Abstract
Pseudomonas veronii 1YdBTEX2, a benzene and toluene degrader, and Pseudomonas veronii 1YB2, a benzene degrader, have previously been shown to be key players in a benzene-contaminated site. These strains harbor unique catabolic pathways for the degradation of benzene comprising a gene cluster encoding an isopropylbenzene dioxygenase where genes encoding downstream enzymes were interrupted by stop codons. Extradiol dioxygenases were recruited from gene clusters comprising genes encoding a 2-hydroxymuconic semialdehyde dehydrogenase necessary for benzene degradation but typically absent from isopropylbenzene dioxygenase-encoding gene clusters. The benzene dihydrodiol dehydrogenase-encoding gene was not clustered with any other aromatic degradation genes, and the encoded protein was only distantly related to dehydrogenases of aromatic degradation pathways. The involvement of the different gene clusters in the degradation pathways was suggested by real-time quantitative reverse transcription PCR.
INTRODUCTION
Members of the Pseudomonas genus are highly versatile and known to colonize diverse habitats such as humans, animals, plants, water, and soil (1), and Pseudomonas veronii strains have been implicated in human pathogenicity (2), heavy metal biosorption (3), and dibenzo-p-dioxin, chloroaromatic, and alkyl methyl ketone degradation (4, 5). P. veronii strains were also pointed out as key catabolic players in a BTEX (benzene, toluene, ethylbenzene, xylenes)-contaminated area where benzene had been the major contaminant (6). Two strains, termed P. veronii 1YB2 and P. veronii 1YdBTEX2, were subjected to a detailed analysis, as they contained catabolism gene variants shown to be highly abundant at the site (6). Interestingly, both strains differed in their substrate specificities, and whereas P. veronii 1YdBTEX2 grows on benzene and toluene, strain 1YB2 grows on benzene only. The difference in substrate specificity was due to small sequence differences in the α-subunit of the initial multicomponent Rieske nonheme iron oxygenase responsible for the activation of the aromatic ring (7).
Even though benzene is the most simple aromatic compound, the biodegradation pathways used by bacteria to degrade this compound have been documented in detail in only a few cases (8). Typically, the degradation of benzene is supposed to occur in analogy to the pathways observed for toluene degradation and to proceed via either initial dioxygenation (9) or two subsequent initial monooxygenations (10). In the case of initial dioxygenation, enzymes involved in the degradation of benzene and toluene are expected to show the highest similarities to enzymes termed benzene/toluene dioxygenases (11, 12). However, P. veronii strains 1YdBTEX2 and1YB2 harbor dioxygenases most closely related to isopropylbenzene dioxygenases (7). In the well-characterized isopropylbenzene-degradative pathways of Pseudomonas putida RE204 and Pseudomonas fluorescens IP01 (13, 14), the genes coding for the oxygenase subunits are organized in operons together with genes encoding a cis-dihydrodiol dehydrogenase and an extradiol dioxygenase of subfamily I.3.A (15). However, P. veronii 1YB2 and 1YdBTEX2 do not express a subfamily I.3.A extradiol dioxygenase (EXDO K2) during growth on benzene but recruit a subfamily I.2.A extradiol dioxygenase (catechol 2,3-dioxygenase [C23O]) (EXDO A) with high similarity to the archetype TOL plasmid-encoded enzyme (6, 16). To our knowledge, such a recruitment has not been reported previously for the degradation of benzene or toluene via a dioxygenolytic route. Since P. veronii 1YdBTEX2 and 1YB2 were indicated to be key catabolic players with unique pathway arrangements, we sequenced the genomes of both strains to elucidate the organization of the benzene and/or toluene catabolic pathways.
MATERIALS AND METHODS
Strains and culture conditions.
P. veronii 1YB2 and P. veronii 1YdBTEX2 were cultured in mineral medium (17) containing 50 mM phosphate buffer (pH 7.4) supplemented with trace elements and growth substrates at concentrations of 2 mM. Benzene or toluene was supplied via the gas phase (2 to 5 mM each) (16), and cultures were incubated at room temperature without agitation.
Genome sequencing and assembly.
Genomes were assembled as previously described (18). Briefly, both genomes were sequenced on an Illumina GAII genome analyzer generating 110-bp paired ends. Approximately 10 million short sequence reads were obtained for each genome. Whole-genome shotgun sequencing data were assembled by using Velvet (19) and Edena (20). Both data sets were then merged by using Minimus (21). In order to further diminish the number of gaps, scaffolds were constructed by using SSPACE (22), and comparisons between contigs and scaffolds were made by using MAUVE (23). Contigs were also compared to the P. fluorescens SBW25 genome (24). These strategies resulted in a possible assembly for a number of contigs. In order to confirm this information and to close gaps between contigs, primers framing these gaps were designed, and PCR products were sequenced by Sanger sequencing.
Phylogenetic comparisons.
Nucleotide sequences of 16S rRNA genes and the DNA gyrase subunit B gene (gyrB) were aligned by using MUSCLE 3.7 (25). Phylogenetic trees were constructed with MEGA5 (26) by using the neighbor-joining (N-J) algorithm (27) with Jukes-Cantor correction and pairwise deletion of gaps and missing data. A total of 100 bootstrap replications were done to test for branch robustness.
