ABSTRACT
Oral ingestion is the major route of infection for the white spot syndrome virus (WSSV). However, the mechanism by which virus particles in the digestive tract invade host cells is unknown. In the present study, we demonstrate that WSSV virions can bind to chitin through one of the major envelope proteins (VP24). Mutagenesis analysis indicated that amino acids (aa) 186 to 200 in the C terminus of VP24 were required for chitin binding. Moreover, the P-VP24186–200 peptide derived from the VP24 chitin binding region significantly inhibited the VP24-chitin interaction and the WSSV-chitin interaction, implying that VP24 participates in WSSV binding to chitin. Oral inoculation experiments showed that P-VP24186–200 treatment reduced the number of virus particles remaining in the digestive tract during the early stage of infection and greatly hindered WSSV proliferation in shrimp. These data indicate that binding of WSSV to chitin through the viral envelope protein VP24 is essential for WSSV per os infection and provide new ideas for preventing WSSV infection in shrimp farms.
IMPORTANCE In this study, we show that WSSV can bind to chitin through the envelope protein VP24. The chitin-binding domain of VP24 maps to amino acids 186 to 200 in the C terminus. Binding of WSSV to chitin through the viral envelope protein VP24 is essential for WSSV per os infection. These findings not only extend our knowledge of WSSV infection but also provide new insights into strategies to prevent WSSV infection in shrimp farms.
INTRODUCTION
White spot syndrome virus (WSSV) is an enveloped double-stranded DNA (dsDNA) virus belonging to the genus Whispovirus, family Nimaviridae (1). WSSV has caused enormous economic losses to the shrimp farming industry since 1993. Infectious WSSV virions are typically 250 to 380 nm in length and 120 to 150 nm in diameter (2, 3), and they contain a genome of ∼300 kb that encodes ∼180 proteins (4, 5).
Ingestion of WSSV-infected sick or dead shrimp has been accepted as the major route of natural infection due to the cannibalistic nature of shrimp (6–11). Therefore the digestive tract of shrimp may be a primary site of infection. The digestive tract of shrimp is composed of the esophagus, stomach, midgut, and hindgut. The esophagus, stomach, and hindgut possess a chitinous lining, which is not present in the midgut (12, 13). Instead, the midgut epithelium is generally lined with the peritrophic membrane (PM), which is a noncellular structure surrounding the food bolus. The PM is composed of regularly arranged chitin fibrils embedded in a matrix of proteins, proteoglycans, and mucopolysaccharides (14–17). Therefore, WSSV must cross the PM in the midgut or the chitinous lining in the other parts of the digestive tract to traverse the basal membranes and reach the host cells. We speculate that the interaction between WSSV and chitin may be important for WSSV infection in the shrimp digestive tract.
More than 40 WSSV structural proteins have been identified using proteomic methods to date (18–20). Among them, VP28, VP26, VP24, and VP19 are the four major viral envelope proteins (20, 21); these proteins form a multiprotein complex together with some low-abundance proteins (22–26). In addition to the importance of this envelope protein complex in virus assembly, it is thought to be an “infectome” that functions in cell recognition during infection (24, 27). However, whether the viral envelope proteins play a role in the initial stage of WSSV invasion is unknown.
In this study, we analyzed the interaction between WSSV particles and chitin. VP24 was found to be a chitin-binding protein (CBP), and its chitin-binding domain (CBD) was identified. Moreover, the VP24-chitin interaction was found to be essential for WSSV per os infection. These findings will extend our knowledge on the pathogenesis of WSSV and help control the disease.
MATERIALS AND METHODS
Animals and virus.
Whiteleg shrimp (Litopenaeus vannamei) with an average body weight of 10 g were purchased from a local market in Xiamen, China, and maintained in water tanks containing seawater with 25% salinity at 25°C with proper aeration. The animals were acclimatized for 2 days and verified to be WSSV free using a WSSV quantitative PCR (qPCR) detection kit (Xiamen Lulong Biotech Co., Ltd., Xiamen, Fujian, China) prior to the experiments.
Red swamp crayfish (Procambarus clarkia) with an average body weight of 20 g were purchased from a local market in Xiamen. Healthy crayfish were used for WSSV propagation, and WSSV virions were purified from moribund crayfish as previously described (28). The viral concentration was quantified by spectrophotometry (29). The viral envelopes and nucleocapsids were prepared according to previously described procedures (20).
Plasmids and recombinant protein expression.
In previous studies, hydrophobicity analysis of the amino acid (aa) sequences of VP24, VP26 and VP28 showed that strong hydrophobic regions are present in the N termini of these proteins (30, 31). Full-length expression of VP24, VP26, and VP28 resulted in the production of insoluble proteins (32–34). Therefore, in this study, N-terminal hydrophobic region-truncated VP24, VP26, and VP28 mutants were expressed to obtain soluble proteins. The N-terminal hydrophobic region-truncated VP28 (aa 31 to 204) and full-length VP19 were constructed in pET-His. The N-terminal hydrophobic region-truncated VP24 (aa 26 to 208) and VP26 (aa 36 to 204) were constructed in pET-V5, which was generated by replacing the His tag in pET-28a (Novagen) with a V5 tag. A series of VP24 deletion mutants were generated by inverse PCR using pET-V5-VP24 (aa 26 to 208) as the template. The primers are listed in Table 1.
TABLE 1.
Primers used in this study
| Name | Primer sequence |
|---|---|
| V5-VP2426–208 | |
| Forward | ATGCGGATCCACCAACATAGAACTTAA |
| Reverse | ATGCGAATTCTTATTTTTCCCCAACCTTAA |
| V5-VP2426–172 | |
| Forward | ATCGGAATTCGAGCTCCGTCGACA |
| Reverse | ATGCGAATTCTTATGTGTTGATCCTATTTTT |
| V5-VP2426–135 | |
| Forward | ATCGGAATTCGAGCTCCGTCGACA |
| Reverse | ATGCGAATTCTTACACTGTTATATCCCTCTT |
| V5-VP2426–98 | |
| Forward | ATCGGAATTCGAGCTCCGTCGACA |
| Reverse | ATGCGAATTCTTAGAGGATTATGTCTCCTT |
| V5-VP2462–208 | |
| Forward | ATGCGGATCCTTTAACTTTGTAAACGGCACAT |
| Reverse | ATCGGGATCCACGCGTAGAATCGA |
| V5-VP2499–208 | |
| Forward | ATGCGGATCCACATCTTTACTTGGAGAC |
| Reverse | ATCGGGATCCACGCGTAGAATCGA |
| V5-VP24136–208 | |
| Forward | ATGCGGATCCGACTCTGTTTCACTGTCTC |
| Reverse | ATCGGGATCCACGCGTAGAATCGA |
| V5-VP2426–180 | |
| Forward | ATGCGGATCCACCAACATAGAACTTAACAAGA |
| Reverse | ATGCGAATTCTTAGTCAAACGTTGCTCCAAAC |
| V5-VP2426–185 | |
| Forward | ATGCGGATCCACCAACATAGAACTTAACAAGA |
| Reverse | ATGCGAATTCTTAATCATCGATGTCTTCGTCA |
| V5-VP2426–195 | |
| Forward | ATGCGGATCCACCAACATAGAACTTAACAAGA |
| Reverse | ATGCGAATTCTTATCGCATACTTAACAGATAA |
| V5-VP2426–200 | |
| Forward | ATGCGGATCCACCAACATAGAACTTAACAAGA |
| Reverse | ATGCGAATTCTTAATTGCCAGGAGAAAATCGC |
| V5-VP2636–204 | |
| Forward | ATGCGGATCCACACGTGTTGGAAGAAGCGTCG |
| Reverse | ATGCGAATTCTTACTTCTTCTTGATTTCGTCCTTG |
| His-VP2831–204 | |
| Forward | ATGCGGATCCAACACTGTGACCAAGACCATCG |
| Reverse | ATGCGGATCCTTACTCGGTCTCAGTGCCAGAGT |
| His-VP19 | |
| Forward | ATGCGGATCCATGGCCACCACGACTAACACTCTT |
| Reverse | ATGCGAATTCTTACTGCCTCCTCTTGGGGTAAG |
The plasmids for recombinant expression were transformed into E. coli BL21(DE3). The cultures were induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 24 h at 18°C. Bacterial cells were harvested and lysed by sonication in 4 ml of binding buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% Triton X-100) supplemented with a protease inhibitor cocktail tablet (Roche). Finally, the lysate was centrifuged at 40,000 × g for 20 min, and the supernatant was collected for analysis.
