ABSTRACT
The immediate early 62 protein (IE62) of varicella-zoster virus (VZV), a major viral trans-activator, initiates the virus life cycle and is a key component of pathogenesis. The IE62 possesses several domains essential for trans-activation, including an acidic trans-activation domain (TAD), a serine-rich tract (SRT), and binding domains for USF, TFIIB, and TATA box binding protein (TBP). Transient-transfection assays showed that the VZV IE62 lacking the SRT trans-activated the early VZV ORF61 promoter at only 16% of the level of the full-length IE62. When the SRT of IE62 was replaced with the SRT of equine herpesvirus 1 (EHV-1) IEP, its trans-activation activity was completely restored. Herpes simplex virus 1 (HSV-1) ICP4 that lacks a TAD very weakly (1.5-fold) trans-activated the ORF61 promoter. An IE62 TAD-ICP4 chimeric protein exhibited trans-activation ability (10.2-fold), indicating that the IE62 TAD functions with the SRT of HSV-1 ICP4 to trans-activate viral promoters. When the serine and acidic residues of the SRT were replaced with Ala, Leu, and Gly, trans-activation activities of the modified IE62 proteins IE62-SRTΔSe and IE62-SRTΔAc were reduced to 46% and 29% of wild-type activity, respectively. Bimolecular complementation assays showed that the TAD of IE62, EHV-1 IEP, and HSV-1 VP16 interacted with Mediator 25 in human melanoma MeWo cells. The SRT of IE62 interacted with the nucleolar-ribosomal protein EAP, which resulted in the formation of globular structures within the nucleus. These results suggest that the SRT plays an important role in VZV viral gene expression and replication.
IMPORTANCE The immediate early 62 protein (IE62) of varicella-zoster virus (VZV) is a major viral trans-activator and is essential for viral growth. Our data show that the serine-rich tract (SRT) of VZV IE62, which is well conserved within the alphaherpesviruses, is needed for trans-activation mediated by the acidic trans-activation domain (TAD). The TADs of IE62, EHV-1 IEP, and HSV-1 VP16 interacted with cellular Mediator 25 in bimolecular complementation assays. The interaction of the IE62 SRT with nucleolar-ribosomal protein EAP resulted in the formation of globular structures within the nucleus. Understanding the mechanisms by which the TAD and SRT of IE62 contribute to the function of this essential regulatory protein is important in understanding the gene program of this human pathogen.
INTRODUCTION
Varicella-zoster virus (VZV), a member of the Alphaherpesvirinae, is a ubiquitous human pathogen that causes varicella (chicken pox) in primary lytic infection and zoster (shingles) during reactivation from latent infection (1). VZV has a linear DNA genome of approximately 125 kbp that encodes at least 70 open reading frames (ORFs) (1). Herpesviruses with genomes of group D, such as equine herpesvirus 1 (EHV-1), bovine herpesvirus 1 (BHV-1), pseudorabies virus (PRV), and varicella-zoster virus (VZV), have a fixed long region (UL) covalently linked to a short (S) genomic region comprised of a pair of inverted repeat sequences that bracket a unique short segment (US) (2–4). Two alphaherpesviruses, VZV and EHV-1, are members of genus Varicellovirus and have very similar genomic structures (2, 4). The VZV immediate early 62 protein (IE62) of 1,310 amino acids (aa) is the major viral trans-activator and is essential for viral growth (1, 5, 6). IE62 is encoded by duplicated ORFs 62 and 71 and, as a tegument protein, is delivered to newly infected cell nuclei, where it initiates VZV replication by transactivating viral immediate early, early, and late genes (7, 8). IE62 binds several viral proteins, such as ORF4 (9), ORF9 (10), ORF47 (11), and ORF63 (12), and cellular transcription factors, including upstream stimulatory factor (USF), TATA-binding protein (TBP), TFIIB, and Sp1 (13–16).
The acidic trans-activation domain (TAD) of IE62 shows some compositional similarity (containing primarily aliphatic and acidic residues) to the 81-aa HSV-1 VP16 TAD (17). The acidic TAD is not present in ICP4 of herpes simplex virus 1 (HSV-1) (18–20). The IE62 TAD targets a Mediator complex lacking CDK8 and interacts directly with aa 402 to 590 of the Med25 subunit, and site-specific TAD mutations abolish this interaction (16, 17). Lester and DeLuca showed that four components of the Mediator complex, such as MED7, MED8, TRAP220, and CRSP77, were copurified with HSV-1 ICP4 (21). Mediator complexes contain over 30 polypeptides organized as subunits to form head, middle, tail, and CDK submodules (22–24). Mediators are believed to transduce signals between RNA polymerase II (Pol II) and transcriptional activators, such as Sp1, p53, the adenovirus E1A protein, the HSV-1 activator VP16 acidic trans-activation domain (TAD), and the general transcription apparatus (17, 23, 25). In the current model of activated transcription, Mediator is recruited to the promoter by the TADs of activators. This interaction allows further recruitment of general transcription factors and Pol II, resulting in preinitiation-complex assembly and subsequent activated transcription (24).
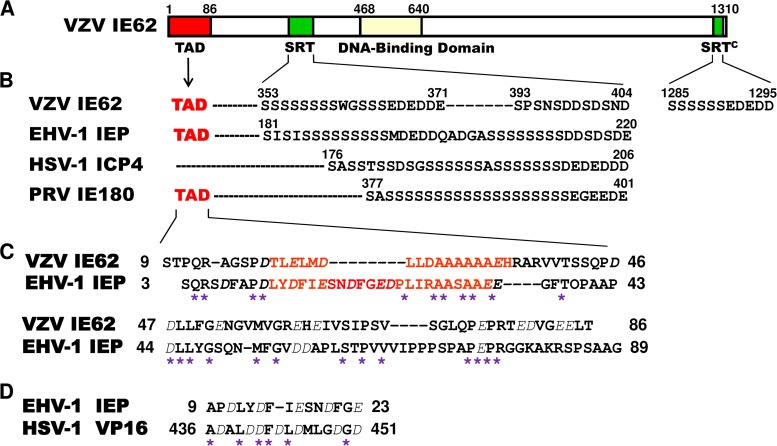
A comparison of the amino acid sequences of the serine-rich tract (SRT) of VZV IE62, EHV-1 IEP, HSV-1 ICP4, and pseudorabies virus (PRV) IE180 showed that the SRTs of these alphaherpesviruses are very similar (26) (Fig. 1). The serine-rich region is followed by five to seven acidic residues. The SRT at aa 353 to 404 of VZV IE62 contains two serine-rich regions and two acidic regions (Fig. 1). The second SRT, consisting of aa 1285 to 1295 of VZV IE62, is located in the C terminus (SRTc) (Fig. 1). The SRT of HSV-1 ICP4 functionally colocalizes and interacts with the nucleolar-ribosomal protein EAP (Epstein-Barr virus [EBV]-associated protein), and a late viral function is required for ICP4-EAP association (27, 28). The SRT of EHV-1 IEP also interacts with EAP, which is important for viral gene expression (29) and is essential for viral growth (30). EBV, an oncogenic herpesvirus, encodes two small RNAs (EBERs) that are expressed at high levels during latent transformation of human B lymphocytes. EBERs are associated with EAP, which is a 15-kDa nucleolar-ribosomal protein (31), later found to be L22, a ribosomal component (32).
FIG 1.
Comparison of the amino acid sequences of the serine-rich region (SRT) and acidic regions of VZV IE62, EHV-1 IEP, HSV-1 ICP4, and PRV IE180. (A) The top diagram represents the 1,310-aa IE62 of VZV. TAD, acidic trans-activation domain. All numbers refer to the number of amino acids from the N terminus of each protein. (B) The similarity between the sequences, with the run of serines followed by five to seven acidic residues, is evident. (C) Comparison of the deduced TAD domains of VZV IE62 and EHV-1 IEP. Dashes represent gaps introduced for maximal alignment. Acidic amino acids are italicized. Potential α-helix regions are in red. Asterisks indicated conserved amino acids. (D) Comparison of EHV-1 IEP residues 9 to 23 to residues 436 to 451 of HSV-1 VP16.
In the work presented here, we examined the molecular mechanism by which the TAD and SRT of IE62 function in activating transcription. Our data indicate that the SRT of VZV IE62 is necessary for TAD-mediated trans-activation. Bimolecular complementation (BiMC) assays show that the TAD of IE62 interacts with cellular Mediator 25, and the results suggest that the TAD and SRT play an important role in VZV gene programming.
MATERIALS AND METHODS
Cell culture.
Human melanoma MeWo cells were kindly provided by Jeffrey Cohen (18). Mouse fibroblast L-M, rabbit kidney RK13, and MeWo cells were maintained at 37°C in complete Eagle's minimum essential medium (EMEM) supplemented with 100 U/ml of penicillin, 100 μg/ml of streptomycin, nonessential amino acids, and 5% fetal bovine serum.
Mammalian expression plasmids.
Plasmids were constructed and maintained in Escherichia coli (E. coli) HB101 or JM109 by standard methods (33). Plasmids pSVIE and pSVIR2 have been described previously (34, 35). To generate plasmid pSVTAD-IR2, an IEP TAD-IR2N minigene that contains aa 1 to 89 of the EHV-1 IEP TAD [TAD(1–89)] and IR2P(1–105) was synthesized by Integrated DNA Technologies, Inc. (IDT;Coralville, IA). The 584-bp fragment of the IEP TAD-IR2N was cloned into the KpnI and BamHI sites of pSVIR2-KH (29), and the plasmid was designated pSVTAD-IR2. To generate plasmid pSVTS-IR2, an IEP TS-IR2N minigene that contains IEP TAD, SRT, and IR2P(1–105) was synthesized (IDT), and the 784-bp fragment of the IEP-TS-IR2N was cloned into the KpnI and BamHI sites of pSVIR2-KH; the plasmid was designated pSVTS-IR2N. Plasmid pSVTS-IR2N was digested with XbaI to delete a 36-bp XbaI fragment and self-ligated to generate pSVTS-IR2. The minigene IEP-TAD2, in which some amino acids have different codons from the IEP TAD, was synthesized, and a 279-bp fragment of the IEP-TAD2 was cloned into the PmeI and XbaI sites of pSVTS-IR2 to generate plasmid pSVTT-IR2.
