ABSTRACT
Hantaviruses, which belong to the genus Hantavirus in the family Bunyaviridae, infect mammals, including humans, causing either hemorrhagic fever with renal syndrome (HFRS) or hantavirus cardiopulmonary syndrome (HCPS) in humans with high mortality. Hantavirus encodes a nucleocapsid protein (NP) to encapsidate the genome and form a ribonucleoprotein complex (RNP) together with viral polymerase. Here, we report the crystal structure of the core domains of NP (NPcore) encoded by Sin Nombre virus (SNV) and Andes virus (ANDV), which are two representative members that cause HCPS in the New World. The constructs of SNV and ANDV NPcore exclude the N- and C-terminal portions of full polypeptide to obtain stable proteins for crystallographic study. The structure features an N lobe and a C lobe to clamp RNA-binding crevice and exhibits two protruding extensions in both lobes. The positively charged residues located in the RNA-binding crevice play a key role in RNA binding and virus replication. We further demonstrated that the C-terminal helix and the linker region connecting the N-terminal coiled-coil domain and NPcore are essential for hantavirus NP oligomerization through contacts made with two adjacent protomers. Moreover, electron microscopy (EM) visualization of native RNPs extracted from the virions revealed that a monomer-sized NP-RNA complex is the building block of viral RNP. This work provides insight into the formation of hantavirus RNP and provides an understanding of the evolutionary connections that exist among bunyaviruses.
IMPORTANCE Hantaviruses are distributed across a wide and increasing range of host reservoirs throughout the world. In particular, hantaviruses can be transmitted via aerosols of rodent excreta to humans or from human to human and cause HFRS and HCPS, with mortalities of 15% and 50%, respectively. Hantavirus is therefore listed as a category C pathogen. Hantavirus encodes an NP that plays essential roles both in RNP formation and in multiple biological functions. NP is also the exclusive target for the serological diagnoses. This work reveals the structure of hantavirus NP, furthering the knowledge of hantavirus RNP formation, revealing the relationship between hantavirus NP and serological specificity and raising the potential for the development of new diagnosis and therapeutics targeting hantavirus infection.
INTRODUCTION
Hantavirus causes severe infectious diseases in human and animals (1, 2). In addition to 24 representative species, increasing numbers of antigenically and genetically distinct hantaviruses have recently been registered as new members of the genus Hantavirus (3, 4). The reservoir hosts of hantaviruses in nature are primarily rodents, shrews, moles, and bats, and the transmission of hantaviruses via aerosols of rodent excreta to humans causes two human diseases: (i) hemorrhagic fever with renal syndrome (HFRS), which is primarily caused by Hantaan virus (HTNV) in Asia, by Puumala virus (PUUV) and Dobrava virus (DOBV) in Europe, and by Seoul virus (SEOV) worldwide, and (ii) hantavirus cardiopulmonary syndrome (HCPS), which is caused by Sin Nombre virus (SNV) in North America and by Andes virus (ANDV) in Latin America (2). In particular, person-to-person transmission was reported during an HCPS endemic in South America (5). The worldwide occurrence of HFRS and HCPS outbreaks and the emergence of new hantaviruses emphasize the importance of furthering our understanding of hantavirus biology.
Hantavirus belongs to the genus Hantavirus of the family Bunyaviridae, which is the largest segmented negative-sense single-strand RNA (−ssRNA) virus family and is subdivided into the genera Orthobunyavirus, Hantavirus, Nairovirus, Phlebovirus, and Tospovirus (6, 7). As in other bunyaviruses, the genome of hantavirus is trisegmented, including large (L), middle (M), and small (S) segments. L segment encodes an RNA-dependent RNA polymerase (RdRp), M segment encodes a precursor of glycoproteins (Gn and Gc), and S segment encodes a nucleocapsid protein (NP). Throughout the replication cycle of hantavirus, genomic RNA must be encapsidated by NP to form a ribonucleoprotein (RNP) complex together with RdRp. The RNP either further serves as an active template for RNA synthesis or is packaged into the virion (8–10). Since 2010, a series of structural studies, including studies on the structures of NPs encoded by phleboviruses (Rift Valley fever virus [RVFV] [11–13] and severe fever with thrombocytopenia syndrome virus [SFTSV] [14]), nairovirus (Crimean-Congo hemorrhagic fever virus [CCHFV]) (6, 15, 16), and orthobunyaviruses (Bunyamwera virus [BUNV], etc.) (17–21), has advanced the understanding of the structures and functions of bunyavirus NPs.
Hantavirus NP has the second highest molecular mass (approximately 46 kDa) among all bunyavirus NPs and plays multiple roles in virus proliferation. Hantavirus NPs form contacts with other viral proteins, e.g., Gn (22) and RdRp (23), to regulate virus assembly and proliferation. Moreover, key studies revealed that hantavirus NP simultaneously binds both m7G caps and viral RNA (vRNA) at distinct sites, suggesting that hantavirus NP has key functions in cap snatching and viral mRNA synthesis (24). Furthermore, hantavirus NPs have been reported to regulate the functions of host eIFs (25) and ribosomal proteins (26, 27) to enhance the translation of viral mRNA. In particular, hantavirus NP not only is a target for host innate immunity against infection via its association with MxA-mediated antiviral response (28) but also has been found to interfere with host innate immune responses by downregulating apoptosis (29), inhibiting interferon (IFN) signaling responses (30, 31), and blocking tumor necrosis factor alpha (TNF-α)-induced activation of NF-κB (32, 33).
However, the structural information on hantavirus NP remains very limited. Previous results revealed that hantavirus NP undergoes trimerization (34, 35) and that NP trimers specifically recognize a conserved panhandle structure that is formed by the complementary base pairing of the 5′ and 3′ ends of the hantaviral genome (36, 37). Further results showed that the N-terminal domains of the ANDV (38) and SNV NPs (34) possess a coiled-coil architecture that is comprised of two antiparallel helices stabilized by conserved hydrophobic residues, while the C-terminal domain (residues 370 to 429) forms two helices; both domains are essential for NP oligomerization (35).
Because of the major impact of hantavirus on public health, efforts have been concentrated on developing diagnostics and therapeutics in recent decades. Hantavirus NP is the major antigen that elicits early serological responses in infected humans and has been used as a biomarker to develop antibodies for epidemiological surveillance in regions where various hantavirus species cocirculate (4, 39). NP has also been shown to elicit protection against hantavirus infection in animals and therefore was considered for use as a component of a vaccine (40, 41). Our interest in understanding the structures and functions of −ssRNA virus-encoded NPs, as well as in discovering relevant information to aid in the development of antihantavirus reagents, prompted us to initiate a crystallographic investigation of hantavirus NP.
MATERIALS AND METHODS
Protein production.
Codon-optimized cDNAs for full-length SNV NP and Andes virus NP were synthesized in GENEWIZ. SNV NPcore (residues 111 to 398) was subcloned into the pGEX-6p-1 (GE Healthcare) vector with BamHI and XhoI restriction sites using the cloning primers 5′-CCGGGATCCGAGCCAAGTGGGCAAACAG-3′ (forward) and 5′-GGCCTCGAGTTATCCAAGATGGAAGTG-3′ (reverse). Andes virus NPcore (residues 111 to 398) was subcloned into the pET-32a (Novagen) vector with BamHI and XhoI restriction sites using the cloning primers 5′-CGGGATCCGAGCCTACCGGTCAGACAG-3′ (forward) and 5′-GCCCTCGAGTTACAGGTGGAAGTTATCCAC-3′ (reverse). The accuracy of the inserts was verified by sequencing.
The recombinant plasmid of SNV NPcore was transformed into Escherichia coli strain BL21(DE3) and overexpressed as a glutathione S-transferase (GST) fusion protein. The cells were cultured at 37°C in 800 ml LB medium containing 100 μg/ml ampicillin. Once the optical density at 600 nm (OD600) reached 0.6, the culture was transferred to 16°C, and protein was induced by incubation with 0.5 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) (catalog no. A100487; Sangon Biotech, Shanghai, China) for an additional 20 h. Harvested cells were resuspended in lysis buffer containing 20 mM Tris-HCl (pH 8.5) (catalog no. A100826; Sangon Biotech), 500 mM NaCl (catalog no. A100241; Sangon Biotech), 5% (vol/vol) glycerol (catalog no. A100854; Sangon Biotech), 1 μg/ml DNase I (catalog no. B002138; Sangon Biotech), and 1 μg/ml RNase (catalog no. B500474; Sangon Biotech) and homogenized with a low-temperature ultrahigh-pressure cell disrupter (JNBIO). The lysate was centrifuged at 25,000 × g for 30 min at 4°C to remove cell debris. The supernatant was then loaded twice onto a GST column pre-equilibrated with lysis buffer, and the GST tag was removed by digestion with PreScission protease (catalog no. 27-0843-01; GE Healthcare) on a column overnight at 4°C. Eluted SNV NPcore protein was further purified by Superdex-200 gel filtration chromatography column (catalog no. 17-5175-01; GE Healthcare). SDS-PAGE analysis revealed over 95% purity of the final purified recombinant protein. The purified protein was then concentrated to 3 mg/ml in a buffer containing 20 mM Tris-HCl (pH 8.5), 200 mM NaCl, and 5% (vol/vol) glycerol.
The recombinant plasmid of ANDV NPcore was transformed into E. coli strain BL21(DE3) and overexpressed as a Trx tag and 6× His tag fusion protein. The cells were cultured at 37°C in 800 ml LB medium containing 100 μg/ml ampicillin. Once the OD600 reached 0.6, the culture was transferred to 20°C, and protein was induced by incubating with 0.2 mM IPTG for an additional 20 h. Harvested cells were resuspended in lysis buffer containing 20 mM HEPES (pH 7.5) (catalog no. A100511; Sangon Biotech), 500 mM NaCl and homogenized with a low-temperature ultrahigh-pressure cell disrupter (JNBIO). The lysate was centrifuged at 25,000 × g for 30 min at 4°C to remove cell debris. The fusion protein was first purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography and elution with lysis buffer, and the Trx tag and 6× His tag were removed by digestion with thrombin (catalog no. 27-0846-01; GE Healthcare) overnight at 4°C. Eluted Andes virus NP (residues 111 to 398) protein was further purified by Resource Q (catalog no. 17-1177-01; GE Healthcare) by using a Superdex-200 gel filtration chromatography column (GE Healthcare). SDS-PAGE analysis revealed over 90% purity of the final purified recombinant protein. The purified protein was then concentrated to 4 mg/ml in a buffer containing 20 mM HEPES (pH 7.5), 200 mM NaCl.