Chitin-binding assay.
For the WSSV-chitin binding experiments, 10 μl of chitin beads (New England BioLabs) preequilibrated with TMN buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2 mM MgCl2) was mixed with 109 virions, the envelope fraction (from 109 virions) or nucleocapsids (from 109 virions) in 100 μl of TMN buffer and incubated for 1 h at room temperature (RT) with gentle rotation. Subsequently, the beads were washed five times with wash buffer 1 (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 2 mM MgCl2). For the protein-chitin binding experiments, 10 μl of chitin beads preequilibrated with binding buffer was mixed with 100 μl of bacterial lysate supernatant containing the indicated recombinant proteins and incubated for 1 h at RT. Then the beads were washed five times with washing buffer 2 (50 mM Tris-HCl [pH 8.0], 250 mM NaCl, 0.5% Triton X-100). The bound virions, envelopes, nucleocapsids, or proteins were dissociated from the beads by boiling in SDS-PAGE sample buffer for 10 min and analyzed by Western blotting.
Western blot analysis.
The samples were separated on 14% SDS-PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore). The membranes were blocked by incubation in Bløk-CH reagent (Millipore) for 1 h at RT. Immunoblotting was performed by incubating the membranes with different primary antibodies diluted in Bløk-CH reagent for 1 h at RT. Each membrane was then incubated with a secondary antibody (alkaline phosphatase-conjugated goat anti-mouse IgG [Pierce]) for 1 h at RT. After three more washes with TBST (50 mM Tris-Cl, 150 mM NaCl, 0.05% Tween 20 [pH 7.5]), the alkaline phosphatase signal was detected using the NBT/BCIP (nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate) substrate (Roche).
Anti-VP28 and anti-VP26 monoclonal antibodies were produced by the Shanghai Immune Biotech Company (China); anti-VP24, anti-VP19, and anti-VP51 monoclonal antibodies were produced by Abmart (China).
Oligochitosan blocking assay.
Oligochitosan (chitosan oligosaccharide lactate, Sigma) was dissolved in TMN buffer (for the blocking experiment with WSSV virions) or binding buffer (for the blocking experiment with V5-VP24) to concentrations of 5, 7.5, and 10 mg/ml. WSSV (109 virions in 100 μl of TMN buffer) or 100 μl of lysis supernatant containing V5-VP24 was incubated with 300 μl of oligochitosan at different concentrations for 3 h at RT with gentle rotation. The final amounts of oligochitosan in the reaction mixtures were 0, 1.5, 2.25, and 3 mg. Finally, the chitin-binding assay was performed as described above.
Peptide blocking assay.
The P-VP24186–200 (TNRHYLLSMRFSPGN) peptide corresponding to aa 186 to 200 of VP24 and the same-size control peptide P-VP24148–162 (GREFSANKFVLYFKP) from the non-chitin-binding region (aa 148 to 162 of VP24) were synthesized by Shanghai Science Peptide Biological Technology Co., Ltd. The peptides were dissolved in TMN buffer (for the blocking experiments with WSSV virions) or binding buffer (for the blocking experiments with V5-VP24) to concentrations of 2, 3, and 4 mg/ml. Ten microliters of chitin beads was incubated with 300 μl of peptides for 1 h at RT with gentle rotation. Subsequently, WSSV (109 virions in 100 μl of TMN buffer) or 100 μl of lysis supernatant containing V5-VP24 was added to the mixture and incubated for another hour. The peptide amounts in the reaction mixtures were 0, 0.6, 0.9, and 1.2 mg. Finally the chitin-binding assay was performed as described above.
Oral inoculation procedure.
A 1-cm-long flexible silicone tube (diameter, 1.5 mm; wall thickness, 0.3 mm) was connected to a 100-μl plastic pipette tip. L. vannamei shrimp were placed flat on a sponge (mouth up). The pipette tip with the silicone tube was infused into the oral cavity, and the viral inoculum was delivered into the lumen of the esophagus.
In vivo protection assay with peptides by oral delivery.
The effects of peptides on viral infection were assessed in an in vivo challenge experiment. L. vannamei shrimp were randomly divided into four groups (30 individuals per group). For the positive-control group, each shrimp was orally inoculated with 5 × 108 WSSV in 30 μl of normal saline. For the negative-control group, each shrimp was infused with 30 μl of normal saline. For the experimental groups, the shrimp were inoculated orally with 5 × 108 WSSV plus 120 μg of P-VP24186–200 or P-VP24148–162 in 30 μl of normal saline. In each group, 24 shrimp were used to determine the viral load in the digestive tract and hemolymph, and 6 shrimp were used for cryosectioning.
Quantification of viral load in the digestive tract and hemolymph.
At 4 h postinoculation (hpi), 12 shrimp were randomly selected from each group for analysis. The intact digestive tract (including the esophagus, stomach, midgut, and hindgut) of each shrimp was weighed and homogenized in 10 volumes (wt/vol) of normal saline. The viral load in each sample was measured using quantitative PCR (qPCR) according to the instructions of the WSSV-qPCR detection kit (Xiamen Lulong Biotech Co., Ltd., Xiamen, Fujian, China). The amplification reactions were performed as follows: denaturing at 95°C for 2 min, followed by 40 cycles of 94°C for 10 s and 60°C for 30 s. Additionally, 12 shrimp in each group were used to analyze the viral load in the hemolymph. At 24, 48, 72, and 96 hpi, 20 μl of hemolymph was withdrawn from each shrimp, and the viral copies in the hemolymph were quantitatively measured by qPCR. The data obtained from the qPCR analysis were subjected to one-way analysis of variance (ANOVA) using SPSS software 16.0. P values of <0.05 were considered statistically significant.
Cryosectioning and immunofluorescence analysis.
Six shrimp were analyzed from each group 4 hpi. The midgut of each animal was collected and cut into sections 1 cm in length. The samples were placed in optimal cutting temperature compound, transferred to liquid nitrogen, and stored at −80°C prior to sectioning. The midgut samples were cross-sectioned into 5-μm-thick slices using a Leica CM1850 cryostat. The slices were sequentially fixed with 4% paraformaldehyde for 15 min, probed with the anti-P28 antibody, and incubated with the Alexa Fluor 488 donkey anti-mouse IgG (H+L) secondary antibody (Life Technologies).
In vivo protection assay with oligochitosan by oral delivery.