Minigene 62TS that contains VZV IE62 TAD and SRT was synthesized, and the 489-bp fragment of the 62TS was cloned into the KpnI and XbaI sites of pSVTS-IR2 to generate plasmid pSV62TS-IR2. To generate plasmid pSV62T-IR2, pSV62TS-IR2 was digested with PmeI and XbaI, and a synthetic DNA linker 62 (coding strand, 5′-CAAGACGACTACT-3′; noncoding strand, 5′-CAAGACGACTACT-3′) was inserted. Minigene IE62-TAD2, in which some amino acids have different codons from the IEP TAD, was synthesized, and the 270-bp fragment of the IEP-STAD was cloned into the PmeI and XbaI sites of pSV62TS-IR2 to generate plasmid pSVTT-IR2. Minigene IE62-SRTΔSe, in which 10 of 16 serines of the SRT were replaced with Ala, Leu, and Gly, was synthesized, and a 204-bp fragment of the IE62-SRTΔSe construct was cloned into the PmeI and XbaI sites of pSV62TS-IR2 to generate plasmid pSV62TSΔSe-IR2. Minigene IE62-SRTΔAc, in which 8 of 10 acidic residues of the SRT were replaced with Ala, Leu, and Gly, was synthesized, and a 204-bp fragment of the IE62-SRTΔAc construct was cloned into the PmeI and XbaI sites of pSV62TS-IR2 to generate plasmid pSV62TSΔAc-IR2. To generate pSVIR2-62TS, a 489-bp fragment of the minigene 62TS was cloned into the XhoI and HindIII sites of pSVIR2-H2 (29). To generate plasmid pSV62TS2-IR2, a 630-bp IE62 DNA fragment (aa 1 to 210) was amplified from pCMV-IE62 (pOka strain) by PCR with the primers IE62SRT-F (forward) and IE62SRT-R2 (reverse), and the fragment was cloned into the KpnI and PmeI sites of pSV62TS-IR2. To generate plasmid pSV62TS3-IR2, a 1,060-bp IE62 DNA fragment (aa 1 to 350) was amplified from pCMV-IE62 by PCR with the primers IE62SRT-F (forward) and IE62SRT-R1 (reverse) (see Table S1 in the supplemental material for sequences of the primers named above), and the fragment was cloned into the KpnI and PmeI sites of pSV62TS-IR2.
Plasmid pcDNA-IE62 (pCMV-IE62; pOka strain), which expresses the full-length VZV IE62, was provided by Ann M. Arvin (Stanford University School of Medicine) (36). To generate plasmid pCMV-IE62ΔS (SRT), a 1,050-bp N-terminal IE62 DNA fragment (IE62 aa 1 to 350) was amplified from pCMV-IE62 by PCR with primers IE62N-F1 (forward) and IE62N-R1 (reverse), which contain an NheI sequence (underlined in the primer sequences in Table S1). The fragment was cloned into the EcoRI and XbaI sites of the pSVSPORT1, and the plasmid was designated pIE62-N. A 500-bp IE62 DNA fragment (IE62 aa 411 to 571) was amplified from pCMV-IE62 by PCR with primers IE62N-F2 (forward) and IE62N-R2 (reverse) that contain an EcoRV sequence (underlined in the primer sequences in Table S1), and the fragment was cloned into the EcoRI and KpnI sites of the pIE62-N. This plasmid was designated pIE62-NM. A 1,650-bp fragment of pIE62-NM was cloned into the KpnI and NheI sites of the pCMV-IE62 to generate pCMV-IE62ΔS. The minigene IE62ΔSc (IE62 aa 1167 to 1310), in which 4 of 6 serines and 3 of 5 acidic residues of the SRTc were replaced with Ala, Leu, and Gly, was synthesized, and the 430-bp fragment of the IE62ΔSc was cloned into the BsmI and XbaI sites of pCMV-IE62 to generate plasmid pCMV-IE62ΔSc. To generate plasmid pCMV-IE62ΔSSc, the 430-bp fragment of the IE62ΔSc construct was cloned into the BsmI and XbaI sites of pCMV-IE62ΔS. To generate plasmid pCMV-IE62ΔS-IEP SRT, a 150-bp IEP SRT DNA fragment (IEP aa 176 to 221) was amplified from pSVIE by PCR with primers YSRT-F3 (forward) and YSRT-R3 (reverse) (see Table S1 in the supplemental material for sequences) that contain the EcoRV sequence (underlined in Table S1). The fragment was cloned into the EcoRI and EcoRI sites of the pIE62-NM, and the plasmid was designated pIE62NM-IEP SRT. A 1,800-bp fragment of pIE62NM-IEP SRT was cloned into the KpnI and NheI sites of the pCMV-IE62 to generate pCMV-IE62ΔS-IEP SRT.
To generate plasmid pCMV-IE62-SRTΔSe, a 200-bp IE62 SRTΔSe DNA was amplified from the minigene IE62-SRTΔSe by PCR with primers SRT-F5 (forward) and SRT-R5 (reverse) (see Table S1 in the supplemental material for sequences) and synthesized; the 200-bp fragment was cloned into the EcoRV and EcoRI sites of pIE62-NM, and the plasmid was designated pIE62-NSΔSe. A 1,750-bp fragment of pIE62-NSΔSe was cloned into the KpnI and NheI sites of pCMV-IE62 to generate pCMV-IE62-SRTΔSe. To generate plasmid pCMV-IE62-SRTΔAc, a 200-bp IE62 SRTΔSe DNA fragment was amplified from the minigene IE62-SRTΔAc by PCR with primers SRT-F6 (forward) and SRT-R5 (reverse) (see Table S1 in the supplemental material for sequences)and synthesized; the 200-bp fragment was cloned into the EcoRV and EcoRI sites of pIE62-NM, and the plasmid was designated pIE62-NSΔAc. A 1,750-bp fragment of pIE62-NSΔAc was cloned into the KpnI and NheI sites of the pCMV-IE62 to generate pCMV-IE62-SRTΔAc.
To generate plasmid pSV-ICP4, a 4.0-kb HSV-1 ICP4 DNA fragment was amplified from pK1-2 (provided by Neal A. DeLuca [37]) by PCR with primers (forward, 5′-GGGAATTCCGCCGTCGCAGCCGTATCCCCGGAGGATCGCCCCGCATCG-3′; reverse, 5′-GGACTAGTTTATTGCGTCTTCGGGTCTCACAAGCGCCCCGCCCCGTCC-3′) that contain EcoRI and SpeI sites, respectively, and the fragment was cloned into the EcoRI and SpeI sites of the pSVSPORT1. To generate plasmid pSV-62TAD-ICP4, a 370-bp N-terminal ICP4 DNA was amplified from pSV-ICP4 by PCR with primers TAD-P4-F (forward) and TAD-P4-R (reverse) that contain KpnI and PmeI sequences (underlined, respectively, in the primer sequences in Table S1 in the supplemental material), and the fragment was cloned into the EcoRI and PvuII sites of the pSVIR2. This plasmid was designated pICP4-N. The minigene IE62 TAD-SRT that contains the IE62 TAD and SRT was synthesized, and a 270-bp fragment of IE62 TAD-SRT was cloned into the KpnI and PmeI sites of pICP4-N; the plasmid was designated pSV-62TAD-ICP4.
YFP fusion plasmids.
Plasmid pCS2-YFP-venus-FL (pYFP) (where YFP is yellow fluorescent protein and FL is full-length) was provided by Ronald N. Harty (University of Pennsylvania) (38). To generate pNYFP, a 540-bp N-terminal YFP DNA fragment was amplified from pYFP by PCR with primers NYFP-F (forward) and NYFP-R (reverse) (see Table S1 in the supplemental material for sequences), and the fragment was cloned into the EcoRI and NotI sites of pYFP. To generate pCYFP, a 210-bp C-terminal YFP DNA was amplified from pYFP by PCR with primers CYFP-F (forward) and CYFP-R (reverse) (see Table S1), and the fragment was cloned into the EcoRI and NotI sites of pYFP. To generate pYn-62TAD, a 279-bp IE62 TAD(1–86) DNA fragment was amplified from pCMV-IE62 by PCR with primers YIE62-F (forward) and YIE62-R2 (reverse) (see Table S1), and the fragment was cloned into the KpnI and AflII sites of the pNYFP. To generate pYn-IEP TAD, a 290-bp IE62 TAD DNA was amplified from pSVIE by PCR with primers IETAD1-F (forward) and IETAD89-R2 (reverse) (see Table S1), and the fragment was cloned into the KpnI and NotI sites of the pNYFP. To generate pYn-16TAD, a 290-bp VP16 TAD(411–490) DNA fragment was amplified from pGEX-VP16TAD by PCR with primers YVP16-F (forward) and VP16-R2 (reverse) (see Table S1), and the fragment was cloned into the KpnI and NotI sites of the pNYFP. To generate pYn-EAP, a 384-bp EAP DNA fragment was amplified from the minigene IDT-EAP (IDT) by PCR with primers YEAP-F (forward) and YEAP-R (reverse), and the fragment was cloned into the KpnI and NotI sites of the pNYFP.
To generate plasmid pYc-62S-IR2, pYn-62S was digested with KpnI and AflII, a synthetic DNA linker, YH (coding strand, 5′-CCGTTTAAACGGAAGCTTC-3′; noncoding strand, 5′-TTAAGAAGCTTCCGTTTAAACGGGTAC-3′), was inserted, and the plasmid was designated pYn-PH. A 200-bp fragment of pCYFP was cloned into the EcoRI and KpnI sites of pYn-PH to generate plasmid pYc-PH. A 4.1-kbp fragment of the pSV62TS-IR2 was cloned into the PmeI and HindIII sites of pYc-PH to generate plasmid pYc-62SIR2. A 4.1-kbp fragment of pSVTS-IR2 was cloned into the PmeI and HindIII sites of pYc-PH to generate plasmid pYc-SIR2. To generate plasmid pYc-IR2, pYc-62SIR2 was digested with PmeI and XbaI, and a synthetic DNA linker, IR2-PX (coding strand, 5′-AAACGCTGGTGCGT-3′; noncoding strand, 5′-CTAGACGCACCAGCGTTT-3′), was inserted. To generate pYc-ETIF, a 1.35-kbp ETIF DNA fragment was amplified from plasmid pCETIF (39) by PCR with primers YETIF-F (forward) and YETIF-R (reverse) (see Table S1 in the supplemental material for sequences), and a 1.35-kbp ETIF fragment was cloned into the XbaI and HindIII sites of the pYc-IR2.
