ABSTRACT
Between November 2013 and February 2014, China reported three human cases of H10N8 influenza virus infection in the Jiangxi province, two of which were fatal. Using hybridoma technology, we isolated a panel of H10- and N8-directed monoclonal antibodies (MAbs) and further characterized the binding reactivity of these antibodies (via enzyme-linked immunosorbent assay) to a range of purified virus and recombinant protein substrates. The H10-directed MAbs displayed functional hemagglutination inhibition (HI) and neutralization activity, and the N8-directed antibodies displayed functional neuraminidase inhibition (NI) activity against H10N8. Surprisingly, the HI-reactive H10 antibodies, as well as a previously generated, group 2 hemagglutinin (HA) stalk-reactive antibody, demonstrated NI activity against H10N8 and an H10N7 strain; this phenomenon was absent when virus was treated with detergent, suggesting the anti-HA antibodies inhibited neuraminidase enzymatic activity through steric hindrance. We tested the prophylactic efficacy of one representative H10-reactive, N8-reactive, and group 2 HA stalk-reactive antibody in vivo using a BALB/c challenge model. All three antibodies were protective at a high dose (5 mg/kg). At a low dose (0.5 mg/kg), only the anti-N8 antibody prevented weight loss. Together, these data suggest that antibody targets other than the globular head domain of the HA may be efficacious in preventing influenza virus-induced morbidity and mortality.
IMPORTANCE Avian H10N8 and H10N7 viruses have recently crossed the species barrier, causing morbidity and mortality in humans and other mammals. Although these reports are likely isolated incidents, it is possible that more cases may emerge in future winter seasons, similar to H7N9. Furthermore, regular transmission of avian influenza viruses to humans increases the risk of adaptive mutations and reassortment events, which may result in a novel virus with pandemic potential. Currently, no specific therapeutics or vaccines are available against the H10N8 influenza virus subtype. We generated a panel of H10- and N8-reactive MAbs. Although these antibodies may practically be developed into therapeutic agents, characterizing the protective potential of MAbs that have targets other than the HA globular head domain will provide insight into novel antibody-mediated mechanisms of protection and help to better understand correlates of protection for influenza A virus infection.
INTRODUCTION
Recently, avian influenza A viruses of the H10 subtype have been reported to infect seals and humans and have generated concern over their pandemic potential. Three human cases of H10N8 virus have been reported in China so far, two of which were fatal (1–3). Furthermore, an avian H10N7 strain was found to be the etiological agent responsible for the massive die-off harbor seals in the Baltic Sea, an epidemic that killed more than 10% of the local seal population (4–6). The receptor binding profile of H10 viruses is currently debated (7–12), but the subtype has been proven to cause productive infections in humans (13, 14). Currently, the only treatment option for patients infected with an H10 subtype influenza virus is the use of antiviral inhibitors that target the viral neuraminidase (NA). Stalk-reactive monoclonal antibodies (MAbs) are actively being explored as a possible therapeutic approach to infections with avian viruses but remain in clinical development. Several stalk-reactive antibodies recognize and neutralize the H10 subtype (15–19), but no data regarding the protective efficacy of stalk MAbs against this subtype have been published so far. We generated a panel of antibodies against H10N8, including anti-H10 and anti-N8 antibodies. These antibodies were then characterized in terms of breadth, functionality, and mechanism of protection and were compared both in vitro and in vivo to a stalk-reactive antibody that also recognizes H10 subtype viruses.
MATERIALS AND METHODS
Cells, viruses, and proteins.
Madin-Darby canine kidney (MDCK) cells were grown in complete Dulbecco's modified Eagle medium (DMEM; Life Technologies) supplemented with antibiotics (100 U/ml penicillin-100 μg/ml streptomycin [Pen-Strep]; Gibco), 10% fetal bovine serum (FBS; HyClone), and 10 ml of 1 M HEPES (Life Technologies). Sf9 insect cells were grown in TNM-FH insect medium (Gemini Bioproducts) supplemented with antibiotics (Pen-Strep) and 10% FBS, and High Five cells (BTI-TN-5B1-4 subclone; Vienna Institute of Biotechnology) (20) were grown in serum-free SFX-insect cell medium (HyClone). SP2/0 mouse myeloma cells (originated from SP2/0-Ag14; ATCC CRL-1581) were passaged and maintained in complete DMEM supplemented with antibiotics (Pen-Step) prior to fusion with primary mouse splenocytes. Monoclonal, immortalized B cells (obtained from the hybridoma fusion) were initially grown in Clonacell-HY Medium E (Stemcell Technologies) and gradually switched to less enriched, serum-free hybridoma medium (Hybridoma-SFM; Life Technologies) for high-volume production.
The influenza viruses A/mallard/IA/10BM01929/10 (H10N7), A/Northern shoveler/Alaska/7MP1708/07 (H3N8), and JD13 (A/Puerto Rico/8/1934 [PR8, H1N1] internal genes and hemagglutinin [HA] and neuraminidase [NA] from A/Jiangxi-Donghu/346/13 [H10N8]) (21) were grown in 8- to 10-day-old embryonated chicken eggs, and titers were determined on MDCK cells in the presence of TPCK (tolylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin. The latter JD13 reassortant virus was rescued and characterized as previously described (21). To create purified virus preps, allantoic fluid containing virus was harvested and subjected to low-speed centrifugation (at a relative centrifugal force of 3,000 for 30 min at 4°C) to remove cellular debris. Viruses were pelleted through a 30% sucrose cushion (30% sucrose in NTE buffer [100 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA]; pH 7.4) by ultracentrifugation (Beckman L7-65 ultracentrifuge with SW-28 rotor at 25,000 rpm for 2 h). Once all of the supernatant was aspirated, virus pellets were resuspended in phosphate-buffered saline (PBS).
The recombinant proteins used—A/Jiangxi-Donghu/346/13 H10 and N8, A/harbor seal/Germany/1/14 H10, A/mallard/Interior Alaska/10BM01929/10 H10, A/mallard/Sweden/50/02 N8, and A/chicken/Netherlands/14015531/14 N8—were expressed in High Five cells and purified from cell culture supernatants as described previously (22, 23). In brief, cultures were infected with recombinant baculoviruses at a multiplicity of infection of 10. Supernatants were then harvested by low-speed centrifugation at 72 h postinfection and purified by using Ni-nitrilotriacetic acid resin (Qiagen) according to a published protocol (22).
Generation and screening of MAbs.