Crystallization.
The SNV NPcore and ANDV NPcore proteins were stored in the respective solutions described above, and initial crystallization conditions were screened by the hanging-drop vapor-diffusion method using commercial crystal screening kits at 16°C. Crystals were obtained by mixing 1 μl of the protein solution with an equal volume of a reservoir solution and equilibrating the mixed drop against 300 μl of reservoir solution.
Small crystals of SNV NPcore in long-axis form first appeared after 1 day in 200 mM lithium sulfate monohydrate and 20% (wt/vol) PEG 3350. The final optimized crystals in short-axis form were grown in 200 mM HEPES (pH 7.5), 200 mM lithium sulfate monohydrate, 23% (wt/vol) PEG 3350, and 3% d-sorbitol. Needle crystals grew to a final size of 10 μm by 10 μm by 220 μm within 3 days at 20°C. Crystals were harvested and cryoprotected in the well solution containing an additional 20% (vol/vol) glycerol and cooled in a dry nitrogen stream at 100 K for X-ray data collection.
Small crystals of ANDV NPcore first appeared after 1 day in 100 mM HEPES (pH 7.5), 200 mM l-proline, and 10% (wt/vol) PEG3350. The final optimized crystals were grown in 100 mM HEPES (pH 7.5), 200 mM l-proline, 9% (wt/vol) polyethylene glycol (PEG) 3350, and 3% (vol/vol) d-(+)-glucose monohydrate. Rod-like crystals grew to a final size of 50 μm by 50 μm by 220 μm within 2 days at 20°C. Crystals were harvested and cryoprotected in the well solution containing an additional 20% (vol/vol) glycerol and cooled in dry nitrogen stream at 100 K for X-ray data collection. All chemicals used the in crystallization process were purchased from Sigma-Aldrich.
X-ray data collection and processing and structure determination.
The selenomethionine single-wavelength anomalous dispersion (SAD) data set for SNV NPcore in the P6122 space group was collected at 3.0 Å using a wavelength corresponding to the Se peak at the BL17U beamline in Shanghai Synchrotron Radiation Facility (SSRF). Other native data sets were also collected at BL17U. All data sets were processed using the HKL2000 package (42). Seven of eight selenium atoms in one asymmetric unit were located and refined, and the SAD data phases were calculated and substantially improved by solvent flattening using the PHENIX program (43). A model was manually built into the modified experimental electron density using COOT (44) and further refined in PHENIX. Other structures were solved by using the molecular replacement method with the structure of SNV NPcore in long-axis form as the initial searching model and refined in PHENIX. Model geometry was verified using the program MolProbity (45). Structural figures were drawn using the program PyMOL (46). Data collection and the final refinement statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statisticsa
| Parameter | SNV NP |
ANDV NP (native) | |
|---|---|---|---|
| Native in C2221 | Se-SAD peak in P6122 | ||
| PDB code | 5E05 | 5E06 | 5E04 |
| Data collection statistics | |||
| Cell parameters | |||
| a (Å) | 49.7 | 49.3 | 52.5 |
| b (Å) | 74.9 | 49.3 | 99.7 |
| c (Å) | 154.4 | 419.9 | 137.4 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 | 90, 90, 90 |
| Space group | C2221 | P6122 | P212121 |
| Wavelength used (Å) | 0.9798 | 0.9798 | 1.0000 |
| Resolution (Å) | 50.0–2.3 (2.34–2.30) | 50.0–3.0 (3.05–3.00) | 50.00–2.25 (2.28–2.25) |
| No. of all reflections | 93,734 (4,634) | 108,042 (4,416) | 207,719 (4,505) |
| No. of unique reflections | 13,234 (662) | 7,053 (368) | 33,842 (1,204) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 90.8 (83.4) |
| Average I/σ(I) | 31.0 (6.0) | 45 (7.8) | 23.2 (2.9) |
| Rmergeb (%) | 11.8 (47.5) | 10.4 (32.3) | 11.2 (36.2) |
| Refinement statistics | |||
| Resolution | 50.0–2.3 | 50.0–3.0 | 50.00–2.25 |
| No. of reflections used [σ(F) > 0] | 13,200 | 6,937 | 33,764 |
| Rworkc (%) | 20.3 | 22.4 | 21.5 |
| Rfreed (%) | 23.1 | 28.9 | 25.2 |
| RMSD bond distance (Å) | 0.008 | 0.010 | 0.008 |
| RMSD bond angle (°) | 1.097 | 1.289 | 1.211 |
| Average B value (Å2): | |||
| For protein atoms | 33.5 | 76.2 | 40.3 |
| For ligand atoms | 42.7 | 52.1 | |
| No. of: | |||
| Protein atoms | 2,151 | 2,155 | 4,352 |
| Ligand atoms | 5 | ||
| Solvent atoms | 45 | 92 | |
| Ramachandran plot (% resolution): | |||
| In favored regions | 98.0 | 91.0 | 92.0 |
| In allowed regions | 1.6 | 7.5 | 8.0 |
| In outlier regions | 0.4 | 1.5 | 0.0 |
Numbers in parentheses are the corresponding values for the highest-resolution shell.
Rmerge = ΣhΣl | Iih − 〈Ih〉 |/ΣhΣI 〈Ih〉, where 〈Ih〉 is the mean of the observations Iih of reflection h.
Rwork = Σ(||Fp(obs)| − |Fp(calc)||)/Σ|Fp(obs)|.
Rfree is an R factor for a preselected subset (5%) of reflections that was not included in refinement.
EM of natural hantavirus RNPs.
HTNV (strain HV114) (47) virions released by infected Vero cells were produced and purified following a previously described procedure (48). The Vero cells were infected with HTNV at a multiplicity of infection (MOI) of 0.01. Supernatants were harvested at 7 days postinfection (p.i.), clarified at 4,500 rpm for 5 min at 4°C, and further filtered through a 0.45-μm filter to remove cell debris. The filtered supernatants were mixed with 0.5 M NaCl and 10% (wt/vol) PEG 6000 (Sigma), stirred on ice for 20 min, and incubated overnight at 4°C. Virions were precipitated by centrifugation at 3,000 rpm for 30 min at 4°C. The pellets were resuspended in cold TEN buffer (0.01 M Tris-HCl [pH 7.4], 1 mM EDTA, and 100 mM NaCl) and loaded on a 5 to 25% continuous Optiprep iodixanol density gradient (Sigma). The gradient was prepared by placing 5 layers (bottom to top) of 0.8 ml of 25%, 20%, 15%, 10%, and 5% (vol/vol) Optiprep iodixanol density gradient dissolved in TEN buffer with protease inhibitor (Roche). Each layer was frozen in dry ice before a new one was added. These tubes were kept frozen and placed at 4°C overnight before use. During melting, a continuous gradient is formed. The virus preparation was centrifuged at 28,000 × g for 1.5 h. Fractions of 500 μl were collected from the bottom of the gradient and treated with 1 M sucrose to release RNPs. RNPs were further purified by gradient ultracentrifugation by using the same gradient as that used to purify virions. The purified RNPs were adsorbed to electron microscopy (EM) grids covered by Formvar and carbon and made hydrophilic by glow discharge. Adsorbed RNPs were washed by floating the grids on two drops of TEN buffer before negative staining with 2% (wt/vol) uranyl acetate for 30 s. The samples were subsequently imaged on a JEOL JEM 1011 electron microscope operating at 100 kV.
Pulldown assays.
We performed pulldown assays to detect the impact of the residues located at the interprotomer interface on NP-NP homotypic contacts following a previously described procedure (10) by using wild-type (wt) and mutated NPCT, NPlinker, and wt NPcore as probes. The wt and mutated SNV NPCT were expressed and purified as GST-fused proteins. NPlinker and relevant mutations were expressed and purified with a C-terminal His tag to exclude the influence of the recombinant tag on NP-NP interactions. Equal amounts (100 μg) of GST-NPCT and His-NPlinker were bound to 50 μl glutathione-Sepharose or Ni-NTA beads, respectively, in 300 μl buffer. Identical amount (100 μg) wt NPcore with a Flag tag was subsequently added, and the mixture was incubated overnight at 4°C. The beads were then washed three times, and the complexes were eluted from the glutathione-Sepharose or Ni-NTA beads with reduced glutathione (GSH) solution or the buffer containing additional 200 mM imidazole. We then moved the supernatant to new Eppendorf tubes and added SDS loading buffer. We then incubated the supernatants in boiling water for 5 min and subjected the samples to SDS-PAGE followed by Western blot analysis with the antibody against the Flag tag.
Docking and molecular dynamics simulations.
Docking studies were performed using the software Hex 8.0 (49). The C-terminal region was placed near the binding surface of the main region. The correlation type was set to shape plus electron. The sampling method was set to range angles. Postprocessing was accomplished using optimized potentials for liquid simulations (OPLS) minimization. The steric san level was set to 18, and the final search level was set to 25. A total of 2000 solutions were clustered into 100 clusters. The best cluster was selected as the final docking result.
