Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) infection modulates the host cell cycle to create an environment optimal for its viral-DNA replication during the lytic life cycle. We report here that KSHV vIRF4 targets the β-catenin/CBP cofactor and blocks its occupancy on the cyclin D1 promoter, suppressing the G1-S cell cycle progression and enhancing KSHV replication. This shows that KSHV vIRF4 suppresses host G1-S transition, possibly providing an intracellular milieu favorable for its replication.
TEXT
Once Kaposi's sarcoma-associated herpesvirus (KSHV), a causative agent of Kaposi's sarcoma (KS) and two lymphotropic diseases (1, 2), enters the cell, it manipulates the host environment in order to establish a productive infection and ensure viral replication. The canonical Wnt/β-catenin pathway is one signaling pathway that is perturbed by gammaherpesviruses (3). β-Catenin, the major effector protein in the canonical Wnt signaling pathway, is normally retained in the cytoplasm in an inactive state through its interaction with a large protein complex (4–6). This complex maintains a low basal level of β-catenin through constant proteasome-mediated degradation (4, 6). Upon Wnt activation, glycogen synthase kinase 3β (GSK-3β)-dependent phosphorylation of β-catenin is inhibited, resulting in the stabilization of β-catenin, followed by its translocation to the nucleus. Once in the nucleus, β-catenin binds T cell-specific factor (TCF)/lymphoid enhancer-binding factor-1 (LEF-1) DNA-binding factors to activate transcription of numerous target genes involved in cell proliferation and survival (7–9).
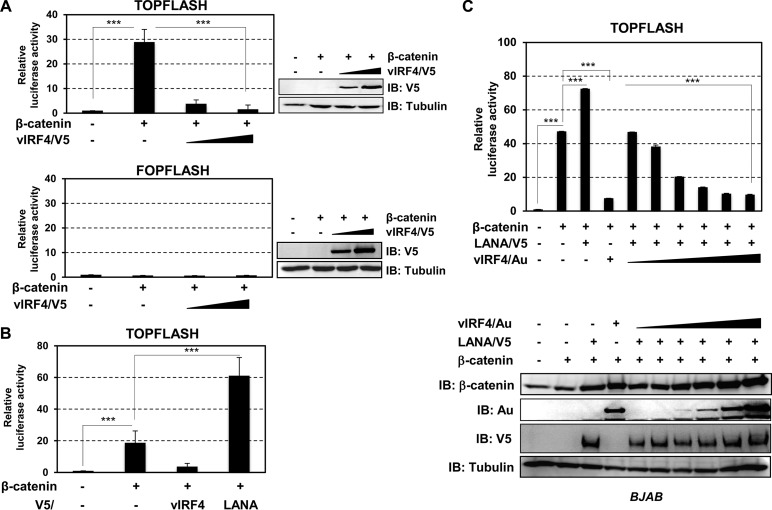
A number of studies have broadened our understanding of the mechanism by which herpesviruses modulate Wnt/β-catenin signaling (3, 10). For instance, KSHV latency-associated nuclear antigen (LANA)-mediated intracellular redistribution of GSK-3β accumulates nuclear β-catenin to transcribe high levels of TCF target genes, such as cyclin D1 and c-Myc, resulting in cell proliferation for KSHV persistency (3, 11–13). In a previous study, we found that KSHV viral interferon regulatory factor 4 (vIRF4) robustly downregulated c-Myc expression, generating a favorable environment for viral lytic replication (14). Furthermore, our microarray data also showed that vIRF4 downregulated expression of cyclin D1, a key regulator in G1/S phase (14). To further define the potential action of KSHV vIRF4 in β-catenin-mediated signal transduction, we first determined whether vIRF4 regulates the β-catenin/TCF transcription complex. A wild-type (WT) TCF binding site (TOPFLASH)-luciferase reporter plasmid and a β-catenin plasmid were transfected together with increasing amounts of vIRF4 into 293T cells. FOPFLASH, which instead uses a TCF/LEF binding site-defective mutant reporter plasmid, was used as a control. Comparison of luciferase activities showed that vIRF4 significantly suppressed β-catenin/TCF-mediated transcriptional activity (Fig. 1A), while LANA dramatically increased β-catenin/TCF-mediated transcriptional activity (Fig. 1B). Given that the activation of the TOPFLASH promoter is primarily determined by the LANA-mediated stabilization of β-catenin, we postulated that vIRF4 might compete with LANA. Indeed, while LANA expression led to increases in the β-catenin level and β-catenin/TCF transactivation, vIRF4 effectively decreased LANA-mediated β-catenin/TCF transactivation (Fig. 1C). However, this vIRF4 effect was independent of β-catenin stability (Fig. 1C, bottom), suggesting that vIRF4 is a negative regulator of β-catenin-mediated signal transduction in a protein stability-independent manner.