The effects of oligochitosan on viral infection were assessed in an in vivo challenge experiment. L. vannamei shrimp were randomly divided into four groups (12 individuals per group). For the positive-control group, each shrimp was orally inoculated with 5 × 108 virions in 30 μl of normal saline. For the negative-control group, each shrimp was infused with 30 μl of normal saline. For the experimental group, the shrimp were orally inoculated with 5 × 108 virions in 30 μl of normal saline containing 10 mg/ml oligochitosan. The virions were preincubated with oligochitosan in normal saline for 3 h prior to administration to the animals. At 4 hpi, the viral load in the digestive tract of each shrimp was evaluated by qPCR as described above. The data obtained from the qPCR analysis were subjected to one-way analysis of variance (ANOVA) using SPSS software 16.0. P values of <0.05 were considered statistically significant.
RESULTS
WSSV binds to chitin.
To investigate the interaction between WSSV and chitin, purified intact virions, viral envelopes, or nucleocapsids were incubated with chitin beads. The proteins bound to the chitin beads were analyzed by Western blotting with an antibody against the WSSV major envelope protein VP28 or the capsid protein VP51. Purified WSSV virions, envelopes, and nucleocapsids were also analyzed by Western blotting. As shown in Fig. 1, intact virions (A) and the viral envelopes (C) bound to the chitin beads, whereas viral nucleocapsids (B) failed to bind to the chitin beads. This result suggests that the binding of WSSV to the chitin beads was dependent on the envelope protein(s). Additionally, the interaction between the intact virions and the chitin beads was inhibited in a dose-dependent manner by oligochitosan (Fig. 1D).
FIG 1.
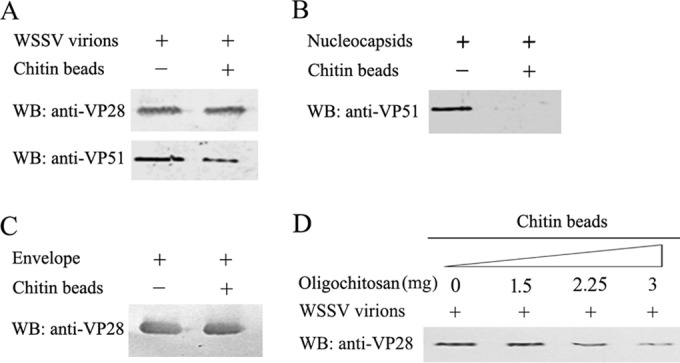
Chitin-binding assays for WSSV particles, envelope and nucleocapsid. WSSV particles (109) (A), nucleocapsids (from 109 virions) (B), and envelopes (from 109 virions) (C) were incubated separately with 10 μl of chitin beads. The chitin-bound fractions were analyzed by Western blotting (WB) with an anti-VP28 antibody (1:10,000) and anti-VP51 antibody (1:3,000). For the oligochitosan blocking assay (D), WSSV particles (109) was preincubated with 0, 1.5, 2.25, and 3 mg of oligochitosan in 400 μl of TMN buffer for 3 h. Then 10 μl of chitin beads was added to the mixture. After further incubation for 1 h, the beads were washed and the chitin-bound complexes were analyzed by Western blotting with an anti-VP28 antibody.
VP24 is a CBP.
Because VP28, VP26, VP24, and VP19 are the four most abundant WSSV envelope proteins (26), we speculated that one or some of them might bind to chitin. To test this hypothesis, recombinant His-VP19 (aa 1 to 121), His-VP28 (aa 31 to 204), V5-VP24 (aa 26 to 208), and V5-VP26 (aa 36 to 204) were expressed in E. coli. Bacterial lysates containing the recombinant proteins were incubated with chitin beads, and the protein(s) bound to the beads was analyzed by Western blotting. As shown in Fig. 2, only VP24 was associated with the chitin beads (B), suggesting that VP24 was a CBP and VP28, VP26, and VP19 were not. This finding was supported by the results of the oligochitosan blocking experiment, in which the binding of V5-VP24 with the chitin beads was significantly blocked by oligochitosan in a dose-dependent manner (Fig. 2C).
FIG 2.
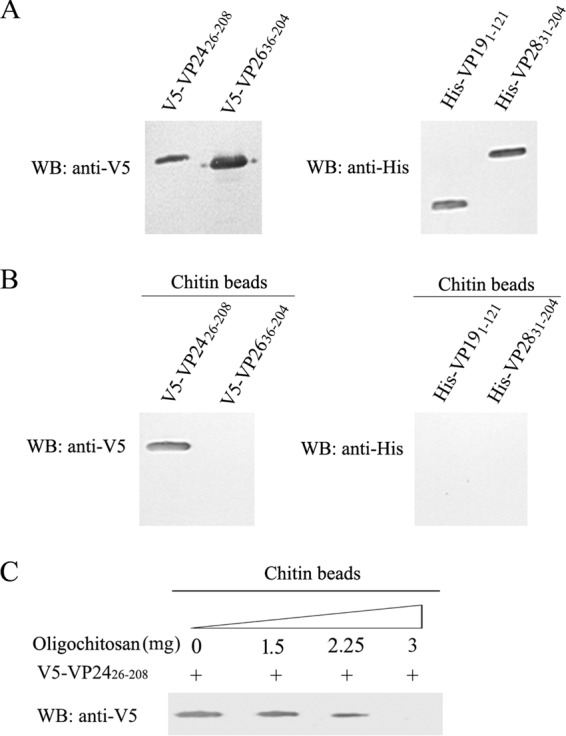
Chitin-binding assays for VP24, VP26, VP19, and VP28. (A) V5-tagged VP24, V5-tagged VP26, His-tagged VP19, and His-tagged VP28 were expressed separately in E. coli BL21. The bacterial lysates were analyzed by Western blotting (WB) to evaluate the expression of His-VP19 (aa 1 to 121), His-VP28 (aa 31 to 204), V5-VP24 (aa 26 to 208), and V5-VP26 (aa 36 to 204). (B) One hundred microliters of bacterial lysate containing recombinant proteins was incubated with 10 μl of chitin beads, and the chitin-bound proteins were analyzed by Western blotting with an anti-V5 or anti-His antibody. (C) For the oligochitosan blocking experiments, a bacterial lysate containing V5-VP24 was preincubated with 0, 1.5, 2.25, and 3 mg of oligochitosan separately for 1 h at RT. Then 10 μl of chitin beads were added to the mixture. After incubation for 1 h with gentle rotation, the beads were washed five times, and the chitin-bound complexes were analyzed by Western blotting with an anti-V5 antibody.
The C-terminal region (aa 186 to 200) of VP24 is required for chitin binding.
