Abstract
Foodborne outbreaks of human noroviruses (HuNoVs) are frequently associated with leafy greens. Because there is no effective method to eliminate HuNoV from postharvest leafy greens, understanding virus survival under preharvest conditions is crucial. The objective of this study was to evaluate the survival of HuNoV and its surrogate viruses, murine norovirus (MNV), porcine sapovirus (SaV), and Tulane virus (TV), on preharvest lettuce and spinach that were subjected to abiotic stress (physical damage, heat, or flood). We also examined the bacteria culturable from the phyllosphere in response to abiotic stress and in relation to viral persistence. Mature plants were subjected to stressors 2 days prior to inoculation of the viruses on leaves. We quantified the viral RNA, determined the infectivity of the surrogates, and performed bacterial counts on postinoculation days (PIDs) 0, 1, 7, and 14. For both plant types, time exerted significant effects on HuNoV, MNV, SaV, and TV RNA titers, with greater effects being seen for the surrogates. Infectious surrogate viruses were undetectable on PID 14. Only physical damage on PID 14 significantly enhanced HuNoV RNA persistence on lettuce, while the three stressors differentially enhanced the persistence of MNV and TV RNA. Bacterial counts were significantly affected by time and plant type but not by the stressors. However, bacterial counts correlated significantly with HuNoV RNA titers on spinach and with the presence of surrogate viruses on both plant types under various conditions. In conclusion, abiotic stressors and phyllosphere bacterial density may differentially influence the survival of HuNoV and its surrogates on lettuce and spinach, emphasizing the need for the use of preventive measures at the preharvest stage.
INTRODUCTION
Human noroviruses (HuNoVs) are the leading cause of acute gastroenteritis and foodborne illnesses in the United States, causing 19 million to 21 million cases, 570 to 800 deaths, and $777 million in health care-associated costs annually (1). These single-stranded RNA viruses belong to the Caliciviridae family and are 28 to 35 nm in diameter and nonenveloped. Although these viruses cause self-limiting gastroenteritis, more serious infections can develop in high-risk groups, such as the elderly and immunocompromised populations (2). The virus is mainly transmitted to humans via the fecal-oral route either by contact with an infected person or fomites or by ingestion of contaminated food and water (2). Leafy greens are frequently associated with HuNoV outbreaks (1) and are globally recognized to be a high priority in terms of the microbial safety of fresh produce (3). Contamination of leafy greens with HuNoVs can occur at any stage along the farm-to-fork chain (3) through a number of sources, including fecally contaminated water used for irrigation, improperly treated sewage sludge used for fertilization, and food harvesters or food handlers who use improper hygiene practices (4, 5). Because HuNoVs have a low infectious dose (∼18 to 1,000 viral particles) (6) and postharvest leafy greens are not processed in ways that effectively remove/inactivate HuNoVs (4, 7), contamination of leafy greens with HuNoVs constitutes a serious risk to consumers. Therefore, there is a need to understand the factors influencing the survival/persistence characteristics of HuNoVs on preharvest leafy greens to develop proper measures to prevent contamination.
Leafy greens are grown in various regions of the world under a wide range of climatic conditions to fulfill the demands of both domestic and export markets (3). In the field, plants are exposed to various biotic and abiotic stress factors, such as phytopathogen infections, physical damage, waterlogging, and rapid and wide fluctuations in temperature (8, 9). Depending on the duration and the type of the stressor and the plant's ability to adapt to stress, the stressor may affect the plant's growth and productivity (10). For example, severe flooding causes many nutrient deficiencies in the affected vegetable because its roots cannot take up nutrients from the oxygen-deficient flooded soils, resulting in crop losses. However, short-term flooding (<48 h) due to poor field drainage does not reduce crop yields (11). In addition, stressors may alter the microenvironment on the leaf surfaces (9, 12), which may then influence the survival of human pathogens on the phyllosphere in cases of fecal contamination. For example, heat-stressed plants leach carbohydrates more easily than plants held at lower temperatures (13), and carbohydrates and proteins are exuded as a result of physical damage, such as open cuts, and can be used as nutrient sources by enteric bacterial pathogens (9, 12). Leached or exposed cell wall carbohydrates may act as binding sites for HuNoVs, which have been shown to bind cell wall carbohydrates in lettuce (14). In addition to abiotic stress factors, the plant phyllosphere is colonized by diverse microorganisms that may promote or impede the ability of enteric bacterial pathogens to persist on the leaf surface (5). However, little is known about whether any of these factors affect the survival of HuNoV on preharvest leafy greens, in part because HuNoV cannot be routinely propagated in cell culture; hence, its infectivity cannot be quantified.
Surrogate culturable viruses are often used as proxies to estimate HuNoV infectivity and survival in the environment. Murine norovirus (MNV), porcine sapovirus (SaV), and Tulane virus (TV) are the only three enteric viruses belonging to the Caliciviridae family which can be propagated in cell culture and as such have been used as HuNoV surrogates. These HuNoV calicivirus surrogates differ from each other in terms of their susceptible hosts, disease symptoms, cell surface receptor binding, and physiochemical properties but share various degrees of similarity to HuNoV (15–17). For example, like HuNoV, TV binds to carbohydrates of histo-blood group antigens (HBGAs), whereas MNV and SaV bind to sialic acids on ganglioside and on glycoproteins, respectively (18, 19). Like HuNoV, SaV and TV cause gastroenteritis in the corresponding host species (20, 21), whereas MNV causes a modest intestinal pathology in wild-type mice and severe to lethal systemic disease in immunocompromised mice (22). In addition, like HuNoV, SaV and TV have the potential for zoonotic infections (20, 23), and human SaVs are emerging as foodborne pathogens (24). In spite of the lack of a surrogate virus with characteristics that comprehensively mimic the various characteristics of HuNoV, a comparison of these surrogate viruses to each other and to HuNoV on leafy greens is needed to understand their survival characteristics with respect to those of each other and to those of HuNoV. Taken together, the main objective of this study was to evaluate the survival of HuNoV and its surrogates, MNV, SaV, and TV, on preharvest lettuce and spinach plants subjected to physical damage, heat, or flood stressors. We also examined the bacteria culturable from the phyllosphere in response to abiotic stress and in relation to viral persistence.
MATERIALS AND METHODS
Preparation of viruses.
Cell culture propagation of MNV, SaV, and TV was performed as described previously (17, 25, 26) using the following cell lines: a mouse leukemic macrophage cell line (RAW 264.7, ATCC TIB-71), an LLC-porcine kidney cell line (LLC-PK1, ATCC CL-101), and an LLC-monkey kidney cell line (LLC-MK2, ATCC CCL-7), respectively. Briefly, SaV was propagated in minimum essential medium (MEM; Life Technologies, Grand Island, NY) without fetal bovine serum (FBS), while TV and MNV were propagated in M199 cell culture medium and high-glucose Dulbecco modified Eagle medium (Life Technologies) containing 5 and 10% FBS (Thermo Scientific, Rockford, IL), respectively. All viruses were released from the cells by applying three cycles of freezing-thawing. The viruses were separated from the cell debris by centrifugation at 4°C and 2,095 × g for 30 min. The supernatants containing the viruses were aliquoted and stored at −80°C until further use. The HuNoV strain used in this study belonged to GII.4/HS194/2009/US and was genetically characterized in our laboratory (27). Briefly, a stool sample containing HuNoV was diluted (1:10) in 0.01 M phosphate-buffered saline (PBS; pH 7.4), vortexed, and centrifuged at 4°C and 2,095 × g for 30 min. The supernatant was filtered through 0.45-μm-pore-size nitrocellulose filters, aliquoted, and stored at −80°C until further use. Virus titers were assayed using reverse transcription (RT)-quantitative real-time PCR (qPCR) and infectivity assays for surrogate viruses (as described below).
Plant cultivation.
Seeds of romaine lettuce cultivar Tall Guzmaine Elite (Siegers Seed Co., Holland, MI) and baby's leaf hybrid spinach (Burpee, Warminster, PA) were grown in 200-cell trays containing Fafard superfine germinating mix (Conrad Fafard, Aguwam, MA) under greenhouse conditions with temperatures of ∼23°C during the daytime and 15°C during the nighttime and with a 12-h photoperiod (28). At 2 weeks of age, the seedlings were transferred to 15-cm-diameter pots containing sterile soil (Wooster sandy loam) and were fertilized biweekly using Osmocote slow-release fertilizer. The spinach and lettuce plants were watered twice a day using an overhead irrigation sprinkler hose until they reached ages of 6 and 8 weeks, respectively.
Plant treatments.