FIG 1.
vIRF4 suppresses β-catenin-mediated TOPFLASH activity. (A) Effect of vIRF4 on β-catenin-mediated TOPFLASH activity. TOPFLASH (containing a wild-type TCF/LEF binding site) and FOPFLASH (containing a TCF/LEF binding site-defective mutant) reporters were transfected into 293T cells with vIRF4 and β-catenin as indicated. (B) Comparison of the effects of vIRF4 and LANA on β-catenin-mediated TOPFLASH activity. 293T cells were transfected with either β-catenin/vIRF4 or β-catenin/LANA, together with TOPFLASH reporter plasmid. (C) Effects of vIRF4 on the activation of LANA-mediated TOPFLASH activity. BJAB cells were transfected with β-catenin/LANA, along with vIRF4, in a dosage-dependent manner. Equal amounts of total proteins were analyzed by immunoblotting (IB) with an anti-β-catenin antibody, anti-Au, anti-V5, or anti-tubulin antibody. ***, P < 0.00001. The error bars indicate standard deviations.
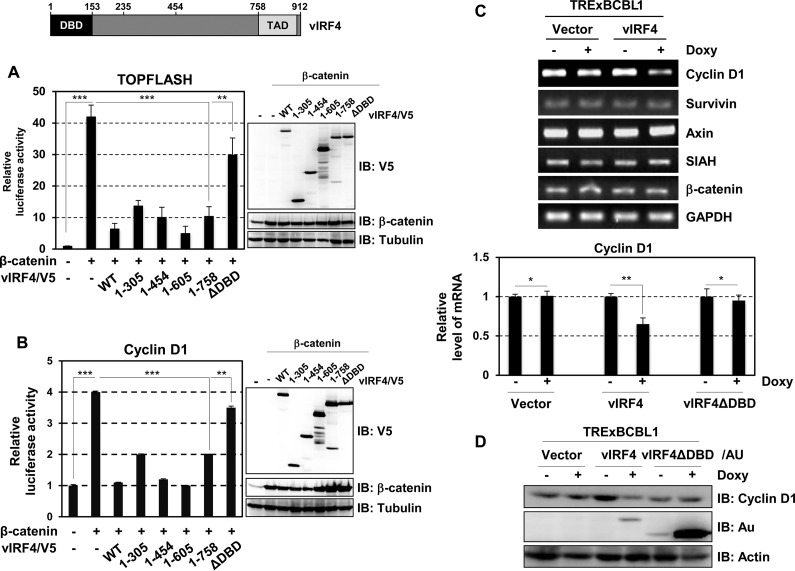
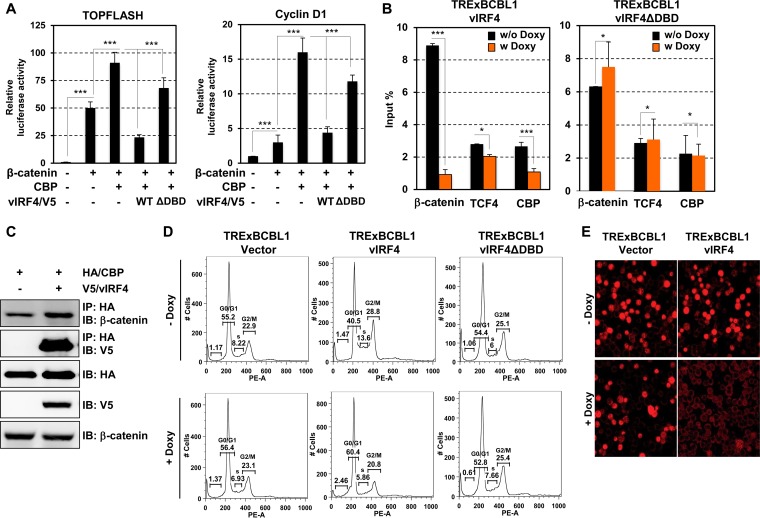
Mutational analysis showed that the DNA-binding domain (DBD) (residues 1 to 153) of vIRF4 was responsible for inhibiting β-catenin/TCF transcriptional activity (Fig. 2A). As cyclin D1 is a target gene for β-catenin/TCF-mediated transactivation (12, 15), the β-catenin-mediated activation of cyclin D1 promoter activity was detectably reduced by WT vIRF4, but not by the ΔDBD mutant (Fig. 2B). Interestingly, vIRF4 expression effectively downregulated cyclin D1 expression without affecting the expression of other β-catenin/TCF target genes, survivin, SIAH, and axin (Fig. 2C and D). β-Catenin binds either CBP or p300, along with the basal transcription machinery, to generate a transcriptionally active complex and selectively activate β-catenin/TCF-mediated transactivation (16, 17). In line with this, knocking down CBP using short interfering RNA decreases cyclin D1 but does not affect other β-catenin/TCF-dependent genes (18). Thus, we investigated whether vIRF4 has the ability to deregulate CBP-dependent β-catenin