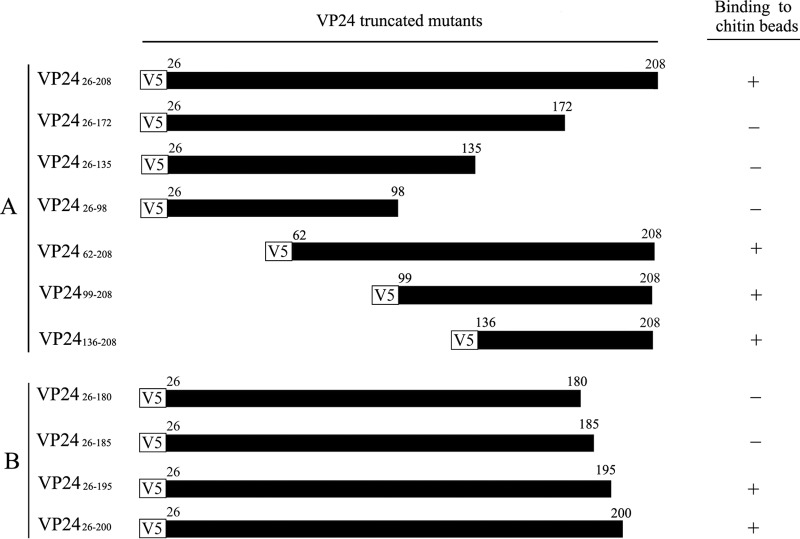
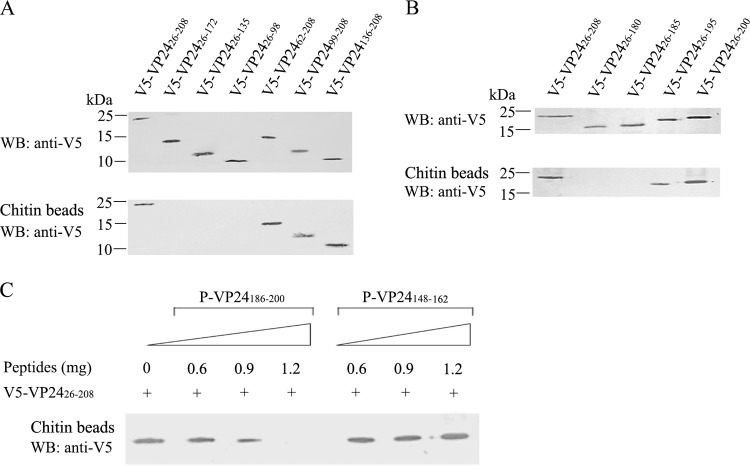
To identify the CBD of VP24, we constructed a series of VP24 deletion mutants (see Fig. 4A, left panel) and analyzed their interactions with chitin beads. As shown in Fig. 3A (upper panel), V5-tagged VP2426–208, VP2426–172, VP2426–135, VP2426–98, VP2462–208, VP2499–208, and VP24136–208 were expressed well with the expected molecular weight. Among them, VP2426–208, VP2462–208, VP2499–208, and VP24136–208 bound to the chitin beads, whereas VP2426–172, VP2426–135, and VP2426–98 did not (Fig. 3A, lower panel). These results suggest that the CBD is located in the C-terminal region (aa 173 to 208) of VP24 (Fig. 4A).
FIG 4.
Schematic diagram of the VP24 mutants and summary of their interaction with chitin. The V5 tag is indicated by a white box. VP24 and its mutants are indicated by black boxes. “+” indicates that the protein can bind to chitin, and “−” indicates that the protein does not bind to chitin.
FIG 3.
Chitin-binding assays for VP24 mutants. (A and B) Chitin-binding assay. VP24 mutants were expressed separately in E. coli BL21, and the expression of the recombinant proteins was analyzed by Western blotting with an anti-V5 antibody (upper panels in panels A and B). One hundred microliters of bacterial lysate containing recombinant proteins was incubated with 10 μl of chitin beads, and the chitin-bound proteins were analyzed by Western blotting with an anti-V5 antibody (lower panels in panels A and B). A bacterial lysate containing VP2426–208 was used as a positive control (lane 1, lower panels in panels A and B). (C) The P-VP24186–200 peptide inhibits the VP24-chitin interaction. Ten microliters of chitin beads was preincubated with 0, 0.6, 0.9, and 1.2 mg of P-VP24186–200 or P-VP24148–162 (control peptide) in 300 μl of binding buffer for 1 h. Then 100 μl of bacterial lysate containing V5-VP2426–208 was added to the mixture. After incubation for 1 h with gentle rotation, the beads were washed five times, and the chitin-bound proteins were analyzed by Western blotting with an anti-V5 antibody.
To more precisely map the chitin-binding site of VP24, four VP24 mutants (VP2426–180, VP2426–185, VP2426–195, and VP2426–200) were generated (Fig. 4B, left panel), and their interactions with chitin beads were examined. As shown in Fig. 3B, deletion of aa 180 to 208 or aa 185 to 208 of VP24 completely abolished the chitin-binding ability of this protein. Moreover, VP2426–195 showed a weaker chitin-binding ability than VP2426–200. The interactions between the VP24 mutants and the chitin beads are summarized in Fig. 4. These results indicate that aa 186 to 200 of VP24 are essential for chitin binding.
The peptide blocking analysis showed that the P-VP24186–200 peptide corresponding to aa 186 to 200 of VP24 was able to inhibit the binding of VP24 to the chitin beads in a dose-dependent manner, whereas the control peptide P-VP24148–162 did not affect the VP24-chitin interaction (Fig. 3C). These results are consistent with the results of the mutagenesis experiments. Therefore, we conclude that the CBD of VP24 is located within aa 186 to 200. However, this CBD shares no homology to typical CBDs, such as the cysteine-containing CBD and arthropod cuticular protein CBD (35, 36) (data not shown).
VP24-chitin interaction is essential for WSSV per os infection.
Among the four most abundant WSSV envelope proteins, only VP24 could interact with chitin (Fig. 2). Therefore, we speculated that VP24 might be required for WSSV-chitin binding. To investigate this possibility, chitin beads were preincubated with different amounts of the P-VP24186–200 or P-VP24148–162 peptides and then subjected to binding assays with the WSSV virions. As shown in Fig. 5, the WSSV-chitin interaction was significantly reduced by P-VP24186–200 in a dose-dependent manner. The interaction was greatly inhibited when 1.2 mg of P-VP24186–200 was added to the reaction mixture. In contrast, the control peptide P-VP24148–162 did not affect the WSSV-chitin interaction. These data demonstrate that VP24 participates in the WSSV-chitin interaction.
FIG 5.
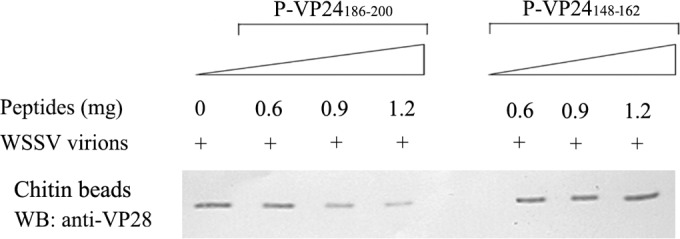
The P-VP24186–200 peptide inhibits the WSSV-chitin interaction. Ten microliters of chitin beads were preincubated with 0, 0.6, 0.9, and 1.2 mg of P-VP24186–200 or P-VP24148–162 (control peptide) in 300 μl of TMN buffer for 1 h, and then WSSV (109 virions in 100 μl TMN buffer) was added to the mixture and incubated for 1 h with gentle rotation. The beads were washed five times, and the chitin-bound fraction was analyzed by Western blotting (WB) with an anti-VP28 antibody.
