ABSTRACT
Upon release of HIV-1 particles from the infected cell, the viral protease cleaves the Gag polyprotein at specific sites, triggering maturation. During this process, which is essential for infectivity, the capsid protein (CA) reassembles into a conical core. Maturation inhibitors (MIs) block HIV-1 maturation by interfering with protease-mediated CA-spacer peptide 1 (CA-SP1) processing, concomitantly stabilizing the immature CA-SP1 lattice; virions from MI-treated cells retain an immature-like CA-SP1 lattice, whereas mutational abolition of cleavage at the CA-SP1 site results in virions in which the CA-SP1 lattice converts to a mature-like form. We previously reported that propagation of HIV-1 in the presence of MI PF-46396 selected for assembly-defective, compound-dependent mutants with amino acid substitutions in the major homology region (MHR) of CA. Propagation of these mutants in the absence of PF-46396 resulted in the acquisition of second-site compensatory mutations. These included a Thr-to-Ile substitution at SP1 residue 8 (T8I), which results in impaired CA-SP1 processing. Thus, the T8I mutation phenocopies PF-46396 treatment in terms of its ability to rescue the replication defect imposed by the MHR mutations and to impede CA-SP1 processing. Here, we use cryo-electron tomography to show that, like MIs, the T8I mutation stabilizes the immature-like CA-SP1 lattice. These results have important implications for the mechanism of action of HIV-1 MIs; they also suggest that T8I may provide a valuable tool for structural definition of the CA-SP1 boundary region, which has thus far been refractory to high-resolution analysis, apparently because of conformational flexibility in this region of Gag.
IMPORTANCE HIV-1 maturation involves dissection of the Gag polyprotein by the viral protease and assembly of a conical capsid enclosing the viral ribonucleoprotein. Maturation inhibitors (MIs) prevent the final cleavage step at the site between the capsid protein (CA) and spacer peptide 1 (SP1), apparently by binding at this site and denying the protease access. Additionally, MIs stabilize the immature-like CA-SP1 lattice, preventing release of CA into the soluble pool. We previously found that T8I, a mutation in SP1, rescues a PF-46396-dependent CA mutant and blocks CA-SP1 cleavage. In this study, we imaged T8I virions by cryo-electron tomography and showed that T8I mutants, like MI-treated virions, contain an immature CA-SP1 lattice. These results lay the groundwork needed to understand the structure of the CA-SP1 interface region and further illuminate the mechanism of action of MIs.
INTRODUCTION
The production of HIV-1 particles is driven primarily by Pr55Gag, the Gag precursor protein, in concert with cellular factors. Pr55Gag is composed of several major domains and spacer peptides organized from the N terminus to the C terminus as follows: matrix (MA), capsid (CA), spacer peptide 1 (SP1), nucleocapsid (NC), spacer peptide 2 (SP2), and p6. During Gag translation, an infrequent ribosomal frameshifting event leads to the synthesis of the larger GagPol polyprotein, Pr160GagPol, which additionally contains the viral protease (PR), reverse transcriptase (RT), and integrase (IN) (1, 2).
As the immature virion buds from the infected cell, the PR is activated and dissects the Gag and GagPol precursor polyproteins. The Gag cleavage sites are processed in a specific order (3, 4) (Fig. 1). Cleavage starts at the SP1-NC site, detaching the viral nucleoprotein complex (vRNP; NC plus genomic RNA) from the residual Gag shell. This is followed by cleavage at the MA-CA site, separating CA from the membrane-bound MA layer, and, finally, by cleavage between CA and SP1. Upon its liberation from the Gag precursor, CA is released into a soluble pool from which a conical capsid is assembled (here, we use the term “capsid” to denote the assembled CA protein shell and the term “core” for the capsid plus whatever it may contain). Although both the immature and mature CA lattices are predominantly hexameric, the strain induced by curvature in the immature lattice is accommodated by gaps in the lattice (5–7), whereas the mature capsid is organized on the basis of fullerene geometry, in which a hexameric lattice is closed by 12 vertices thought to be occupied by CA pentamers (8).
FIG 1.
Schematic diagram of the HIV-1 Gag cleavage and maturation process. WT virions mature through the 4 stages shown on the top row, with ∼95% of them assembling a capsid, 80% to 85% of which are conical (top row, right diagram). (In the remaining ∼5%, which lack a core, all of CA stays in the soluble pool [top row, third diagram].) In MI-treated virions (11, 15), much of the CA (typically, 50% or so [42]) remains in an immature-like lattice (bottom row, left diagram). Therefore, the size of the immature-like lattices is reduced compared to that of the immature ones. In CA5 virions, in which CA-SP1 cleavage is completely blocked, the CA shell progresses to a mature-like conformation (bottom row, right diagram). (Bottom left) Diagram showing the SP1 sequence, with the Thr at residue 8 of SP1 that is mutated to Ile in the T8I mutant underlined, and the secondary cleavage site marked with an arrowhead. Note that SP2 and p6 (distal to NC [4, 43]) are not included in these diagrams.