One day prior to abiotic stress treatments, spinach plants (at 6 weeks of age) and lettuce plants (at 8 weeks of age) were placed inside 22-by-11-in. plastic trays (6 pots per tray; Hummert, Topeka, KS), and ∼2 liters of water, which was not allowed to come into contact with the leaves, was added to each tray. On the next day, the plants were subjected to the following stressors, as described previously (8, 9, 29): (i) mechanical damage, whereby 6 outer leaves were bent in half widthwise to crack the central vein while maintaining an adequate water supply to the plants (∼2 liters/tray/day); (ii) heat stress, whereby the plants were placed inside a growth chamber at temperatures of 36°C during the daytime and 15°C during the nighttime for 2 days while maintaining an adequate water supply as needed (∼3 liters/tray/day); or (iii) flood stress, whereby the plants were subjected to overwatering by maintaining ∼6 liters of water per tray for 2 days. Physical damage was applied only to outer leaves to mimic field damage, and heat and flooding stressors were applied only for 2 days to avoid bolting (caused by extended heat) and root suffocation (caused by extended flooding), which result in unmarketable plants. In addition, heat-stressed plants received additional water to avoid wilting and the drought effect. Physically damaged plants, flooded plants, and control plants (2 liters/tray/day) with no signs of physical damage to the leaves were all held for 2 days at the greenhouse at temperatures of ∼23°C during the daytime and 15°C during the nighttime and with a 12-h photoperiod. According to Ge et al. (29), normal irrigation for lettuce plants is defined as 250 ml per pot per day. We used a similar approach for calculating the volume of water per tray containing 6 pots (i.e., ∼1.5 liters/tray/day). We added an extra 0.5 liter per tray to take into consideration evaporation and the age difference between our plants (6 to 8 weeks old) and those of Ge et al. (4 weeks old) (29). Following the stress treatments, the soil water content was measured by obtaining 100-g soil samples from random plant pots. The soil samples were heated in an oven at 60°C overnight, and the calculated weight of water was divided by the weight of the dried soil to obtain the gravimetric soil water content (grams of water per gram of soil) as described previously (8). The soil water content was significantly different between flooded plants (0.88 ± 0.04 g of water/g of soil) and control and physically damaged plants (0.53 ± 0.03 g of water/g of soil) and heat-stressed plants (0.67 ± 0.03 g of water/g of soil).
Contamination of plant leaves with viruses.
Following abiotic stress treatment, viral contamination of plant leaves was performed inside a biohazard hood. The suspension of feces containing HuNoV and the cell lysates of MNV, SaV, and TV were buffer exchanged to sterile water (to mimic contamination through irrigation water) using Amicon 100K Ultra-15 centrifugal devices (Millipore, Billerica, MA) that were centrifuged at 4,000 × g for 0.5 h at 4°C as described previously (30). The titer of HuNoV RNA used in subsequent experiments was 6.3 ± 0.1 log10 genomic equivalent (GE)/ml. The infectivity titers of MNV, SaV, and TV used were 6 ± 0.1, 5.5 ± 0.2, and 6 ± 0.3 log10 tissue culture infectious dose affecting 50% of the cultures (TCID50)/ml, corresponding to RNA titers of 8.2 ± 0.2, 10.3 ± 0.2, and 9.8 ± 0.1 log10 GE/ml, respectively. The infectivity titers were not significantly different from each other. However, the HuNoV, MNV, SaV, and TV RNA titers were all significantly different from each other, with the exception of the SaV and TV RNA titers. Therefore, the y axes for the infectivity graphs for surrogate viruses start at the infectivity assay detection limit of 0.3 log10 TCID50/ml, while those for the graphs for RNA titers start at different titers for different viruses but span 4 log10 units for all viruses.
Viruses were spot inoculated onto stressed lettuce and spinach outer leaves (1 ml per leaf over an area of ∼30 cm2). Spot inoculation was chosen as the method for viral application because it has been shown to be a more efficient method than immersion for application of virus to leaves (31). Control plants included plants not subjected to any of the three stressors and inoculated with viruses (negative treatment control) and stressed plants inoculated with sterile water (negative virus control). Specifically, the four viruses were inoculated on different leaves (marked with a pen) of each plant. One milliliter of each virus solution was inoculated as ∼50-μl droplets per leaf. Different plants were used for replication for each time point. The viral droplets were allowed to dry in a biosafety level 2 (BSL2) hood for 2 h before all the plants were transferred to biosafety level 2 plant growth chambers under controlled conditions (12-h photoperiod, 20°C daytime and 15°C nighttime temperatures, 80% relative humidity). Extra care was taken, especially with physically bent leaves, not to lose any viral droplets from the leaves at the time of inoculation. This was achieved by limiting the area inoculated to a circle about 6 cm in diameter and allowing time (usually 2 h) for the applied viral droplets to dry. Water (2 liters/tray containing 6 plants/day) was added to the trays daily as described above. Sampling of leaves was done on postinoculation days (PIDs) 0 (immediately following the 2-h dry period), 1, 7, and 14.
Elution of viruses from leaves.
On each harvesting day, for each sampling time point, three marked leaves from each of three different plants inoculated with each virus and receiving each treatment were aseptically detached from the plants. Each leaf was weighed, cut into small pieces, and individually suspended in elution buffer. The viruses were eluted from the leaf samples using MEM supplemented with a 1% antibiotic-antimycotic cocktail (Invitrogen, Carlsbad, CA, USA) and 2% heat-inactivated (60°C for 1 h) fetal bovine serum (Thermo Scientific HyClone) as described previously (28). Briefly, the samples were shaken vigorously (vortexing for 1 min, followed by shaking at 250 rpm for 10 min at 4°C), and the resulting solutions were transferred to sterile 50-ml Falcon tubes and centrifuged at 2,095 × g for 10 min to remove bacterial cells and plant debris. The supernatants were ultracentrifuged at 112,700 × g for 1.5 h to concentrate the viruses. The resulting pellets were suspended back in the original inoculum volume of 1 ml in sterile 0.01 M PBS (pH 7.4) supplemented with the 1% antibiotic-antimycotic cocktail (Invitrogen). The latter was used for RNA and infectivity assays, as described below.
Infectivity assays.
Viruses were titrated for the determination of their TCID50s using their respective cell lines cultured in 96-well plates, as described previously (17, 32–34). Briefly, 1- to 2-day-old confluent cell monolayers (except for RAW 264.7 cell monolayers, which were used at ∼50% confluence) in 96-well plates were infected in quadruplicate with serially diluted samples (diluted 1:10 in the respective cell culture medium supplemented with 1% antibiotic-antimycotic cocktail) and incubated at 37°C. The plates were inspected daily for a cytopathic effect (CPE), whereby final observations were performed on day 5 for MNV and TV. For SaV, on day 5, virus-infected cells in the wells with the highest dilution showing an isolated CPE and negative-control wells were indistinguishable. Therefore, an immunohistochemistry protocol described previously (17) was used to stain virus-infected cells to determine the TCID50 of SaV on day 5. The CPE for SaV and TV manifested as rounding of the cells, followed by their detachment from the cell monolayer, while that for MNV was observed as a shrinking of the cells, detachment, and a loss of their translucent appearance. The wells with infected cells were scored positive, and the viral titers were estimated following the Reed-Muench equation for the calculation of TCID50 (35). Leaf samples processed with the negative virus control were used to assess bacterial contamination or the cytotoxic effects of plant debris on cells. No bacterial contamination or cytotoxic effects were observed on any of the cell lines. The minimum viral concentrations that had to be on the leaf for virus to be detected were 1.3 ± 0.1, 2.1 ± 0.3, and 1.9 ± 0.1 log10 TCID50/ml for MNV, SaV, and TV, respectively. An average of 0.6 ± 0.1, 1.2 ± 0.2, and 0.9 ± 0.2 log10 TCID50/ml was lost during the recovery process for MNV, SaV, and TV, respectively. The infectivity assay's detection limit was 0.3 log10 TCID50/ml.
RNA extraction and quantification by RT-qPCR.
Viral RNA was extracted from 250 μl of the processed leaf samples using an RNeasy minikit (Qiagen, Valencia, CA, USA). In addition to negative virus control plant samples, a sterile water sample was extracted with every run to serve as an RNA extraction control. Prior to RNA extraction, samples were treated with RNase A (0.5 μg/μl; Invitrogen) for 1 h at 37°C. The RNA was eluted in 50 μl nuclease-free water and stored at −20°C. A one-step TaqMan virus-specific RT-qPCR was used to estimate the virus RNA titers in plant samples as described previously for HuNoV, MNV, SaV, and TV (17, 21, 36). All reactions were performed by taking 2 μl of each RNA sample and mixing it with 18 μl of master mix prepared using a Qiagen one-step RT-PCR kit (Qiagen). The master mix contained 1× PCR buffer, 400 μM deoxynucleoside triphosphates, 200 nM each primer, 100 nM TaqMan probe, 0.8 μl of enzyme mix, and 0.4 units/μl of RNasin (Promega, Madison, WI). The amplification cycle for HuNoV, MNV, SaV, and TV consisted of a reverse transcription step (50°C for 30 min) and 1 cycle at 95°C for 15 min, followed by 45 cycles consisting of two steps: 15 s at 95°C and 1 min at either 56°C for HuNoV, 60°C for MNV, 57.5°C for SaV, or 60°C for TV. RT-qPCR was performed with a MasterCycler RealPlex system (Eppendorf, Germany). Negative samples (nuclease-free water) and positive samples (virus with a known threshold cycle [CT] value) were included with each run. Each sample was tested in duplicate. An internal RNA control was spiked into randomly selected samples to check for the presence of RT-PCR inhibitors as described previously (17). None of the RNA samples showed any PCR inhibitors.
To avoid biases in virus infectivity and RNA titer calculation due to the significant differences in the weights of lettuce (5.23 ± 0.14 g) and spinach (1.82 ± 0.13 g) leaves, the viral inoculum used was fixed to 1 ml per replicate leaf sample and the droplets were spread over an approximately similar surface area (a 6-cm-diameter circle; total area, ∼30 cm2) on all leaves. Since our processing protocol involved a concentration step and suspension back into the original inoculum volume, the viruses' infectivity and RNA titers were reported in TCID50 and GE per milliliter of recovered virus instead of per gram of leaf tissue.