Six- to eight-week-old female BALB/c mice were intranasally infected with a sublethal dose (105 PFU) of A/mallard/Interior Alaska/10BM01929/10 (H10N7), followed 6 weeks later by an intranasal infection (105 PFU) with JD13 (H10N8) virus. Approximately 6 weeks after the second infection, one mouse was boosted with a unilateral, intraperitoneal injection of 100 μg of formalin-inactivated, purified JD13 (H10N8) virus adjuvanted with 10 μg of poly(I·C). At 3 days postboost, the mouse was sacrificed, and its spleen was sterilely removed. The spleen was flushed forcefully with serum-free DMEM (with antibiotics [Pen-Strep]) using a 10-ml syringe with a 20-gauge needle, followed by repeated pulverization with flat-ending forceps. Splenocytes and SP2/0 myeloma cells (in log phase) were combined in a 5:1 ratio, and cell fusion was mediated via slow, dropwise addition of 1 ml of polyethylene glycol (molecular weight, 4,000). The splenocyte/SP2 mixture was resuspended in 25 ml of complete DMEM (supplemented with antibiotics [Pen-Strep], FBS, and HEPES) and left to incubate for 24 h. After this incubation, the cells were spun down, resuspended in 10 ml of complete DMEM, mixed with a proprietary bottle of 90 ml of semisolid Clonacell-HY Medium D (Stemcell Technologies), and dispensed onto tissue culture dishes (10 ml each) using a 10-ml syringe with a 15-gauge Luer Stub adapter (Becton Dickinson). Individual colonies were picked 10 days later and transferred into 96-well plates containing Clonacell-HY Medium E. Five days after transfer to 96-well plates, hybridoma supernatants were screened by ELISA for reactivity to purified, baculovirus-expressed H10 or N8 from A/Jiangxi-Donghu/346/13 (full ELISA protocol described below). Positive clones were isotyped using a Pierce rapid antibody isotyping kit (Life Technologies); only the MAbs isotyped to the IgG heavy-chain subclasses were selected for further expansion and purification. All animal procedures were performed in accordance with the Icahn School of Medicine at Mount Sinai Institutional Animal Care and Use Committee (IACUC).
Expansion and purification of MAbs.
Hybridoma cultures were initially expanded in Clonacell-HY Medium E but gradually switched to serum-free hybridoma medium until a final volume of 300 to 500 ml was achieved. When cells appeared no longer viable (∼10 days after the final expansion step), the cultures were harvested by low-speed centrifugation (30 min, 5,500 × g), and the supernatants were passed through 0.22-μm-pore size sterile filtration units (Millipore). Filtered supernatants were passed through a gravity flow column packed with protein G-Sepharose 4 Fast Flow beads (GE Healthcare). After washing with 3 column volumes (450 ml) of sterile PBS (pH 7.4), antibody was eluted with 45 ml of 0.1 M glycine-HCl buffer (pH 2.7), and the eluate was immediately neutralized with 5 ml of 2 M Tris-HCl buffer (pH 10). The MAb was further concentrated and buffer exchanged against PBS (pH 7.4) using Amicon Ultra centrifugal filter units (10 kDa cutoff; Millipore). The final protein concentration was determined using a NanoDrop device (Thermo Scientific) and the A280 method.
ELISA.
Ninety-six-well, flat-bottom, nonsterile Immulon 4 HBX plates (Thermo Scientific) were coated overnight with either 2 μg/ml (50 μl/well) of purified protein or 5 μg/ml (50 μl/well) of purified virus in coating buffer (carbonate-bicarbonate buffer; pH 9.4) at 4°C. The coating buffer was discarded, and the plates were blocked with 3% milk in PBS containing 0.1% Tween 20 (TPBS; 100 μl/well) for 1 h at room temperature. In the case of hybridoma screening, 50 μl of undiluted supernatant from each hybridoma clone was added directly to wells as the primary antibody step. In the case of endpoint-titer enzyme-linked immunosorbent assays (ELISAs), MAbs were added at a starting concentration of 30 μg/ml and serially diluted 1:3 in 1% milk TPBS so that the final volume in each well was 100 μl. The plates were then incubated for 1 h at room temperature. After three washes with TPBS (100 μl/well for each wash), the plates were incubated for another hour at room temperature with secondary antibody solution (horseradish peroxidase [HRP]-labeled anti-mouse antibody [1:3,000; GE Healthcare] in 1% milk TPBS, 100 μl/well) and developed using SigmaFast OPD (o-phenylenediamine dihydrochloride; 100 μl per well [Sigma]) after another round of three washes. The plates were developed for 10 min, stopped with 3 M HCl (50 μl/well), and read at an optical density of 490 nm with a Synergy H1 hybrid multimode microplate reader (BioTek). An endpoint titer was defined as the final concentration at which the antibody signal remained greater than 3 standard deviations above the average of the blank wells.
PRNAs.
Plaque reduction neutralization assays (PRNAs) were performed according to a protocol described by Tan et al. (24). Briefly, MAbs at different dilutions were incubated with 100 PFU of JD13 (H10N8) virus for 1 h at room temperature. These dilutions were then plaqued on MDCK cell monolayers in six-well plates. Cells were overlaid with minimum essential medium (MEM) containing TPCK-treated trypsin at 1 μg/ml, 0.64% agarose (Oxoid), and the corresponding MAb dilutions. After 3 days of incubation, the cells were fixed with 4% paraformaldehyde, and the plaques were stained using a polyclonal anti-H10N8 mouse serum (1:1,000), an anti-mouse secondary antibody conjugated to HRP (Sigma) and Trueblue reagent (KPL). The plaques were counted in each MAb dilution, and the percent inhibition for each MAb and dilution was calculated based on a no-antibody control. The data were analyzed by using Prism software (GraphPad), and the concentration at which a MAb inhibited 50% of plaques (IC50) was calculated using a nonlinear regression.
Since anti-NA antibodies do not typically decrease plaque number but are known to restrict virus growth, we also assessed the decrease in plaque size upon incubation with antibody. To analyze average plaque diameter, 10 plaques were randomly selected in each well, and the plaque diameter was measured in three random directions for each plaque using the program ImageJ. The average diameter of each plaque was calculated from these values, and these 10 collective averages were used to calculate a single average plaque diameter for each well. Data points were analyzed using Prism software.
NA inhibition assay.
NA inhibition (NI) assays were performed as described in detail earlier (25). In brief, 96-well, flat-bottom, nonsterile Immulon 4 HBX plates (Thermo Scientific) were coated with 150 μl of fetuin (Sigma) at a concentration of 50 μg/μl and refrigerated at 4°C overnight. Individual MAbs were serially diluted 1:2 in PBS from a starting concentration of 300 μg/ml in separate 96-well, sterile, flat-bottom tissue culture plates (final volume, 75 μl/well). Virus stocks were diluted in PBS containing 1% BSA to 2× the 50% effective concentration (EC50; based on an NA assay) and added to the MAb dilution plates (75 μl/well). The plates were briefly tapped (for mixing) and incubated at room temperature for 1 h 40 min. During this time, the coating buffer was discarded from the fetuin-coated ELISA plates, and the wells were blocked for at least 1 h at room temperature with 200 μl of blocking solution (PBS containing 5% BSA). Immediately before the antibody-virus incubation period of 1 h and 40 min expired, the blocked plates were washed six times using TPBS, and 100-μl portions of the antibody-virus mixture were transferred in parallel to each well of the fetuin-coated plates. After 2 h of incubation at 37°C, the plates were washed six times using TPBS (225 μl/well), and a secondary solution of peanut agglutinin conjugated to HRP (PNA-HRP; Sigma) at a concentration of 5 μg/ml in PBS was added (100 μl/well). After incubation for 1 h and 45 min in the dark, the plates were again washed six times using TBPS (225 μl/well) and developed with 100 μl of SigmaFast OPD. The developing process was stopped after 7 min with 3 M HCl, and the reaction mixture was read at an absorbance of 490 nm with a Synergy H1 hybrid multimode microplate reader (BioTek). Data points were analyzed using Prism software (GraphPad) and the EC50 was defined as concentration at which 50% of the NA activity was inhibited compared to the negative control.
NI assay in the presence of detergent.
