ABSTRACT
CD4 T cells provide protection against cytomegalovirus (CMV) and other persistent viruses, and the ability to quantify and characterize epitope-specific responses is essential to gain a more precise understanding of their effector roles in this regard. Here, we report the first two I-Ad-restricted CD4 T cell responses specific for mouse CMV (MCMV) epitopes and use a major histocompatibility complex class II (MHC-II) tetramer to characterize their phenotypes and functions. We demonstrate that MCMV-specific CD4 T cells can express high levels of granzyme B and kill target cells in an epitope- and organ-specific manner. In addition, CD4 T cell epitope vaccination of immunocompetent mice reduced MCMV replication in the same organs where CD4 cytotoxic T lymphocyte (CTL) activity was observed. Together, our studies show that MCMV epitope-specific CD4 T cells have the potential to mediate antiviral defense by multiple effector mechanisms in vivo.
IMPORTANCE CD4 T cells mediate immune protection by using their T cell receptors to recognize specific portions of viral proteins, called epitopes, that are presented by major histocompatibility complex class II (MHC-II) molecules on the surfaces of professional antigen-presenting cells (APCs). In this study, we discovered the first two epitopes derived from mouse cytomegalovirus (MCMV) that are recognized by CD4 T cells in BALB/c mice, a mouse strain commonly used to study the pathogenesis of this virus infection. Here, we report the sequences of these epitopes, characterize the CD4 T cells that recognize them to fight off MCMV infection, and show that we can use the epitopes to vaccinate mice and protect against MCMV.
INTRODUCTION
Cytomegalovirus (CMV)/human herpesvirus 5 (HHV-5) (the prototypic betaherpesvirus) infection is endemic in humans and wild mice and establishes a lifelong infection in the absence of acute disease in healthy hosts. Adaptive immunity is a critical component of CMV defenses, restricting primary infection and dampening reactivation, helping to maintain the largely benign host-virus equilibrium. However, if immunity is naive or compromised (e.g., in transplant recipients or congenital infection), CMV can cause serious disease (1, 2). In immunocompetent mice, CD8 T cells help to control acute CMV infection and establish latency (3, 4), and their adoptive transfer prevents disease in mice and humans with weakened immunity (5–8). Although much less well studied, CD4 T cells also contribute to defense against CMV. Their rapid expansion and numbers correlate with reduced disease in transplant and HIV patients (9–11), and correspondingly, delayed induction of CMV-specific CD4 T cells is associated with increased congenital infection (12) and prolonged viral shedding in infants (13). Notably, no protective correlates were seen with CMV-specific CD8 T cell responses in several of these studies. In mice, CD4 T cells are absolutely required to control mouse cytomegalovirus (MCMV) replication in the salivary glands—the key site of viral dissemination, where CD8 T cells can exert no control (14, 15)—and also contribute to immune control in several other organs. Adoptive transfer of MCMV-specific transgenic CD4 T cells provides some protection in immunocompromised mice (16), and cotransferring CMV-specific CD4 T cells reduces viral load and promotes “help” for CMV-specific CD8 T cell responses in patients receiving cellular immunotherapy (17).
In addition to traditional helper functions, CD4 T cells can also mediate direct antiviral activity in some cases. In chronic human CMV (HCMV), Epstein-Barr virus (EBV), and HIV infections, CD4 T cells displaying a terminally differentiated effector phenotype and/or expressing canonical cytolytic molecules (e.g., granzymes and perforin) are present in peripheral blood (18–20). The ability of these CD4 T cells to directly kill cells in an antigen-specific fashion has been demonstrated in most cases after their isolation and subsequent expansion/manipulation in culture (21–23). Notably, however, a few studies have shown that HCMV-specific CD4 T cells can mediate killing directly ex vivo in cell culture assays (24–26). CD4 cytotoxic T lymphocytes (CTLs) can be induced in virus-infected mice within a relatively short time (weeks) (27–32), albeit ex vivo assays have normally been used to define their killing capacity and human CD4 CTLs have been isolated and studied largely from persons who have been chronically infected for several years. Studies in both mice and humans suggest that perforin and granzyme are key mediators of CD4 T cell-cytolytic activity, but tumor necrosis factor (TNF) family ligands, such as FasL and TRAIL, likely can also contribute (24, 25, 27, 29, 32). Notably, despite the fact that several studies have assessed the phenotype and/or function of virus-specific CD4 CTLs that develop in CMV-infected humans, almost nothing is known about their role in the context of MCMV infection.
Although CD4 T cells have the capacity to mediate antiviral defense via cytolysis in some cases, the relative importance of this CTL activity, as well as the factors regulating their differentiation, remains largely unclear. We hypothesized that epitope-specific CD4 CTLs might be induced during MCMV infection, given what has been observed in CMV-infected humans. Consistent with this hypothesis, we now report the identification of the first MCMV epitope-specific CD4 T cell responses restricted by major histocompatibility complex class II (MHC-II) (I-Ad) in BALB/c mice, a model of CMV infection utilized for more than 50 years. An MHC-II tetramer comprised of the m78417–431 epitope was constructed and was utilized to enrich and characterize the phenotype and function of these cells. We demonstrate that MCMV epitope-specific CD4 T cells can mediate the killing/loss of peptide-loaded target cells in vivo and that this effector function varies dramatically depending on the tissue where they reside. Finally, epitope vaccination protected against MCMV challenge in immunocompetent mice, the first evidence that CD4 T cells can mediate nonredundant, early defense against CMV infection. Altogether, this study significantly furthers our understanding of how CMV-specific CD4 T cells function during natural infection and highlights the importance of considering their contributions in the context of vaccination against this persistent virus.
MATERIALS AND METHODS
Mice and virus.
BALB/c mice were purchased from Jackson Laboratories (Bar Harbor, ME) and bred under specific-pathogen-free conditions at the La Jolla Institute for Allergy and Immunology (LJI). All experiments were performed in 8- to 12-week-old mice in accordance with the guidelines established by the AAALAC and the LJI IACUC. Viral stocks derived from the bacterial artificial chromosome (BAC)-derived Smith strain of MCMV (33) or a stock obtained from the ATCC (VR-1399) were used, and no significant differences were seen in the results obtained with either. Intraperitoneal infection was performed with 2 × 104 PFU of salivary gland-derived (SG) or 2 × 105 PFU of mouse embryonic fibroblast (MEF)-derived (TC) viral stocks. MCMV replication levels in organs were determined by plaque assay in 3T3 cells as described previously (34).
IFN-γ ELISPOT assay and ICCS.
