Abstract
Recent genome-scale studies have begun to establish the scope and magnitude of the impacts of carbohydrate source and availability on the regulation of gene expression in bacteria. The effects of sugars on gene expression are particularly profound in a group of lactic acid bacteria that rely almost entirely on their saccharolytic activities for energy production and growth. For Streptococcus mutans, the major etiologic agent of human dental caries, sucrose is the carbohydrate that contributes in the most significant manner to establishment, persistence, and virulence of the organism. However, because this organism produces multiple extracellular sucrolytic enzymes that can release hexoses from sucrose, it has not been possible to study the specific effects of sucrose transport and metabolism on gene expression in the absence of carbohydrates that by themselves can elicit catabolite repression and induce expression of multiple genes. By employing RNA deep-sequencing (RNA-Seq) technology and mutants that lacked particular sucrose-metabolizing enzymes, we compared the transcriptomes of S. mutans bacteria growing on glucose, fructose, or sucrose as the sole carbohydrate source. The results provide a variety of new insights into the impact of sucrose transport and metabolism by S. mutans, including the likely expulsion of fructose after sucrose internalization and hydrolysis, and identify a set of genes that are differentially regulated by sucrose versus fructose. The findings significantly enhance our understanding of the genetics and physiology of this cariogenic pathogen.
INTRODUCTION
Nearly all isolates of the primary etiologic agent of human dental caries, Streptococcus mutans, encode at least five secreted enzymes that act on sucrose in the extracellular environment, including three glucosyltransferases (GtfB, GtfC, and GtfD) that incorporate the glucose moiety of sucrose into high-molecular-mass α1,3- and α1,6-linked homopolymers of glucose that promote biofilm formation (1, 2); a fructosyltransferase (Ftf) that catalyzes the incorporation of fructose into β2,1- and β2,6-linked homopolymers of fructose (fructans) that serve as extracellular stores of carbohydrate within the biofilm matrix (3, 4); and a fructan hydrolase (FruA) that releases fructose from fructans, sucrose, and other β-fructosides (4, 5). While these exoenzymes contribute significantly to the virulence of S. mutans, most of the sucrose presented to S. mutans is rapidly internalized by a high-affinity, high-capacity sugar-phosphotransferase system (PTS) (6, 7). A single open reading frame (ORF), scrA, encodes the enzyme II permease of a sucrose-specific PTS, which internalizes and concomitantly phosphorylates sucrose. Transcribed divergently from scrA are two genes that encode a sucrose-6-PO4 (S-6-P) hydrolase (scrB), which cleaves S-6-P into glucose-6-PO4 (G-6-P) and fructose, and a transcriptional regulator (scrR) that represses the expression of both scrA and scrB (6, 8, 9). In addition, the multiple-sugar metabolism (msm) ABC transporter and a trehalose-PTS enzyme II of S. mutans have also been shown to be capable of internalizing sucrose from the environment (10–12), albeit much less efficiently. There has been extensive research establishing that the abilities of S. mutans to ferment sucrose and to convert it to polysaccharides are critical factors that directly contribute to human tooth decay (13–15).
It appears that S. mutans became associated and evolved closely with humans around the time that humans adopted agriculture and increased their intake of carbohydrate-rich foodstuffs. Consistent with this idea, a preliminary survey of the fossil record indicates an absence of S. mutans in Neolithic and Mesolithic populations but its appearance in the post-Mesolithic period, through the industrial revolution, and to the present day (16). Likewise, evolutionary genomic analysis of over 50 sequenced isolates of S. mutans from around the globe led to the conclusion that there was a large expansion in the population and genetic complexity of this species some 5,000 to 10,000 years ago (17). Phylogenetic analyses have also pointed to a strong influence on the genomic composition of S. mutans associated with widespread consumption of refined sucrose during the industrial revolution (18). In particular, it has been suggested that many of the genes responsible for enhanced cariogenicity in S. mutans were introduced into this bacterium via horizontal gene transfer, including the three glucosyltransferase genes, which encode enzymes that produce biofilm-promoting, adhesive glucans (17–19). Therefore, characterization of the effects of sucrose on gene expression and physiology of S. mutans is critical not only to gain a fuller appreciation of how this carbohydrate enhances the cariogenicity of the organism but also to gain insights into niche adaptations that occurred during coevolution with a human host, which can be highly specific to S. mutans and thus lead to new therapeutic or preventive strategies.
We previously reported the construction of a number of otherwise isogenic sucrase-deficient mutants derived from the cariogenic human isolate S. mutans UA159 and the systematic assessment of the contributions to sucrose metabolism by various enzymes and pathways (20). Growth phenotypes of these mutants indicated that gtfBCD, ftf, and the scrAB pathway constitute the majority of the sucrose-catabolizing capacity. Importantly, we also observed sucrose-dependent induction of the fructan hydrolase operon (fruAB) (5) and a fructose-PTS operon (levDEFGX or EIILev), with both being mediated by an unusual four-component signal transduction system (LevQRST) (21) and modulated by the sucrose-specific PTS enzyme II (ScrA) (20). Specifically, in a mutant that lacked all known extracellular sucrolytic activities, addition of sucrose resulted in increased expression of a levD::cat promoter fusion. This was an unusual finding, because prior studies had determined that fructose and mannose appeared to be the primary activators of the fruA and levD promoters by LevQRST (22). Despite some progress in understanding how sucrose influences gene expression in S. mutans, many questions remain (23). However, sucrose-specific impacts on S. mutans gene regulation can be studied only in a genetic background that is devoid of all extracellular enzymes capable of modifying sucrose; otherwise, the observed results will be due to combined effects of sucrose, liberated hexoses, and hydrolysis products of sucrose-derived exopolysaccharides. To assess the global impact of sucrose metabolism on bacterial gene regulation, we applied next-generation RNA sequencing technology (RNA-Seq) (24) to mRNAs from selected sucrase-deficient mutants of S. mutans (20). The exceptional depth and resolution of RNA-Seq allowed for a comprehensive examination of sucrose and the sucrose-PTS in the transcriptome.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Streptococcus mutans strains were maintained on brain heart infusion (BHI; Difco Laboratories, Detroit, MI) agar plates. To select for transformants bearing antibiotic resistance gene cassettes, antibiotics were added to the agar plates at the following concentrations: erythromycin, 10 μg/ml; kanamycin, 1 mg/ml; and spectinomycin, 1 mg/ml. Bacterial cells used for extraction of RNA were grown in tryptone (3%)-yeast extract (0.1%) (TY) medium supplemented with 0.5% glucose, fructose, or sucrose (Thermo Fisher Scientific, Waltham, MA). Bacterial cultures were incubated aerobically without agitation in the presence of 5% CO2 at 37°C until they reached mid-exponential phase (optical density at 600 nm [OD600] ≈ 0.5) and were then harvested by centrifugation at 4°C for 5 min, treated with RNAprotect bacterial reagent (Qiagen, Germantown, MD), and immediately stored at −80°C. To explore whether cells expelled internalized carbohydrate, bacterial strains were grown to mid-exponential phase in tryptone-vitamin (TV) medium (25) supplemented with 0.5% galactose, and then various amounts of sucrose were added. Cells were incubated for 30 min before the supernatants of the cultures were collected by centrifugation and filter sterilized through a 0.22-μm filter, and the culture supernatants were assayed for the presence of fructose by use of a fructose assay kit (Sigma, St. Louis, MO).
Standard molecular cloning techniques (26) and transformation assays established for S. mutans (27) were utilized to create all the strains required in this study (Table 1). To construct mutants deficient in extracellular sucrolytic activities, strain MMZ950 (gtfBC::Tc gtfDM9stop ftfM9stop) (20) was used in a transformation assay as the recipient of the suicide plasmid pBGE, which replaced the gtfA gene with an erythromycin resistance marker, and a PCR product that replaced the fruAB coding sequence with a nonpolar kanamycin marker. The resultant strain, MMZ1009 (gtfABCD ftf fruAB), was validated by allele-specific PCR (28) for point mutations in gtfD and ftf that were established previously through site-directed mutagenesis (20). Another mutant, designated MMZ1170, which lacks the scrA gene in addition to harboring all the mutations of strain MMZ1009, was constructed by transforming strain MMZ952 (gtfA::Em gtfBCD ftf fruAM12stop) (20) simultaneously with two PCR products: one containing an scrA::Km allelic replacement marker (conferring kanamycin resistance) and the other containing a fruBM1stop mutation that replaced the start codon of fruB with a stop codon, as detailed elsewhere (20, 22). While the scrA::Km fragment was amplified using strain MMZ993 (20) as a template, the fruBM1stop-containing fragment was generated via recombinant PCR using the following four primers: fruB-5′, fruBM1stop-3′, fruBM1stop-5′, and fruBdelta113-3′ (see Table S4 in the supplemental material) (22). Similarly, all point mutations (in gtfD, ftf, and fruB) in strain MMZ1170 were confirmed by allele-specific PCR followed by sequencing. Lastly, a PCR product containing a manL::Sp replacement marker (conferring spectinomycin resistance) (29) was introduced into strain MMZ1009 via transformation, resulting in strain MMZ1196 (gtfABCD ftf fruAB manL).