Quantification of culturable bacterial population associated with leaf surfaces.
To quantify the culturable aerobic bacteria on the surface of leaves, outer leaves were sampled and processed as described previously (37). Briefly, the leaves were weighed and aseptically cut into small pieces before they were transferred into sterile Whirl-Pak bags (Nasco, Salida, CA, USA). Each leaf sample received 10 ml of the elution buffer (1% peptone supplemented with 1% Tween 90), and the mixture was shaken vigorously (by vortexing for 1 min, followed by shaking at 250 rpm for 10 min at 4°C) to detach the bacteria. The resulting suspensions were collected, placed into new tubes, and serially diluted in 0.01 M PBS. A 100-μl aliquot from multiple dilutions per sample was plated onto R2A medium (Thermo Scientific) supplemented with cycloheximide (100 μg/ml; Sigma) to prevent fungal growth (38). The plates were incubated at room temperature for 3 to 4 days. The bacterial CFU were enumerated from dilutions showing 30 to 300 colonies.
Statistical analysis.
GraphPad Prism (version 5) software (GraphPad Software, USA) was used for statistical analyses. The entire data set for viruses and bacteria was log10 transformed. Significant differences in mean infectivity, RNA titers, and bacterial counts were determined by using two-way analysis of variance (ANOVA) followed by the Bonferroni posttest. The factors analyzed included time, stress treatment, and plant type. Each greenhouse experiment was repeated twice with triplicate plants per time point per treatment per plant type. Differences in means were considered significant when the P value was <0.05. Data are expressed as the mean ± standard error (SE). Pearson product-moment correlation analysis was used to determine significant correlations (P < 0.05) between the infectivity and RNA titers of each virus and between the viral titers and bacterial counts over time within each plant.
RESULTS
Abiotic stress treatments did not affect the infectivity of MNV, SaV, or TV on PID 1 or 7.
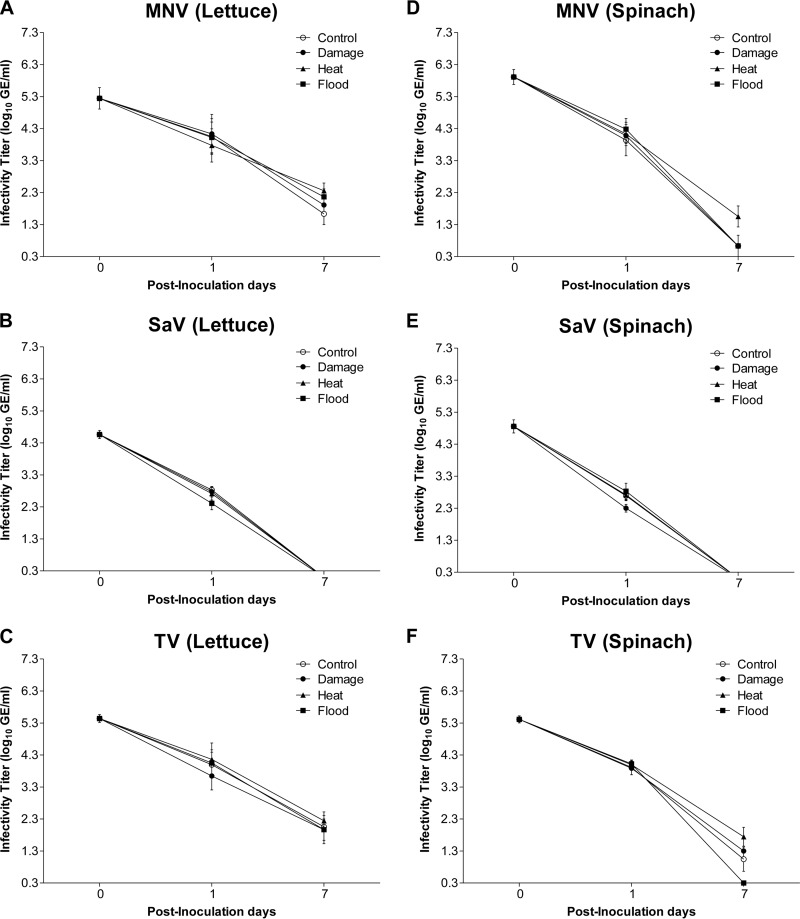
On PID 1, the infectivity titers of MNV, SaV, and TV inoculated on lettuce and spinach plants subjected to abiotic stressors (physical damage, heat, or flood) showed no significant differences from those of the corresponding viruses inoculated on plants held under control conditions (Fig. 1A to F). A similar result was obtained for PID 7. On both plant types, MNV, SaV, and TV showed significant decreases in infectivity titers on PID 0 compared to those on PID 1 and PID 7 and on PID 1 compared to those on PID 7 under control and all stress treatments (Table 1). Infectious viruses could not be detected for SaV on PID 7 and for all surrogate viruses on PID 14.
FIG 1.
Infectivity titers (log10 TCID50 per milliliter) for MNV, SaV, and TV on lettuce (A to C) and spinach (D to F) subjected to the following stressors: physical damage, heat, and flood. No significant differences between stress and control treatments on each PID were detected for any of the viruses.
TABLE 1.
Results of statistical analyses comparing viruses (infectivity and RNA titers) or bacterial counts between different PID
| Plant | PID data compareda | Statistically significant difference byb: |
||||
|---|---|---|---|---|---|---|
| RNA titer |
Bacterial count | |||||
| HuNoV | MNV | SaV | TV | |||
| Lettuce | 0 and 1 | None | CDHF | None | None | None |
| 1 and 7 | None | None | None | None | H | |
| 7 and 14 | C | CDF | CDHF | CD | None | |
| 0 and 7 | None | CDHF | None | H | CDH | |
| 0 and 14 | C | CDHF | CDHF | CDHF | CHF | |
| 1 and 14 | None | CDHF | CDHF | CDHF | H | |
| Spinach | 0 and 1 | F | CF | None | None | F |
| 1 and 7 | None | HF | None | DF | None | |
| 7 and 14 | None | C | None | C | H | |
| 0 and 7 | F | CDHF | CF | DF | F | |
| 0 and 14 | C | CDHF | CDHF | CF | CDHF | |
| 1 and 14 | None | CDF | CDHF | CDF | CH | |
For infectivity data, control, damage, heat, and flood treatments were all significantly different between PID 0 and PID 1, PID 0 and PID 7, and PID 1 and PID 7 for MNV, SaV, and TV.
C, D, H, and F, control, damage, heat, and flood treatments, respectively. The presence of a letter (C, D, H, or F) indicates a significant difference for that treatment between the two PIDs compared, while “none” indicates no significant differences between the two PIDs compared. Statistical analyses were performed by two-way ANOVA.
There were no significant differences in infectivity titers between the corresponding viruses on PIDs 0, 1, and 7 when infectivities for lettuce were compared to those for spinach, with the exception of the damage treatment for MNV and the flood stress treatment for MNV and TV on PID 7 (Table 2). Two-way ANOVA with time and stress treatments as factors revealed that time exerted significant effects on MNV, SaV, and TV infectivity for both lettuce and spinach (Table 3). Neither the treatment nor the interaction between the two factors had any significant effects (Table 3). Two-way ANOVA with plant and stress treatments as factors revealed that plant type exerted significant effects on PID 7 for MNV and TV (Table 4).
TABLE 2.
Results of statistical analyses comparing viruses (infectivity and RNA titers) or bacterial counts between lettuce and spinach
| PID | Statistically significant difference bya: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Infectivity |
RNA titer |
Bacterial count | ||||||
| MNV | SaV | TV | HuNoV | MNV | SaV | TV | ||
| 0 | None | None | None | None | None | None | None | HF |
| 1 | None | None | None | F | None | CHF | None | None |
| 7 | DF | — | F | None | F | CDHF | F | CDH |
| 14 | — | — | — | None | None | None | None | None |
C, D, H, and F, control, damage, heat, and flood treatments, respectively. The presence of a letter (C, D, H, or F) indicates a significant difference between the two plants compared for that group, while “none” indicates no significant differences. Statistical analyses were performed by two-way ANOVA. —, no data were available because infectivity titers were below the method's detection limit.
TABLE 3.
Results of statistical analyses of effect of each factor (time and stress treatment) and their interaction on the total variance of lettuce and spinach infectivity for RNA and bacterial count data sets
| Plant | Factor | % by which the factor accounted for total variance of data seta |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Infectivity |
RNA |
Bacterial count | |||||||
| MNV | SaV | TV | HuNoV | MNV | SaV | TV | |||
| Lettuce | Interaction | 0.9 | 0.2 | 0.3 | 12.9 | 3.0 | 1.9 | 5.6 | 6.2 |
| Stress | 0.2 | 0.1 | 0.4 | 6.4 | 0.7 | 0.7 | 1.7 | 1.0 | |
| Time | 61.9 | 96.3 | 76.4 | 11.9 | 73.3 | 62.9 | 61.1 | 33.9 | |
| Spinach | Interaction | 0.7 | 0.3 | 1.3 | 14.9 | 6.2 | 1.6 | 10.1 | 8.3 |
| Stress | 0.5 | 0.1 | 0.9 | 3.3 | 3.9 | 2.1 | 5.2 | 0.5 | |
| Time | 88 | 95 | 88.6 | 13.7 | 64.4 | 48.4 | 41.6 | 45.0 | |
Numbers in bold indicate a significant effect (P < 0.05) for that factor. Statistical analyses were performed by two-way ANOVA.