High-resolution structures have been obtained for the individual Gag domains MA, CA, NC, and p6 (1, 2). However, owing to its large size and the flexible nature of the interdomain linker regions, the structure of full-length Pr55Gag has not been defined. Of particular importance to the present study is the region where CA connects to SP1. Peptides corresponding to this region adopt a helical conformation in vitro (9, 10), and cryo-electron tomography (cryo-ET) studies have suggested that SP1 forms a six-helix bundle connecting the CA lattice to the less-ordered NC/RNA layer (7, 11, 12). However, its conformation(s) in ordered lattices remains poorly resolved (12). This is a point of great interest, as the CA-SP1 boundary region is thought to be the binding site for HIV-1 maturation inhibitors (MIs; see below).
By generating cleavage-preventing point mutations at salient sites in Gag, it has been shown that initiation of disassembly of the immature CA lattice requires cleavage on both sides of CA-SP1 (i.e., at the MA-CA and SP1-NC sites), while assembly of a core also requires cleavage between CA and SP1 (13). An immature-like “thick” (∼10-nm) conformation of the CA shell is found in mutants in which the MA-CA or the SP1-NC cleavage events are prevented (13). Immature-like lattices are approximately the same thickness (10 nm) as immature lattices, but they have less contrasted substructure. Strikingly, when CA-SP1 cleavage was completely blocked by mutagenesis (e.g., in the CA5 mutant [14]), the CA shell was found in a mature-like “thin” (∼8-nm) conformation (11, 13, 15) (mature-like lattices are approximately the same thickness as mature lattices [8 nm], but appear at the current resolution to be more textured), implying that, under these conditions, the immature-like CA lattice can progressively convert to a mature-like lattice without disassembling (15). However, such viral particles are not infectious.
PR inhibitors act by binding the enzyme and preventing it from cleaving its target sites in Pr55Gag and Pr160GagPol. Members of a different class of compounds, MIs, prevent maturation by binding to the partially processed Gag lattice and blocking the conversion of CA-SP1 to mature CA (16, 17). Two chemically distinct maturation inhibitors have been reported: bevirimat (BVM) (16–18) and PF-46396 (PF96) (19, 20). Virions produced from MI-treated cells display a morphology characterized by an eccentric electron-dense aggregate, presumably composed of NC plus viral RNA (21), and an incomplete shell of CA-SP1 underlying the MA layer (16). The morphology of MI-treated virions is somewhat reminiscent of that displayed by virions in which CA-SP1 cleavage has been blocked by mutations, as in CA5 (14), with two notable differences: (i) while MIs partially prevent CA-SP1 cleavage, the CA5 mutant completely abolishes it; (ii) the residual Gag shell in MI-treated particles is in an immature-like (thick) state, whereas that observed in virions defective for CA-SP1 cleavage is in a mature-like (thin) state (11, 15). Therefore, we concluded that, in addition to blocking CA-SP1 cleavage partially (but sufficiently), MIs also stabilize the immature-like CA shell (11, 15). Clinical trials performed with bevirimat demonstrated that the compound is safe and effective (22, 23); however, polymorphisms, located predominantly between SP1 residues 6 and 8, reduced the susceptibility of HIV-1 to the compound in a significant percentage of treated patients (24–26).
Propagation of HIV-1 in culture in the presence of PF96 led to the selection of resistance mutations around the CA-SP1 cleavage site (20), where resistance to bevirimat maps (27). Resistance mutations also arose far upstream in CA in the major homology region (MHR), a highly conserved retroviral sequence known to be important for virus assembly (20). Replication of the MHR mutants was markedly PF96 dependent (20). The replication defect exhibited by the PF96-dependent MHR mutants (e.g., CA-P157S) could be rescued not only by PF96 but also by second-site substitutions in Gag that arose spontaneously during propagation of these mutants in the absence of compound. One such mutation was a Thr-to-Ile substitution at residue 8 of SP1 (T8I). Notably, on its own, the T8I mutant was severely replication defective and displayed an accumulation of CA-SP1. Thus, in two important respects, the T8I mutation phenocopies the effect of PF96 binding: (i) it interferes with CA-SP1 processing, and (ii) it rescues the assembly defect imposed by the CA-P157S MHR mutation.
These observations led us to hypothesize that the T8I mutation may, like MIs, stabilize the immature CA-SP1 lattice. By coupling T8I with the cleavage-defective mutant CA5 and examining the resulting virus particles by cryo-ET, we demonstrate that T8I does indeed stabilize the immature CA-SP1 lattice. These results extend the parallels between MI binding and the T8I mutation and further suggest that the T8I mutation may offer a valuable tool for resolving the structure of the highly flexible SP1 region.
MATERIALS AND METHODS
Plasmids.
