ABSTRACT
The cell-transforming activity of human adenovirus 5 (hAd5) E1A is mediated by the N-terminal half of E1A, which interacts with three different major cellular protein complexes, p300/CBP, TRRAP/p400, and pRb family members. Among these protein interactions, the interaction of pRb family proteins with conserved region 2 (CR2) of E1A is known to promote cell proliferation by deregulating the activities of E2F family transcription factors. The functional consequences of interaction with the other two protein complexes in regulating the transforming activity of E1A are not well defined. Here, we report that the E1A N-terminal region also interacted with the cellular proto-oncoprotein c-MYC and the homolog of enhancer of yellow 2 (ENY2). Our results suggested that these proteins interacted with an essential E1A transforming domain spanning amino acid residues 26 to 35 which also interacted with TRRAP and p400. Small interfering RNA (siRNA)-mediated depletion of TRRAP reduced c-MYC interaction with E1A, while p400 depletion did not. In contrast, depletion of TRRAP enhanced ENY2 interaction with E1A, suggesting that ENY2 and TRRAP may interact with E1A in a competitive manner. The same E1A region additionally interacted with the constituents of a deubiquitinase complex consisting of USP22, ATXN7, and ATXN7L3 via TRRAP. Acute short hairpin RNA (shRNA)-mediated depletion of c-MYC reduced the E1A transforming activity, while depletion of ENY2 and MAX did not. These results suggested that the association of c-MYC with E1A may, at least partially, play a role in the E1A transformation activity, independently of MAX.
IMPORTANCE The transforming region of adenovirus E1A consists of three short modules which complex with different cellular protein complexes. The mechanism by which one of the transforming modules, CR2, promotes cell proliferation, through inactivating the activities of the pRb family proteins, is better understood than the activities of the other domains. Our analysis of the E1A proteome revealed the presence of the proto-oncoprotein c-MYC and of ENY2. We mapped these interactions to a critical transforming module of E1A that was previously known to interact with the scaffolding molecule TRRAP and the E1A-binding protein p400. We showed that c-MYC interacted with E1A through TRRAP, while ENY2 interacted with it independently. The data reported here indicated that depletion of c-MYC in normal human cells reduced the transforming activity of E1A. Our result raises a novel paradigm in oncogenic transformation by a DNA viral oncogene, the E1A gene, that may exploit the activity of a cellular oncogene, the c-MYC gene, in addition to inactivation of the tumor suppressors, such as pRb.
INTRODUCTION
Adenovirus E1A is an extensively investigated viral oncogene product that is exploited to elucidate mechanisms of cell proliferation and oncogenic transformation. The E1A gene region codes for two major protein isoforms, S-E1A and L-E1A. Of the two protein isoforms, the smaller isoform (S-E1A) (referred to here as E1A protein) is widely used to study mechanisms of fibroblast and epithelial cell transformation. Mutational dissection of the transforming activity of S-E1A has indicated that the N-terminal half is essential for transformation of primary epithelial cells in cooperation with the activated H-Ras oncogene (reviewed in reference 1). In contrast, the C-terminal half inhibits this function. The transforming activity of the E1A N-terminal region is linked to three short amino acid regions of E1A that interact with three major cellular protein complexes, p300/CBP, TRRAP/p400, and pRb/p107/p130 (reviewed in reference 2). The transforming activity imparted by the interaction of these protein complexes is opposed as a consequence of the interaction of three other protein complexes, FOXK1/K2, DYRK1A/1B/HAN11, and CtBP1/2, with the C-terminal half of E1A (reviewed in references 1 and 3).
Although the interaction of cellular protein complexes with the N-terminal region is well characterized, the functional consequences of their interactions remain to be fully understood. Among the E1A N-terminal protein interactions, the functional significance of the interaction of the pRb family proteins with conserved region 2 (CR2) of E1A (residues 120 to 140) is better understood. CR2 of E1A and the homologous domains of human papillomavirus (E7) and simian virus 40 (T antigen [Ag]) interact with the pRb family members, leading to the activation of the E2F transcriptional pathway by relieving the repression of the S-phase genes (4). The N-terminal 80-amino-acid region of E1A, which includes a conserved domain (CR1), interacts with p300/CBP lysine acetyl transferases (5). In cells infected with human adenovirus 5 (hAd5), E1A expression resulted in quantitative sequestration of these acetyl transferases by E1A, resulting in massive deacetylation of histone H3 Lys18 (H3K18Ac) (6), and also targeted the E1A-pRB repressive complex to a battery of cellular genes that normally inhibit viral replication (7). However, the precise role of E1A-p300/CBP interaction in cell transformation still remains unclear. Some of our results presented here (Fig. 1) suggested that the primary function of p300/CBP interaction with E1A might be to antagonize the activities of the E1A C-terminal region in cell transformation. The mechanism of such action remains to be investigated.
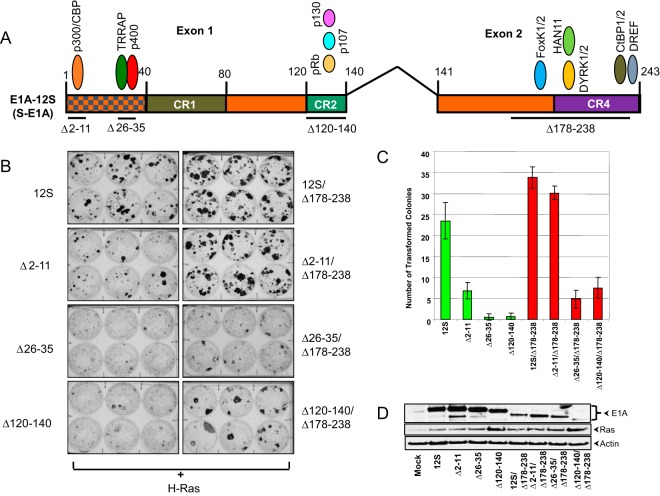
FIG 1.
Domain-specific transforming activities of E1A. (A) Domain map of S-E1A and the interaction of major cellular proteins. The exons of E1A 12S mRNA and the conserved regions (CR) of S-E1A are indicated. The deleted amino acid regions in the E1A mutant constructs are underlined. (B) BRK cells were transfected with plasmids that express E1A mutants with the indicated deletions and activated H-Ras. The transformation assays were carried out as described previously (26). The transfected cells were stained 10 days after transfection and photographed. (C) Quantification of the transformed colonies. (D) Expression of E1A and Ras in BRK cells. BRK cells were transfected with the indicated plasmids, and 48 h after transfection, the cells were analyzed by Western blotting using a mixture of E1A Abs (M58 and Ab 1-80) and Abs specific for Ras and actin.
The N-terminal 80-amino-acid region also interacts with TRRAP (8–10) and p400 (11). An E1A mutant lacking residues 26 to 35, dl1102, was deficient in interaction with both TRRAP and p400. Proteomic analyses have shown that the scaffolding protein TRRAP forms two different multiprotein complexes that contain either of the histone acetylases TIP60 (12, 13) and GCN5 (14, 15). The TIP60 complex also contained p400 (12, 13, 16), raising the possibility that E1A-associated p400 might be a constituent of the TRRAP complex. The presence of TIP60 as a constituent of the E1A-associated TRRAP/p400 complex has not been addressed. The possibility that p400 might interact with E1A independently of TRRAP also exists. The E1A domain spanning residues 26 to 35 was also shown to be defective in interaction with TRRAP and GCN5 (10), suggesting that E1A interacts with the TRRAP/GCN5 complex (also known as the SAGA complex) (reviewed in reference 15).