activity. Indeed, the synergistic activation of either TOPFLASH or cyclin D1 promoter activity by β-catenin and CBP was readily abolished by WT vIRF4, but not the ΔDBD mutant (Fig. 3A). By using chromatin immunoprecipitation (ChIP) assays, we then monitored whether vIRF4 altered the occupancy of TCF/β-catenin/CBP on the cyclin D1 promoter. This showed that the cyclin D1 promoter occupancy of β-catenin and CBP was dramatically reduced upon vIRF4 expression, whereas that of TCF4 was not altered (Fig. 3B). In contrast, the vIRF4ΔDBD mutant did not affect the occupancy of β-catenin and CBP on the cyclin D1 promoter (Fig. 3B). Finally, we found that vIRF4 specifically interacted with the CBP that was capable of binding endogenous β-catenin (Fig. 3C). These data collectively indicate that vIRF4 interaction reduces the accessibility of β-catenin and CBP on the cyclin D1 promoter.
FIG 2.
The DNA-binding domain region of vIRF4 is necessary for modulation of TOPFLASH and cyclin D1 promoter activity. (A and B) Mapping of the region of vIRF4 responsible for β-catenin-mediated TOPFLASH or cyclin D1 activity. 293T cells were cotransfected with the indicated vIRF4 constructs, along with a β-catenin and TOPFLASH (A) or cyclin D1 promoter (B) construct. The cell lysates were used for luciferase assay, followed by IB with the indicated antibodies to determine expression levels of WT vIRF4 and mutants. In the vIRF4 diagram (top), TAD represents the transcriptional activation domain. (C and D) Effects of vIRF4 on the expression of cyclin D1 and other β-catenin/TCF-regulated genes. TRExBCBL1-Vector, TRExBCBL1-vIRF4, and TRExBCBL1-vIRF4ΔDBD cells were mock treated or Doxy treated for 24 h. The cell lysates were used for either quantitative reverse transcription (qRT)-PCR analyses with cyclin D1-, survivin-, SIAH-, axin-, β-catenin-, and GAPDH-specific primers (C) or IB with anti-cyclin D1, anti-Au, or anti-actin antibodies (D). *, P < 0.05; **, P < 0.0001; ***, P < 0.00001. The error bars indicate standard deviations.