TABLE 1.
Bacterial strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| UA159 | Wild type | University of Alabama |
| MMZ950 | gtfBC::Tc gtfDM9stop ftfM9stop | 20 |
| MMZ952 | gtfA::Em gtfBCD ftf fruAM12stop | MMZ950 |
| MMZ993 | gtfABCD ftf fruA scrA::Km | MMZ952 |
| MMZ998 | gtfABCD ftf fruA PlevD-cat | MMZ952 |
| MMZ1002 | gtfABCD ftf fruA scrA PlevD-cat | 20 |
| MMZ1009 | gtfA::Em gtfBCD ftf fruAB::Km | MMZ950 |
| MMZ1170 | gtfABCD ftf fruA fruBM1stop scrA::Km | MMZ952 |
| MMZ931 | scrB::Km | 20 |
| MMZ932 | scrA::Em | 20 |
| MMZ1196 | gtfABCD ftf fruAB manL::Sp | MMZ1009 |
RNA extraction and deep sequencing (RNA-Seq).
Total RNA was extracted from bacterial cells by use of an RNeasy minikit (Qiagen) according to protocols described elsewhere (30). To remove 16S and 23S rRNAs, 10 μg of total RNA was first treated twice by use of a MICROBExpress bacterial mRNA enrichment kit (Ambion Life Technologies, Grand Island, NY), followed by ethanol precipitation. The final product was resuspended in 25 μl of nuclease-free water. The quality of enriched mRNA samples was analyzed using an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA).
cDNA libraries were created from the enriched mRNA samples by using an NEBNext Ultra directional RNA library prep kit for Illumina and NEBNext multiplex oligonucleotides for Illumina (New England BioLabs, Ipswich, MA), following the instructions from the supplier. Deep sequencing was performed by the NextGen DNA Sequencing Core Laboratory of ICBR at the University of Florida (Gainesville, FL). Read mapping was performed on a Galaxy server hosted by the research computing center at the University of Florida, using Map with Bowtie for Illumina (version 1.1.2). Mapped reads were then counted using htseq-count for both sense and antisense transcripts. A detailed description of the entire procedure and parameters employed is provided in Text S1 in the supplemental material.
Differential expression analysis.
Data containing read counts for each gene, excluding all rRNA and tRNA genes, were uploaded into the Web-based analysis tool DEB (31), and then Degust (32), for differential analysis. Results obtained from both websites were comparable. Since the software hosted at the Degust website allowed for simultaneous comparison of data from more than two different experiments, the results obtained by the Degust method for cells grown under glucose, sucrose, and fructose conditions are presented.
To confirm some of the differential analysis results obtained using RNA-Seq, a conventional real-time quantitative reverse transcription-PCR (qRT-PCR) was employed to measure changes in the mRNA level of each ORF. For first-strand cDNA synthesis, 1 μg of total RNA without rRNA depletion treatment was used in reverse transcription reaction mixtures in the presence of random hexamers and SuperScript III reverse transcriptase (Thermo Fisher). Subsequently, gene-specific primers for each target ORF were used in quantitative PCRs performed using a CFX96 Touch real-time PCR detection system (Bio-Rad, Hercules, CA) following the instructions from the supplier. A housekeeping gene (gyrA) was used as an internal control for calculating fold changes in cDNA levels according to the ΔΔCq method (33). The sequences of all primers used for real-time PCRs are listed in Table S4 in the supplemental material.
RESULTS
Expulsion of fructose by S. mutans exposed to sucrose.
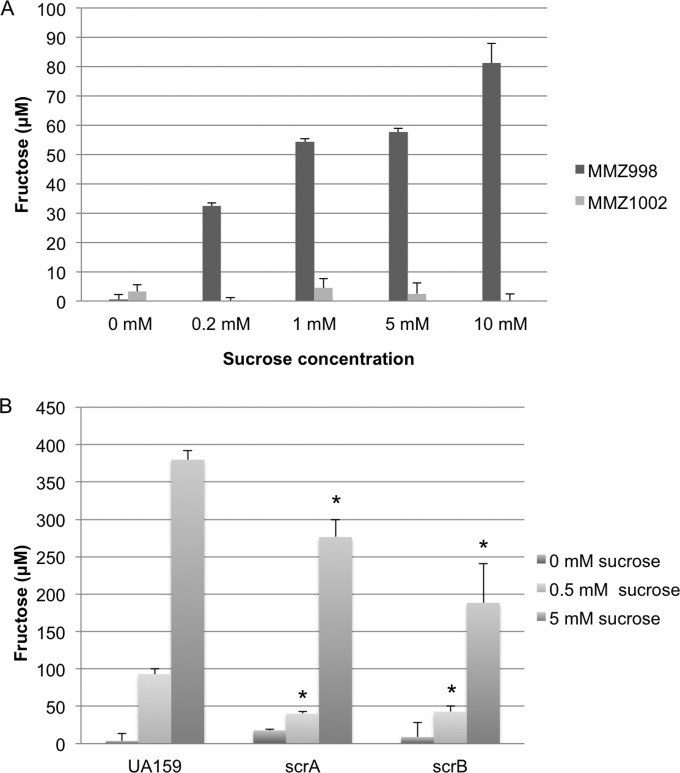
We previously demonstrated the ability of sucrose to activate a promoter-reporter fusion in S. mutans, constructed using the levD promoter and a promoterless cat gene in a strain that lacked the extracellular sucrases GtfBCD, Ftf, and FruA (20). The levDEFG genes encode the A, B, C, and D domains of a fructose/mannose-type enzyme II (EIILev) of the PTS. Interestingly, activation of the levD promoter required the presence of an intact LevQRST four-component system that was previously shown to activate levD in response to fructose (21, 22); sucrose-mediated activation of levD also required the presence of an intact scrA gene, encoding EIIScr (20). Since the recombinant strain MMZ998 (gtfABCD ftf fruA) used in these assays lacked all secreted sucrases, we postulated that free fructose may be released into the medium as a result of hydrolysis by ScrB (S-6-P hydrolase), as fructose has been known to activate levD via the LevQRST and other fructose-specific pathways (23). Here we measured free fructose levels in culture supernatants of strain MMZ998 and its derivative MMZ1002, which is otherwise isogenic to MMZ998 but lacks the scrA gene, after adding various concentrations of sucrose to cells growing exponentially in TV medium containing galactose. As shown in Fig. 1A, 30 min after the addition of sucrose, free fructose was detected in MMZ998 culture supernatants, at concentrations ranging from 30 to 80 μM that directly correlated with the amount of sucrose added to the cultures. Similar levels of fructose have been shown to be sufficient to activate the LevQRST pathway and expression of the genes for EIILev in S. mutans (21). In contrast, no fructose was detected in the MMZ1002 cultures as a result of pulsing with sucrose. Furthermore, similar results were obtained when the experiment was repeated using strains bearing similar sucrase gene deletions but lacking the reporter gene fusion present in MMZ998 and MMZ1002 (MMZ952 and MMZ993, respectively) (Table 1; see Fig. S1 in the supplemental material); thus, the presence of the reporter gene did not influence the results. Therefore, it appears that free fructose is released into the extracellular environment by S. mutans exposed to sucrose and that the sucrose must be internalized by the sucrose-PTS. Since the strains we tested have no other means of metabolizing sucrose (strains MMZ993 and MMZ1002 show drastically reduced growth on sucrose-based media, and further deletion of scrB completely abolishes growth) (20), we propose that PTS-mediated internalization of sucrose, subsequent hydrolysis by S-6-P hydrolase (ScrB) to yield G-6-P and fructose, and expulsion of fructose into the supernatant fluid comprise the source of fructose.
FIG 1.
Sucrose pulsing leads to release of fructose by S. mutans in an ScrA-dependent fashion. (A) Strains MMZ998 (gtfABCD ftf fruA; dark gray bars) and MMZ1002 (gtfABCD ftf fruA scrA; light gray bars) were cultivated in TV medium supplemented with 0.5% galactose to the exponential growth phase, followed by pulsing for 30 min with sucrose at final concentrations ranging from 0 to 10 mM. Supernatants of the bacterial cultures were collected, and free fructose was measured. Results are the averages for three independent cultures, with error bars representing the standard deviations. (B) Strain UA159 and its scrA and scrB mutants were similarly pulsed with a final concentration of 0, 0.5, or 5 mM sucrose before measurement of fructose in supernatants. Asterisks indicate statistically significant differences in relation to the parental strain UA159, according to Student's t test (P < 0.005).