The pNL4-3 molecular clone (28) and the T8I (20) and CA5 (14; kindly provided by H.-G. Kräusslich) derivatives have been reported previously. The CA5/T8I double mutant was constructed by site-directed mutagenesis using the Quikchange method (Stratagene) according to the manufacturer's instructions.
Infectivity and CA-SP1 processing assays.
Single-cycle infectivity assays were performed by using the TZM-bl indicator cell line (obtained from J. Kappes through the NIH AIDS Reagent Program [29]) as previously described (30). Briefly, 293T cells were transfected with wild-type (WT) pNL4-3 or derivatives containing the indicated Gag mutations. Virus stocks were harvested, filtered, normalized for RT activity, and used to infect TZM-bl cells. Two days postinfection, luciferase activity was measured. To monitor CA-SP1 accumulation (27, 31), HeLa cells transfected with the indicated HIV-1 molecular clones were metabolically labeled with [35S]methionine-cysteine ([35S]Met-Cys) for 2 h at 1 day posttransfection. Virus-containing supernatants were harvested, filtered, and subjected to ultracentrifugation at 75,000 × g for 45 to 60 min. Virus pellets were resuspended in lysis buffer (31) and characterized by SDS-PAGE. CA and CA-SP1 bands were quantified by phosphorimager analysis using Quantity One software (Bio-Rad).
Cryo-ET and subtomogram averaging.
These operations were performed essentially as previously described (32). In brief, paraformaldehyde-fixed virus was mixed (2:1) with 10-nm-diameter colloidal gold particles (Aurion, Wageningen, The Netherlands), applied to Quantifoil R2/2 holey carbon grids, and plunge-frozen in a Vitrobot cryostation (FEI, Hillsboro, OR). Grids were then transferred to a cryo-holder (type 626; Gatan, Warrendale, PA), and single-axis tilt series were recorded on a Tecnai-12 electron microscope (FEI) equipped with an energy filter (GIF 2002; Gatan). The microscope was operated at 120 keV in zero-loss mode with an energy slit width of 20 eV. Images were acquired using SerialEM (33) and recorded using a 2,048-pixel-by-2,048-pixel charge-coupled-device (CCD) camera (Gatan). Tilt series data were acquired at 2° intervals from ∼−66° to ∼66° at an electron dose of ∼1.1 e−/Å2 per projection (total cumulative dose, ∼75 e−/Å2). The magnification used was ×38,500 (0.78 nm/pixel), and the nominal defocus was −4 μm [first contrast transfer function zero at (3.7 nm)−1]. Tilt series images were aligned and reconstructed using the Bsoft package (34), and virions were extracted and denoised by 20 iterations of anisotropic nonlinear diffusion (35). The in-plane resolution of the tomograms was 5.0 to 5.5 nm for individual virions as calculated by the NLOO-2D (noise-compensated leave one out in two dimensions) method (36).
Subtomograms containing structures of interest (subvolumes containing patches of Gag-related lattice, 39 nm on a side) were located manually in the denoised virions and extracted from the corresponding raw reconstructions of the virion. Initial orientations of the patches were defined by vectors from the virion centers directed radially outwards that were thus approximately perpendicular to the viral envelope. A density map calculated by averaging all selected patches was then generated, cylindrically symmetrized, and used as a reference for translationally aligning all subtomograms. Subtomogram alignment was done taking into account the missing wedge of information (37), performed with routines from Bsoft (38) modified as needed, and wrapped into Python scripts. The procedure was repeated two more times, using the average from the preceding cycle as a reference for the next cycle. As a result of this process, subtomograms were translationally but not rotationally aligned, and therefore the Gag-related lattices were not yet in register. For the next steps, the viral membrane and MA layers were masked off to maximize the influence of the Gag-related lattice. One subtomogram was selected, C6 symmetry was applied, and this subvolume was used as a reference to rotationally and translationally align the other patches. This alignment procedure was iterated five times, using as reference the average of the top ∼5% to 10% of the particles (as ranked by correlation coefficients) from the previous round. Classification and averaging were then performed by maximum likelihood analysis as implemented in the Xmipp package (39). Approximately the top 33% of the initially selected subtomograms were used to calculate the final average. The percentage of data excluded in subtomogram averaging usually ranges from 45% to 60% (12, 40). In this study, the results obtained with 70% and 50% exclusion were very similar but we elected to use the former analysis because it made the CA repeat slightly clearer. The resolutions of the subtomogram averages, as given by the Fourier shell correlation (FSC), are given in Table 1.
TABLE 1.
Subtomogram averaging information
| Strain | No. of tomograms | No. of virions | No. of selected subtomograms | No. of averaged subtomograms | Avg resolution (nm) (FSCa cutoff, 0.5) | Spacing of hexagonal lattice (nm) |
|---|---|---|---|---|---|---|
| T8I | 9 | 49 | 730 | 248 | 4.4 | 7.8 |
| CA5 | 3 | 40 | 1,097 | 385 | 3.9 | NAa |
| CA5-T8I | 7 | 169 | 6,035 | 2,079 | 3.7 | 7.8 |
FSC, Fourier shell correlation; NA, not applicable.