FIG 3.
vIRF4 inhibits β-catenin/CBP-mediated cyclin D1 expression and G1-S cell cycle transition. (A) Effect of vIRF4 in a TCF/β-catenin/CBP-dependent manner in either TOPFLASH reporter activity (left) or cyclin D1 reporter activity (right). 293T cells were transfected with β-catenin, CBP, and/or vIRF4. (B) TCF/β-catenin/CBP occupancy on the cyclin D1 promoter. TRExBCBL1-vIRF4 or TRExBCBL1-vIRF4ΔDBD cells were mock treated or Doxy treated for 24 h. β-Catenin, TCF4, or CBP antibodies were used for ChIP assay, and ChIP DNAs were subjected to real-time PCR using primers for the cyclin D1 promoter regions. (C) vIRF4-CBP interaction. 293T cells were transiently transfected with V5-tagged vIRF4 and/or hemagglutinin (HA)-tagged CBP, followed by immunoprecipitation (IP) with an anti-HA antibody and IB with an anti-β-catenin and an anti-V5 antibody. (D) Enhancing G1 accumulation of the cells expressing vIRF4. TRExBCBL1-Vector, TRExBCBL1-vIRF4, or TRExBCBL1-vIRF4ΔDBD cells were stimulated with Doxy (1 μg/ml) and then assessed for PI staining. The data represent the means (plus standard deviations) of the combined results from three independent experiments. *, P < 0.05; ***, P < 0.00001. (E) Inhibition of newly synthesized cellular DNA by vIRF4. TRExBCBL1-Vector and TRExBCBL1-vIRF4 cells were treated with Doxy (1 μg/ml) for 24 h, and the cells were incorporated with BrdU, followed by immunostaining with anti-BrdU antibody for confocal analysis.
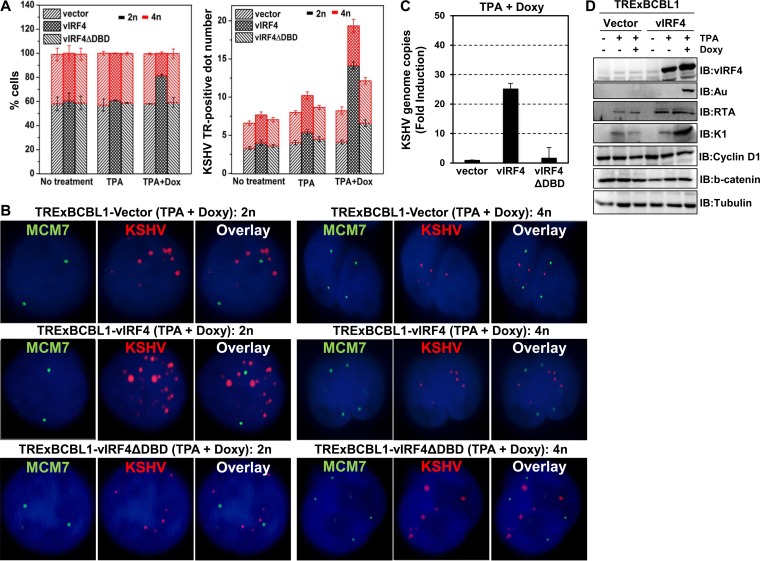
Since cyclin D1 is central to the coordination of the G1-S transition (19), tetracycline-inducible TRExBCBL1-Vector and TRExBCBL1-vIRF4 cells were treated or not with 1 μg/ml doxycycline (Doxy) for 24 h, followed by propidium iodide (PI) staining to determine the cell cycle profile via flow cytometry. This showed detectable increase of the G1 phase upon vIRF4 expression but no increase in vIRF4ΔDBD mutant expression (Fig. 3D). To support this, we performed a bromodeoxyuridine (BrdU) incorporation cell proliferation assay, which allowed us to detect cells undergoing DNA synthesis during S phase. BrdU effectively integrated into newly synthesized DNAs in Doxy-treated TRExBCBL1-Vector cells; however, the BrdU-positive cell population was notably decreased in Doxy-treated TRExBCBL1-vIRF4 cells (Fig. 3E). To examine vIRF4's effects on KSHV viral-DNA synthesis and cellular-DNA synthesis simultaneously, we performed small-molecule DNA fluorescence in situ hybridization (smDNA FISH) using probes containing either the chromosome regions of minichromosome maintenance 7 (MCM7) or KSHV terminal-repeat regions (20, 21). This showed that Doxy-induced vIRF4 expression in TRExBCBL1-vIRF4 cells led to fewer tetraploid (4n) cells (18.5% ± 1.09%) than with untreated or tetradecanoylphorbol acetate (TPA)-treated TRExBCBL1-vIRF4 cells (39% ± 0.34%) (Fig. 4A and B). In contrast, no detectable changes in the tetraploid cell population were detected in TRExBCBL1-Vector and TRExBCBL1-vIRF4ΔDBD cells under any conditions. Finally, the