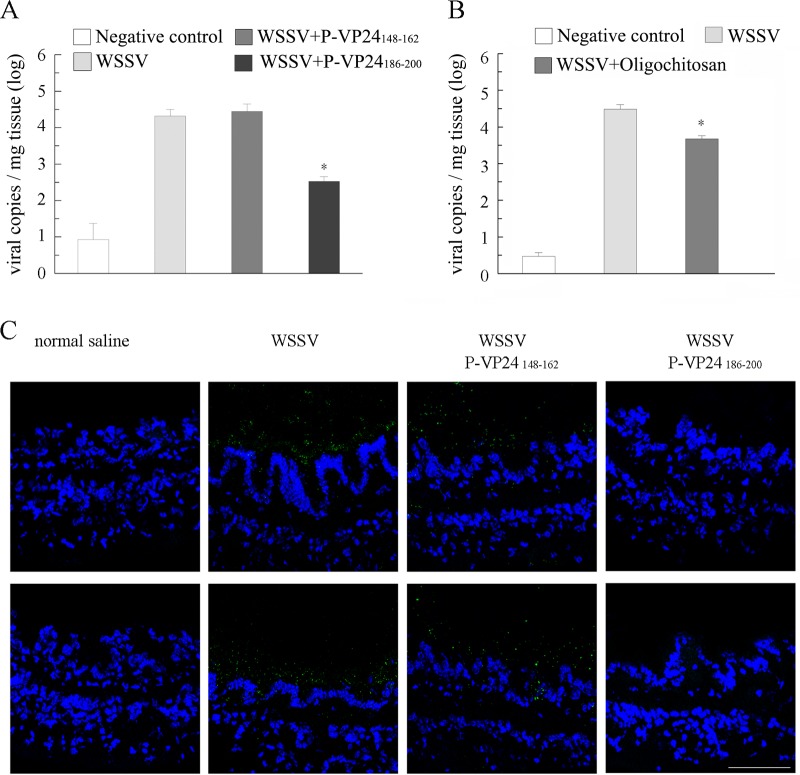
Because the digestive tract of shrimp is lined with a chitinous structure, we analyzed whether P-VP24186–200 could inhibit the binding of WSSV virions to the inner surface of the shrimp digestive tract during per os infection. P-VP24186–200 or the control peptide P-VP24148–162 was delivered into the oral cavity of the shrimp together with the purified WSSV virions. The clearance time of ingested food from the entire digestive system has been quantified as 4 h for L. vannamei (37). Therefore, the entire digestive tract was collected from 12 shrimp from each group at 4 hpi, and the amount of viral genomic DNA in each sample was determined via qPCR. As shown in Fig. 6A, the viral copy numbers in the P-VP24148–162-treated group and WSSV-positive-control group were 2.8 × 104 copies/mg tissue and 2.1 × 104 copies/mg tissue at 4 hpi, respectively. The viral load in the P-VP24186–200-treated group was significantly lower (3.4 × 102 copies/mg tissue). Because oligochitosan could also block the VP24-chitin interaction (Fig. 2), the effect of oligochitosan on WSSV infection was investigated. The viral copy numbers in the WSSV-positive-control group and oligochitosan-treated group were 3.0 × 104 copies/mg tissue and 4.8 × 103 copies/mg tissue at 4 hpi, respectively (Fig. 6 B). These data suggest that P-VP24186–200 and oligochitosan both impede the binding of WSSV virions to the inner surface of the shrimp digestive tract, with P-VP24186–200 blocking the interaction more efficiently than oligochitosan.
FIG 6.
The P-VP24186–200 peptide and oligochitosan prevent WSSV from binding to the inner surface of the digestive tract. (A and B) Shrimp were orally inoculated with WSSV, WSSV plus peptide P-VP24186–200, WSSV plus peptide P-VP24148–162, WSSV plus oligochitosan, or an equal volume of normal saline (negative control). The intact digestive tract of each shrimp was collected at 4 hpi, and the viral load in each sample was analyzed via qPCR. (C) Shrimp were orally inoculated with WSSV, WSSV plus peptide P-VP24186–200, WSSV plus peptide P-VP24148–162, or an equal volume of normal saline. The midgut of each shrimp was collected, cross-sectioned, and probed with an anti-VP28 antibody (green). The nucleus was stained with DAPI (4′,6-diamidino-2-phenylindole [blue]). The inner surface of the midgut is facing upwards. Bar, 75 μm. Two different images are shown for each treatment.
The midguts of shrimp inoculated with WSSV, WSSV plus P-VP24186–200, WSSV plus P-VP24148–162, or normal saline were collected at 4 hpi, cross-sectioned, and probed with the anti-VP28 antibody. As shown in Fig. 6C, virions were distributed along the inner surface of the midgut in the WSSV-infected shrimp and shrimp inoculated with WSSV plus P-VP24148–162. In contrast, virions were rarely observed in the midguts of shrimp inoculated with WSSV plus P-VP24186–200. These data indicate that the VP24-chitin interaction is required for the binding of WSSV to the inner surface of the shrimp digestive tract.
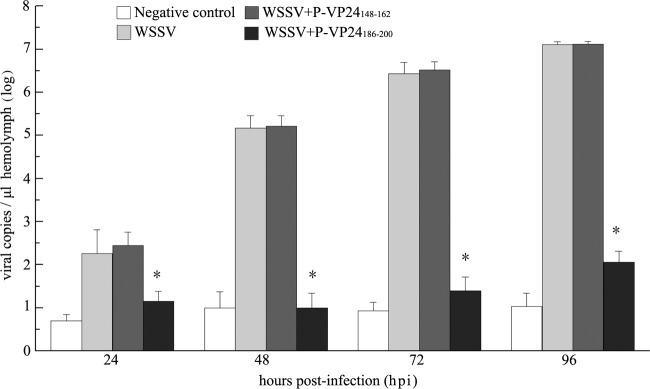
To investigate the importance of the VP24-chitin interaction during WSSV per os infection, P-VP24186–200 or the control peptide P-VP24148–162 was delivered into the oral cavity of the shrimp together with purified WSSV virions as described above. The amounts of viral genomic DNA in the hemolymph of the infected animals were measured by qPCR 24, 48, 72, and 96 hpi. As shown in Fig. 7, the viral copy numbers in the P-VP24148–162-treated group and the positive-control group increased rapidly with the progress of infection, reaching 1.3 × 107 copies/μl hemolymph at 96 hpi. In contrast, the viral load in the P-VP24186–200-treated group showed a very limited increase (1.2 × 102 copies/μl hemolymph at 96 hpi) that was slightly higher than that in the uninfected control. These results suggest that WSSV infection through the digestive tract is greatly hindered by P-VP24186–200 treatment.
FIG 7.
The P-VP24186–200 peptide inhibits WSSV per os infection. Shrimp were orally inoculated with WSSV, WSSV plus peptide P-VP24186–200, WSSV plus peptide P-VP24148–162, or an equal volume of normal saline (negative control). Hemolymph was collected from each shrimp at 24, 48, 72, and 96 hpi. The viral copy number in each sample was determined via qPCR. The columns represent the log base 10 of the mean WSSV copy number of 12 shrimp in each group, and the standard deviations were calculated. Asterisks indicate significant differences (P < 0.05) between the WSSV-positive-control group and P-VP24186–200-treated group.
DISCUSSION
Ingestion of WSSV-infected sick or dead shrimp is a major route of the natural infection of WSSV in shrimp farms (6, 7, 38). The inner surfaces of the shrimp esophagus, stomach, and hindgut are covered with a chitinous lining, whereas the midgut epithelium is separated from the food bolus by the PM, which is composed of chitin fibrils (13, 17). The digestive system of insects is similar to that of shrimp. Studies of insects suggested that the chitinous structures lining the inner surfaces of the digestive tract might serve as physical infectious barriers; pathogens that infect the digestive system through the oral route need to cross these barriers before they can reach host cells (39, 40). The methods by which WSSV crosses the chitinous barriers in the digestive tract of its hosts are unknown.