TABLE 4.
Summary of statistical analyses of effect of each factor (plant and stress treatment) and their interaction on the total variance of lettuce and spinach infectivity for RNA and bacterial count data sets
| PID | Factor | % by which the factor accounted for total variance of data seta |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Infectivity |
RNA titer |
Bacterial count | |||||||
| MNV | SaV | TV | HuNoV | MNV | SaV | TV | |||
| 1 | Interaction | 0.9 | 14.0 | 1.0 | 12.3 | 1.0 | 0.6 | 2.0 | 6.7 |
| Plant | 0.3 | 0.3 | 0.1 | 7.2 | 6.6 | 42 | 6.8 | 24.9 | |
| Stress | 0.6 | 4.1 | 2.6 | 11.6 | 5.6 | 3.9 | 13.4 | 4.6 | |
| 7 | Interaction | 3.1 | — | 7.2 | 4.0 | 6.4 | 1.6 | 6.0 | 9 |
| Plant | 53.9 | — | 45.8 | 3.7 | 15.2 | 65 | 25.6 | 59 | |
| Stress | 15.8 | — | 15.8 | 16.1 | 3.6 | 0.6 | 2.2 | 2.0 | |
| 14 | Interaction | — | — | — | 7.3 | 8.7 | 2.8 | 4.0 | 3.0 |
| Plant | — | — | — | 1.2 | 1.0 | 13.2 | 0.2 | 10.6 | |
| Stress | — | — | — | 17 | 30.2 | 6.4 | 31.9 | 11.8 | |
Numbers in bold indicate a significant effect (P < 0.05) for that factor. Statistical analyses were performed by two-way ANOVA. —, no data were available because infectivity titers were below the method's detection limit.
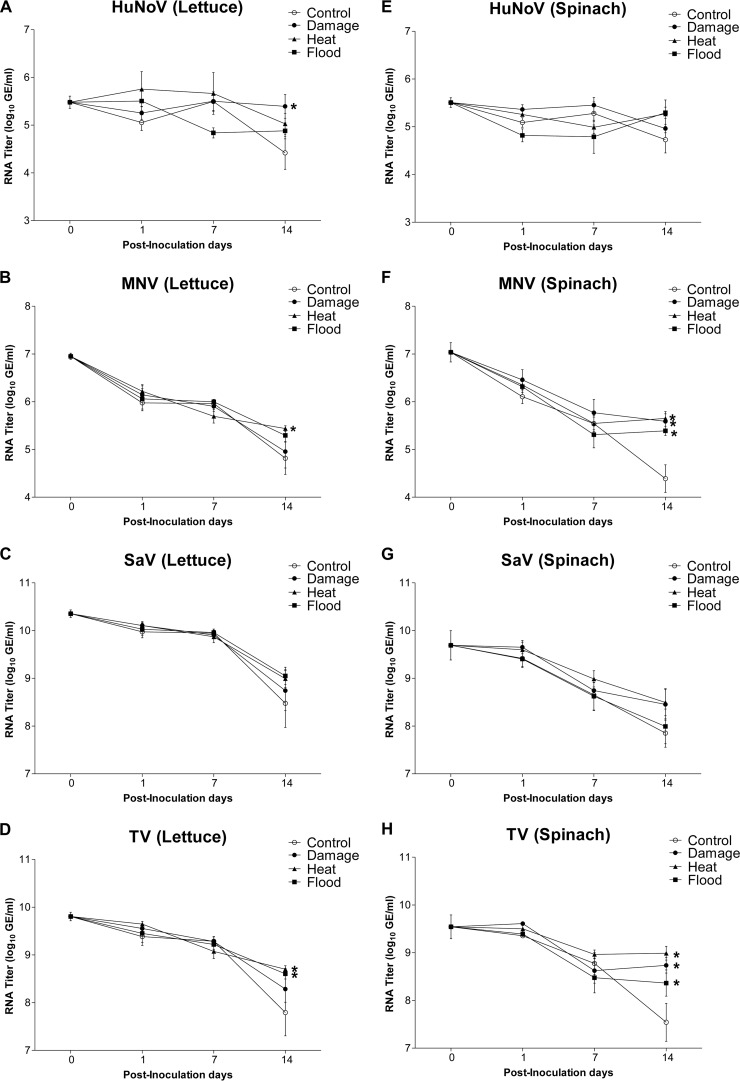
Abiotic stress treatments differentially affect the RNA titers of HuNoV, MNV, and TV only on PID 14.
On either PID 1 or PID 7, the HuNoV RNA titers on lettuce and spinach plants subjected to abiotic stressors (physical damage, heat, or flood) were not significantly different from those of the corresponding viruses inoculated on plants held under control conditions (Fig. 2A and E). The trend for all surrogate viruses mimicked this trend (Fig. 2B to D and F to H). On PID 14, the persistence of HuNoV RNA on lettuce subjected to physical damage was significantly enhanced compared to the persistence of HuNoV RNA on lettuce plants held under control conditions (Fig. 2A). In contrast, MNV RNA titers were significantly enhanced by heat treatment, while TV RNA titers were significantly enhanced by heat or flood treatment (Fig. 2B and D). For spinach, none of the stressors showed any significant effects on HuNoV RNA titers (Fig. 2E). In contrast, all stressors significantly enhanced MNV and TV RNA persistence when the RNA titers were compared to the RNA titers of the corresponding viruses inoculated on control plants (Fig. 2F and H). SaV RNA titers were not affected by any of the stressors (Fig. 2C and G).
FIG 2.
RNA titers (log10 GE per milliliter) for HuNoV, MNV, SaV, and TV on lettuce (A to D) and spinach (E to H) subjected to the following stressors: physical damage, heat, and flood. *, significant differences between stress and control treatments on each PID for each of the viruses.
On both plant types, the RNA titers of HuNoV on PID 14 were not significantly different from those on PID 1 (Table 1). In contrast, all surrogate viruses showed significant decreases in RNA titers on PID 14 compared to those on PID 1 under control and all stress conditions (Table 1). In general, there were no significant differences in HuNoV RNA titers between lettuce and spinach plants with any of the treatments except the flood treatment (Table 2). MNV and TV RNA titers showed a similar trend (Table 2).
Two-way ANOVA with time and stress treatments as factors revealed that time exerted significant effects on HuNoV, MNV, SaV, and TV RNA titers for both lettuce and spinach, with greater effects being seen with the surrogate viruses (Table 3), whereas stress treatment and the interaction between the two factors showed significant effects only for MNV and TV on spinach (Table 3). Two-way ANOVA with plant and stress treatments as factors revealed that stress exerted a significant effect on HuNoV RNA titers on PID 14 (Table 4). Both MNV and TV showed a similar trend. In contrast, plant type exerted a significant effect on MNV and TV RNA titers on PID 7, SaV RNA titers on all PIDs, and HuNoV RNA titers only on PID 1 (Table 4).
Significant correlations between the entire infectivity data set and the RNA titer data set were found for MNV, SaV, and TV on lettuce and spinach (r = 0.67, 0.66, and 0.55, respectively, for the infectivity data set and 0.82, 0.54, and 0.9, respectively, for the RNA titer data set).
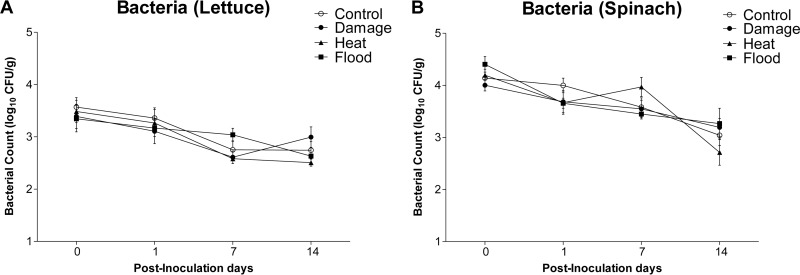
Stress treatment did not affect the bacterial counts on the phyllosphere of lettuce or spinach leaves.
On PIDs 0, 1, 7, and 14, both lettuce and spinach plants subjected to physical damage, heat, or flood stress exhibited no significant differences in their bacterial counts compared to those on control plants (Fig. 3A and B). When the results for PID 0 were compared to those for PID 14, it was revealed that the bacterial counts on control and all stressed lettuce and spinach plants decreased significantly, with the exception of those on lettuce subjected to the physical damage treatment (Table 1). Comparison of the bacterial counts on lettuce to those on spinach revealed significantly higher counts on spinach on PID 0 and PID 7 for certain groups (Fig. 3 and Table 2). However, by PID 14, there were no significant differences between the bacterial counts on lettuce and those on spinach for the control or any of the stress treatments (Table 2). Two-way ANOVA with time and stress treatment as factors revealed that time had a significant effect on the total variance of the data set on both lettuce and spinach (Table 3). Two-way ANOVA with plant and stress treatments as factors revealed that plant type exerted a significant effect on the total variance of the data set on all PIDs (Table 4).
FIG 3.