The complete set of protein and coding sequences from P. veronii 1YdBTEX2 and 1YB2; P. fluorescens SBW25, A506, Pf0-1, and F113; P. protegens CHA0 and Pf-5; P. brassicacearum NFM421; and P. aeruginosa PAO1 were obtained from the NCBI ftp website (http://www.ncbi.nlm.nih.gov/Ftp). Comparisons between pairs of strains were performed independently at the protein and coding sequence levels through an in-house Perl script detecting the reciprocal best hit (RBH). For every RBH pair (proteins and coding sequences), the identity score was calculated considering the percent identity and the percent coverage of the query (alignment length × % identity/query length) (28, 29). If a sequence reported no RBH, the identity was scored as zero. Two data matrices containing the identity score for all protein or coding sequence comparisons among the 10 genomes were then generated (59,527 proteins). Similarity matrices were generated by using the Bray-Curtis coefficient (30), and dendrograms were created by group-average agglomerative hierarchical clustering. All statistical routines were carried out by using PRIMER 6 (v.6.1.6; Primer-E, Plymouth Marine Laboratory, United Kingdom) (31).
Partial purification and identification of benzene dihydrodiol dehydrogenase.
Cells of P. veronii 1YdBTEX2 and P. veronii 1YB2 (100 ml of culture) were harvested and disrupted with a French press (Aminco, Silver Spring, MD). Benzene dihydrodiol dehydrogenase was partially purified by using a fast protein liquid chromatography system (Amersham Biosciences, Freiburg, Germany). Cell extracts (1 ml) containing 1.4 to 2.0 mg of protein were applied directly to the column, and proteins were eluted with a linear gradient of 0 to 0.5 M NaCl over 33 ml at a flow rate of 0.5 ml/min. Fraction volumes were 0.5 ml. The activity of benzene dihydrodiol dehydrogenase was determined spectrophotometrically by measuring the 3,5-cyclohexadiene-1,2-diol (benzene dihydrodiol) (0.1 mM)-dependent reduction of NAD+ (0.1 mM) in Tris-HCl (100 mM) (pH 7.5) at 340 nm (εNADH = 6,220 M−1 · cm−1). Protein concentrations were determined by the Bradford method (32).
Aliquots of active fractions were subjected to SDS-PAGE on a Bio-Rad Miniprotean II instrument as described previously (33), and major protein bands with a molecular mass of ∼50 kDa obtained from P. veronii 1YdBTEX2 were analyzed by N-terminal amino acid sequencing performed with an Applied Biosystems model 494 A Procise HT sequencer, as previously described (34). Proteins from P. veronii 1YdBTEX2 and 1YB2 annotated by the RAST server (35) were searched for the presence of the determined N-terminal sequence.
Real-time quantitative PCR.
Primers annealing inside the genes of interest and targeting genes encoding the α-subunit of the isopropylbenzene dioxygenase (ipbAfw [GCTGGTTCAATGATGACTTCTC] and ipbArev [GATGAAAGACCAGACTTCGAC]), benzene dihydrodiol dehydrogenase (DHfw [GTGATTTTCTGCTCGAAGCTG] and DHrev [ATCCCAGGTCATCGCCGTAC]), EXDO A (ExdoA-1fw [GTTGACAAATTCTCCGTGGT] and ExdoA-1rev [TCTGCATACAACTCGAAGTGA]), and EXDO A2 (ExdoA-2fw [GCGTGTACTCGATATGGACA] and ExdoA-2rev [TGGGTCAGCTGATACAGACA]) were designed. After PCR, fragments were cloned into the pGEM-T Easy vector (Promega). Vectors with inserts were transformed into Escherichia coli JM109 cells, and positive clones were confirmed by PCR. The appropriate plasmids were purified by using the QIAprep Spin Miniprep kit (Qiagen, Germany).
For RNA preparation, strain 1YdBTEX2 was grown with benzene or toluene, strain 1YB2 was grown with benzene as the carbon source (inducing conditions), and both strains were grown on glucose (2 g/liter) as a control (noninducing conditions). RNA extractions were performed as previously described (36), and cDNA was prepared from 800 ng of RNA by using 0.3 μg of random primers and 400 U of Superscript III reverse transcriptase (RT) (Invitrogen, Life Technologies, Germany), according to the instructions of the providers.
The primers described above were used for the amplification of cDNA. Real-time PCRs were performed as previously described (37), using a LightCycler 480 real-time PCR system programmed for a hold at 95°C for 10 min and completion of 50 cycles of 94°C for 15 s, 56°C for 30 s, and 72°C for 30 s. Results were analyzed by using LightCycler 480 software (Roche Applied Science). Standard curves were generated, and a melting step was added to confirm amplification specificity.
Nucleotide sequence accession numbers.
The genome sequences of P. veronii 1YdBTEX2 and 1YB2 have been deposited in GenBank under accession numbers AOUH00000000 (18) and JGYI00000000, respectively.
RESULTS
General characteristics and phylogenetic analyses of P. veronii 1YdBTEX2 and 1YB2.
The Pseudomonas strain YdBTEX2 and 1YB2 genomes were sequenced, and ∼9.5 million and 3.2 million reads were assembled, giving 63 and 115 contigs, respectively (18). The genomes contained 5,981 and 6,884 candidate protein-encoding genes, respectively, where >5,100 of the encoded proteins are shared between the strains (nucleotide sequence identity of ≥80%) (see Table S1 in the supplemental material for comparison with finished genomes of strains of the P. fluorescens group).
