ABSTRACT
The HIV-1 Vif protein inactivates the cellular antiviral cytidine deaminase APOBEC3F (A3F) in virus-infected cells by specifically targeting it for proteasomal degradation. Several studies identified Vif sequence motifs involved in A3F interaction, whereas a Vif-binding A3F interface was proposed based on our analysis of highly similar APOBEC3C (A3C). However, the structural mechanism of specific Vif-A3F recognition is still poorly understood. Here we report structural features of interaction interfaces for both HIV-1 Vif and A3F molecules. Alanine-scanning analysis of Vif revealed that six residues located within the conserved Vif F1-, F2-, and F3-box motifs are essential for both A3C and A3F degradation, and an additional four residues are uniquely required for A3F degradation. Modeling of the Vif structure on an HIV-1 Vif crystal structure revealed that three discontinuous flexible loops of Vif F1-, F2-, and F3-box motifs sterically cluster to form a flexible A3F interaction interface, which represents hydrophobic and positively charged surfaces. We found that the basic Vif interface patch (R17, E171, and R173) involved in the interactions with A3C and A3F differs. Furthermore, our crystal structure determination and extensive mutational analysis of the A3F C-terminal domain demonstrated that the A3F interface includes a unique acidic stretch (L291, A292, R293, and E324) crucial for Vif interaction, suggesting additional electrostatic complementarity to the Vif interface compared with the A3C interface. Taken together, these findings provide structural insights into the A3F-Vif interaction mechanism, which will provide an important basis for development of novel anti-HIV-1 drugs using cellular cytidine deaminases.
IMPORTANCE HIV-1 Vif targets cellular antiviral APOBEC3F (A3F) enzyme for degradation. However, the details on the structural mechanism for specific A3F recognition remain unclear. This study reports structural features of interaction interfaces for both HIV-1 Vif and A3F molecules. Three discontinuous sequence motifs of Vif, F1, F2, and F3 boxes, assemble to form an A3F interaction interface. In addition, we determined a crystal structure of the wild-type A3F C-terminal domain responsible for the Vif interaction. These results demonstrated that both electrostatic and hydrophobic interactions are the key force driving Vif-A3F binding and that the Vif-A3F interfaces are larger than the Vif-A3C interfaces. These findings will allow us to determine the configurations of the Vif-A3F complex and to construct a structural model of the complex, which will provide an important basis for inhibitor development.
INTRODUCTION
Human cells have evolved intrinsic defense systems against retroviruses, which include the APOBEC3 (A3) family of polynucleotide cytidine deaminases (reviewed in references 1, 2, 3, and 4]). The A3 family comprises seven members that contain either one (A3A, A3C, and A3H) or two (A3B, A3D, A3F, and A3G) Zn2+ coordination domains (Z domains) with conserved HXE(X)23–28CXXC motifs (5, 6). Based on amino acid sequence homology, each domain is classified into three domain types: Z1 (A3A and the C-terminal domains [CTDs] of A3B and A3G), Z2 (A3C, both domains of A3D and A3F, and the N-terminal domains [NTDs] of A3B and A3G); and Z3 (A3H) (5, 6). Z domain categorization is closely correlated with distinct structural and functional features, as well as evolutionary diversification of the domains in mammals.
HIV-1 inactivates A3 antiviral functions in infected cells through expression of the virion infectivity factor (Vif) protein. The most potent A3 proteins, A3F, A3G, and A3H (haplotype II), play central roles in cellular defense systems against HIV-1 (7–11). In the absence of Vif, the A3 proteins are packaged into progeny virions and block virus replication in newly infected cells (reviewed in reference 3). The molecular mechanisms of replication inhibition are primarily dependent on or independent of deaminase activities (12–20). However, during vif-proficient HIV-1 infection, Vif interacts with A3 proteins and recruits them to a cullin 5 (CUL5)-based E3 ubiquitin ligase complex for proteasomal degradation (21), which accordingly prevents A3 packaging and allows subsequent viral replication in the newly infected cells. The E3 complex consists of CUL5, elongin B (ELOB), elongin C (ELOC), a RING-box subunit 2 (RBX2), and core binding factor-β (CBF-β) (22, 23). CBF-β interaction with HIV-1 Vif is a prerequisite for intracellular Vif stability and specific A3-Vif degradation. The specificity of A3 degradation is determined by HIV-1 Vif interactions with A3 proteins. A3C, -D, -F, -G, and -H (haplotype II) are vulnerable to HIV-1 Vif-mediated degradation, whereas A3A, B, and H (haplotype I) are resistant. A3C, the CTDs of A3D and A3F, the A3G NTD, and A3H contain residues responsible for interaction with Vif (reviewed in references 24 and 25).
The Vif protein is encoded by all lentiviruses, with the exception of equine infectious anemia virus (26). Vif's antagonism of host A3 restriction is presumably conserved among the viruses, although their amino acid sequence similarities are low. Many studies have focused on the Vif of HIV-1 group M, and mutational analyses have identified several functional Vif motifs. The N-terminal half of Vif is mainly involved in the interaction with A3G and A3F through the G box (YRHHY [residues 40 to 44]) and the F1 (DRMR [residues 14 to 17]) and F2 (TGERDW [residues 74 to 79]) boxes, respectively (27–30). The FG box has been reported as a critical sequence motif for binding to both A3F and A3G (30). The C-terminal half of Vif contains three conserved motifs: (i) a Zn2+ coordination motif that mediates the interaction with CUL5 and stabilizes the overall conformation; (ii) the BC box, which binds ELOC; and (iii) the PPLP motif, which is important for Vif dimerization (31). Additionally, Dang et al. have reported that EDRWN (residues 171 to 175) sequences encoded in the C-terminal half are highly conserved in HIV-1 Vif and that the first four residues (EDRW) contribute to formation of a critical motif for binding to A3F (32). Thus, the EDRWN sequences are designated the “F3 box” in this study. As all of these A3F-binding Vif motifs have been identified in the context of A3F interaction, the details of their involvement in A3C binding remain unclear. Moreover, it is still poorly understood how three discontinuous Vif sequence motifs are involved in the A3F binding.
Previously, we determined a high-resolution crystal structure of A3C and identified 10 residues critical for Vif interaction by extensive structure-guided mutagenesis (33). Additionally, we reported that 10 equivalent residues of A3D and A3F are also involved in the Vif interaction (33). These data suggested that the conformation of the Vif interaction interface is highly conserved in homologous Z2-typed A3s, including A3C and the CTDs of A3D and A3F. In contrast to this conservation, it has also been demonstrated that one position in A3F (E324) that is adjacent to the interface is critical for the Vif interaction, although the equivalent A3C residue (E141) is uninvolved (34). These results suggested that a local conformation of the interaction interface might differ between A3C and A3F. However, such different structural features of their interfaces remained unclear because A3F residues critical for the Vif-A3F interaction have not wholly been determined.
In this study, we performed structure-based analyses of the interaction interfaces on Vif and A3F and determined the wild-type (WT) A3F CTD crystal structure to elucidate the entire Vif-binding A3F region. We found that three discontinuous Vif sequence motifs cluster to form the A3C/F-binding interface, which consists of flexible loop structure regions. Moreover, our results demonstrate that hydrophobic and electrostatic interface surfaces are key structural features for the Vif-A3F interaction. Notably, critical residues in the electrostatic regions involved in the interaction differ between Vif-A3C and Vif-A3F. These data allow us to understand the precise structural configurations of the A3F-Vif interaction.
MATERIALS AND METHODS
Plasmids and transfection.
For the pET-41 glutathione S-transferase (GST)-A3F CTD plasmid for expression in bacteria, a DNA fragment of the A3F CTD open reading frame (ORF) region (positions 187 to 373) was amplified by PCR using the primers A3F_187-(+)_KpnI and A3F_Stop_XhoI-(–) (data not shown). The amplified fragment was cloned into pET-41a(+) (Merck Millipore), which has an N-terminal GST tag and an enterokinase (EK) cleavage site, as previously described for the construction of the A3C expression plasmid (33). To achieve efficient cleavage by EK, a tripeptide spacer (MNP) was inserted between the EK cleavage site and the A3F CTD. To make recombinant bacmid DNA for a baculovirus expression system, pFastBac-GST-A3F plasmids containing either WT or catalytic motif mutations were constructed by replacing the A3G ORF portion of pFastBac-GST-A3G (12) with a fragment of the full-length A3F ORF. The mammalian expression plasmids of A3C (pcDNA A3C-MH), A3F (pcDNA A3F-MH), or A3G (pcDNA A3G-MH) containing a C-terminal myc-His (MH) tag were described previously (33). Mutant A3F expression plasmids were constructed by oligonucleotide-directed PCR using each primer. For analysis of A3F-binding motifs in HIV-1 Vif, single alanine substitutions were introduced into pcDNA-HVif (35) by PCR with each primer set (the list of primers is available upon request). For all mutants, the sequences of both the insert and the boundary regions were verified by DNA sequencing. The pNL4-3 WT and pNL4-3ΔVif plasmids have been described previously (36, 37).
Cells, transfection, and virus purification.
Human embryonic kidney cells (293T) and TZM-bl cells were obtained from the AIDS Research and Reference Reagent Program, NIAID, NIH (38–42) and maintained in Dulbecco's modified Eagle medium (DMEM) (Sigma-Aldrich) supplemented with 10% (vol/vol) fetal bovine serum (FBS) plus penicillin (100 U/ml) and streptomycin (100 μg/ml) (Life Technologies). For the Vif-mediated degradation assay, the A3C, A3F, or A3G expression plasmid (1 μg) plus pcDNA-HVif or the pcDNA 3.1(–) vector control (3 μg) was cotransfected into 6-well plates using FuGENE HD (Promega) according to the manufacturer's instructions. At 48 h after transfection, cell lysates were prepared with Laemmli buffer (Bio-Rad) containing 2.5% (vol/vol) 2-mercaptoethanol (2-ME). Transfection for the immunoprecipitation assay was performed as previously described (43).