In this study, we analyzed the interaction of WSSV with chitin and showed that WSSV virions were capable of binding to chitin beads through their envelopes (Fig. 1). Further investigation demonstrated that VP24 was the only CBP among the four most abundant WSSV envelope proteins (Fig. 2). A peptide corresponding to the CBD of VP24 inhibited both VP24-chitin binding and WSSV-chitin binding in a dose-dependent manner (Fig. 3 and 5), implying that VP24 played an important role in the WSSV-chitin interaction. However, P-VP24186–200 treatment could not completely abolish the WSSV-chitin interaction in our experiment (Fig. 5). Therefore, some other low-abundance envelope protein(s) may also participate in WSSV-chitin binding. Because we used N-terminal hydrophobic region-truncated VP26 and VP28 in our experiments, we could not totally exclude the possibility that VP26 and VP28 could bind to chitin in their natural forms.
To elucidate the role of VP24-chitin binding in WSSV per os infection, L. vannamei shrimp were orally inoculated with WSSV together with P-VP24186–200 or the control peptide P-VP24148–162. The P-VP24186–200 peptide corresponding to the VP24 CBD significantly reduced the number of virions in the digestive tract 4 hpi (Fig. 6A and C). Moreover, viral replication in the animals was greatly inhibited within 96 hpi by P-VP24186–200 (Fig. 7). The clearance time for ingested food from the entire digestive tract of the shrimp has been reported to be 4 h (37). Therefore, we deduce that the VP24-chitin interaction is essential for the binding of WSSV to the inner surface of the shrimp digestive tract, and failure in the attachment process will lead to unsuccessful infection.
According to research with insects, the chitin lining on the inner surface of the digestive tract is impermeable (39). The pore on the shrimp PM is only 20 nm in diameter (17), which is much smaller than WSSV particles. Therefore, WSSV cannot simply cross through the chitinous barriers by penetration. It is possible that WSSV recruits a host chitinase to degrade the chitin on the binding site after attachment to the chitin layer, thereby allowing WSSV to break though the chitin barriers and infect the underlying epithelium.
Interestingly, VP24 was recently found to interact with the Penaeus monodon CBP PmCBP, which exists in different shrimp species (27) and is widely expressed in many tissues, including the stomach and midgut (41). In nature, CBPs are the major group of proteins that associate with chitin and participate in the formation, maintenance, and regulation of the functions of these extracellular structures (42). Other classes of CBPs function as lectins and chitinases (43, 44). Although the function of PmCBP remains unclear, depletion of PmCBP by double-stranded RNA (dsRNA) interference (27) or neutralization of PmCBP with recombinant protein (41) leads to an increase in the shrimp survival rate after WSSV challenge. Because PmCBP has been found on the cell surface and can interact with six WSSV envelope proteins, including VP24, it is believed to be a good receptor candidate for WSSV (27, 41). Combined with the results of the present study, we speculate that VP24 is a key factor involved in WSSV infection. The VP24-chitin interaction makes it possible for the virus to bind and cross the chitinous barriers in the digestive tract. Then the six envelope proteins that bind PmCBP may attach to PmCBP on the cell surface and mediate WSSV entry.
VP24 was previously identified as the core of a WSSV envelope protein complex called the “infectome.” However, the location of VP24 on the WSSV membrane has been predicted differently in different models (24, 27). In an early model, VP24 was proposed to be embedded in the WSSV envelope with only its C-terminal end projecting toward the inside the virion. However, insufficient experimental evidence has been reported in support of this hypothesis. In 2014, Chen et al. provided a new model of the WSSV infectome in which the infectome contained two groups: one group that bound PmCBP and the other group that did not bind PmCBP. VP24 serves as a link between these two groups, and the C terminus of VP24 is thought to be located in the exterior of the viral envelope because VP24 can interact with CBP and other proteins. In our study, VP24 was found to bind chitin through its C terminus (aa 186 to 200); therefore, it should be exposed outside the viral envelope, which supports Chen's model. However, additional evidence is required to clarify the exact location of VP24 on the envelope.
Notably, the findings of this study provide new ideas for WSSV control strategies in shrimp aquaculture. Numerous studies have explored different vaccination strategies to protect shrimp from WSSV infection, including inactivated WSSV vaccines (45), recombinant protein-based vaccines (46, 47), DNA-based vaccines (48–50), and RNA-based vaccines (51–57). VP28, which is the most abundant WSSV envelope protein, is the major target for vaccine design, and oral vaccination is thought to be the most practical route for vaccine delivery (58). Here, we showed that WSSV per os infection could be efficiently inhibited by P-VP24186–200, which might abolish the interaction between WSSV and chitin in the shrimp digestive tract (Fig. 5, 6, and 7). Therefore, P-VP24186–200 may be used as an orally delivered drug to prevent WSSV infection. Because oligochitosan hampered the WSSV-chitin interaction (Fig. 1 and 2), it is also an attractive choice. Although oligochitosan impeded the binding of WSSV to the inner surface of the digestive tract, it was not as efficient as P-VP24186–200 (Fig. 6B). We speculate that the sticky nature of the oligochitosan solution may prolong the virion's stay in the digestive tract and facilitate viral infection.
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Program of China (973 Program, no. 2012CB114401), National Natural Science Foundation of China (no. 31272698 and no. 41376173), and the China Agriculture Research System (no. CARS-47).