Bacterial counts (log10 CFU per gram) over time for lettuce (A) and spinach (B) subjected to the following stressors: physical damage, heat, and flood. No significant differences between stress and control treatments were detected on any PID.
Phyllosphere bacterial counts correlated significantly with HuNoV, MNV, SaV, and TV RNA titers under various conditions.
On control spinach, only HuNoV RNA titers correlated significantly with bacterial counts (Table 5). On control spinach, both SaV infectivity titers and RNA titers showed significant correlations with bacterial counts (Table 5). Various significant correlations between the infectivity and RNA titers of the different surrogate viruses and bacterial counts were found under different conditions. For example, the bacterial count on heat-stressed lettuce and flood-damaged spinach consistently correlated significantly with the infectivity and RNA titers of all surrogate viruses but not with the HuNoV RNA titer on the corresponding stressed plants (Table 5).
TABLE 5.
r values obtained by Pearson product-moment correlation analyses performed on virus infectivity or RNA titer and bacterial counts on lettuce or spinach plants over time
| Plant | Bacterial source |
r valuea |
||||||
|---|---|---|---|---|---|---|---|---|
| Infectivity |
RNA titer |
|||||||
| MNV | SaV | TV | HuNoV | MNV | SaV | TV | ||
| Lettuce | Control | 0.75 | 0.6 | 0.68 | 0.07 | 0.3 | 0.2 | 0.17 |
| Damage | 0.8 | 0.4 | 0.6 | −0.26 | 0.27 | −0.03 | 0.09 | |
| Heat | 0.74 | 0.64 | 0.78 | −0.1 | 0.72 | 0.58 | 0.57 | |
| Flood | 0.61 | 0.3 | 0.42 | 0.37 | 0.51 | 0.51 | 0.41 | |
| Spinach | Control | 0.33 | 0.63 | 0.5 | 0.44 | 0.67 | 0.83 | 0.86 |
| Damage | 0.37 | 0.62 | 0.52 | 0.39 | 0.15 | 0.55 | 0.24 | |
| Heat | −0.01 | 0.27 | 0.15 | −0.01 | 0.22 | 0.45 | 0.42 | |
| Flood | 0.69 | 0.80 | 0.74 | 0.31 | 0.60 | 0.73 | 0.66 | |
Numbers in bold indicate a significant correlation (P < 0.05).
DISCUSSION
The preharvest contamination of leafy greens with HuNoV is of great concern because of the reported low infectious dose of the virus (18 to 1,000 particles) and the lack of an effective postharvest decontamination method to remove/inactivate HuNoV (4). However, the duration of HuNoV survival on preharvest leafy greens is not known. The survival of these viruses may differ between preharvest and postharvest environments, and survival on the leaf surfaces may differ from that within the leaf tissues. Multiple studies that have investigated the postharvest survival of HuNoV on fresh produce have shown that this virus or its calicivirus surrogates (MNV, SaV, and TV) can survive during the chilling storage period and well beyond the produce shelf life (17, 30, 39–42). For example, HuNoV inoculated onto cut lettuce pieces persisted at 4°C for the entire 14-day study period with only a 1- to 1.5-log-unit reduction in RNA titers (39).
In contrast, few studies have investigated the preharvest survival of HuNoV or its surrogate viruses on leafy greens. A number of studies reported that HuNoV and its surrogates can internalize inside lettuce and spinach plants under normal conditions (28, 43, 44) and in lettuce under certain biotic and extreme abiotic stress conditions (45). However, only one study investigated the preharvest survival of HuNoV on leaf surfaces, reporting that the virus can persist on the leaves of mature spinach plants and showing no significant differences in viral RNA titers during the 7-day study period (46).
Our results expand on those of the latter study and show that HuNoV RNA can persist on mature lettuce and spinach plant leaves for at least 14 days. In contrast, MNV, SaV, and TV showed increased inactivation over time (according to their infectivity and RNA titers), consistent with the findings of previous studies utilizing MNV and TV on preharvest spinach (46) and SaV on lettuce (28). These findings are also consistent with those from a recent systematic review showing that, on the basis of RNA data, HuNoV is more stable than its surrogates (47). Both MNV and TV inoculated on spinach adaxial surfaces exhibited a 1-log-unit reduction in infectious titer every 2 to 3 days (46). This finding suggests what has been shown in our study that, by day 14, the virus in an initial inoculum of 6 log10 TCID50/ml is completely inactivated or is present at levels below the detection limit of current methods. In addition, plant type (lettuce or spinach) had stronger effects on surrogate viruses (infectivity and RNA titer) than on HuNoV, again suggesting that HuNoV is more stable than its surrogates.
Under the experimental conditions applied, HuNoV, SaV, MNV, and TV are negatively charged, because their isoelectric points (pI; 4.15, 5.4, 4.7, and 4.8, respectively [17, 48–50]) are less than the pH of their suspension matrix (water, pH 7). This information suggests that viral charge does not explain the different survival patterns between HuNoV and its surrogates. When subjected to heat treatment at 56°C, MNV and TV behaved similarly (51) and both MNV and SaV showed behavior similar to that of HuNoV (17, 52), suggesting that the capsids of the four viruses have similar stabilities at 56°C. Therefore, capsid stability does not explain the difference between HuNoV and its surrogates observed under the greenhouse conditions used in this study (23°C).
A recent study has shown that HuNoV VLP, TV, and MNV preferentially aggregate inside lettuce stomata, suggesting that the three viruses exhibit similar patterns of attachment to lettuce leaves (53). However, specific binding was not investigated in that study. HuNoV is known to bind to HBGAs and was found to attach specifically to cell wall carbohydrates in lettuce leaves (14). Of the three surrogates used, only TV mimics HuNoV binding to HBGAs; however, it is not known whether TV can bind plant carbohydrates. The specific binding of HuNoV to plant carbohydrates may explain its higher degree of stability in comparison to that of the surrogates. However, the specific binding of surrogates to plant carbohydrates needs to be investigated.
In our study, plant type and abiotic stress (with the exception of SaV) made significant contributions to the total variance of the results for the different viruses, suggesting that different viruses may be indirectly affected by the changes in the leaf microenvironment induced by stress on different plants. In addition, phyllosphere bacteria may play a role in the differential survival of HuNoV in comparison to that of SaV, MNV, and TV since bacterial counts more often correlated with the RNA titers of the surrogates than with the RNA titer of HuNoV under the various abiotic stress conditions. The latter result suggests that the titer of these surrogates may be influenced by bacterial density to a greater extent than the titer of HuNoV. Further investigation is needed to understand the microbial as well as the metabolic changes happening on the surfaces of lettuce and spinach leaves with and without abiotic stress and in relation to virus survival.
Although infectious surrogate viruses were undetectable on PID 14, on PID 7 the plants were market ready and had detectable infectious viruses. Similar to our results, it was reported that approximately 1 log unit of the viral infectivity titer is lost during the processing step (31, 46). This loss could be due to drying, the elution step, and/or the release of various inactivating/damaging factors from plant tissues during the sample processing steps. Therefore, since no current methods allow the complete recovery of viruses from the leaves, potentially infectious viruses may still be present on leaf samples that test negative (i.e., viruses may be present at levels below the test method's detection limit). The 14-day study period was chosen because by 7 and 14 days plants can be harvested and because of the importance of the period between a contamination event and harvest time. In addition, the 14-day study period used in our study is similar to or longer than the study periods reported for studies dealing with the survival of HuNoV (7 days) or TV and MNV (14 days) on leaf surfaces (45, 46). Collectively, because HuNoV RNA showed a higher degree of stability than MNV, SaV, and TV RNAs and RNA titers correlated significantly with infectivity titers for all surrogates, our results suggest that infectious HuNoVs contaminating leafy greens close to the harvest time may persist until the vegetables are harvested.
Production of leafy greens on the farm exposes the plants to several abiotic stressors of various durations. We followed the same methods for application of the heat, flood, and physical damage stressors reported in previous studies that addressed the effects of these abiotic stressors on human-pathogenic bacteria, such as Escherichia coli O157:H7 and Salmonella enterica serovar Typhimurium and evaluated the same parameters (8, 9, 29). The heat (36°C) and flood stressors were applied for a short duration (2 days), whereas physical damage was limited to bending of the outer leaves only (mimicking the common damage due to animals, farmers, equipment, etc., that occurs in the field). More importantly, extended periods of heat or flood result in bolting and root suffocation, making the plants unfit for sale (11, 46). As such, we focused on the end product, i.e., plant growth and productivity, rather than investigate how plants respond to the stressors because any visible deformities or stunted growth renders leafy greens unmarketable and the question of whether HuNoV survives on the leaf or not does not pose any further risk to consumers. The fresh weights of the leaves of the control lettuce and spinach plants on any sampling date were not significantly different from those of stressed plants for any of the stressors (data not shown), indicating that the stressors applied in our study did not affect the overall growth of the plants.