The existing literature on the role in cell transformation of the E1A domain that is involved in interaction with the TRRAP-associated protein complexes is somewhat mixed. Some have reported that deletion of E1A residues 26 to 35 did not significantly affect E1A-Ras transformation (17, 18). However, subsequent studies showed that the same E1A mutant was deficient in E1A-Ras transformation (8, 9, 11). It has been reported that the interaction of p400 with this domain resulted in enhanced stability of c-MYC, suggesting that E1A may contribute to cell transformation by exploiting the enhanced c-MYC protein levels (19). However, the specific role of p400 in c-MYC stability has not been addressed. Here, we show that the E1A domain spanning residues 26 to 35 is essential for E1A-Ras cooperative transformation and that this domain recruits c-MYC through TRRAP. We further show that the transforming activity of E1A is dependent, at least in part, on c-MYC, while the DNA-binding heterodimeric partner MAX is not required.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
HeLa, 293, HNK (human normal kidney), and BRK (baby rat kidney) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. KB cell suspensions were maintained in Joklik modified minimal essential medium (MEM) containing 5% horse serum (Sigma). The construction of hAd5 12S (Flag- and hemagglutinin [HA]-tagged S-E1A [FH-12S]) is described in Komorek et al. (20). hAd5 dl1102 (21) was a gift from J. Mymryk. The plasmids pME18S-Flag-c-MYC (from R. Eisenman), pEGFP-c-MYC, and pCMV-F-AMY-1 (MYCBP) (from Hiroyoshi Ariga) were gifts from different investigators. Plasmid pcDNA4/TO/myc-His-ENY2 was purchased from Abgent. Plasmids pCβF-TRRAP (from M. Cole) and pLPC-12S (from S. Lowe) were gifts from various investigators.
Cell transformation assays.
BRK (baby rat kidney) cells for cell transformation assays were plated in 6-well plates 1 day prior to transfection. Various E1A mutant plasmids (1 μg), along with pcDNA3 (1 μg), were transfected using jetPEI (Polyplus) according to the manufacturer's specifications. On the day after transfection, the medium was changed, and 50 μg/ml of G418 was added. Every fourth day, the medium was changed, along with the G418. On the 10th day after transfection, cells were stained with crystal violet, and the transformed colonies were counted and photographed. The transformation assays with HNK (human normal kidney) cells were carried out using various retroviral and lentiviral vectors. Low-passage-number HNK cells were purchased from Diagnostic Hybrids, Inc., and grown in DMEM supplemented with 10% fetal bovine serum. For the preparation of lentiviruses, 293T cells were plated in 25-cm2 flasks and transfected with different small hairpin RNA (shRNA) expression vectors along with the packaging plasmids (pCMVΔ8.2ΔVPR and pCMV-VSV-G). For the preparation of retroviruses, Phoenix A cells were transfected with pBABE-E1A-12S-177-9 (Δ178–238 mutant) or H-Ras using the jetPEI transfection reagent. Forty-eight hours after transfection, the supernatants containing the retrovirus or lentivirus were collected and used for infecting HNK cells in 25-cm2 flasks along with Polybrene (8 μg/ml). After 2 h at 37°C, the medium was changed, and on the next day, cells were fed with fresh medium containing 1 μg/ml of puromycin. After 10 days of selection, transformed colonies were stained with crystal violet and counted.
siRNA transfection and shRNA transduction.
On-Target plus smart pool human c-MYC, MYCBP, TRRAP, and p400 small interfering RNAs (siRNAs) were purchased from Dharmacon-Fisher Scientific. Dilution and transfection of siRNAs were performed according to the manufacturer's instructions. Cells were transfected with siRNA at a final concentration of 50 nM using DharmaFECT1 and infected with hAd5 36 h after transfection, and the knockdown of the target gene was determined by Western blotting. The following Mission shRNA vectors were purchased from Sigma Corp.: pLKO.1-puro-c-MYC-shRNA (TRCN0000174055 [c-MYC-1] and TRCN0000039640 [c-MYC-2]), pLKO.1-puro-ENY2-shRNA (TRCN0000274401 [ENY2-1] and TRCN0000274467 [ENY2-2]), and pLKO.1-puro-MAX-shRNA (TRCN0000231550). The lentiviral control vector pLKO.1-puro-GFP-shRNA (ID30323) was purchased from Addgene. The lentiviral vectors were transfected into 293T cells, and the viruses were collected at 48 h after transfection.
Immunoprecipitation and Western blotting.
Cells were collected 17 h after viral infection or 24 h posttransfection (Xtreme HP transfection reagent [Roche] or Lipofectamine 2000 [Life Technologies]), and cell extracts were prepared. The cell lysates were passed through a 26-gauge needle attached to a 1-ml syringe 10 times and treated with DNase for immunoprecipitation of TRRAP and p400. The lysates were precleared using protein A-agarose fast flow (P3476 Sigma) and immunoprecipitated with the antibodies indicated in the figures, as described previously (22). The bound proteins were resolved by NuPAGE Novex (Life Technologies) 3 to 8% Tris-acetate gels with Tris-acetate running buffer (for TRRAP and p400), 10% Bis-Tris gels with MOPS (morpholinepropanesulfonic acid) running buffer (for c-MYC), or 4 to 12% Bis-Tris gel with MES (morpholineethanesulfonic acid) running buffer (for all other proteins) and analyzed by Western blotting. The following antibodies were used for immunoprecipitation and Western blot analysis: Flag antibody (Ab) (F1804; Sigma), anti-HA Ab (clone 12CA5 11666606001; Roche), anti-USP22 Ab (NBP1-49644; Novus), anti-ENY2 Ab (15778-1-AP; Proteintech), and anti-MAX (A302-866A) and anti-ATXN7L3 (A302-800A) Abs (both from Bethyl Laboratories). Anti-E1A Ab (clone M73 05-599) and anti-ATXN7 Ab (04-1573) were purchased from Millipore. Anti-TRRAP Ab (ab183517), anti-p400 Ab (ab5201), and anti-DRYK1A Ab (ab54944) were purchased from Abcam. Anti-CtBP1 Ab (612042), anti-E1A (M58) Ab (554155), and anti-glutathione S-transferase (GST) Ab (554805) were purchased from BD Biosciences, and anti-p300 Ab (SC584), anti-actin Ab (SC1615), anti-MYC Ab (clone 9E10 SC-40), anti-Rb (C15) Ab (SC-50), anti-Rb (IF8) Ab (SC-102), anti-MYCBP Ab (SC-102030), and anti-GCN5 Ab (SC-6303) were purchased from Santa Cruz Biotechnology. All secondary horseradish peroxidase (HRP)-conjugated antibodies were purchased from Santa Cruz Biotechnology, except the secondary antibody used for c-MYC (mouse TrueBlot ultra [anti-mouse Ig HRP 18-8817-33; Rockland Antibodies]) to reduce the IgG heavy chain background.
Tandem affinity purification (TAP) and MS.
The E1A-associated cellular protein complexes were prepared from KB cells infected with hAd5 mutants that express epitope-tagged (Flag and HA) S-E1A (FH-12S) or L-E1A (FH-13S). The protein complexes were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described previously (20).