Molecular dynamics (MD) simulations were performed starting from the best docking pose of the C-terminal region. The N-terminal region was manually placed in the relative position based on the model structure of BUNV NP (PDB code 4IJS) (17). The parameterization of the protein was performed with the leap module of Amber 12 (50) using the ff99SB force field. Hydrogens were added. N termini were capped with acetyl, and C termini were capped with methylamine. Counterions were added to neutralize the whole system. Subsequently, the system was solvated in a cubic TIP3P explicit water box with a 10-Å buffer. Before MD simulations, the system was first minimized with 2,000 steps at the steepest descent minimization, followed by 2,000 steps at conjugate gradient minimization. Then, the system was heated from 0 K to 300 K during 200 ps in a constant-volume MD simulation using Langevin dynamics with a collision frequency gamma ln of 2.0. Then, 50 ns of constant-pressure MD simulations were performed at 300 K and a pressure of 1 atm. The SHAKE (51) algorithm was used to treat hydrogen-containing bonds. During this simulation, a harmonic force constraint of 5 kcal/mol · Å was applied to the main region. In contrast, no constraint was applied to the N-terminal or C-terminal regions. During the simulations, the particle mesh Ewald method was employed to calculate long-range electrostatic interactions. An 8-Å cutoff value was used for the nonbonded interactions, and a time step of 2 fs was used for the simulations. The MD trajectories were analyzed using the module ccptraj in AMBERTOOLS. Root mean square deviation (RMSD) was calculated for the backbone of each residue during the production stage. In particular, 100 snapshots were extracted for the last 5 ns of the production phase of the simulation and were used to evaluate the binding free energy between the N- and C-terminal regions and the main region of the protein using an MM/GBSA (52) approach.
EMSA.
Electrophoretic mobility shift assays (EMSA) were performed to evaluate potential differences between the SNV NPcore and RNA probe, following the conventional method, by using a commercial EMSA kit (chemiluminescent EMSA kit; Beyotime Institute of Biotechnology, Haimen, China). Biotin-labeled ssRNA (5′-UAAAGCUGGAAUGAGCACCC-3′, derived from Sin Nombre virus S segment viral genomic RNA) was synthesized by TaKaRa Biotechnology (Dalian, China). Each reaction mixture contained 0.1 nM wt NP or the relevant mutant and 0.15 nM RNA probe. After incubation for 1 h at 37°C, the reaction mixture was loaded on a 10% native polyacrylamide gel in 0.5% Tris-borate-EDTA, and then the protein-RNA complex was transferred and UV-cross-linked for 15 min onto nylon membranes (FFN12; Beyotime Institute of Biotechnology). Bands were visualized using a probe biotin-labeling kit (BeyoECL Plus; Beyotime Institute of Biotechnology).
Minigenome replicon activity.
The construction of a minigenome replicon with sequences from HTNV strain HV114 and the measurement of the activity of the chloramphenicol acetyltransferase (CAT) reporter gene were performed as previously described (53–55). Briefly, a minigenome cassette containing the CAT reporter gene flanked by the M-segment noncoding regions of the vRNA was cloned into an expression vector driven by a T7 bacteriophage RNA polymerase promoter (pT7-MCAT), which was used to transcribe the minigenome cassette containing the CAT reporter gene and the M-segment noncoding regions of the viral RNA (vRNA). The addition of RdRp (L) and NP proteins forms the ribonucleoprotein complex (RNP). The activity of the minigenome replicon was detected by measuring the activity of the reporter gene CAT. The quantitative determination of CAT was measured using a CAT enzyme-linked immunosorbent assay (ELISA) kit (Roche, Germany) following the manufacturer's protocol. Cells were lysed in lysis buffer for 30 min on ice. Aliquots of 200 ml of cell extracts were added to anti-CAT-coated microplate modules and incubated for 1 h at 37°C. Then, 200 ml of anti-CAT-digoxigenin (DIG) was pipetted into each of the modules, the mixtures were incubated for 1 h at 37°C, and subsequently, 200 ml of anti-DIG-peroxidase (POD) was added and incubated at 37°C for 1 h. Finally, POD substrate was added for a 20-min incubation. The absorptions of the samples were measured at 405 nm.
Protein structure accession numbers.
Structures have been deposited in PDB under the following accession numbers: ANDV NP, 5E04; SNV NP in short-axis form, 5E05; and SNV NP in long-axis form, 5E06.
RESULTS
Purification and characterization of SNV and ANDV NPs.
We selected two representative viruses, SNV and ANDV, that cause HCPS in the New World to study the structure of hantavirus NP. We initially expressed full-length SNV and ANDV NPs in Escherichia coli, but both had poor solution properties with heavy random nucleic acid binding and extensive inhomogeneous high-ordered multimerization.
Previous results demonstrated that the minimal 77 residues in the N-terminal coiled-coil domain (NPNCC) (34, 38) and the last 50 residues in the C-terminal tail (NPCT) (35) play critical roles in NP oligomerization. We speculated that these two regions may interfere with the solubility and stability needed for protein crystallization. A subset of constructs was therefore generated (Fig. 1A). Most of these constructs displayed solution properties similar to those of the full-length protein. The one exception was the construct that was comprised of residues 1 to 398 from SNV NP. However, purified SNV NP 1 to 398 presented clear degradation, and mass spectrum analysis demonstrated that NPNCC was spontaneously lost. This observation suggests that the linkage between SNV NPNCC and the core domain of NP (NPcore) is not stable, which is consistent with previous analyses in which residues 76 to 93 were found to display intrinsic disorder (34). Finally, a construct that spanned residues E111 to G398 of the SNV and ANDV NPs was successfully expressed, purified as a monomeric protein without nucleic acid binding, and crystallized (Fig. 1A and B).
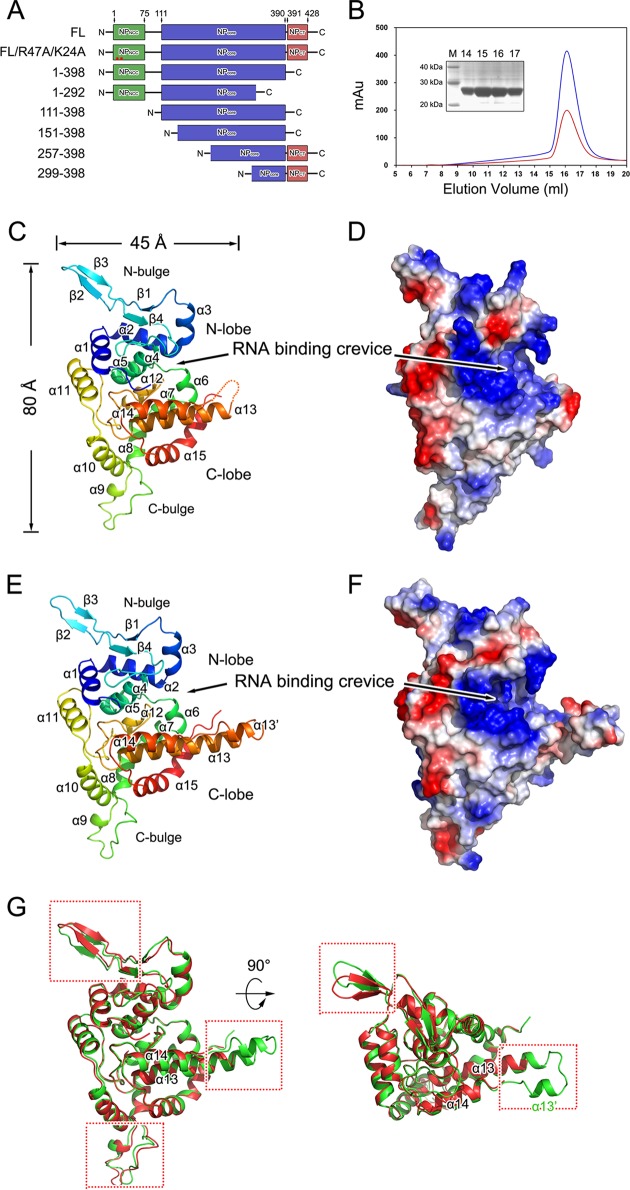
FIG 1.
Structures of SNV NP and ANDV NPs. (A) Schematic diagram of the construction of SNV NP. Previously suggested NPNCC, NPcore, and NPCT structures are in green, blue, and red, respectively. The residue numbers for each end are at the top. (B) Size exclusion chromatogram (SEC) of the construct covering residues E111 to G398, which was run on a Superdex-200 column. The blue and red lines indicate absorbance at 280 and 260 nm, respectively. The inset shows a 12% SDS-PAGE gel of the major peak in SEC. Ribbon representation of SNV (C) and ANDV (E) NPs with rainbow coloring from the N (blue) to the C terminus (red). Each structural portion and secondary structural element is labeled. (D and F) Electrostatic surfaces of SNV and ANDV NPs. Positive and negative charges are in blue and red, respectively. RNA-binding crevices, indicated by large positively charged residues, are noted. (G) Comparison of SNV and ANDV NPcore. The overall structures of SNV and ADNV NPcore are aligned and are shown in two perpendicular views. The SNV and ANDV NPcore molecules are shown as cartoon diagrams in red and green, respectively. Structural elements with obvious differences are highlighted by red frames.
Structure of SNV and ANDV NPcore.
SNV NPcore was crystallized in two different space groups: a long-axis form in the P6122 space group and a short-axis form in the C2221 space group (Table 1). The long-axis form was the first to be crystallized, but it diffracted less than 4.0 Å. Only one of the crystals (out of over 200) that was screened out with a 3.0-Å resolution to solve the structure using the single-wavelength anomalous dispersion method. To obtain more stable protein for improved crystallization and diffraction, we employed fluorescence-based thermal shift assays to search for potential hits that could increase the melting temperature (Tm). Among all tested compounds in an additive kit from Hampton Research, d-sorbitol displayed the most prominent hit, showing a 6°C shift in the Tm. By supplying d-sorbitol as an additive compound, we consequently obtained a short-axis crystal form with the best resolution, 2.3 Å, and the structure was solved by molecular replacement (MR) using a refined long-axis structure as a search model. Although the c axis in the long-axis form is rather large (420 Å), the final model suggested that there is no potential oligomerized architecture that is relayed by crystallographic symmetry. ANDV NPcore was crystallized in the P212121 space group and solved with MR by using the SNV NPcore structure as the searching model (Table 1).