To generate pYc-Med25, a Med25 minigene that contains Mediator 25 VBD (aa 391 to 590) was synthesized, and a 570-bp fragment of Med25 was cloned into the KpnI and NotI sites of the pCYFP. To generate pYn-62SRT, a 180-bp IE62 SRT (aa 351 to 410) DNA fragment was amplified from pCMV-IE62 by PCR with primers Y62SRT-F (forward) and Y62SRT-R3 (reverse) (see Table S1 in the supplemental material for sequences), and the fragment was cloned into the KpnI and AflII sites of the pNYFP. To generate pYn-SRT, a 180-bp IEP SRT (aa 176 to 221) DNA fragment was amplified from pSVIE by PCR with primers YSRT-F (forward) and YSRT-R (reverse) (see Table S1), and the PCR fragment was cloned into the KpnI and AflII sites of the pNYFP. To generate pYn-IE62, pYn-SRT was digested with KpnI and NotI, and a synthetic DNA linker, Yn-NX (coding strand, 5′-AGCTAGCTTAGGTACCTTATCTAGAGC-3′; noncoding strand, 5′-GGCCGCTCTAGATAAGGTACCTAAGCTAGCTGTAC-3′) that contains NheI, KpnI, and XbaI (underlined) was inserted. This plasmid, designated pYn-NX, has a KpnI cloning site that cannot be recleaved because the synthetic linker Yn-NX lacks the KpnI consensus sequence at the end of the linker Yn-NX (TGTAC in the above sequence). The 4-kbp fragment of pCMV-IE62 was cloned into the NheI and XbaI sites of pYn-NX to generate pYn-IE62. The 1,550-bp fragment of pIE62-NM was cloned into the KpnI and NheI sites of the pYn-IE62 to generate pYn-IE62ΔSRT. To generate pYc-IE62, a 200-bp Yc DNA fragment of the plasmid pYc was cloned into the BglII and KpnI sites of the pYn-IE62.
Luciferase reporter and the secreted alkaline phosphatase (SEAP) plasmids.
To generate plasmid pORF61-Luc, a ORF61-Pro minigene that contains the VZV ORF61 promoter region located at bp −363 to −1 relative to the first ATG of the OFR61 open reading frame (VZV pOka strain) was synthesized (IDT) and cloned into the BglII and HindIII sites of pGL3-basic (Promega). To generate plasmid pgI-Luc, minigene gI-Pro that contains the VZV glycoprotein I (gI) promoter region located at bp −360 to −1 relative to the first ATG of the open reading frame (VZV pOka strain) was synthesized and cloned into the BglII and HindIII sites of pGL3-basic (Promega). Plasmid pICP22-SEAP has been described previously (40).
Luciferase reporter and the SEAP assays.
The luciferase reporter assay was performed by using Lipofectamine LTX with Plus reagent (Invitrogen, San Diego, CA) according to the manufacturer's protocol. L-M cells were seeded at 50% confluence in 24-well plates and transfected with 0.12 pmol of the reporter vector and 0.08 pmol of effectors in each well. Lipofectamine LTX (5.4 μl) was diluted with 114 μl of Opti-MEM (Invitrogen). DNA and 1.95 μl of Plus reagent were mixed with 114 μl of Opti-MEM. The total amount of DNA was adjusted to the same amount with pSVSPORT1 DNA. The solutions were combined and incubated at room temperature for 5 min, and one-third volume was transferred into each of three wells of L-M cells. At 40 h posttransfection, luciferase activity was measured with a luciferase assay kit (Promega, Madison, WI) and a Polarstar Optima plate reader (BMG Labtech, Inc., Cary, NC). SEAP activity in culture medium was determined as previously described (29) by a chemiluminescence method using a Great Escape SEAP kit (Clontech, Palo Alto, CA).
Generation of anti-IE62 trans-activation domain (TAD) polyclonal antibody (pAb).
Two peptides, PPMQRSTPQRAGSP (TAD1; aa 4 to 17) and SGLQPEPRTEDVGE (TAD2; aa 70 to 83), of the IE62 TAD of VZV were synthesized, conjugated to keyhole limpet hemocyanin (KLH), and used for immunization in rabbit and generation of polyclonal anti-IE62 TAD antibodies (GenScript Corp., Piscataway, NJ). Anti-IE62 TAD polyclonal antibodies (IE62T1 and IE62T2) were affinity purified and verified by Western blot analysis (see Fig. 5E).
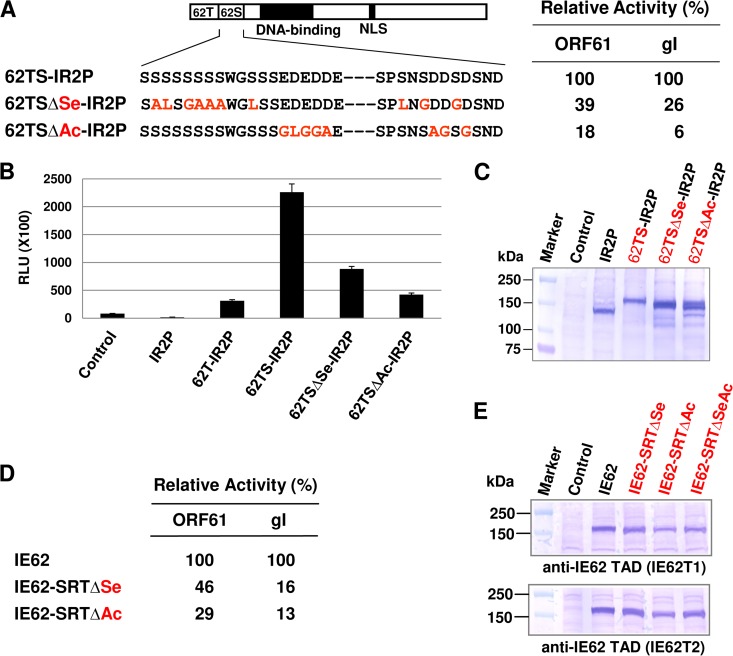
FIG 5.
Both the serine-rich region and the acidic residues of IE62 SRT are necessary for TAD-mediated trans-activation. (A) The top diagram represents IE62 TAD-SRT-IR2P chimeric fusion proteins. IE62 SRT mutants with multiple amino acid substitutions were generated and analyzed by a luciferase reporter assay. Ten of 16 serines and 8 of 10 acidic residues were replaced with Ala, Leu, and Gly. The table on the right shows a summary of the luciferase reporter assays. ORF61 and gI represent VZV ORF61 and glycoprotein I (gI) promoter-luciferase reporter plasmids, respectively. (B) Luciferase reporter assays were performed as described in the legend of Fig. 2. Data are averages and are representative of three independent experiments. RLU, relative luminescence units. (C) The IE62 SRT mutants were confirmed by Western blot analyses and DNA sequencing (data not shown). The SRT mutants were detected with the IEP antibody OC33. Luciferase reporter assays (D) and Western blot analyses (E) were performed with two IE62 mutants, IE62-SRTΔSe and IE62-SRTΔAc, represented in the scheme in panel A. Data are averages and are representative of three independent experiments. The IE62 mutants were detected with the anti-IE62 TAD polyclonal antibodies IE62T1 and IE62T2 (see Materials and Methods).
Western blot analysis.
The preparation of cytoplasmic and nuclear extracts of transfected cells and Western blot analysis were performed as previously described (41). Blots were incubated with a polyclonal antibody to IEP OC33 (42), a monoclonal antibody to ICP4 58S (ATCC), a polyclonal antibody to IE62 (sc-17525; Santa Cruz Biotechnology, Santa Cruz, CA), or a polyclonal antibody to green fluorescent protein (GFP) (sc-8334; Santa Cruz Biotech) for 2 h. The blots were washed three times for 10 min each in Tris-buffered saline plus Tween (TBST) and incubated with the secondary antibody (anti-rabbit IgG Fc-alkaline phosphatase [AP] conjugate [Promega]) for 1 h. Proteins were visualized by incubating the membranes containing blotted protein in AP conjugate substrate (AP conjugate substrate kit; Bio-Rad) according to the manufacturer's directions.
Transfection and confocal microscopy.
Transfection and confocal microscopy were carried out as previously described (43), with slight modifications. RK13 cells were seeded on two-chamber glass slides (Nalgene Nunc International, Naperville, IL) and transfected with the indicated plasmids using Lipofectamine LTX and Plus reagent (Invitrogen). After 48 h, cells were washed twice with phosphate-buffered saline (PBS; pH 7.2) and fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. Cells were washed four times with PBS and affixed to glass slides with Prolong Gold Antifade with 4′,6′-diamidino-2-phenylindole (DAPI; Invitrogen). Slides were viewed by a Leica TCS SP5 confocal microscope (Leica Microsystems, Inc., Buffalo Grove, IL).
RESULTS
The SRT of the VZV IE62 is important for its trans-activation activity.