Comparison of the 16S rRNA gene (Fig. 1A) and a 798-base sequence stretch of the DNA gyrase subunit B-encoding gene gyrB (Fig. 1B) with the respective genes of P. veronii CIP104663T, P. fluorescens IAM12022T, and other genome-sequenced P. fluorescens group organisms (24, 38–42) as well as comparison of the similarity of total predicted proteins (see Fig. S1 in the supplemental material) support the notion that both 1YB2 and 1YdBTEX2 belong to the P. veronii species.
FIG 1.
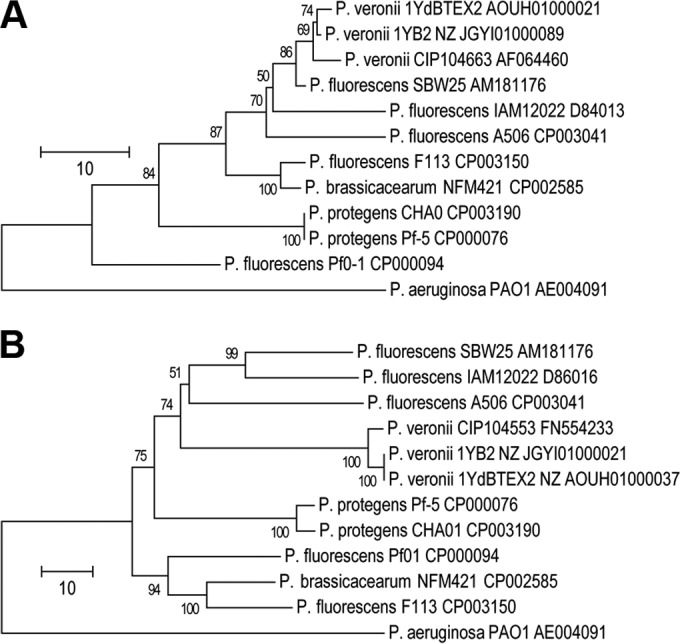
Phylogenetic relationships among Pseudomonas fluorescens group organisms. Phylogenetic trees are based on 1,513- to 1,515-base 16S rRNA gene fragments (A) and 798-base DNA gyrase subunit B gene (gyrB) fragments (B) of P. aeruginosa PAO1; P. veronii 1YdBTEX2, 1YB2, and CIP104663T; P. fluorescens A506, SBW25, F113, Pf0-1, and IAM12022T; P. brassicacearum NFM421; and P. protegens CHA01 and Pf-5. Phylogenetic trees were constructed with MEGA5 (26) using the N-J algorithm (27) with Jukes-Cantor correction and pairwise deletion of gaps and missing data. Bootstrap values of >50% from 100 neighbor-joining trees are indicated to the left of the nodes. The bar indicates nucleotide differences per site.
Genetic analysis of determinants of the benzene catabolic pathway.
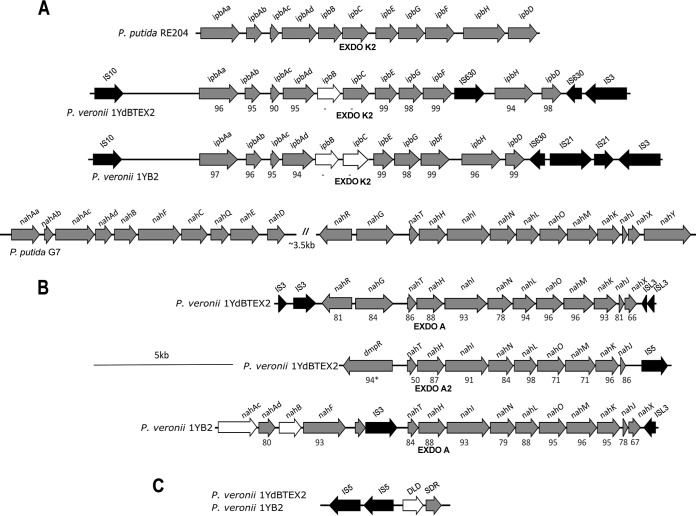
In previous studies, some of the isopropylbenzene dioxygenase-encoding genes (ipb) (7) and the genes encoding an EXDO A protein involved in the degradation of benzene by both strains (6, 16) were identified. Genome sequencing indicated the presence of a complete isopropylbenzene catabolism operon in both strains (GenBank accession numbers NZ_AOUH01000041 and NZ_JGYI01000039) (Fig. 2A), with a gene organization similar to the one found for plasmid pRE4 of P. putida RE204 (accession number AF006691) (14). The presence of putative transposases flanking the ipb operons in both 1YB2 and 1YdBTEX2 indicates that they were probably acquired via horizontal gene transfer. Curiously, the activity of the putatively encoded isopropylcatechol 2,3-dioxygenase (EXDO K2) has never been observed in any strain (16). In accordance with the absence of any activity, the extradiol dioxygenase-encoding gene of 1YB2 showed the presence of an internal stop codon probably resulting in a nonfunctional gene product. In 1YdBTEX2, the extradiol dioxygenase-encoding gene was obviously functional; however, sequence analysis revealed the presence of stop codons in both isopropylbenzene dihydrodiol dehydrogenase-encoding genes (ipbB genes) upstream of the extradiol dioxygenase-encoding gene, probably resulting in nonfunctional gene products in both strains.
FIG 2.