Two preparations each of the WT, T8I, and CA5, and three of CA5/T8I, were imaged by cryo-EM, and the results obtained were consistent in each case. Cryo-ET was performed using one preparation for WT and T8I, two preparations for CA5, and three preparations for CA5/T8I.
RESULTS AND DISCUSSION
The T8I mutation impairs CA-SP1 processing and inhibits HIV-1 infectivity.
To measure the infectivity of the T8I mutant in a single-round assay, 293T cells were transfected with WT molecular clone pNL4-3 (28) or the mutants CA5 (14), T8I (20), and CA5/T8I. Infectivity was measured in the TZM-bl indicator cell line (29) (Fig. 2A). These results indicated that the infectivity of T8I was approximately 15% of that of WT. As shown previously (14, 30), the CA5 mutant, which has two substitutions that completely block CA-SP1 processing, was noninfectious, as was a CA5/T8I double mutant (Fig. 2A). Effects of the CA5, T8I, and CA5/T8I mutations on CA-SP1 processing were confirmed by metabolic radiolabeling (Fig. 2B). T8I virions showed an approximately 70% accumulation of CA-SP1, whereas CA5 and CA5/T8I mutants were completely blocked for CA-SP1 processing; only CA-SP1 was detected, and no mature CA was detected. In contrast, WT virions showed only ∼5% to 10% accumulation of CA-SP1. No differences in the amounts of uncleaved Gag or any other CA-containing cleavage products were found.
FIG 2.
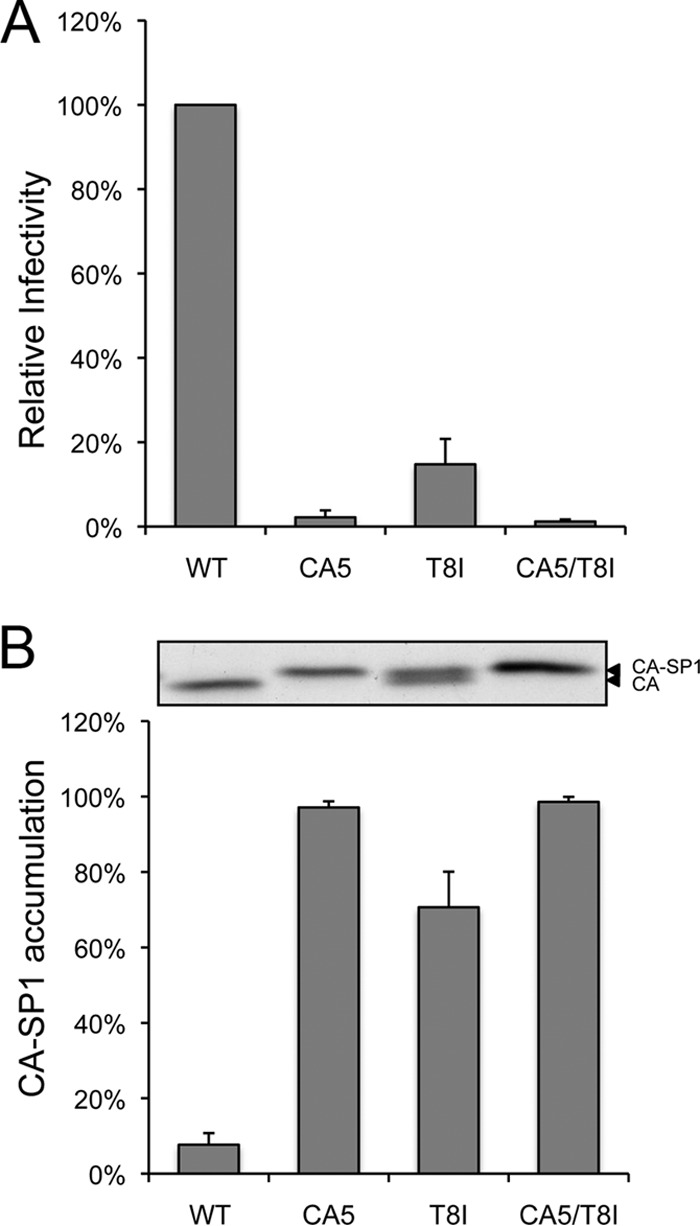
Effect of CA5, T8I, and CA5/T8I mutations on virus infectivity and CA-SP1 processing. (A) Viruses were produced in 239T cells by transfection with WT or mutant pNL4-3 molecular clones. Virus-containing supernatants were normalized for RT activity and used to infect TZM-bl cells. The luciferase signal was normalized to the corresponding RT values. Error bars indicate standard deviations from 3 independent experiments. (B) HeLa cells were transfected with the pNL4-3 WT, CA5, T8I, and CA5/T8I and were metabolically labeled with [35S]Met-Cys. Released virions were collected by ultracentrifugation, and virion-associated CA and CA-SP1 were analyzed by SDS-PAGE and quantified by phosphorimager analysis. A representative gel image is shown at the top, and quantification of the percentage of CA-SP1 relative to the total amount of CA plus CA-SP1 is presented in the graph. Error bars indicate standard deviations of the results from five independent experiments.