To evaluate the packaging efficiency of WT A3F or mutants into HIV-1ΔVif, 293T cells (T75 flask) were cotransfected with 13.5 μg of pNL4-3ΔVif and 13.5 μg of pcDNA A3F-MH (WT or mutant) or pcDNA3.1(–) (vector control) using FuGENE HD. At 48 h after transfection, virion-associated proteins were prepared from the transfectant supernatants and separated from cellular debris by centrifugation at 750 × g for 10 min and then filtered through a 0.22-μm-pore-size membrane (Merck Millipore). Virus particles were concentrated by centrifugation through a 20% (wt/vol) sucrose cushion at 111,000 × g for 1.5 h in an SW32Ti rotor (Beckman Coulter).
Protein expression and purification.
Purification of recombinant proteins from the Escherichia coli expression system was performed as previously reported with slight modifications (33). Briefly, Rosetta2(DE3)pLysS bacterial cells (Merck Millipore) transformed with pET-41 GST-A3F CTD were grown in Luria-Bertani medium containing kanamycin (50 μg/ml) and chloramphenicol (34 μg/ml) at 37°C to an optical density at 600 nm (OD600) of ∼0.6. The cells were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and 1 μM ZnSO4 at 20°C for 20 h. The bacterial pellets were harvested and resuspended in lysis buffer (10 mM CaCl2, 5 mM 2-ME, 10% [vol/vol] glycerol, 1% [vol/vol] Triton X-100, 1 M NaCl, 4 mM MgCl2, 40 μg/ml RNase A [Qiagen], 0.75 μg/ml DNase I [TaKaRa Bio], 50 mM Tris HCl [pH 8.0]). The lysed cells were sonicated and then subjected to centrifugation and filtration. The soluble fraction was applied to a glutathione Sepharose 4 FF column (GE Healthcare) for affinity purification. The column was washed with lysis buffer and then wash buffer (10 mM CaCl2, 5 mM 2-ME, 10% [vol/vol] glycerol, 500 mM NaCl, and 50 mM Tris HCl [pH 8.0]). The GST-A3F CTD fraction was obtained using elution buffer (4 mM CaCl2, 1 mM 2-ME, 2% [vol/vol] glycerol, 500 mM NaCl, 40 mM reduced l-glutathione, 30 mM Tris HCl [pH 7.4]). The recombinant EK (Merck Millipore) was added to the eluted fraction to remove the GST tag by incubation at 20°C for 20 h. The EK reaction sample was dialyzed in dialysis buffer (150 mM NaCl, 500 mM l-arginine HCl, 5 mM 2-ME, 50 mM Tris HCl [pH 8.0]) at 4°C. High salt or l-arginine helped the A3F CTD stay soluble without precipitation. The A3F CTD was further purified using Superdex 75 gel filtration chromatography (GE Healthcare) equilibrated with gel filtration buffer (150 mM NaCl, 300 mM l-arginine HCl, 5 mM 2-ME, 50 mM Tris HCl [pH 8.0]).
To obtain full-length A3F recombinant proteins containing an N-terminal GST tag (GST-A3F) for in vitro deaminase assays, we used the baculovirus expression system because of technical difficulties in expressing enzymatically active full-length A3F enzymes using the E. coli expression system, similar to our previous report for A3G (12). GST-A3F proteins were expressed according to the protocol supplied by the manufacturer for the Bac-to-Bac baculovirus expression system (Life Technologies) and purified as previously described for GST-A3G (12). The final protein concentrations were determined by UV absorbance at 280 nm or Coomassie (Bradford) protein assay kit (Thermo Scientific).
Protein crystallization.
The purified A3F CTD protein was dialyzed to crystallization buffer [50 mM NaCl, 300 mM l-arginine HCl, 1 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 10 mM Na HEPES (pH 7.0)] and concentrated to 3 mg/ml using an Amicon Ultra-0.5 (Merck Millipore). Needle-like crystals of A3F CTD were obtained by the hanging-drop vapor-diffusion method. The optimized crystallization solution contained 8.5% (wt/vol) polyethylene glycol 20000 (PEG 20000), 2% (vol/vol) 1,4-dioxane, 300 mM l-arginine HCl, and 85 mM Na Bicine (pH 9.0). The hanging drops were incubated at 20°C.
Data collection and processing.
The crystal was soaked in a cryoprotectant solution (8.5% [wt/vol] PEG 20000, 2% [vol/vol] 1,4-dioxane, 300 mM l-arginine HCl, 15% [vol/vol] glycerol, 85 mM Na Bicine [pH 9.0]) for a few seconds and flash-cooled in nitrogen gas. Diffraction data sets were collected at 93 K using synchrotron radiation (λ = 0.98 Å) on beamline BL-5A at the Photon Factory, KEK, in Tsukuba, Japan. The data sets were indexed, integrated, and scaled using the HKL-2000 suite (44).
Analysis of structure data and construction of structure model.
The A3F CTD structure was determined using the molecular replacement method starting with the A3C crystal structure (Protein Data Bank [PDB] ID no. 3VOW) (33) using the program Phaser (45) implemented in the CCP4 suite (46). The structure was then refined using the program REFMAC5 (47) and manually fitted with COOT (48). Root-mean-square deviation (RMSD) values of reported structures from our crystal structure were calculated using the “cealign” command in PyMOL version 1.6 (49). The electrostatic potential of our structure was visualized with the APBS tools implemented in PyMOL version 1.6.
Immunoprecipitation, immunoblotting, and antibodies.
The coimmunoprecipitation and immunoblotting assays were performed as previously described (33, 43). Anti-His monoclonal antibody (MAb) (1:2,500 dilution), anti-His rabbit serum (D291-3 and PM002; Medical & Biological Laboratories Co.), anti-Vif MAb [319] (1:103 dilution), and β-tubulin rabbit polyclonal antibody (1:103 dilution) (ab66643 and ab6046; Abcam) were purchased. Anti-p24 (CA) antibody (1:105 dilution) was obtained from the AIDS Research and Reference Reagent Program (4250).
In vitro DNA cytidine deamination assay.
The deamination assay conditions were adapted from our previous report (12). A total of 12 μl of each reaction mixture containing 150 nM a 40-nucleotide (nt) single-stranded DNA (ssDNA) substrate (5′-ATTATTATTATTATTATTCTTATTTAGGATTTATTTATTA) labeled at its 5′ end with 6-carboxyfluorescein (6-FAM) (Eurofins) and various concentrations of A3F recombinant proteins in a deamination buffer (15 mM Tris HCl [pH 8.0], 55 mM NaCl, 1 mM TCEP, and 3% [vol/vol] glycerol) was used. The deamination reactions were performed at 37°C with GST-A3F (WT, E67Q, E251Q, or E67Q E251Q) for 8 h and the A3F CTD (WT or E251Q) for 2 h. The deamination reaction was stopped by heat denaturation at 94°C for 5 min, followed by incubation at 37°C for 1 h with 0.2 U/μl uracil-DNA glycosylase (UDG) (New England BioLabs) and UDG buffer. The mixture was treated with 0.15 M NaOH at 37°C for 30 min. GEL loading buffer II (Life Technologies) was added to the mixture, and the mixture was heated at 94°C for 5 min. The final products were subjected to 20% denaturing urea-polyacrylamide gel electrophoresis (PAGE). The gels were scanned by Typhoon (GE Healthcare) and analyzed using ImageQuant (GE Healthcare).
HIV-1 infectivity assay.
The effects of WT A3F or mutants on the infectivity of HIV-1ΔVif were analyzed as previously described (33). Briefly, 293T cells (6-well plates) were cotransfected with 1 μg of pNL4-3 WT or pNL4-3ΔVif plus 0.5 μg of pcDNA A3F-MH or pcDNA 3.1(–) (vector control). At 48 h after transfection, virus-containing supernatants were harvested and filtered. The amount of HIV-1 p24 antigen in the supernatants was determined by enzyme-linked immunosorbent assay (ELISA) (ZeptoMetrix). Virus infectivity was determined by single-cycle replication assays using TZM-bl cells inoculated with viral supernatant containing 10 ng of p24 antigen. Relative infectivity as relative light units (RLU) was measured using the Bright-Glo luciferase assay system kit (Promega) and an ARVO MX luminescence counter (PerkinElmer). The percentage of relative infectivity was calculated relative to the vector control, which was set at 100%.
Structure prediction of Vif containing F3 box.
The crystal structure of HIV-1 Vif in complex with CBF-β, ELOB/C, and the N-terminal domain of CUL5 (PDB ID no. 4N9F) (50) was used as a template. The determined Vif structure, which terminates at D172, lacks structural information on a part of the F3-box (EDRWN [residues 171 to 175]) motif. To predict potential configurations of the F3-box motif in the Vif structure, five residues (RWNKP [residues 173 to 177]) were extended in silico. Using Modeler 9v8 (51), 100 structures of the pentapeptide-containing Vif were obtained and optimized through 300 steps of the variable target function method, followed by gradient-based refinement using molecular dynamics (MD) with simulated annealing using the “refine.slow” option. The structure with the lowest DOPE score was selected and refined by energy minimization using the AMBER9 software package (52) with ff10 and the cationic dummy atom approach proposed by Pang et al. (53). Ten thousand steps of minimization by the steepest descent method and another 10,000 steps by the conjugated gradient method were performed in the gas phase. Finally, the same number of steps for minimization was performed using the generalized born method (IGB = 5). The cutoff distance for the long-range potential energy function was set to 15.0 Å.
Molecular docking and modeling of the A3F CTD-Vif-CBF-β trimeric complex.
To predict the complex structure of the A3F CTD, Vif, and CBF-β, we first performed a docking simulation of A3F CTD and Vif using Rosetta version 3.4 (54) based on our crystal structure of the A3F CTD and the crystal structure of the Vif–CBF-β–ELOB/C–N-terminal CUL5 complex (PDB ID no. 4N9F) (50). Five thousand complex structures were obtained, and the structure with the lowest energy was selected. The docking simulation was performed under distance constraints of Cα atoms between Vif M16 and A3F CTD F290 by a flat harmonic function. Next, optimized extension of the five residues (RWNKP [residues 173 to 177]) was executed concurrently with modeling of the trimeric complex structure of A3F CTD, Vif containing the 5 additional residues, and CBF-β using Modeler 9v8 (51). We then generated 100 structures of the trimeric complex using three templates: our A3F CTD structure, the Vif-CBF-β structure in the E3 ligase complex, and the predicted complex model of A3F CTD and Vif. Finally, we selected and refined the model structure using the same procedure as for the F3-box-containing Vif. Information on the structural models of extended Vif and predicted A3F CTD-Vif complex is available upon request.