Organization of gene clusters involved in benzene degradation by P. veronii 1YdBTEX2 and 1YB2. Gene clusters of P. putida RE204 and P. putida G7 were used for comparison. (A) Isopropylbenzene gene clusters (ipb) of P. putida RE204 (14) (GenBank accession number AF006691; locus tags AAC03436 to AAC03446), P. veronii 1YdBTEX2 (accession number NZ_AOUH01000041; locus tags H736_RS0125995 to H736_RS0126080), and P. veronii 1YB2 (accession number NZ_JGYI01000039; locus tags Y055_RS13435 to Y055_RS13520). (B) Upper and lower naphthalene catabolism gene clusters (nah) located on plasmid Nah7 of P. putida G7 (accession number NC_007926; locus tags NAH7_34 to NAH7_56) (43), a gene cluster in P. veronii 1YdBTEX2 encoding enzymes for salicylate degradation (accession number NZ_AOUH01000059), a gene cluster in P. veronii 1YdBTEX2 harboring a second EXDO A (EXDO A2)-encoding gene (accession number NZ_AOUH01000042; locus tags H736_RS0126300 to H736_RS0126240), and the truncated nah gene cluster in P. veronii 1YB2 (accession number NZ_JGYI01000001; locus tags Y055_RS00010 to Y055_RS00080). (C) Gene encoding benzene dihydrodiol dehydrogenase and its genetic environment (accession numbers NZ_AOUH01000056 [locus tags H736_RS0127880 to H736_RS0127895] and NZ_JGYI01000072 [locus tags Y055_RS18330 to Y055_RS18315]). White arrows indicate interrupted genes, and black arrows indicate mobile genetic elements. SDR indicates the gene encoding benzene dihydrodiol dehydrogenase of the short-chain dehydrogenase/reductase family, and DLD indicates the gene encoding a truncated putative dihydrolipoamide dehydrogenase. Putative mobile genetic elements were annotated by using TnpPred (52), and putative families of transposases are indicated. The percent sequence identity between the encoded proteins of P. putida RE204 and P. veronii 1YdBTEX2 or 1YB2 (A) and between the encoded proteins of P. putida G7 and P. putida 1YdBTEX2 or 1YB2 (B) are given below the gene clusters.
Sequence comparisons show that in P. veronii 1YdBTEX2, EXDO A is encoded by a salicylate catabolism gene cluster (GenBank accession number NZ_AOUH01000059), which is recruited for benzene degradation. The presence of a complete salicylate catabolism gene cluster (nahGTHINLOMKJ) (Fig. 2) allows the strain to grow on this substrate. Typically, salicylate catabolism gene clusters are localized in the proximity of naphthalene catabolism gene clusters, as is the case in P. putida G7 (43), where both the nahAaAbAcAdBCDEF genes encoding enzymes for the transformation of naphthalene to salicylate and the genes responsible for salicylate oxidation are localized on plasmid NAH7 (Fig. 2B). However, there was no indication of the presence of a naphthalene operon in P. veronii 1YdBTEX2. In contrast, in P. veronii 1YB2, the EXDO A-encoding gene is localized near a truncated naphthalene gene cluster (Fig. 2B) composed of only the nahAaAbBF genes where the naphthalene dihydrodiol dehydrogenase-encoding gene (nahB) is interrupted (GenBank accession number NZ_JGYI01000001), which explains the failure of the strain to grow on naphthalene. The absence of a nahG gene encoding salicylate hydroxylase explains the failure of strain 1YB2 to grow on salicylate.
P. veronii 1YdBTEX2 harbors a second extradiol dioxygenase (EXDO A2), which exhibits 84% sequence identity with EXDO A and which is localized in a gene cluster comprising only genes for downstream processing of catechol ring cleavage products (GenBank accession number NZ_AOUH01000042) (Fig. 2). The absence of any gene encoding enzymes for the formation of catechol may indicate the function of this operon in the processing of catechol derived from different sources by gene products encoded elsewhere on the genome.
Identification of the benzene dihydrodiol dehydrogenase recruited for benzene degradation.
Evidently, no functional dihydrodiol dehydrogenase was observed in any of the above-mentioned gene clusters, and screening of the genomes did not reveal any open reading frame that could encode an enzyme with high similarity to those transforming isopropylbenzene dihydrodiol, naphthalene dihydrodiol, or benzene dihydrodiol in either strain. However, benzene-grown cells of P. veronii 1YdBTEX2 and 1YB2 exhibited benzene dihydrodiol dehydrogenase activities of 590 and 860 U/g protein, respectively. The benzene dihydrodiol dehydrogenases of P. veronii 1YdBTEX2 and 1YB2 were partially purified from benzene-grown cells by anion-exchange chromatography, and activities of both cell extracts eluted at an NaCl concentration of 0.38 ± 0.01 M. In each case, 32 to 40% of the applied activity was recovered, reaching specific activities of 4,800 U/g protein.
Proteins in the active fractions were subjected to N-terminal sequencing. Only one open reading frame of each genome (GenBank accession number WP_046487322 for 1YB2 and accession number WP_017849553 for 1YdBTEX2) matched the observed MQRFQNKVVLVTGAA N-terminal sequence. Exploring the gene organization, the benzene dihydrodiol dehydrogenase-encoding genes in both strains are localized in identical gene clusters, devoid of other genes encoding aromatic degradation enzymes (accession numbers NZ_AOUH01000056 and NZ_JGYI01000072). Importantly, the benzene dihydrodiol dehydrogenases of both 1YdBTEX2 and 1YB2 are only distantly related to previously described dehydrogenases involved in aromatic degradation pathways (Fig. 3).