To perform the NI assay in the presence of detergent, all steps remained identical to those listed above, except the following: a final concentration of ca. 2% of Triton X-100 (Fisher Bioreagents) was first added directly to virus stock aliquots once thawed, and the virus preparations were allowed to shake gently at 37°C for 1 h. During this time, antibodies were diluted in PBS with 2% Triton X-100. Prior to incubation with MAbs, the virus preparation was diluted to 2× the EC50 (the EC50 was recalculated in the presence of 2% Triton X-100) in PBS containing 1% BSA and 2% Triton X-100. In addition, antibodies were diluted using PBS with 2% Triton X-100 and the virus. The NI assay was then performed as detailed above.
Evaluation of the prophylactic and therapeutic efficacy in mice.
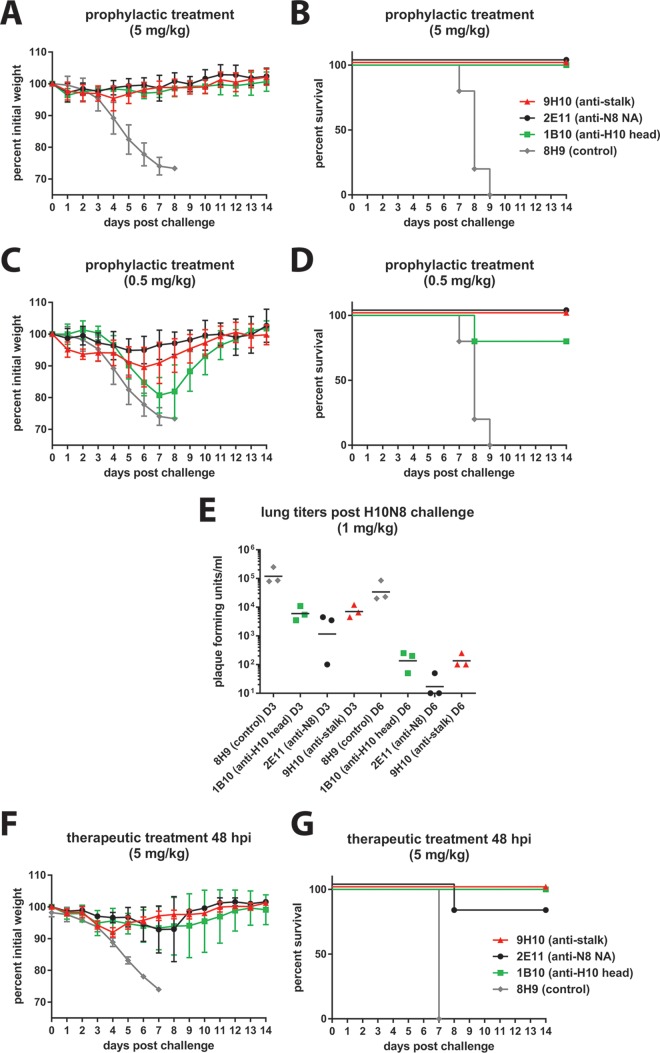
In prophylactic studies, groups of five female BALB/c mice (The Jackson Laboratory) aged 6 to 8 weeks received a 5-, 1-, or 0.5-mg/kg dose of purified mouse monoclonal antibody intraperitoneally (anti-H10 head MAb 1B10 [IgG2b], anti-stalk MAbs 9H10 [IgG2a], anti-N8 antibody 2E11 [IgG2b], and control MAb 8H9 [anti-H6]). At 2 h after treatment, the mice were anesthetized using a ketamine-xylazine mixture and infected with 5× the 50% lethal dose (LD50) of JD13 (H10N8) virus diluted in PBS (pH 7.4). The LD50 of this virus in BALB/c mice has been previously determined and characterized in detail (21). In a therapeutic setting, mice received a 5-mg/kg dose of each antibody intraperitoneally 48 h after infection. Mice that received a 1-mg/kg antibody dose prophylactically were sacrificed either on day 3 or 6 postinfection for lung titer analysis. At these time points, the lungs were harvested and homogenized using a BeadBlaster 24 (Benchmark) homogenizer, and viral lung titers were measured by plaquing lung homogenate on MDCK cells. In all other groups, mice were monitored daily for survival and weight loss until day 14 postinfection. Mice that lost more than 25% of their initial body weights were euthanized. All animal procedures were performed in accordance with the Icahn School of Medicine at Mount Sinai IACUC.
RESULTS
Anti-H10 and anti-N8 antibodies show high cross-reactivity within their respective subtypes.
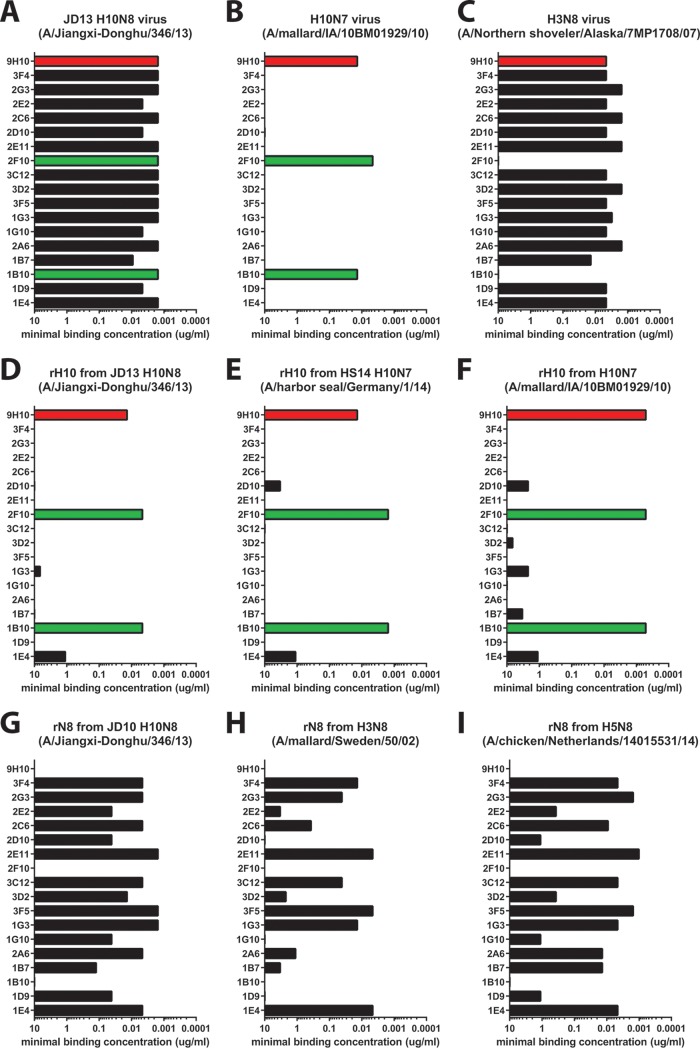
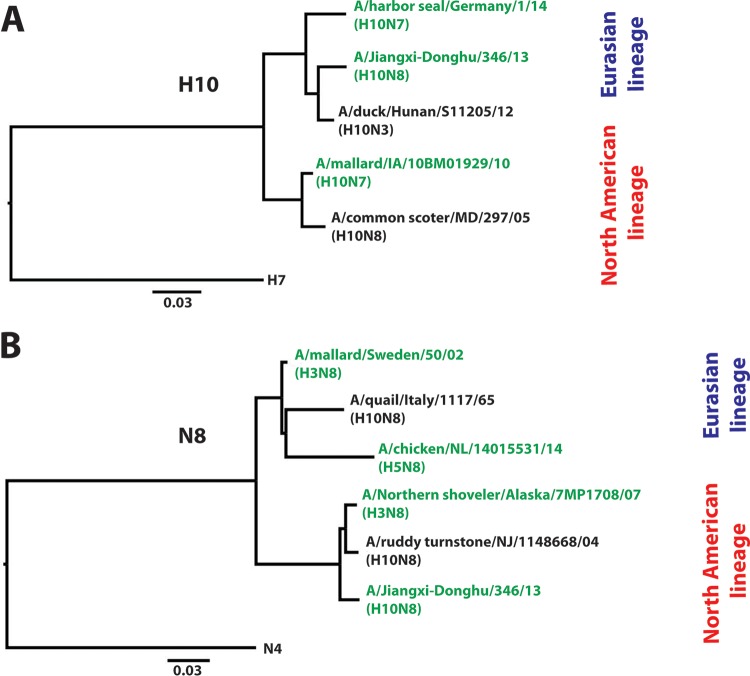
Using traditional hybridoma technology, we isolated a total of 17 MAbs with specific reactivity to H10N8 virus strain JD13. In all further analyses, we decided to include stalk-reactive MAb 9H10, an antibody which has recently been reported to have activity against the H10 subtype, as an additional control. All 18 antibodies tested showed high reactivity to purified JD13 (H10N8) virus in a quantitative ELISA (Fig. 1A). Two of the newly isolated MAbs (1B10 and 2F10) also showed high reactivity to a purified H10N7 (A/mallard/IA/10BM01929/10) preparation (Fig. 1B). Conversely, the remaining 15 MAbs we generated strongly reacted to an H3N8 (A/Northern shoveler/Alaska/7MP1708/07) preparation. These results indicate that only two of the isolated MAbs, 1B10 and 2F10, are reactive to H10 HA, whereas the remaining MAbs are reactive to the N8 NA (Fig. 1C). MAb 9H10 reacted strongly with both preparations, which was expected since it displays broad reactivity to the group 2 HA stalk domain. To corroborate these findings, we also tested the reactivity of all 18 MAbs to recombinant H10 hemagglutinin and N8 neuraminidase from JD13 (H10N8) virus. The results confirmed the findings from the purified, whole-virus