SNV and ANDV NPcore fold into a compact body that exhibits two horn-like extensions, which include 15 or 16 α-helices and 4 β-strands (Fig. 1C to F). The body of SNV NPcore contains two relatively separate portions: an N lobe (G114 to P218) and a C lobe (V219 to L397). The N lobe is comprised of a helical core that contains α1 to α5 and a bulge structure (N bulge) (N152 to M181), which folds into four β-strands in an antiparallel manner. The C lobe is predominately formed by α-helices (α6 to α15) and possesses a C-terminal bulge (C bulge) (A242 to N261) that is comprised of a short α-helix (α9) and a long extended structure. The RNA-binding site was indicated as a large positively charged crevice located at the interface of the N and C lobes.
The overall structures of SNV and ANDV NPcore show high structural similarities, whereas N and C bulges present obvious differences (Fig. 1G). In particular, the linkage between M342 and S358 of SNV NP is completely missing in the final model but forms an additional α13′ in ANDV NP (Fig. 1G). The structural variations between SNV and ANDV NPcore are likely to be caused by crystal packing and indicate the mobility of these parts during the formation of the high-ordered RNP. A previous analysis suggested that residues 370 to 428 of SNV NP form two helical structures (helix 1 includes residues 373 to 387; helix 2 includes residues 404 to 420) and further organize the C-terminal tail for trimerization (35). In the structures of SNV and ADNV NPcore, residues 370 to 390, which are suggested to create the first helix of the C-terminal domain (35), fold into a bent α15 to constitute the C lobe, while the extended region (residues 390 to 397) presents flexible architecture, suggesting that the orientation of the last helix (residues P405 to E420) with NPcore is alterable. Therefore, the composition of the C-terminal tail of hantavirus NP (NPCT) should be refined to include residues 390 to 429. A previous result from a mammalian two-hybrid system demonstrated that G389P reduced 50% of NP homotypic interactions but G399P did not (35). It is reasoned that G389 is located at the turning point of the bent α15, and therefore the mutation G398P restricts the correct folding of NP, whereas G399 lies in the intrinsic flexible region that connects NPcore and NPCT, and thus its replacement with a proline has only a very slight influence. Similarly, the residues following K77 in NPNCC were shown to be structurally disordered (34), and the residues beyond G114 were also missing electron density, indicating that the orientation of NPNCC-NPcore is also variable.
Comparison with other −ssRNA viruses encoding NPs.
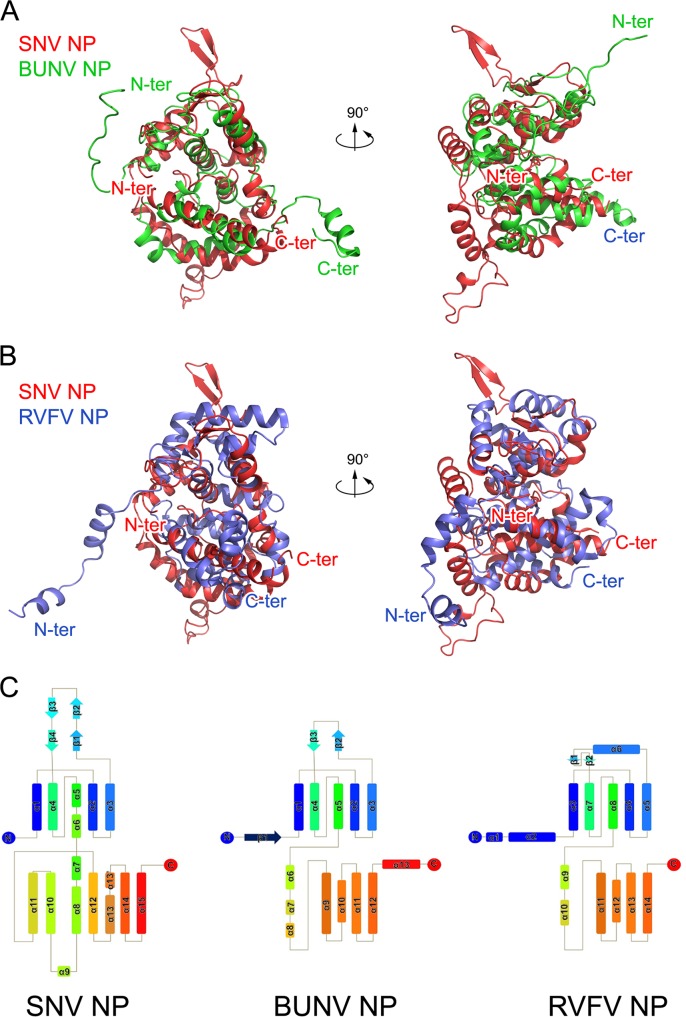
Virus-encoded NPs present large structural varieties (8, 9). We first performed structural comparisons to determine whether hantavirus NP has novel protein folding among members of the viral NP family. The comparison of SNV and ANDV NPcore with structures in the Protein Data Bank (PDB) using the DALI server (56) yielded two clusters of structural homologues. The most significant matches, based on the highest Z scores and the lowest RMSD values, included the structures of NPs encoded by orthobunyaviruses, including BUNV (PDB code 4IJS) (17), Leanyer virus (PDB code 4J1J) (57), Schmallenberg virus (PDB code 4JNG) (18), and La Crosse virus (PDB code 4BGP) (20). The Z score between SNV NPcore and orthobunyavirus NPs is high, 8.8, and the RMSD of 175 aligned residues compared against all 232 residues of orthobunyavirus NP is low, 3.4 Å, indicating that close structural similarities exist between these proteins (Fig. 2A). Although sequence identity based on the structural alignment of SNV NPcore with orthobunyavirus NPs is not visible (data not shown), their topologies are remarkably similar (Fig. 2C). The large variations appear at the N and C bulges, which exist in the structure of the hantavirus NPcore and flank the body part of NPcore, but do not have counterparts in orthobunyavirus NP. Actually, orthobunyavirus NP also has two β-strands in its N lobe, but they attach to other structural components to form a compact architecture (Fig. 2A and C).
FIG 2.
Comparison of hantavirus NP with orthobunyavirus and phlebovirus NPs. (A and B) The structures of BUNV (orthobunyavirus) NP (A) and RVFV (phlebovirus) NP (B) are aligned with that of SNV NP, and they are shown in two perpendicular views. Polypeptides of SNV, BUNV, and RVFV NPs are in red, green, and blue, respectively. The N- and C-terminal ends of each molecule are indicated. (C) Topologies of SNV, BUNV, and RVFV NPs. Helices and strands are, respectively, presented as rectangles and arrows and are rainbow colored, where blue and red indicate N- and C-terminal ends.
Additionally, hantavirus NPcore also exhibits homology with NPs encoded by phleboviruses, including RVFV (PDB code 4H5L) (11–13), SFTSV (PDB code 4J4X) (14, 19), and Toscana virus (PDB code 4CSG) (58). The Z score between SNV NPcore and the phlebovirus NP is approximately 5.8, and the RMSD of 148 aligned residues is 4.5 Å, suggesting that they have partial structural homology (Fig. 2B and C).
Interprotomer interactions.
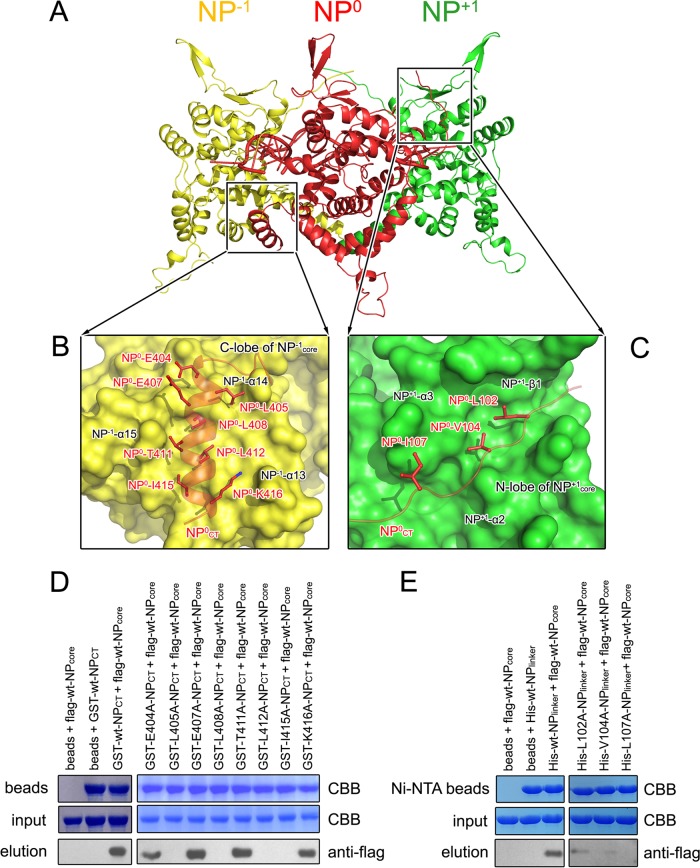
Oligomerization is a key feature of viral NP, and it is well known that the N-terminal coiled-coil portion and C-terminal portion are critical for hantavirus NP oligomerization. We therefore carried out further experiments to obtain detailed information on oligomerization based on core region structure. Considering the high structural similarity between orthobunyavirus and hantavirus NPs, it is reasonable to propose that hantavirus may also utilize the core region for RNA binding and the N-terminal and C-terminal portions for oligomerization. By comparing with orthobunyavirus and phlebovirus NP multimers, a model for SNV NPcore oligomer was built (Fig. 3A). Interprotomer interactions were found to occur in two separate regions: (i) NP0CT, with the C lobe of a neighboring protomer on the left (NP−1), and (ii) the linker NP0core-NP0NCC, with the N lobe of a different adjacent protomer on the right (NP+1) (Fig. 3A and B). To visualize the interprotomer interactions, we employed molecular dynamic stimulations to analyze the contacts between either the structure of NPCT or the linker of NPcore-NPNCC with NPcore. The model with the best docking scores (−331) and lowest binding energy (−53 kcal/mol) was selected, and the final complexes were quite stable, remaining intact during 50 ns of molecular dynamics simulation.