GST pulldown assay.
GST and GST-E1A proteins were expressed in One Shot BL21star (DE3) chemically competent E. coli (Life Technologies) and purified as described previously (23, 24). The whole-cell protein lysate of HeLa cells was prepared and used for the pulldown assay as described previously (24).
RESULTS
Defining the transforming activity of E1A N-terminal domains.
The N-terminal half of E1A (coded by exon 1) consists of the N-terminal region and two conserved regions, CR1 and CR2 (25). These E1A domains have been shown to mediate cell transformation through the interaction of three major protein complexes, p300/CBP, TRRAP-p400/TRRAP-GCN5, and the pRb family proteins (reviewed in references 1 and 2). Although it is generally accepted that the interactions with p300/CBP and pRb family proteins are required for the transforming activities of E1A, the functional significance of the interaction of TRRAP-containing protein complexes remains ambiguous (11). Here, we carried out a study to better define the transforming functions of the E1A N-terminal domains. We generated E1A (S-E1A) deletion mutants with deletions of amino acid residues 2 to 11, 26 to 35, and 120 to 140 (Δ2–11, Δ26–35, and Δ120–140, respectively) that individually abolish interaction with p300/CBP, TRRAP-p400/TRRAP-GCN5, and the Rb family proteins in the presence or absence of the C-terminal region (Fig. 1A). Primary baby rat kidney cells were transfected with various E1A deletion mutants along with the activated H-Ras oncogene and assayed for the formation of transformed foci (Fig. 1B). As expected, all three N-terminal mutants were generally defective in transformation when expressed in the presence of the C-terminal region. Among the three mutants, mutant Δ2–11 induced slightly higher numbers of foci than the other two (Δ26–35 and Δ120–140). In contrast, when the E1A mutant constructs contained a deletion of the C-terminal region (residues 178 to 238 [Δ178–238]), mutant Δ2–11 did not cause a significant reduction in focus formation. Even though mutant Δ120–140 was strongly defective, it still induced a detectable number of foci. However, mutant Δ26–35 was strongly defective in the absence or presence of the C-terminal region. These results suggested that the E1A region encompassing amino acid residues 26 to 35 might be the most critical transforming domain of E1A. These results further suggested that the N-terminal region encompassing residues 2 to 11 (which interacts with p300/CBP) might play a role in relieving the transformation suppression activity of the C-terminal region (26).
Proteomic analysis of E1A.
A number of previous immunoprecipitation studies using the well-defined hAd5 E1A mutant dl1102 (18) have identified the interaction of TRRAP (8, 10) and p400 (11) with the E1A domain encompassing residues 26 to 35. During our unbiased proteomic analyses of cellular proteins associated with L-E1A and S-E1A (20, 22), we detected associations of several proteins (in addition to TRRAP, p400, and GCN5) that we reasoned might be constituents of the TRRAP/p400 and TRRAP-GCN5 complexes. These proteins included Tip49, Tip50, BAF53, c-MYC, MYCBP, and ENY2. Among these proteins, we chose to further investigate the interaction of c-MYC and ENY2 (homolog of enhancer of yellow 2).
c-MYC interacts with the E1A transforming domain.
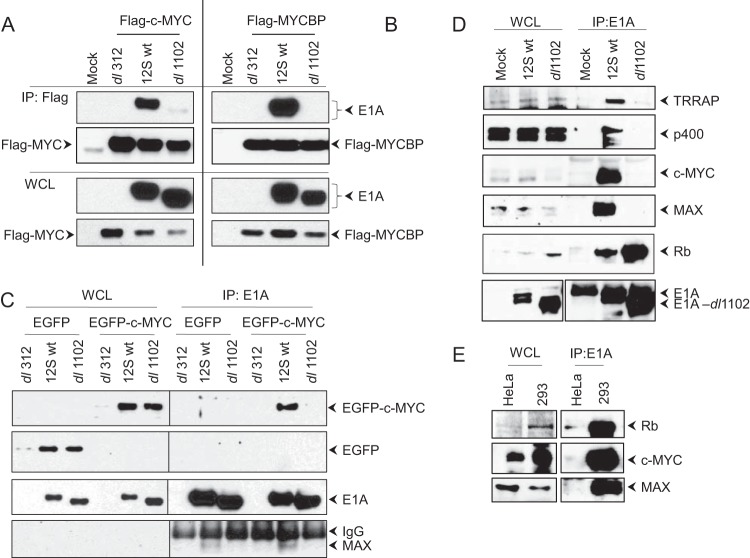
We observed the presence of a single trypsin-generated c-MYC peptide (residues 428 to 436) in three different LC-MS analyses of E1A proteomes. These analyses also revealed the presence of two different peptides of MYCBP, a protein that has previously been reported to bind with the N terminus of c-MYC (27). We carried out coimmunoprecipitation (co-IP) and Western blot analyses to substantiate the interaction of c-MYC and MYCBP with E1A. HeLa cells were transiently transfected with Flag-tagged c-MYC (Fig. 2A), Flag-tagged MYCBP (Fig. 2B), or enhanced green fluorescent protein (EGFP)-tagged c-MYC (Fig. 2C). The transfected cells were infected with hAd5 12S wild type (wt) (S-E1A) or dl1102 (Δ26–35), and the proteins were immunoprecipitated with the Flag Ab (Fig. 2A and B) or E1A Ab (M73) (Fig. 2C) and probed with E1A or EGFP Abs in Western blots. These analyses indicated strong interaction of exogenously expressed Flag-MYC, Flag-MYCBP, and EGFP-MYC with wt S-E1A and not with dl1102. Interestingly, the immunoprecipitates from cells that expressed wt S-E1A also contained endogenous MAX (Fig. 2C, lowermost gel). Next, we determined the interaction of endogenous c-MYC and MAX with E1A in HeLa cells infected with hAd5 E1A 12S wt or dl1102. The Western blots (Fig. 2D) were probed with Abs specific to different E1A-interacting proteins. This analysis also revealed potent interaction of c-MYC and MAX with wt S-E1A and not with dl1102. As expected, there was no detectable interaction of TRRAP or p400 with dl1102, while pRb interacted with wt E1A as well as dl1102. Similarly, there was strong interaction of c-MYC and MAX with E1A in hAd5-transformed 293 cells compared to their interactions in control HeLa cells (Fig. 2E). Together, these results indicated potent interaction of c-MYC and the MYC-binding proteins MAX and MYCBP with the E1A region encompassing residues 26 to 35 that also interacts with TRRAP and p400.
FIG 2.
Interaction of c-MYC with E1A. (A to C) Interaction of exogenously expressed c-MYC and MYCBP. HeLa cells were transfected with the expression vectors for Flag-MYC, EGFP, EGFP-MYC, and Flag-MYCBP. Twenty-four hours after transfection, the cells were infected with hAd5 dl312 (E1A null), 12S wt, or dl1102 (100 PFU/cell). The whole-cell lysates (WCL) were immunoprecipitated (IP) with the Flag Ab (A and B) or the E1A Ab (M73) (C). The blots were probed with the E1A Ab (M73) (A to C) or the EGFP Ab (C). (D and E) Interaction of endogenous c-MYC and MAX. (D) HeLa cells were infected with hAd5 12S wt or mutant dl1102, and the WCLs were immunoprecipitated with the E1A Ab (M73). The Western blots were probed with the Abs for pRb (IF8), c-MYC, MAX, p400, and TRRAP. (E) Interaction of c-MYC with E1A in transformed cells. The WCLs of HeLa or 293 cells were immunoprecipitated with the E1A Ab (M73), and the Western blots were probed for pRb (C15), c-MYC, and MAX.
c-MYC interacts through TRRAP.