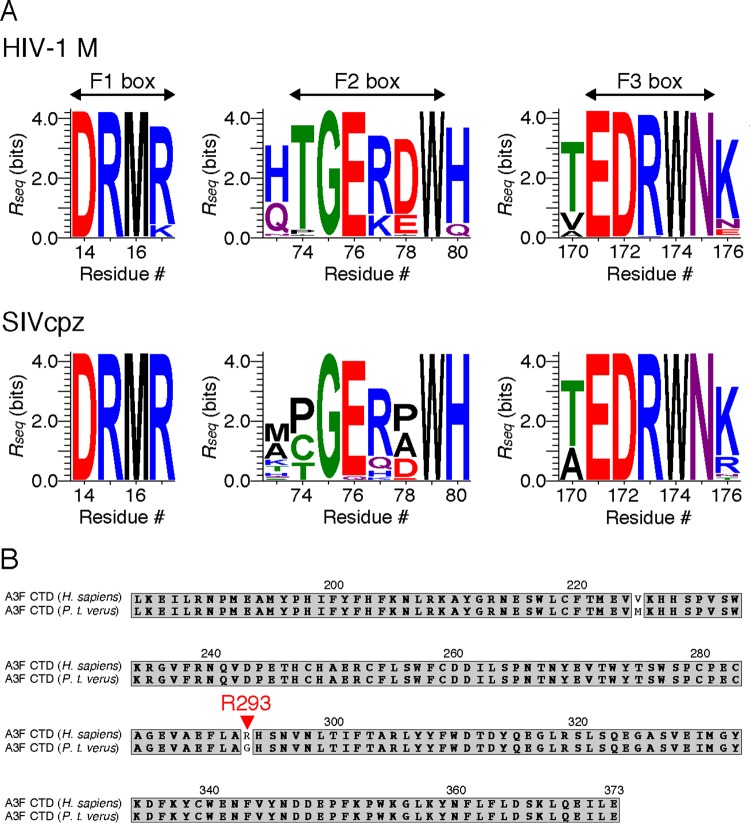
Analysis of amino acid sequence conservation of A3F-binding Vif motifs.
Amino acid sequences of HIV-1 and chimpanzee simian immunodeficiency virus (SIVcpz) Vif were obtained from the Los Alamos HIV sequence database (http://www.hiv.lanl.gov/) in June 2014. For HIV-1 Vif, we selected one sequence from each patient as well as sequences belonging to the major subtypes (A, B, C, D, F, G, H, J, and K), CRF01_AE, or CRF02_AG. A total of 2,935 HIV-1 Vif sequences were aligned with MUSCLE (55) implemented in MEGA5 (Molecular Evolutionary Genetic Analysis; http://www.megasoftware.net/), and conservation at each alignment position was examined using the WebLogo 3.4 program (56). The amino acid sequence conservation at a particular position in the alignment Rseq was defined using the following equation:
| (1) |
where pn is the observed frequency of each type of amino acid at the position. Thus, a larger value of Rseq indicates higher conservation of the sequence. The y axis represents Rseq at each residue number in the F1, F2, and F3 boxes as the total height of logos and pn as the relative height of a logo within the total height. Conservation at each amino acid positions of 27 SIVcpz Vif sequences was also analyzed by the same methods for HIV-1.
Protein structure accession number.
The PDB accession number for the A3F CTD structure and diffraction data is 3WUS.
RESULTS
Three discontinuous HIV-1 Vif sequence motifs are structurally neighbored to form an A3F/A3C interaction interface.
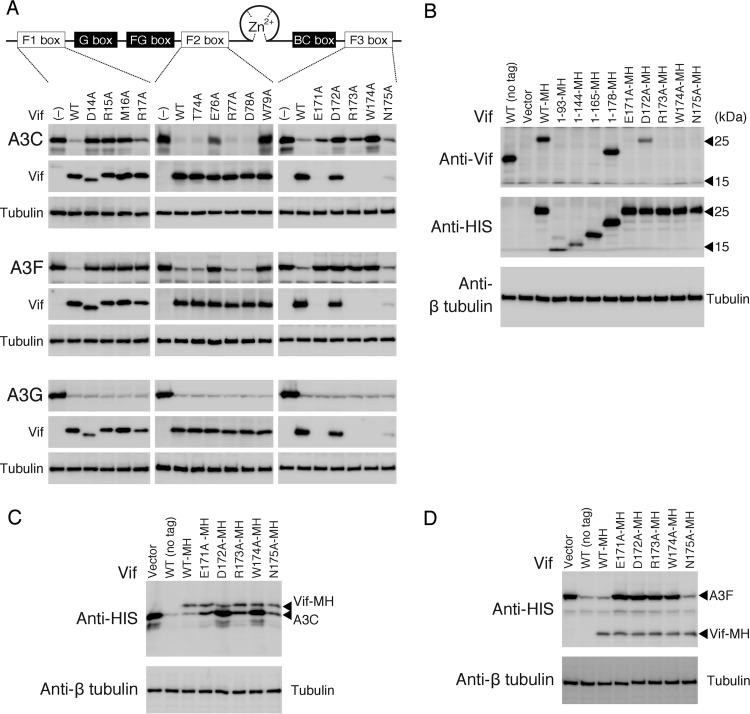
Evidence on highly conserved sequences between A3C and the A3F CTD prompted us to assess a possibility that the A3F-binding motifs of the HIV-1 Vif F1, F2, and F3 boxes might be also required for the A3C interaction. First, we introduced alanine substitutions into the Vif motifs and analyzed the effects of each mutation on the degradation of A3C, A3F, and A3G (Fig. 1). The results showed that Vif mutations at D14, R15, M16, W79, D172, and W174 fully disrupted both A3C and A3F degradation, suggesting that these Vif residues are required to degrade both A3C and A3F. Unexpectedly, Vif R17A, E76A, E171A, and R173A showed slightly different degradation effects between A3C and A3F. Based on three independent experiments, the levels (percentages) of A3C and A3F in the presence of the Vif mutants were evaluated compared with those in the absence of Vif. The A3C levels were 29.4 (R17A), 31.8 (E76A), 23.8 (E171A), and 35.1% (R173A), whereas the A3F levels were 94.2 (R17A), 81.6 (E76A), 88.3 (E171A), and 75.6% (D173A). Alanine substitutions at T74, R77, D78, and N175 did not affect A3F or A3C degradation. In contrast, all Vif mutants in the A3F-binding motifs induced A3G degradation as efficiently as WT Vif, suggesting a functional dissociation between the A3F and A3G interaction motifs. Because the epitope of an anti-Vif monoclonal antibody (MAb), 319 (57), used in this study overlapped the F3 box (Fig. 1B), the Vif F3-box mutants (except for Vif D172A) could not be detected by immunoblotting using this MAb (Fig. 1). However, the F3-box mutants with an MH tag at the C-terminal end (Fig. 1C and D) showed a similar pattern of A3C/A3F degradation to those without a tag (Fig. 1). These data suggest that four Vif residues (R17, E76, E171, and R173) are involved in differential degradation between A3C and A3F.
FIG 1.
Critical residues of HIV-1 Vif in the conserved motifs required for A3 interaction. (A) A diagram of HIV-1 Vif functional domains is shown at the top. Three discontinuous motifs, DRMR (F1 box), TGERDW (F2 box), and EDRWN (F3 box), and the YRHHY motif (G box) are important for binding to A3F and A3G, respectively. The FG box, a Zn2+ coordination motif, and the BC box are critical for A3F/A3G interaction, CUL5 selection, and ELOC binding, respectively. Alanine-substituted Vif mutants were made in the F1, F2, and F3 boxes. A3C, A3F, or A3G with a C-terminal MH tag was expressed in the presence of WT or mutant Vif in 293T cells. A3 and Vif were detected by Western blotting with anti-HIS MAb and anti-Vif MAb, respectively. The loading control was β-tubulin. −, no Vif expression. Note that four Vif F3-box mutants (E171A, R173A, W174A, and N175A) are not detected by the anti-Vif MAb due to lack of the MAb epitope by the alanine substitution. A representative experiment is shown (n = 3). (B) Reactivity of anti-Vif MAb 319 (Abcam) against a series of C-terminal-truncated Vif proteins and the F3-box mutant with a C-terminal MH tag. 293T cells were transfected with each Vif expression plasmid. The intracellular Vif was immunoblotted with the anti-Vif MAb or anti-HIS MAb. Only WT Vif (WT-MH) and the Vif 1–178 truncation mutant (1–178-MH) were detected by the anti-Vif MAb, whereas all the MH-tagged Vif proteins were detected using the anti-HIS MAb. All four of the mutations (E171A, R173A, W174A, and N175A) within the F3-box motif abolished Vif recognition by the anti-Vif MAb. (C) Effects of the F3-box mutations on A3C and (D) A3F degradation using a C-terminal MH-tagged version of Vif.