FIG 3.
Interprotomer interactions. (A) A model of SNV NP oligomerization was built according to its high structural homology with orthobunyavirus NP. Three adjacent SNV NP molecules, including NPCT and the linker NPcore-NPNCC, are shown as cartoons in red, yellow, and green. (B and C) Contact of NPCT of the NP0 protomer with the C lobe of the NP−1 protomer and the contact of the linker NPcore-NPNCC with the N lobe of the NP+1 protomer. (D and E) Pulldown assays to verify the effects of mutations on NP homotypic interactions in the region of NPCT-NPcore (D) and the linker NPcore-NPNCC with NPcore (E).
The C-terminal interprotomer interaction occurred between NP0CT and the C lobe of the NP−1 protomer (Fig. 3B). The residues E404, L405, E407, L408, R411, L412, I415, and K416 of the NP0 protomer, particularly their hydrophobic side chains, occupied a hydrophobic groove formed by residues from the last three α-helices (α13, α14, and α15) in the C lobe of NP−1 protomer. Moreover, according to the length of the linker region connecting NPcore and NPNCC, this region interacts with the N lobe of NP+1 protomer on the right (Fig. 3C). Residues A137, M140, and L141 of α2, T148 of α3, and I157 to F159 of β1 in the NP+1 protomer form a relatively hydrophobic pocket to accommodate the hydrophobic side chains L102, V104, and I107 of the NP0 protomer, mainly contributing to the N-terminal interaction. Additionally, a subset of hydrogen bonds connecting the main atoms of G99 to I107 with the residues of NPcore helps stabilize this N-terminal interaction.
To further verify the impacts of the residues at the NP0CT-NP−1 interface, we replaced these residues with alanines and evaluated whether these mutations disrupted the homotypic interaction using pulldown assays (Fig. 3D). We first expressed the wild-type (wt) and mutated polypeptides of G398-R418 of SNV NP as GST-fused proteins and bound identical amounts of GST-fused proteins onto the beads. Subsequently, Flag-tagged wt SNV NPcore was used as a probe to test the impact of the SNV NPCT substitutions on interprotomer interactions. The results showed that four substitutions, L405A, L408A, L412A, and I415A, led to the most substantial attenuation of NP-NP contacts, indicating the critical roles of these hydrophobic residues in interprotomer interactions. The substitution mutants of E404 and K416 retained over 50% of their interprotomer binding abilities. Other mutations, i.e., E407A and T411A, did not obviously affect the intermolecular interactions.
To examine the N-terminal interaction, we constructed the polypeptide G99-G398 from SNV NP (named NPlinker), which included a C-terminal His tag to exclude the influence of the recombinant tag on NP-NP interactions, and replaced L102, V104, and I107 with alanines. Identical amounts of His-tagged SNV NPlinker with and without mutations were bound to Ni-NTA beads and probed using a Flag-tagged wt SNV NPcore monomer to assess the impacts of the substitutions (Fig. 3E). The results showed that all three substitutions, L102A, V104A, and I107A, eliminated interactions with the wt SNV NPcore, supporting their essential roles in forming NP homotypic contacts. Consistent with our structural observations, a previous work demonstrated that residues 100 to 125 (particularly residue W119) and 404 to 429 were essential for NP-NP interactions in a yeast expression system (59). Collectively, these results supported the model that both SNV NPCT and the linker of NPcore-NPNCC contribute to the oligomerization of hantavirus NP.
RNA-binding crevice.
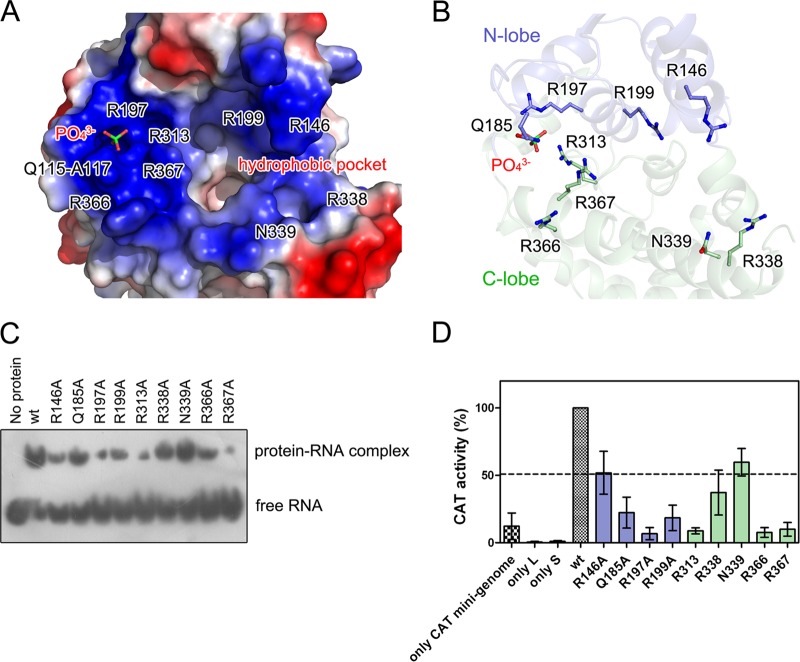
The RNA-binding crevice of hantavirus NP is clamped by the N and C lobes (Fig. 1D and F). The basic residues of the viral NP are known to provide positive charges that enable interactions with the phosphate moieties of nucleic acids. The side chains of a subset of positively charged residues from α1 to α4 in the N lobe, as well as α6/α14/α15 and the linkage of α12 and α13, predominantly contribute to the constitution of the RNA-binding crevice, including the residues R146, Q185, R197, R199, R313, R338, N339, R366, and R367 (Fig. 4A and B). All of these residues are highly conserved, indicating that an identical RNA binding mechanism exists throughout the Hantavirus genus.
FIG 4.
RNA-binding crevice of hantavirus NP. (A) Electric potential surface of the RNA-binding crevice in SNV NPcore. The residues that provide positive charges are labeled. Bound PO43+ is shown as colored sticks. The deep pocket is indicated. To show the bound PO43+, the surfaces of residues P182 to E192 were removed. (B) Localization of positively charged residues that orchestrate RNA binding in SNV NP. SNV NPcore is shown as a semitransparent cartoon diagram, in which the N and C lobes are in blue and green, respectively. The basic residues and a bound PO43+ are displayed as colored sticks. (C) Binding affinities of the recombinant wt and mutational SNV NPcore with probe RNA. (D) Impacts of positively charged residues on SNV replication. The histogram shows the activity of the chloramphenicol acetyltransferase (CAT) reporter gene in SNV NP mutants, which are colored as in panel B and are normalized to wt SNV NP. Each experiment was performed in three replicates, and error bars indicate standard deviations.
A phosphate anion was found to tightly bind into a positively charged pocket (Fig. 4A). This region is constituted by the side chains of R197/R313/R367 and by the nitrogen atoms of the residues Q115-A117 (Fig. 4A and B). Moreover, the aromatic side chain W119 helps stabilize the conformation of this pocket to anchor the phosphate anion. The phosphate anion's presence highlights the idea that these residues are potentially involved in RNA binding. Interestingly, W119 was reported to significantly impact NP-NP interactions (59), suggesting a dual function of W119 in RNP formation.
To investigate the influence of these positively charged residues to genome encapsidation, we employed an electrophoretic mobility shift assay (EMSA) to test the binding affinity of SNV NPcore and relevant mutations with RNA (Fig. 4C). The results of EMSA showed that the replacements of R197, R313, and R367 with alanine residues resulted in the most significant decrease in binding to probe RNA, while the mutations R146A, R199A, and R366A resulted in modest attenuation to the binding with probe RNA. In a contrast, the replacement of Q185, R338, and N339 with alanines had only a slight impact on RNA binding.
To further verify the impact of the residues in RNA-binding crevice on hantavirus replication, we evaluated the effect of substitutions for these residues on a reported hantavirus replicon system (53–55). Results showed that replacing R197, R313, R366, and R367 each with an alanine significantly attenuated replicon activity to less than 10% of that of wt NP (Fig. 4D). Additionally, the mutations Q185A and R199A of NP diminished replicon activity to approximately 20% that of wt NP. The mutations R146A, R338A, and N339A resulted in modest attenuation of replicon activity, retaining approximately 50% to 60% of the activity. The impacts of most of these positively charged residues on replicon activity are consistent with their influences on RNA binding. Notably, R366A showed modest attenuation of RNA binding but clearly reduced replicon activity; R146A obviously diminished RNA binding but retained 50% of replicon activity. This discrepancy needs further structural evidence for the hantavirus NP-RNA or NP-RdRp complex to be verified. All of these results support the idea that these positively charged residues play essential roles in hantavirus proliferation.
A distinct feature of the NPs encoded by orthobunyavirus and phlebovirus compared with other viral NPs is that orthobunyavirus and phlebovirus NPs sequester several RNA bases within a deep hydrophobic pocket inside the RNA-binding crevice (11, 17). Similarly, we found a remarkably deep hydrophobic pocket in the RNA-binding crevice of the SNV and ANDV NPcore (Fig. 4A). This pocket is constituted by a subset of hydrophobic residues, including M220, F307, S310, F331, and Y364, and is very similar to the hydrophobic pocket in RVFV NP, which sequesters nucleotide B2 (11), and to that in BUNV NP, which binds nucleotides 6 and 7 (17).
Visualization of the native SNV RNP.