FIG 3.

Evolutionary relationships among members of the short-chain dehydrogenase/reductase enzyme family. The evolutionary histories were inferred by using the neighbor-joining method and the p-distance model. All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons. Phylogenetic analyses were conducted with MEGA5 (26). Bootstrap values of >50% from 100 neighbor-joining trees are indicated to the left of the nodes. The bar indicates amino acid differences per site. R. jostii, Rhodococcus jostii; B. sartisoli, Burkholderia sartisoli; R. globerulus, Rhodococcus globerulus; B. xenovorans, Burkholderia xenovorans; L. grayi, Listeria grayi; P. abietaniphila, Pseudomonas abietaniphila; A. piechaudii, Achromobacter piechaudii; P. stutzeri, Pseudomonas stutzeri; E. litoralis, Erythrobacter litoralis; M. extorquens, Methylobacterium extorquens; Y. ruckeri, Yersinia ruckeri; S. proteamaculans, Serratia proteamaculans.
Expression of genes of the benzene catabolic pathway.
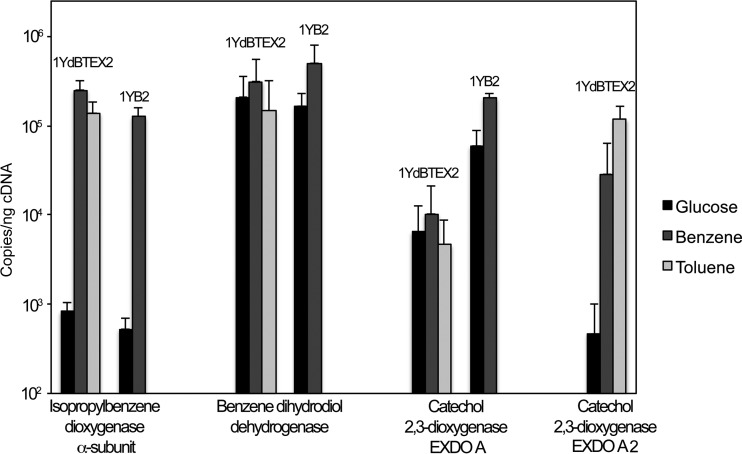
According to the data presented above, benzene degradation in both strains is most likely catalyzed by proteins of at least three catabolism gene clusters (see also Fig. S2 in the supplemental material). In addition, 1YdBTEX2 harbors two distinct extradiol dioxygenases of the EXDO A subfamily (EXDO A and EXDO A2). To analyze whether both dioxygenases were recruited and to determine whether all three gene clusters described above were transcribed in response to benzene and/or toluene, transcript levels of key genes of the above-mentioned gene clusters were quantified. This analysis comprised the quantification of expression of genes encoding the α-subunit of isopropylbenzene dioxygenase (ipbAa), the novel benzene dihydrodiol dehydrogenase, and EXDO A and EXDO A2 (1YdBTEX2 only) during growth on benzene (P. veronii 1YB2) and on benzene and toluene (P. veronii 1YdBTEX2) compared to transcript levels observed under noninducing conditions (growth on glucose).
The novel type of benzene dihydrodiol dehydrogenase was expressed constitutively in both strains with high transcript levels of 1.5 × 105 to 3.1 × 105 copies/ng cDNA (Fig. 4). EXDO A was also expressed constitutively but at levels of 4.6 × 103 to 10.3 × 103 copies/ng cDNA in the case of P. veronii 1YdBTEX2 and at a higher level of 5.9 × 103 to 20 × 103 copies/ng cDNA in the case of P. veronii 1YB2. In contrast, IpbAa was inducible in both strains, with transcript levels being increased by roughly 2 orders of magnitude after growth on benzene or toluene (1.5 × 105 to 2.5 × 105 copies/ng cDNA) compared to levels in glucose-grown cells (5.3 × 102 to 8.2 × 102 copies/ng cDNA). Similarly, EXDO A2 (present only in 1YdBTEX2) was inducible, with transcript levels increasing from 5.3 × 102 copies/ng cDNA to 2.9 × 104 and 1.2 × 105 copies/ng cDNA after growth on benzene and toluene, respectively (Fig. 4).
FIG 4.
Expression levels of catabolism genes involved in benzene or toluene degradation by P. veronii 1YdBTEX2 and 1YB2. Accumulation of transcripts of catabolism genes during growth on benzene for both P. veronii 1YdBTEX2 and 1YB2 and on toluene only for P. veronii 1YdBTEX2 was measured by real-time quantitative reverse transcription-PCR. Glucose was used as a noninducing negative control.
DISCUSSION
Considering that P. veronii 1YdBTEX2 and 1YB2 were isolated from a benzene-contaminated site, it was not surprising that they were able to degrade a variety of aromatic hydrocarbons. The use of at least three different catabolism gene clusters for benzene degradation, including a novel benzene dihydrodiol dehydrogenase, was upon first view unexpected, as such a mosaic kind of pathway is more typically observed for other genera such as Sphingomonas (44). However, this highlights the plasticity of the Pseudomonas genome in performing more efficient degradation in the face of dramatic environmental challenges.