ELISAs (Fig. 1D and G). Furthermore, we tested all MAbs for binding to recombinant H10 and N8 proteins from more divergent viruses to test cross-reactivity (Fig. 1E, F, H, and I). We found strong cross-reactivity for the anti-H10 antibodies 1B10 and 2F10 that spanned both the North American and Eurasian lineages (Fig. 2A). However, neither 1B10 nor 2F10 reacted with the H3N8 virus (and do not display HI activity [see Fig. 5]), suggesting that they are more specifically directed to the HA globular head domain. Furthermore, we found strong N8 cross-reactivity within the North American lineage (Fig. 1A, C, and G). In addition more than half of the anti-N8 MAbs were also able to bind to the more distant Eurasian N8 lineage (Fig. 1H and I and 2B), including the N8 from the highly pathogenic Fujian H5 clade 2.3.4.4, which has recently spread in Asia, Europe, and North America (Fig. 1H) (26–32).
FIG 1.
Anti-H10 and anti-N8 antibodies display heterologous cross-reactivity. The minimal binding concentrations of anti-H10 (green) and anti-N8 (black) MAbs and stalk-reactive MAb 9H10 (red) to either purified whole virus (coated at 5 μg/ml) or recombinant, baculovirus-expressed proteins (coated at 2 μg/ml) as measured by ELISA are shown. All 18 MAbs showed high reactivity to purified JD13 (H10N8) virus (A), whereas only the HA-directed MAbs displayed reactivity to an H10N7 virus (A/mallard/Interior Alaska/10BM01929/10) (B) and only the NA-directed MAbs displayed reactivity to an H3N8 virus (A/Northern shoveler/Alaska/7MP1708/07) (C). These findings were confirmed by quantitative ELISAs to purified, recombinant H10 and N8 proteins from either the matched JD13 (H10N8) virus or the divergent strains (D to I) from both the Eurasian and North American lineages.
FIG 2.
H10N8 MAbs display cross-lineage, heterologous binding activity. Phylogenetic trees of representative H10 (A) and N8 (B) sequences from the Eurasian and North American lineages are shown. Both H10 HA and N8 NA cluster into one Eurasian and one North American lineage. Scale bars represent a 3% difference in amino acid identity. The anti-H10 antibodies generated, 1B10 and 2F10, displayed high cross-reactivity to viruses from both lineages (tested strains are highlighted in green). The N8-directed MAbs also displayed remarkable cross-lineage binding reactivity as well (tested strains are highlighted in green). Trees were generated in Clustal Omega and visualized in FigTree.
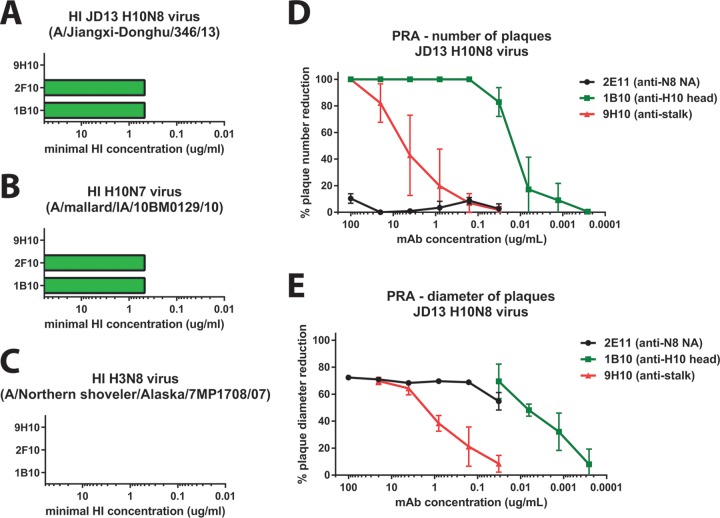
FIG 5.
Activity of selected MAbs in HI and PRNAs. HA-reactive antibodies were tested in an HI assay against JD13 H10N8 (A), H10N7 virus (A/mallard/Interior Alaska/10BM01929/10) (B), and an H3N8 virus (A/Northern shoveler/Alaska/7MP1708/07) (C). MAbs 1B10 and 2F10 showed HI activity against H10N8 and H10N7 viruses but lacked reactivity to H3N8. Stalk-reactive MAb 9H10 showed no HI activity against the tested viruses. (D) A PRNA with anti-H10 head antibodies 1B10 (anti-head), 2E11 (anti-NA) and 9H10 (anti-stalk) was performed. Both anti-head and anti-stalk antibodies were effective in reducing the number of plaques, with the anti-head antibody 1B10 displaying 320 times greater potency than that of the anti-stalk MAb 9H10. Anti-NA antibody 2E11 was not able to reduce plaque numbers at any tested concentration. (E) Reduction of plaque size (as measured by plaque diameter) by the three tested MAbs. Using this readout, the anti-NA antibody 2E11 displays robust reactivity.
Anti-HA head, anti-HA stalk, and anti-NA MAbs show NI activity.