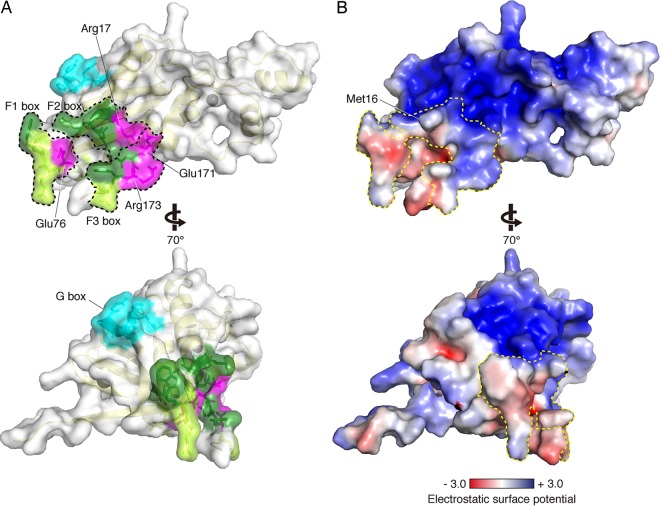
Next, to rationalize how these distal residues of the F-box motifs are involved in the A3C/A3F interaction, we mapped residues critical for degradation of A3C, A3F, and A3G onto the HIV-1 Vif structure. As the crystal structure (PDB ID no. 4N9F) (50) terminates at residue D172 and lacks a part of the F3-box motif, the Vif structure was modeled in silico onto the crystal structure with an extension of the pentapeptide RWNKP (residues 173 to 177) and used for mapping. As shown in Fig. 2A, three distal F-box motifs are assembled at one surface region on the Vif structure, providing a common interaction interface between A3C and A3F. Notably, this F-box interface region consists of the residues located in flexible loop regions, except M16 and R17, and is spatially distal from the G box to bind with A3G. In the F-box surface, a hydrophobic side chain of M16 projects outward at the middle position of the interface surface. In addition, three residues (R17, E171, and R173) that are differentially critical for A3C and A3F degradation form a cluster at the interface brim comprising a positively charged stretch (Fig. 2B), with only E76 separate from the three. Outside this positively charged brim, most of the F-box area does not contain either strongly positive or negative electrostatic polarization. These results suggest that electrostatic and hydrophobic interactions are the fundamental driving force of the A3F-Vif interaction. Of note, critical residues of the FG box are buried inside the Vif molecule and are closely positioned toward the CBF-β contact site, suggesting that the FG box likely plays a role in maintaining the interface conformation necessary for interaction with the CBF-β scaffold (50). Therefore, for example, the side chains of L64 and L66, whose mutations abolish both Vif-A3F and Vif-A3G interactions (30), are projected toward the hydrophobic inner core of Vif.
FIG 2.
Residues involved in each A3 interaction on the Vif structure. (A and B) Based on the crystal structure of Vif (PDB ID no. 4N9F), five residues (RWNKP [residues 173 to 177]) were extended using Modeler. (A) Residues critical only for A3F degradation (R17, E76, E171, and R173) and commonly required for A3C/A3F degradation (D14, R15, M16, W79, D172, and W174) are mapped in magenta and green, respectively. Amino acid residues for which alanine substitution showed no change in A3C/A3F degradation levels are colored in light green. The G box is shown in cyan. The Vif secondary structure is traced in light yellow. Each area of the F1, F2, and F3 boxes is enclosed by the black dotted lines. A gray sphere indicates a zinc atom coordinated in Vif. (B) Electrostatic potential representations of Vif in the same orientation as those in panel A. The surface area is colored according to the calculated potentials from −3.0 kT/e (red) to +3.0 kT/e (blue). The enclosed area with the yellow dotted line indicates the F1, F2, and F3 boxes.
Four additional A3F residues are required for Vif interaction.
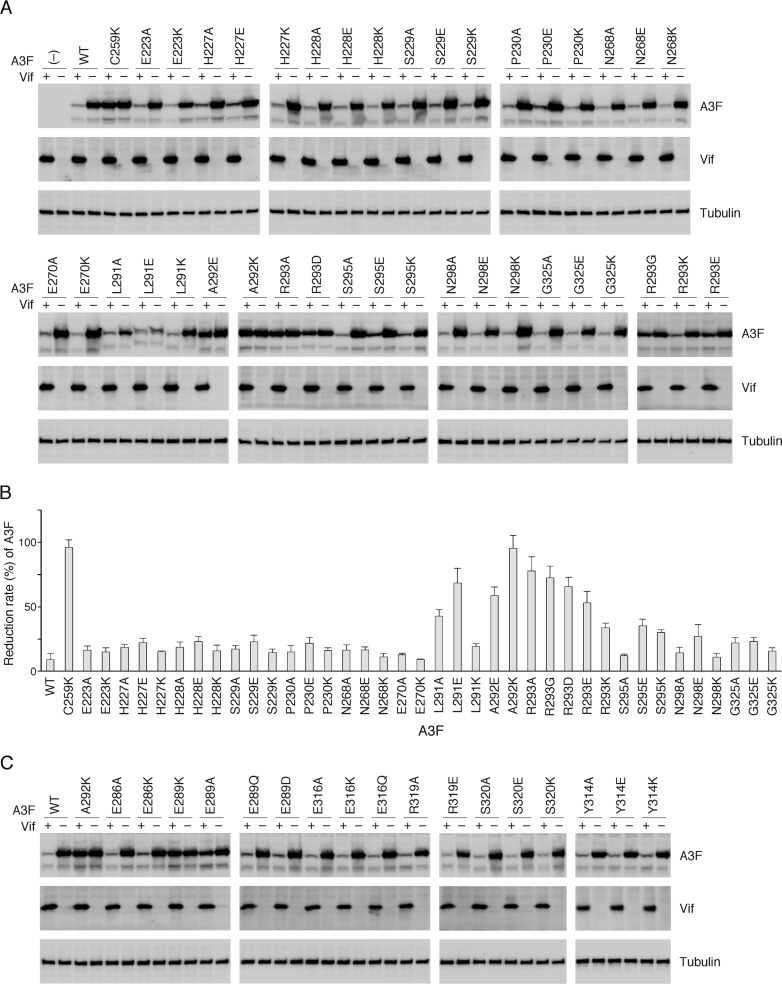
Because of the enlarged positively charged region on Vif that is required for the A3F interaction, but not A3C's, there is the potential for involvement of additional A3F residues that may be critical for binding with the Vif surface. Our previous studies demonstrated that 10 residues between helices α2 and α3 of A3C (residues equivalent to L255, F258, C259, I262, L263, S264, Y269, E289, F290, and H294 in A3F) constitute the Vif interaction interface. This conformation is fundamentally conserved among A3C, -D, and -F (33). In contrast to A3C, A3F has another key residue (E324) that is required for interaction with Vif (33). Bohn et al. reported that an additional 13 residues in the vicinity of the Vif interaction interface are presumably involved in the Vif-A3F interaction, according to predictions based on electrostatic calculations of the A3F CTD surface (58). To assess whether these additional A3F residues are actually involved in the Vif interaction, we tested the effects of a mutation at each residue on changes in susceptibility to Vif-mediated degradation. Single mutations substituted with alanine, negatively charged glutamate, or positively charged lysine were introduced into the full-length A3F expression plasmid. After cotransfection of the A3F and HIV-1 Vif expression plasmids into 293T cells, the steady-state levels of A3F were determined by immunoblotting analysis (Fig. 3A). WT A3F was efficiently degraded by Vif (Vif sensitive), whereas the expression levels of A3F C259K did not change even in the presence of Vif (Vif resistant) as previously described (33). For each A3F mutant, the amount of A3F was quantified, and the percentage of A3F in the presence of Vif relative to the absence of Vif was obtained as a reduction rate (Fig. 3B). The results indicated that two A3F mutants (A292K and R293A) were highly resistant to Vif-mediated degradation (95% and 78%, respectively), and three mutants (L291E, A292E, and R293E) were moderately resistant (68%, 59%, and 53%, respectively). In contrast, single-substitution mutations at eight residues (E223, H227, H228, S229, P230, N268, E270, and G325) did not significantly change the Vif resistance level, whereas S295E, S295K, and N298E resulted in resistance levels of 35%, 30%, and 27%, respectively. These results indicated that L291, A292, and R293, are involved in Vif-mediated A3F degradation, whereas S295 and N298 contribute negligibly to degradation. For the R293 residue, mutations to aspartate or glycine increased the Vif resistance level to 66% (R293D) or 72% (R293G) in comparison with R293E (53%) and R293K (36%) (Fig. 3B), suggesting requirements for side-chain size and a positive charge at this position. Notably, single mutations of the four residues (E286, E316, R319, and S320) that were previously predicted by Siu et al. (59) did not result in changes in the Vif sensitivity (Fig. 3C), suggesting that these four residues do not play a major role in the A3F-Vif interaction. This discrepancy is likely attributable to the use of different assay systems; our assay is a cell-based system using 293T cells that express CBF-β, whereas the in vitro Biolayer interferometry assay for A3F-Vif binding performed by Siu et al. used recombinant A3F and Vif proteins in the absence of CBF-β.
FIG 3.
Identification of critical residues in A3F for Vif-mediated degradation. WT A3F and mutants were assayed for intracellular degradation in the presence (+) or absence (–) of HIV-1 Vif in 293T cells. The steady-state levels of A3F were analyzed by Western blotting using an anti-His MAb. (A) Thirteen residues predicted by Bohn et al. (58) were analyzed. (B) Based on three independent analyses for each mutant, the percentage of A3F in the presence of Vif relative to that in the absence of Vif was calculated as the Vif resistance level. (C) Additional A3F residues identified by Siu et al. (59) and the Y314 residue were also analyzed. Vif expression levels were determined by immunoblotting with an anti-Vif MAb. A3F C259K and A3F E289K were used as a control of Vif-resistant A3F as previously reported by our group. The loading control was β-tubulin.
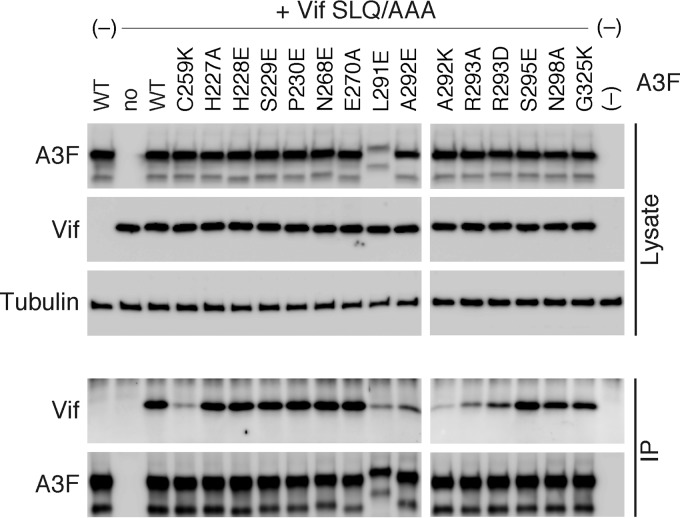
Next, to verify whether these residues were intrinsically involved in the Vif interaction, a coimmunoprecipitation (co-IP) assay of Vif SLQ/AAA was performed using MH-tagged WT A3F or various A3F mutants, as previously reported (33). As shown in Fig. 4, Vif was immunoprecipitated with WT A3F, whereas significantly reduced amounts of Vif were pulled down with the control A3F C259K mutant, which is incapable of binding to Vif (33). All co-IP complexes of the Vif-resistant A3F mutants (L291E, A292E, A292K, R293A, and R293D) contained reduced amounts of Vif (Fig. 4), indicating that residues L291, A292, and R293 do play critical roles in the Vif interaction. In contrast, three equivalent A3C residues (L108, A109, and R110) were shown to be dispensable for efficient interaction (33). Furthermore, in conjunction with previous observations that A3F residue E324 is involved in Vif binding and the analogous position E141 of A3C is not (33, 34), these results suggest that Vif interaction with the A3F interface occurs over a larger area, including 4 additional residues (L291, A292, R293, and E324) compared with the equivalent A3C residues.