REFERENCES
- 1.Mayo MA. 2002. A summary of taxonomic changes recently approved by ICTV. Arch Virol 147:1655–1663. doi: 10.1007/s007050200039. [DOI] [PubMed] [Google Scholar]
- 2.Wongteerasupaya C, Vickers J, Sriurairatana S, Nash G, Akarajamorn A, Boonsaeng V, Panyim S, Tassanakajon A, Withyachumnarnkul B, Flegel T. 1995. A non-occluded, systemic baculovirus that occurs in cells of ectodermal and mesodermal origin and causes high mortality in the black tiger prawn Penaeus monodon. Dis Aquat Organ 21:69–77. doi: 10.3354/dao021069. [DOI] [Google Scholar]
- 3.Wang CH, Lo CF, Leu JH, Chou CM, Yeh PY, Chou HY, Tung MC, Chang CF, Su MS, Kou GH. 1995. Purification and genomic analysis of baculovirus associated with white spot syndrome (WSBV) of Penaeus monodon. Dis Aquat Organ 23:239–242. doi: 10.3354/dao023239. [DOI] [Google Scholar]
- 4.Yang F, He J, Lin X, Li Q, Pan D, Zhang X, Xu X. 2001. Complete genome sequence of the shrimp white spot bacilliform virus. J Virol 75:11811–11820. doi: 10.1128/JVI.75.23.11811-11820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Hulten MC, Witteveldt J, Peters S, Kloosterboer N, Tarchini R, Fiers M, Sandbrink H, Lankhorst RK, Vlak JM. 2001. The white spot syndrome virus DNA genome sequence. Virology 286:7–22. doi: 10.1006/viro.2001.1002. [DOI] [PubMed] [Google Scholar]
- 6.Chou HY, Huang CY, Lo CF, Kou GH. 1998. Studies on transmission of white spot syndrome associated baculovirus (WSBV) in Penaeus monodon and P. japonicus via waterborne contact and oral ingestion. Aquaculture 164:263–276. doi: 10.1016/S0044-8486(98)00192-6. [DOI] [Google Scholar]
- 7.Wu JL, Namikoshi A, Nishizawa T, Mushiak K, Teruya K, Muroga K. 2001. Effects of shrimp density on transmission of penaeid acute viremia in Penaeus japonicus by cannibalism and the waterborne route. Dis Aquat Organ 47:129–135. doi: 10.3354/dao047129. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, White BL, Redman RM, Lightner DV. 1999. Per os challenge of Litopenaeus vannamei postlarvae and Farfantepenaeus duorarum juveniles with six geographic isolates of white spot syndrome virus. Aquaculture 170:179–194. doi: 10.1016/S0044-8486(98)00425-6. [DOI] [Google Scholar]
- 9.Soto MA, Lotz JM. 2001. Epidemiological parameters of white spot syndrome virus infections in Litopenaeus vannamei and L. setiferus. J Invertebr Pathol 78:9–15. doi: 10.1006/jipa.2001.5035. [DOI] [PubMed] [Google Scholar]
- 10.Soto MA, Shervette VR, Lotz JM. 2001. Transmission of white spot syndrome virus (WSSV) to Litopenaeus vannamei from infected cephalothorax, abdomen, or whole shrimp cadaver. Dis Aquat Organ 45:81–87. doi: 10.3354/dao045081. [DOI] [PubMed] [Google Scholar]
- 11.Escobedo-Bonilla CM, Wille M, Sanz VA, Sorgeloos P, Pensaert MB, Nauwynck HJ. 2005. In vivo titration of white spot syndrome virus (WSSV) in specific pathogen-free Litopenaeus vannamei by intramuscular and oral routes. Dis Aquat Organ 66:163–170. doi: 10.3354/dao066163. [DOI] [PubMed] [Google Scholar]
- 12.Hackman R. 1987. Chitin and the fine structure of cuticles. Chitin and benzoylphenyl ureas. Ser Entomol 38:1–32. [Google Scholar]
- 13.Felgenhauer BE. 1992. Internal anatomy of the Decapoda: an overview, p 45–75. In Harrison FW, Humes AG (ed), Microscopic anatomy of invertebrates, vol 10 Wiley-Liss, Inc, New York, NY. [Google Scholar]
- 14.Merzendorfer H, Zimoch L. 2003. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. J Exp Biol 206:4393–4412. doi: 10.1242/jeb.00709. [DOI] [PubMed] [Google Scholar]
- 15.Tellam RL, Wijffels G, Willadsen P. 1999. Peritrophic matrix proteins. Insect Biochem Mol Biol 29:87–101. doi: 10.1016/S0965-1748(98)00123-4. [DOI] [PubMed] [Google Scholar]
- 16.Eisemann C, Binnington K. 1994. The peritrophic membrane: its formation, structure, chemical composition and permeability in relation to vaccination against ectoparasitic arthropods. Int J Parasitol 24:15–26. doi: 10.1016/0020-7519(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 17.Martin GG, Simcox R, Nguyen A, Chilingaryan A. 2006. Peritrophic membrane of the penaeid shrimp Sicyonia ingentis: structure, formation, and permeability. Biol Bull 211:275–285. doi: 10.2307/4134549. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Lin Q, Chen J, Wu JL, Lim TK, Loh SS, Tang X, Hew CL. 2007. Shotgun identification of the structural proteome of shrimp white spot syndrome virus and iTRAQ differentiation of envelope and nucleocapsid subproteomes. Mol Cell Proteomics 6:1609–1620. doi: 10.1074/mcp.M600327-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Tsai JM, Wang HC, Leu JH, Hsiao HH, Wang AH, Kou GH, Lo CF. 2004. Genomic and proteomic analysis of thirty-nine structural proteins of shrimp white spot syndrome virus. J Virol 78:11360–11370. doi: 10.1128/JVI.78.20.11360-11370.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie X, Xu L, Yang F. 2006. Proteomic analysis of the major envelope and nucleocapsid proteins of white spot syndrome virus. J Virol 80:10615–10623. doi: 10.1128/JVI.01452-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang C, Zhang L, Zhang J, Xiao L, Wu Q, Chen D, Li JK. 2001. Purification and characterization of white spot syndrome virus (WSSV) produced in an alternate host: crayfish, Cambarus clarkii. Virus Res 76:115–125. doi: 10.1016/S0168-1702(01)00247-7. [DOI] [PubMed] [Google Scholar]
- 22.Lin Y, Xu L, Yang F. 2010. Tetramerization of white spot syndrome virus envelope protein VP33 and its interaction with VP24. Arch Virol 155:833–838. doi: 10.1007/s00705-010-0650-z. [DOI] [PubMed] [Google Scholar]
- 23.Jie Z, Xu L, Yang F. 2008. The C-terminal region of envelope protein VP38 from white spot syndrome virus is indispensable for interaction with VP24. Arch Virol 153:2103–2106. doi: 10.1007/s00705-008-0221-8. [DOI] [PubMed] [Google Scholar]
- 24.Chang YS, Liu WJ, Lee CC, Chou TL, Lee YT, Wu TS, Huang JY, Huang WT, Lee TL, Kou GH, Wang AH, Lo CF. 2010. A 3D model of the membrane protein complex formed by the white spot syndrome virus structural proteins. PLoS One 5:e10718. doi: 10.1371/journal.pone.0010718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Li Z, Hew C-L. 2007. Characterization of a novel envelope protein WSV010 of shrimp white spot syndrome virus and its interaction with a major viral structural protein VP24. Virology 364:208–213. doi: 10.1016/j.virol.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 26.Li Z, Xu L, Li F, Zhou Q, Yang F. 2011. Analysis of white spot syndrome virus envelope protein complexome by two-dimensional blue native/SDS PAGE combined with mass spectrometry. Arch Virol 156:1125–1135. doi: 10.1007/s00705-011-0954-7. [DOI] [PubMed] [Google Scholar]
- 27.Huang PY, Leu JH, Chen LL. 2014. A newly identified protein complex that mediates white spot syndrome virus infection via chitin-binding protein. J Gen Virol 95:1799–1808. doi: 10.1099/vir.0.064782-0. [DOI] [PubMed] [Google Scholar]
- 28.Xie X, Li H, Xu L, Yang F. 2005. A simple and efficient method for purification of intact white spot syndrome virus (WSSV) viral particles. Virus Res 108:63–67. doi: 10.1016/j.virusres.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Q, Qi YP, Yang F. 2007. Application of spectrophotometry to evaluate the concentration of purified white spot syndrome virus. J Virol Methods 146:288–292. doi: 10.1016/j.jviromet.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 30.van Hulten MC, Westenberg M, Goodall SD, Vlak JM. 2000. Identification of two major virion protein genes of white spot syndrome virus of shrimp. Virology 266:227–236. doi: 10.1006/viro.1999.0088. [DOI] [PubMed] [Google Scholar]