Enzyme-linked immunospot (ELISPOT) assays were performed as described previously (35). For CD4 T cell intracellular cytokine staining (ICCS) of spleen, liver, or lung cells, 1 × 106 cells were incubated with 5 μg/ml of m53285–299 or m78417–431 15-mer peptides for 8 h or treated with phorbol myristate acetate (PMA) (100 ng/ml) and ionomycin (500 ng/ml) for 5 h in the presence of brefeldin A (2 μg/ml). The cells were then surface stained, fixed, and permeabilized using BD Cytofix/Cytoperm buffer and stained for intracellular cytokines. The antibodies used were Alexa-Fluor 700 CD3, efluor450 CD11a, and peridinin chlorophyll protein (PerCP)-efluor710 CD49d (all from eBioscience); brilliant violet 570 (BV570) CD4 and BV605 TNF-α (clone MP6-XT22) (both from Biolegend); and V500 CD44, phycoerythrin (PE)-Cy7 gamma interferon (IFN-γ) (clone XMG1.2), PE-CF594 interleukin 2 (IL-2) (clone JES6-5H4), allophycocyanin (APC) IL-10 (clone JES65-16E3), and PE IL-17A (clone TC11-18H10) (all from BD Biosciences). Samples were acquired on a BD LSR II cytometer, and data were analyzed using FlowJo software (FlowJo).
In vivo peptide restimulation.
On day 8 of infection, mice were injected intravenously (i.v.) with 100 μg of m78417–431 peptide. After 3 h, spleens, livers, and lungs were harvested. Single-cell suspensions from the spleen and liver were prepared as described previously (36). The lungs were perfused prior to harvest, cut into small pieces, and incubated in RPMI medium supplemented with collagenase D (1 mg/ml), DNase I (10 μg/ml), and 5 mM CaCl2 for 30 min at 37°C. The digested organ was passed through a 70-μm cell strainer and further processed similarly to the spleen. The resulting cell suspensions were subjected to m78 tetramer enrichment as described below in medium containing 10 μg/ml brefeldin A. The enriched fraction was analyzed for cell surface and intracellular marker/cytokine expression as described above for ICCS.
Peptide–MHC-II tetramer enrichment and flow cytometry.
Biotin-labeled m78 (I-Ad) monomers were generated by the NIH Tetramer Core Facility (Emory, AL) and were tetramerized by addition of streptavidin-APC (Life Technologies) according to their protocol. Single-cell suspensions prepared from various organs were subjected to m78-specific CD4 T cell tetramer enrichment as described by the Pepper et al. (37). Briefly, the cells were incubated for 1 h at 25°C with m78-APC tetramer, followed by addition of anti-APC magnetic beads (Miltenyi Biotec). Samples were enriched for bead-bound cells on magnetic columns. Cells from the enriched fraction (or flowthrough) were incubated with various antibodies to determine the phenotype of m78-specific CD4 T cells: PE-CF594 CD69 (H1.253) (BD Biosciences), fluorescein isothiocyanate (FITC) KLRG-1 (2F1), PE-Cy7 CD28 (clone 37.51), APC-efluor780 PE BTLA (clone 8F4), and CD127 (clone A7R34) (all from eBioscience) and BV605 PD-1 (clone 29F.1A12), PECy7 CD27 (clone LG-3A10), APC-Cy7 CD43 (1B11), and PE HVEM (all from Biolegend).
In vivo CD4 T cell cytotoxicity assay.
Splenocytes were harvested from naive BALB/c mice and incubated with either 5 μg/ml each of m53285–299 and m78417–431 peptides (targets) or dimethyl sulfoxide (DMSO) for 1 h at 37°C and then washed and labeled with carboxyfluorescein succinimidyl ester (CFSE) (Molecular Probes) in phosphate-buffered saline (PBS)-0.1% bovine serum albumin (BSA) for 10 min at 37°C. The peptide-labeled targets were incubated with 1 μM CFSE (CFSEhi) and the DMSO-pulsed cells with 500 nM CFSE (CFSElo), washed, and mixed at a 1:1 ratio, and ∼1 × 107 cells were injected i.v. into naive or MCMV-infected (day 8) mice. After 16 h, the mice were sacrificed, spleen and liver mononuclear cells were isolated and stained with efluor450 anti-MHC-II (clone M5/114.15.2; eBioscience), and the relative numbers of MHC-II+ CFSEhi and CFSElo cells were analyzed. Percent killing was calculated as follows: 100 − ([percent CFSEhi in infected mice/percent CFSElo in infected mice]/[percent CFSEhi in naive mice/percent CFSElo in naive mice] × 100). For analyses in mice depleted of CD4 (clone GK1.5) or CD8 (clone 2.43) T cells, 150 μg of antibody was injected intraperitoneally (i.p.) at days 5 and 7 of infection prior to injecting CFSE-labeled targets on day 8, and >97% depletion was achieved in all mice. For analysis of granzyme B expression, total or tetramer-enriched spleen or liver cells were ex vivo fixed, permeabilized, and stained with PE anti-granzyme B (clone GB12; Invitrogen).
Peptide immunization.
BALB/c mice were immunized a single time subcutaneously (s.c.) with 50 μg of m53285–299 or m78417–431 peptide emulsified in complete Freund's adjuvant (CFA) (Difco, Detroit, MI). Mock-immunized mice received DMSO emulsified in CFA. Three weeks later, the mice were challenged with TC MCMV, and 4 days later, they were sacrificed for analysis of viral replication.
Statistical analysis.
Statistical significance was analyzed by a Mann-Whitney test or a Kruskal-Wallis test as indicated in the figure legends. Unless otherwise indicated, the data represent means and standard errors of the mean (SEM).
RESULTS
Identification of MCMV epitope-specific CD4 T cell responses.
To identify MHC class II (I-Ad)-restricted MCMV peptide epitopes, defined MCMV open reading frames (38) were used for algorithm-based predictions using tools available in the Immune Epitope Database (http://www.iedb.org), and the 234 highest ranked 15-mer peptides were subjected to further screening. CD4 T cells isolated from day 8 MCMV-infected mice were first incubated with pools of 10 peptides, and ELISPOT analysis was performed to assess their IFN-γ production, followed by deconvolution of potentially positive pools (Fig. 1A). Out of the 234 peptides, two (m53285–299 and m78417–431) were found to activate CD4 T cells above the threshold value of 100 spot-forming cells (SFC)/106 CD4 T cells (Fig. 1B). Although both m53- and m78-specific IFN-γ-producing CD4 T cells were readily measurable by ELISPOT assay, detecting them by ICCS and flow cytometry was challenging, with m78 comprising ∼0.15% of total splenic CD4 T cells and m53 even less (∼0.06%) at day 8 of infection (Fig. 1C). These two CD4 T cell responses were virtually undetectable at day 28 when assessed by either assay, indicating they display largely canonical expansion and contraction kinetics and do not “inflate” over time (Fig. 1D), as some MCMV-specific CD8 and CD4 T cells are known to do (39, 40). To further assess whether these predicted epitopes might selectively identify inflationary MCMV-specific CD4 T cells, all 234 peptides were tested again in day 28-infected mice. However, unlike the I-Ab-restricted m09133–47-specific CD4 T cells we have previously shown to selectively expand during the persistent phase of MCMV infection in C57BL/6 mice (39), no analogous inflationary response was identified using this pool of potential epitopes (data not shown).