Only two previous studies examined the role of abiotic stress on the survival of HuNoV on leafy greens. The first study investigated the role of sunlight (UVA/UVB) on the survival of the surrogate viruses MNV and TV on spinach for 7 days and reported that it had a negative effect only when viruses were inoculated on the adaxial surfaces (46). The other study reported that extreme drought decreased the rate of MNV and TV internalization in lettuce plants (45). However, both studies relied on surrogates and did not assess the survival of HuNoV on plants subjected to abiotic stress. In contrast, more studies have investigated the effects of abiotic stressors on the survival of enteric bacterial pathogens on leafy greens. For example, physical damage, such as cutting, bruising, or bending of the leaves, promoted the survival and growth of E. coli O157:H7 on postharvest spinach (54) and pre- and postharvest lettuce (9, 54, 55). Heat and drought applied to lettuce plants individually or in combination for 2 days did not promote the internalization of E. coli O157:H7 from soil into leaves (8). Similarly, the application of flooding or drought stress to lettuce plants for 2 days did not enhance the internalization of Salmonella Typhimurium from leaf surfaces into internal tissues (29).
Although the findings of these studies on enteric bacterial pathogens cannot be directly compared to those of our study, it can be inferred that certain stressors, such as physical damage, can promote the survival of human bacterial pathogens on leafy greens. In our study, physical damage significantly affected the persistence of HuNoV RNA on lettuce leaves only on PID 14. The latter effect was not detected for any of the surrogate viruses on physically damaged lettuce or for HuNoV on physically damaged spinach but was detected for most surrogate viruses on physically damaged spinach. Therefore, surrogate viruses can behave differently from HuNoV, depending on the plant type and condition. This may be because spinach and lettuce respond differently when subjected to physical damage, thus exerting differential effects on different enteric viruses.
In our study, browning occurred at the damaged sites on lettuce leaves, while no such effect was noted on spinach leaves. This observation is consistent with the findings of previous studies that showed that ascorbic acid, which causes inhibition of enzyme-derived browning, has a higher antioxidant capacity on spinach leaves than lettuce leaves (54, 56). Since HuNoV was found to specifically bind to cell wall carbohydrates of lettuce leaves (14), exposed carbohydrates at damaged sites on lettuce may enhance the persistence of HuNoV, whereas other factors at damaged sites on spinach (e.g., antioxidants [57, 58]) may interfere with HuNoV RNA persistence but not MNV and TV RNA persistence.
Furthermore, the effects of all stressors on viruses were observed only on PID 14, indicating that the short-term stress (2 days in this study) had relatively long-term effects on virus survival. A previous study, however, implemented an extreme weather situation where drought or flooding was applied for 14 days (45). That study showed a negative effect of drought on virus internalization. However, leafy greens subjected to drought or flood for a long duration are unmarketable (11), and their contamination with HuNoV does not pose an increased risk to consumers. In contrast, previous studies, such as the study of Ge et al. (29), tripled the normal amount of water to achieve flooding conditions, and Erickson et al. (59) decreased the water level to 50% below normal to achieve stress from reduced amounts of water. Consistent with these studies, we used overwatering, maintaining triple the normal volume of water for 2 days, to simulate short-term flooding scenarios, and we showed a significant difference in soil water content between flooded and control soils. Collectively, since tissue damage is common not only in the field but also during harvest and postharvest, contamination with HuNoV on damaged lettuce leaves may pose a greater risk to consumers. Also, our results suggest that the occurrence of high temperatures or flooding for short periods (2 days) does not enhance HuNoV persistence in the case of accidental contamination.
Bacteria on the phyllosphere have been shown to influence the survival and growth of human bacterial pathogens (37, 60, 61). Previous studies have shown that the application of physical damage changes the microenvironment of the leaf due to nutrient leakage and sugar exudation, which in turn may affect the survival of human-pathogenic bacteria on the surfaces of leaves (9, 12, 54). However, no previous studies have explored whether phyllosphere bacteria have any role in the persistence of HuNoV on plants. In our study, the overall range of bacterial counts obtained on PID 0 (3.5 and 4.5 log10 CFU/g for lettuce and spinach, respectively) was similar to previously reported ranges of culturable bacteria on plants cultivated under greenhouse conditions (62, 63).
Consistent with the findings of previous studies, our results showed that plant type significantly impacts bacterial density (64, 65). Furthermore, the culturable bacteria on both lettuce and spinach showed a trend toward a decrease over time with respect to the count on PID 0. The latter may be due to changes in the amount of water available on leaf surfaces, as bacterial density can be affected by the leaf moisture content (63) and the plants in our study were watered through secondary pots (i.e., they were watered by the capillary action of the roots).
The response of the phyllosphere bacterial density to stress is largely unknown for leafy greens (63). In our study, the applied stressors did not affect the bacterial counts on lettuce or spinach. However, since culturable bacterial counts represent a small fraction of the total bacteria on the plant's phyllosphere, it is possible that the effect of abiotic stressors on these bacteria can be missed when only culturable counts are examined. In addition, the short period of stress application (2 days in this study) might not have significant effects on bacterial counts, which are known to change over the long growing season (65, 66). On the other hand, the structure of phyllosphere bacterial communities can be influenced by a number of environmental factors (63, 67, 68), which may explain the various significant correlations between the different viruses and the bacterial counts on lettuce and spinach observed under different conditions. In our study, both the levels of bacteria and the virus infectivity and RNA titers showed significant positive correlations under multiple conditions for multiple viruses. The latter indicates that the decrease in the levels of bacteria correlates with the decrease in viral titers.
Time may be one of the factors driving the various significant correlations between the viral titers and bacterial counts observed, as these were significantly impacted by time. Furthermore, other biological factors, such as leaf leachates (12, 69), may impact the survival of both viruses and bacteria on leaves. However, since virus-like particles of HuNoV were previously reported to adhere to certain locations on the plant phyllosphere (14, 70, 71) where native bacteria are reported to adhere, such as the leaf veins and stomata (72), it is possible that the HuNoVs contaminating plant leaves occupy niches similar to those that native bacteria occupy and therefore directly interact with those bacteria. A recent study by our group has shown an enhanced persistence of SaV on postharvest lettuce leaves that were infected by a plant-pathogenic bacterium preharvest (73), suggesting that plant bacteria may have a direct influence on the survival of viruses on the leaves.
Further research is needed to examine the lettuce and spinach bacterial community structure and their effects on HuNoV survival on the phyllosphere. This is especially important, as (i) HuNoVs are known to bind to multiple carbohydrate moieties (14), (ii) bacteria are known to express extracellular polysaccharides (5), and (iii) recent reports suggest that HuNoV can bind to bacteria found in the intestinal tract (74, 75). Collectively, our data suggest that the culturable bacterial density in the phyllosphere may influence the persistence of HuNoV and its surrogates on leafy greens.
In conclusion, our results suggest that (i) HuNoV RNA may persist under preharvest conditions on lettuce and spinach until harvest; (ii) abiotic stress, especially physical damage and the bacterial density in the phyllosphere of spinach, may influence the HuNoV RNA titer; and (iii) the persistence of no one surrogate virus mimicked the persistence of HuNoV over time and with the different abiotic stressors.
ACKNOWLEDGMENTS
We thank Mahesh Khatri and Issmat Kassem for their critical review of the manuscript.
Funding Statement
Salaries and research support were provided by state and federal funds provided to the Ohio Agricultural Research and Development Center (OARDC), The Ohio State University.