- 31.van Hulten MC, Goldbach RW, Vlak JM. 2000. Three functionally diverged major structural proteins of white spot syndrome virus evolved by gene duplication. J Gen Virol 81:2525–2529. doi: 10.1099/0022-1317-81-10-2525. [DOI] [PubMed] [Google Scholar]
- 32.Tang X, Wu J, Sivaraman J, Hew CL. 2007. Crystal structures of major envelope proteins VP26 and VP28 from white spot syndrome virus shed light on their evolutionary relationship. J Virol 81:6709–6717. doi: 10.1128/JVI.02505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie X, Yang F. 2006. White spot syndrome virus VP24 interacts with VP28 and is involved in virus infection. J Gen Virol 87:1903–1908. doi: 10.1099/vir.0.81570-0. [DOI] [PubMed] [Google Scholar]
- 34.Xie X, Yang F. 2005. Interaction of white spot syndrome virus VP26 protein with actin. Virology 336:93–99. doi: 10.1016/j.virol.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 35.Rebers JE, Willis JH. 2001. A conserved domain in arthropod cuticular proteins binds chitin. Insect Biochem Mol Biol 31:1083–1093. doi: 10.1016/S0965-1748(01)00056-X. [DOI] [PubMed] [Google Scholar]
- 36.Chen LL, Lu LC, Wu WJ, Lo CF, Huang WP. 2007. White spot syndrome virus envelope protein VP53A interacts with Penaeus monodon chitin-binding protein (PmCBP). Dis Aquat Organ 74:171–178. doi: 10.3354/dao074171. [DOI] [PubMed] [Google Scholar]
- 37.Marte CL. 1980. The food and feeding habit of Penaeus monodon Fabricius collected from Makato River, Aklan, Philippines (Decapoda Natantia). Crustaceana 38:225–236. doi: 10.1163/156854080X00139. [DOI] [Google Scholar]
- 38.Kiran R, Rajendran K, Jung S, Oh M. 2002. Experimental susceptibility of different life-stages of the giant freshwater prawn, Macrobrachium rosenbergii (de Man), to white spot syndrome virus (WSSV). J Fish Dis 25:201–207. doi: 10.1046/j.1365-2761.2002.00357.x. [DOI] [Google Scholar]
- 39.Lemaitre B, Miguel-Aliaga I. 2013. The digestive tract of Drosophila melanogaster. Annu Rev Genet 47:377–404. doi: 10.1146/annurev-genet-111212-133343. [DOI] [PubMed] [Google Scholar]
- 40.Wang P, Granados RR. 1997. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc Natl Acad Sci U S A 94:6977–6982. doi: 10.1073/pnas.94.13.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen KY, Hsu TC, Huang PY, Kang ST, Lo CF, Huang WP, Chen LL. 2009. Penaeus monodon chitin-binding protein (PmCBP) is involved in white spot syndrome virus (WSSV) infection. Fish Shellfish Immunol 27:460–465. doi: 10.1016/j.fsi.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 42.Tetreau G, Dittmer NT, Cao X, Agrawal S, Chen Y, Muthukrishnan S, Haobo J, Blissard GW, Kanost MR, Wang P. 2014. Analysis of chitin-binding proteins from Manduca sexta provides new insights into evolution of peritrophin A-type chitin-binding domains in insects. Insect Biochem Mol Biol 62:127–141. doi: 10.1016/j.ibmb.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen Z, Jacobs-Lorena M. 1999. Evolution of chitin-binding proteins in invertebrates. J Mol Evol 48:341–347. doi: 10.1007/PL00006478. [DOI] [PubMed] [Google Scholar]
- 44.Marques MRF, Barracco MA. 2000. Lectins, as non-self-recognition factors, in crustaceans. Aquaculture 191:23–44. doi: 10.1016/S0044-8486(00)00417-8. [DOI] [Google Scholar]
- 45.Namikoshi A, Wu JL, Yamashita T, Nishizawa T, Nishioka T, Arimoto M, Muroga K. 2004. Vaccination trials with Penaeus japonicus to induce resistance to white spot syndrome virus. Aquaculture 229:25–35. doi: 10.1016/S0044-8486(03)00363-6. [DOI] [Google Scholar]
- 46.Witteveldt J, Vlak JM, van Hulten MC. 2004. Protection of Penaeus monodon against white spot syndrome virus using a WSSV subunit vaccine. Fish Shellfish Immunol 16:571–579. doi: 10.1016/j.fsi.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Kulkarni A, Rombout JH, Singh IS, Sudheer NS, Vlak JM, Caipang CM, Brinchmann MF, Kiron V. 2013. Truncated VP28 as oral vaccine candidate against WSSV infection in shrimp: an uptake and processing study in the midgut of Penaeus monodon. Fish Shellfish Immunol 34:159–166. doi: 10.1016/j.fsi.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 48.Rajesh Kumar S, Ishaq Ahamed VP, Sarathi M, Nazeer Basha A, Sahul Hameed AS. 2008. Immunological responses of Penaeus monodon to DNA vaccine and its efficacy to protect shrimp against white spot syndrome virus (WSSV). Fish Shellfish Immunol 24:467–478. doi: 10.1016/j.fsi.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 49.Ning JF, Zhu W, Xu JP, Zheng CY, Meng XL. 2009. Oral delivery of DNA vaccine encoding VP28 against white spot syndrome virus in crayfish by attenuated Salmonella typhimurium. Vaccine 27:1127–1135. doi: 10.1016/j.vaccine.2008.11.075. [DOI] [PubMed] [Google Scholar]
- 50.Rout N, Kumar S, Jaganmohan S, Murugan V. 2007. DNA vaccines encoding viral envelope proteins confer protective immunity against WSSV in black tiger shrimp. Vaccine 25:2778–2786. doi: 10.1016/j.vaccine.2006.12.056. [DOI] [PubMed] [Google Scholar]
- 51.Wu Y, Lü L, Yang L-S, Weng S-P, Chan S-M, He J-G. 2007. Inhibition of white spot syndrome virus in Litopenaeus vannamei shrimp by sequence-specific siRNA. Aquaculture 271:21–30. doi: 10.1016/j.aquaculture.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mejia-Ruiz CH, Vega-Pena S, Alvarez-Ruiz P, Escobedo-Bonilla CM. 2011. Double-stranded RNA against white spot syndrome virus (WSSV) vp28 or vp26 reduced susceptibility of Litopenaeus vannamei to WSSV, and survivors exhibited decreased susceptibility in subsequent re-infections. J Invertebr Pathol 107:65–68. doi: 10.1016/j.jip.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 53.Xu J, Han F, Zhang X. 2007. Silencing shrimp white spot syndrome virus (WSSV) genes by siRNA. Antiviral Res 73:126–131. doi: 10.1016/j.antiviral.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 54.Kim CS, Kosuke Z, Nam YK, Kim SK, Kim KH. 2007. Protection of shrimp (Penaeus chinensis) against white spot syndrome virus (WSSV) challenge by double-stranded RNA. Fish Shellfish Immunol 23:242–246. doi: 10.1016/j.fsi.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 55.Robalino J, Browdy CL, Prior S, Metz A, Parnell P, Gross P, Warr G. 2004. Induction of antiviral immunity by double-stranded RNA in a marine invertebrate. J Virol 78:10442–10448. doi: 10.1128/JVI.78.19.10442-10448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarathi M, Simon MC, Ahmed VP, Kumar SR, Hameed AS. 2008. Silencing VP28 gene of white spot syndrome virus of shrimp by bacterially expressed dsRNA. Mar Biotechnol 10:198–206. doi: 10.1007/s10126-007-9052-y. [DOI] [PubMed] [Google Scholar]
- 57.Sarathi M, Simon MC, Venkatesan C, Hameed AS. 2008. Oral administration of bacterially expressed VP28dsRNA to protect Penaeus monodon from white spot syndrome virus. Mar Biotechnol 10:242–249. doi: 10.1007/s10126-007-9057-6. [DOI] [PubMed] [Google Scholar]
- 58.Witteveldt J, Cifuentes CC, Vlak JM, van Hulten MC. 2004. Protection of Penaeus monodon against white spot syndrome virus by oral vaccination. J Virol 78:2057–2061. doi: 10.1128/JVI.78.4.2057-2061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]