We also performed the same sucrose-pulsing assays using the wild-type strain UA159 and its otherwise isogenic mutants lacking scrA or scrB. This was done to ensure that the behavior of MMZ993 and MMZ1002 was not an anomaly associated with the presence of multiple mutations in the sucrase genes. As shown in Fig. 1B, treatment with increasing concentrations of sucrose led to the release of larger amounts of fructose into the culture supernatants by all three strains than by MMZ998. Importantly, when strains were treated with the same amounts of sucrose, the ScrA- and ScrB-deficient strains released far lower levels of fructose than did the parental strain. Therefore, it appears that when S. mutans is presented with sucrose under the conditions tested, the sucrose-PTS rapidly internalizes the sucrose, the S-6-P is cleaved by ScrB, and at least some of the fructose that is liberated by ScrB is released from the cells. The knowledge that fructose would be present in the environment under these conditions was therefore factored into the design of the RNA-Seq experiments.
RNA-Seq to identify genes regulated by ScrA-mediated sucrose metabolism.
We have established that fruA and fruB are cotranscribed and that FruB is secreted into the supernatant fluid (data not shown). FruB is annotated as a potential β-fructosidase, although we have been unable to detect any activity of this protein on sucrose or fructan polymers (our unpublished data). However, there is low but significant sequence similarity between FruB and ScrB, and the best alignments of FruB in BLAST searches are with β-fructosidases. For the purpose of studying genes and enzymatic pathways that are directly affected by ScrA-mediated sucrose metabolism, we created a new mutant (MMZ1009) (Table 1) that lacks fruB, in addition to other known sucrase-encoding genes (gtfABCD, ftf, and fruA). MMZ1009 was cultivated in tryptone broth containing 0.1% yeast extract (TY) supplemented with 0.5% glucose or sucrose as the carbohydrate source, and cells were harvested at exponential phase (OD600 = 0.5). S. mutans MMZ1009 was also cultured in TY-fructose to allow for detection of genes that were differentially expressed as a result of fructose released into the medium from the ScrAB pathway. Total RNA was extracted from cells and treated with MICROBExpress to deplete rRNAs, and cDNA libraries were then constructed and subjected to high-throughput sequencing (see Materials and Methods). To further elucidate the influence of ScrA on regulation of gene expression, an scrA mutation was introduced into the background of MMZ1009 to create strain MMZ1170 (Table 1). Since MMZ1170 lacks the ability to grow efficiently on sucrose-based medium, RNA-Seq was performed only on MMZ1170 grown on glucose. As an additional control, in recognition that trace amounts of glucose present in medium components might have an impact on the results, a mutation of the major glucose-PTS permease EIIMan (by deleting manL) was established in the background of MMZ1009, creating strain MMZ1196. RNA-Seq data obtained for MMZ1196 growing in TY-glucose or TY-sucrose, together with those for MMZ1170, were then used to help interrogate the transcriptomic data for MMZ1009 growing in glucose or sucrose.
The cutoff for designating a gene as being differentially expressed was a change in mRNA level of at least 1.5-fold and a false discovery rate (FDR) of <0.001 to generate the lists of genes in Table 2 (fold changes of >2) and in Table S1 in the supplemental material (fold changes of >1.5). Overall, a substantial portion of the MMZ1009 transcriptome was significantly altered by the presence of sucrose compared with the presence of glucose (Table 2; see Table S1). Importantly, the presence or absence of scrA (MMZ1009 versus MMZ1170) did not influence gene expression when the strains were cultivated in medium with glucose as the primary carbohydrate source. On the other hand, loss of manL (EIIABMan) resulted in significant changes in the transcriptome when cells were grown on glucose but had almost no impact on gene expression patterns in cells grown on sucrose (see Tables S2 and S3). This important finding indicates that there is essentially no overlap in the scrA and manL regulons. The following three sections focus on differential gene regulation in strain MMZ1009, with carbohydrate as the environmental variable of interest (Table 2; see Table S1). Focusing on genes showing the highest levels of differential expression and those that are critical to interpretation of the transcriptomic data, a subset of these results was confirmed using qRT-PCR (Table 3).
TABLE 2.
Genes differentially expressed in MMZ1009 (fold changes of >2) growing in TY medium supplemented with glucose (Glc), sucrose (Scr), or fructose (Fru)
| Gene ID | Log2 (Scr/Glc) | Log2 (Fru/Glc) | FDR | Gene product description |
|---|---|---|---|---|
| SMU.29 | −1.04 | −0.81 | 6.11E−04 | Putative SAICAR synthase |
| SMU.31 | −1.00 | −0.72 | 7.76E−04 | Hypothetical protein |
| SMU.84 | 0.34 | 1.31 | 7.67E−06 | Putative tRNA pseudouridine synthase A TruA |
| SMU.85 | 0.30 | 1.26 | 3.20E−05 | Putative phosphomethylpyrimidine kinase ThiD |
| SMU.86 | 0.21 | 1.32 | 2.70E−04 | Hypothetical membrane protein |
| SMU.87 | 0.26 | 1.22 | 5.24E−05 | Hypothetical protein |
| SMU.100 | 0.73 | 1.88 | 8.31E−04 | PTS, nigerose-specific enzyme IIB NigB |
| SMU.102 | 0.92 | 1.73 | 3.18E−06 | PTS, nigerose-specific enzyme IID NigD |
| SMU.104 | 0.76 | 1.60 | 7.26E−06 | Putative alpha-glucosidase NigE |
| SMU.125 | 0.21 | 1.10 | 1.48E−05 | Hypothetical protein |
| SMU.148 | −1.94 | −2.60 | 4.21E−06 | Putative alcohol-acetaldehyde dehydrogenase AdhE |
| SMU.149 | −1.17 | −1.92 | 7.92E−04 | Putative transposase |
| SMU.179 | −1.27 | −1.52 | 1.43E−05 | Hypothetical protein |
| SMU.180 | −1.62 | −1.80 | 4.36E−07 | Putative oxidoreductase IlvC |
| SMU.402 | −2.06 | −2.89 | 1.55E−06 | Pyruvate formate-lyase Pfl |
| SMU.493 | −0.23 | −1.44 | 1.88E−04 | Formate acetyltransferase (pyruvate formate-lyase 2) |
| SMU.495 | −0.24 | −1.24 | 2.33E−04 | Glycerol dehydrogenase GldA |
| SMU.498 | 1.79 | −0.35 | 9.31E−04 | Putative late competence protein ComF |
| SMU.500 | −0.62 | −1.23 | 1.13E−06 | Putative ribosome-associated protein |
| SMU.510c | 0.57 | 1.30 | 6.40E−05 | Hypothetical protein |
| SMU.564 | −0.39 | −1.03 | 4.87E−04 | Hypothetical protein |
| SMU.600c | 0.92 | 1.03 | 1.12E−04 | Hypothetical protein |
| SMU.602 | 1.14 | 0.68 | 1.58E−04 | Putative sodium-dependent transporter |