FIG 4.
Effects of A3F mutations on the Vif-binding ability of A3F. Shown are the results from coimmunoprecipitation of HIV-1 Vif SLQ/AAA with A3F-MH (WT) and mutants. Total cell lysates (Lysate) and immunoprecipitated complexes (IP) were analyzed by immunoblotting with anti-Vif or anti-His MAbs, and β-tubulin was used as the loading control.
Virion packaging and anti-HIV-1 activity of Vif-resistant A3F.
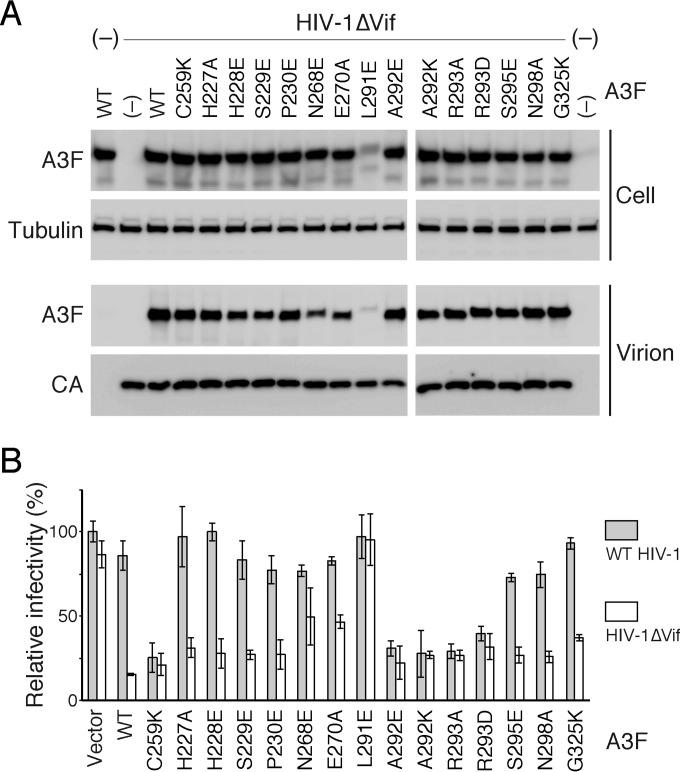
The incorporation of A3 proteins into virions is a prerequisite to confer anti-HIV-1 activity in newly infected cells. To evaluate the incorporation efficiency of Vif-resistant A3F, we analyzed the amounts of WT and mutant A3F in cells and vif-deficient HIV-1 (HIV-1ΔVif) virions by immunoblot assay (Fig. 5A). As previously reported (33), WT A3F and Vif-resistant control A3F C259K were detected in virions at similar levels. Intracellular levels of A3F L291E were relatively low, although the mutant was packaged into virions. The other A3F mutants, including Vif-resistant A292E, A292K, R293A, and R293D, were efficiently packaged into virions in a manner that was dependent upon intracellular levels, although incorporation of A3F N268E and E270A was slightly less efficient. These results suggest that mutations at residues L291, A292, and R293 that are critical for the Vif interaction do not severely alter the incorporation efficiency of A3F proteins into HIV-1ΔVif.
FIG 5.
Packaging efficiency and antiviral activity of WT and mutant A3F. (A) 293T cells were cotransfected with pNL4-3ΔVif (HIV-1ΔVif) and each A3F expression plasmid. The amounts of A3F in cells and virions were determined by Western blotting. Virion-associated p24 (CA) levels were detected using anti-p24 antibody. The loading control was β-tubulin. (B) The antiviral effects of WT and mutant A3F on WT HIV-1 or HIV-1ΔVif were assessed in a single-round replication assay using TZM-bl cells. The relative viral infectivity of WT HIV-1 in the absence of A3F (Vector) was set to 100%.
We next measured the anti-HIV-1 activity of the Vif-resistant A3F in a single-round replication assay using TZM-bl reporter cells (Fig. 5B). Consistent with our previous report (33), Vif-resistant control A3F C259K suppressed both WT and HIV-1ΔVif infectivity, although WT A3F inhibited only vif-deficient virus infectivity. Analogous to the controls, we found that the Vif-resistant A3F mutants (A292E, A292K, R293A, and R293D) showed similar levels of antiviral activity against WT and HIV-1ΔVif, whereas the Vif-sensitive A3F mutants did not reduce the infectivity of WT HIV-1 as efficiently as HIV-1ΔVif infectivity. The antiviral activity of A3F mutant L291E could not be detected in this study, partly because the inefficient intracellular expression and/or the intracellular instability of the mutant protein caused less packaging. These results support our previous findings that disruption of A3F-Vif interactions prevents A3F degradation and exerts antiviral activity against WT HIV-1.
Crystal structure determination of the A3F CTD and comparative analysis of the Vif-interaction interfaces of A3C and A3F.
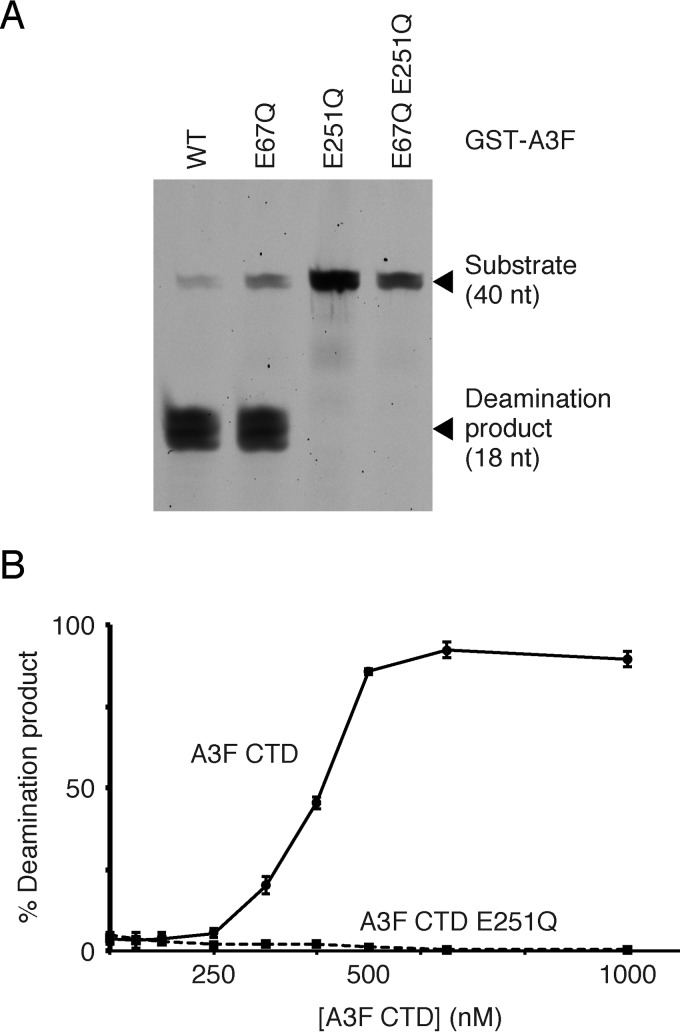
Two crystal structures of the A3F CTD are currently available, although they are both modified from WT A3F sequences: one, called A3F185–373-11X (PDB ID no. 4IOU), has 11 amino acid substitutions (58), whereas the other (PDB ID no. 4J4J) has a 23-amino-acid replacement that included swapping of the whole α1 helix architecture of the A3F CTD with A3G residues 197 to 221 (59). Both substitutions were used to improve protein solubility. Therefore, we attempted to purify an enzymatically active A3F CTD protein without mutations for crystallization and structure determination. Because A3 family proteins tend to form irreversible aggregates during the purification and concentration steps, l-arginine was used to increase protein solubility, as previously described for A3C (33). Our A3F CTD consisted of the entire CTD region (residues 187 to 373) and three additional residues (MNP) at the N-terminal end. A deamination assay using recombinant proteins of full-length A3F expressed in the baculovirus expression system demonstrated that only the CTD of A3F was responsible for its enzymatic activity (Fig. 6A), which was consistent with a previous report of an E. coli mutation assay (60). Additionally, we found that the highly purified A3F CTD from the E. coli expression system possessed catalytic activity in the UDG-based deamination assay (Fig. 6B). These data suggested that the integrity of the A3F CTD architecture was fundamentally maintained.
FIG 6.
Deaminase activity of WT and mutant A3F. (A) The A3F domain(s) responsible for enzymatic activity are shown. Recombinant GST-A3F proteins were expressed in the baculovirus expression system and purified. The deaminase activity of full-length WT and mutant A3F proteins (E67Q, E251Q, and E67Q E251Q) containing an N-terminal GST tag was assayed using the UDG-dependent assay. The positions of the TTCA-containing DNA oligonucleotide substrate (40 nucleotides [nt]) and the deamination product (18 nt) are indicated. (B) Deaminase activity of the A3F CTD. The recombinant proteins were expressed in the E. coli expression system. The relative amounts of deamination product (percentage) versus the A3F CTD concentration (nanomolar) are shown. Error bars represent the standard deviations from three independent measurements. The A3F CTD E251Q was used as a deaminase-deficient control.
