ABSTRACT
Susceptibility or resistance to prion infection in humans and animals depends on single prion protein (PrP) amino acid substitutions in the host, but the agent's modulating role has not been well investigated. Compared to disease incubation times in wild-type homozygous ARQ/ARQ (where each triplet represents the amino acids at codons 136, 154, and 171, respectively) sheep, scrapie susceptibility is reduced to near resistance in ARR/ARR animals while it is strongly enhanced in VRQ/VRQ carriers. Heterozygous ARR/VRQ animals exhibit delayed incubation periods. In bovine spongiform encephalopathy (BSE) infection, the polymorphism effect is quite different although the ARR allotype remains the least susceptible. In this study, PrP allotype composition in protease-resistant prion protein (PrPres) from brain of heterozygous ARR/VRQ scrapie-infected sheep was compared with that of BSE-infected sheep with a similar genotype. A triplex Western blotting technique was used to estimate the two allotype PrP fractions in PrPres material from BSE-infected ARR/VRQ sheep. PrPres in BSE contained equimolar amounts of VRQ- and ARR-PrP, which contrasts with the excess (>95%) VRQ-PrP fraction found in PrP in scrapie. This is evidence that transmissible spongiform encephalopathy (TSE) agent properties alone, perhaps structural aspects of prions (such as PrP amino acid sequence variants and PrP conformational state), determine the polymorphic dependence of the PrPres accumulation process in prion formation as well as the disease-associated phenotypic expressions in the host.
IMPORTANCE Transmissible spongiform encephalopathies (TSEs) are fatal neurodegenerative and transmissible diseases caused by prions. Amino acid sequence variants of the prion protein (PrP) determine transmissibility in the hosts, as has been shown for classical scrapie in sheep. Each individual produces a separate PrP molecule from its two PrP gene copies. Heterozygous scrapie-infected sheep that produce two PrP variants associated with opposite scrapie susceptibilities (136V-PrP variant, high; 171R-PrP variant, very low) contain in their prion material over 95% of the 136V PrP variant. However, when these sheep are infected with prions from cattle (bovine spongiform encephalopathy [BSE]), both PrP variants occur in equal ratios. This shows that the infecting prion type determines the accumulating PrP variant ratio in the heterozygous host. While the host's PrP is considered a determining factor, these results emphasize that prion structure plays a role during host infection and that PrP variant involvement in prions of heterozygous carriers is a critical field for understanding prion formation.
INTRODUCTION
Transmissible spongiform encephalopathies (TSEs), or prion diseases, are fatal neurological diseases occurring in some mammalian species, including humans. The TSE agent or prion is characterized by the pivotal role of the host prion protein (PrP) that in disease appears aggregated and structurally abnormal and is named PrPSc, where “Sc” refers to scrapie in small ruminants, which was recognized in the 18th century in Spanish Merino sheep (1). In healthy situations PrP is a cellular membrane protein (PrPC) and fully susceptible to proteases, while its PrPSc isoform is partially resistant to digestion with proteinase K (PK), usually leading to an N-terminally shortened protein called PrPres that still retains the associated infectivity (2–4).
From many studies it is obvious that TSEs occur in distinct phenotypic forms that are recognized as TSE or prion disease types, such as classical scrapie in sheep and goat, Creutzfeldt-Jakob disease in humans, chronic wasting disease in cervids, and bovine spongiform encephalopathy (BSE) in cattle (5–15). In the experimental situation, these types can be considered strains when they are subpassaged to homogeneity in rodent bioassays (16–20). Susceptibility (and resistance) to animal and human prion diseases, either under infectious or spontaneous conditions, is dependent on single amino acid substitutions in the host's PrP sequence. In most species such substitutions are naturally occurring polymorphisms (7, 10, 21–24).