Cryo-ET analysis reveals that the T8I mutation stabilizes the immature Gag lattice.
HIV-1 virions imaged by cryo-ET can be classified according to core morphology as conical (Fig. 3A and D) or nonconical (Fig. 3B and E) or having no core (Fig. 3C and F) and by the presence (Fig. 3D to F) or absence (Fig. 3A to C) of a partial CA-SP1 lattice (an “inner shell”) underneath and somewhat offset from the viral envelope (i.e., the bilayer plus MA layer).
FIG 3.

Cryo-ET analysis of WT virions and T8I, CA5, and CA5/T8I mutants. Tomographic central sections (A to F) and distributions in percentages of HIV virions (G) classified according to core morphology and the presence or absence of an inner shell of density. The panels show chosen representative images from the HIV WT (A) and the T8I mutant (B to F) used in this study. CA-SP1 inner shells are labeled with white arrowheads. The numbers in bold correspond to the majority species for each sample and help to identify which panels represent the samples analyzed in the study. Scale bar, 50 nm.
Most (∼80%) WT virions possess a conical core, and none of them contain an inner shell (Fig. 3G; for an example, see Fig. 3A). In contrast, most virions produced in the presence of MIs lack conical cores—they are present in only ∼6% of BVM-treated virions and ∼13% of PF96-treated virions—but contain an inner shell (found in ∼82% of BVM-treated virions and in ∼56% of PF96-treated virions) (11, 15). As previously demonstrated, this inner shell is in the immature-like/thick conformation in MI-treated virions (11, 15). Additionally, MI-treated virions that lack a core often contain an electron-dense “eccentric condensate” similar to those observed when virions are produced in the presence of allosteric IN inhibitors (ALLINIs) or in class II IN mutants (41). The eccentric condensates observed in ALLINI-treated preparations have been demonstrated to consist of the vRNP (21).
As with MIs, the T8I mutation reduces the proportion of virions with conical cores—in this case, to ∼31% (representative examples of T8I mutants are shown in Fig. 4A to D). Moreover, a significant (∼32%) fraction of the T8I particles contain an inner shell whose extent can vary but that is on average only about half that of the original Gag shell (Fig. 3G and 4D to F). Also, as with MI-treated virions, the T8I inner shells are mostly in the immature-like “thick” conformation (Fig. 4D to F), although the Gag shell appears to be in the mature-like “thin” conformation in ∼10% of cases (Fig. 4 G and H). A small fraction of these virions (3% of T8I particles containing a CA-SP1 shell) present a mosaic of thick and thin CA-SP1 shells. An alternative explanation for the occasional mature-like shell in T8I virions is that they could represent malformed cores.
FIG 4.
Tomographic sections of T8I, CA5, and CA5/T8I mutants. (A to H) Tomographic sections of T8I virions. Panels A to D illustrate the distribution of the morphologies seen in T8I virions as follows: A, conical core; B and D, nonconical core; C, no core; A to C, no inner shell; D, contains inner shell. Panels E and F illustrate T8I virions with inner CA-SP1 shells; the inner shell is in an immature-like conformation in ∼90% of cases (E and F; see also panel D), and it is in a mature-like conformation in ∼10% of cases (G and H). (I to L) Tomographic sections of CA5 mutants. I, nonconical core and no inner shell; J and K, no core, but containing an inner shell in mature-like conformation; L, tomographic section from one of the rare (∼3%) CA5 virions containing a mosaic immature/mature inner shell of CA-SP1. (M to P) Tomographic sections of CA5/T8I virions with no core and an immature-like CA-SP1 inner shell. White arrowheads, immature-like CA-SP1 shell; black arrowheads, mature-like CA-SP1 shell; white arrows, eccentric condensates. Scale bar, 50 nm.
One difference between T8I-treated virions and the MI-treated virions is that T8I produces more virions that lack a core (∼28% for T8I compared to ∼1% in BVM-treated virions and ∼2% in PF96-treated virions [15]). As in BVM- and PF-96-treated virions, most (∼90%) of these T8I particles contain RNPs packed in eccentric condensates (see, e.g., Fig. 4E and F). The basis for the difference in the percentages of particles lacking a core that are observed with T8I-treated virions versus MI-treated virions is currently unknown.