The WT A3F CTD was crystallized, and its structure was successfully determined at a resolution of 2.54 Å with two molecules in the asymmetric unit (chains A and B) (Table 1). As expected, the WT A3F CTD has a canonical core structure composed of six α-helices (α1 to α6) over five β-strands (β1 to β5). The WT A3F CTD shared high structural similarity to other polynucleotide cytidine deaminases (25) with Cα RMSDs of 3.2 Å and 2.9 Å over A3A (PDB ID no. 2M65) (61) and the A3G CTD (PDB ID no. 3IR2) (62), respectively. At the catalytic center, one zinc ion is tetrahedrally coordinated directly to H249, C280, and C283 and indirectly to the catalytic glutamate E251 via a water molecule. As shown in Fig. 7A, the superimposition of the A3F CTD core over A3C (PDB ID no. 3VOW) revealed a highly conserved structure with a Cα RMSD of 2.2 Å. However, in the Vif interaction interfaces, the distance between α2 and α3 was larger in the A3F CTD than A3C, whereas the α3-α4 distance in the A3F CTD was narrower than in A3C. Furthermore, we compared the WT A3F CTD with two previously reported structures: A3F185–373-11X (PDB ID no. 4IOU) and the other A3F CTD (PDB ID no. 4J4J). WT A3F CTD (3WUS) also resembles A3F185–373-11X and the other A3F CTD with overall Cα RMSDs of 1.5 and 2.3 Å, respectively. However, comparative analyses of electrostatic potentials suggest that WT A3F CTD obviously displays a bipolar distribution of two different surface charges: a large negatively charged patch around the Vif-binding interface and a positively charged patch along the putative nucleic acid-binding surface. In contrast, the two previous structures exhibit relatively dispersed distribution of the charges, especially around the nucleic acid-binding surface. These observations suggest that the reduction of the positive charges of nucleic acid-binding surface by introducing some mutations (e.g., Y196D, W310D, K355D, and F363D in A3F185–373-11X) may improve the solubility of A3 proteins.
TABLE 1.
Data collection and refinement statistics
| Parameter | Value(s) for APOBEC3F CTDa |
|---|---|
| Data collection | |
| Space group | P3121 |
| Cell dimensions, a, c (Å) | 117.3, 79.0 |
| Wavelength (Å) | 1.000 |
| Resolution (Å) | 50–2.54 (2.58–2.54) |
| Rmerge (%)b | 10.7 (77.6) |
| I/σ〈I〉 | 36.1 (3.39) |
| Completeness (%) | 99.7 (98.7) |
| Redundancy | 10.9 (8.9) |
| Refinement | |
| Resolution (Å) | 50–2.54 |
| No. of reflections | 18,743 |
| Rwork/Rfree (%)c | 20.7/26.0 |
| No. of atoms | 3,139 |
| Protein | 3,130 |
| Zn2+ | 2 |
| Water | 7 |
| Avg B factors (Å2) | 54.8 |
| RMSD | |
| Bond lengths (Å) | 0.0161 |
| Bond angles (°) | 1.863 |
Values in parentheses are for the highest-resolution shell.
Rmerge = ΣhklΣi |Ii(hkl) − 〈I(hkl)〉|/Σhkl Σi Ii(hkl), where Ii(hkl) is the ith observation of reflection hkl and áI(hkl)〉 is the weighted mean of all observations (after rejection of outliers).
Rwork = Σhkl||Fobs| − |Fcalc||/Σhkl|Fobs|. Rfree was calculated using 5% of the data that were randomly chosen and excluded from the refinement.
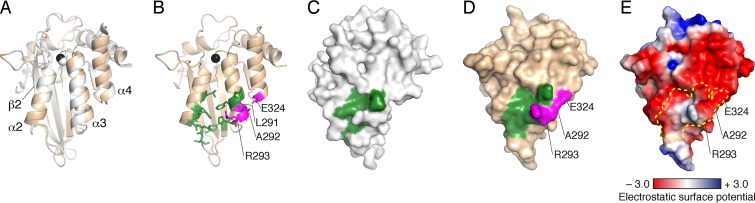
FIG 7.
Crystal structure of the A3F CTD and the Vif interaction interface. (A) Ribbon representation of the superimposed structures of the A3F CTD and A3C (PDB ID no. 3VOW). The backbone traces of the A3F CTD and A3C are colored in wheat and gray, respectively. Catalytic zinc ions of A3F and A3C are shown as black and gray spheres, respectively. (B) Ribbon representation of the A3F CTD alone. Critical residues of the Vif interaction interface that are analogous to A3C are shown in green. Four residues (L291, A292, R293, and E324) shown in magenta are uniquely required for the A3F-Vif interaction. (C) Surface representation of A3C with the Vif interaction interface shown in green. (D) Surface representation of A3F CTD with the Vif interaction interface with the same coloring and labeling as in panel B. (E) Electrostatic potentials of A3F CTD. The surface area is colored according to the calculated potentials from −3.0 kT/e (red) to +3.0 kT/e (blue). The enclosed area with the yellow dotted line indicates the Vif-binding interface.
We mapped 14 critical residues for the Vif interaction onto the A3F CTD structure (Fig. 7B): 10 residues (L255, F258, C259, I262, L263, S264, Y269, E289, F290, and H294) that are equivalent to A3C residues necessary for the Vif interaction are shown in green, and four residues (L291, A292, R293, and E324) whose analogous A3C residues are not essential for the Vif interaction are colored in magenta. As previously described, the former 10 residues cluster in one region between the α2 and α3 helices on the A3F CTD (33). The surface representations of A3C and the A3F CTD (Fig. 7C and D) indicate that these equivalent residues form a similar shallow cavity on the molecules. Additionally, four unique residues (shown in magenta) for the A3F-Vif interaction extend the Vif-binding interface to the bottom region between the α3 and α4 helices. These data suggest that A3F requires an extended surface for interaction with HIV-1 Vif. Furthermore, analysis of the electrostatic potentials on the A3F surface implies that the four unique residues reside in a highly negatively charged portion of the Vif interaction interface of A3F (Fig. 7E).
DISCUSSION
The principal contribution of this study is the first characterization of structural features for HIV-1 Vif-A3F/C interaction and the determination of the WT A3F CTD crystal structure. The F-box residues of HIV-1 Vif are structurally neighbored to form the A3F/C interaction interface despite their distal separation in the primary sequence. Interestingly, a 4-residue-larger Vif interface is required for A3F degradation but not absolutely for A3C. Coincidentally, our mutational analysis of A3F revealed that the Vif-binding interface of A3F includes 4 additional residues in conjunction with our previously identified 10 residues that are highly conserved between A3C and A3F (33). These suggest that the A3F interface consists of an extended area compared with A3C. In addition, the extended portion of the A3F interface is electrostatically complementary to the region of the Vif interface that differentially contributes to the interaction between A3C and A3F. These data provide important insights into structural configurations of the A3F-Vif interaction, because no information is available concerning the structure of the A3F-Vif (or A3F-Vif-CBF-β) complex to date.
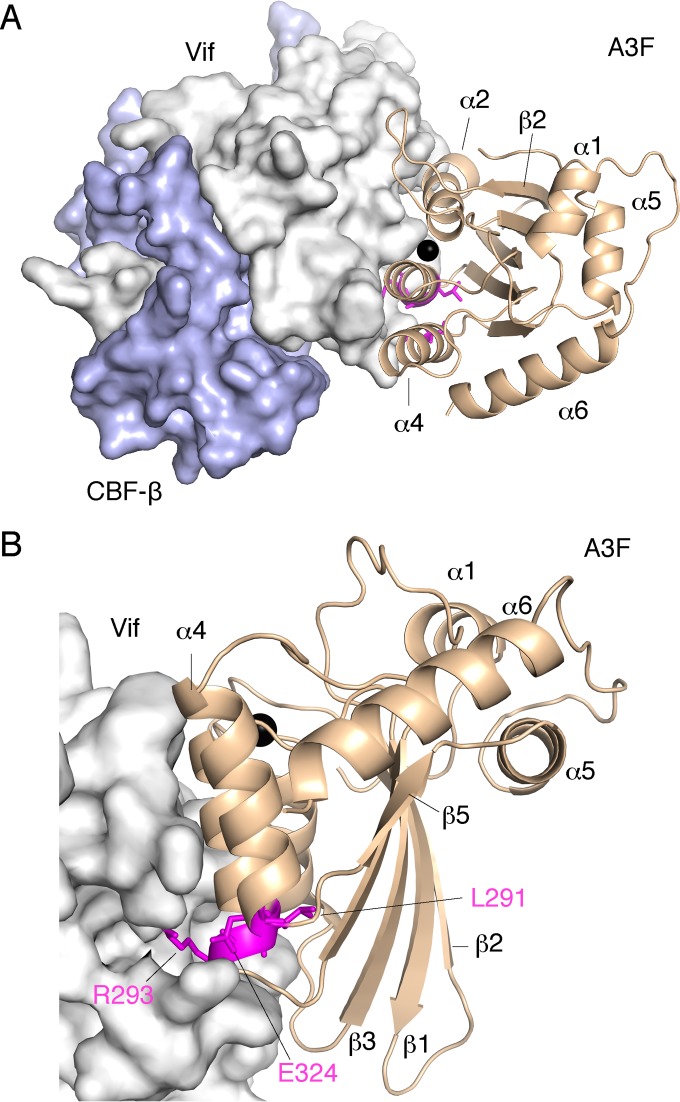
The A3F-binding interface is clearly separate from the YRHHY (residues 40 to 44) position required for A3G binding (Fig. 2). Detailed analysis of the A3F-binding interface conformation using a Vif structural model with a minimum extension of RWNKP (residues 173 to 177) exhibited two unique characteristics: (i) one-half of the region responsible for A3F binding, especially a portion consisting of R17, E171, and R173, represents a highly positively charged surface, and (ii) a hydrophobic side chain of M16 projects out of the surface (Fig. 2). Interestingly, the electrostatic distribution of the Vif interface is inversely correlated with the electrostatic distribution of the negatively charged A3F interface. Additionally, our analysis of Vif mutagenesis demonstrates that the Vif interface is more extended for A3F than for A3C (Fig. 1 and 2), which coincides with the evidence that four additional residues of A3F are critical for the Vif interaction. These data suggest that the Vif surface composed of R17, E171, and R173 might electrostatically interact with the extended interface region, including A3F residues A292, R293, and E324. In a parallel manner, the hydrophobic M16 of Vif might directly interact with the hydrophobic A3F cavity between the α2 and α3 helices because these hydrophobic interactions are critical for the degradation of A3F as well as A3C. Therefore, we sought to predict a model of the A3F CTD and Vif complex using docking simulations, with Vif M16 fixed in the hydrophobic A3F cavity (Fig. 8; see Movie S1 in the supplemental material). The model structure suggests that Vif R17, E171, and R173 and A3F A292, R293, and E324 tend to cluster around the bottom region between the A3F α3 and α4 helices, whereas Vif W174 is likely located at the A3F CTD cavity between the α2 and α3 helices. These observations support our proposal that the electrostatic complementarities between the A3F-unique residues and concurrent anchoring of Vif hydrophobic residues into A3F are required for the A3F-Vif interaction. Moreover, our detailed analyses of the structural model suggest (i) the model is plausible, (ii) two Vif residues (M16 and W174) are clustered to form hydrophobic interaction with the shallow cavity (L255, F258, C259, I262, Y269, F290 and H294) of A3F CTD α2/α3, (iii) three hydrophilic residues (E289, R293 and E324) clustered at the A3F CTD α3/α4 region are positioned closely to four Vif residues (D14, R15, E76, and R173), and (iv) four Vif residues (R17, E171, D172, and N175) of the F-box motifs are proximal to each other and are likely to form intramolecular hydrogen bonds and/or salt bridges. These residues (except for N175) involved in their interaction in our structural model were actually identified by our experiments (Fig. 1, 3, and 4) as critical residues for the A3F-Vif interaction, supporting our structural complex model. In contrast, we identified a new potential interaction that was not investigated: Vif W79 (F2 box) is close to A3F Y314. Therefore, we further assessed whether the Y314 plays a critical role in the Vif-mediated degradation. Three additional A3F mutants, A3F Y314A, Y314E, and Y314K, were constructed, and their Vif sensitivity was evaluated. The results demonstrated that the three A3F mutants (Y314A, Y314E, and Y314K) exhibited Vif sensitivity, suggesting there was no involvement of the A3F Y314 residue in their interaction. The mechanistic reason for this discordance regarding Y314 is not clear yet. Further experiments are required for refinement of our structural model.