The inactivation of a gene encoding an extradiol dioxygenase as observed for the EXDO K2-encoding gene in the isopropylbenzene catabolism gene cluster of 1YB2 was previously reported for other gene clusters. The degradation of 1,2,4-trichlorobenzene by Ralstonia sp. strain PS12 is initiated by a Rieske nonheme iron oxygenase with high similarity to isopropylbenzene dioxygenases (45). However, the further degradation of central chlorocatechols is achieved by an intradiol cleavage chlorocatechol pathway, whereas extradiol dioxygenases are typically inactivated during chlorocatechol turnover (46) and, thus, have no function. To avoid the energy-costing synthesis of an enzyme without function, the respective gene is inactivated in strain PS12 (47). Similarly, it may be speculated that inactivation of the EXDO K2-encoding gene avoids the unnecessary synthesis of the respective gene products, which in turn may be of a selective advantage.
The degradation of benzene via an extradiol cleavage pathway requires a 2-hydroxymuconic semialdehyde dehydrogenase (48); however, the corresponding genes are absent from gene clusters encoding enzymes for biphenyl/isopropylbenzene/toluene degradation via a dioxygenolytic route, and only a 2-hydroxymuconic semialdehyde hydrolase (encoded by ipbD) (Fig. 2) is typically encoded (9, 14). Thus, a 2-hydroxymuconic semialdehyde dehydrogenase has to be recruited from elsewhere. In P. veronii 1YdBTEX2, a 2-hydroxymuconic semialdehyde dehydrogenase-encoding gene (nahI) (Fig. 2) was localized in a salicylate catabolic cluster together with a gene encoding an EXDO A-type extradiol dioxygenase (Fig. 2). In P. veronii 1YB2, EXDO A- and 2-hydroxymuconic semialdehyde dehydrogenase-encoding genes were localized in a truncated naphthalene gene cluster (Fig. 2). Thus, recruitment of a gene cluster encoding an extradiol dioxygenase and a 2-hydroxymuconic semialdehyde dehydrogenase would be sufficient to allow growth on benzene and make expression of the extradiol dioxygenase encoded in the isopropylbenzene dioxygenase gene cluster dispensable. However, the reasons why dihydrodiol dehydrogenase encoded by this gene cluster is also inactivated and why such an activity is recruited from elsewhere in both strains are not so obvious. Interestingly, in Pseudomonas putida ML2, one of the few bacterial strains isolated based on its ability to grow on benzene (49) and characterized in detail regarding enzymes involved in benzene degradation, the dehydrogenation of cis-benzene dihydrodiol is catalyzed by BedD, a cis-benzene dihydrodiol dehydrogenase with high similarity to glycerol dehydrogenases (50), whereas genes encoding enzymes related to toluene cis-dihydrodiol dehydrogenases of the short-chain dehydrogenase family were absent. Curiously, degradation of intermediate catechol proceeds via an intradiol cleavage pathway (see Fig. S2 in the supplemental material) (8), which belongs to the core genomes of most Pseudomonas strains (51). Similarly to the situation for strain ML2, dehydrogenation of intermediate cis-benzene dihydrodiol in 1YB2 and 1YdBTEX2 is catalyzed by enzymes that are not closely related to toluene cis-dihydrodiol dehydrogenases. Thus, in P. putida ML2 as well as P. veronii 1YB2 and 1YdBTEX2, degradation of benzene, except for the initial dioxygenation step, is not catalyzed by enzymes similar to those involved in toluene degradation via a dioxygenolytic pathway. Whether this is due to the poor activity of enzymes of the toluene dioxygenase pathway remains to be elucidated.
Supplementary Material
ACKNOWLEDGMENTS
We give special thanks to Robert Geffers and Michael Jarek from the sequencing facility at the Helmholtz Centre for Infection Research and to Iris Plumeier and Agnes Waliczek for technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03026-15.
REFERENCES
- 1.Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW. 2011. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- 2.Cheuk W, Woo PC, Yuen KY, Yu PH, Chan JK. 2000. Intestinal inflammatory pseudotumour with regional lymph node involvement: identification of a new bacterium as the aetiological agent. J Pathol 192:289–292. doi:. [DOI] [PubMed] [Google Scholar]
- 3.Vullo DL, Ceretti HM, Daniel MA, Ramirez SA, Zalts A. 2008. Cadmium, zinc and copper biosorption mediated by Pseudomonas veronii 2E. Bioresour Technol 99:5574–5581. doi: 10.1016/j.biortech.2007.10.060. [DOI] [PubMed] [Google Scholar]
- 4.Hong HB, Nam IH, Murugesan K, Kim YM, Chang YS. 2004. Biodegradation of dibenzo-p-dioxin, dibenzofuran, and chlorodibenzo-p-dioxins by Pseudomonas veronii PH-03. Biodegradation 15:303–313. doi: 10.1023/B:BIOD.0000042185.04905.0d. [DOI] [PubMed] [Google Scholar]
- 5.Onaca C, Kieninger M, Engesser KH, Altenbuchner J. 2007. Degradation of alkyl methyl ketones by Pseudomonas veronii MEK700. J Bacteriol 189:3759–3767. doi: 10.1128/JB.01279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Junca H, Pieper DH. 2004. Functional gene diversity analysis in BTEX contaminated soils by means of PCR-SSCP DNA fingerprinting: comparative diversity assessment against bacterial isolates and PCR-DNA clone libraries. Environ Microbiol 6:95–110. [DOI] [PubMed] [Google Scholar]