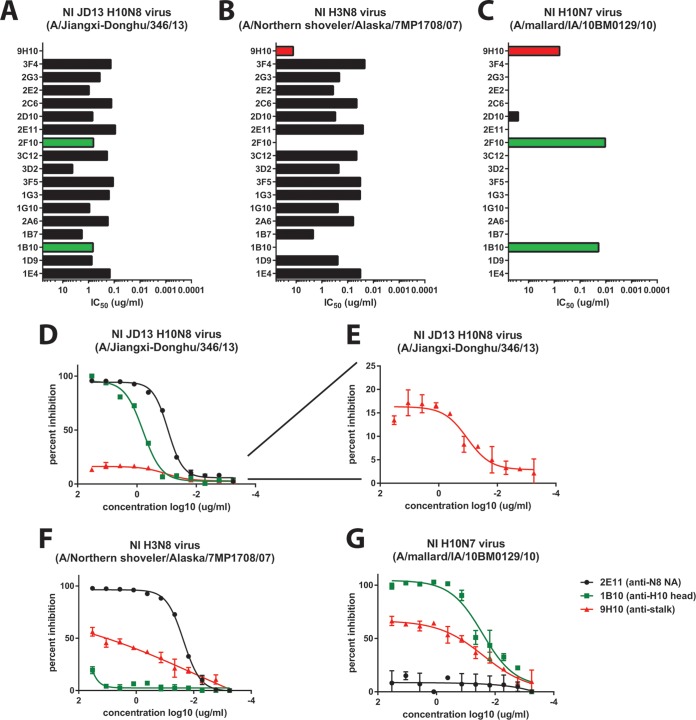
Neuraminidase inhibition is mediated by antibodies that bind to the NA and prevent its ability to enzymatically cleave sialic acids via direct binding to the active site or by steric hindrance. We tested the isolated MAbs (including 9H10) for their activity in a fetuin-based neuraminidase inhibition assay (33). First, we tested the antibodies against JD13 (H10N8) virus and found strong NI activity for all tested NA-reactive antibodies (Fig. 3A). Interestingly, the two H10-reactive antibodies showed strong NI activity as well (Fig. 3A). To determine whether this NI activity was dependent on the subtype of the HA, we tested all MAbs for NI activity against H3N8 (Fig. 3B). All N8-reactive MAbs maintained their activity against this virus, but the two H10-reactive MAbs showed no NI (Fig. 3B). Surprisingly, stalk-reactive MAb 9H10 also showed low—but significant—NI activity against H3N8 (Fig. 3B). Finally, we wanted to test NI activity of the MAb panel against an H10N7 virus. Only the two H10-reactive MAbs showed high activity (Fig. 3C) in this assay. 9H10 showed good NI activity against this subtype as well (Fig. 3C). To follow up on these findings we selected three antibodies—an H10-reactive MAb (1B10), a stalk-reactive MAb (9H10), and an N8-reactive MAb (2E11)—to further examine. Upon reexamination of the NI data against JD13 (H10N8) we found that, of the three selected antibodies, MAb 2E11 (N8) had the strongest NI activity, followed closely by MAb 1B10 (H10) (Fig. 3D). However, MAb 9H10 (stalk) also showed activity with a plateau at ca. 15% maximum inhibition (Fig. 3D and E); this indicates that the antibody has measurable NI activity against the virus but that this value cannot be reported as an IC50, since 50% maximum inhibition is never reached. MAb 2E11 also strongly inhibited H3N8, followed by 9H10, whereas H10-reactive MAb 1B10 displayed no NI activity against this strain (Fig. 3F). Finally, MAb 1B10 showed strong activity against H10N7, followed by 9H10, whereas N8-reactive MAb 2E11 displayed no activity against this virus (Fig. 3G). We postulate that the NI activity of anti-H10 head and anti-HA stalk MAbs is most likely mediated by steric hindrance (Fig. 4). To test this hypothesis, we performed the NI assay in the presence of the detergent Triton X-100, which historically has been used to disrupt the lipid bilayer of virions for the further extraction of the viral glycoproteins during the production of split influenza virus vaccines (34). Although HA and NA are in close contact in the virion, the Triton X-100 treatment solubilizes these proteins, and they dissociate from each other as long as the detergent is present. In support of our hypothesis, the NI activity of HA-directed antibodies was no longer observable upon addition of Triton X-100, while the NI activity of the NA-directed antibodies remained robust (Fig. 4D).
FIG 3.
Anti-HA head, anti-HA stalk, and anti-NA MAbs show NI activity. (A to C) All 18 MAbs generated (and 9H10) were tested in a classic neuraminidase inhibition (NI) assay, in which fetuin-coated ELISA plates were used as the target substrate. (A) N8 antibodies were able to significantly inhibit the NA activity of JD13 (H10N8) virus. (B) All anti-N8 antibodies were also able to significantly inhibit the NA activity of an H3N8 virus. Surprisingly, the anti-H10 antibodies displayed NI activity against JD13 (A) and an H10N7 strain (A/mallard/Interior Alaska/10BM01929/10) (C), and the anti-HA stalk antibodies displayed low—but significant—NI activity to both the H3N8 (A/Northern shoveler/Alaska/7MP1708/07) (B) and H10N7 (C) strains tested. To investigate this phenomenon, the NI data from one representative H10-reactive (1B10), N8-reactive (2E11), and the HA stalk-reactive antibody (9H10) were plotted and fit to nonlinear regression curves (D to G). At this resolution, it is apparent that 9H10 displays NI activity to H10N8 as well; this inhibition displays sigmoidal kinetics, but plateaus at only 15% maximal inhibition and so is not captured by the IC50 graph in panel A.
FIG 4.