FIG 1.
Identification of MCMV-specific CD4 T cells. (A) BALB/c mice were infected with MCMV, and day 8 postinfection, CD4 T cells from the spleens were purified for screening of 24 peptide pools (10 peptides/pool; 10 μg/ml each peptide). The mean numbers of SFC per 106 CD4 T cells from four independent experiments are shown. Responses were considered positive if the stimulation index (SI) exceeded twice the mean of the negative-control wells (effectors plus APCs without peptide) and the net numbers of spots were above the threshold of 100 SFC/106 CD4 cells in each individual experiment (indicated by the dashed line). Positive pools (*) and pools showing an SI of >2 in at least two experiments (arrows) were deconvoluted. (B) Out of all individually tested peptides, two 15-mers were identified (*) that elicited a significant IFN-γ response. They were m53285–99 (IAHQRITLTARCLRL) and m78417–31 (SQQKMTSLPMSVFYS). (C) Splenocytes from day 8 MCMV-infected BALB/c mice were restimulated with m78 or m53 peptide epitopes, and intracellular cytokine staining for IFN-γ and TNF-α was performed. (D) ELISPOT analysis for IFN-γ production by CD4 T cells from day 28 (d28) MCMV-infected mice after restimulation with m53- and m78- peptide epitopes (means of the results of two independent experiments are shown). The data represent means and SEM.
Phenotype of m78-specific CD4 T cells during acute and persistent MCMV infections.
CD4 T cells play a critical role in controlling MCMV in mucosal organs, such as the salivary gland and lung (15). One hurdle in studying the identified MCMV epitope-specific CD4 T cells was their very low frequencies. Consequently, an MHC-II tetramer was generated using the m78417–431 epitope, and mononuclear cells isolated from the spleen, liver, lungs, and salivary glands were subjected to tetramer enrichment (37). A representative plot from each organ showing the successful enrichment of m78-specific CD4 T cells at day 8 after infection is depicted in Fig. 2A. An MHC-II tetramer generated using the m53285–299 epitope was unfortunately not functional. The expression of various activation markers, adhesion molecules, and stimulatory/inhibitory cosignaling molecules of the TNF receptor (TNFR) and CD28 families was also assessed on m78-specific CD4 T cells and compared to levels expressed by naive CD4 T cells (CD44lo) isolated from the same tissues at both days 8 and 28 (Fig. 2B to D). The m78-specific CD4 T cells displayed roughly similar phenotypes in most cases, although some organ-specific differences in CD27, BTLA, and KLRG-1 expression were observed. At day 28, spleen and liver m78-specific CD4 T cells exhibited reduced expression of several surface markers, such as CD69, CD43, BTLA, PD-1, and CD27, compared to the day 8 levels. Notably, surface marker expression by m78-specific CD4 T cells was similar to that of the total MCMV-specific CD4 T cell response as gauged by CD44hi CD11ahi CD49d+ expression (reference 41 and data not shown), at both peak expansion and early memory time points.
FIG 2.
Phenotypic analysis of m78-specific CD4 T cells after MHC-II tetramer enrichment. (A) Tetramer (tet) enrichment of m78-specific CD4 T cells from spleen, liver, lung, and salivary glands (bottom) compared to nonspecific CLIP tetramer (top) from day 8 MCMV-infected BALB/c mice. (B and C) Phenotype of m78-specific tetramer-enriched CD4 T cells (open black histograms) compared to that of CD44lo naive CD4 T cells (filled gray histogram) from spleens and livers (B) and lungs and salivary glands (C) of day 8 MCMV-infected mice. (D) Phenotype of m78-specific CD4 T cells from spleens and livers of day 28 MCMV-infected mice. Representative fluorescence-activated cell sorter (FACS) plots of the results of 2 or 3 independent experiments with groups of 4 mice each for both time points are shown. For liver, lung, and salivary glands, cells were pooled from multiple mice prior to enrichment due to low numbers.
Cytokine expression of MCMV-specific CD4 T cells.
As an initial attempt to gauge the magnitude and characteristics of the total MCMV-specific CD4 T cell response in various organs, cells isolated from naive and infected mice were restimulated ex vivo with PMA/ionomycin and their production of IFN-γ was assessed. At day 8 of infection, an ∼4% increase in the proportion of IFN-γ-producing CD4 T cells was observed in MCMV-infected spleens (∼8% in MCMV-infected and ∼4% in naive mice) (Fig. 3A). Roughly similar enhanced proportions of IFN-γ-producing CD4 T cells were also observed in the liver (∼6%) and lung (∼3%) at day 8 of infection (Fig. 3A). Although the absolute numbers of IFN-γ+ CD4 T cells resident in these nonlymphoid organs were much lower than in the spleen, the numbers increased after infection in all the tissues examined (Fig. 3A).
FIG 3.
Cytokine expression by MCMV-specific CD4 T cells. (A) Frequencies and absolute numbers of IFN-γ-producing total CD4 T cells in spleens, livers, and lungs of naive and day 8 MCMV-infected BALB/c mice after PMA/ionomycin stimulation. The data are representative of the results of 2 separate experiments with 4 or 5 mice per group analyzed by the Mann-Whitney test. (B) Representative FACS plots showing the CD11a and CD49d profiles of total CD4 T cells from the spleens, livers, and lungs of naive and day 8 infected mice. (C) Total CD4 T cells (gray contour plots) overlaid with m78-specific tetramer-enriched CD4 T cells (black dots) from the spleens and livers of day 8 infected mice analyzed for CD11a and CD49d expression. (D) Representative FACS plots showing cytokine production by CD44hi CD11ahi CD49+ MCMV-specific splenic CD4 T cells from day 8 MCMV-infected mice and CD44lo CD4 T cells from naive mice after PMA/ionomycin restimulation. (E) Frequencies and absolute numbers of CD44hi CD11ahi CD49+ MCMV-specific CD4 T cells producing IFN-γ after PMA/ionomycin stimulation of naive (gray bars) and day 8 MCMV-infected (black bars) mice. The data are representative of 2 separate experiments with 4 or 5 mice per group analyzed by the Mann-Whitney test. For analysis of the liver, invariant NK T cells were excluded by CD1d tetramer staining. Ag, antigen. (F) Frequencies of IFN-γ-producing m78-specific CD4 T cells at day 8 of infection were determined after in vivo m78 peptide restimulation followed by tetramer enrichment and intracellular cytokine staining. The data are representative of 2 independent experiments with 4 mice per group. The data represent means and SEM; *, P < 0.05; **, P < 0.005.