| SMU.609 | 0.06 | 1.18 | 8.68E−05 | Putative 40K cell wall protein precursor |
| SMU.625 | 2.33 | 0.95 | 1.78E−04 | Putative competence protein ComEA |
| SMU.626 | 2.53 | 0.46 | 3.90E−04 | Putative competence protein ComEC |
| SMU.633 | 0.15 | 1.92 | 1.27E−07 | Putative thioesterase |
| SMU.870 | 1.93 | 1.89 | 2.96E−08 | Transcription regulator for fructose metabolism FruR |
| SMU.871 | 2.02 | 1.97 | 6.93E−09 | Fructose-1-phosphate kinase FruK |
| SMU.872 | 1.90 | 1.92 | 5.15E−08 | PTS, fructose-specific enzyme IIABC FruI |
| SMU.877 | −1.17 | −1.64 | 2.67E−07 | Alpha-galactosidase AgaL |
| SMU.878 | −1.22 | −1.77 | 1.37E−06 | Multiple-sugar-binding ABC transporter MsmE |
| SMU.879 | −1.27 | −1.90 | 3.71E−06 | Multiple-sugar-binding ABC transporter MsmF |
| SMU.880 | −1.30 | −1.84 | 9.72E−06 | Multiple-sugar-binding ABC transporter MsmG |
| SMU.881 | −1.29 | −1.68 | 3.11E−07 | Sucrose phosphorylase GtfA |
| SMU.882 | −1.15 | −1.60 | 1.43E−05 | Multiple-sugar-binding ABC transporter MsmK |
| SMU.883 | −1.10 | −1.34 | 6.13E−07 | Dextran glucosidase DexB |
| SMU.913 | 0.97 | 1.38 | 3.20E−05 | Putative NADP-specific glutamate dehydrogenase |
| SMU.921 | 0.07 | 1.05 | 7.92E−04 | MarR-type transcriptional regulator RcrR |
| SMU.925 | −0.07 | 1.03 | 5.13E−04 | Hypothetical protein |
| SMU.935 | 0.58 | −2.30 | 6.13E−04 | Putative amino acid ABC transporter |
| SMU.940c | −0.55 | −3.33 | 1.65E−08 | Putative hemolysin III |
| SMU.941c | −0.45 | −3.36 | 6.91E−08 | Hypothetical protein |
| SMU.954 | 0.14 | 1.73 | 2.37E−05 | Putative pyridoxal kinase |
| SMU.955 | 0.34 | 1.50 | 7.92E−04 | Hypothetical membrane protein |
| SMU.997 | −0.07 | 1.18 | 2.26E−04 | Putative inorganic ion ABC transporter |
| SMU.1077 | −1.10 | −1.30 | 8.26E−06 | Phosphoglucomutase Pgm |
| SMU.1175 | 1.68 | 2.35 | 7.67E−06 | Putative sodium/amino acid (alanine) symporter |
| SMU.1339 | 0.05 | −1.48 | 5.19E−04 | Bacitracin synthetase BacD |
| SMU.1340 | 0.06 | −1.64 | 4.13E−04 | Putative surfactin synthetase BacA2 |
| SMU.1341c | 0.03 | −1.49 | 1.58E−04 | Putative gramicidin S synthetase |
| SMU.1342 | −0.01 | −1.55 | 2.18E−04 | Putative bacitracin synthetase 1 BacA1 |
| SMU.1343c | 0.04 | −1.43 | 3.97E−05 | Putative polyketide synthase |
| SMU.1398 | −0.65 | −1.13 | 1.20E−04 | Transcriptional regulator IrvR |
| SMU.1410 | −1.53 | −2.49 | 2.11E−04 | Putative reductase |
| SMU.1411 | −1.55 | −2.36 | 1.84E−05 | Hypothetical protein |
| SMU.1535 | −0.97 | −1.15 | 1.91E−06 | Glycogen phosphorylase PhsG |
| SMU.1536 | −1.09 | −1.45 | 6.88E−07 | Bacterial glycogen synthase GlgA |
| SMU.1537 | −1.20 | −1.48 | 9.79E−06 | Putative glycogen biosynthesis protein GlgD |
| SMU.1538 | −1.04 | −1.42 | 3.06E−06 | Glucose-1-phosphate adenylyltransferase GlgC |
| SMU.1539 | −0.92 | −1.20 | 1.30E−05 | 1,4-Alpha-glucan branching enzyme GlgB |
| SMU.1550c | 0.21 | 1.01 | 3.21E−04 | Hypothetical membrane protein |
| SMU.1551c | 0.22 | 1.00 | 5.06E−05 | Putative ABC transporter |
| SMU.1552c | 0.23 | 1.10 | 2.33E−04 | Hypothetical protein |
| SMU.1564 | −0.80 | −1.25 | 9.72E−06 | Glycogen phosphorylase GlgP |
| SMU.1565 | −0.54 | −1.26 | 5.19E−04 | Putative 4-alpha-glucanotransferase MalQ |
| SMU.1568 | −0.86 | −1.16 | 2.26E−04 | Maltose/maltodextrin ABC transporter MalX |
| SMU.1569 | −0.83 | −1.21 | 3.12E−04 | Maltose/maltodextrin ABC transporter MalF |
| SMU.1570 | −0.96 | −1.28 | 1.05E−04 | Maltose/maltodextrin ABC transporter MalG |
| SMU.1571 | −0.98 | −1.24 | 3.83E−04 | Putative ABC transporter MsmK-like protein |
| SMU.1700c | 0.99 | 1.81 | 9.79E−06 | CidB |
| SMU.1702c | 0.03 | 2.67 | 6.63E−09 | Putative phosphatase |
| SMU.1703c | −0.18 | 1.49 | 4.94E−05 | Hypothetical membrane protein |
| SMU.1927 | −0.88 | −1.19 | 1.20E−04 | Putative ABC transporter |
| SMU.1928 | −0.96 | −1.27 | 1.89E−04 | Putative ABC transporter PsaB |
| SMU.1956c | 2.43 | 2.51 | 3.21E−06 | LevX |
| SMU.1957 | 2.42 | 2.50 | 1.65E−08 | PTS, fructose-specific enzyme IID LevG |
| SMU.1958c | 2.43 | 2.68 | 6.91E−08 | PTS, fructose-specific enzyme IIC LevF |
| SMU.1960c | 2.24 | 2.35 | 6.23E−07 | PTS, fructose-specific enzyme IIB LevE |
| SMU.1961c | 2.50 | 2.47 | 3.34E−07 | PTS, fructose-specific enzyme IIA LevD |
| SMU.1980c | 1.45 | 0.18 | 5.81E−04 | Hypothetical protein |
| SMU.1981c | 1.05 | −0.09 | 2.01E−04 | Competence protein ComGF |
| SMU.1982c | 1.23 | −0.07 | 5.34E−04 | Hypothetical protein |
| SMU.1985 | 1.35 | 0.13 | 1.89E−04 | ABC transporter ComYB |
| SMU.1987 | 1.33 | −0.11 | 2.94E−04 | ABC transporter, late competence protein ComYA |
| SMU.2046c | −0.79 | −1.03 | 1.45E−04 | Metal-dependent hydrolase |
| SMU.2047 | −0.82 | −1.12 | 9.79E−06 | PTS, maltose-specific IIABC PtsG |
| SMU.2127 | −1.37 | −1.70 | 4.40E−07 | Putative succinate semialdehyde dehydrogenase |
TABLE 3.
qRT-PCR confirmation of selected genes in strain MMZ1009 growing in TY medium supplemented with glucose (Glc), sucrose (Scr), or fructose (Fru)a
| Gene | Log2 (Scr/Glc) |
Log2 (Fru/Glc) |
||
|---|---|---|---|---|
| RNA-Seq | qRT-PCR | RNA-Seq | qRT-PCR | |
| scrA | 3.38 | 3.78 ± 0.31 | −0.19 | 0.24 ± 0.26 |
| levD | 2.50 | 2.40 ± 0.23 | 2.47 | 0.49 ± 0.14 |
| fruR | 1.93 | 2.37 ± 0.20 | 1.89 | 2.27 ± 0.04 |
| comEA | 2.33 | 2.03 ± 0.95 | 0.95 | 0.64 ± 0.33 |
| comYA | 1.33 | 1.77 ± 0.65 | −0.11 | −0.75 ± 0.30 |
| scrB | 0.59 | 1.16 ± 0.06 | −0.85 | −0.51 ± 0.35 |
| cidB | 0.99 | 1.01 ± 0.24 | 1.81 | 3.04 ± 0.17 |
| SMU.1175 | 1.68 | 0.83 ± 0.23 | 2.35 | 1.55 ± 0.27 |
| SMU.101 | 0.98 | 0.62 ± 0.30 | NS | 1.39 ± 0.25 |
| rcrR | 0.07 | 0.42 ± 0.09 | 1.05 | 1.23 ± 0.28 |
| gtfD | NS | 0.21 ± 0.27 | NS | −0.49 ± 0.22 |
| bacD | 0.05 | 0.09 ± 0.28 | −1.48 | −1.14 ± 0.29 |
| scrK | NS | −0.02 ± 0.19 | NS | 0.35 ± 0.37 |
| SMU.1349 | NS | −0.14 ± 0.11 | NS | 0.51 ± 0.28 |
| SMU.1927 | −0.88 | −0.49 ± 0.24 | −1.19 | −0.72 ± 0.17 |
| ptsH | −0.54 | −0.68 ± 0.25 | −0.48 | −0.61 ± 0.09 |
| ptsI | −0.57 | −0.73 ± 0.09 | NS | −0.55 ± 0.14 |
| ftf | −0.77 | −0.86 ± 0.15 | −0.83 | −0.66 ± 0.35 |
| ptsG | −0.82 | −0.99 ± 0.37 | −1.12 | −1.00 ± 0.04 |
| lytS | −0.57 | −1.04 ± 0.22 | −0.55 | 0.01 ± 0.04 |
| SMU.1077 (pgm) | −1.10 | −1.08 ± 0.07 | −1.30 | −1.35 ± 0.03 |
| purC | −1.04 | −1.21 ± 0.41 | −0.81 | −0.98 ± 0.13 |
| glgD | −1.20 | −1.53 ± 0.56 | −1.48 | −0.96 ± 0.44 |
| SMU.180 (ilvC) | −1.62 | −1.63 ± 0.56 | −1.80 | −1.60 ± 0.24 |
| pfl | −2.06 | −2.11 ± 0.17 | −2.89 | −2.70 ± 0.15 |
Changes in transcript levels were determined using gyrA as an internal control, and qRT-PCR results are presented as averages ± standard deviations. NS, not significant.