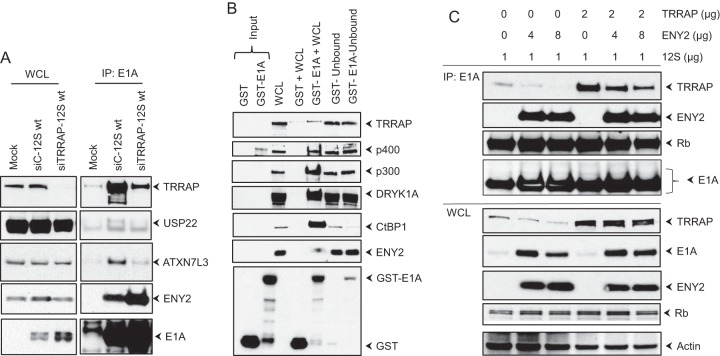
Since the transforming domain of E1A spanning residues 26 to 35 formed complexes with c-MYC, MYCBP, TRRAP, and p400, we wanted to determine whether c-MYC interacted with E1A through any of the other proteins. HeLa cells were depleted of TRRAP, p400, or MYCBP by transient siRNA transfection and infected with hAd5 FH-12S (Flag- and HA-tagged 12S) wt. The proteins were immunoprecipitated with the E1A Ab (M73) and analyzed by Western blotting (Fig. 3). The presence of c-MYC in the E1A protein complex was greatly diminished in TRRAP-depleted cells. In contrast, there was no significant reduction in the levels of c-MYC present in the E1A complex in cells that were depleted of p400 or MYCBP. These results suggested that c-MYC interacted with E1A through TRRAP. Additionally, there was a pronounced reduction in the interaction of p400 with E1A in cells that were depleted of TRRAP, suggesting that p400 may also interact with E1A via TRRAP.
FIG 3.
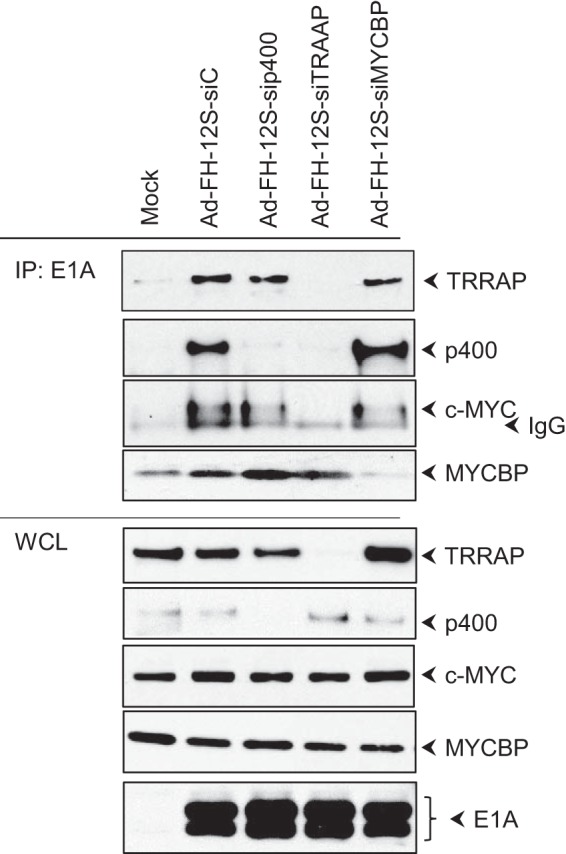
Mode of c-MYC interaction with E1A. (A) HeLa cells were transfected with siRNAs directed against TRRAP, p400, or MYCBP or with control siRNA. Thirty-six hours after transfection, cells were infected with hAd5 12S wt, and WCLs were immunoprecipitated with the E1A Ab (M73). The Western blots were probed with Abs against TRRAP, p400, c-MYC, and MYCBP.
ENY2 and USP22 interact with the E1A transforming domain.
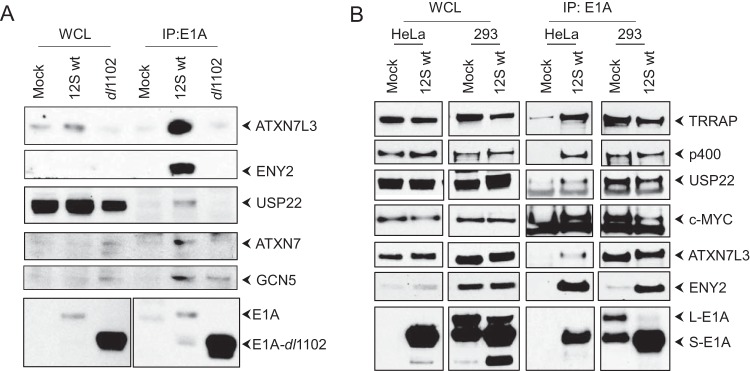
Our proteomics analysis indicated the interaction of ENY2 (three different peptides) with E1A. ENY2 has been reported to be a component of a deubiquitinase (DUB) complex consisting of ATXN7, ATXN7L3, and the ubiquitin-specific peptidase USP22 (28). The USP22 DUB complex has been reported to be a distinct subcomplex of the larger TRRAP-GCN5 (or SAGA) protein complex (28, 29). The TRRAP-GCN5 complex has been previously reported to interact with the E1A sequences spanning residues 26 to 35 (10). Therefore, we carried out co-IP studies to determine whether the transforming domain also interacted with the USP22 complex. KB cells were infected with hAd5 12S wt or dl1102, proteins were immunoprecipitated with the E1A Ab, and the Western blot was probed for the components of the ENY2 complex (Fig. 4A). These results showed the interaction of GCN5, ATXN7, ATXN7L3, ENY2, and USP22 with wt S-E1A. The E1A mutant dl1102 was deficient in interaction with these proteins, suggesting that the transforming domain spanning residues 26 to 35 also interacted with the ENY2-USP22 DUB complex. The interaction of GCN5 with dl1102 was only partially reduced, suggesting that GCN5 may interact with additional E1A regions. We note that a previous study reported that the E1A N-terminal 29-amino-acid region interacted with the yeast GCN5 (30).
FIG 4.
Interaction of ENY2-USP22 complex with E1A. (A) KB cells were infected with hAd5 12S wt or mutant dl1102, and the WCLs were immunoprecipitated with the E1A Ab (M73). The Western blots were probed with Abs for ENY2, ATXN7L3, ATXN7, USP22, GCN5, or E1A (M73). (B) 293 or HeLa cells were either mock infected or infected with hAd5 12S wt, and the E1A-interacting proteins were analyzed as described for panel A.