Distinct subsets of CD4 T cells can be identified in large part based on their cytokine production, and whether MCMV-specific CD4 T cells were capable of producing additional cytokines (e.g., TNF-α, IL-2, IL-10, and IL-17) was determined after PMA/ionomycin stimulation. The polyclonal MCMV-specific CD4 T cell response was estimated by CD44hi CD11ahi CD49d+ surface marker expression (41). Representative flow plots depicting the expression profiles of CD11a and CD49d of total CD4 T cells from spleens, livers, and lungs of naive and day 8 MCMV-infected mice are shown in Fig. 3B. In naive mice, ∼5 to 20% of total CD4 T cells are CD11ahi CD49d+, varying by organ, whereas 8 days after infection, an ∼2- to 3-fold increase in the proportion of cells displaying this phenotype was observed. The CD44hi CD11ahi CD49d+ compartment is not comprised solely of MCMV-specific cells in infected mice, as these “antigen-experienced” cells also exist at measurable levels in naive mice (Fig. 3B). However, >95% of m78-specific cells show this phenotype (Fig. 3C), indicating that almost all MCMV-specific CD4 T cells will likely be contained in this CD4 T cell compartment, as they are for several other viruses, but not all (41). The majority (>50%) of CD44hi CD11ahi CD49d+ CD4 T cells in the spleens of MCMV-infected mice were found to display a Th1-like phenotype, producing IFN-γ and/or TNF-α, much greater than the ∼15% observed in naive mice. A substantial proportion of these cells also produced IL-2 (∼40% compared to ∼15% in naive mice), and a small, but readily identifiable, proportion also produced IL-10 (∼5%). No IL-17-producing CD4 T cells were observed at day 8 of infection following this polyclonal restimulation (Fig. 3D). Although a similar enhanced proportion of CD44hi CD11ahi CD49d+ CD4 T cells were found to produce IFN-γ in the spleen, liver, and lungs (∼15 to 30%) after MCMV infection, again, their absolute numbers were much higher in the spleen than in the other nonlymphoid organs (Fig. 3E).
The ability of m78-specific CD4 T cells to produce various effector cytokines was then assessed following their restimulation in vivo by injecting the m78 peptide epitope directly in day 8 infected mice, followed by tetramer enrichment. The m78-specific CD4 T cells were found by this method to primarily differentiate into IFN-γ-producing Th1-like cells, with virtually no expression of TNF-α, IL-2, or IL-17 detected (data not shown). Similar to what was observed for the polyclonal MCMV-specific CD4 T cell response, ∼20% of all splenic m78-specific cells produced IFN-γ in the spleen, but interestingly, ∼60% of all liver-resident m78-specific CD4 T cells were found to be IFN-γ+ after in vivo peptide restimulation (Fig. 3F).
CMV-specific CD4 T cells display CTL activity in vivo and confer protection after peptide immunization.
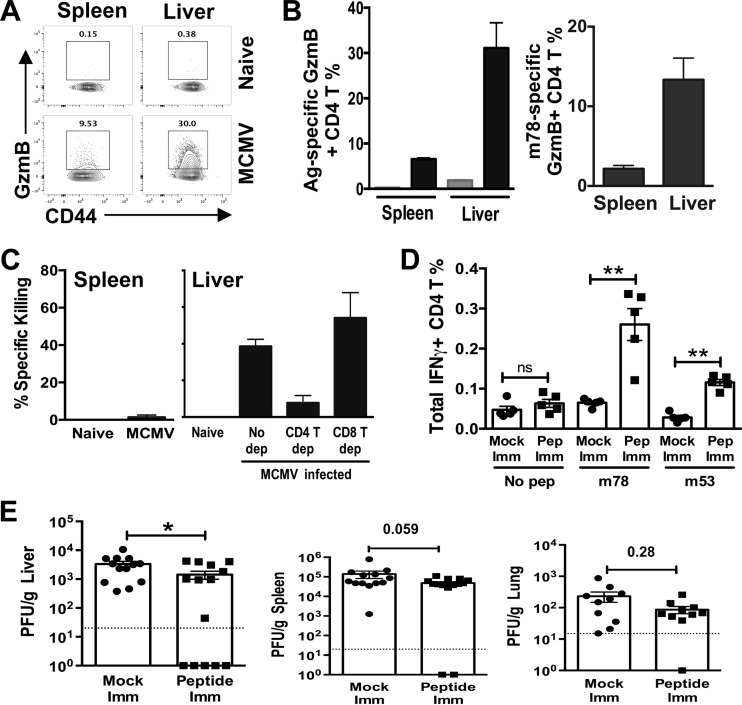
In addition to helping antibody and CD8 T cell responses, CD4 T cells can mediate direct antiviral activity through their production of effector cytokines (e.g., IFN-γ and TNF-α). Although few studies exist in the context of MCMV infection, production of IFN-γ by CD4 T cells has been reported to be a key component of their antiviral effector functions (14, 42). Notably, CD4 T cells displaying a CTL-like phenotype circulate in persons infected with HCMV, and we wished to examine if this was also true for MCMV and also whether MCMV epitope-specific CD4 T cells might be capable of mediating direct cytolysis in vivo. Expression of granzyme B (encoded by gzmB) by CD4 T cells is a marker commonly used to identify potential CD4 CTLs, as it is a key effector molecule for killing target cells. gzmB expression was assessed in both CD44hi and m78-specific CD4 T cells in both spleen and liver at day 8 after MCMV infection. Only a small proportion of CD44hi CD4 T cells expressed gzmB in the spleen (∼3 to 5%), whereas ∼30% of CD4 T cells in the liver expressed gzmB. When m78-specific CD4 T cells were analyzed, similar preferential expression levels of gzmB were seen in the liver and the spleen (spleen, ∼2%, and liver, ∼16%) (Fig. 4A and B). To assess whether this expression of gzmB correlated with the potential in vivo cytolytic activity of MCMV epitope-specific CD4 T cells, target cells were pulsed with m53 and m78 peptides and injected into day 8 MCMV-infected mice. Approximately 40% loss of MHC-II-positive target cells was observed in the liver, whereas essentially no loss of target cells was seen in the spleen (Fig. 4C), entirely consistent with the relative expression of gzmB by MCMV-specific CD4 T cells in the two organs and strongly suggesting the cells can mediate direct cytolysis in vivo. As a comparison, target cells loaded with MCMV epitopes targeted by CD8 T cells (IE1168–176 and m164257–265) showed >90% loss/lysis in both organs (data not shown). To demonstrate that CD4 T cells were responsible for the apparent killing of epitope-loaded target cells, mice were antibody depleted of either CD4 or CD8 T cells. As expected, target cell cytolysis was markedly (5-fold) reduced in the livers of CD4 T cell-depleted mice, with CD8 T cell depletion showing no effect (Fig. 4C).