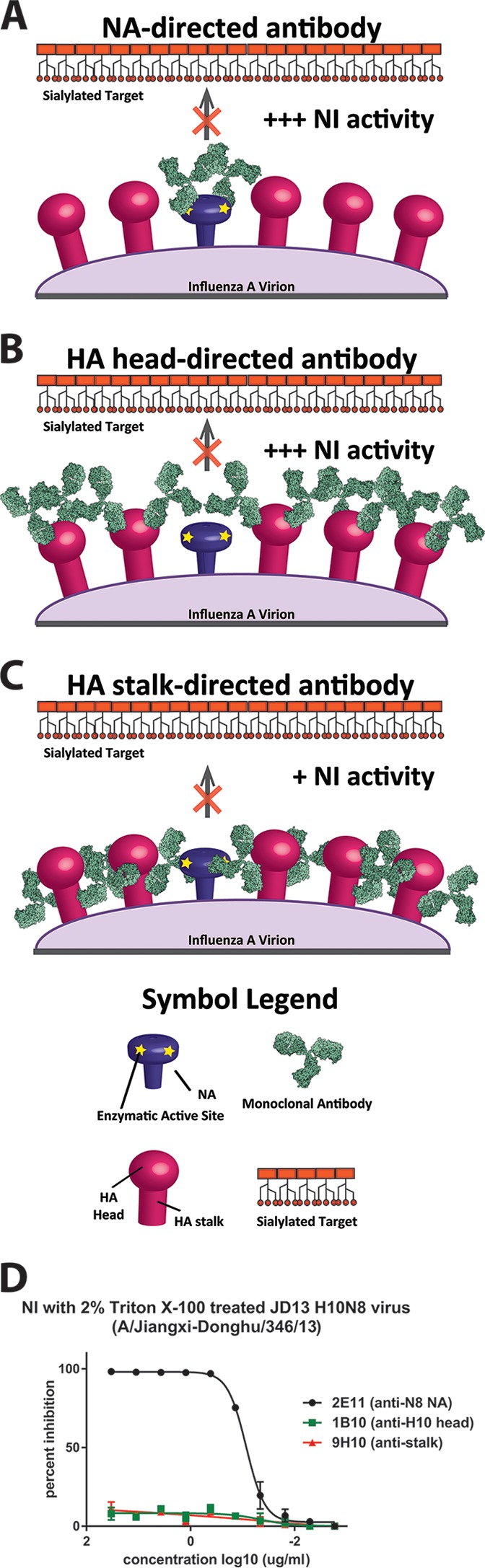
Theoretical mechanism for anti-HA antibody mediated NI activity. (A) Anti-NA antibodies are able to result in NI activity by directly binding to and blocking the enzymatic active site of the NA. (B and C) However, it is conceivable that antibodies against the HA head and stalk domain may, by steric hindrance, shield the NA active site from the substrate. Because stalk-directed MAbs bind lower on the HA glycoprotein, this steric hindrance effect may not be as robust (C), possibly explaining the lower NI seen with 9H10 in Fig. 3. (D) This scenario may be further complicated by the HA/NA ratio, extent of NA clustering on the virus surface, differences in specific NA activity and differences in strengths and kinetics of individual MAb binding profiles. Detergent treatment (Triton X-100) of the virus releases HA and NA from the viral membrane and disassociates them from each other. Both HA head and stalk MAbs lost all NI activity under these conditions, while the NA antibody retained its activity.
In vitro neutralizing activity of divergent anti-H10N8 antibodies.
As a first step, we assessed the hemagglutination inhibition (HI) activity of the three HA-reactive antibodies. The HI titer is used as a correlate of protection and is usually measured to assess influenza virus vaccine efficacy; furthermore, HI active antibodies are usually potently neutralizing. The two H10 specific MAbs 1B10 and 2F10 both showed high HI activity against H10N8 strain JD13 and against H10N7 strain A/mallard/IA/10BM01929/10 but expectedly lacked activity against an H3N8 strain (Fig. 5A to C). The stalk-reactive antibody 9H10 completely lacked HI activity against all three strains which is not surprising given that its membrane-proximal footprint on the HA is far from the receptor binding site (15).
Next, we measured the in vitro neutralizing activity of MAbs 9H10 (anti-HA stalk), 1B10 (anti-HA head), and 2E11 (anti-NA) in a plaque reduction neutralization assay. The traditional way to read this assay is to assess the reduction in the number of plaques per well caused by the antibody. Using this analysis, we found that the anti-head MAb strongly neutralized the JD13 (H10N8) virus (IC50 = 0.014 μg/ml) (Fig. 5D). Anti-stalk MAb 9H10 neutralized as well but had an ∼320-fold higher IC50 (4.503 μg/ml). Anti-NA MAb did not show neutralizing activity as assessed by plaque count (Fig. 5D).
However, we observed that the plaques in the presence of MAb 2E11 had a smaller plaque phenotype. We therefore chose to analyze percent reduction in plaque diameter. Both HA head- and stalk-reactive antibodies reduced plaque size at concentrations at which they also reduced plaque numbers (Fig. 5E). The NA-reactive MAb 2E11 also showed strong activity in this assay and was able to reduce plaque size by ca. 75% at concentrations between 100 and 0.16 μg/ml. This activity began to drop only at the lowest concentration of 2E11 tested (0.032 μg/ml) (Fig. 5E).
In vivo protective activity of anti-HA head, anti-HA stalk, and anti-NA MAbs.
Finally, we investigated the protective effect of anti-H10 head, anti-N8, and anti-stalk MAbs in an in vivo challenge model. We used the same three representative MAbs selected earlier (1B10, 2E11, and 9H10) in a passive-transfer challenge experiment with JD13 (H10N8) virus in mice. Mice received MAb at 5 or 0.5 mg/kg 2 h prior to infection. Upon challenge, weight loss and survival were monitored. Treatment of mice with 5 mg/kg completely protected mice from weight loss and mortality, whereas mice treated with an irrelevant anti-H6 specific MAb lost weight rapidly and succumbed to infection by day 9 (Fig. 6A and B). At 0.5 mg/kg, however, we observed marked differences between MAbs. To our surprise, the anti-N8 MAb 2E11 was still completely protective and inhibited weight loss and mortality (Fig. 6C and D). Stalk-reactive MAb 9H10 protected from mortality, but mice showed an intermediate weight loss phenotype with a maximum weight loss of 10% on day 6 but fast recovery after day 6. HI-active, neutralizing anti-head MAb 1B10 performed less favorably. The MAb did not protect from weight loss at 0.5 mg/kg, and a maximum average weight loss of ca. 20% was reached on day 7 postinfection. In addition, the survival rate for 1B10 was only 80% compared to 100% for the other two MAbs. To follow up on these findings, we repeated this prophylactic study using a 1-mg/kg antibody dose and harvested lungs on days 3 and day 6 postinfection to determine viral lung titers. Although all MAbs were able to significantly reduce viral lung titers on both days, the anti-N8 MAb 2E11 showed the greatest overall reduction compared to the two HA-reactive MAbs, with two of three lung isolates showing undetectable levels of virus on day 6 postinfection (Fig. 6E). Finally, we also wanted to examine the therapeutic activity of the three MAbs. This time we infected mice and then treated them with 5-mg/kg doses of the respective MAbs at 48 h postinfection. All three antibodies showed therapeutic activity with 100% survival for the anti-H10 head and the anti-stalk MAbs and 80% survival for the anti-N8 MAb (Fig. 6F and G).
FIG 6.
Anti-HA head, anti-HA stalk, and anti-NA antibodies are protective against H10N8 in vivo. To test prophylactic efficacy, female BALB/c mice (5 per group) were administered either 5 or 0.5 mg/kg of a representative anti-H10 head (1B10), anti-H10 stalk (9H10), anti-N8 (2E11), or an irrelevant negative-control antibody (8H9, an anti-H6 head antibody) 2 h prior to a 5× LD50 challenge with JD13 (H10N8) virus. All antibodies (excluding the negative control) were protective against morbidity (A) and mortality (B) at the higher dose. At the lower dose, mice in the anti-N8 MAb group displayed the least weight loss (C), and all groups (except that receiving the anti-H10 head antibody) demonstrated 100% survival (D). The experimental results shown in panels A to D were performed in parallel and share the negative-control group. Prophylactic studies were repeated at a dose of 1 mg/kg, and mice were sacrificed at days 3 and 6 for lung titer analysis. (E) In line with previous weight loss results, the anti-N8 MAb was most effective in reducing titers at both time points. To test therapeutic efficacy, mice were administered 5 mg/kg of each antibody 48 h after a 5× LD50 challenge with JD13 (H10N8) virus. 1B10 and 9H10 were 100% protective, while 2E11 was 80% protective (F to G).