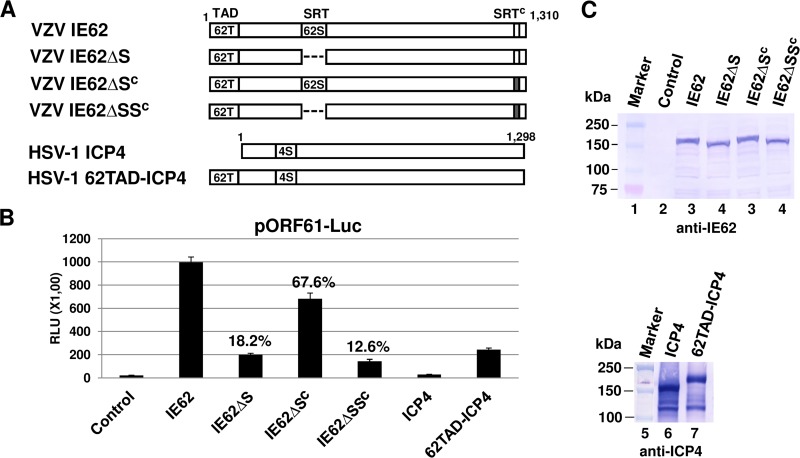
The SRT of alphaherpesviruses is well conserved, and the SRT of HSV-1 ICP4 and EHV-1 IEP interacts with the nucleolar-ribosomal protein EAP (28, 43). It has been suggested that the SRT plays a role in viral gene regulation. To investigate whether the SRT of the VZV IE62 functions in activating transcription, SRT deletion mutants of the IE62 were generated (Fig. 2A) and confirmed by DNA sequencing (see Fig. S1 and S2 in the supplemental material for sequences) and Western blot analysis (Fig. 2C). The sequence data confirm that the SRT deletion mutants are in frame and that the sizes of the proteins reflected the deletions (Fig. 2C). In a luciferase reporter assay, the intact IE62 strongly trans-activated the early VZV ORF61 promoter (Fig. 2B, IE62). However, the IE62 lacking the SRT activated the ORF61 promoter to only 18.2% of the level of the full-length IE62 (Fig. 2B, IE62ΔS). When 11 amino acids (SSSSSSEDEDD) of the C-terminal SRT (SRTc) of IE62 were mutated to SAGAGSALGDD, its trans-activation activity was reduced to 67.6% of that of full-length IE62 (Fig. 2B, IE62ΔSc). The IE62 lacking both the SRT and SRTc activated the ORF61 promoter to only 12.6% of the level of the full-length IE62 (Fig. 2B, IE62ΔSSc), indicating that both the SRT and SRTc are important for IE62's trans-activation activity. HSV-1 ICP4, which lacks the activation domain similar to the acidic transcriptional activation domain (TAD) of VZV IE62 (18, 19, 44), minimally trans-activated the ORF61 promoter (Fig. 2B, ICP4). Interestingly, when the IE62 TAD was fused to the N terminus of HSV-1 ICP4 (Fig. 2B, 62TAD-ICP4), the chimeric fusion protein increased its trans-activation activity from 1.4-fold for ICP4 to 11.6-fold, indicating that the SRT is important for trans-activation ability.
FIG 2.
The SRT of IE62 is important for its trans-activation activity. (A) Schematic diagram of SRT-deletion mutants. 62T, IE62 TAD; 62S, IE62 SRT; Sc, IE62 SRTc; 4S, HSV-1 ICP4 SRT. (B) Luciferase reporter assay. L-M cells were cotransfected with 0.12 pmol of the reporter plasmid pORF61-Luc and 0.08 pmol of effector plasmid (pSV-SPORT1, pCMV-IE62, pCMV-IE62ΔSRT, pCMV-IE62ΔSc, pCMV-IE62ΔSSc, pSV-ICP4, or pSV62TAD-ICP4). The firefly luciferase signals were normalized to the internal secreted alkaline phosphatase (SEAP) transfection control. Data are averages and are representative of three independent experiments. RLU, relative luminescence units. The percentages represent the relative activity of the mutant IE62 compared to that of the wild-type control. (C) Western blot analysis of the chimeric fusion proteins. Western blot analysis was performed by using anti-ICP4 monoclonal antibody 58S (ATCC) or an anti-IE62 pAb (sc-17525; Santa Cruz Biotechnology, Santa Cruz, CA).
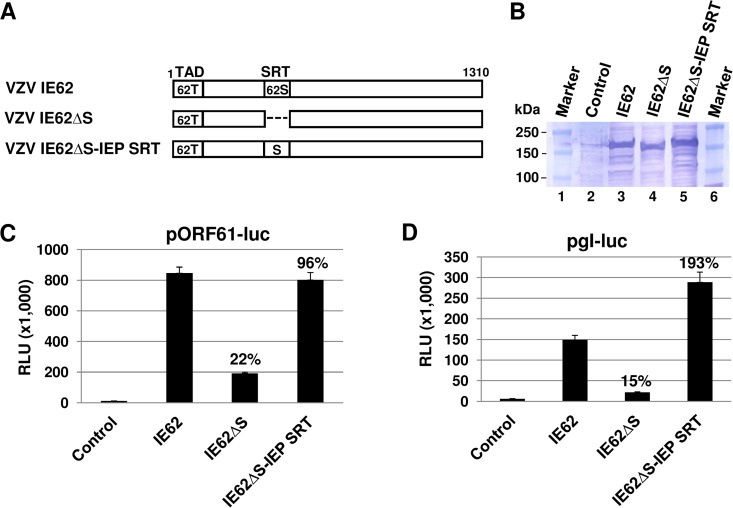
The SRT of EHV-1 IEP can functionally replace the SRT of IE62.
Since the SRT of alphaherpesviruses is well conserved, the SRT of IE62 was replaced with the SRT of EHV-1 IEP (Fig. 3A). The chimeric IE62 protein was confirmed by DNA sequencing (see Fig. S1 in the supplemental material for sequences) and Western blot analysis (Fig. 3B). As expected, IE62ΔSRT (IE62ΔS) trans-activated the ORF61 promoter to about 22% of the level of the full-length IE62 (Fig. 3C). The chimeric IE62ΔS-IEPSRT protein trans-activated the early ORF61 promoter to about 96% of the level of the full-length IE62. The IE62ΔS-IEPSRT trans-activated the late gI promoter by 193% of the level of the full-length IE62 (Fig. 3D, fourth bar). These results demonstrated that the chimeric IE62ΔS-IEPSRT protein restored trans-activation activity equal to that of the wild-type (wt) IE62.
FIG 3.
The SRT of IE62 can be replaced with the SRT of EHV-1 IEP. (A) Schematic diagram of IE62 SRT substitution mutants. 62T, IE62 TAD; 62S, IE62 SRT. (B) Western blot analysis of the SRT deletion and insertion mutants. Western blot analysis was performed by using anti-IE62 polyclonal antibody vc-20 (Santa Cruz Biotechnology). Control, cells transfected with empty vector. Luciferase reporter assays were performed with pORF61-Luc (C) and pgI-Luc (D). L-M cells were cotransfected with 0.12 pmol of the reporter plasmid pORF61-Luc or pgI-Luc and 0.08 pmol of effector plasmids. The firefly luciferase signals were normalized to the internal secreted alkaline phosphatase (SEAP) transfection control. The percentages represent the relative activities of the mutants IE62 compared to the activity of the wild-type control. Data are averages and are representative of three independent experiments. RLU, relative luminescence units.
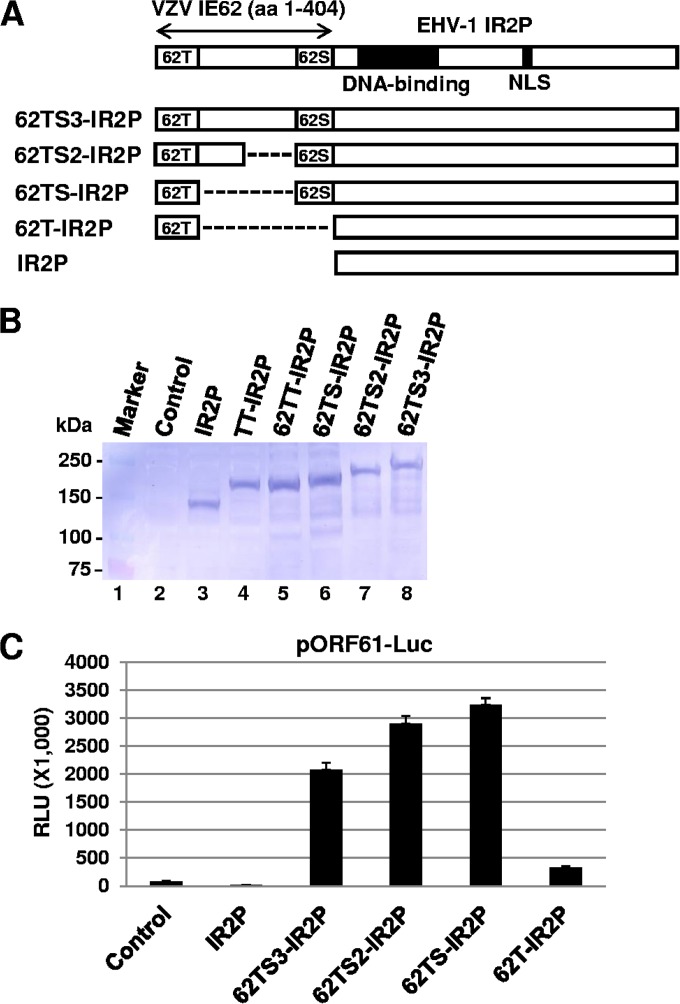
The distance between the TAD and SRT is not important for the trans-activation ability of IE62.
To determine the effect of the 264-aa distance between the acidic trans-activation domain (TAD) and SRT on the trans-activation activity of IE62, N-terminal sequences (aa 1 to 404) of IE62 were fused to the N terminus of the EHV-1 IR2 protein that lacks the TAD and SRT, resulting in the generation of plasmid pSV62TS3-IR2P (Fig. 4A). The construction of the chimeric protein 62TS3-IR2P was confirmed by DNA sequencing (data not shown) and Western blot analysis (Fig. 4B). When the plasmid pSV62TS3-IR2P was cotransfected with the ORF61 promoter luciferase reporter plasmid into L-M cells, strong luciferase activity was observed (Fig. 4C, 62TS3-IR2P). In contrast, the IR2 protein by itself downregulated the ORF61 promoter (Fig. 4C, IR2P), which is consistent with our previous results (29, 41). Plasmids pSV62TS2-IR2P and pSV62TS-IR2P, in which the 264-aa sequences between the TAD and SRT were partially and completely deleted, respectively, strongly trans-activated the ORF61 promoter (Fig. 4C, 62TS2-IR2P and 62TS-IR2P, respectively). The plasmid pSV62T-IR2P that expresses IE62 TAD(1–86)-IR2P very weakly trans-activated the ORF61 promoter (Fig. 4C, 62T-IR2P). These results indicated that the distance between the TAD and SRT of the IE62 is not important for its trans-activation ability and that the 264-aa sequences between the TAD and SRT may not be required for trans-activation activity.
FIG 4.