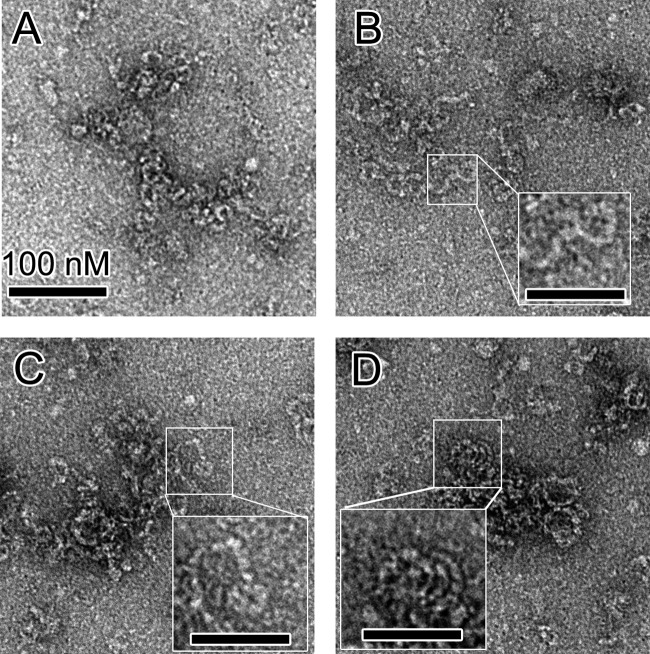
The native bunyavirus RNPs prefer to adopt a linear state without clear helical architecture, suggesting that a monomer-sized NP-RNA complex is the building block of bunyavirus RNP (11, 12, 17, 20). To study the high-ordered architecture of hantavirus RNP, we extracted native RNPs from HTNV (strain HV114) virions and visualized them using negative-stain EM (Fig. 5).
FIG 5.
EM images of native hantavirus RNPs extracted from virions. (A, B, and C) Most native HNTV RNPs displayed relaxed and linear architectures with widths of approximately 10 nm. (D) In some visualizations, select regions of the native RNPs displayed apparent helical characteristics (inset). Bars, 100 nm (A) and 50 nm (B, C, and D).
Native RNP from the HTNV virion exhibited a linear structure without remarkable helical symmetry, which was similar to observations of phlebovirus and orthobunyavirus RNPs. The width of the virion-extracted HTNV RNP is approximately 80 to 100 Å and was consistent with the size of a hantavirus NP monomer (Fig. 1C), suggesting that the monomer-sized NP-RNA complex is likely the building block of the linear hantavirus RNP. Interestingly, in some views of EM images, a relaxed helical architecture with a radius of approximately 250 to 300 Å was observed (Fig. 5D), suggesting that the further formation of a high-ordered helical structure would be facilitated during the packaging of RNP into mature virions. This observation strengthens the unique features of bunyavirus RNP relative to other −ssRNA virus RNPs.
DISCUSSION
Comparison of the mechanism of hantavirus RNP formation with other −ssRNA viruses.
NPs encoded by most −ssRNA viruses possess N and C lobes that face each other to form a positively charged crevice for RNA binding but use diverse structural components to create interprotomer interactions (8, 9). The exceptions are the NPs encoded by Lassa fever virus (LAFV; family Arenaviridae) (10, 60, 61) and CCHFV (genus Nairovirus, family Bunyaviridae) (6). Although LAFV and CCHFV belong to different virus families and their NPs were found to have additional biological functions, their structural regions used for genome encapsidation are highly similar and distinct from other −ssRNA virus NPs (8, 9). The molecular weight of hantavirus NP is almost 2-fold more than that of orthobunyavirus and phlebovirus NPs, but the core region of hantavirus NP exhibits bona fide structural homologies with orthobunyavirus NP and partially with phlebovirus NP (Fig. 2). They all use extensions in their N- and/or C-terminal ends for oligomer formation and encapsidate RNA in a positively charged groove within their cores. This structural information suggests that evolutionary relationships exist between orthobunyavirus, phlebovirus, and hantavirus within the family Bunyaviridae and also implies that nairovirus may have a closer connection with arenavirus.
Notably, a number of works have shown that hantavirus NP exists as trimer and that trimerization is essential for hantavirus replication (34–36). Combining the structural information on hantavirus NPcore with other structural information on hantavirus NP shows that the linkages NPNCC-NPcore and NPcore-NPCT have highly flexible architecture, suggesting that the orientations of NPNCC-NPcore and NPcore-NPCT can undergo substantial shifting during the oligomerization of hantavirus NP and further RNP formation. Moreover, the linear architecture of the native RNP extracted from the hantavirus virion supports the idea that the monomer-sized NP-RNA complex could be the building block of hantavirus RNP (Fig. 6). Furthermore, conformational shifts between the core region and N- and/or C-terminal extensions are also observed during orthobunyavirus and phlebovirus NP oligomerization, and all orthobunyavirus, phlebovirus, and hantavirus RNPs present flexible arrangements in vitro without clear symmetrical structures (11–13, 17, 20, 57). Collectively, this evidence indicates that trimerization is one, but not the most essential, oligomerization state of hantavirus NP and suggests that the structural mobility of N- and/or C-terminal regions of NP during RNP formation is conserved among all bunyaviruses.
FIG 6.
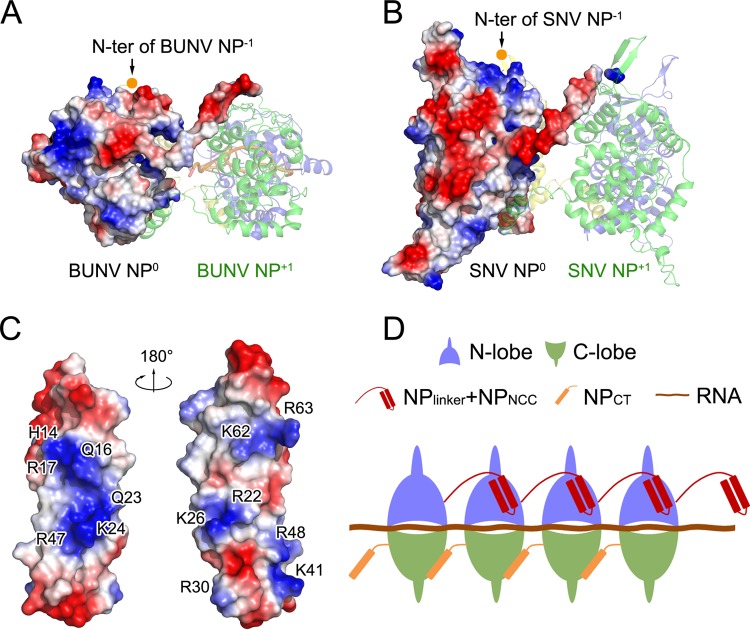
Implication of the role of hantavirus NPNCC in NP oligomerization. (A and B) Comparison of the BUNV NP-RNA tetramer (PDB code 4IJS) and a model of the SNV NP oligomer. For each tetramer, NP0 was covered with an electrostatic surface potential, while the other three protomers are presented as colored cartoon diagrams. (C) Electrostatic surface potential of SNV NPNCC (PDB code 2IC6) in two views. The basic residues on the surface are labeled. (D) Schematic model of the linear hantavirus RNP formed using the monomer-sized NP RNA as a building block.
Role of NPNCC in hantavirus RNP formation.
Previous results revealed that hantavirus NPNCC is essential for NP oligomerization (34, 35). Unlike orthobunyavirus NP, hantavirus NPNCC is unique to hantavirus NP and thus cannot be fitted into the oligomeric model that was generated based on the structure of an orthobunyavirus NP oligomer. In the model of a hantavirus NP oligomer, a highly negatively charged region can be observed on the surface of NPcore on the side close to the adjacent protomer on the right, but BUNV NP does not display this feature (Fig. 6A and B). Structural studies of SNV and ANDV NPNCC have shown that a set of basic residues constitute positively charged regions on NPNCC (Fig. 6C). We speculate that NPNCC interacts with NPcore via contacts between these positive and negative charges and thus helps stabilize the interprotomer of NP for the formation of RNP (Fig. 6D). This warrants further structural investigation of the oligomer of full-length hantavirus NP.
Implications for the diagnostics of and therapeutics against hantavirus.
Hantavirus infections cause various diseases and pathogenicities in humans depending on the hantavirus species (62). Therefore, early diagnoses of suspected infections, as well as the surveillance of infected animals, are crucial for the control of hantavirus infection. The current widely used strategy for hantavirus serological screening is first carried out using either ELISA or an immunochromatographic (ICG) strip test that recognizes 103 amino acids from the N-terminal region of NP, and positive sera must be further examined for serotyping by using an NP that lacks the N-terminal domain antigen (4).
Among representative viruses from three phylogenetic clusters of hantavirus, a total of 123 invariant residues are concentrated in the N lobe and the C-terminal half of the C lobe of NPcore, as well as NPCT. The side of NPcore that was facing the inner side of RNP, in particular the RNA-binding crevice, possesses the highest degree of conservation among all types of hantavirus (Fig. 7A). In sharp contrast, the residues on the side of hantavirus NPcore that face the outer side of RNP, in particular the C bulge, are variable. Because this region is exposed to solvent during the virus life cycle, it is conceivable that it could be used as a target to develop clinically useful agents for hantavirus early diagnosis and serotyping. Consistently, epitopes on SNV and ANDV NPs, including residues 244 to 263 and 274 to 286, which are located within the most variable regions of the C bulge and the C lobe, exhibit the most significant serotype specificity and have led to the generation of monoclonal antibodies (MAbs) that can be used to distinguish these two viruses (4) (Fig. 7B).
FIG 7.
