′-subunit may be associated with RNA elongation by Mg2+ binding in the active center. Therefore, the RNA polymerase β/β′-subunits of phage CR5 may play roles similar to those of two Pseudomonas phages in RNA transcription and elongation.
To evaluate the lysis activity of phage CR5 against C. sakazakii by a bacterial challenge assay for food application, it was added to infant formula milk containing C. sakazakii clinical or/and food isolates (ATCC 29544 and/or food isolate 31-3). Surprisingly, phage CR5 controlled both isolates completely up to 10 h at an MOI of only 105, which is much lower than that of previously reported C. sakazakii phages (20); this suggests that phage CR5 is a good candidate for a biocontrol agent against both clinical and food isolates in infant formula milk. However, these food applications using CR5 phage represent a model experiment under specific food conditions to control C. sakazakii in infant formula. For real industrial applications, a cheap phage purification and concentration method and an optimized food application method should be developed. In addition, although the U.S. FDA approved the phage application in foods as a natural food preservative or biocontrol agent, customers may not prefer using phage in some sensitive foods, including milk formula. Therefore, food applications to control food-borne pathogens using phage need to be considered carefully. This study provides insights into the characteristics and genome of phage CR5 for further development of a novel phage biocontrol agent against C. sakazakii in foods.
Funding Statement
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Republic of Korea government (MSIP) (no. NRF-2014R1A2A1A10051563).
REFERENCES
- 1.Farmer JJ III, Asbury MA, Hickman FW, Brenner DJ, the Enterobacteriaceae Study Group. 1980. Enterobacter sakazakii: a new species of “Enterobacteriaceae” isolated from clinical specimens. Int J Syst Bacteriol 30:569–584. doi: 10.1099/00207713-30-3-569. [DOI] [Google Scholar]
- 2.Iversen C, Lehner A, Mullane N, Bidlas E, Cleenwerck I, Marugg J, Fanning S, Stephan R, Joosten H. 2007. The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov. and descriptions of Cronobacter sakazakii comb. nov. Cronobacter sakazakii subsp. sakazakii, comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies 1. BMC Evol Biol 7:64. doi: 10.1186/1471-2148-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drudy D, Mullane NR, Quinn T, Wall PG, Fanning S. 2006. Enterobacter sakazakii: an emerging pathogen in powdered infant formula. Clin Infect Dis 42:996–1002. doi: 10.1086/501019. [DOI] [PubMed] [Google Scholar]
- 4.Nazarowec-White M, Farber JM. 1997. Enterobacter sakazakii: a review. Int J Food Microbiol 34:103–113. doi: 10.1016/S0168-1605(96)01172-5. [DOI] [PubMed] [Google Scholar]
- 5.Tsai H, Liao C, Huang Y, Lee P, Ren HP. 2013. Cronobacter infections not from infant formula, Taiwan. Emerg Infect Dis 19:167–169. doi: 10.3201/eid1901.120774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai KK. 2001. Enterobacter sakazakii infections among neonates, infants, children, and adults: case reports and a review of the literature. Medicine (Baltimore) 80:113–122. doi: 10.1097/00005792-200103000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Calendar R. 2006. The bacteriophages, 2nd ed Oxford University Press, New York, NY. [Google Scholar]