DISCUSSION
Recent fatal human cases of H10N8 virus infections have caused increased interest in avian influenza viruses of the H10 subtype (1, 35). Here, we showed that the antibody response toward H10N8 induced protective antibodies against both the HA and the NA surface glycoproteins. The isolated H10 antibodies displayed a broad binding profile spanning both the North American and the Eurasian lineages of H10. Of note, JD13 (H10N8), which caused human fatalities in China in 2013, belongs to the North American lineage (1), whereas the H10N7 strain that caused a deadly epidemic in Baltic harbor seals in 2014 belongs to the Eurasian lineage (4–6). The two isolated MAbs would very likely be effective against both strains. The anti-N8 antibodies generated by our fusion also showed a very broad binding profile; over half of the isolated MAbs exhibited binding to both the North American and the Eurasian N8 lineages. Importantly, we found strong reactivity to potential pandemic viruses like H3N8, which caused a severe outbreak in harbor seals in New England in 2012, as well as against the N8 NA of the newly emerging, highly pathogenic H5N8 viruses (26, 30, 31, 36, 37). The H3N8 subtype is also important, since it was speculated to be the cause of a human pandemic in 1889 (38). This suggests that the isolated MAbs could be used as therapeutics against emerging viruses of the H10 or N8 subtype and that H10N8 vaccines could have broad efficacy against those viruses as well.
Another interesting finding of this study is the fact that antibodies directed against the HA can have NI activity. Although there is evidence for this phenomenon in the literature for HA-head reactive antibodies, we report this effect for broadly reactive anti-stalk antibodies for the first time (33). We hypothesize that this activity is mediated by steric hindrance. Antibody bound to HA might block access of the NA to its substrate. The NA is slightly shorter than the HA, and it is conceivable that the Fc part of MAbs bound to HA can form a shield over the NA molecules. This effect seems to be stronger for anti-head MAbs than for stalk MAbs. Anti-stalk MAbs bind closer to the viral membrane, and thus the effect of steric hindrance may be less pronounced. Furthermore, the NI activity of anti-HA MAbs might be modulated by the HA/NA ratio on the virus surface and by the specific activity of the NA. The World Health Organization recommends the use of viruses with mismatched HAs to measure NI titers in serum (http://www.who.int/csr/resources/publications/influenza/whocdscsrncs20025rev.pdf). However, anti-stalk MAbs with broad binding activity might interfere with this assay since they are able to bind to mismatched HAs as well. Anti-stalk MAbs might therefore contribute to NI titers tested in polyclonal serum. A solution for this problem might be the use of recombinant NA in NI assays.
In our study, we had the opportunity to test an anti-HA head, an anti-HA stalk, and an anti-NA antibody side by side. It was expected that the HA head-directed antibody, with its high in vitro neutralization activity and HI activity, would be superior in protecting animals from virus challenge. However, both anti-HA stalk and anti-NA antibodies outperformed the anti-head antibody at low concentrations in regard to protective efficacy. The in vitro neutralizing activities of the three MAbs are strikingly different. In a plaque reduction neutralization assay the anti-head MAb was ∼320-fold more active than the anti-stalk MAbs. Although the anti-NA MAb did not affect plaque number, the plaque size was markedly reduced; this is concordant with the major enzymatic role of NA, which is to free emerging influenza virus particles from the surfaces of infected cells. It is possible that these results do not reflect polyclonal responses or do not apply to other influenza virus subtypes. However, these findings shine a new light on the HI assay, which is currently used as a correlate of protection for seasonal and pandemic influenza virus vaccines.
ACKNOWLEDGMENTS
We thank Jonathan Runstadler (MIT) for providing the H3N8 and H10N7 viruses used in this study.
Funding was provided in part by the NIH Centers of Excellence in Influenza Virus Research and Surveillance (CEIRS; contract HHSN272201400008C) and U19 AI109946-01.
REFERENCES
- 1.Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, Zou S, Yang L, Chen T, Dong L, Bo H, Zhao X, Zhang Y, Lan Y, Bai T, Dong J, Li Q, Wang S, Li H, Gong T, Shi Y, Ni X, Li J, Fan J, Wu J, Zhou X, Hu M, Wan J, Yang W, Li D, Wu G, Feng Z, Gao GF, Wang Y, Jin Q, Liu M, Shu Y. 2014. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2014. Avian influenza A (H10N8). World Health Organization, Geneva, Switzerland: http://www.wpro.who.int/china/mediacentre/factsheets/h10n8/en/. [Google Scholar]
- 3.Krammer F. 2015. Emerging influenza viruses and the prospect of a universal influenza virus vaccine. Biotechnol J 10:690–701. doi: 10.1002/biot.201400393. [DOI] [PubMed] [Google Scholar]
- 4.Bodewes R, Bestebroer TM, van der Vries E, Verhagen JH, Herfst S, Koopmans MP, Fouchier RA, Pfankuche VM, Wohlsein P, Siebert U, Baumgärtner W, Osterhaus AD. 2015. Avian influenza A(H10N7) virus-associated mass deaths among harbor seals. Emerg Infect Dis 21:720–722. doi: 10.3201/eid2104.141675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krog JS, Hansen MS, Holm E, Hjulsager CK, Chriél M, Pedersen K, Andresen LO, Abildstrøm M, Jensen TH, Larsen LE. 2015. Influenza A(H10N7) virus in dead harbor seals, Denmark. Emerg Infect Dis 21:684–687. doi: 10.3201/eid2104.141484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zohari S, Neimanis A, Harkonen T, Moraeus C, Valarcher J. 2014. Avian influenza A(H10N7) virus involvement in mass mortality of harbour seals (Phoca vitulina) in Sweden, March through October 2014. Euro Surveill 19:pii20967. [DOI] [PubMed] [Google Scholar]
- 7.Deng G, Shi J, Wang J, Kong H, Cui P, Zhang F, Tan D, Suzuki Y, Liu L, Jiang Y, Guan Y, Chen H. 2015. Genetics, receptor binding, and virulence in mice of H10N8 influenza viruses isolated from ducks and chickens in live poultry markets in China. J Virol 89:6506–6510. doi: 10.1128/JVI.00017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vachieri SG, Xiong X, Collins PJ, Walker PA, Martin SR, Haire LF, Zhang Y, McCauley JW, Gamblin SJ, Skehel JJ. 2014. Receptor binding by H10 influenza viruses. Nature 511:475–477. doi: 10.1038/nature13443. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, de Vries RP, Tzarum N, Zhu X, Yu W, McBride R, Paulson JC, Wilson IA. 2015. A human-infecting H10N8 influenza virus retains a strong preference for avian-type receptors. Cell Host Microbe 17:377–384. doi: 10.1016/j.chom.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang H, Carney PJ, Chang JC, Villanueva JM, Stevens J. 2015. Structure and receptor binding preferences of recombinant hemagglutinins from avian and human H6 and H10 influenza A virus subtypes. J Virol 89:4612–4623. doi: 10.1128/JVI.03456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M, Zhang W, Qi J, Wang F, Zhou J, Bi Y, Wu Y, Sun H, Liu J, Huang C, Li X, Yan J, Shu Y, Shi Y, Gao GF. 2015. Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus. Nat Commun 6:5600. doi: 10.1038/ncomms6600. [DOI] [PubMed] [Google Scholar]