We then determined whether the components ENY2 and USP22 of the DUB complex interacted with E1A in hAd5-transformed 293 cells. HeLa and 293 cells were either mock infected or infected with hAd5 12S wt, and the proteins were immunoprecipitated with the E1A Ab and analyzed by Western blotting (Fig. 4B). In 293 and HeLa cells infected with hAd5 12S wt, there was proficient interaction of ATXN7L3, USP22, and ENY2. Similar levels of interaction of USP22 and ATXN7L3 with E1A (i.e., endogenous E1A) were observed in uninfected 293 cells. However, the interaction of ENY2 with endogenous E1A was reduced in uninfected 293 cells. These results suggested that endogenous E1A (L-E1A and S-E1A) expressed in 293 cells interacted with reduced levels of ENY2 compared to the relative levels of USP22 and ATXN7L3. However, the expression of S-E1A (through viral infection) in 293 cells substantially enhanced the interaction with ENY2. It is unclear whether there is any difference in the relative affinities of ENY2 for L-E1A versus S-E1A or whether transiently overexpressed E1A via viral infection may have enhanced affinity for ENY2.
ENY2 interacts independently of TRRAP and USP22 interacts through TRRAP.
To determine the mode of interaction of ENY2 and USP22 with E1A, we carried out coimmunoprecipitation analysis in HeLa cells that were depleted of TRRAP by transient transfection of siRNA and infected with hAd5 12S wt (Fig. 5A). While the interactions of USP22 and ATXN7L3 with S-E1A were reduced in TRRAP-depleted cells, the interaction of ENY2 was enhanced. These results suggested that USP22 and ATXN7L3 interacted with E1A through TRRAP, while ENY2 interacted independently of TRRAP. However, it should be noted that our results did not rule out additional interaction of ENY2 (as a constituent of the USP22 DUB subcomplex) with TRRAP. Since the interaction of ENY2 was enhanced in TRRAP-depleted cells, it appears that ENY2 and TRRAP may interact with E1A competitively.
FIG 5.
Mode of interaction of ENY2 with E1A. (A) Effect of TRRAP depletion. HeLa cells were transfected with siRNAs directed against TRRAP or control siRNA, the WCLs were immunoprecipitated with the E1A Ab (M73), and the Western blots were probed with Abs against TRRAP, USP22, ATXN7L3, ENY2, and E1A (M73). (B) GST pulldown assay for E1A-interacting proteins. The WCLs from HeLa cells were incubated with GST or GST-E1A recombinant proteins, and the interacting proteins were purified by using the glutathione-agarose affinity beads. The proteins bound to the beads were analyzed in the flowthrough fractions by Western blotting using Abs against various E1A-interacting proteins. (C) Competitive interaction of ENY2 and TRRAP. HeLa cells were transfected with E1A 12S wt (pLPC-12S) with or without TRRAP and two different concentrations of ENY2. The interactions of TRRAP, ENY2, and pRb with E1A were analyzed by immunoprecipitation with the E1A Ab (M73) and Western blotting.
We then determined whether ENY2 interacted with E1A in vitro by pulldown with the GST-E1A (S-E1A) fusion protein. The whole-cell extract from uninfected HeLa cells was incubated with GST or GST-E1A, and the interacting proteins were purified by affinity chromatography on glutathione-agarose beads. Western blot analysis of the proteins bound to GST or GST-E1A revealed significantly more interaction of ENY2 with GST-E1A than with GST (Fig. 5B). These results provided additional support for the idea that ENY2 interacts with E1A avidly, similar to other in vitro interactions of p400, p300, DYRK1A, and CtBP1 with GST-E1A. Our analysis was not sensitive enough to determine whether TRRAP interacted with GST-E1A.
To determine whether ENY2 interaction with E1A may alter the level of TRRAP interaction, we transfected two different concentrations of an ENY2 expression vector, with or without the TRRAP expression vector, along with a plasmid that expressed E1A 12S wt. The proteins were immunoprecipitated with the E1A Ab (M73) and analyzed by Western blotting (Fig. 5C). The transfected ENY2 decreased the levels of interaction of endogenously and exogenously expressed TRRAP. Interestingly, cells transfected with ENY2 contained significantly increased levels of E1A, suggesting that ENY2 may enhance the expression of E1A.
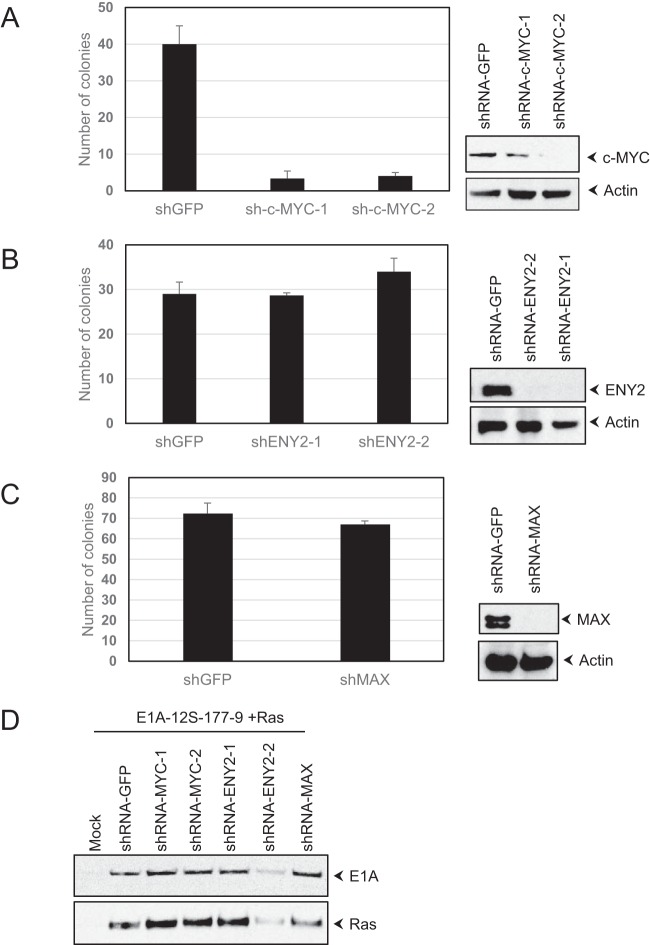
c-MYC is required for E1A-mediated cell transformation.
Having established the interaction of c-MYC and ENY2 with E1A, we decided to determine the roles of these proteins, as well as MAX, in E1A-mediated cell transformation. Human normal kidney (HNK) cells were infected with retro and lentiviruses that express an E1A hyper-transforming mutant (Δ178–238 mutant) (26), the activated H-Ras oncogene, and specific shRNAs targeted against c-MYC, ENY2, MAX, or GFP and assayed for focus formation by selection with puromycin (Fig. 6). The expression of shRNA directed against c-MYC drastically reduced the frequency of transformation compared to the rate in cells that expressed the GFP shRNA (Fig. 6A). In contrast, the expression of shRNA directed against ENY2 (Fig. 6B) or MAX (Fig. 6C) did not cause a reduction in the frequency of transformation. These results suggested that c-MYC may play a direct role in E1A-mediated transformation, independently of the interaction with MAX.
FIG 6.