(i) Regulation of the scr locus by sucrose.
Previous research suggested that scrA and scrB are likely coregulated by the GalR/LacI-type transcription regulator ScrR, which binds to the operator sequences located between the divergently transcribed scrA and scrBR genes (34). The RNA-Seq results confirmed that transcription of the scrA and scrB genes was significantly higher in cells growing in the presence of sucrose, although the level of increase in scrA transcripts (14.0- ± 3-fold, as measured by qRT-PCR) (Table 3) was larger than that for scrB transcripts (2.2- ± 0.1-fold). Also, the transcript levels of scrB showed an interesting pattern in response to carbohydrates (sucrose > glucose > fructose), similar to what was previously reported (8). Note that there was little change in the expression of the gene encoding an ATP-dependent fructokinase (scrK) (34), which would be predicted to be involved in phosphorylation of the fructose liberated by ScrB and which is situated immediately downstream of scrA, with any of the three carbohydrates tested.
(ii) The presence of sucrose alters expression levels of genes for fructose-PTS and glycolytic enzymes.
As presented above, pulsing of cells containing an intact ScrA protein with sucrose resulted in release of free fructose into the culture medium at concentrations that are able to activate particular genes involved in fructose transport and metabolism. When strain MMZ1009 was treated with sucrose, two fructose-PTS operons, fruRKI and EIILev (levDEFGX), were upregulated. Both of these operons were also upregulated to similar degrees in cells growing on fructose. While both FruI and EIILev are inducible fructose-PTS permeases (21, 35), the expression of EIILev was previously shown to be activated by pulsing with sucrose (20). In addition to the two fructose-PTS permease operons, the expression of a recently characterized operon (SMU.100 to SMU.105) that encodes components of a nigerose-specific EII complex (PTSBio), an α-glucosidase and a putative transcriptional regulator, was elevated in cells growing on sucrose but was induced to an even greater degree in cells growing on fructose.
In contrast, a maltose-specific PTS (ptsG) (36), a maltose/maltodextrin-specific ABC transporter operon (SMU.1564 to SMU.1571), and the multiple-sugar metabolism (msm) pathway genes (SMU.877 to SMU.883) showed decreases in expression levels in cells growing on sucrose or on fructose; the msm operon was also shown to be regulated in this way by use of microarrays (23). Consistent with the absence of glucose in the growth medium, there were significant reductions in expression for genes encoding a glucose kinase (Glk; SMU.542) and a phosphoglucomutase (Pgm; SMU.1077). Transcripts for the general components enzyme I (EI) and HPr of the PTS (ptsI and ptsH), pyruvate-formate lyase (pfl), formate acetyltransferase (pfl2), acetate kinase (ackA), the glg operon gene products (responsible for bacterial glycogen biosynthesis) (SMU.1535 to SMU.1539), and the catabolite control protein (ccpA) were also downregulated. Reduced expression of these genes is perhaps reflective of a significant downshift as well as a change in carbon flux in bacterial glycolysis (24).
(iii) Sucrose and fructose differentially affect expression of genes for competence and stress tolerance.
While the effects of sucrose and fructose on the expression of some of the PTS-encoding operons appear to be similar in both their direction and magnitude, there were several loci in the genome where transcriptional activities were differentially affected by growth on fructose or sucrose. First, in comparison to growth on glucose, growth on sucrose led to increased expression of genes shown to be required for competence development (comYA-comGF operon; SMU.1987 to SMU.1981c) (37), as well as that of comEA (SMU.625/626) and comF (SMU.498) (38). At the same time, fructose-grown cells did not show similar changes in expression of the same group of genes.
Conversely, growth on fructose led to altered expression of several stress-related loci. For example, an operon structure recently indicated to be involved in controlling tolerance of acid and oxidative stress, development of genetic competence, and (p)ppGpp metabolism, namely, rcrRPQ-relPRS (SMU.921 to SMU.928), showed increased expression in cells growing on fructose, but not in cells growing on sucrose, compared to that in glucose-grown cells. The expression of the cidB (SMU.1700c) and SMU.1702c/1703c operons was also notably higher in fructose-grown cells (3-fold for cidB and 6- and 3-fold, respectively, for SMU.1702c/1703c); CidB has been implicated in oxidative stress tolerance, control of autolysis, and biofilm maturation (39). Interestingly, the LytST two-component system, which has been shown to play important roles in oxidative stress tolerance, showed reduced transcript levels in cells growing on sucrose or fructose compared to those in cells growing on glucose. Also, a set of putative bacitracin/surfactin/gramicidin-synthesizing genes, whose expression has been shown to be affected by mutation of lytS, displayed reduced expression only in cells cultured in fructose (40). Lastly, among the hypothetical genes showing the greatest changes in mRNA levels only in cells growing on fructose were SMU.84 to SMU.87, which are situated immediately downstream of the genes encoding two major heat shock proteins (DnaK and DnaJ), putative hemolysin III (SMU.940c; downregulated 10-fold) and hypothetical protein (SMU.941c; downregulated 10-fold) genes, two MarR-type transcription regulator genes (SMU.632 and SMU.124; each upregulated 2-fold), and a putative thioesterase gene (SMU.633; upregulated 4-fold). Enhanced expression of these stress-related genes/operons points to less-than-favorable metabolic conditions associated with growth on fructose, possibly consistent with the active release of fructose by this bacterium when growing on sucrose.
Antisense transcriptome of S. mutans.
The technology used for the construction of cDNA libraries in this study retains only the first strand of cDNA for sequencing, allowing differentiation of RNA strandedness at the step of read counting. This directional approach led to the quantification of antisense RNA transcripts from the genome of S. mutans for the first time. Listed in Table 4 are some of the most abundant antisense RNAs identified in this study. In fact, very few of these RNAs were found to have been differentially expressed in response to the carbohydrate source. One exception was SMU.253 (dacA), encoding a putative d-alanyl–d-alanine carboxypeptidase, which showed a 4-fold decrease (confirmed by RT-PCR) in the antisense transcript in strain MMZ1196 (gtfABCD ftf fruAB manL) growing on sucrose compared with that in the same strain growing on glucose. In contrast, the sense transcript of dacA was present in quantities that were comparable to that of the antisense transcript, and there was no change in the dacA mRNA level as a function of carbohydrate source.
TABLE 4.
Top 50 genes with significant levels of antisense transcript
| Gene ID | Description of gene or gene product | Relative abundancea |
|---|---|---|
| SMU.959c | Hypothetical protein | 123,352 |
| SMU.1907 | Hypothetical protein | 109,215 |
| SMU.847c | Hypothetical protein | 88,958 |
| SMU.1928 | ABC transporter | 28,623 |
| SMU.803c | ABC transporter | 25,019 |
| SMU.1812 | Transposase | 20,845 |
| SMU.1605 | MDR permease | 17,571 |
| SMU.1399 | Hypothetical protein | 13,601 |
| SMU.1678 | Hypothetical protein | 13,449 |
| SMU.590c | Transposase | 12,947 |
| SMU.1898 | sunT ortholog | 11,742 |
| SMU.1592 | pepQ | 10,231 |
| SMU.1677 | murE | 10,033 |
| SMU.640c | GntR family regulator | 9,505 |
| SMU.1831 | aspG | 8,022 |
| SMU.1900 | Hypothetical protein | 7,850 |
| SMU.527 | Hypothetical protein | 7,811 |
| SMU.1078c | ABC transporter | 7,365 |
| SMU.121 | DinF efflux pump | 7,363 |
| SMU.956 | Clp protease | 7,311 |
| SMU.666 | argD | 6,856 |
| SMU.401c | Hypothetical protein | 6,840 |
| SMU.610 | spaP | 6,796 |
| SMU.2081 | Hypothetical protein | 6,718 |
| SMU.412c | Hit-like protein | 6,363 |
| SMU.1748 | akh | 6,312 |
| SMU.1856c | Hypothetical protein | 6,233 |
| SMU.1195 | Possible permease | 6,137 |
| SMU.694c | Putative ferredoxin | 6,099 |
| SMU.910 | gtfD | 6,004 |
| SMU.1357 | Transposase | 5,926 |
| SMU.365 | gltA | 5,919 |
| SMU.493 | pfl2 | 5,790 |
| SMU.1881c | sunT | 5,779 |
| SMU.78 | fruA | 5,748 |
| SMU.1929 | HtpX protease | 5,716 |
| SMU.985 | bglA | 5,575 |
| SMU.1004 | gtfB | 5,508 |
| SMU.1844 | scrR | 5,455 |
| SMU.1005 | gtfC | 5,431 |
| SMU.1927 | ABC transporter | 5,426 |
| SMU.1480 | Hypothetical protein | 5,347 |
| SMU.877 | agaL | 5,320 |
| SMU.2080 | Hypothetical protein | 5,238 |
| SMU.1992 | Tyrosyl-tRNA synthetase | 5,210 |
| SMU.30 | purL | 5,201 |
| SMU.123 | DNA polymerase III, alpha subunit | 5,073 |
| SMU.637c | Hypothetical protein | 5,014 |
| SMU.1155 | Hypothetical protein | 4,977 |
Relative abundances of antisense transcripts were calculated based on counts obtained from three independent samples.