FIG 4.
MCMV-specific CD4 T cells demonstrate CTL activity and can protect after peptide immunization. (A and B) GzmB expression in CD44hi CD4 T cells (A) or m78-specific CD4 T cells (B) in livers and spleens of day 8 MCMV-infected (black bars) or naive (gray bar) BALB/c mice. The results are representative of 3 independent experiments with at least 4 mice per experiment, and invariant NK T cells were excluded by CD1d tetramer staining. (C) The percent specific killing by CD4 T cells was assessed in spleens and livers of day 8 MCMV-infected and naive mice by the ability of CD4 T cells to kill m78 and m53 peptide-loaded MHC-II+ target cells. The killing results are representative of four independent experiments with at least 4 mice per group, and CD4 and CD8 T cell depletion was performed twice in groups of 3 mice with similar results. Statistical analysis was performed using the Kruskal-Wallis test (P = 0.014). (D) ICCS for IFN-γ production by CD4 T cells in day 4 MCMV-challenged mice in mock-immunized (imm) (circles) or vaccinated (squares) mice restimulated with no peptide (No pep) or the individual epitopes. The data are representative of the results of 2 separate experiments with 5 mice per group analyzed by the Mann-Whitney test. (E) Mice were immunized with m78 and m53 epitopes (squares) or mock immunized (circles) 3 weeks prior to MCMV challenge, and replication was determined 4 days postinfection. The results are a composite of 3 separate experiments with 5 mice per experiment analyzed by the Mann-Whitney test. The dotted lines denote the detection limit. The data represent means and SEM; *, P < 0.05; **, P < 0.005; ns, not significant.
The importance of CD4 T cells in controlling chronic virus infection has made them a focus in the context of vaccine development (43, 44). To assess whether these cells have the capacity to provide protection against CMV in a vaccine setting, immunocompetent mice received a single vaccination with m53 and m78 peptides in CFA, followed by MCMV challenge and assessment of viral replication 4 days later. We have observed that CD4 T cells normally provide no protection at this early time point in the spleen or liver during normal MCMV infection (determined after depletion with anti-CD4 antibody), while by day 8 of infection, their depletion results in dramatically higher titers in both organs (data not shown). Therefore, we focused on the day 4 time point to determine whether more rapid CD4 T cell-dependent control could be induced after epitope vaccination. Significantly enhanced proportions of IFN-γ-producing m78 and m53 CD4 T cells were seen in immunized mice following infection (Fig. 4D), and ∼1,000 m78-specific CD4 T cells could be tetramer enriched from spleens of vaccinated mice prior to infection (data not shown), indicating the vaccination regimen was successful. In turn, vaccinated mice showed significantly reduced MCMV replication in the liver at day 4 of infection, with spleens and lungs trending toward protection but not reaching significance (Fig. 4E). Notably, CD4 T epitope vaccination did not boost the MCMV epitope-specific CD8 T cell response (data not shown), indicating protection was likely due to directly enhancing CD4 T effector function.
DISCUSSION
Here, we identify the first MCMV epitope-specific CD4 T cells induced in BALB/c mice, extending our previous work identifying MCMV epitopes targeted by CD4 T cells in C57BL/6 mice (39). BALB/c mice have been used for decades to show the utility of CD8 T cell immunotherapy for protection against CMV-induced disease in the context of bone marrow transplantation (45). The identification of these epitopes will facilitate studying how MCMV-specific CD4 T cells contribute in this transplant model, in addition to providing valuable new tools to assess their function(s) during normal infection and vaccination settings. BALB/c mice do not encode the Ly49H-activating receptor expressed by C57BL/6 mice, which binds the viral m157 protein and results in a potent NK cell response that is not representative of what occurs in most MCMV infections of outbred/wild mice (46, 47). As NK cells play a role in shaping MCMV T cell responses (48, 49), the ability to study MCMV epitope-specific CD4 T cell responses in both BALB/c and C57BL/6 mice will provide valuable new insight.
Virus epitope-specific CD4 T cells are often induced at very low frequency, which is the case for the m78 and m53 responses identified here, and this poses a challenge for their study. Fortunately, an MHC-II tetramer was successfully generated for analysis of CD4 T cells targeting the m78417–431 epitope, allowing their characterization from several tissues after performing tetramer enrichment, which increases detection sensitivity by ∼100-fold (37). When the cell surface marker phenotype of m78-specific CD4 T cells was compared to that of the total antigen-experienced CD4 T cell compartment in MCMV-infected mice (based on their CD44hi CD11ahi CD49d+ phenotype [41]), they were quite similar for most markers examined, despite the fact that the m78 epitope likely identifies only ∼3 to 5% of the entire MCMV-specific CD4 T cell response at times of peak expansion. This result strongly suggests that the apparent CD4 CTL activity mediated by m78 and m53 CD4 T cells in liver compared to spleen, as well as the enhanced protection in the organ after vaccination, is not likely to be unique to these two particular MCMV epitope-specific CD4 T cell populations. In addition, to estimate the cytokine-producing potential of m78 CD4 T cells, we performed in vivo restimulation with this epitope, which may be more representative of how these cells respond to natural encounter of MCMV antigens in vivo (50). Interestingly, after peptide epitope injection into mice, only ∼15% of m78-specific CD4 T cells present in the spleen produced IFN-γ, and no expression of IL-2 or TNF was observed. Consistently, a similar percentage of IFN-γ+ m78 CD4 T cells was observed after PMA/ionomycin restimulation at the same time (data not shown). In contrast, ∼40 to 50% of CD44hi CD11ahi CD49d+ splenic CD4 T cells produced IFN-γ, IL-2, and/or TNF after PMA/ionomycin stimulation ex vivo, with many producing more than one of the cytokines. Whether this represents an inherent difference in the cytokine-producing potential of m78-specific cells from those of other MCMV-specific CD4 T cells or whether it is the result of technical differences when restimulating CD4 T cells by the two different methods remains to be determined.