- 7.Witzig R, Junca H, Hecht HJ, Pieper DH. 2006. Assessment of toluene/biphenyl dioxygenase gene diversity in benzene-polluted soils: links between benzene biodegradation and genes similar to those encoding isopropylbenzene dioxygenases. Appl Environ Microbiol 72:3504–3514. doi: 10.1128/AEM.72.5.3504-3514.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan H-M, Fong KP-Y. 1993. Molecular analysis of the plasmid-borne bed gene cluster from Pseudomonas putida ML2 and cloning of the cis-benzene dihydrodiol dehydroxygenase gene. Can J Microbiol 39:357–362. doi: 10.1139/m93-052. [DOI] [PubMed] [Google Scholar]
- 9.Zylstra GJ, McCombie WR, Gibson DT, Finette BA. 1988. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microbiol 54:1498–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shields MS, Montgomery SO, Chapman PJ, Cuskey SM, Pritchard PH. 1989. Novel pathway of toluene catabolism in the trichloroethylene-degrading bacterium g4. Appl Environ Microbiol 55:1624–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duarte M, Jauregui R, Vilchez-Vargas R, Junca H, Pieper DH. 2014. AromaDeg, a novel database for phylogenomics of aerobic bacterial degradation of aromatics. Database 2014:bau118. doi: 10.1093/database/bau118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson DT, Parales RE. 2000. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Curr Opin Biotechnol 11:236–243. doi: 10.1016/S0958-1669(00)00090-2. [DOI] [PubMed] [Google Scholar]
- 13.Aoki H, Kimura T, Habe H, Yamane H, Kodama T, Omori T. 1996. Cloning, nucleotide sequence, and characterization of the genes encoding enzymes involved in the degradation of cumene to 2-hydroxy-6-oxo-7-methylocta-2,4-dienoic acid in Pseudomonas fluorescens IP01. J Ferment Bioeng 81:187–196. doi: 10.1016/0922-338X(96)82207-0. [DOI] [Google Scholar]
- 14.Eaton RW, Selifonova OV, Gedney RM. 1998. Isopropylbenzene catabolic pathway in Pseudomonas putida RE204: nucleotide sequence analysis of the ipb operon and neighboring DNA from pRE4. Biodegradation 9:119–132. doi: 10.1023/A:1008386221961. [DOI] [PubMed] [Google Scholar]
- 15.Eltis LD, Bolin JT. 1996. Evolutionary relationships among extradiol dioxygenases. J Bacteriol 178:5930–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Junca H, Plumeier I, Hecht HJ, Pieper DH. 2004. Difference in kinetic behaviour of catechol 2,3-dioxygenase variants from a polluted environment. Microbiology 150:4181–4187. doi: 10.1099/mic.0.27451-0. [DOI] [PubMed] [Google Scholar]
- 17.Dorn E, Knackmuss H-J. 1978. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1,2-dioxygenation of catechol. Biochem J 174:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lima-Morales D, Chaves-Moreno D, Jarek M, Vilchez-Vargas R, Jauregui R, Pieper DH. 2013. Draft genome sequence of Pseudomonas veronii strain 1YdBTEX2. Genome Announc 1(3):e00258-13. doi: 10.1128/genomeA.00258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernandez D, Francois P, Farinelli L, Osteras M, Schrenzel J. 2008. De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res 18:802–809. doi: 10.1101/gr.072033.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sommer DD, Delcher AL, Salzberg SL, Pop M. 2007. Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8:64. doi: 10.1186/1471-2105-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 23.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silby MW, Cerdeno-Tarraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM, Zhang XX, Moon CD, Gehrig SM, Godfrey SA, Knight CG, Malone JG, Robinson Z, Spiers AJ, Harris S, Challis GL, Yaxley AM, Harris D, Seeger K, Murphy L, Rutter S, Squares R, Quail MA, Saunders E, Mavromatis K, Brettin TS, Bentley SD, Hothersall J, Stephens E, Thomas CM, Parkhill J, Levy SB, Rainey PB, Thomson NR. 2009. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens.. Genome Biol 10:R51. doi: 10.1186/gb-2009-10-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 28.Moreno-Hagelsieb G, Latimer K. 2008. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24:319–324. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- 29.Sierra R, Rodriguez RL, Chaves D, Pinzon A, Grajales A, Rojas A, Mutis G, Cardenas M, Burbano D, Jimenez P, Bernal A, Restrepo S. 2010. Discovery of Phytophthora infestans genes expressed in planta through mining of cDNA libraries. PLoS One 5:e9847. doi: 10.1371/journal.pone.0009847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:325–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 31.Clarke KR, Warwick RM. 2001. Change in marine communities: an approach to statistical analysis and interpretation, 2nd ed Primer-E, Plymouth, United Kingdom. [Google Scholar]
- 32.Bradford MM. 1976. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Heim S, Ferrer M, Heuer H, Regenhardt D, Nimtz M, Timmis KN. 2003. Proteome reference map of Pseudomonas putida strain KT2440 for genome expression profiling: distinct responses of KT2440 and Pseudomonas aeruginosa strain PAO1 to iron deprivation and a new form of superoxide dismutase. Environ Microbiol 5:1257–1269. doi: 10.1111/j.1462-2920.2003.00465.x. [DOI] [PubMed] [Google Scholar]