FIG 8.
Predicted interaction between the A3F CTD and Vif. (A and B) Two views of their interaction surfaces, from the A3F catalytic groove side (A) and from the A3F CTD α4 helix side (B), are shown. Structures of the A3F CTD and Vif are represented as wheat ribbons and gray surface, respectively. The A3F-unique residues crucial for Vif binding are highlighted with magenta sticks. A zinc ion in the catalytic groove is shown as a black sphere.
The Vif F-box interface consists of the flexible loop structures of the F1, F2, and F3 boxes. This plasticity might be of adaptive advantage for viruses to change substrate recognition interfaces with overall Vif architecture maintained, especially when viruses need to antagonize different species of A3F. Nevertheless, the structural features of the A3F-binding interface on Vif are likely maintained among the major group of HIV-1 (HIV-1 M) and chimpanzee SIV (SIVcpz), as the interface residues critical for the A3F interaction are highly conserved among Vif sequences (Fig. 9). It has been reported that during adaptation of SIVs in chimpanzees, a unique 3′-vif region containing the entire F3 box was created by overprinting associated with the vpx gene loss (63). Such a virus adaptation might be due to high selective pressures that forced HIV-1/SIVcpz Vif to extend the A3F-binding interface to maintain the Vif-mediated antagonizing activity against the A3F protein with high antiviral activity compared with A3C (64, 65). Chimpanzee A3D might also play an important role in the evolution of the HIV-1/SIVcpz Vif capacity for antagonism, as it was reported that A3D gained increased antiviral activity in the chimpanzee-bonobo lineage (66).
FIG 9.
Amino acid sequence conservation of A3F-binding Vif motifs and Vif-binding A3F residues. (A) Conservation at each alignment position in the F1, F2, and F3 boxes. A total of 2,935 HIV-1 sequences and 27 SIVcpz Vif sequences were aligned. The conservation rate at each position (residue) was analyzed using the WebLogo 3.4 program and is shown as Rseq values on the y axis. Amino acids are colored according to chemical properties: polar (green), neutral (purple), basic (blue), acidic (red), and hydrophobic (black). (B) Amino acid sequence alignment of human (Homo sapiens) and chimpanzee (Pan troglodytes verus) A3F CTD. Identical residues are shaded in gray. A red arrowhead indicates residue R293 of human A3F.
Bohn et al. reported the crystal structure of the A3F CTD with 11 amino acid substitutions and predicted a large continuous surface to be required for the Vif interaction, including 11 critical residues that were previously described (33) and 13 additional residues (58). The prediction was based on their analysis encompassing the negatively charged surface of the A3F CTD structure (58). Indeed, within these 13 residues, we verified that 3 residues (L291, A292, and R293) are critical for the Vif interaction. Hence, including the E324 position (33, 34), four A3F residues are uniquely involved in Vif binding that do not represent equivalent roles in A3C. The previously reported A3F motif EFLARH (residues 289 to 294) includes L291, A292, and R293 (67); the interaction of these residues with Vif was initially missed because our previous investigation was based on reported A3C interface residues (33). A recent study of the HIV-1 Vif sensitivity of rhesus macaque A3F strongly supports that A3F E324 is a critical determinant for the Vif interaction (68). MD simulations of the A3F CTD structure have previously suggested that conformational differences in the α3-α4 bottoms, where the four critical residues are located, might be responsible for differential Vif sensitivity between the E141K A3C and E324K A3F mutants (33). In fact, the α3 positions are in a slightly different position between the A3C crystal structure (PDB ID no. 3VOW, chain A) and the A3F CTD determined in this study (chain A), although the positioning of the α2 and α4 helices is similar (Fig. 7A). The α3 residues are shifted toward α4 in the A3F CTD compared with A3C. This shifted positioning causes a narrower α3-α4 space in the A3F CTD than A3C: the distance between the Cαs of A3F CTD α2 (C259)-α4 (E324) is 0.7 Å shorter than in A3C α2 (C76)-α4 (E141), whereas that of A3F CTD α3 (A292)-α4 (E324) is 2.8 Å shorter than in A3C α3 (A108)-α4 (E141). These results suggest that A3F positions L291, A292, R293, and E324 may be more sensitive to mutation than A3C, possibly due to induction of local conformational distortion. Interestingly, the A3F R293A or A3F R293G mutants showed a more Vif-resistant phenotype than R293E or R293D, whereas the A292E mutation attenuated Vif sensitivity and Vif-binding ability (Fig. 3A and 4). These results indicate that the presence of a negatively charged region on the A3F surface is not simply associated with specific Vif binding, but rather the shape and distribution of electric charges at the local A3F surface might be critical for the A3F-Vif interaction. Thereby, mutational disruption of such a charge network at the A3F CTD α3-α4 bottom could be vulnerable to Vif-binding ability.
In this work, A3F R293 was identified as one of the most critical residues for the interaction with HIV-1 Vif. Comparative analysis of A3F amino acid sequences among the family Hominidae showed that the chimpanzee carries an A3F gene with an HIV-1 Vif-resistant genotype, G293 instead of R293 (GenBank Accession DQ373064) (Fig. 9). The chimpanzee G293 genotype is also found in the sequence database of the Great Ape Genome Project (69). Adaptation of SIV in chimpanzees cleared cross-species barriers of host restriction factors and became the source of HIV-1 M, which is currently causing a pandemic in humans (70, 71). Indeed, of the four chimpanzee subspecies, Pan troglodytes troglodytes (P. t. troglodytes) and P. t. schweinfurthii have been shown to harbor natural infections of SIVcpz in Central and Eastern Africa, whereas P. t. verus does not (71). Additionally, phylogenetic analyses of HIV-1 M and SIVcpz show high genetic similarity (71). The amino acid sequences of Vif are also highly similar between HIV-1 M and SIVcpz, with 91% similarity between HIV-1 HXB2 and SIVcpz LB7 strains and high levels of conservation of the F-box motifs (Fig. 9A). These results prompt several questions in terms of the roles of A3F restriction. (i) Do HIV-1 M and/or SIVcpz Vifs antagonize the antiviral A3F function in chimpanzees? (ii) Does A3F exert only a negligible effect on the restriction of SIVcpz in chimpanzees? (iii) Are there any wild-living chimpanzees with the A3F R293 genotype, especially among naturally infected species in Central and Eastern Africa? Additional investigations are required to answer these questions.
In summary, our structure-based analyses of the A3C/F-binding interfaces on Vif revealed that three discontinuous F-box motifs are assembled at one region of the molecule with hydrophobic and positively charged surface patches. The positively charged Vif stretch contributes to formation of an extended interface critical for the A3F interaction, although it is not essential for the A3C binding. Moreover, our determination of the WT A3F CTD crystal structure and identification of four additional A3F residues critical for the Vif interaction demonstrated that the Vif-binding A3F interface includes an additional negatively charged region, which electrostatically intercorrelates with the positively charged extension on the Vif interface. These findings establish a framework for understanding the structural mechanism of specific recognition between A3F and Vif. Furthermore, these results will advance our understanding of host-virus interactions and antagonisms in terms of A3-Vif during the cross-species transmission that led to the current human pandemic of HIV-1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Atsuo Suzuki (Nagoya University) for structure data analysis. The TZM-bl cells were provided by J. C. Kappes and X. Wu through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health, NIH Reagent Program.
This work was supported in part by a grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan, by a grant for HIV/AIDS Research from the Ministry of Health, Labor, and Welfare of Japan (to Y.I.) and by a grant-in-aid for the Japan Society for the Promotion of Science (JSPS) Fellows (to M. Nakashima). This work was performed under the approval of the Photon Factory Program Advisory Committee (proposal no. 2012G642 and 2014G559).
The authors declare that no conflict of interest exists.