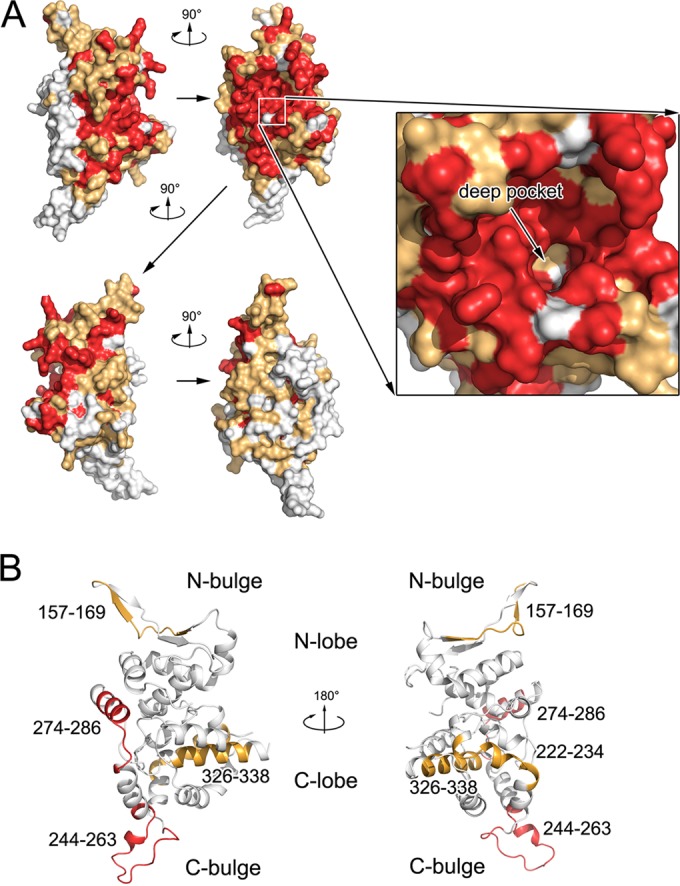
Epitopes in hantavirus NPcore. (A) Sequence conservation mapping on the surface of hantavirus NPs. Primary sequence homologies of representative hantaviruses are defined, and the structure of SNV NPcore is shown in front, side, and back views, covering its molecular surface. The deep pocket inside the RNA-binding crevice is enlarged for detail. Strictly conserved residues are in red, while conserved and variable residues are in gold and white, respectively. (B) Epitopes related to serotype specificity in hantavirus NPcore. Previously reported epitopes on NPcore that can distinguish SNV and ANDV (64) are highlighted in the structure of SNV NPcore. The two regions with the most serotype specificity (i.e., 244 to 263 and 274 to 286) are in red; the other identified epitopes are in gold.
Hantavirus NP is a potential target for the discovery of antihantavirus reagents. The correct formation and function of RNP constitute a key step for the proliferation of all −ssRNA viruses, and NP was recently identified as a druggable target, which was accomplished by either interfering with correct oligomerization (63) or blocking RNA encapsidation (19). In this study of hantavirus NP, we found that the deep hydrophobic pocket inside the RNA-binding crevice is similar to the hydrophobic pocket in RVFV NP, which sequesters nucleotide B2 (11), and to that in BUNV NP, which binds nucleotides 6 and 7 (17). This could lead to the discovery of new compounds that bind to hantavirus NP in this deep hydrophobic pocket and further disturb the encapsidation of the viral genome. Moreover, the NPNCC of one NP protomer binds to a relatively hydrophobic groove of an adjacent NP protomer. A chemical compound or peptide mimetic that could bind to this groove with high affinity is very likely to disrupt RNP formation and thus inhibit hantavirus proliferation.
ACKNOWLEDGMENTS
We thank the staff of beamline BL17U at the Shanghai Synchrotron Radiation Facility for their assistance with diffraction data collection. We also thank Wang X. from Tsinghua University for her help with EM imaging.
This work was supported by the Ministry of Science and Technology 973 Program (2013CB911103 and 2014CB542800), the National Natural Science Foundation of China (81322023, 31300606, 31170678, 31370733, and 31000332), the Importation and Development of High-Caliber Talents Project of Beijing Municipal Institutions (YETP0166), the Tsinghua University Initiative Scientific Research Program (20131089228), and the Tianjin Municipal Natural Science Foundation (14JCQNJC10100 and 10ZCKFSY08800).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Bennett SN, Gu SH, Kang HJ, Arai S, Yanagihara R. 2014. Reconstructing the evolutionary origins and phylogeography of hantaviruses. Trends Microbiol 22:473–482. doi: 10.1016/j.tim.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaheri A, Strandin T, Hepojoki J, Sironen T, Henttonen H, Makela S, Mustonen J. 2013. Uncovering the mysteries of hantavirus infections. Nat Rev Microbiol 11:539–550. doi: 10.1038/nrmicro3066. [DOI] [PubMed] [Google Scholar]
- 3.Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist Å, Schmaljohn CS, Tesh RB. 2010. Bunyaviridae, p 725–741. In King AMQ, Lefkowitz EJ, Adams MJ, Carstens EB (ed), Virus taxonomy: classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomyof Viruses. Elsevier, San Diego, CA. [Google Scholar]
- 4.Yoshimatsu K, Arikawa J. 2014. Serological diagnosis with recombinant N antigen for hantavirus infection. Virus Res 187:77–83. doi: 10.1016/j.virusres.2013.12.040. [DOI] [PubMed] [Google Scholar]
- 5.Martinez VP, Bellomo C, San Juan J, Pinna D, Forlenza R, Elder M, Padula PJ. 2005. Person-to-person transmission of Andes virus. Emerg Infect Dis 11:1848–1853. doi: 10.3201/eid1112.050501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Y, Wang W, Ji W, Deng M, Sun Y, Zhou H, Yang C, Deng F, Wang H, Hu Z, Lou Z, Rao Z. 2012. Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses. Proc Natl Acad Sci U S A 109:5046–5051. doi: 10.1073/pnas.1200808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guu TS, Zheng W, Tao YJ. 2012. Bunyavirus: structure and replication. Adv Exp Med Biol 726:245–266. doi: 10.1007/978-1-4614-0980-9_11. [DOI] [PubMed] [Google Scholar]
- 8.Zhou H, Sun Y, Guo Y, Lou Z. 2013. Structural perspective on the formation of ribonucleoprotein complex in negative-sense single-stranded RNA viruses. Trends Microbiol 21:475–484. doi: 10.1016/j.tim.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 9.Sun Y, Guo Y, Lou Z. 2012. A versatile building block: the structures and functions of negative-sense single-stranded RNA virus nucleocapsid proteins. Protein Cell 3:893–902. doi: 10.1007/s13238-012-2087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qing J, Wang Y, Sun Y, Huang J, Yan W, Wang J, Su D, Ni C, Li J, Rao Z, Liu L, Lou Z. 2014. Cyclophilin A associates with enterovirus-71 virus capsid and plays an essential role in viral infection as an uncoating regulator. PLoS Pathog 10:e1004422. doi: 10.1371/journal.ppat.1004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raymond DD, Piper ME, Gerrard SR, Skiniotis G, Smith JL. 2012. Phleboviruses encapsidate their genomes by sequestering RNA bases. Proc Natl Acad Sci U S A 109:19208–19213. doi: 10.1073/pnas.1213553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raymond DD, Piper ME, Gerrard SR, Smith JL. 2010. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc Natl Acad Sci U S A 107:11769–11774. doi: 10.1073/pnas.1001760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferron F, Li Z, Danek EI, Luo D, Wong Y, Coutard B, Lantez V, Charrel R, Canard B, Walz T, Lescar J. 2011. The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog 7:e1002030. doi: 10.1371/journal.ppat.1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou H, Sun Y, Liu M, Wang Y, Liu C, Wang W, Liu X, Li L, Deng F, Guo Y, Lou Z. 2013. The nucleoprotein of severe fever with thrombocytopenia syndrome virus processes an oligomeric ring to facilitate RNA encapsidation. Protein Cell 4:445–455. doi: 10.1007/s13238-013-3901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter SD, Surtees R, Walter CT, Ariza A, Bergeron E, Nichol ST, Hiscox JA, Edwards TA, Barr JN. 2012. Structure, function, and evolution of the Crimean-Congo hemorrhagic fever virus nucleocapsid protein. J Virol 86:10914–10923. doi: 10.1128/JVI.01555-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, Liu X, Wang X, Dong H, Ma C, Wang J, Liu B, Mao Y, Wang Y, Li T, Yang C, Guo Y. 5 August 2015. Structural and functional diversity of nairovirus-encoded nucleoproteins. J Virol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Wang Q, Pan X, Fernandez de Castro I, Sun Y, Guo Y, Tao X, Risco C, Sui SF, Lou Z. 2013. Bunyamwera virus possesses a distinct nucleocapsid protein to facilitate genome encapsidation. Proc Natl Acad Sci U S A 110:9048–9053. doi: 10.1073/pnas.1222552110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ariza A, Tanner SJ, Walter CT, Dent KC, Shepherd DA, Wu W, Matthews SV, Hiscox JA, Green TJ, Luo M, Elliott RM, Fooks AR, Ashcroft AE, Stonehouse NJ, Ranson NA, Barr JN, Edwards TA. 2013. Nucleocapsid protein structures from orthobunyaviruses reveal insight into ribonucleoprotein architecture and RNA polymerization. Nucleic Acids Res 41:5912–5926. doi: 10.1093/nar/gkt268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiao L, Ouyang S, Liang M, Niu F, Shaw N, Wu W, Ding W, Jin C, Peng Y, Zhu Y, Zhang F, Wang T, Li C, Zuo X, Luan CH, Li D, Liu ZJ. 2013. Structure of severe fever with thrombocytopenia syndrome virus nucleocapsid protein in complex with suramin reveals therapeutic potential. J Virol 87:6829–6839. doi: 10.1128/JVI.00672-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reguera J, Malet H, Weber F, Cusack S. 2013. Structural basis for encapsidation of genomic RNA by La Crosse orthobunyavirus nucleoprotein. Proc Natl Acad Sci U S A 110:7246–7251. doi: 10.1073/pnas.1302298110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong H, Li P, Bottcher B, Elliott RM, Dong C. 2013. Crystal structure of Schmallenberg orthobunyavirus nucleoprotein-RNA complex reveals a novel RNA sequestration mechanism. RNA 19:1129–1136. doi: 10.1261/rna.039057.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganaie SS, Mir MA. 2014. The role of viral genomic RNA and nucleocapsid protein in the autophagic clearance of hantavirus glycoprotein Gn. Virus Res 187:72–76. doi: 10.1016/j.virusres.2013.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng E, Wang Z, Mir MA. 2014. Interaction between hantavirus nucleocapsid protein (N) and RNA dependent RNA polymerase (RdRp) mutants reveals the requirement of an N-RdRp interaction for viral RNA synthesis. J Virol 88:8706–8712. doi: 10.1128/JVI.00405-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mir MA, Sheema S, Haseeb A, Haque A. 2010. Hantavirus nucleocapsid protein has distinct m7G cap- and RNA-binding sites. J Biol Chem 285:11357–11368. doi: 10.1074/jbc.M110.102459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mir MA, Panganiban AT. 2008. A protein that replaces the entire cellular eIF4F complex. EMBO J 27:3129–3139. doi: 10.1038/emboj.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haque A, Mir MA. 2010. Interaction of hantavirus nucleocapsid protein with ribosomal protein S19. J Virol 84:12450–12453. doi: 10.1128/JVI.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng E, Haque A, Rimmer MA, Hussein IT, Sheema S, Little A, Mir MA. 2011. Characterization of the Interaction between hantavirus nucleocapsid protein (N) and ribosomal protein S19 (RPS19). J Biol Chem 286:11814–11824. doi: 10.1074/jbc.M110.210179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanerva M, Melen K, Vaheri A, Julkunen I. 1996. Inhibition of Puumala and Tula hantaviruses in Vero cells by MxA protein. Virology 224:55–62. doi: 10.1006/viro.1996.0506. [DOI] [PubMed] [Google Scholar]