DISCUSSION
A primary challenge for S. mutans, a dental pathogen that relies almost entirely on carbohydrates for energy and survival, is the frequent fluctuation in the sources and availability of sugars in the human oral cavity due to intermittent eating patterns and the diversity in human diets. Thus, sensing the sources and amounts of particular carbohydrates in a rapid manner is essential for persistence and for pathogenesis. We previously identified a four-component system in S. mutans (21) that can sense the presence of specific carbohydrates and alter the expression of at least two operons for carbohydrate transport and catabolism. There are also a number of dedicated transcriptional regulators that regulate the expression of cognate catabolic pathways (e.g., GalR, NagR, ScrR, and LacR) (9, 29, 41, 42). However, the dominant pathway for detecting the presence of particular carbohydrates and for monitoring the flux of carbohydrates through the glycolytic pathway in S. mutans is the sugar-phosphotransferase system (PTS). The most prominent example of PTS-dependent gene regulation in S. mutans is the identification of the regulon controlled by a glucose/mannose-PTS (EIIMan), as previously analyzed using microarrays (43) and confirmed and expanded here using RNA-Seq (see Table S2 in the supplemental material). Studies by our group have also highlighted the critical role that HPr has on gene regulation, often in cooperation with EIIABMan (44). In contrast, the major catabolite control protein (CcpA) that regulates carbon catabolite repression (CCR) in many low-G+C Gram-positive organisms plays a very minor role in regulating CCR in S. mutans (45, 46). The fact that S. mutans has evolved in a way that places PTS permeases in a dominant position to influence gene expression and virulence traits makes it critical to delineate the extent to which the presence of certain carbohydrates affects gene expression at the global transcriptomic level. Utilizing mainly one mutant strain, MMZ1009, that lacks all known secreted sucrolytic enzymes (GtfABCD, Ftf, and FruAB), we have been able to probe changes in the transcriptome of S. mutans in great depth and with great sensitivity as a function of exposure to glucose, sucrose, and fructose. These experiments have facilitated the first description of genes that are specifically regulated in response to the presence of sucrose versus hexoses or degradation products of the exopolymers produced by this bacterium.
One intriguing finding from this study was the detection of free fructose in the culture supernatants of S. mutans MMZ998 (Fig. 1) (and, similarly, MMZ952) when cells were exposed to sucrose, since this strain lacks all extracellular sucrolytic activities. Importantly, fructose was not detectable in strain MMZ1002, which is isogenic to MMZ998 except for the lack of the sucrose-PTS permease, ScrA. The simplest explanation for these observations is that sucrose is internalized by S. mutans and cleaved to G-6-P and fructose by ScrB, but then fructose is expelled from the cells. Not only was fructose detected enzymatically in culture supernatants, but it was also shown that the fructose expelled into the environment could activate the targets of the LevQRST four-component system, which responds to fructose or mannose, in MMZ998 but not in the MMZ1002 strain that lacks ScrA (20). The phenomenon of exporting internalized carbohydrates is often referred to as inducer expulsion, as the metabolite capable of inducing the metabolic pathway is actively transported to the extracellular environment (47). However, in this case, the released fructose actually leads to induction of the PTS permeases (EIILev and FruI) (Table 2) by interacting either extracellularly with the LevQRST apparatus (responsible for EIILev) (22) or intracellularly with the GalR/LacI-type regulator FruR (for fruRKI) (our unpublished data). One possible explanation for this behavior could be that the cells derive some bioenergetic advantage. In particular, introduction of fructose into the glycolytic pathway by the fructokinase ScrK requires ATP (34). If instead the cells expel fructose that is derived intracellularly from S-6-P and reinternalize it via the PTS, it will be phosphorylated at the expense of phosphoenolpyruvate (PEP) by the PTS, allowing cells to conserve ATP for other reactions. Consistent with this interpretation, our RNA-Seq analyses of strain MMZ1009, which is similar to MMZ998 in that it lacks all secreted sucrases, showed that while scrA and scrB were among the genes that were most upregulated in response to sucrose, the transcript levels of scrK remained constant (Table 3). Earlier in vitro work also indicated that the fructokinase activity of the ScrK protein is only slightly activated by fructose (34). During the preparation of this communication, a new study reported the involvement of the glucose-PTS in maltose metabolism by S. mutans, with evidence suggesting a similar phenomenon: active release of glucose after internalization and hydrolysis of maltose and other maltooligosaccharides (48). Together, these studies suggest that sugar expulsion is a more frequent phenomenon in S. mutans than previously recognized.
Primarily as a control experiment to help distinguish the effects on the transcriptome of fructose expulsion from internalization of sucrose via ScrA from changes in gene expression elicited via sucrose itself, we carried out RNA-Seq analysis on MMZ1009 cultivated with fructose as the primary carbohydrate source. In comparison to the presence of glucose, the presence of fructose not only led to upregulation of the fructose-PTS operons, including the fruRKI and levDEFGX operons, but also, surprisingly, increased expression of the recently characterized rcrRPQ and relPRS operons and the cidAB genes. The relPRS locus encodes a (p)ppGpp synthase (RelP) and a two-component system (RelRS) that appears to positively regulate relP transcription; other potential targets of RelRS have not been investigated. RelP is the primary contributor to (p)ppGpp pools during exponential growth and appears to downregulate growth to avoid cellular damage from environmental stress or internally generated stressors (49, 50). Similarly, the product of the cidB gene, a bacteriophage holin-like protein, has a strong influence on bacterial growth under conditions of oxidative stress (39). Importantly, work conducted by colleagues in our laboratory demonstrated that RcrR, a MarR-type (multiple-antibiotic resistance) transcription regulator (51, 52), along with two putative ABC transporters (RcrPQ) and two small peptides encoded immediately downstream of rcrQ (53), plays important roles in bacterial stress tolerance by serving as molecular connections of (p)ppGpp metabolism, acid and oxidative stress tolerance, and genetic competence. Lastly, a set of genes (SMU.84 to SMU.87) located immediately downstream of the Hsp70 heat shock protein-encoding operon (dnaK) and the genes for two putative MarR-type regulators (SMU.632 and SMU.124) also showed increased expression in fructose-grown cells. The finding that fructose alone can affect the expression of these particular genes highlights an important and previously undisclosed interrelationship of carbohydrate source with stress response pathways and the optimization of growth parameters via (p)ppGpp production, which itself broadly influences gene expression (50, 54). Why fructose in particular elicits these effects, more so than sucrose or glucose, is not entirely clear. However, given the dramatic increases in the use of fructose as a sweetening agent in foodstuffs, these observations on the connections between fructose and stress response pathways warrant further investigation.
In addition to the impact on fructose-PTS operons mentioned above, there are other similarities in the transcriptomic profiles between sucrose- and fructose-grown cells, notably the downregulation of the msm pathway, the glycogen biosynthetic operon, and the central glycolytic enzyme genes ptsH, ptsI, pfl, and ackA (Table 3). Downregulation of some of these genes may be associated with reductions in HPr levels in sucrose- or fructose-grown cells or with changes in the levels of glycolytic intermediates, such as fructose-1,6-bisphosphate. Specifically, downregulation of pfl and ackA could indicate reduction in a pathway of pyruvate metabolism that results in acetate production and ATP. These results are consistent with the fact that the dominant pathway for pyruvate metabolism by S. mutans growing with relatively high concentrations of carbohydrates, such as those used for these experiments, is via lactate dehydrogenase (LDH) and that LDH is allosterically activated by the increase in the levels of fructose-1,6-bisphosphate. Similarly, Pfl and the acetate kinase pathway tend to dominate when cells are growing under carbohydrate-limited conditions (55). Another notable example related to the similarity between sucrose- and fructose-specific transcriptomes is the upregulation of the nigerose-PTS operon (EIIBio) compared to its expression in cells growing on glucose. The EIIBio operon was recently identified as a sucrose-inducible pathway that is responsible for transport and metabolism of the α-1,3-glucoside nigerose (56). While sucrose-mediated induction of EIIBio was confirmed here, its upregulation in fructose medium was unexpected, as it was not reported to be induced in a previous study (56). The differences in our results and the prior report could be due in part to the fact that the original study was carried out using strain UA159 in biofilms, whereas planktonic cells were utilized for the present study. If biofilm growth causes a differential response to fructose, this too is interesting and raises the possibility that sensing of cell density through intercellular communication pathways that involve peptides (57, 58) or other pathways (59) can be integrated with catabolism of specific carbohydrates.