In sheep two PrP polymorphisms in the PrP sequence, V136 and R171(where V is valine and R is arginine, according to the single-letter code used by the IUPAC-IUB Joint Commission on Biochemical Nomenclature), provide, respectively, high and very low susceptibilities to natural scrapie compared to the homozygous wild-type variants with A136 and Q171 (where A is alanine and Q is glutamine). Other variants also influence susceptibility, for example, H154 (where H is histidine) (13, 24–30). Together, this evidence has led to policies for eradication of scrapie in sheep breeds by focusing on codons 136, 154, and 171, in which the different alleles are designated, in respective order, ARQ (the wild type), VRQ, AHQ, and ARR (31, 32). When both codon 136 and 171 variants occur in heterozygous sheep, the genotype code is indicated as ARR/VRQ, while homozygous sheep could have genotype ARQ/ARQ (the wild type), ARR/ARR, or VRQ/VRQ (7).
In a previous study we reported that in scrapie-infected ARR/VRQ sheep, the VRQ-PrP in PrPres was highly overrepresented, with 91 to 100% VRQ-PrP product (33, 34). Yet the expression levels of the PrPC alleles in heterozygous animals are considered equal (34, 35), which means that during PrPSc formation in ARR/VRQ scrapie-infected animals, there occurs a selective incorporation of the VRQ-PrP allotype. In vitro assays confirm the relatively high, but not absolute, resistance to conversion of ARR-PrP when this allotype is subjected to scrapie or BSE prions (12, 15, 26, 36). This special property of the ARR-PrP allotype is confirmed under in vivo intracerebral (i.c.) BSE challenge conditions, but the VRQ-PrP allotype, in contrast to its strong link to susceptibility to scrapie, in VRQ/VRQ sheep appeared to confer far more resistance to BSE than that found in ARQ/ARQ sheep (37).
In this paper we investigated whether the level of the VRQ-PrP allotype in PrPres from ARR/VRQ BSE i.c. infected sheep generated by Houston et al. (37) would be comparably high to that found in the same genotype of sheep with natural scrapie. This was accomplished by comparing brain PrPres in scrapie- and BSE-infected ARR/VRQ sheep. A previously developed robust triplex Western blotting (WB) method (38, 39) was used to quantitatively estimate PrP concentrations. In this technique the Q171-PrP fraction (VRQ and ARQ) can be quantitatively estimated using a mixture of two antibodies on the same blot membrane, with one antibody (SAF84) recognizing only the VRQ fraction while the other binds equally well both VRQ-PrP and ARR-PrP. The outcome yielded a clear-cut difference in VRQ contents deposited in the prions of these two different TSE types. This new information is special since it reports on PrP allotype expression for two separate prion types from a mammalian species (sheep) heterozygous for two non-wild-type PrP alleles differing widely in their effects on susceptibility/resistance to prion infection.
MATERIALS AND METHODS
Sheep brain and antibodies.
Brain tissues were available from ARR/VRQ, VRQ/VRQ, ARQ/ARQ, and ARR/ARR sheep clinically affected following intracerebral challenge with cattle BSE and from naturally infected scrapie sheep with genotypes ARR/VRQ, VRQ/VRQ, ARQ/ARQ, and ARQ/VRQ detected in active surveillance monitoring. The details of the different groups of sheep are presented in Table 1. The BSE and classical scrapie diagnosis was carried out on brain stem tissue of each animal by immunohistochemistry and by Western blotting (40–42). Tissues used in the different laboratories were obtained from sheep experiments performed under EU convention ET S 123 in accordance with the rules for ethical animal experimentation carried out in the European Community.
TABLE 1.