REFERENCES
- 1.Hall AJ, Wikswo ME, Pringle K, Gould LH, Parashar UD, Division of Viral Diseases, National Center for Immunization and Respiratory Diseases, CDC . 2014. Vital signs: foodborne norovirus outbreaks—United States, 2009-2012. MMWR Morb Mortal Wkly Rep 63:491–495. [PMC free article] [PubMed] [Google Scholar]
- 2.Siebenga JJ, Vennema H, Zheng DP, Vinje J, Lee BE, Pang XL, Ho EC, Lim W, Choudekar A, Broor S, Halperin T, Rasool NB, Hewitt J, Greening GE, Jin M, Duan ZJ, Lucero Y, O'Ryan M, Hoehne M, Schreier E, Ratcliff RM, White PA, Iritani N, Reuter G, Koopmans M. 2009. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001-2007. J Infect Dis 200:802–812. doi: 10.1086/605127. [DOI] [PubMed] [Google Scholar]
- 3.Gil MI, Selma MV, Suslow T, Jacxsens L, Uyttendaele M, Allende A. 2015. Pre- and postharvest preventive measures and intervention strategies to control microbial food safety hazards of fresh leafy vegetables. Crit Rev Food Sci Nutr 55:453–468. doi: 10.1080/10408398.2012.657808. [DOI] [PubMed] [Google Scholar]
- 4.Bouwknegt M, Verhaelen K, Rzezutka A, Kozyra I, Maunula L, von Bonsdorff CH, Vantarakis A, Kokkinos P, Petrovic T, Lazic S, Pavlik I, Vasickova P, Willems KA, Havelaar AH, Rutjes SA, de Roda Husman AM. 2015. Quantitative farm-to-fork risk assessment model for norovirus and hepatitis A virus in European leafy green vegetable and berry fruit supply chains. Int J Food Microbiol 198:50–58. doi: 10.1016/j.ijfoodmicro.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 5.Matthews KR. 2009. Leafy vegetables, p 165–187. In Sapers GM, Solomon EB, Matthews KR (ed), The produce contamination problem: causes and solutions. Academic Press/Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 6.Teunis PF, Moe CL, Liu P, Miller SE, Lindesmith L, Baric RS, Le Pendu J, Calderon RL. 2008. Norwalk virus: how infectious is it? J Med Virol 80:1468–1476. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- 7.Jung Y, Jang H, Matthews KR. 2014. Effect of the food production chain from farm practices to vegetable processing on outbreak incidence. Microb Biotechnol 7:517–527. doi: 10.1111/1751-7915.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang G, Ma L, Beuchat LR, Erickson MC, Phelan VH, Doyle MP. 2009. Heat and drought stress during growth of lettuce (Lactuca sativa L.) does not promote internalization of Escherichia coli O157:H7. J Food Prot 72:2471–2475. [DOI] [PubMed] [Google Scholar]
- 9.Aruscavage D, Miller SA, Ivey ML, Lee K, LeJeune JT. 2008. Survival and dissemination of Escherichia coli O157:H7 on physically and biologically damaged lettuce plants. J Food Prot 71:2384–2388. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Vinocur B, Altman A. 2003. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- 11.Reiners S. 10 March 2015. Dealing with flooded vegetable fields. Cornell University, Ithaca, NY: http://emergencypreparedness.cce.cornell.edu/disasters/Documents/PDFs/Dealing%20with%20Flooded%20Vegetable%20Fields.pdf. [Google Scholar]
- 12.Aruscavage D, Phelan PL, Lee K, LeJeune JT. 2010. Impact of changes in sugar exudate created by biological damage to tomato plants on the persistence of Escherichia coli O157:H7. J Food Sci 75:M187–M192. doi: 10.1111/j.1750-3841.2010.01593.x. [DOI] [PubMed] [Google Scholar]
- 13.Tukey JHB. 1970. The leaching of substances from plants. Annu Rev Plant Physiol 21:305–324. doi: 10.1146/annurev.pp.21.060170.001513. [DOI] [Google Scholar]
- 14.Esseili MA, Wang Q, Saif LJ. 2012. Binding of human GII.4 norovirus virus-like particles to carbohydrates of romaine lettuce leaf cell wall materials. Appl Environ Microbiol 78:786–794. doi: 10.1128/AEM.07081-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cromeans T, Park GW, Costantini V, Lee D, Wang Q, Farkas T, Lee A, Vinje J. 2014. Comprehensive comparison of cultivable norovirus surrogates in response to different inactivation and disinfection treatments. Appl Environ Microbiol 80:5743–5751. doi: 10.1128/AEM.01532-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kniel KE. 2014. The makings of a good human norovirus surrogate. Curr Opin Virol 4:85–90. doi: 10.1016/j.coviro.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Zhang Z, Saif LJ. 2012. Stability of and attachment to lettuce by a culturable porcine sapovirus surrogate for human caliciviruses. Appl Environ Microbiol 78:3932–3940. doi: 10.1128/AEM.06600-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoelzer K, Fanaselle W, Pouillot R, Van Doren JM, Dennis S. 2013. Virus inactivation on hard surfaces or in suspension by chemical disinfectants: systematic review and meta-analysis of norovirus surrogates. J Food Prot 76:1006–1016. doi: 10.4315/0362-028X.JFP-12-438. [DOI] [PubMed] [Google Scholar]
- 19.Kim DS, Hosmillo M, Alfajaro MM, Kim JY, Park JG, Son KY, Ryu EH, Sorgeloos F, Kwon HJ, Park SJ, Lee WS, Cho D, Kwon J, Choi JS, Kang MI, Goodfellow I, Cho KO. 2014. Both alpha2,3- and alpha2,6-linked sialic acids on O-linked glycoproteins act as functional receptors for porcine sapovirus. PLoS Pathog 10:e1004172. doi: 10.1371/journal.ppat.1004172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang QH, Costantini V, Saif LJ. 2007. Porcine enteric caliciviruses: genetic and antigenic relatedness to human caliciviruses, diagnosis and epidemiology. Vaccine 25:5453–5466. doi: 10.1016/j.vaccine.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sestak K, Feely S, Fey B, Dufour J, Hargitt E, Alvarez X, Pahar B, Gregoricus N, Vinje J, Farkas T. 2012. Experimental inoculation of juvenile rhesus macaques with primate enteric caliciviruses. PLoS One 7:e37973. doi: 10.1371/journal.pone.0037973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kahan SM, Liu G, Reinhard MK, Hsu CC, Livingston RS, Karst SM. 2011. Comparative murine norovirus studies reveal a lack of correlation between intestinal virus titers and enteric pathology. Virology 421:202–210. doi: 10.1016/j.virol.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farkas T, Dufour J, Jiang X, Sestak K. 2010. Detection of norovirus-, sapovirus- and rhesus enteric calicivirus-specific antibodies in captive juvenile macaques. J Gen Virol 91:734–738. doi: 10.1099/vir.0.015263-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svraka S, Vennema H, van der Veer B, Hedlund KO, Thorhagen M, Siebenga J, Duizer E, Koopmans M. 2010. Epidemiology and genotype analysis of emerging sapovirus-associated infections across Europe. J Clin Microbiol 48:2191–2198. doi: 10.1128/JCM.02427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farkas T, Sestak K, Wei C, Jiang X. 2008. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J Virol 82:5408–5416. doi: 10.1128/JVI.00070-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang S, Alhatlani B, Arias A, Caddy SL, Christodoulou C, Cunha JB, Emmott E, Gonzalez-Hernandez M, Kolawole A, Lu J, Rippinger C, Sorgeloos F, Thorne L, Vashist S, Goodfellow I, Wobus CE. 2014. Murine norovirus: propagation, quantification, and genetic manipulation. Curr Protoc Microbiol 33:15K.2.1–15K.2.61. doi: 10.1002/9780471729259.mc15k02s33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takanashi S, Wang Q, Chen N, Shen Q, Jung K, Zhang Z, Yokoyama M, Lindesmith LC, Baric RS, Saif LJ. 2011. Characterization of emerging GII.g/GII.12 noroviruses from a gastroenteritis outbreak in the United States in 2010. J Clin Microbiol 49:3234–3244. doi: 10.1128/JCM.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esseili MA, Wang Q, Zhang Z, Saif LJ. 2012. Internalization of sapovirus, a surrogate for norovirus, in romaine lettuce and the effect of lettuce latex on virus infectivity. Appl Environ Microbiol 78:6271–6279. doi: 10.1128/AEM.01295-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge C, Lee C, Nangle E, Li J, Gardner D, Kleinhenz M, Lee J. 2014. Impact of phytopathogen infection and extreme weather stress on internalization of Salmonella Typhimurium in lettuce. Int J Food Microbiol 168-169:24–31. [DOI] [PubMed] [Google Scholar]
- 30.Esseili MA, Saif LJ, Farkas T, Wang Q. 2015. Feline calicivirus, murine norovirus, porcine sapovirus, and Tulane virus survival on postharvest lettuce. Appl Environ Microbiol 81:5085–5092. doi: 10.1128/AEM.00558-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fino VR, Kniel KE. 2008. Comparative recovery of foodborne viruses from fresh produce. Foodborne Pathog Dis 5:819–825. doi: 10.1089/fpd.2008.0145. [DOI] [PubMed] [Google Scholar]
- 32.Fan Q, Wei C, Xia M, Jiang X. 2013. Inhibition of Tulane virus replication in vitro with RNA interference. J Med Virol 85:179–186. doi: 10.1002/jmv.23340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovac K, Diez-Valcarce M, Raspor P, Hernandez M, Rodriguez-Lazaro D. 2012. Effect of high hydrostatic pressure processing on norovirus infectivity and genome stability in strawberry puree and mineral water. Int J Food Microbiol 152:35–39. doi: 10.1016/j.ijfoodmicro.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 34.Toffan A, Brutti A, De Pasquale A, Cappellozza E, Pascoli F, Cigarini M, Di Rocco M, Terregino C, Arcangeli G. 2014. The effectiveness of domestic cook on inactivation of murine norovirus in experimentally infected Manila clams (Ruditapes philippinarum). J Appl Microbiol 116:191–198. doi: 10.1111/jam.12346. [DOI] [PubMed] [Google Scholar]