Our results are the first we are aware of demonstrating that MCMV epitope-specific CD4 T cells can express high levels of granzyme B and that this expression is highly dependent upon the organ they reside in. Our data strongly suggest these cells can kill antigen-expressing target cells in vivo, and although other potential explanations for the specific loss of the peptide-loaded targets we observed are formally possible, we favor a model where these cells mediate direct cytolysis. Although helper functions of CD4 T cells have been studied for decades, more recently there has been increasing emphasis on deciphering nontraditional roles for CD4 T cells, especially with regard to harnessing these functions for potential vaccine development. In the context of several chronic viruses (e.g., HIV, HCMV, and EBV), CD4 T cells expressing canonical cytolytic molecules routinely circulate in the peripheral blood, but assessing the in vivo cytolytic potential of the cells is a major challenge for these human viral infections (21–23). In mice, there are only a few reports presenting data consistent with CD4 T cells mediating in vivo antiviral CTL activity (27–32), highlighting the complexity and technical challenges associated with developing robust assays/models to assess this. Relatively early during MCMV infection, ∼20% of m78-specific CD4 T cells resident in the liver express gzmB and appear capable of efficiently killing target cells there, whereas the ∼2% that express gzmB in the spleen exhibit virtually no cytolytic activity. This dramatic organ-specific difference in m78 CD4 CTL functions is very intriguing and raises important issues regarding the need to further our understanding of how the tissue environment(s) regulates the priming and/or differentiation of virus-specific CD4 T cells with distinct effector functions. Recent studies show that the transcription factors ThPOK and Runx3 are key regulators of CD4 versus CD8 T cell lineage differentiation decisions. ThPOK maintains the helper identity of CD4 T cells, whereas forced expression of Runx3 induces a cytolytic gene expression program (51, 52). Notably, in the gut mucosa, CD4 T cells lose ThPOK expression and acquire Runx3, suggesting chronic exposure to microbiota antigens can induce CD4 CTLs (53). Whether the expression balance of these two transcription factors may also impact this process during MCMV infection is an interesting question. In turn, determining if “stable” and “inflationary” MCMV-specific CD4 T cells exhibit distinct CTL activities in C57BL/6 mice will be of interest, as inflationary CD4 T cells expand at times when MCMV replication is restricted largely to the salivary gland mucosa (39).
In this study, we show for the first time that immunization with MHC-II (I-Ad)-restricted epitopes protects against MCMV challenge at times when CD4 T cells normally do not contribute to immune defense. As the frequencies of ICCS-detectable, IFN-γ-producing m53-specific CD4 T cells are very low during natural infection (Fig. 1C), it was comforting to also see a readily measurable population of these cells at day 4 following vaccination and MCMV challenge (Fig. 4D). Although a significant reduction in MCMV replication was not observed in the spleen or lung (despite trending lower) and reduced liver replication levels were modest, we chose a relatively simple, single-priming vaccination regimen in these first studies. Whether greater vaccine-mediated protection in the liver is a result of enhanced CD4 CTL activity in the organ is currently being explored. In addition, discovering whether boosting with the peptide epitopes, altering the adjuvant regimen, and/or including other methods to enhance CD4 T cell induction (e.g., anti-CD40 or anti-OX40) will improve protection will be of great interest.
ACKNOWLEDGMENTS
We thank Raphael Zellweger and Elina Zuniga for advice regarding establishing the in vivo CD4 T cell killing assay and Sujan Shresta and Mick Croft for valuable suggestions and feedback. We thank the NIH Tetramer Core Facility at Emory, AL, for generating the m78–MHC-II tetramer used in this study.
This work was supported by National Institutes of Health grant AI101423 to Chris A. Benedict and National Institutes of Health contract HHSN27220140045C to Alessandro Sette.
Funding Statement
The funding agencies had no role in study design, data collection and interpretation, or publication decisions.
REFERENCES
- 1.Staras SA, Dollard SC, Radford KW, Flanders WD, Pass RF, Cannon MJ. 2006. Seroprevalence of cytomegalovirus infection in the United States, 1988-1994. Clin Infect Dis 43:1143–1151. doi: 10.1086/508173. [DOI] [PubMed] [Google Scholar]
- 2.Pereira L, Maidji E, McDonagh S, Tabata T. 2005. Insights into viral transmission at the uterine-placental interface. Trends Microbiol 13:164–174. doi: 10.1016/j.tim.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Reddehase MJ, Mutter W, Munch K, Buhring HJ, Koszinowski UH. 1987. CD8-positive T lymphocytes specific for murine cytomegalovirus immediate-early antigens mediate protective immunity. J Virol 61:3102–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurz S, Steffens HP, Mayer A, Harris JR, Reddehase MJ. 1997. Latency versus persistence or intermittent recurrences: evidence for a latent state of murine cytomegalovirus in the lungs. J Virol 71:2980–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feuchtinger T, Opherk K, Bethge WA, Topp MS, Schuster FR, Weissinger EM, Mohty M, Or R, Maschan M, Schumm M, Hamprecht K, Handgretinger R, Lang P, Einsele H. 2010. Adoptive transfer of pp65-specific T cells for the treatment of chemorefractory cytomegalovirus disease or reactivation after haploidentical and matched unrelated stem cell transplantation. Blood 116:4360–4367. doi: 10.1182/blood-2010-01-262089. [DOI] [PubMed] [Google Scholar]
- 6.Peggs KS, Verfuerth S, Pizzey A, Khan N, Guiver M, Moss PA, Mackinnon S. 2003. Adoptive cellular therapy for early cytomegalovirus infection after allogeneic stem-cell transplantation with virus-specific T-cell lines. Lancet 362:1375–1377. doi: 10.1016/S0140-6736(03)14634-X. [DOI] [PubMed] [Google Scholar]