As previously described (11, 15), almost all (∼82%) CA5 virions contain an uneroded CA-SP1 shell (Fig. 3G and 4J and K). (The Gag shells of immature virions have a sizable gap at the budding site, and we infer that erosion takes place around that site after Gag processing and accounts for the less complete CA-SP1 shells observed after MI treatment or with the T8I mutant). The CA-SP1 shells of CA5 virions are mostly in the mature-like (thin-walled) conformation (Fig. 4J and K), although in a few cases (∼3% of the CA-SP1-containing particles) they present a mosaic of thick and thin regions (Fig. 4L). The mosaic arrangement was previously found in CA5 virions treated with the MI PF96 and was suggested to represent an intermediate step in a displacive in situ transformation of the CA-SP1 shell (15). However, this pseudomaturation process does not generate bona fide capsids and does not produce infectious particles (15). When CA5 virions are produced in the presence of BVM or PF96, the inner shell remains in the immature-like (thick-walled) conformation, consistent with the ability of MIs to prevent the immature-phase-to-mature-phase transition.
To determine whether the T8I mutation is able, like MIs, to stabilize the immature-like CA-SP1 lattice, we combined the T8I and CA5 mutations and examined the resulting virions by cryo-ET. As we had previously seen with MI-treated CA5 virions (15), the CA5/T8I particles exhibited the thick, immature-like Gag shells (Fig. 4M to P).
To enhance the features of the immature-like CA-SP1 shell, subtomogram averaging was performed on T8I, CA5, and CA5/T8I mutant virions (Table 1; Fig. 5). These data confirm that the CA shell from T8I and CA5/T8I mutants is organized as a honeycomb lattice similar to that seen with immature and MI-treated virions (11, 15), although there may be small differences between them that are not detectable at the current resolution. Additionally, SP1 is seen as a faint connecting density, again in agreement with previous results. The CA5 CA-SP1 shell does not exhibit a regular pattern, most likely because it has a flatter surface topography. This is consistent with what is observed in the immature-phase-to-mature-phase conformational change occurring in the CA5 CA-SP1 lattice (15).
FIG 5.
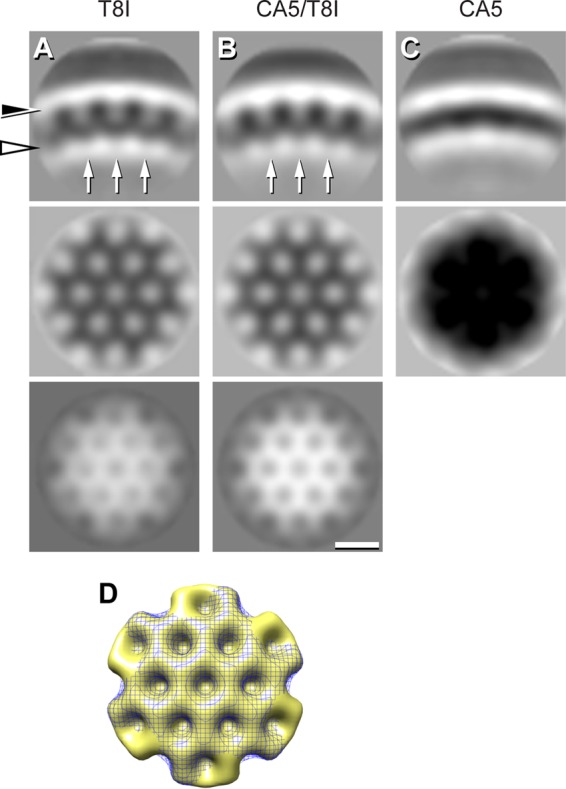
Subtomogram averaging of the CA-SP1 inner shell of density. (A to C) Sections through the three-dimensional (3D) maps. Top row, radial sections; central row, in-plane section at the height of CA (black arrowhead); bottom row, in-plane section at the height of SP1 from the immature-like shells (white arrowhead). The arrows in the top panels label the faint densities corresponding to SP1. To enhance the signal from the CA-SP1 layer, the viral membrane and MA layer were not used during subtomogram aligning and classification; as a result, they are not resolved in the final average. This suggests that the distance between the CA-SP1 shell and the MA is not constant, in agreement with the fact that cleavage between MA and CA has taken place. Scale bar, 10 nm. (D) Top view of surface renderings of the CA-SP1 lattices of CA5/T8I (gold, solid) and of T8I (blue, mesh). To compare with the CA-SP1 lattice from WT protease-defective virions, the reader is referred to two studies by Keller et al. (11, 15).
Summary.