Effect of c-MYC depletion on E1A-Ras cooperative transformation. (A to C) HNK cells were infected with retroviral vectors that express the Δ178–238 E1A mutant and the activated H-Ras oncogene, along with lentiviral vectors that express specific shRNAs targeted against c-MYC (A), ENY2 (B), MAX (C), or GFP. The cells were then selected with puromycin. The foci were stained with crystal violet and counted (graphs). The transduction of retroviral vectors that expressed only E1A-12S-177-9 (Δ178–238) or H-Ras induced low numbers of lightly stained (with crystal violet) colonies that were readily distinguished and discarded during quantification. The activities of shRNA vectors in the downregulation of c-MYC (A), ENY2 (B), and MAX (C) were determined in HeLa cells with different lentiviral vectors and selection with puromycin (blots). (D) Western blot analysis of E1A and Ras proteins. Seventy-two hours after transduction with different retroviral vectors and shRNA lentiviral vectors, HNK cells were analyzed by Western blotting using the E1A Ab (M58) and Ras antibodies.
DISCUSSION
Although three different N-terminal domains of E1A determine its transforming activity, at present, there is a consensus on the understanding of the activity of the CR2 domain, which mediates cell proliferation by disrupting the activities of the pRb family proteins (reviewed in reference 4). Here, we provide evidence that a second essential transforming domain that spans E1A residues 26 to 35 mediates transformation, at least partially, through the cellular oncogene c-MYC. Previously, the activity of this domain was attributed to the recruitment of p400 (11) and/or TRRAP (8–10). Our results suggest that c-MYC recruited via TRRAP may play a critical role in E1A-mediated cell transformation. This conclusion is based on our results that revealed the presence of c-MYC in E1A-containing protein complexes and showed that shRNA-mediated depletion of c-MYC reduced the E1A transforming activity. Although the recruitment of c-MYC by E1A has not been previously reported, such an interaction would be consistent with the previously published results showing that TRRAP is a c-MYC binding factor (31). Our analysis of E1A proteomes additionally identified previously known c-MYC-associated factors, such as BAF53, Tip49, and Tip50 (32, 33). These proteins were also shown to be constituents of a larger protein complex consisting of TRRAP and p400 (also known as the Tip60 complex) (11–13). The mechanistic basis of the role of c-MYC in E1A-mediated transformation remains to be elucidated. However, depletion of MAX did not reduce the transforming activity of E1A, suggesting that the activity may be independent of the MYC-MAX heterodimeric complex (reviewed in reference 34). Cell proliferation and oncogenic activities of c-MYC that are independent of the MYC-MAX axis have been described previously (reviewed in references 35 to 37). For example, in Drosophila melanogaster, a mutant of MYC that is deficient in interaction with MAX retains substantial MYC functions (38). The c-MYC transactivation domain was reported to induce global phosphorylation of the RNA polymerase II C-terminal domain (CTD) and associate with the transcription initiation complex regulating the expression of some of its target genes (39). c-MYC has also been reported to exert translational and DNA replication activities independently of MAX (37). Since our analyses revealed significant levels of MAX in E1A immunoprecipitates, a role of MYC-MAX in certain other E1A activities is also possible. A previous report showed transcriptional activation of a set of c-MYC target genes in E1A-expressing cells (19). What are the roles of TRRAP and p400 in c-MYC-dependent E1A transforming activity? Our results are consistent with a model that TRRAP may serve as a scaffold for the recruitment of various components of the TRRAP complex that includes p400, as well as c-MYC, by E1A to impart a collective effect on cell transformation, with c-MYC playing a critical role. It has been reported that E1A expression increased the stability of c-MYC (19, 40). This phenomenon was attributed to the interaction of E1A with p400 (19). Here, we show that siRNA-mediated depletion of p400 did not significantly affect c-MYC's interaction with E1A. The possibility that the association of c-MYC with the E1A protein complex, along with certain protein-stabilizing cofactors (see below), may contribute to the stability of c-MYC remains to be investigated. E1A has also been reported to induce c-MYC activation and S-phase entry through interacting with p300/CBP via its N-terminal region (41). Thus, it appears that E1A may employ redundant strategies to garner c-MYC to promote cell proliferation and transformation.
In our proteomic analysis, we identified the interaction of ENY2 with E1A. A high-throughput yeast two-hybrid protein interaction analysis using various viral protein baits also identified the interaction between E1A and ENY2 (42). ENY2 (and its fly and yeast homologs), along with the homologs of USP22, ATXN7, and ATXN7L3, have been shown to be constituents of a DUB subcomplex of the SAGA complex that deubiquitinates histone H2B (28, 29, 43, 44). The DUB activity of the USP22 complex is controlled by allosteric interactions of all four proteins (45). Crystal structures of the yeast DUB complex have revealed that the ENY2 homolog (Sus1) was stabilized by interaction between the homologs of USP22 and ATXN7L3 (46, 47). Our Western blot analysis revealed the association of various components of the USP22 DUB complex with E1A (Fig. 4), and depletion of TRRAP reduced the levels of interaction of USP22, ATXN7, and ATXN7L3 with E1A (Fig. 5A). However, our data cannot suggest that the E1A-associated USP22 subcomplex also contained ENY2, since TRRAP depletion enhanced the interaction of ENY2 with E1A (Fig. 5). Our GST-E1A pulldown experiment showed strong in vitro interaction between ENY2 and E1A (Fig. 5B). These results suggest that ENY2 might competitively bind with E1A at the sequences that are also required for TRRAP interaction. Such interaction of ENY2 with E1A may regulate the stability of the SAGA-associated USP22 DUB complex. ENY2 has been reported to be a constituent of at least two other protein complexes that mediate mRNA export and RNA polymerase II loading to nascent mRNA (reviewed in reference 48). The possibility that E1A-associated ENY2 may mediate a role in mRNA transport and/or transcriptional activation remains to be investigated.
The USP22 DUB complex that is associated with the SAGA complex has been linked to transcriptional activation by the nuclear receptors (28) and c-MYC (29) via deubiquitination of histone H2B. The human SAGA-associated USP22 DUB complex, in addition to targeting histone H2B, has been reported to target nonhistone proteins, such as the telomerase maintenance factor TRX1 (49) and the protein deacetylase SIRT1 (50, 51), to enhance their stability. The interaction between USP22 and SIRT1 resulted in deubiquitination of SIRT1 (50, 51). SIRT1 deacetylated different components of the SAGA complex (50) and c-MYC (52). Deacetylation of c-MYC by SIRT1 at K63 resulted in enhanced stability of c-MYC by preventing ubiquitination at K63 (52). In light of these reports, it will be of interest to determine whether the E1A-associated USP22 also recruits SIRT1. It has been previously reported that c-MYC was stabilized as a result of E1A expression through inhibition of the ubiquitination of c-MYC (19). These authors attributed this phenomenon to the interaction of p400 with E1A and not to any specific DUB activity (19). It remains to be investigated whether USP22 plays any significant role in the stability of c-MYC associated with E1A. USP22 associated with the SAGA complex may also contribute to the transcriptional activity of E1A-associated c-MYC by regulating histone H2B ubiquitination. It is interesting to note that E1A also targets (via the N-terminal 25-amino-acid region) the hBre1/RNF20 complex to block the host interferon response by blocking histone H2B monoubiquitination (53). Thus, the significance of E1A targeting two different protein complexes that regulate the ubiquitination status of histone H2B remains to be investigated.