- 7.Reddehase MJ, Jonjic S, Weiland F, Mutter W, Koszinowski UH. 1988. Adoptive immunotherapy of murine cytomegalovirus adrenalitis in the immunocompromised host: CD4-helper-independent antiviral function of CD8-positive memory T lymphocytes derived from latently infected donors. J Virol 62:1061–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter EA, Greenberg PD, Gilbert MJ, Finch RJ, Watanabe KS, Thomas ED, Riddell SR. 1995. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N Engl J Med 333:1038–1044. doi: 10.1056/NEJM199510193331603. [DOI] [PubMed] [Google Scholar]
- 9.Gamadia LE, Remmerswaal EB, Weel JF, Bemelman F, van Lier RA, Ten Berge IJ. 2003. Primary immune responses to human CMV: a critical role for IFN-gamma-producing CD4+ T cells in protection against CMV disease. Blood 101:2686–2692. doi: 10.1182/blood-2002-08-2502. [DOI] [PubMed] [Google Scholar]
- 10.Komanduri KV, Viswanathan MN, Wieder ED, Schmidt DK, Bredt BM, Jacobson MA, McCune JM. 1998. Restoration of cytomegalovirus-specific CD4+ T-lymphocyte responses after ganciclovir and highly active antiretroviral therapy in individuals infected with HIV-1. Nat Med 4:953–956. doi: 10.1038/nm0898-953. [DOI] [PubMed] [Google Scholar]
- 11.Sester M, Sester U, Gartner B, Heine G, Girndt M, Mueller-Lantzsch N, Meyerhans A, Kohler H. 2001. Levels of virus-specific CD4 T cells correlate with cytomegalovirus control and predict virus-induced disease after renal transplantation. Transplantation 71:1287–1294. doi: 10.1097/00007890-200105150-00018. [DOI] [PubMed] [Google Scholar]
- 12.Lilleri D, Fornara C, Revello MG, Gerna G. 2008. Human cytomegalovirus-specific memory CD8+ and CD4+ T cell differentiation after primary infection. J Infect Dis 198:536–543. doi: 10.1086/590118. [DOI] [PubMed] [Google Scholar]
- 13.Tu W, Chen S, Sharp M, Dekker C, Manganello AM, Tongson EC, Maecker HT, Holmes TH, Wang Z, Kemble G, Adler S, Arvin A, Lewis DB. 2004. Persistent and selective deficiency of CD4+ T cell immunity to cytomegalovirus in immunocompetent young children. J Immunol 172:3260–3267. doi: 10.4049/jimmunol.172.5.3260. [DOI] [PubMed] [Google Scholar]
- 14.Lucin P, Pavic I, Polic B, Jonjic S, Koszinowski UH. 1992. Gamma interferon-dependent clearance of cytomegalovirus infection in salivary glands. J Virol 66:1977–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polic B, Hengel H, Krmpotic A, Trgovcich J, Pavic I, Luccaronin P, Jonjic S, Koszinowski UH. 1998. Hierarchical and redundant lymphocyte subset control precludes cytomegalovirus replication during latent infection. J Exp Med 188:1047–1054. doi: 10.1084/jem.188.6.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeitziner SM, Walton SM, Torti N, Oxenius A. 2013. Adoptive transfer of cytomegalovirus-specific effector CD4+ T cells provides antiviral protection from murine CMV infection. Eur J Immunol 43:2886–2895. doi: 10.1002/eji.201343690. [DOI] [PubMed] [Google Scholar]
- 17.Einsele H, Roosnek E, Rufer N, Sinzger C, Riegler S, Loffler J, Grigoleit U, Moris A, Rammensee HG, Kanz L, Kleihauer A, Frank F, Jahn G, Hebart H. 2002. Infusion of cytomegalovirus (CMV)-specific T cells for the treatment of CMV infection not responding to antiviral chemotherapy. Blood 99:3916–3922. doi: 10.1182/blood.V99.11.3916. [DOI] [PubMed] [Google Scholar]
- 18.Brown DM. 2010. Cytolytic CD4 cells: direct mediators in infectious disease and malignancy. Cell Immunol 262:89–95. doi: 10.1016/j.cellimm.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall NB, Swain SL. 2011. Cytotoxic CD4 T cells in antiviral immunity. J Biomed Biotechnol 2011:954602. doi: 10.1155/2011/954602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soghoian DZ, Streeck H. 2010. Cytolytic CD4(+) T cells in viral immunity. Expert Rev Vaccines 9:1453–1463. doi: 10.1586/erv.10.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elkington R, Shoukry NH, Walker S, Crough T, Fazou C, Kaur A, Walker CM, Khanna R. 2004. Cross-reactive recognition of human and primate cytomegalovirus sequences by human CD4 cytotoxic T lymphocytes specific for glycoprotein B and H. Eur J Immunol 34:3216–3226. doi: 10.1002/eji.200425203. [DOI] [PubMed] [Google Scholar]
- 22.Fleischer B. 1984. Acquisition of specific cytotoxic activity by human T4+ T lymphocytes in culture. Nature 308:365–367. doi: 10.1038/308365a0. [DOI] [PubMed] [Google Scholar]
- 23.Hammoud B, Schmueck M, Fischer AM, Fuehrer H, Park SJ, Akyuez L, Schefold JC, Raftery MJ, Schonrich G, Kaufmann AM, Volk HD, Reinke P. 2013. HCMV-specific T-cell therapy: do not forget supply of help. J Immunother 36:93–101. doi: 10.1097/CJI.0b013e31827b87cc. [DOI] [PubMed] [Google Scholar]
- 24.Casazza JP, Betts MR, Price DA, Precopio ML, Ruff LE, Brenchley JM, Hill BJ, Roederer M, Douek DC, Koup RA. 2006. Acquisition of direct antiviral effector functions by CMV-specific CD4+ T lymphocytes with cellular maturation. J Exp Med 203:2865–2877. doi: 10.1084/jem.20052246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suni MA, Ghanekar SA, Houck DW, Maecker HT, Wormsley SB, Picker LJ, Moss RB, Maino VC. 2001. CD4(+)CD8(dim) T lymphocytes exhibit enhanced cytokine expression, proliferation and cytotoxic activity in response to HCMV and HIV-1 antigens. Eur J Immunol 31:2512–2520. doi:. [DOI] [PubMed] [Google Scholar]
- 26.van Leeuwen EM, Remmerswaal EB, Heemskerk MH, ten Berge IJ, van Lier RA. 2006. Strong selection of virus-specific cytotoxic CD4+ T-cell clones during primary human cytomegalovirus infection. Blood 108:3121–3127. doi: 10.1182/blood-2006-03-006809. [DOI] [PubMed] [Google Scholar]
- 27.Brien JD, Uhrlaub JL, Nikolich-Zugich J. 2008. West Nile virus-specific CD4 T cells exhibit direct antiviral cytokine secretion and cytotoxicity and are sufficient for antiviral protection. J Immunol 181:8568–8575. doi: 10.4049/jimmunol.181.12.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hildemann SK, Eberlein J, Davenport B, Nguyen TT, Victorino F, Homann D. 2013. High efficiency of antiviral CD4(+) killer T cells. PLoS One 8:e60420. doi: 10.1371/journal.pone.0060420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jellison ER, Kim SK, Welsh RM. 2005. Cutting edge: MHC class II-restricted killing in vivo during viral infection. J Immunol 174:614–618. doi: 10.4049/jimmunol.174.2.614. [DOI] [PubMed] [Google Scholar]