The distance between the TAD and SRT of IE62 is not important for its trans-activation activity. (A) Schematic diagram of IE62 TAD-SRT-IR2P chimeric fusion proteins. T, trans-activation domain (TAD); S, SRT; 62, IE62; NLS, nuclear localization signal. Western blot analysis (B) and a luciferase reporter assay (C) were performed as described in the legend of Fig. 2. The fusion proteins were detected with the IEP antibody OC33 (42). Data are averages and are representative of three independent experiments. RLU, relative luminescence units.
Both the serine-rich region and the acidic residues of the SRT of IE62 are necessary for TAD-mediated trans-activation.
The SRT of IE62 contains 16 serines and 10 acidic residues. To investigate which residues are important for its trans-activation activity, multiple amino acid substitution mutants were generated and analyzed (Fig. 5). The two amino acid substitution mutants of the IE62 were confirmed by DNA sequencing (data not shown) and Western blot analysis (Fig. 5C). When 10 of the 16 serines of the IE62 SRT were replaced with Ala, Leu, and Gly, its trans-activation activity was reduced to 39% of wt activity for the early ORF61 promoter and 26% for the late gI promoter (Fig. 5A, right table, and B). When 8 of the 10 acidic residues of the IE62 SRT were replaced with Ala, Leu, and Gly, its trans-activation activity was reduced to 18% for the ORF61 promoter and 6% for the gI promoter (Fig. 5A, right table). To investigate whether the SRT mutation is dominant negative over trans-activation by intact IE62, two IE62 mutants, IE62-SRTΔSe and IE62-SRTΔAc, were generated and confirmed by DNA sequencing (data not shown) and Western blot analysis (Fig. 5E). As shown in Fig. 5D, very similar results were obtained with the two IE62 mutants (compare Fig. 5A and D). These results demonstrated that both the serine-rich region and the acidic residues are important for TAD-mediated trans-activation.
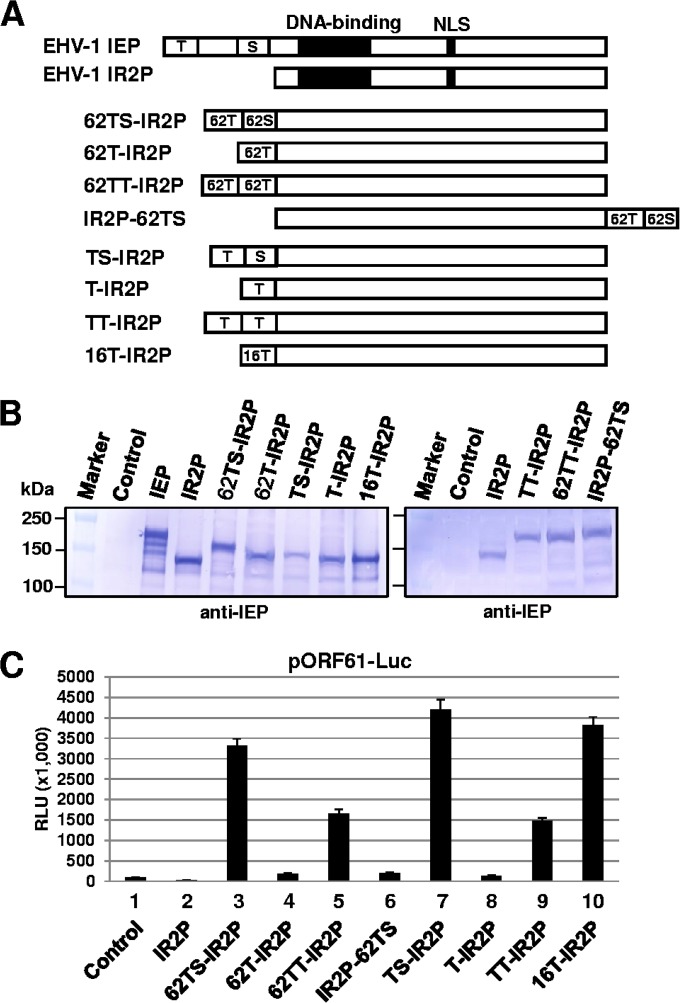
The addition of TAD partially compensates for the SRT deletion.
The data in Fig. 5 showed that the acidic residues within the SRT of the IE62 and IEP are essential for TAD-mediated trans-activation. The TADs of IE62 and IEP contain 15 and 13 acidic residues, respectively. We speculated that the SRT function could be compensated with the acidic residues of the TAD. To address this possibility, various TAD-SRT chimeric constructs were generated and analyzed (Fig. 6). The IR2 protein (IR2P), an early 1,165-aa truncated form of the EHV-1 IEP, lacks the TAD and SRT and negatively regulates viral gene expression (29, 41). The validity of the chimeric fusion constructs was confirmed by DNA sequencing (data not shown). Also, Western blot analysis showed that expression of each chimeric construct in transfected mouse L-M cells generated a chimeric fusion protein of the expected size (Fig. 6B). When the IE62 SRT was replaced with the IE62 TAD, the IE62TAD-TAD-IR2 (62TT-IR2P) chimeric protein trans-activated the ORF61 promoter at 48% of the level of IE62TAD-SRT-IR2P (62TS-IR2P) (Fig. 6C, bar 5). IEPTAD-TAD-IR2 (TT-IR2P) trans-activated the ORF61 promoter at 36% of the level of IEPTAD-SRT-IR2P (TS-IR2P) (Fig. 6C, bar 9). In contrast, 62TAD-IR2P and TAD-IR2 chimeric proteins lacking the SRT were capable of only very weak trans-activation (Fig. 6C, bars 4 and 8), demonstrating that the function of the SRT can be partially compensated and restored by the replacement with TAD. When the IE62 TAD and SRT were moved to the C-terminal end of the IR2P, the IR2P-62TS lost most of its trans-activation activity (Fig. 6C, bar 6), indicating that the positions of the TAD and SRT are important for TAD-mediated trans-activation.
FIG 6.
The TAD compensated and restored the function of the SRT of IE62. (A) Schematic diagram of IE62 TAD-SRT-IR2P chimeric fusion proteins. T, trans-activation domain (TAD); S, SRT; 62, IE62. Western blot analysis (B) and luciferase reporter assays (C) were performed as described in the legend of Fig. 2. Data are averages and are representative of three independent experiments. RLU, relative luminescence units.
Mediator 25 interacts with the TAD of VZV IE62, EHV-1 IEP, and HSV-1 VP16.
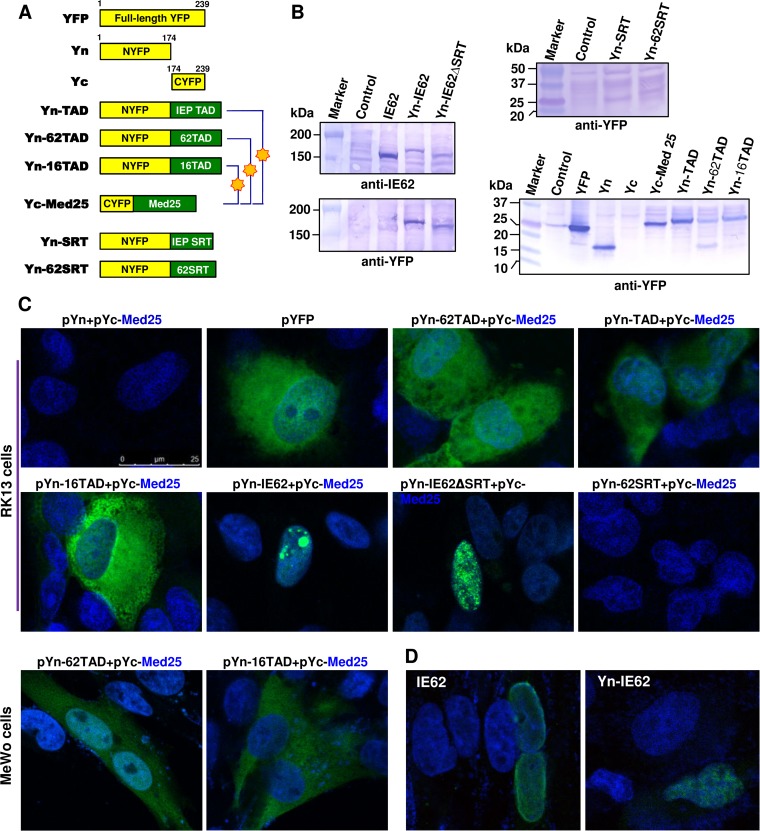
Mediator is required for most RNA polymerase II (Pol II)-mediated transcription since it acts as a bridging molecule between activators and Pol II. The IE62 TAD interacts directly with aa 402 to 590 of the Mediator 25 subunit (17, 25). HSV-1 VP16 TAD functionally interacts with the Mediator 25 VP16-binding domain (VBD) (aa 402 to 590) (45). VZV IE62 TAD shows 27% amino acid sequence identity with EHV-1 IEP TAD (Fig. 1C). The VP16 TAD contains three functionally important hydrophobic amino acids (Leu439, Phe442, and Leu444) (46), among which the Leu439 and Phe442 residues align directly with IEP counterparts within the TAD (Fig. 1D). To confirm the TAD and Med25 interaction in their natural cellular environment, a bimolecular complementation (BiMC) assay was employed. The basic principle of the BiMC assay is as follows: yellow fluorescent protein (YFP) is split into two nonfluorescent fragments, NYFP (Yn) and CYFP (Yc), which are joined in frame to two proteins of interest (P1 and P2). After cotransfection into cells, if the two proteins of interest interact, this interaction allows P1 with P2 to reconstitute YFP, and the resultant fluorescent signal is detected by fluorescence microscopy.