Our results suggest a complex pattern of interaction of cellular proteins with the transforming domain of E1A encompassing residues 26 to 35 (summarized in Fig. 7). In addition to TRRAP (8–10), this domain has been shown to interact with p400 (11). Although E1A interaction with TRRAP and p400 has been known for many years, the mode of their interaction remained unknown. Our GST-E1A pulldown assay suggests an in vitro interaction of p400 with E1A. Whether TRRAP may also interact with E1A in a similar fashion still remains unresolved. Potential independent interactions of TRRAP and p400 with E1A were hinted at in some earlier co-IP studies using mutants with different mutations of the E1A N-terminal region (19, 54). The region comprising E1A residues 10 to 39, when fused to a c-MYC deletion mutant (defective in interaction with TRRAP), was shown to interact with TRRAP and not with Tip49, a component of the p400 complex (9). The possibility of independent interactions of p400 and TRRAP is puzzling, considering that both proteins are generally believed to be in a single complex (i.e., the TRRAP-p400 complex, also known as the Tip60 complex [12, 13, 16]). Our results showed that siRNA-mediated depletion of TRRAP significantly reduced the interaction of p400 with E1A (Fig. 3). It is possible that interaction of TRRAP with E1A may facilitate the stable interaction of p400 with E1A. In addition to the p400 complex, the transforming domain is known to interact with GCN5, a critical component of the TRRAP-SAGA complex (Fig. 4) (10). Our results suggest that the components of the USP22 DUB complex, USP22 and ATXN7L3, also interact with E1A via TRRAP (Fig. 5). These results suggest that the USP22 subcomplex might interact with E1A via the TRRAP-SAGA complex. Since TRRAP depletion enhanced the level of ENY2 interaction with the transforming domain, it is possible that ENY2 may displace TRRAP's interaction with E1A. Since ENY2 is an integral part of the USP22 DUB complex (46, 47), independent interaction of ENY2 with E1A may also specifically disrupt the assembly and activity of the USP22 DUB complex. The competitive interaction of ENY2 with E1A might play a role in the regulation of E1A-mediated cell proliferation and cell transformation. Thus, our studies have illuminated the complexity of the short E1A domain in regulating cell proliferation and transformation.
FIG 7.
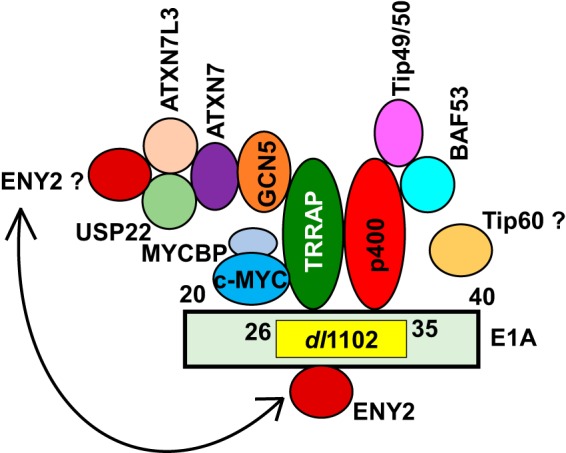
Interaction of cellular proteins with the transforming domain of E1A. The interactions of TRRAP, p400, and GCN5 with the E1A domain spanning residues 26 to 35 were previously known. These interactions were not seen with the mutant dl1102. Our present results revealed the interactions of other proteins shown in the figure via TRRAP and p400. The interaction of Tip60 (indicated by the question mark) was not detected in our LC-MS or by E1A co-IP experiments. Our results suggest that ENY2 may interact with E1A independently of TRRAP. Since ENY2 is an integral part of the USP22 DUB complex, we postulate that ENY2 may additionally interact with E1A via the USP22 DUB complex.
ACKNOWLEDGMENTS
We thank Robert Eisenman and Hiroshi Ariga for the gift of plasmid vectors.
REFERENCES
- 1.Chinnadurai G. 2011. Opposing oncogenic activities of small DNA tumor virus transforming proteins. Trends Microbiol 19:174–183. doi: 10.1016/j.tim.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berk AJ. 2005. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 24:7673–7685. doi: 10.1038/sj.onc.1209040. [DOI] [PubMed] [Google Scholar]
- 3.Yousef AF, Fonseca GJ, Cohen MJ, Mymryk JS. 2012. The C-terminal region of E1A: a molecular tool for cellular cartography. Biochem Cell Biol 90:153–163. doi: 10.1139/o11-080. [DOI] [PubMed] [Google Scholar]
- 4.Nevins JR. 1992. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science 258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 5.Wang HG, Rikitake Y, Carter MC, Yaciuk P, Abraham SE, Zerler B, Moran E. 1993. Identification of specific adenovirus E1A N-terminal residues critical to the binding of cellular proteins and to the control of cell growth. J Virol 67:476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrari R, Pellegrini M, Horwitz GA, Xie W, Berk AJ, Kurdistani SK. 2008. Epigenetic reprogramming by adenovirus E1A. Science 321:1086–1088. doi: 10.1126/science.1155546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrari R, Gou D, Jawdekar G, Johnson SA, Nava M, Su T, Yousef AF, Zemke NR, Pellegrini M, Kurdistani SK, Berk AJ. 2014. Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host Microbe 16:663–676. doi: 10.1016/j.chom.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deleu L, Shellard S, Alevizopoulos K, Amati B, Land H. 2001. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene 20:8270–8275. doi: 10.1038/sj.onc.1205159. [DOI] [PubMed] [Google Scholar]
- 9.Nikiforov MA, Chandriani S, Park J, Kotenko I, Matheos D, Johnsson A, McMahon SB, Cole MD. 2002. TRRAP-dependent and TRRAP-independent transcriptional activation by Myc family oncoproteins. Mol Cell Biol 22:5054–5063. doi: 10.1128/MCB.22.14.5054-5063.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang SE, Hearing P. 2003. The adenovirus E1A oncoprotein recruits the cellular TRRAP/GCN5 histone acetyltransferase complex. Oncogene 22:2836–2841. doi: 10.1038/sj.onc.1206376. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297–307. doi: 10.1016/S0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 12.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463–473. doi: 10.1016/S0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 13.Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. 2003. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem 278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- 14.McMahon SB, Wood MA, Cole MD. 2000. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol Cell Biol 20:556–562. doi: 10.1128/MCB.20.2.556-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sterner DE, Berger SL. 2000. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 64:435–459. doi: 10.1128/MMBR.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyon Y, Cote J. 2004. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev 14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 17.Whyte P, Buchkovich KJ, Horowitz JM, Friend SH, Raybuck M, Weinberg RA, Harlow E. 1988. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature 334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 18.Jelsma TN, Howe JA, Mymryk JS, Evelegh CM, Cunniff NF, Bayley ST. 1989. Sequences in E1A proteins of human adenovirus 5 required for cell transformation, repression of a transcriptional enhancer, and induction of proliferating cell nuclear antigen. Virology 171:120–130. doi: 10.1016/0042-6822(89)90518-7. [DOI] [PubMed] [Google Scholar]