- 35.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 36.Kitajima M, Oka T, Takagi H, Tohya Y, Katayama H, Takeda N, Katayama K. 2010. Development and application of a broadly reactive real-time reverse transcription-PCR assay for detection of murine noroviruses. J Virol Methods 169:269–273. doi: 10.1016/j.jviromet.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Velasco G, Tydings HA, Boyer RR, Falkinham JO III, Ponder MA. 2012. Characterization of interactions between Escherichia coli O157:H7 with epiphytic bacteria in vitro and on spinach leaf surfaces. Int J Food Microbiol 153:351–357. doi: 10.1016/j.ijfoodmicro.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 38.Mercier J, Lindow SE. 2000. Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl Environ Microbiol 66:369–374. doi: 10.1128/AEM.66.1.369-374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Escudero BI, Rawsthorne H, Gensel C, Jaykus LA. 2012. Persistence and transferability of noroviruses on and between common surfaces and foods. J Food Prot 75:927–935. doi: 10.4315/0362-028X.JFP-11-460. [DOI] [PubMed] [Google Scholar]
- 40.Fallahi S, Mattison K. 2011. Evaluation of murine norovirus persistence in environments relevant to food production and processing. J Food Prot 74:1847–1851. doi: 10.4315/0362-028X.JFP-11-081. [DOI] [PubMed] [Google Scholar]
- 41.Lamhoujeb S, Fliss I, Ngazoa SE, Jean J. 2008. Evaluation of the persistence of infectious human noroviruses on food surfaces by using real-time nucleic acid sequence-based amplification. Appl Environ Microbiol 74:3349–3355. doi: 10.1128/AEM.02878-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattison K, Karthikeyan K, Abebe M, Malik N, Sattar SA, Farber JM, Bidawid S. 2007. Survival of calicivirus in foods and on surfaces: experiments with feline calicivirus as a surrogate for norovirus. J Food Prot 70:500–503. [DOI] [PubMed] [Google Scholar]
- 43.Dicaprio E, Ma Y, Purgianto A, Hughes J, Li J. 2012. Internalization and dissemination of human norovirus and animal caliciviruses in hydroponically grown romaine lettuce. Appl Environ Microbiol 78:6143–6152. doi: 10.1128/AEM.01081-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei J, Jin Y, Sims T, Kniel KE. 2011. Internalization of murine norovirus 1 by Lactuca sativa during irrigation. Appl Environ Microbiol 77:2508–2512. doi: 10.1128/AEM.02701-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DiCaprio E, Purgianto A, Li J. 2015. Effects of abiotic and biotic stresses on the internalization and dissemination of human norovirus surrogates in growing romaine lettuce. Appl Environ Microbiol 81:4791–4800. doi: 10.1128/AEM.00650-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirneisen KA, Kniel KE. 2013. Norovirus surrogate survival on spinach during preharvest growth. Phytopathology 103:389–394. doi: 10.1094/PHYTO-09-12-0231-FI. [DOI] [PubMed] [Google Scholar]
- 47.Knight A, Haines J, Stals A, Li D, Uyttendaele M, Knight A, Jaykus LA. 2015. A systematic review of human norovirus survival reveals a greater persistence of human norovirus RT-qPCR signals compared to those of cultivable surrogate viruses. Int J Food Microbiol 216:40–49. doi: 10.1016/j.ijfoodmicro.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 48.Bolton SL, Kotwal G, Harrison MA, Law SE, Harrison JA, Cannon JL. 2013. Sanitizer efficacy against murine norovirus, a surrogate for human norovirus, on stainless steel surfaces when using three application methods. Appl Environ Microbiol 79:1368–1377. doi: 10.1128/AEM.02843-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samandoulgou I, Fliss I, Jean J. 2015. Zeta potential and aggregation of virus-like particle of human norovirus and feline calicivirus under different physicochemical conditions. Food Environ Virol 7:249–260. doi: 10.1007/s12560-015-9198-0. [DOI] [PubMed] [Google Scholar]
- 50.Vega E, Smith J, Garland J, Matos A, Pillaii SD. 2005. Variability of virus attachment patterns to butterhead lettuce. J Food Prot 68:2112–2117. [DOI] [PubMed] [Google Scholar]
- 51.Hirneisen KA, Kniel KE. 2013. Comparing human norovirus surrogates: murine norovirus and Tulane virus. J Food Prot 76:139–143. doi: 10.4315/0362-028X.JFP-12-216. [DOI] [PubMed] [Google Scholar]
- 52.Li D, Baert L, Xia M, Zhong W, Van Coillie E, Jiang X, Uyttendaele M. 2012. Evaluation of methods measuring the capsid integrity and/or functions of noroviruses by heat inactivation. J Virol Methods 181:1–5. doi: 10.1016/j.jviromet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 53.DiCaprio E, Purgianto A, Ma Y, Hughes J, Dai X, Li J. 2015. Attachment and localization of human norovirus and animal caliciviruses in fresh produce. Int J Food Microbiol 211:101–108. doi: 10.1016/j.ijfoodmicro.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 54.Khalil RK, Frank JF. 2010. Behavior of Escherichia coli O157:H7 on damaged leaves of spinach, lettuce, cilantro, and parsley stored at abusive temperatures. J Food Prot 73:212–220. [DOI] [PubMed] [Google Scholar]
- 55.Brandl MT. 2008. Plant lesions promote the rapid multiplication of Escherichia coli O157:H7 on postharvest lettuce. Appl Environ Microbiol 74:5285–5289. doi: 10.1128/AEM.01073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Altunkaya A, Gökmen V. 2008. Effect of various inhibitors on enzymatic browning, antioxidant activity and total phenol content of fresh lettuce (Lactuca sativa). Food Chem 107:1173–1179. doi: 10.1016/j.foodchem.2007.09.046. [DOI] [Google Scholar]
- 57.Su X, D'Souza DH. 2011. Grape seed extract for control of human enteric viruses. Appl Environ Microbiol 77:3982–3987. doi: 10.1128/AEM.00193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su X, Sangster MY, D'Souza DH. 2010. In vitro effects of pomegranate juice and pomegranate polyphenols on foodborne viral surrogates. Foodborne Pathog Dis 7:1473–1479. doi: 10.1089/fpd.2010.0583. [DOI] [PubMed] [Google Scholar]
- 59.Erickson MC, Liao J, Payton AS, Webb CC, Ma L, Zhang G, Flitcroft I, Doyle MP, Beuchat LR. 2013. Fate of Escherichia coli O157:H7 and Salmonella in soil and lettuce roots as affected by potential home gardening practices. J Sci Food Agric 93:3841–3849. doi: 10.1002/jsfa.6321. [DOI] [PubMed] [Google Scholar]
- 60.Cooley MB, Chao D, Mandrell RE. 2006. Escherichia coli O157:H7 survival and growth on lettuce is altered by the presence of epiphytic bacteria. J Food Prot 69:2329–2335. [DOI] [PubMed] [Google Scholar]
- 61.Johnston MA, Harrison MA, Morrow RA. 2009. Microbial antagonists of Escherichia coli O157:H7 on fresh-cut lettuce and spinach. J Food Prot 72:1569–1575. [DOI] [PubMed] [Google Scholar]
- 62.Williams TR, Marco ML. 2014. Phyllosphere microbiota composition and microbial community transplantation on lettuce plants grown indoors. mBio 5:e01564-14. doi: 10.1128/mBio.01564-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams TR, Moyne AL, Harris LJ, Marco ML. 2013. Season, irrigation, leaf age, and Escherichia coli inoculation influence the bacterial diversity in the lettuce phyllosphere. PLoS One 8:e68642. doi: 10.1371/journal.pone.0068642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jackson CR, Randolph KC, Osborn SL, Tyler HL. 2013. Culture dependent and independent analysis of bacterial communities associated with commercial salad leaf vegetables. BMC Microbiol 13:274. doi: 10.1186/1471-2180-13-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson IP, Bailey MJ, Fenlon JS, Fermor TR, Lilley AK, Lynch JM, McCormack PJ, McQuilken MP, Purdy KJ, Rainey PB, Whipps JM. 1993. Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar beet (Beta vulgaris). Plant Soil 150:177–191. doi: 10.1007/BF00013015. [DOI] [Google Scholar]
- 66.Mew TW, Kennedy BW. 1982. Seasonal variation in populations of pathogenic pseudomonads on soybean leaves. Phytopathology 72:103–105. doi: 10.1094/Phyto-72-103. [DOI] [Google Scholar]
- 67.Lopez-Velasco G, Carder PA, Welbaum GE, Ponder MA. 2013. Diversity of the spinach (Spinacia oleracea) spermosphere and phyllosphere bacterial communities. FEMS Microbiol Lett 346:146–154. doi: 10.1111/1574-6968.12216. [DOI] [PubMed] [Google Scholar]
- 68.Vorholt JA. 2012. Microbial life in the phyllosphere. Nat Rev Microbiol 10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 69.Blakeman JP. 1973. The chemical environment of leaf surfaces with special reference to spore germination of pathogenic fungi. Pesticide Sci 4:575–588. doi: 10.1002/ps.2780040415. [DOI] [Google Scholar]
- 70.Gandhi KM, Mandrell RE, Tian P. 2010. Binding of virus-like particles of Norwalk virus to romaine lettuce veins. Appl Environ Microbiol 76:7997–8003. doi: 10.1128/AEM.01566-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wei J, Jin Y, Sims T, Kniel KE. 2010. Manure- and biosolids-resident murine norovirus 1 attachment to and internalization by romaine lettuce. Appl Environ Microbiol 76:578–583. doi: 10.1128/AEM.02088-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baldotto LE, Olivares FL. 2008. Phylloepiphytic interaction between bacteria and different plant species in a tropical agricultural system. Can J Microbiol 54:918–931. doi: 10.1139/W08-087. [DOI] [PubMed] [Google Scholar]
- 73.Esseili MA, Chin A, Saif LJ, Miller SA, Qu F, Lewis Ivey ML, Wang Q. 2015. Postharvest survival of porcine sapovirus, a human norovirus surrogate, on phytopathogen-infected leafy greens. J Food Prot 78:1472–1480. doi: 10.4315/0362-028X.JFP-14-518. [DOI] [PubMed] [Google Scholar]
- 74.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinje J, Tibbetts SA, Wallet SM, Karst SM. 2014. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miura T, Sano D, Suenaga A, Yoshimura T, Fuzawa M, Nakagomi T, Nakagomi O, Okabe S. 2013. Histo-blood group antigen-like substances of human enteric bacteria as specific adsorbents for human noroviruses. J Virol 87:9441–9451. doi: 10.1128/JVI.01060-13. [DOI] [PMC free article] [PubMed] [Google Scholar]