- 35.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vilchez-Vargas R, Geffers R, Suarez-Diez M, Conte I, Waliczek A, Kaser VS, Kralova M, Junca H, Pieper DH. 2013. Analysis of the microbial gene landscape and transcriptome for aromatic pollutants and alkane degradation using a novel internally calibrated microarray system. Environ Microbiol 15:1016–1039. doi: 10.1111/j.1462-2920.2012.02752.x. [DOI] [PubMed] [Google Scholar]
- 37.Camarinha-Silva A, Wos-Oxley ML, Jauregui R, Becker K, Pieper DH. 2012. Validating T-RFLP as a sensitive and high-throughput approach to assess bacterial diversity patterns in human anterior nares. FEMS Microbiol Ecol 79:98–108. doi: 10.1111/j.1574-6941.2011.01197.x. [DOI] [PubMed] [Google Scholar]
- 38.Jousset A, Schuldes J, Keel C, Maurhofer M, Daniel R, Scheu S, Thuermer A. 2014. Full-genome sequence of the plant growth-promoting bacterium Pseudomonas protegens CHA0. Genome Announc 2(2):e00322-14. doi: 10.1128/genomeA.00322-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loper JE, Hassan KA, Mavrodi DV, Davis EW II, Lim CK, Shaffer BT, Elbourne LD, Stockwell VO, Hartney SL, Breakwell K, Henkels MD, Tetu SG, Rangel LI, Kidarsa TA, Wilson NL, van de Mortel JE, Song C, Blumhagen R, Radune D, Hostetler JB, Brinkac LM, Durkin AS, Kluepfel DA, Wechter WP, Anderson AJ, Kim YC, Pierson LS III, Pierson EA, Lindow SE, Kobayashi DY, Raaijmakers JM, Weller DM, Thomashow LS, Allen AE, Paulsen IT. 2012. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet 8:e1002784. doi: 10.1371/journal.pgen.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortet P, Barakat M, Lalaouna D, Fochesato S, Barbe V, Vacherie B, Santaella C, Heulin T, Achouak W. 2011. Complete genome sequence of a beneficial plant root-associated bacterium, Pseudomonas brassicacearum.. J Bacteriol 193:3146. doi: 10.1128/JB.00411-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GS, Mavrodi DV, DeBoy RT, Seshadri R, Ren Q, Madupu R, Dodson RJ, Durkin AS, Brinkac LM, Daugherty SC, Sullivan SA, Rosovitz MJ, Gwinn ML, Zhou L, Schneider DJ, Cartinhour SW, Nelson WC, Weidman J, Watkins K, Tran K, Khouri H, Pierson EA, Pierson LS III, Thomashow LS, Loper JE. 2005. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol 23:873–878. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Redondo-Nieto M, Barret M, Morrisey JP, Germaine K, Martinez-Granero F, Barahona E, Navazo A, Sanchez-Contreras M, Moynihan JA, Giddens SR, Coppoolse ER, Muriel C, Stiekema WJ, Rainey PB, Dowling D, O'Gara F, Martin M, Rivilla R. 2012. Genome sequence of the biocontrol strain Pseudomonas fluorescens F113. J Bacteriol 194:1273–1274. doi: 10.1128/JB.06601-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yen KM, Gunsalus IC. 1982. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A 79:874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Armengaud J, Timmis KN, Wittich RM. 1999. A functional 4-hydroxysalicylate/hydroxyquinol degradative pathway gene cluster is linked to the initial dibenzo-p-dioxin pathway genes in Sphingomonas sp. strain RW1. J Bacteriol 181:3452–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beil S, Happe B, Timmis KN, Pieper DH. 1997. Genetic and biochemical characterization of the broad-spectrum chlorobenzene dioxygenase from Burkholderia sp. strain PS12: dechlorination of 1,2,4,5-tetrachlorobenzene. Eur J Biochem 247:190–199. doi: 10.1111/j.1432-1033.1997.00190.x. [DOI] [PubMed] [Google Scholar]
- 46.Bartels I, Knackmuss H-J, Reineke W. 1984. Suicide inactivation of catechol 2,3-dioxygenase from Pseudomonas putida mt-2 by 3-halocatechols. Appl Environ Microbiol 47:500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beil S, Timmis KN, Pieper DH. 1999. Genetic and biochemical analyses of the tec operon suggest a route for evolution of chlorobenzene degradation genes. J Bacteriol 181:341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harayama S, Mermod N, Rekik M, Lehrbach PR, Timmis KN. 1987. Roles of the divergent branches of the meta-cleavage pathway in the degradation of benzoate and substituted benzoates. J Bacteriol 169:558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan H-M, Mason JR. 1990. Cloning and expression of the plasmid-encoded benzene dioxygenase genes from Pseudomonas putida ML2. FEMS Microbiol Lett 72:259–264. doi: 10.1111/j.1574-6968.1990.tb03899.x. [DOI] [PubMed] [Google Scholar]
- 50.Fong KPY, Goh CBH, Tan HM. 1996. Characterization and expression of the plasmid-borne bedD gene from Pseudomonas putida ML2, which codes for a NAD(+)-dependent cis-benzene dihydrodiol dehydrogenase. J Bacteriol 178:5592–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perez-Pantoja D, Donoso R, Junca H, Gonzalez B, Pieper DH. 2009. Phylogenomics of aerobic bacterial degradation of aromatics, p 1356–1397. In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 52.Riadi G, Medina-Moenne C, Holmes DS. 2012. TnpPred: a Web service for the robust prediction of prokaryotic transposases. Comp Funct Genomics 2012:678761. doi: 10.1155/2012/678761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.