In conclusion, cryo-ET and subtomogram averaging confirm and extend the hypothesis that the T8I mutation has effects on maturation similar to those of MIs. This mutation rescues the replication defect conferred by PF96-dependent MHR mutations (20), impedes CA-SP1 processing, and results in the stabilization of immature-like CA-SP1 shells. Given that residue 8 of SP1 lies outside the PR recognition sequence (which involves several amino acids on either side of the cleavage site), this mutation may disrupt CA-SP1 processing by stiffening SP1, which may need to be flexible for efficient CA-SP1 processing. The striking parallels between the effects of MI binding and of the T8I mutation suggest that MIs may likewise disrupt CA-SP1 processing not only by binding to the cleavage site and preventing access by PR but also by restricting the conformational flexibility around the CA-SP1 junction. The ability of the T8I mutation to stabilize the immature-like Gag shell further suggests that this mutant will be a useful tool for resolving the structure of the highly disordered SP1 domain of Gag in the context of virus particles and for understanding the effect of MI binding on Gag structure.
ACKNOWLEDGMENTS
We thank members of the Freed and Steven laboratories for helpful discussions, B. Heymann and D. Winkler for support with imaging resources, and members of the Freed laboratory for critical reviews of the manuscript.
Funding Statement
Funding was also provided by the Intramural AIDS Targeted Antiviral Program.
REFERENCES
- 1.Freed EO. 2015. HIV-1 assembly, release and maturation. Nat Rev Microbiol 13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sundquist WI, Kräusslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2:a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Konvalinka J, Kräusslich HG, Müller B. 2015. Retroviral proteases and their roles in virion maturation. Virology 479–480:403–417. [DOI] [PubMed] [Google Scholar]
- 4.Lee S-K, Potempa M, Swanstrom R. 2012. The choreography of HIV-1 proteolytic processing and virion assembly. J Biol Chem 287:40867–40874. doi: 10.1074/jbc.R112.399444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Kräusslich HG. 2009. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A 106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuller SD, Wilk T, Gowen BE, Kräusslich HG, Vogt VM. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol 7:729–738. doi: 10.1016/S0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 7.Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J 26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S, Hill CP, Sundquist WI, Finch JT. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 9.Datta SA, Temeselew LG, Crist RM, Soheilian F, Kamata A, Mirro J, Harvin D, Nagashima K, Cachau RE, Rein A. 2011. On the role of the SP1 domain in HIV-1 particle assembly: a molecular switch? J Virol 85:4111–4121. doi: 10.1128/JVI.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morellet N, Druillennec S, Lenoir C, Bouaziz S, Roques BP. 2005. Helical structure determined by NMR of the HIV-1 (345–392)Gag sequence, surrounding p2: implications for particle assembly and RNA packaging. Protein Sci 14:375–386. doi: 10.1110/ps.041087605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keller PW, Adamson CS, Heymann JB, Freed EO, Steven AC. 2011. HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J Virol 85:1420–1428. doi: 10.1128/JVI.01926-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schur FK, Hagen WJ, Rumlova M, Ruml T, Muller B, Kräusslich HG, Briggs JA. 2015. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 A resolution. Nature 517:505–508. [DOI] [PubMed] [Google Scholar]
- 13.de Marco A, Muller B, Glass B, Riches JD, Kräusslich HG, Briggs JA. 2010. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog 6:e1001215. doi: 10.1371/journal.ppat.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Kräusslich HG. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol 72:2846–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keller PW, Huang RK, England MR, Waki K, Cheng N, Heymann JB, Craven RC, Freed EO, Steven AC. 2013. A two-pronged structural analysis of retroviral maturation indicates that core formation proceeds by a disassembly-reassembly pathway rather than a displacive transition. J Virol 87:13655–13664. doi: 10.1128/JVI.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li F, Goila-Gaur R, Salzwedel K, Kilgore NR, Reddick M, Matallana C, Castillo A, Zoumplis D, Martin DE, Orenstein JM, Allaway GP, Freed EO, Wild CT. 2003. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Natl Acad Sci U S A 100:13555–13560. doi: 10.1073/pnas.2234683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou J, Yuan X, Dismuke D, Forshey BM, Lundquist C, Lee KH, Aiken C, Chen CH. 2004. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J Virol 78:922–929. doi: 10.1128/JVI.78.2.922-929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanamoto T, Kashiwada Y, Kanbara K, Gotoh K, Yoshimori M, Goto T, Sano K, Nakashima H. 2001. Anti-human immunodeficiency virus activity of YK-FH312 (a betulinic acid derivative), a novel compound blocking viral maturation. Antimicrob Agents Chemother 45:1225–1230. doi: 10.1128/AAC.45.4.1225-1230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blair WS, Cao J, Fok-Seang J, Griffin P, Isaacson J, Jackson RL, Murray E, Patick AK, Peng Q, Perros M, Pickford C, Wu H, Butler SL. 2009. New small-molecule inhibitor class targeting human immunodeficiency virus type 1 virion maturation. Antimicrob Agents Chemother 53:5080–5087. doi: 10.1128/AAC.00759-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waki K, Durell SR, Soheilian F, Nagashima K, Butler SL, Freed EO. 2012. Structural and functional insights into the HIV-1 maturation inhibitor binding pocket. PLoS Pathog 8:e1002997. doi: 10.1371/journal.ppat.1002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontana J, Jurado KA, Cheng N, Ly NL, Fuchs JR, Gorelick RJ, Engelman AN, Steven AC. 15 July 2015. Distribution and redistribution of HIV-1 nucleocapsid protein in immature, mature, and integrase-inhibited virions: a role for integrase in maturation. J Virol doi: 10.1128/JVI.01522-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salzwedel K, Martin DE, Sakalian M. 2007. Maturation inhibitors: a new therapeutic class targets the virus structure. AIDS Rev 9:162–172. [PubMed] [Google Scholar]
- 23.Smith PF, Ogundele A, Forrest A, Wilton J, Salzwedel K, Doto J, Allaway GP, Martin DE. 2007. Phase I and II study of the safety, virologic effect, and pharmacokinetics/pharmacodynamics of single-dose 3-o-(3′,3′-dimethylsuccinyl)betulinic acid (bevirimat) against human immunodeficiency virus infection. Antimicrob Agents Chemother 51:3574–3581. doi: 10.1128/AAC.00152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adamson CS, Sakalian M, Salzwedel K, Freed EO. 2010. Polymorphisms in Gag spacer peptide 1 confer varying levels of resistance to the HIV-1 maturation inhibitor bevirimat. Retrovirology 7:36. doi: 10.1186/1742-4690-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCallister S, Lalezari J, Richmond G, Thompson M, Harrigan R, Martin D, Salzwedel K, Allaway G. 2008. HIV-1 Gag polymorphisms determine treatment response to bevirimat (PA-457). Antivir Ther 13 (Suppl 3):A10. [Google Scholar]
- 26.Van Baelen K, Salzwedel K, Rondelez E, Van Eygen V, De Vos S, Verheyen A, Steegen K, Verlinden Y, Allaway GP, Stuyver LJ. 2009. Susceptibility of human immunodeficiency virus type 1 to the maturation inhibitor bevirimat is modulated by baseline polymorphisms in Gag spacer peptide 1. Antimicrob Agents Chemother 53:2185–2188. doi: 10.1128/AAC.01650-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adamson CS, Ablan SD, Boeras I, Goila-Gaur R, Soheilian F, Nagashima K, Li F, Salzwedel K, Sakalian M, Wild CT, Freed EO. 2006. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (bevirimat). J Virol 80:10957–10971. doi: 10.1128/JVI.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Checkley MA, Luttge BG, Soheilian F, Nagashima K, Freed EO. 2010. The capsid-spacer peptide 1 Gag processing intermediate is a dominant-negative inhibitor of HIV-1 maturation. Virology 400:137–144. doi: 10.1016/j.virol.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waheed AA, Ono A, Freed EO. 2009. Methods for the study of HIV-1 assembly. Methods Mol Biol 485:163–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontana J, Steven AC. 2013. At low pH, influenza virus matrix protein M1 undergoes a conformational change prior to dissociating from the membrane. J Virol 87:5621–5628. doi: 10.1128/JVI.00276-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Heymann JB, Cardone G, Winkler DC, Steven AC. 2008. Computational resources for cryo-electron tomography in Bsoft. J Struct Biol 161:232–242. doi: 10.1016/j.jsb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frangakis AS, Hegerl R. 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol 135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 36.Cardone G, Grunewald K, Steven AC. 2005. A resolution criterion for electron tomography based on cross-validation. J Struct Biol 151:117–129. doi: 10.1016/j.jsb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 37.Frank J. 2006. Electron tomography: methods for three-dimensional visualization of structures in the cell. Springer, New York, NY. [Google Scholar]
- 38.Heymann JB, Belnap DM. 2007. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol 157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Scheres SH, Melero R, Valle M, Carazo JM. 2009. Averaging of electron subtomograms and random conical tilt reconstructions through likelihood optimization. Structure 17:1563–1572. doi: 10.1016/j.str.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zanetti G, Briggs JA, Grünewald K, Sattentau QJ, Fuller SD. 25 August 2006, posting date Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engelman A. 1999. In vivo analysis of retroviral integrase structure and function. Adv Virus Res 52:411–426. doi: 10.1016/S0065-3527(08)60309-7. [DOI] [PubMed] [Google Scholar]
- 42.Lanman J, Lam TT, Emmett MR, Marshall AG, Sakalian M, Prevelige PE Jr. 2004. Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat Struct Mol Biol 11:676–677. doi: 10.1038/nsmb790. [DOI] [PubMed] [Google Scholar]
- 43.de Marco A, Heuser AM, Glass B, Kräusslich HG, Muller B, Briggs JA. 2012. Role of the SP2 domain and its proteolytic cleavage in HIV-1 structural maturation and infectivity. J Virol 86:13708–13716. doi: 10.1128/JVI.01704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]