Funding Statement
This work was funded by the Japan Society for the Promotion of Science (JSPS) under grant 15J12567 (Masaaki Nakashima). This work was funded by JSPS under KAKENHI grant 15H04740 (Yasumasa Iwatani) and by the Japan Agency for Medical Research and Development (AMED) (Yasumasa Iwatani). This work was performed under the approval of the Photon Factory Program Advisory Committee (proposal no. 2012G642, 2014G559).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02369-15.
REFERENCES
- 1.Albin JS, Harris RS. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol Med 12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desimmie BA, Delviks-Frankenberry KA, Burdick RC, Qi D, Izumi T, Pathak VK. 2014. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol 426:1220–1245. doi: 10.1016/j.jmb.2013.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goila-Gaur R, Strebel K. 2008. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 5:51. doi: 10.1186/1742-4690-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malim MH, Emerman M. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 5.LaRue RS, Andresdottir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jonsson SR, Landau NR, Lochelt M, Malik HS, Malim MH, Munk C, O'Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu XF, Yuhki N, Harris RS. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol 83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM. 2012. Functions and regulation of the APOBEC family of proteins. Semin Cell Dev Biol 23:258–268. doi: 10.1016/j.semcdb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang Y, Siew LM, Wang X, Han Y, Lampen R, Zheng YH. 2008. Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J Biol Chem 283:11606–11614. doi: 10.1074/jbc.M707586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ooms M, Brayton B, Letko M, Maio SM, Pilcher CD, Hecht FM, Barbour JD, Simon V. 2013. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe 14:411–421. doi: 10.1016/j.chom.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 10.Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J 23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol 78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iwatani Y, Takeuchi H, Strebel K, Levin JG. 2006. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J Virol 80:5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwatani Y, Chan DS, Wang F, Maynard KS, Sugiura W, Gronenborn AM, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res 35:7096–7108. doi: 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809. doi: 10.1016/S0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 15.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 16.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishop KN, Holmes RK, Malim MH. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J Virol 80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr Biol 15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 20.Chaurasiya KR, McCauley MJ, Wang W, Qualley DF, Wu T, Kitamura S, Geertsema H, Chan DS, Hertz A, Iwatani Y, Levin JG, Musier-Forsyth K, Rouzina I, Williams MC. 2014. Oligomerization transforms human APOBEC3G from an efficient enzyme to a slowly dissociating nucleic acid-binding protein. Nat Chem 6:28–33. doi: 10.1038/nchem.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 22.Jager S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, Mahon C, Kane J, Franks-Skiba K, Cimermancic P, Burlingame A, Sali A, Craik CS, Harris RS, Gross JD, Krogan NJ. 2012. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature 481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Du J, Evans SL, Yu Y, Yu XF. 2012. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 24.Kitamura S, Ode H, Iwatani Y. 2011. Structural features of antiviral APOBEC3 proteins are linked to their functional activities. Front Microbiol 2:258. doi: 10.3389/fmicb.2011.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aydin H, Taylor MW, Lee JE. 2014. Structure-guided analysis of the human APOBEC3-HIV restrictome. Structure 22:668–684. doi: 10.1016/j.str.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Oberste MS, Gonda MA. 1992. Conservation of amino-acid sequence motifs in lentivirus Vif proteins. Virus Genes 6:95–102. doi: 10.1007/BF01703760. [DOI] [PubMed] [Google Scholar]
- 27.Tian C, Yu X, Zhang W, Wang T, Xu R, Yu XF. 2006. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J Virol 80:3112–3115. doi: 10.1128/JVI.80.6.3112-3115.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrofelbauer B, Senger T, Manning G, Landau NR. 2006. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J Virol 80:5984–5991. doi: 10.1128/JVI.00388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell RA, Pathak VK. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J Virol 81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Z, Zhang W, Chen G, Xu R, Yu XF. 2008. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J Mol Biol 381:1000–1011. doi: 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- 31.Donahue JP, Vetter ML, Mukhtar NA, D'Aquila RT. 2008. The HIV-1 Vif PPLP motif is necessary for human APOBEC3G binding and degradation. Virology 377:49–53. doi: 10.1016/j.virol.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dang Y, Davis RW, York IA, Zheng YH. 2010. Identification of 81LGxGxxIxW89 and 171EDRW174 domains from human immunodeficiency virus type 1 Vif that regulate APOBEC3G and APOBEC3F neutralizing activity. J Virol 84:5741–5750. doi: 10.1128/JVI.00079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kitamura S, Ode H, Nakashima M, Imahashi M, Naganawa Y, Kurosawa T, Yokomaku Y, Yamane T, Watanabe N, Suzuki A, Sugiura W, Iwatani Y. 2012. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol 19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 34.Albin JS, LaRue RS, Weaver JA, Brown WL, Shindo K, Harjes E, Matsuo H, Harris RS. 2010. A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. J Biol Chem 285:40785–40792. doi: 10.1074/jbc.M110.173161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen K-L, Llano M, Akari H, Miyagi E, Poeschla EM, Strebel K, Bour S. 2004. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology 319:163–175. doi: 10.1016/j.virol.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 36.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59:284–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kao S, Khan MA, Miyagi E, Plishka R, Buckler-White A, Strebel K. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J Virol 77:11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O'Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol 74:8358–8367. doi: 10.1128/JVI.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi Y, McClure MO, Pizzato M. 2008. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J Virol 82:12585–12588. doi: 10.1128/JVI.01726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. 2009. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol 83:8289–8292. doi: 10.1128/JVI.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwatani Y, Chan DS, Liu L, Yoshii H, Shibata J, Yamamoto N, Levin JG, Gronenborn AM, Sugiura W. 2009. HIV-1 Vif-mediated ubiquitination/degradation of APOBEC3G involves four critical lysine residues in its C-terminal domain. Proc Natl Acad Sci U S A 106:19539–19544. doi: 10.1073/pnas.0906652106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. 2004. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr 60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Schrodinger LLC. 2010. The PyMOL molecular graphics system, version 1.6. Schrodinger, LLC, New York, NY. [Google Scholar]
- 50.Guo Y, Dong L, Qiu X, Wang Y, Zhang B, Liu H, Yu Y, Zang Y, Yang M, Huang Z. 2014. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature 505:229–233. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 51.Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 52.Case DA, Darden TA, Cheatham TEI, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. 2006. AMBER 9. University of California, San Francisco, CA. [Google Scholar]
- 53.Pang YP, Xu K, Yazal JE, Prendergas FG. 2000. Successful molecular dynamics simulation of the zinc-bound farnesyltransferase using the cationic dummy atom approach. Protein Sci 9:1857–1865. [PMC free article] [PubMed] [Google Scholar]
- 54.Schueler-Furman O, Wang C, Baker D. 2005. Progress in protein-protein docking: atomic resolution predictions in the CAPRI experiment using RosettaDock with an improved treatment of side-chain flexibility. Proteins 60:187–194. doi: 10.1002/prot.20556. [DOI] [PubMed] [Google Scholar]
- 55.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simon JH, Southerling TE, Peterson JC, Meyer BE, Malim MH. 1995. Complementation of vif-defective human immunodeficiency virus type 1 by primate, but not nonprimate, lentivirus vif genes. J Virol 69:4166–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bohn MF, Shandilya SM, Albin JS, Kouno T, Anderson BD, McDougle RM, Carpenter MA, Rathore A, Evans L, Davis AN, Zhang J, Lu Y, Somasundaran M, Matsuo H, Harris RS, Schiffer CA. 2013. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure 21:1042–1050. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siu KK, Sultana A, Azimi FC, Lee JE. 2013. Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat Commun 4:2593. doi: 10.1038/ncomms3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holmes RK, Koning FA, Bishop KN, Malim MH. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem 282:2587–2595. [DOI] [PubMed] [Google Scholar]
- 61.Byeon IJ, Ahn J, Mitra M, Byeon CH, Hercik K, Hritz J, Charlton LM, Levin JG, Gronenborn AM. 2013. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat Commun 4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shandilya SM, Nalam MN, Nalivaika EA, Gross PJ, Valesano JC, Shindo K, Li M, Munson M, Royer WE, Harjes E, Kono T, Matsuo H, Harris RS, Somasundaran M, Schiffer CA. 2010. Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure 18:28–38. doi: 10.1016/j.str.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Etienne L, Hahn BH, Sharp PM, Matsen FA, Emerman M. 2013. Gene loss and adaptation to hominids underlie the ancient origin of HIV-1. Cell Host Microbe 14:85–92. doi: 10.1016/j.chom.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu Q, Chen D, Konig R, Mariani R, Unutmaz D, Landau NR. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J Biol Chem 279:53379–53386. doi: 10.1074/jbc.M408802200. [DOI] [PubMed] [Google Scholar]
- 65.Hultquist JF, Lengyel JA, Refsland EW, LaRue RS, Lackey L, Brown WL, Harris RS. 2011. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol 85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Duggal NK, Malik HS, Emerman M. 2011. The breadth of antiviral activity of Apobec3DE in chimpanzees has been driven by positive selection. J Virol 85:11361–11371. doi: 10.1128/JVI.05046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith JL, Pathak VK. 2010. Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif. J Virol 84:12599–12608. doi: 10.1128/JVI.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Land AM, Shaban NM, Evans L, Hultquist JF, Albin JS, Harris RS. 2014. APOBEC3F determinants of HIV-1 Vif sensitivity. J Virol 88:12923–12927. doi: 10.1128/JVI.02362-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O'Connor TD, Santpere G, Cagan A, Theunert C, Casals F, Laayouni H, Munch K, Hobolth A, Halager AE, Malig M, Hernandez-Rodriguez J, Hernando-Herraez I, Prufer K, Pybus M, Johnstone L, Lachmann M, Alkan C, Twigg D, Petit N, Baker C, Hormozdiari F, Fernandez-Callejo M, Dabad M, Wilson ML, Stevison L, Camprubi C, Carvalho T, Ruiz-Herrera A, Vives L, Mele M, Abello T, Kondova I, Bontrop RE, Pusey A, Lankester F, Kiyang JA, Bergl RA, Lonsdorf E, Myers S, Ventura M, Gagneux P, Comas D, et al. 2013. Great ape genetic diversity and population history. Nature 499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, Sharp PM, Hahn BH. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 71.Sharp PM, Hahn BH. 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med 1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.