- 30.Stuller KA, Flano E. 2009. CD4 T cells mediate killing during persistent gammaherpesvirus 68 infection. J Virol 83:4700–4703. doi: 10.1128/JVI.02240-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yauch LE, Prestwood TR, May MM, Morar MM, Zellweger RM, Peters B, Sette A, Shresta S. 2010. CD4+ T cells are not required for the induction of dengue virus-specific CD8+ T cell or antibody responses but contribute to protection after vaccination. J Immunol 185:5405–5416. doi: 10.4049/jimmunol.1001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown DM, Dilzer AM, Meents DL, Swain SL. 2006. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J Immunol 177:2888–2898. doi: 10.4049/jimmunol.177.5.2888. [DOI] [PubMed] [Google Scholar]
- 33.Jordan S, Krause J, Prager A, Mitrovic M, Jonjic S, Koszinowski UH, Adler B. 2011. Virus progeny of murine cytomegalovirus bacterial artificial chromosome pSM3fr show reduced growth in salivary glands due to a fixed mutation of MCK-2. J Virol 85:10346–10353. doi: 10.1128/JVI.00545-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verma S, Wang Q, Chodaczek G, Benedict CA. 2013. Lymphoid-tissue stromal cells coordinate innate defense to cytomegalovirus. J Virol 87:6201–6210. doi: 10.1128/JVI.00113-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiskopf D, Yauch LE, Angelo MA, John DV, Greenbaum JA, Sidney J, Kolla RV, De Silva AD, de Silva AM, Grey H, Peters B, Shresta S, Sette A. 2011. Insights into HLA-restricted T cell responses in a novel mouse model of dengue virus infection point toward new implications for vaccine design. J Immunol 187:4268–4279. doi: 10.4049/jimmunol.1101970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verma S, Loewendorf A, Wang Q, McDonald B, Redwood A, Benedict CA. 2014. Inhibition of the TRAIL death receptor by CMV reveals its importance in NK cell-mediated antiviral defense. PLoS Pathog 10:e1004268. doi: 10.1371/journal.ppat.1004268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pepper M, Linehan JL, Pagan AJ, Zell T, Dileepan T, Cleary PP, Jenkins MK. 2010. Different routes of bacterial infection induce long-lived TH1 memory cells and short-lived TH17 cells. Nat Immunol 11:83–89. doi: 10.1038/ni.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rawlinson WD, Farrell HE, Barrell BG. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol 70:8833–8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arens R, Wang P, Sidney J, Loewendorf A, Sette A, Schoenberger SP, Peters B, Benedict CA. 2008. Cutting edge: murine cytomegalovirus induces a polyfunctional CD4 T cell response. J Immunol 180:6472–6476. doi: 10.4049/jimmunol.180.10.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munks MW, Cho KS, Pinto AK, Sierro S, Klenerman P, Hill AB. 2006. Four distinct patterns of memory CD8 T cell responses to chronic murine cytomegalovirus infection. J Immunol 177:450–458. doi: 10.4049/jimmunol.177.1.450. [DOI] [PubMed] [Google Scholar]
- 41.McDermott DS, Varga SM. 2011. Quantifying antigen-specific CD4 T cells during a viral infection: CD4 T cell responses are larger than we think. J Immunol 187:5568–5576. doi: 10.4049/jimmunol.1102104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walton SM, Mandaric S, Torti N, Zimmermann A, Hengel H, Oxenius A. 2011. Absence of cross-presenting cells in the salivary gland and viral immune evasion confine cytomegalovirus immune control to effector CD4 T cells. PLoS Pathog 7:e1002214. doi: 10.1371/journal.ppat.1002214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Streeck H, D'Souza MP, Littman DR, Crotty S. 2013. Harnessing CD4(+) T cell responses in HIV vaccine development. Nat Med 19:143–149. doi: 10.1038/nm.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kurtz JR, Petersen HE, Frederick DR, Morici LA, McLachlan JB. 2014. Vaccination with a single CD4 T cell peptide epitope from a Salmonella type III-secreted effector protein provides protection against lethal infection. Infect Immun 82:2424–2433. doi: 10.1128/IAI.00052-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ebert S, Lemmermann NA, Thomas D, Renzaho A, Reddehase MJ, Holtappels R. 2012. Immune control in the absence of immunodominant epitopes: implications for immunotherapy of cytomegalovirus infection with antiviral CD8 T cells. Med Microbiol Immunol 201:541–550. doi: 10.1007/s00430-012-0268-8. [DOI] [PubMed] [Google Scholar]
- 46.Brizic I, Lenac Rovis T, Krmpotic A, Jonjic S. 2014. MCMV avoidance of recognition and control by NK cells. Semin Immunopathol 36:641–650. doi: 10.1007/s00281-014-0441-9. [DOI] [PubMed] [Google Scholar]
- 47.Scalzo AA, Yokoyama WM. 2008. Cmv1 and natural killer cell responses to murine cytomegalovirus infection. Curr Top Microbiol Immunol 321:101–122. [DOI] [PubMed] [Google Scholar]
- 48.Andrews DM, Estcourt MJ, Andoniou CE, Wikstrom ME, Khong A, Voigt V, Fleming P, Tabarias H, Hill GR, van der Most RG, Scalzo AA, Smyth MJ, Degli-Esposti MA. 2010. Innate immunity defines the capacity of antiviral T cells to limit persistent infection. J Exp Med 207:1333–1343. doi: 10.1084/jem.20091193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitrovic M, Arapovic J, Jordan S, Fodil-Cornu N, Ebert S, Vidal SM, Krmpotic A, Reddehase MJ, Jonjic S. 2012. The NK cell response to mouse cytomegalovirus infection affects the level and kinetics of the early CD8(+) T-cell response. J Virol 86:2165–2175. doi: 10.1128/JVI.06042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cabrera-Perez J, Condotta SA, James BR, Kashem SW, Brincks EL, Rai D, Kucaba TA, Badovinac VP, Griffith TS. 2015. Alterations in antigen-specific naive CD4 T cell precursors after sepsis impairs their responsiveness to pathogen challenge. J Immunol 194:1609–1620. doi: 10.4049/jimmunol.1401711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naito T, Taniuchi I. 2010. The network of transcription factors that underlie the CD4 versus CD8 lineage decision. Int Immunol 22:791–796. doi: 10.1093/intimm/dxq436. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Wildt KF, Castro E, Xiong Y, Feigenbaum L, Tessarollo L, Bosselut R. 2008. The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity 29:876–887. doi: 10.1016/j.immuni.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reis BS, Rogoz A, Costa-Pinto FA, Taniuchi I, Mucida D. 2013. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4(+) T cell immunity. Nat Immunol 14:271–280. doi: 10.1038/ni.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]