average copy numbers of KSHV were significantly higher in diploid (2n) cells expressing vIRF4 (14.16 ± 0.47 copies) than in diploid cells expressing vector or vIRF4ΔDBD (5.17 ± 0.89 copies) (Fig. 4A and B). Furthermore, induction of vIRF4 during lytic replication leads to a robust increase in the viral copy number, as well as viral-protein expression in KSHV-infected B cells (Fig. 4C and D). Taken together, vIRF4 expression leads to a decrease of G1 phase and an increase in the KSHV DNA copy number.
FIG 4.
vIRF4 expression leads to a decrease in cellular-DNA replication and an increase in KSHV replication. (A and B) vIRF4-dependent cellular and KSHV replication at the single-cell level. In order to perform the smDNA FISH analysis, we generated oligonucleotide probes complementary to the noncoding regions of chromosomal MCM7 and the terminal-repeat regions of KSHV and labeled the probes with Alexa 594 and Cy5, respectively. TRExBCBL1-Vector, TRExBCBL1-vIRF4, and TRExBCBL1-vIRF4ΔDBD cells were treated or not with Doxy (1 μg/ml), together with TPA (20 ng/ml) treatment. The images were acquired with a Zeiss Axiovert Illumination optical-sectioning system. Sixty z-stacks were taken automatically at 0.3-μm intervals at an exposure time of 0.5 s. (C) Effects of vIRF4-mediated G1-S transition on KSHV viral-DNA copy numbers. The reactivated KSHV contained supernatants from TRExBCBL1-Vector, TRExBCBL1-vIRF4, and TRExBCBL1-vIRF4ΔDBD mutant cells that were collected and filtered at 72 h after Doxy and TPA treatment. Virion-associated DNA was purified, and KSHV DNA levels were determined by quantitative PCR (qPCR) with specific primers against the KSHV late gene (ORF26). Shown are the relative rates of KSHV DNA copy number/μl for Vector versus vIRF4 (or the vIRF4ΔDBD mutant) expressed in TRExBCBL1 cell lines. (D) Effects of vIRF4-mediated G1-S transition on KSHV viral-protein expression. TREx BCBL1-Vector and -vIRF4 cells were treated with TPA alone or together with doxycycline for 24 h. The cells were lysed, and IB was subsequently performed with the indicated antibodies. The error bars indicate standard deviations.
Most herpesviruses block the G1-S transition early in the lytic cycle, possibly to avoid competition with host cell DNA synthesis for the limited supply of free nucleotides and to provide nuclear spaces for progeny viral-DNA accumulation (22). Collectively, this study demonstrates a novel strategy for KSHV to suppress host G1-S transition by using the lytic protein vIRF4 to prevent host cell DNA replication and by enhancing viral-DNA replication.
ACKNOWLEDGMENTS
This work was partly supported by NIH grants CA82057, CA31363, CA115284, CA180779, DE023926, HL110609, AI073099, and AI116585, the Hastings Foundation, and the Fletcher Jones Foundation (J.U.J.); grants GM 065367 and PHY 082613 and the Howard Hughes Medical Institute (T.H.); the 21C Frontier Microbial Genomics and Applications Center Program, and a KRIBB Initiative Program grant (KGM4541521) (M.H.K.).