- 8.Sulakvelidze A, Alavidze Z, Morris JG Jr. 2001. Bacteriophage therapy. Antimicrob Agents Chemother 45:649–659. doi: 10.1128/AAC.45.3.649-659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y-D, Chang H-I, Park J-H. 2011. Complete genomic sequence of virulent Cronobacter sakazakii phage ESSI-2 isolated from swine feces. Arch Virol 156:721–724. doi: 10.1007/s00705-011-0934-y. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y-D, Park J-H, Chang H-I. 2011. Genomic sequence analysis of virulent Cronobacter sakazakii bacteriophage ES2. Arch Virol 156:2105–2108. doi: 10.1007/s00705-011-1096-7. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y-D, Kim J-Y, Park J-H, Chang H. 2012. Genomic analysis of bacteriophage ESP2949-1, which is virulent for Cronobacter sakazakii.. Arch Virol 157:199–202. doi: 10.1007/s00705-011-1147-0. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y-D, Park J-H. 2012. Complete genome of temperate phage ENT39118 from Cronobacter sakazakii.. J Virol 86:5400–5401. doi: 10.1128/JVI.00345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin H, Lee J-H, Kim Y, Ryu S. 2012. Complete genome sequence of Cronobacter sakazakii bacteriophage CR3. J Virol 86:6367–6368. doi: 10.1128/JVI.00636-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J-H, Choi Y, Shin H, Lee J, Ryu S. 2012. Complete genome sequence of Cronobacter sakazakii temperate bacteriophage phiES15. J Virol 86:7713–7714. doi: 10.1128/JVI.01042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbasifar R, Kropinski AM, Sabour PM, Ackermann H-W, Lingohr EJ, Griffiths MW. 2012. Complete genome sequence of Cronobacter sakazakii bacteriophage vB_CsaM_GAP161. J Virol 86:13806–13807. doi: 10.1128/JVI.02546-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abbasifar R, Kropinski AM, Sabour PM, Ackermann H-W, Alanis Villa A, Abbasifar A, Griffiths MW. 2012. Genome sequence of Cronobacter sakazakii myovirus vB_CsaM_GAP31. J Virol 86:13830–13831. doi: 10.1128/JVI.02629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abbasifar R, Kropinski AM, Sabour PM, Ackermann H-W, Alanis Villa A, Abbasifar A, Griffiths MW. 2013. The genome of Cronobacter sakazakii bacteriophage vB_CsaP_GAP227 suggests a new genus within the Autographivirinae.. Genome Announc 1(1):e00122-12. doi: 10.1128/genomeA.00122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Endersen L, Guinane CM, Johnston C, Neve H, Coffey A, Ross RP, McAuliffe O, O'Mahony J. 2015. Genome analysis of Cronobacter phage vB_CsaP_Ss1 reveals an endolysin with potential for biocontrol of Gram-negative bacterial pathogens. J Gen Virol 96:463–477. doi: 10.1099/vir.0.068494-0. [DOI] [PubMed] [Google Scholar]
- 19.Abbasifar R, Griffiths MW, Sabour PM, Ackermann HW, Vandersteegen K, Lavigne R, Noben JP, Alanis Villa A, Abbasifar A, Nash JH, Kropinski AM. 2014. Supersize me: Cronobacter sakazakii phage GAP32. Virology 460–461:138–146. doi: 10.1016/j.virol.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Kim KP, Klumpp J, Loessner MJ. 2007. Enterobacter sakazakii bacteriophages can prevent bacterial growth in reconstituted infant formula. Int J Food Microbiol 115:195–203. doi: 10.1016/j.ijfoodmicro.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 21.Zuber S, Boissin-Delaporte C, Michot L, Iversen C, Diep B, Brussow H, Breeuwer P. 2008. Decreasing Enterobacter sakazakii (Cronobacter spp.) food contamination level with bacteriophages: prospects and problems. Microbiol Biotechnol 1:532–543. doi: 10.1111/j.1751-7915.2008.00058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tóthová L, Celec P, Bábíčková J, Gajdošová J, Al-Alami H, Kamodyova N, Drahovská H, Liptáková A, Turňa J, Hodosy J. 2011. Phage therapy of Cronobacter-induced urinary tract infection in mice. Med Sci Monit 17:BR173–BR178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M, Ryu S. 2011. Characterization of a T5-like coliphage SPC35 and differential development of resistance to SPC35 in Salmonella Typhimurium and Escherichia coli.. Appl Environ Microbiol 77:2042–2050. doi: 10.1128/AEM.02504-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fauquet C, Mayo MA, Maniloff J, Desselberger U, Ball LA (ed). 2005. Virus taxonomy: eighth report of the International Committee on the Taxonomy of Viruses, 1st ed Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 25.Kim K, Kim K-P, Choi J, Lim J-A, Lee J, Hwang S, Ryu S. 2010. Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii.. Appl Environ Microbiol 76:5188–5198. doi: 10.1128/AEM.02498-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilcox SA, Toder R, Foster JW. 1996. Rapid isolation of recombinant lambda phage DNA for use in fluorescence in situ hybridization. Chromosome Res 4:397–398. doi: 10.1007/BF02257276. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 29.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Zdobnov EM, Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 33.Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, Parkhill J, Rajandream M-A. 2008. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartmann I, Carranza P, Lehner A, Stephan R, Eberl L, Riedel K. 2010. Genes involved in Cronobacter sakazakii biofilm formation. Appl Environ Microbiol 76:2251–2261. doi: 10.1128/AEM.00930-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans TJ, Trauner A, Komitopoulou E, Salmond GP. 2010. Exploitation of a new flagellatropic phage of Erwinia for positive selection of bacterial mutants attenuated in plant virulence: towards phage therapy. J Appl Microbiol 108:676–685. doi: 10.1111/j.1365-2672.2009.04462.x. [DOI] [PubMed] [Google Scholar]
- 36.Lee JH, Shin H, Kim H, Ryu S. 2011. Complete genome sequence of Salmonella bacteriophage SPN3US. J Virol 85:13470–13471. doi: 10.1128/JVI.06344-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dömötör D, Becságh P, Rákhely G, Schneider G, Kovács T. 2012. Complete genomic sequence of Erwinia amylovora phage PhiEaH2. J Virol 86:10899. doi: 10.1128/JVI.01870-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. 1995. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J Mol Biol 253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- 39.Hagens S, Loessner MJ. 2007. Application of bacteriophages for detection and control of foodborne pathogens. Appl Microbiol Biotechnol 76:513–519. doi: 10.1007/s00253-007-1031-8. [DOI] [PubMed] [Google Scholar]
- 40.Thomas JA, Rolando MR, Carroll CA, Shen PS, Belnap DM, Weintraub ST, Serwer P, Hardies SC. 2008. Characterization of Pseudomonas chlororaphis myovirus 201varphi2-1 via genomic sequencing, mass spectrometry, and electron microscopy. Virology 376:330–338. doi: 10.1016/j.virol.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mesyanzhinov VV, Robben J, Grymonprez B, Kostyuchenko VA, Bourkaltseva MV, Sykilinda NN, Krylov VN, Volckaert G. 2002. The genome of bacteriophage phiKZ of Pseudomonas aeruginosa.. J Mol Biol 317:1–19. doi: 10.1006/jmbi.2001.5396. [DOI] [PubMed] [Google Scholar]
- 42.Monson R, Foulds I, Foweraker J, Welch M, Salmond GP. 2011. The Pseudomonas aeruginosa generalized transducing phage phiPA3 is a new member of the phiKZ-like group of ‘jumbo’ phages, and infects model laboratory strains and clinical isolates from cystic fibrosis patients. Microbiology 157:859–867. doi: 10.1099/mic.0.044701-0. [DOI] [PubMed] [Google Scholar]
- 43.Zaychikov E, Martin E, Denissova L, Kozlov M, Markovtsov V, Kashlev M, Heumann H, Nikiforov V, Goldfarb A, Mustaev A. 1996. Mapping of catalytic residues in the RNA polymerase active center. Science 273:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]
- 44.Kucerova E, Clifton SW, Xia XQ, Long F, Porwollik S, Fulton L, Fronick C, Minx P, Kyung K, Warren W, Fulton R, Feng D, Wollam A, Shah N, Bhonagiri V, Nash WE, Hallsworth-Pepin K, Wilson RK, McClelland M, Forsythe SJ. 2010. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS One 5:e9556. doi: 10.1371/journal.pone.0009556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim K, Jang SS, Kim SK, Park JH, Heu S, Ryu S. 2008. Prevalence and genetic diversity of Enterobacter sakazakii in ingredients of infant foods. Int J Food Microbiol 122:196–203. doi: 10.1016/j.ijfoodmicro.2007.11.072. [DOI] [PubMed] [Google Scholar]