- 29.Park SW, Han MG, Park C, Ju YR, Ahn BY, Ryou J. 2013. Hantaan virus nucleocapsid protein stimulates MDM2-dependent p53 degradation. J Gen Virol 94:2424–2428. doi: 10.1099/vir.0.054312-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cimica V, Dalrymple NA, Roth E, Nasonov A, Mackow ER. 2014. An innate immunity-regulating virulence determinant is uniquely encoded by the Andes virus nucleocapsid protein. mBio 5:e01088-13. doi: 10.1128/mBio.01088-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine JR, Prescott J, Brown KS, Best SM, Ebihara H, Feldmann H. 2010. Antagonism of type I interferon responses by new world hantaviruses. J Virol 84:11790–11801. doi: 10.1128/JVI.00916-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor SL, Frias-Staheli N, Garcia-Sastre A, Schmaljohn CS. 2009. Hantaan virus nucleocapsid protein binds to importin alpha proteins and inhibits tumor necrosis factor alpha-induced activation of nuclear factor kappa B. J Virol 83:1271–1279. doi: 10.1128/JVI.00986-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor SL, Krempel RL, Schmaljohn CS. 2009. Inhibition of TNF-alpha-induced activation of NF-kappaB by hantavirus nucleocapsid proteins. Ann N Y Acad Sci 1171(Suppl 1):E86–E93. doi: 10.1111/j.1749-6632.2009.05049.x. [DOI] [PubMed] [Google Scholar]
- 34.Boudko SP, Kuhn RJ, Rossmann MG. 2007. The coiled-coil domain structure of the Sin Nombre virus nucleocapsid protein. J Mol Biol 366:1538–1544. doi: 10.1016/j.jmb.2006.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaukinen P, Kumar V, Tulimaki K, Engelhardt P, Vaheri A, Plyusnin A. 2004. Oligomerization of Hantavirus N protein: C-terminal alpha-helices interact to form a shared hydrophobic space. J Virol 78:13669–13677. doi: 10.1128/JVI.78.24.13669-13677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mir MA, Panganiban AT. 2004. Trimeric hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. J Virol 78:8281–8288. doi: 10.1128/JVI.78.15.8281-8288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mir MA, Panganiban AT. 2005. The hantavirus nucleocapsid protein recognizes specific features of the viral RNA panhandle and is altered in conformation upon RNA binding. J Virol 79:1824–1835. doi: 10.1128/JVI.79.3.1824-1835.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Boudreaux DM, Estrada DF, Egan CW, St Jeor SC, De Guzman RN. 2008. NMR structure of the N-terminal coiled coil domain of the Andes hantavirus nucleocapsid protein. J Biol Chem 283:28297–28304. doi: 10.1074/jbc.M804869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koma T, Yoshimatsu K, Pini N, Safronetz D, Taruishi M, Levis S, Endo R, Shimizu K, Yasuda SP, Ebihara H, Feldmann H, Enria D, Arikawa J. 2010. Truncated hantavirus nucleocapsid proteins for serotyping Sin Nombre, Andes, and Laguna Negra hantavirus infections in humans and rodents. J Clin Microbiol 48:1635–1642. doi: 10.1128/JCM.00072-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geldmacher A, Schmaler M, Kruger DH, Ulrich R. 2004. Yeast-expressed hantavirus Dobrava nucleocapsid protein induces a strong, long-lasting, and highly cross-reactive immune response in mice. Viral Immunol 17:115–122. doi: 10.1089/088282404322875511. [DOI] [PubMed] [Google Scholar]
- 41.Rigo MM, Antunes DA, Cibulski SP, Sinigaglia M, Chies JA, Vieira GF. 2011. Immunogenic epitopes of hantaviruses' N protein are restricted to conserved regions. Front Biosci (Landmark Ed) 17:1582–1588. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode, p 307–326. In Carter CW Jr, Sweet RM (ed), Macromolecular crystallography, part A, vol 276 Academic Press, New York, NY. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Lovell SC, Davis IW, Arendall WB III, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. 2003. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 46.DeLano W. 2002. The PyMOL molecular graphics system. DeLano Scientific, LLC, San Carlos, CA: http://www.pymol.org. [Google Scholar]
- 47.Xiao SY, Liang M, Schmaljohn CS. 1993. Molecular and antigenic characterization of HV114, a hantavirus isolated from a patient with haemorrhagic fever with renal syndrome in China. J Gen Virol 74:1657–1659. doi: 10.1099/0022-1317-74-8-1657. [DOI] [PubMed] [Google Scholar]
- 48.Novoa RR, Calderita G, Cabezas P, Elliott RM, Risco C. 2005. Key Golgi factors for structural and functional maturation of bunyamwera virus. J Virol 79:10852–10863. doi: 10.1128/JVI.79.17.10852-10863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macindoe G, Mavridis L, Venkatraman V, Devignes MD, Ritchie DW. 2010. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res 38:W445–W449. doi: 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Case DA, Cheatham TE III, Darden T, Gohlke H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. 2005. The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryckaert JP, Ciccotti G, Berendsen HJC. 1977. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comp Phys 23:327–341. doi: 10.1016/0021-9991(77)90098-5. [DOI] [Google Scholar]
- 52.Rastelli G, Del Rio A, Degliesposti G, Sgobba M. 2010. Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J Comput Chem 31:797–810. [DOI] [PubMed] [Google Scholar]
- 53.Flick K, Hooper JW, Schmaljohn CS, Pettersson RF, Feldmann H, Flick R. 2003. Rescue of Hantaan virus minigenomes. Virology 306:219–224. doi: 10.1016/S0042-6822(02)00070-3. [DOI] [PubMed] [Google Scholar]
- 54.Brown KS, Ebihara H, Feldmann H. 2012. Development of a minigenome system for Andes virus, a New World hantavirus. Arch Virol 157:2227–2233. doi: 10.1007/s00705-012-1401-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Li XH, Jiang H, Huang CX, Wang PZ, Mou DL, Sun L, Xu Z, Wei X, Bai XF. 2008. Expression of L protein of Hantaan virus 84FLi strain and its application for recovery of minigenomes. APMIS 116:1089–1096. doi: 10.1111/j.1600-0463.2008.01011.x. [DOI] [PubMed] [Google Scholar]
- 56.Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niu F, Shaw N, Wang YE, Jiao L, Ding W, Li X, Zhu P, Upur H, Ouyang S, Cheng G, Liu ZJ. 2013. Structure of the Leanyer orthobunyavirus nucleoprotein-RNA complex reveals unique architecture for RNA encapsidation. Proc Natl Acad Sci U S A 110:9054–9059. doi: 10.1073/pnas.1300035110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olal D, Dick A, Woods VL Jr, Liu T, Li S, Devignot S, Weber F, Saphire EO, Daumke O. 2014. Structural insights into RNA encapsidation and helical assembly of the Toscana virus nucleoprotein. Nucleic Acids Res 42:6025–6037. doi: 10.1093/nar/gku229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshimatsu K, Lee BH, Araki K, Morimatsu M, Ogino M, Ebihara H, Arikawa J. 2003. The multimerization of hantavirus nucleocapsid protein depends on type-specific epitopes. J Virol 77:943–952. doi: 10.1128/JVI.77.2.943-952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. 2011. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci U S A 108:2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hastie KM, Liu T, Li S, King LB, Ngo N, Zandonatti MA, Woods VL Jr, de la Torre JC, Saphire EO. 2011. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc Natl Acad Sci U S A 108:19365–19370. doi: 10.1073/pnas.1108515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmaljohn C, Hjelle B. 1997. Hantaviruses: a global disease problem. Emerg Infect Dis 3:95–104. doi: 10.3201/eid0302.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kao RY, Yang D, Lau LS, Tsui WH, Hu L, Dai J, Chan MP, Chan CM, Wang P, Zheng BJ, Sun J, Huang JD, Madar J, Chen G, Chen H, Guan Y, Yuen KY. 2010. Identification of influenza A nucleoprotein as an antiviral target. Nat Biotechnol 28:600–605. doi: 10.1038/nbt.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tischler ND, Rosemblatt M, Valenzuela PD. 2008. Characterization of cross-reactive and serotype-specific epitopes on the nucleocapsid proteins of hantaviruses. Virus Res 135:1–9. doi: 10.1016/j.virusres.2008.01.013. [DOI] [PubMed] [Google Scholar]