Along these lines, there is clear evidence from this study that carbohydrate sources can have a strong influence on gene networks that are governed by intercellular signal molecules. In particular, some of the more dramatic differences in gene expression patterns between sucrose- and fructose-grown MMZ1009 cells were noted in genes associated with the development of genetic competence and uptake of exogenous DNA. Natural genetic competence occurs transiently in early-exponential-phase cultures of S. mutans, and only in a subpopulation of cells (60). Competence development, which involves peptide signals for activation, is also sensitive to other environmental conditions, e.g., pH (61). Growth of MMZ1009 on sucrose, but not on fructose or glucose, caused increased expression of several competence-associated genes, mostly late competence genes, such as the comY operon, which encodes proteins required for uptake and processing of DNA (37). Other investigators have shown that competence can be enhanced when cells are cultivated in biofilms, and these studies often included sucrose in the growth medium (57). The interpretation of enhanced competence in biofilms has been that the diffusion of signal peptides is limited, so the effective concentration of the inducing signals is increased. However, the results presented here can be interpreted to mean that in an environment where sucrose is present, bacteria may have a greater chance of acquiring foreign DNA as a nutrient source or to enhance genome diversification (62). The effects of carbohydrate on competence signaling and genetic transformation are under investigation in our laboratory.
Despite the fact that strain MMZ1009 lacks most of the extracellular sucrase genes, including gtfBCD and ftf, it is possible to extract some information about the expression of secreted sucrases from the RNA-Seq data. Specifically, the gtfD mutation in MMZ1009 was created by mutating the codon for the 9th methionine to a TAG stop codon, presumably without affecting promoter activity or the stability of the transcript; this was also the case for ftf. The gtfBC genes were inactivated by allelic exchange with a tetracycline (Tc) resistance marker that replaced some of each coding sequence. However, since the first 708 bp of the gtfB sequence and the last 1.26 kbp of the gtfC gene were intact, it was possible to make comparisons of the expression levels of these genes in response to carbohydrate. We do recognize, however, that other inputs regulate these genes in wild-type S. mutans exposed to sucrose, as reported elsewhere (63–65). Consistent with previous results (64), there were no changes detected in gtfD gene expression in response to the carbohydrates tested. On the other hand, the transcript levels of ftf were slightly downregulated in cells growing on sucrose or fructose relative to that in cells growing on glucose, in contrast to reports that showed an enhancement of ftf expression when sucrose was present (65). Also, read counts for the first 708 bp of the gtfB mRNA and the last 1.26 kbp of gtfC remained similar under all conditions tested (see Fig. S2 in the supplemental material for details), which is different from previous findings indicating enhanced expression of gtfB by the presence of sucrose (64, 65). By acting on sucrose, the Gtfs and Ftf have the capacity to influence bacterial gene expression through production of exopolysaccharides and the effects of these polymers on interbacterial interactions and signal sensing, by releasing free hexoses, and by generating products that can be acted on by dextranase and fructanase, which in turn can act as inducers or repressors of gene expression. For example, adhesive glucans have been shown to significantly affect local biofilm structure and pH homeostasis (66), potentially affecting bacterial gene regulation mediated by diffusible peptides or cell density (57, 59); each of these outcomes is capable of significantly altering the bacterial transcriptome, including the levels of gtfBC and ftf transcripts. Overall, though, the loss of the exoenzymes, but not the transcripts for these enzymes, in our sucrase-deficient mutants enabled us to discriminate direct effects on the expression of these genes associated with the presence and metabolism of sucrose from secondary effects associated with polysaccharide synthesis and hydrolysis. Collectively, the findings support the idea that sucrose or fructose does not in fact lead to induction or repression of the gtf or ftf genes, with the caveat that the proteins themselves must not be able to regulate their own transcription.
Past research on S. mutans has revealed profound impacts by sucrose and other carbohydrates on the physiology and genetics of this opportunistic pathogen. Using a batch culture model and a genetic background that excluded the influence of the by-products of sucrose metabolism, our transcriptomic analysis helped to address some of the fundamental questions regarding the role of sugars, and sucrose in particular, in the behavior of a primary dental pathogen. The findings point to an even broader impact of carbohydrates than previously appreciated, affecting not only energy metabolism and acid production but potentially also stress tolerance, cell-to-cell communication, and bacterial evolution by lateral gene transfer.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02681-15.
REFERENCES
- 1.Bowen WH, Koo H. 2011. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res 45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamashita Y, Bowen WH, Burne RA, Kuramitsu HK. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect Immun 61:3811–3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiroza T, Kuramitsu HK. 1988. Sequence analysis of the Streptococcus mutans fructosyltransferase gene and flanking regions. J Bacteriol 170:810–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burne RA, Chen YY, Wexler DL, Kuramitsu H, Bowen WH. 1996. Cariogenicity of Streptococcus mutans strains with defects in fructan metabolism assessed in a program-fed specific-pathogen-free rat model. J Dent Res 75:1572–1577. doi: 10.1177/00220345960750080801. [DOI] [PubMed] [Google Scholar]
- 5.Burne RA, Schilling K, Bowen WH, Yasbin RE. 1987. Expression, purification, and characterization of an exo-β-d-fructosidase of Streptococcus mutans. J Bacteriol 169:4507–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato Y, Poy F, Jacobson GR, Kuramitsu HK. 1989. Characterization and sequence analysis of the scrA gene encoding enzyme IIScr of the Streptococcus mutans phosphoenolpyruvate-dependent sucrose phosphotransferase system. J Bacteriol 171:263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Postma PW, Lengeler JW, Jacobson GR. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev 57:543–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiratsuka K, Wang B, Sato Y, Kuramitsu H. 1998. Regulation of sucrose-6-phosphate hydrolase activity in Streptococcus mutans: characterization of the scrR gene. Infect Immun 66:3736–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang B, Kuramitsu HK. 2003. Control of enzyme IIScr and sucrose-6-phosphate hydrolase activities in Streptococcus mutans by transcriptional repressor ScrR binding to the cis-active determinants of the scr regulon. J Bacteriol 185:5791–5799. doi: 10.1128/JB.185.19.5791-5799.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell RR, Aduse-Opoku J, Sutcliffe IC, Tao L, Ferretti JJ. 1992. A binding protein-dependent transport system in Streptococcus mutans responsible for multiple sugar metabolism. J Biol Chem 267:4631–4637. [PubMed] [Google Scholar]
- 11.Tao L, Sutcliffe IC, Russell RR, Ferretti JJ. 1993. Transport of sugars, including sucrose, by the msm transport system of Streptococcus mutans. J Dent Res 72:1386–1390. doi: 10.1177/00220345930720100701. [DOI] [PubMed] [Google Scholar]
- 12.Poy F, Jacobson GR. 1990. Evidence that a low-affinity sucrose phosphotransferase activity in Streptococcus mutans GS-5 is a high-affinity trehalose uptake system. Infect Immun 58:1479–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burt BA. 1993. Relative consumption of sucrose and other sugars: has it been a factor in reduced caries experience? Caries Res 27(Suppl 1):S56–S63. [DOI] [PubMed] [Google Scholar]
- 14.Sheiham A. 1983. Sugars and dental decay. Lancet i:282–284. [DOI] [PubMed] [Google Scholar]
- 15.Newbrun E. 1982. Sucrose in the dynamics of the carious process. Int Dent J 32:13–23. [PubMed] [Google Scholar]
- 16.Gibbons A. 2013. How sweet it is: genes show how bacteria colonized human teeth. Science 339:896–897. doi: 10.1126/science.339.6122.896. [DOI] [PubMed] [Google Scholar]
- 17.Cornejo OE, Lefebure T, Pavinski Bitar PD, Lang P, Richards VP, Eilertson K, Do T, Beighton D, Zeng L, Ahn SJ, Burne RA, Siepel A, Bustamante CD, Stanhope MJ. 2013. Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans.. Mol Biol Evol 30:881–893. doi: 10.1093/molbev/mss278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshino T, Fujiwara T, Kawabata S. 2012. Evolution of cariogenic character in Streptococcus mutans: horizontal transmission of glycosyl hydrolase family 70 genes. Sci Rep 2:518. doi: 10.1038/srep00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Argimon S, Alekseyenko AV, DeSalle R, Caufield PW. 2013. Phylogenetic analysis of glucosyltransferases and implications for the coevolution of mutans streptococci with their mammalian hosts. PLoS One 8:e56305. doi: 10.1371/journal.pone.0056305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng L, Burne RA. 2013. Comprehensive mutational analysis of sucrose-metabolizing pathways in Streptococcus mutans reveals novel roles for the sucrose PTS permease. J Bacteriol 195:833–843. doi: 10.1128/JB.02042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng L, Wen ZT, Burne RA. 2006. A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans.. Mol Microbiol 62:187–200. doi: 10.1111/j.1365-2958.2006.05359.x. [DOI] [PubMed] [Google Scholar]