- 12.Ramos I, Mansour M, Wohlbold TJ, Ermler ME, Hirsh A, Runstadler JA, Fernandez-Sesma A, Krammer F. 2015. Hemagglutinin receptor binding of a human isolate of influenza A(H10N8) virus. Emerg Infect Dis 21:1197–1201. doi: 10.3201/eid2107.141755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arzey GG, Kirkland PD, Arzey KE, Frost M, Maywood P, Conaty S, Hurt AC, Deng YM, Iannello P, Barr I, Dwyer DE, Ratnamohan M, McPhie K, Selleck P. 2012. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg Infect Dis 18:814–816. doi: 10.3201/eid1805.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beare AS, Webster RG. 1991. Replication of avian influenza viruses in humans. Arch Virol 119:37–42. doi: 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- 15.Tan GS, Lee PS, Hoffman RM, Mazel-Sanchez B, Krammer F, Leon PE, Ward AB, Wilson IA, Palese P. 2014. Characterization of a broadly neutralizing monoclonal antibody that targets the fusion domain of group 2 influenza a virus hemagglutinin. J Virol 88:13580–13592. doi: 10.1128/JVI.02289-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friesen RH, Lee PS, Stoop EJ, Hoffman RM, Ekiert DC, Bhabha G, Yu W, Juraszek J, Koudstaal W, Jongeneelen M, Korse HJ, Ophorst C, Brinkman-van der Linden EC, Throsby M, Kwakkenbos MJ, Bakker AQ, Beaumont T, Spits H, Kwaks T, Vogels R, Ward AB, Goudsmit J, Wilson IA. 2014. A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci U S A 111:445–450. doi: 10.1073/pnas.1319058110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin Ö, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 20.Krammer F, Schinko T, Palmberger D, Tauer C, Messner P, Grabherr R. 2010. Trichoplusia ni cells (High Five) are highly efficient for the production of influenza A virus-like particles: a comparison of two insect cell lines as production platforms for influenza vaccines. Mol Biotechnol 45:226–234. doi: 10.1007/s12033-010-9268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wohlbold TJ, Hirsh A, Krammer F. 2015. An H10N8 influenza virus vaccine strain and mouse challenge model based on the human isolate A/Jiangxi-Donghu/346/13. Vaccine 33:1102–1106. doi: 10.1016/j.vaccine.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margine I, Palese P, Krammer F. 2013. Expression of functional recombinant hemagglutinin and neuraminidase proteins from the novel H7N9 influenza virus using the baculovirus expression system. J Vis Exp 2013:e51112. doi: 10.3791/51112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krammer F, Margine I, Tan GS, Pica N, Krause JC, Palese P. 2012. A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates. PLoS One 7:e43603. doi: 10.1371/journal.pone.0043603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan GS, Krammer F, Eggink D, Kongchanagul A, Moran TM, Palese P. 2012. A pan-h1 anti-hemagglutinin monoclonal antibody with potent broad-spectrum efficacy in vivo. J Virol 86:6179–6188. doi: 10.1128/JVI.00469-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wohlbold TJ, Nachbagauer R, Xu H, Tan GS, Hirsh A, Brokstad KA, Cox RJ, Palese P, Krammer F. 2015. Vaccination with adjuvanted recombinant neuraminidase induces broad heterologous, but not heterosubtypic, cross-protection against influenza virus infection in mice. mBio 6:e02556. doi: 10.1128/mBio.02556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ip HS, Torchetti MK, Crespo R, Kohrs P, DeBruyn P, Mansfield KG, Baszler T, Badcoe L, Bodenstein B, Shearn-Bochsler V, Killian ML, Pedersen JC, Hines N, Gidlewski T, DeLiberto T, Sleeman JM. 2015. Novel Eurasian highly pathogenic avian influenza A H5 viruses in wild birds, Washington, USA, 2014. Emerg Infect Dis 21:886–890. doi: 10.3201/eid2105.142020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan S, Zhou L, Wu D, Gao X, Pei E, Wang T, Gao Y, Xia X. 2014. A novel highly pathogenic H5N8 avian influenza virus isolated from a wild duck in China. Influenza Other Respir Viruses 8:646–653. doi: 10.1111/irv.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu H, Peng X, Xu L, Jin C, Cheng L, Lu X, Xie T, Yao H, Wu N. 2014. Novel reassortant influenza A(H5N8) viruses in domestic ducks, eastern China. Emerg Infect Dis 20:1315–1318. doi: 10.3201/eid2008.140339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong J, Kang HM, Lee EK, Song BM, Kwon YK, Kim HR, Choi KS, Kim JY, Lee HJ, Moon OK, Jeong W, Choi J, Baek JH, Joo YS, Park YH, Lee HS, Lee YJ. 2014. Highly pathogenic avian influenza virus (H5N8) in domestic poultry and its relationship with migratory birds in South Korea during 2014. Vet Microbiol 173:249–257. doi: 10.1016/j.vetmic.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Ku KB, Park EH, Yum J, Kim JA, Oh SK, Seo SH. 2014. Highly pathogenic avian influenza A(H5N8) virus from waterfowl, South Korea, 2014. Emerg Infect Dis 20:1587–1588. doi: 10.3201/eid2009.140390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YJ, Kang HM, Lee EK, Song BM, Jeong J, Kwon YK, Kim HR, Lee KJ, Hong MS, Jang I, Choi KS, Kim JY, Lee HJ, Kang MS, Jeong OM, Baek JH, Joo YS, Park YH, Lee HS. 2014. Novel reassortant influenza A(H5N8) viruses, South Korea, 2014. Emerg Infect Dis 20:1087–1089. doi: 10.3201/eid2006.140233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anonymous. 2014. Avian influenza outbreak in Yorkshire: strain identified as H5N8. Vet Rec 175:495–496. doi: 10.1136/vr.g6947. [DOI] [PubMed] [Google Scholar]
- 33.Couzens L, Gao J, Westgeest K, Sandbulte M, Lugovtsev V, Fouchier R, Eichelberger M. 2014. An optimized enzyme-linked lectin assay to measure influenza A virus neuraminidase inhibition antibody titers in human sera. J Virol Methods 210C:7–14. doi: 10.1016/j.jviromet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 34.Gross PA, Ennis FA, Gaerlan PF, Denning CR, Setia U, Davis WJ, Bisberg DS. 1981. Comparison of new triton X-100- and tween-ether-treated split-treated vaccines in children. J Clin Microbiol 14:534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.García-Sastre A, Schmolke M. 2014. Avian influenza A H10N8: a virus on the verge? Lancet 383:676–677. doi: 10.1016/S0140-6736(14)60163-X. [DOI] [PubMed] [Google Scholar]
- 36.Anthony SJ, St Leger JA, Pugliares K, Ip HS, Chan JM, Carpenter ZW, Navarrete-Macias I, Sanchez-Leon M, Saliki JT, Pedersen J, Karesh W, Daszak P, Rabadan R, Rowles T, Lipkin WI. 2012. Emergence of fatal avian influenza in New England harbor seals. mBio 3:e00166-12. doi: 10.1128/mBio.00166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee DH, Torchetti MK, Winker K, Ip HS, Song CS, Swayne DE. 2015. Intercontinental spread of Asian-origin H5N8 to North America through Beringia by migratory birds. J Virol 89:6521–6524. doi: 10.1128/JVI.00728-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Worobey M, Han GZ, Rambaut A. 2014. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc Natl Acad Sci U S A 111:8107–8112. doi: 10.1073/pnas.1324197111. [DOI] [PMC free article] [PubMed] [Google Scholar]