NYFP-IE62 TAD (Yn-62TAD), NYFP-IEP TAD (Yn-TAD), NYFP-VP16 TAD (Yn-16TAD), and CYFP-Mediator 25 (Yc-Med25) fusion expression vectors were generated and confirmed by Western blot analyses (Fig. 7B) and DNA sequencing (data not shown). RK13 cells were cotransfected with expression constructs shown at the top of each panel and examined by confocal microscopy. A strong, reproducible YFP signal was observed in the cells expressing Yn-62TAD and Yc-Med25, which is indicative of a 62TAD-Med25 interaction (Fig. 7C). A majority of the YFP signal was detected in the cytoplasm, which is very similar to the expression of full-length YFP (by itself) shown in Fig. 7C. IEP TAD and VP16 TAD also strongly interacted with Med25. However, no signal between Yn-62SRT and Yc-Med25 was detected. In human melanoma MeWo cells, the IE62 TAD and VP16 TAD interacted with Med25 in a pattern similar to those of RK13 cells (Fig. 7C). However, no signal between Yn-62SRT and Yc-Med25 was detected. Full-length IE62 was also fused to the NYFP (Fig. 7A and B). To investigate whether the addition of YFP affects IE62 localization, the distribution of IE62 with or without NYFP was examined in MeWo and RK13 cells. Both the IE62 and Yn-IE62 localized in the nuclei of MeWo (Fig. 7D) and RK13 (data not shown). A strong YFP signal, which appeared as distinct nuclear punctae, was observed in the nuclei of cells expressing Yn-full-length IE62 and Yc-Med25 (Fig. 7C), indicating that the TAD of IE62 interacts with Med25 in the nucleus. In contrast, very small nuclear punctae were observed in the nuclei of cells expressing Yn-full-length IE62ΔSRT and Yc-Med25, suggesting that the IE62 SRT is involved in forming large globular structures within the nucleus.
FIG 7.
Mediator 25 interacts with the TAD of EHV-1 IEP, VZV IE62, and HSV-1 VP16. (A) Schematic diagram and names of constructs. The numbers refer to the number of amino acids from the N terminus of each protein. Blue, DAPI; YFP, enhanced yellow fluorescence protein; Yn, NYFP (1 to 173); Yc, CYFP (aa 174 to 239). Stars indicate fluorescent signals. (B) The YFP-TAD fusion proteins were confirmed by Western blot analyses with anti-IE62 polyclonal antibody (sc-17525; Santa Cruz Biotechnology) and anti-YFP polyclonal antibody (sc-8334; Santa Cruz Biotechnology). (C) Bimolecular complementation assays were performed to detect TAD domain and Mediator 25 interaction. Rabbit kidney RK13 and human melanoma MeWo cells were cotransfected with the constructs shown above each panel and examined by confocal microscopy at 48 h posttransfection. (D) To detect IE62 or Yn-IE62, the MeWo cells transfected with pCMV-IE62 or pYn-IE62 were fixed at 48 h posttransfection, stained with IE62-specific antibody IE62T1 or anti-YFP polyclonal antibody sc-8334 (Santa Cruz Biotechnology), respectively, and with fluorescein isothiocyanate-conjugated secondary antibody.
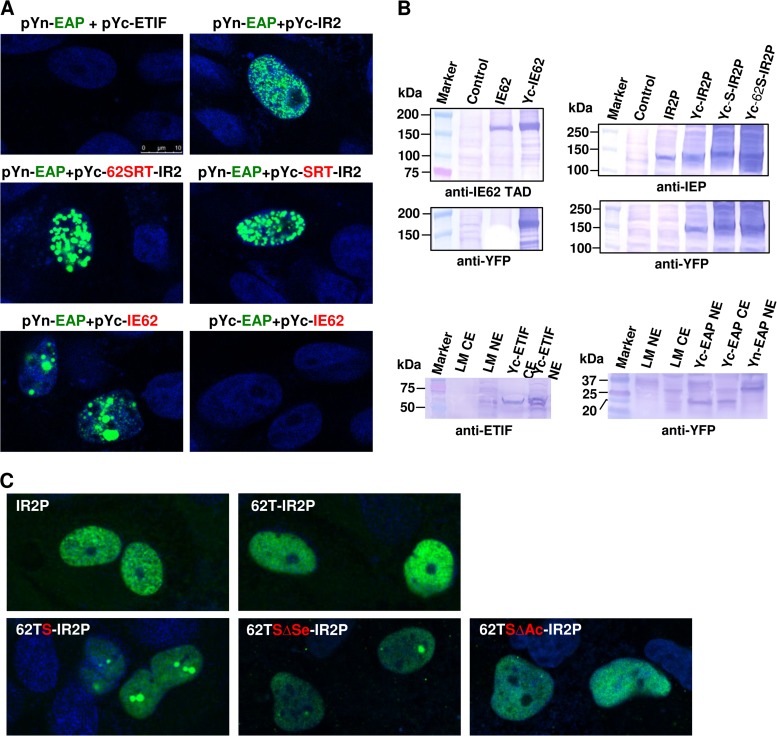
The SRT IE62 may be involved in the formation of large globular structures.
To determine whether the IE62 SRT interacts with a nucleolar-ribosomal EAP, bimolecular complementation (BiMC) assays were performed in MeWo cells. EAP was fused to the NYFP (Yn). The IE62 SRT-IR2 chimeric protein was fused to the CYFP (Yc) because Yc-SRT was unstable. Yc-ETIF, Yc-62S-IR2, Yc-S-IR2, and Yn-Med25 fusion expression vectors were confirmed by Western blot analyses (Fig. 8B) and DNA sequencing (data not shown). ETIF is the EHV-1 homolog of HSV-1 VP16 (39). EAP did not interact with Yc-ETIF but weakly interacted with IR2P (Fig. 8A). The EAP-IR2P interaction was localized in discrete nuclear structures. When the IE62 SRT was fused to the N terminus of IR2P, much larger globular structures were observed in the nuclei (Fig. 8A). The EAP-IE62 interaction was also localized in discrete nuclear structures (Fig. 8A). Very similar results were obtained with RK13 cells (see Fig. S3 in the supplemental material for BiMC data).
FIG 8.
The interaction of the IE62 SRT with EAP results in the formation of globular structures. (A) Bimolecular complementation (BiMC) assay to detect the interaction of cellular EAP and SRT-IR2P. MeWo cells were cotransfected with the constructs shown above each panel and examined by confocal microscopy at 48 h posttransfection. (B) The YFP fusion proteins were confirmed by Western blot analyses of mouse L-M cells with anti-IEP polyclonal antibody OC33, anti-ETIF monoclonal antibody (65), anti-IE62 TAD polyclonal antibody IE62T1, and anti-YFP polyclonal antibody (sc-8334; Santa Cruz Biotechnology). L-M NE and L-M CE refer to control cells transfected with empty vector, where NE is nuclear extract and CE is cytoplasmic extract. (C) The addition of SRT at the N terminus of IR2P results in the formation of large globular structures. RK13 cells were transfected with the constructs shown above each panel. The cells were then fixed at 48 h posttransfection and stained with IR2P-specific antibody OC33 and fluorescein isothiocyanate-conjugated secondary antibody. 62TS-IR2P containing the SRT was detected as globular structures. At least 100 cells were analyzed to determine the percentages of cells containing the large globular structures.
To examine the effect of the SRT on the expression of IE62, IE62 TAD-SRT chimeric fusion proteins (Fig. 6) were used in immunofluorescence (IF) assays. The plasmids expressing the chimeric fusion proteins were transfected into RK13 cells, and IF assays were performed with anti-IR2P pAb OC33 (Fig. 8C). IR2P and IE62TAD-IR2P (62T-IR2P) proteins were dispersed throughout the nucleoplasm, whereas IE62TAD-SRT-IR2P (62TS-IR2P) was tightly aggregated in dense nuclear structures (Fig. 8C). To further investigate whether the SRT is involved in forming globular structures within the nucleus, the two SRT mutants 62TSΔS-IR2P and 62TSΔA-IR2P (Fig. 5) were examined by IF assay. Interestingly, large globular structures were not detected in cells transfected with the acidic residue mutant 62TSΔA-IR2P (Fig. 8C). The large globular structures were detected in 42% of the cells transfected with the serine residue mutant 62TSΔS-IR2P, and the number of globular structures within the cell was also decreased. Very similar results, including percentages of cells with globular structures, were obtained with the EHV-1 IEP SRT mutants (data not shown). These results suggest that the SRT may be involved in forming globular structures within the nucleus.
DISCUSSION
To begin understanding the functions performed by the SRT of the VZV IE62 protein, we analyzed the effect of its deletion on the ability of IE62 to function as a trans-activator. We observed that the deletion of IE62 SRT resulted in the loss of most of its trans-activation ability, indicating that the SRT is necessary for TAD-mediated trans-activation. It has been shown that two transactivation domains, one at the N terminus and one at the C terminus, are largely responsible for the activation functions of ICP4 (47). However, the activation domains of ICP4 are not conserved with the TAD of VZV IE62 (18, 19, 44). Our studies showed that an IE62 TAD-ICP4 chimeric protein in which the TAD of IE62 was inserted in the ICP4 ORF exhibited greater than an 8-fold increase in trans-activation activity, indicating that the IE62 TAD enhances the ability of ICP4 to trans-activate viral promoters. The SRT of HSV-1 ICP4 has been shown to be important for the induction of early and leaky late gene expression, particularly in an ICP4 mutant lacking the C-terminal sequences (48, 49). Residues in the SRT region are potential sites for phosphorylation and are involved in the functions of ICP4 (20, 50). VZV encodes two serine/threonine kinases, ORF47 and ORF66, that phosphorylate IE62 (11, 51). The ORF66 phosphorylated IE62 at residues S686 and S722 (51). The ORF47 potentially phosphorylates at S16 (52). Purified CDK1/cyclin B1 phosphorylated IE62 at residues T10, S245, and T680 in vitro (53). These results indicated that none of the phosphorylation sites of IE62 resides within the SRT(353–404) of the IE62, suggesting that the serine-rich region of the IE62 may not be a site for phosphorylation. We cannot completely exclude the possibility that the serine-rich region could be a determinant for phosphorylation of IE62 as seen in HSV-1 CP4.
With the Alphaherpesvirinae subfamily, members of the Varicellovirus such as VZV, EHV-1, BHV-1, and PRV have similar genomic structures, classified as type group D (2–4). These viruses harbor one major immediate early gene that is diploid and maps near the termini of the inverted repeat component within the short region. The IE62 gene of VZV is clearly a counterpart to the EHV-1 IE, the IE180 gene of PRV, and the BICP4 of BHV-1 (Fig. 1) (26, 54). Comparison of the amino acid sequences of these Varicellovirus IE proteins showed that the major regulatory protein of VZV, the only human virus in this genus, is closely related to the IE protein of the bovine, equine, and porcine viruses. Furthermore, all of these IE proteins including the VZV IE62 contain very similar TADs that are required for the IE protein to activate viral promoters (Fig. 1) (26, 54) and, as shown in this study, can be interchanged without significant loss of function. In contrast, the ICP4 of HSV-1 lacks a TAD and is functionally different from and more distantly related to the four IE proteins of these Varicellovirus members.