- 19.Tworkowski KA, Chakraborty AA, Samuelson AV, Seger YR, Narita M, Hannon GJ, Lowe SW, Tansey WP. 2008. Adenovirus E1A targets p400 to induce the cellular oncoprotein Myc. Proc Natl Acad Sci U S A 105:6103–6108. doi: 10.1073/pnas.0802095105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komorek J, Kuppuswamy M, Subramanian T, Vijayalingam S, Lomonosova E, Zhao LJ, Mymryk JS, Schmitt K, Chinnadurai G. 2010. Adenovirus type 5 E1A and E6 proteins of low-risk cutaneous beta-human papillomaviruses suppress cell transformation through interaction with FOXK1/K2 transcription factors. J Virol 84:2719–2731. doi: 10.1128/JVI.02119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egan C, Jelsma TN, Howe JA, Bayley ST, Ferguson B, Branton PE. 1988. Mapping of cellular protein-binding sites on the products of early-region 1A of human adenovirus type 5. Mol Cell Biol 8:3955–3959. doi: 10.1128/MCB.8.9.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vijayalingam S, Chinnadurai G. 2013. Adenovirus L-E1A activates transcription through mediator complex-dependent recruitment of the super elongation complex. J Virol 87:3425–3434. doi: 10.1128/JVI.03046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyd JM, Subramanian T, Schaeper U, La Regina M, Bayley S, Chinnadurai G. 1993. A region in the C-terminus of adenovirus 2/5 E1a protein is required for association with a cellular phosphoprotein and important for the negative modulation of T24-ras mediated transformation, tumorigenesis and metastasis. EMBO J 12:469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian T, Vijayalingam S, Lomonosova E, Zhao LJ, Chinnadurai G. 2007. Evidence for involvement of BH3-only proapoptotic members in adenovirus-induced apoptosis. J Virol 81:10486–10495. doi: 10.1128/JVI.00808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Avvakumov N, Kajon AE, Hoeben RC, Mymryk JS. 2004. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology 329:477–492. doi: 10.1016/j.virol.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian T, La Regina M, Chinnadurai G. 1989. Enhanced ras oncogene mediated cell transformation and tumorigenesis by adenovirus 2 mutants lacking the C-terminal region of E1a protein. Oncogene 4:415–420. [PubMed] [Google Scholar]
- 27.Taira T, Maeda J, Onishi T, Kitaura H, Yoshida S, Kato H, Ikeda M, Tamai K, Iguchi-Ariga SM, Ariga H. 1998. AMY-1, a novel C-MYC binding protein that stimulates transcription activity of C-MYC. Genes Cells 3:549–565. doi: 10.1046/j.1365-2443.1998.00206.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, Georgieva SG, Schule R, Takeyama K, Kato S, Tora L, Devys D. 2008. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell 29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. 2008. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell 29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shuen M, Avvakumov N, Walfish PG, Brandl CJ, Mymryk JS. 2002. The adenovirus E1A protein targets the SAGA but not the ADA transcriptional regulatory complex through multiple independent domains. J Biol Chem 277:30844–30851. doi: 10.1074/jbc.M201877200. [DOI] [PubMed] [Google Scholar]
- 31.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363–374. doi: 10.1016/S0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 32.Wood MA, McMahon SB, Cole MD. 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol Cell 5:321–330. doi: 10.1016/S1097-2765(00)80427-X. [DOI] [PubMed] [Google Scholar]
- 33.Park J, Wood MA, Cole MD. 2002. BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Mol Cell Biol 22:1307–1316. doi: 10.1128/MCB.22.5.1307-1316.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eilers M, Eisenman RN. 2008. Myc's broad reach. Genes Dev 22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cascon A, Robledo M. 2012. MAX and MYC: a heritable breakup. Cancer Res 72:3119–3124. doi: 10.1158/0008-5472.CAN-11-3891. [DOI] [PubMed] [Google Scholar]
- 36.Gallant P, Steiger D. 2009. Myc's secret life without Max. Cell Cycle 8:3848–3853. doi: 10.4161/cc.8.23.10088. [DOI] [PubMed] [Google Scholar]
- 37.Cole MD, Cowling VH. 2008. Transcription-independent functions of MYC: regulation of translation and DNA replication. Nat Rev Mol Cell Biol 9:810–815. doi: 10.1038/nrm2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steiger D, Furrer M, Schwinkendorf D, Gallant P. 2008. Max-independent functions of Myc in Drosophila melanogaster. Nat Genet 40:1084–1091. doi: 10.1038/ng.178. [DOI] [PubMed] [Google Scholar]
- 39.Cowling VH, Cole MD. 2007. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol 27:2059–2073. doi: 10.1128/MCB.01828-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lohr K, Hartmann O, Schafer H, Dobbelstein M. 2003. Mutual interference of adenovirus infection and myc expression. J Virol 77:7936–7944. doi: 10.1128/JVI.77.14.7936-7944.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baluchamy S, Sankar N, Navaraj A, Moran E, Thimmapaya B. 2007. Relationship between E1A binding to cellular proteins, c-myc activation and S-phase induction. Oncogene 26:781–787. doi: 10.1038/sj.onc.1209825. [DOI] [PubMed] [Google Scholar]
- 42.Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis AR, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabasi AL, Beroukhim R, Kieff E, Cusick ME, Hill DE, Munger K, Marto JA, Quackenbush J, Roth FP, et al. . 2012. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487:491–495. doi: 10.1038/nature11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, Workman JL. 2008. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J 27:394–405. doi: 10.1038/sj.emboj.7601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. 2009. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics Chromatin 2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, Devys D, Tora L. 2011. The tightly controlled deubiquitination activity of the human SAGA complex differentially modifies distinct gene regulatory elements. Mol Cell Biol 31:3734–3744. doi: 10.1128/MCB.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samara NL, Datta AB, Berndsen CE, Zhang X, Yao T, Cohen RE, Wolberger C. 2010. Structural insights into the assembly and function of the SAGA deubiquitinating module. Science 328:1025–1029. doi: 10.1126/science.1190049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohler A, Zimmerman E, Schneider M, Hurt E, Zheng N. 2010. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell 141:606–617. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kopytova DV, Krasnov AN, Orlova AV, Gurskiy DY, Nabirochkina EN, Georgieva SG, Shidlovskii YV. 2010. ENY2: couple, triple…more? Cell Cycle 9:479–481. doi: 10.4161/cc.9.3.10610. [DOI] [PubMed] [Google Scholar]
- 49.Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, Chang S, Dent SY. 2009. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell 35:352–364. doi: 10.1016/j.molcel.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Armour SM, Bennett EJ, Braun CR, Zhang XY, McMahon SB, Gygi SP, Harper JW, Sinclair DA. 2013. A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex. Mol Cell Biol 33:1487–1502. doi: 10.1128/MCB.00971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin Z, Yang H, Kong Q, Li J, Lee SM, Gao B, Dong H, Wei J, Song J, Zhang DD, Fang D. 2012. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell 46:484–494. doi: 10.1016/j.molcel.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 52.Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Luscher B, Larsson LG, Hermeking H. 2012. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci U S A 109:E187–E196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fonseca GJ, Thillainadesan G, Yousef AF, Ablack JN, Mossman KL, Torchia J, Mymryk JS. 2012. Adenovirus evasion of interferon-mediated innate immunity by direct antagonism of a cellular histone posttranslational modification. Cell Host Microbe 11:597–606. doi: 10.1016/j.chom.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 54.Samuelson AV, Narita M, Chan HM, Jin J, de Stanchina E, McCurrach ME, Fuchs M, Livingston DM, Lowe SW. 2005. p400 is required for E1A to promote apoptosis. J Biol Chem 280:21915–21923. doi: 10.1074/jbc.M414564200. [DOI] [PubMed] [Google Scholar]