We thank all the members of the Jung laboratory for their discussions.
REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Hayward SD, Liu J, Fujimuro M. 2006. Notch and Wnt signaling: mimicry and manipulation by gamma herpesviruses. Sci STKE 2006:re4. [DOI] [PubMed] [Google Scholar]
- 4.MacDonald BT, Tamai K, He X. 2009. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohn AD, Moon RT. 2005. Wnt and calcium signaling: beta-catenin-independent pathways. Cell Calcium 38:439–446. doi: 10.1016/j.ceca.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 6.Polakis P. 2007. The many ways of Wnt in cancer. Curr Opin Genet Dev 17:45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Brantjes H, Barker N, van Es J, Clevers H. 2002. TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling. Biol Chem 383:255–261. [DOI] [PubMed] [Google Scholar]
- 8.Mosimann C, Hausmann G, Basler K. 2009. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol 10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- 9.Espada J, Calvo MB, Diaz-Prado S, Medina V. 2009. Wnt signalling and cancer stem cells. Clin Transl Oncol 11:411–427. doi: 10.1007/s12094-009-0380-4. [DOI] [PubMed] [Google Scholar]
- 10.Angelova M, Zwezdaryk K, Ferris M, Shan B, Morris CA, Sullivan DE. 2012. Human cytomegalovirus infection dysregulates the canonical Wnt/beta-catenin signaling pathway. PLoS Pathog 8:e1002959. doi: 10.1371/journal.ppat.1002959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Martin HJ, Liao G, Hayward SD. 2007. The Kaposi's sarcoma-associated herpesvirus LANA protein stabilizes and activates c-Myc. J Virol 81:10451–10459. doi: 10.1128/JVI.00804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujimuro M, Wu FY, ApRhys C, Kajumbula H, Young DB, Hayward GS, Hayward SD. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat Med 9:300–306. doi: 10.1038/nm829. [DOI] [PubMed] [Google Scholar]
- 13.Hagen T. 2009. Characterization of the interaction between latency-associated nuclear antigen and glycogen synthase kinase 3beta. J Virol 83:6312–6317. doi: 10.1128/JVI.01671-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HR, Doganay S, Chung B, Toth Z, Brulois K, Lee S, Kanketayeva Z, Feng P, Ha T, Jung JU. 2014. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 4 (vIRF4) targets expression of cellular IRF4 and the Myc gene to facilitate lytic replication. J Virol 88:2183–2194. doi: 10.1128/JVI.02106-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein EA, Assoian RK. 2008. Transcriptional regulation of the cyclin D1 gene at a glance. J Cell Sci 121:3853–3857. doi: 10.1242/jcs.039131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy L, Wei Y, Labalette C, Wu Y, Renard CA, Buendia MA, Neuveut C. 2004. Acetylation of beta-catenin by p300 regulates beta-catenin-Tcf4 interaction. Mol Cell Biol 24:3404–3414. doi: 10.1128/MCB.24.8.3404-3414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takemaru KI, Moon RT. 2000. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol 149:249–254. doi: 10.1083/jcb.149.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teo JL, Ma H, Nguyen C, Lam C, Kahn M. 2005. Specific inhibition of CBP/beta-catenin interaction rescues defects in neuronal differentiation caused by a presenilin-1 mutation. Proc Natl Acad Sci U S A 102:12171–12176. doi: 10.1073/pnas.0504600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tashiro E, Tsuchiya A, Imoto M. 2007. Functions of cyclin D1 as an oncogene and regulation of cyclin D1 expression. Cancer Sci 98:629–635. doi: 10.1111/j.1349-7006.2007.00449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chavez SL, Loewke KE, Han J, Moussavi F, Colls P, Munne S, Behr B, Reijo Pera RA. 2012. Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat Commun 3:1251. doi: 10.1038/ncomms2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flemington EK. 2001. Herpesvirus lytic replication and the cell cycle: arresting new developments. J Virol 75:4475–4481. doi: 10.1128/JVI.75.10.4475-4481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]