Our findings in this study showed that the interaction of IE62 SRT with EAP resulted in the formation of globular structures and that both the serine-rich region and acidic residues of the SRT were important for TAD-mediated trans-activation. The Arvin laboratory (55) showed that formation of VZV DNA replication compartments started between 4 and 6 h, involved the recruitment of ORF29 to putative IE62 prereplication sites, and resulted in large globular nuclear compartments where newly synthesized viral DNA accumulated. IE62 was detected at 2 h postinfection and appeared as distinct nuclear punctae (55). By 4 h, the diameter of the punctae had increased to form globular structures (55).
As mentioned before, the SRT of HSV-1 ICP4 functionally colocalizes and interacts with the nucleolar-ribosomal protein EAP (28, 53). The SRT of EHV-1 IEP, shown to be essential for virus replication (30), also interacts with the EAP (43). Ribosomal protein EAP (L22) contains a globular domain that sits on the surface of the large ribosomal subunit and an extended loop that penetrates its core (56). These results suggest that the SRT may be involved in forming globular structures of the IE62 within the nucleus. It is possible that the SRT may allow the globular structures to grow larger by stabilizing the fused proteins. The globular structures could be a consequence of phosphorylation of the SRT or also of overexpression in the in vitro approach. Ribosomal assembly is a highly interactive process in which binding of many proteins to the rRNA occurs in a stepwise manner. EAP (L22) is an “early-binding protein” but does not bind with high affinity to naked 23S rRNA; the initial step of EAP incorporation in the ribosome is strongly stimulated by the prior binding of several other proteins, particularly L4 (57). EAP binds specifically to 23S rRNA and makes multiple contacts with different domains of the 23S rRNA in the assembled 50S subunit and ribosome (58–60). These results suggest that EAP binds to the SRT and also makes multiple contacts with the other domains of IE62 and IEP.
A majority of the YFP signal resulting from the interactions of the TADs of IE62, IEP, and VP16 with Med25 was detected in the cytoplasm, the site of localization of full-length YFP in control cells (Fig. 7). In contrast, distinct nuclear punctae were observed in the nuclei of the cells expressing Yn-full-length IE62 and Yc-Med25. VZV IE62 that contains a nuclear localization signal is targeted to subcellular nuclear compartments during very early infection (55). These results suggest that localization of Med25 is dependent on its interacting partners.
The data presented in this study revealed that the deletion of the SRT of IE62 and IEP resulted in a significant loss of their trans-activation activity and that the function of the SRT of IE62 can be compensated by an additional TAD. Surprisingly, the VP16 TAD-IR2P chimeric protein robustly trans-activated viral promoters, indicating that the VP16 TAD is able to strongly activate transcription without the SRT (Fig. 6). Both the IE62 TAD and VP16 TAD interact with the VP16-binding domain (VBD; Med25 aa 390 to 553) (17, 45) of Mediator 25 which is the same domain with an activator-interacting domain (ACID; Med25 aa 391 to 543) (61). However, the interaction between the IE62 TAD and Mediator 25 was considerably weaker than that involving the VP16 (17, 25). Our bimolecular complementation (BiMC) assay also showed that Med25 interacted more strongly with the VP16 TAD than the IE62 TAD and the IEP TAD. The VP16 TAD can be divided into two subdomains (VP16N and VP16C), and each subdomain contains α-helices (62, 63). The net charges of the VP16N and VP16C are −11.8 and −8.9, respectively (Table 1). The TADs of IE62 and IEP contain 15 and 13 acidic residues, respectively. Nuclear magnetic resonance (NMR) data (17) with the IE62 TAD (net charge, −9.8) indicated that the predicted single α-helix is formed only transiently, if at all. Prediction of secondary structures using the Predict-Protein server (http://predictprotein.org) indicated the potential for the presence of a single extended α-helix involving aa 19 to 35 of the IEP TAD (net charge, −7.0) (Fig. 1C) (64). These results suggest that the weak interaction between the Mediator 25 and the IE62 TAD may be due to the presence of a single α-helix within the IE62 TAD. The SRTs of IE62, IEP, and ICP4 contain 10, 10, and 8 acidic residues, respectively. Interestingly, the three SRT peptides are acidic (net charges, −8.0 to −10.0) (Table 1). These results demonstrated that acidic residues in the TAD and SRT are important for TAD-mediated trans-activation and Mediator interaction.
TABLE 1.
The net charges of the TAD and SRT peptides
| Peptide | Net chargea |
|---|---|
| VP16 TAD | −20.7 |
| VP16 TADN | −11.8 |
| VP16 TADC | −8.9 |
| IE62 TAD | −9.8 |
| IEP TAD | −7.0 |
| IE62 SRT | −10.0 |
| IEP SRT | −10.0 |
| ICP4 SRT | −8.0 |
The net charges were calculated by Innovagen's peptide property calculator (http://www.innovagen.se).
The precise mechanism by which the TAD and SRT activate viral gene expression remains unknown. Taken together, the results of this study suggest that the TAD-Mediator 25 and/or SRT-EAP association stimulates gene expression by enhancing the recruitment of other cellular components of the RNA polymerase II machinery to the core promoter of IE62-regulated genes. Ongoing efforts to generate and characterize mutant forms of the SRT and TAD should give more insight into their importance in herpesvirus biology.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ann M. Arvin for providing plasmid pcDNA-IE62 and Ronald N. Harty for providing plasmid pCS2-YFP-Venus-FL. We thank Ann M. Arvin and Nikolaus Osterrieder for critical reading of the manuscript and providing comments.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02096-15.
REFERENCES
- 1.Cohen JI, Straus S, Arvin A. 2007. Varicella-zoster virus replication, pathogenesis and management, p 2773–2818. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Cohen JI. 1999. Genomic structure and organization of varicella-zoster virus. Contrib Microbiol 3:10–12. doi: 10.1159/000060313. [DOI] [PubMed] [Google Scholar]
- 3.Henry BE, Robinson RA, Dauenhauer SA, Atherton SS, Hayward GS, O'Callaghan DJ. 1981. Structure of the genome of equine herpesvirus type 1. Virology 115:97–114. doi: 10.1016/0042-6822(81)90092-1. [DOI] [PubMed] [Google Scholar]
- 4.Roizman B, Pellett P. 2001. The family Herpesviridae: a brief introduction, p 2381–2397. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 5.Moriuchi M, Moriuchi H, Straus SE, Cohen JI. 1994. Varicella-zoster virus (VZV) virion-associated transactivator open reading frame 62 protein enhances the infectivity of VZV DNA. Virology 200:297–300. doi: 10.1006/viro.1994.1190. [DOI] [PubMed] [Google Scholar]
- 6.Ruyechan W. 2004. Mechanism(s) of activation of varicella zoster virus promoters by the VZV IE62 protein. Recent Res Dev Virol 6:145–172. [Google Scholar]
- 7.Kinchington P, Hougland J, Arvin A, Ruyechan W, Hay J. 1992. The varicella-zoster virus immediate-early protein IE62 is a major component of virus particles. J Virol 66:359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinchington PR, Fite K, Seman A, Turse SE. 2001. Virion association of IE62, the varicella-zoster virus (VZV) major transcriptional regulatory protein, requires expression of the VZV open reading frame 66 protein kinase. J Virol 75:9106–9113. doi: 10.1128/JVI.75.19.9106-9113.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spengler ML, Ruyechan WT, Hay J. 2000. Physical interaction between two varicella zoster virus gene regulatory proteins, IE4 and IE62. Virology 272:375–381. doi: 10.1006/viro.2000.0389. [DOI] [PubMed] [Google Scholar]
- 10.Cilloniz C, Jackson W, Grose C, Czechowski D, Hay J, Ruyechan WT. 2007. The varicella-zoster virus (VZV) ORF9 protein interacts with the IE62 major VZV transactivator. J Virol 81:761–774. doi: 10.1128/JVI.01274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng TI, Keenan L, Kinchington PR, Grose C. 1994. Phosphorylation of varicella-zoster virus open reading frame (ORF) 62 regulatory product by viral ORF 47-associated protein kinase. J Virol 68:1350–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynch JM, Kenyon TK, Grose C, Hay J, Ruyechan WT. 2002. Physical and functional interaction between the varicella zoster virus IE63 and IE62 proteins. Virology 302:71–82. doi: 10.1006/viro.2002.1555. [DOI] [PubMed] [Google Scholar]
- 13.Peng H, He H, Hay J, Ruyechan WT. 2003. Interaction between the varicella zoster virus IE62 major transactivator and cellular transcription factor Sp1. J Biol Chem 278:38068–38075. doi: 10.1074/jbc.M302259200. [DOI] [PubMed] [Google Scholar]
- 14.Rahaus M, Desloges N, Yang M, Ruyechan WT, Wolff MH. 2003. Transcription factor USF, expressed during the entire phase of varicella-zoster virus infection, interacts physically with the major viral transactivator IE62 and plays a significant role in virus replication. J Gen Virol 84:2957–2967. doi: 10.1099/vir.0.19335-0. [DOI] [PubMed] [Google Scholar]
- 15.Ruyechan WT, Peng H, Yang M, Hay J. 2003. Cellular factors and IE62 activation of VZV promoters. J Med Virol 70:S90–S94. doi: 10.1002/jmv.10328. [DOI] [PubMed] [Google Scholar]