- 22.Zeng L, Das S, Burne RA. 2011. Genetic analysis of the functions and interactions of components of the LevQRST signal transduction complex of Streptococcus mutans.. PLoS One 6:e17335. doi: 10.1371/journal.pone.0017335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ajdic D, Pham VT. 2007. Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J Bacteriol 189:5049–5059. doi: 10.1128/JB.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng L, Choi SC, Danko CG, Siepel A, Stanhope MJ, Burne RA. 2013. Gene regulation by CcpA and catabolite repression explored by RNA-Seq in Streptococcus mutans.. PLoS One 8:e60465. doi: 10.1371/journal.pone.0060465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burne RA, Wen ZT, Chen YY, Penders JE. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J Bacteriol 181:2863–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 27.Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods 49:193–205. doi: 10.1016/S0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- 28.Cha RS, Zarbl H, Keohavong P, Thilly WG. 1992. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Methods Appl 2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- 29.Moye ZD, Burne RA, Zeng L. 2014. Uptake and metabolism of N-acetylglucosamine and glucosamine by Streptococcus mutans.. Appl Environ Microbiol 80:5053–5067. doi: 10.1128/AEM.00820-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn SJ, Lemos JA, Burne RA. 2005. Role of HtrA in growth and competence of Streptococcus mutans UA159. J Bacteriol 187:3028–3038. doi: 10.1128/JB.187.9.3028-3038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao JQ, Yu F. 2011. DEB: a web interface for RNA-seq digital gene expression analysis. Bioinformation 7:44–45. doi: 10.6026/97320630007044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 34.Sato Y, Yamamoto Y, Kizaki H, Kuramitsu HK. 1993. Isolation, characterization and sequence analysis of the scrK gene encoding fructokinase of Streptococcus mutans.. J Gen Microbiol 139:921–927. doi: 10.1099/00221287-139-5-921. [DOI] [PubMed] [Google Scholar]
- 35.Wen ZT, Browngardt C, Burne RA. 2001. Characterization of two operons that encode components of fructose-specific enzyme II of the sugar:phosphotransferase system of Streptococcus mutans.. FEMS Microbiol Lett 205:337–342. doi: 10.1111/j.1574-6968.2001.tb10969.x. [DOI] [PubMed] [Google Scholar]
- 36.Webb AJ, Homer KA, Hosie AH. 2007. A phosphoenolpyruvate-dependent phosphotransferase system is the principal maltose transporter in Streptococcus mutans.. J Bacteriol 189:3322–3327. doi: 10.1128/JB.01633-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merritt J, Qi F, Shi W. 2005. A unique nine-gene comY operon in Streptococcus mutans.. Microbiology 151:157–166. doi: 10.1099/mic.0.27554-0. [DOI] [PubMed] [Google Scholar]
- 38.Niu G, Okinaga T, Zhu L, Banas J, Qi F, Merritt J. 2008. Characterization of irvR, a novel regulator of the irvA-dependent pathway required for genetic competence and dextran-dependent aggregation in Streptococcus mutans.. J Bacteriol 190:7268–7274. doi: 10.1128/JB.00967-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahn SJ, Rice KC, Oleas J, Bayles KW, Burne RA. 2010. The Streptococcus mutans Cid and Lrg systems modulate virulence traits in response to multiple environmental signals. Microbiology 156:3136–3147. doi: 10.1099/mic.0.039586-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahn SJ, Qu MD, Roberts E, Burne RA, Rice KC. 2012. Identification of the Streptococcus mutans LytST two-component regulon reveals its contribution to oxidative stress tolerance. BMC Microbiol 12:187. doi: 10.1186/1471-2180-12-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng L, Das S, Burne RA. 2010. Utilization of lactose and galactose by Streptococcus mutans: transport, toxicity, and carbon catabolite repression. J Bacteriol 192:2434–2444. doi: 10.1128/JB.01624-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ajdic D, Ferretti JJ. 1998. Transcriptional regulation of the Streptococcus mutans gal operon by the GalR repressor. J Bacteriol 180:5727–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abranches J, Candella MM, Wen ZT, Baker HV, Burne RA. 2006. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans.. J Bacteriol 188:3748–3756. doi: 10.1128/JB.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moye ZD, Zeng L, Burne RA. 2014. Fueling the caries process: carbohydrate metabolism and gene regulation by Streptococcus mutans. J Oral Microbiol 6:24878–24892. doi: 10.3402/jom.v6.24878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeng L, Burne RA. 2010. Seryl-phosphorylated HPr regulates CcpA-independent carbon catabolite repression in conjunction with PTS permeases in Streptococcus mutans. Mol Microbiol 75:1145–1158. doi: 10.1111/j.1365-2958.2009.07029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deutscher J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr Opin Microbiol 11:87–93. doi: 10.1016/j.mib.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 47.Reizer J, Panos C. 1980. Regulation of beta-galactoside phosphate accumulation in Streptococcus pyogenes by an expulsion mechanism. Proc Natl Acad Sci U S A 77:5497–5501. doi: 10.1073/pnas.77.9.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato Y, Okamoto-Shibayama K, Azuma T. 2015. Glucose-PTS involvement in maltose metabolism by Streptococcus mutans. Bull Tokyo Dent Coll 56:93–103. doi: 10.2209/tdcpublication.56.93. [DOI] [PubMed] [Google Scholar]
- 49.Lemos JA, Lin VK, Nascimento MM, Abranches J, Burne RA. 2007. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol Microbiol 65:1568–1581. doi: 10.1111/j.1365-2958.2007.05897.x. [DOI] [PubMed] [Google Scholar]
- 50.Lemos JA, Nascimento MM, Lin VK, Abranches J, Burne RA. 2008. Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J Bacteriol 190:5291–5299. doi: 10.1128/JB.00288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seaton K, Ahn S-J, Burne RA. 2015. Regulation of competence and gene expression in Streptococcus mutans by the RcrR transcriptional regulator. Mol Oral Microbiol 30:147–159. doi: 10.1111/omi.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seaton K, Ahn SJ, Sagstetter AM, Burne RA. 2011. A transcriptional regulator and ABC transporters link stress tolerance, (p)ppGpp, and genetic competence in Streptococcus mutans. J Bacteriol 193:862–874. doi: 10.1128/JB.01257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahn SJ, Kaspar J, Kim JN, Seaton K, Burne RA. 2014. Discovery of novel peptides regulating competence development in Streptococcus mutans. J Bacteriol 196:3735–3745. doi: 10.1128/JB.01942-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gaca AO, Colomer-Winter C, Lemos JA. 2015. Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol 197:1146–1156. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moye ZD, Zeng L, Burne RA. 2014. Modification of gene expression and virulence traits in Streptococcus mutans in response to carbohydrate availability. Appl Environ Microbiol 80:972–985. doi: 10.1128/AEM.03579-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ajdic D, Chen Z. 2013. A novel phosphotransferase system of Streptococcus mutans is responsible for transport of carbohydrates with alpha-1,3 linkage. Mol Oral Microbiol 28:114–128. doi: 10.1111/omi.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol 183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol 78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okinaga T, Niu G, Xie Z, Qi F, Merritt J. 2010. The hdrRM operon of Streptococcus mutans encodes a novel regulatory system for coordinated competence development and bacteriocin production. J Bacteriol 192:1844–1852. doi: 10.1128/JB.01667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perry D, Kuramitsu HK. 1981. Genetic transformation of Streptococcus mutans. Infect Immun 32:1295–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo Q, Ahn SJ, Kaspar J, Zhou X, Burne RA. 2014. Growth phase and pH influence peptide signaling for competence development in Streptococcus mutans. J Bacteriol 196:227–236. doi: 10.1128/JB.00995-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perry D, Wondrack LM, Kuramitsu HK. 1983. Genetic transformation of putative cariogenic properties in Streptococcus mutans. Infect Immun 41:722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Burne RA. 2001. Regulation of the gtfBC and ftf genes of Streptococcus mutans in biofilms in response to pH and carbohydrate. Microbiology 147:2841–2848. doi: 10.1099/00221287-147-10-2841. [DOI] [PubMed] [Google Scholar]
- 64.Shemesh M, Tam A, Feldman M, Steinberg D. 2006. Differential expression profiles of Streptococcus mutans ftf, gtf and vicR genes in the presence of dietary carbohydrates at early and late exponential growth phases. Carbohydr Res 341:2090–2097. doi: 10.1016/j.carres.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 65.Wexler DL, Hudson MC, Burne RA. 1993. Streptococcus mutans fructosyltransferase (ftf) and glucosyltransferase (gtfBC) operon fusion strains in continuous culture. Infect Immun 61:1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR III, Heydorn A, Koo H. 2012. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog 8:e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.