- 16.Yang M, Peng H, Hay J, Ruyechan WT. 2006. Promoter activation by the varicella-zoster virus major transactivator IE62 and the cellular transcription factor USF. J Virol 80:7339–7353. doi: 10.1128/JVI.00309-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto S, Eletsky A, Szyperski T, Hay J, Ruyechan WT. 2009. Analysis of the varicella-zoster virus IE62 N-terminal acidic transactivating domain and its interaction with the human mediator complex. J Virol 83:6300–6305. doi: 10.1128/JVI.00054-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen JI, Heffel D, Seidel K. 1993. The transcriptional activation domain of varicella-zoster virus open reading frame 62 protein is not conserved with its herpes simplex virus homolog. J Virol 67:4246–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shepard A, Imbalzano A, DeLuca N. 1989. Separation of primary structural components conferring autoregulation, transactivation, and DNA-binding properties to the herpes simplex virus transcriptional regulatory protein ICP4. J Virol 63:3714–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia K, DeLuca NA, Knipe DM. 1996. Analysis of phosphorylation sites of herpes simplex virus type 1 ICP4. J Virol 70:1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lester JT, DeLuca NA. 2011. Herpes simplex virus 1 ICP4 forms complexes with TFIID and mediator in virus-infected cells. J Virol 85:5733–5744. doi: 10.1128/JVI.00385-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casamassimi A, Napoli C. 2007. Mediator complexes and eukaryotic transcription regulation: an overview. Biochimie 89:1439–1446. doi: 10.1016/j.biochi.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. 2005. The mammalian Mediator complex and its role in transcriptional regulation. The trends Biochem Sci 30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Malik S, Roeder RG. 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci 30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Yang M, Hay J, Ruyechan WT. 2008. Varicella-zoster virus IE62 protein utilizes the human mediator complex in promoter activation. J Virol 82:12154–12163. doi: 10.1128/JVI.01693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundy FJ, Baumann RP, O'Callaghani DJ. 1989. DNA sequence and comparative analyses of the equine herpesvirus type 1 immediate early gene. Virology 172:223–236. doi: 10.1016/0042-6822(89)90124-4. [DOI] [PubMed] [Google Scholar]
- 27.Leopardi R, Ward PL, Ogle WO, Roizman B. 1997. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the UL 13 protein kinase. J Virol 71:1133–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leopardi R, Roizman B. 1996. Functional interaction and colocalization of the herpes simplex virus 1 major regulatory protein ICP4 with EAP, a nucleolar-ribosomal protein. Proc Natl Acad Sci U S A 93:4572–4576. doi: 10.1073/pnas.93.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SK, Kim S, Dai G, Zhang Y, Ahn BC, O'Callaghan DJ. 2011. Identification of functional domains of the IR2 protein of equine herpesvirus 1 required for inhibition of viral gene expression and replication. Virology 417:430–442. doi: 10.1016/j.virol.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buczynski KA, Kim SK, O'Callaghan DJ. 2005. Initial characterization of 17 viruses harboring mutant forms of the immediate-early gene of equine herpesvirus 1. Virus Genes 31:229–239. doi: 10.1007/s11262-005-1801-2. [DOI] [PubMed] [Google Scholar]
- 31.Toczyski D, Steitz JA. 1991. EAP, a highly conserved cellular protein associated with Epstein-Barr virus small RNAs (EBERs). EMBO J 10:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toczyski DP, Matera AG, Ward DC, Steitz JA. 1994. The Epstein-Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc Natl Acad Sci U S A 91:3463–3467. doi: 10.1073/pnas.91.8.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- 34.Smith R, Caughman G, O'Callaghan D. 1992. Characterization of the regulatory functions of the equine herpesvirus 1 immediate-early gene product. J Virol 66:936–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith RH, Holden VR, O'Callaghan DJ. 1995. Nuclear localization and transcriptional activation activities of truncated versions of the immediate-early gene product of equine herpesvirus 1. J Virol 69:3857–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen N, Sommer M, Che X, White K, Ruyechan WT, Arvin AM. 2010. Varicella-zoster virus immediate-early protein 62 blocks interferon regulatory factor 3 (IRF3) phosphorylation at key serine residues: a novel mechanism of IRF3 inhibition among herpesviruses. J Virol 84:9240–9253. doi: 10.1128/JVI.01147-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLuca NA, Schaffer PA. 1987. Activities of herpes simplex virus type 1 (HSV-1) ICP4 genes specifying nonsense peptides. Nucleic Acids Res 15:4491–4511. doi: 10.1093/nar/15.11.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Stone S, Harty RN. 2011. Characterization of filovirus protein-protein interactions in mammalian cells using bimolecular complementation. J Infect Dis 204:S817–S824. doi: 10.1093/infdis/jir293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SK, O'Callaghan DJ. 2001. Molecular characterizations of the equine herpesvirus 1 ETIF promoter region and translation initiation site. Virology 286:237–247. doi: 10.1006/viro.2001.0988. [DOI] [PubMed] [Google Scholar]
- 40.Bedadala GR, Pinnoji RC, Hsia S-CV. 2007. Early growth response gene 1 (Egr-1) regulates HSV-1 ICP4 and ICP22 gene expression. Cell Res 17:546–555. doi: 10.1038/cr.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim SK, Ahn BC, Albrecht RA, O'callaghan DJ. 2006. The unique IR2 protein of equine herpesvirus 1 negatively regulates viral gene expression. J Virol 80:5041–5049. doi: 10.1128/JVI.80.10.5041-5049.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harty RN, O'Callaghan DJ. 1991. An early gene maps within and is 3′ coterminal with the immediate-early gene of equine herpesvirus 1. J Virol 65:3829–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SK, Buczynski KA, Caughman GB, O'Callaghan DJ. 2001. The equine herpesvirus 1 immediate-early protein interacts with EAP, a nucleolar-ribosomal protein. Virology 279:173–184. doi: 10.1006/viro.2000.0725. [DOI] [PubMed] [Google Scholar]
- 44.Xiao W, Pizer LI, Wilcox KW. 1997. Identification of a promoter-specific transactivation domain in the herpes simplex virus regulatory protein ICP4. J Virol 71:1757–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milbradt AG, Kulkarni M, Yi T, Takeuchi K, Sun Z-YJ, Luna RE, Selenko P, Näär AM, Wagner G. 2011. Structure of the VP16 transactivator target in the Mediator. Nat Struct Mol Biol 18:410–415. doi: 10.1038/nsmb.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Regier JL, Shen F, Triezenberg SJ. 1993. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci U S A 90:883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner LM, Bayer A, DeLuca NA. 2013. Requirement of the N-terminal activation domain of herpes simplex virus ICP4 for viral gene expression. J Virol 87:1010–1018. doi: 10.1128/JVI.02844-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paterson T, Everett R. 1988. Mutational dissection of the HSV-1 immediate-early protein Vmw175 involved in transcriptional transactivation and repression. Virology 166:186–196. doi: 10.1016/0042-6822(88)90160-2. [DOI] [PubMed] [Google Scholar]
- 49.Sampath P, DeLuca NA. 2008. Binding of ICP4, TATA-binding protein, and RNA polymerase II to herpes simplex virus type 1 immediate-early, early, and late promoters in virus-infected cells. J Virol 82:2339–2349. doi: 10.1128/JVI.02459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia K, Knipe DM, DeLuca NA. 1996. Role of protein kinase A and the serine-rich region of herpes simplex virus type 1 ICP4 in viral replication. J Virol 70:1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eisfeld AJ, Turse SE, Jackson SA, Lerner EC, Kinchington PR. 2006. Phosphorylation of the varicella-zoster virus (VZV) major transcriptional regulatory protein IE62 by the VZV open reading frame 66 protein kinase. J Virol 80:1710–1723. doi: 10.1128/JVI.80.4.1710-1723.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kenyon T, Lynch J, Hay J, Ruyechan W, Grose C. 2001. Varicella-zoster virus ORF47 protein serine kinase: characterization of a cloned, biologically active phosphotransferase and two viral substrates, ORF62 and ORF63. J Virol 75:8854–8858. doi: 10.1128/JVI.75.18.8854-8858.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leisenfelder SA, Kinchington PR, Moffat JF. 2008. Cyclin-dependent kinase 1/cyclin B1 phosphorylates varicella-zoster virus IE62 and is incorporated into virions. J Virol 82:12116–12125. doi: 10.1128/JVI.00153-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwyzer M, Vlček, Fraefel C, Menekse O, Fraefel C, Pačes V. 1993. Promoter, spliced leader, and coding sequence for BICP4, the largest of the immediate-early proteins of bovine herpesvirus 1. Virology 197:349–357. doi: 10.1006/viro.1993.1596. [DOI] [PubMed] [Google Scholar]
- 55.Reichelt M, Brady J, Arvin AM. 2009. The replication cycle of varicella-zoster virus: analysis of the kinetics of viral protein expression, genome synthesis, and virion assembly at the single-cell level. J Virol 83:3904–3918. doi: 10.1128/JVI.02137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zengel JM, Jerauld A, Walker A, Wahl MC, Lindahl L. 2003. The extended loops of ribosomal proteins L4 and L22 are not required for ribosome assembly or L4-mediated autogenous control. RNA 9:1188–1197. doi: 10.1261/rna.5400703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nierhaus KH. 1991. The assembly of prokaryotic ribosomes. Biochimie 73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 58.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 59.Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107:679–688. doi: 10.1016/S0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 60.Worbs M, Wahl MC, Lindahl L, Zengel JM. 2002. Comparative anatomy of a regulatory ribosomal protein. Biochimie 84:731–743. doi: 10.1016/S0300-9084(02)01410-4. [DOI] [PubMed] [Google Scholar]
- 61.Eletsky A, Ruyechan WT, Xiao R, Acton TB, Montelione GT, Szyperski T. 2011. Solution NMR structure of MED25 (391-543) comprising the activator-interacting domain (ACID) of human mediator subunit 25. J Struct Funct Genomics 12:159–166. doi: 10.1007/s10969-011-9115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cress WD, Triezenberg SJ. 1991. Critical structural elements of the VP16 transcriptional activation domain. Science 251:87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- 63.Jonker HR, Wechselberger RW, Boelens R, Folkers GE, Kaptein R. 2005. Structural properties of the promiscuous VP16 activation domain. Biochemistry 44:827–839. doi: 10.1021/bi0482912. [DOI] [PubMed] [Google Scholar]
- 64.Smith RH, Zhao Y, O'Callaghan DJ. 1994. The equine herpesvirus type 1 immediate-early gene product contains an acidic transcriptional activation domain. Virology 202:760–770. doi: 10.1006/viro.1994.1398. [DOI] [PubMed] [Google Scholar]
- 65.Lewis JB, Thompson YG, Feng X, Holden VR, O'callaghan D, Caughman GB. 1997. Structural and antigenic identification of the ORF12 protein (αTIF) of equine herpesvirus 1. Virology 230:369–375. doi: 10.1006/viro.1997.8477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.