Sheep genotypes, TSE type tissues, laboratory origin, and breeda
| TSE type | Genotype | No. of cases | Lab sourced | Breed |
|---|---|---|---|---|
| BSEb | ARR/VRQ | 4 | Roslin-UEDINc | Cheviot |
| VRQ/VRQ | 5 | Roslin-UEDINc | Cheviot | |
| ARQ/ARQ | 3 | INRA-Tours2nd | Suffolk | |
| ARR/ARR | 3 | INRA-Tours | Poll Dorset | |
| Natural scrapie | ARR/VRQ | 7 | CVI-Wageningen UR | Texel crossbreed |
| VRQ/VRQ | 2 | CVI-Wageningen UR | Texel crossbreed | |
| ARQ/ARQ | 4 | CVI-Wageningen UR | Texel crossbreed | |
| ARQ/VRQ | 4 | CVI-Wageningen UR | Texel crossbreed |
Scrapie brain stem tissues were from natural field cases, BSE brain stem or midbrain tissues were either from intracerebral infections with bovine BSE in VRQ/VRQ, ARR/VRQ, and ARR/ARR sheep or, in the case of INRA-Tours2nd, obtained by i.c. passage from bovine BSE-infected ARQ/ARQ sheep to ARQ/ARQ sheep.
Intracerebral infection.
Houston and Hunter, unpublished data.
INRA, Institut National de la Recherche Agronomique; UEDIN, University of Edinburgh; CVI, Central Veterinary Institute.
Monoclonal antibodies used were L42, Sha31, and SAF84 (43–45) with respective linear ovine PrP epitope sequences consisting of residues 148 to 153, 148 to 155, and 166 to 172, as determined using Pepscan epitope mapping technology (46), and IgG class numbers a2, 1, and b2. Though L42 and Sha31 share nearly the same linear epitope, they were raised with very different antigens, with L42 being a linear peptide derived from ovine PrP and Sha31 derived from PK-digested nondenatured scrapie-associated fibrils from Syrian hamsters. Molecular Probes Zenon Alexa Fluor mouse labeling kits for mouse IgG1 (Alexa 647), IgG2a (Alexa 647), and IgG2b (Alexa 488) were from ThermoFisher. For molecular mass estimation a Pre-Stained SeeBlue Standards kit (LC5625; ThermoFisher) was used. Ovine recombinant ARQ-PrP was a gift from Human Rezaei (Institut National de la Recherche Agronomique [INRA], Jouy-en Jozas, France) (47).
PrPres preparation and quantification of allotype expression with mixed-antibody Western blotting.
PrPres was prepared from 10% (wt/vol) brain stem homogenates prepared in lysis buffer, digested with PK at 37°C, and further partially purified by precipitation with 1-propanol as described previously (38). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of denatured samples in loading buffer (with lithium-dodecyl sulfate and β-mercaptoethanol) was performed in 17-well gels (33). Detection of PrPres on blot membranes was carried out in our triplex Western blotting system, but for this study a mixture of only two primary antibodies instead of three was used. The antibodies were labeled with Zenon Alexa Fluor kits before application on the blot. Immunochemical quantification of PrPres was subsequently performed by fluorimetric detection monitored in a three-laser-beam imager (Typhoon Trio variable-mode imager; Amersham Biosciences) (38). For estimation of the ARR- and VRQ-PrP fractions in PrPres, a mixture of two antibodies was applied, one of which (SAF84) binds only if the 171Q polymorphism is present (VRQ-PrP or ARQ-PrP) while the other binds equally well to VRQ-, ARQ-, and ARR-PrP (33, 38, 39). Two different mixtures with SAF84 were used: SAF84 with L42 (L42/SAF84 combination) and SAF84 with Sha31 (Sha31/SAF84 combination). SAF84 detection was carried out with a Zenon labeling Alexa 488 kit, and L42 or Sha31 was detected with a Zenon labeling Alexa 647 kit (see above for kit specifications). The VRQ-PrP and ARQ-PrP fractions in PrPres samples were calculated as follows (33, 38, 39). When the SAF84/L42 antibody combination was used, the fraction of the 171Q-PrP product [Fr(171Q-PrP); the VRQ- or ARQ-PrP levels] in scrapie or BSE was obtained by applying the formula Fr(171Q-PrP) = ratiox/ratioQ/Q, where ratiox is the SAF84/L42 ratio of an unknown sample, and ratioQ/Q is the SAF84/L42 ratio determined for Q/Q homozygous material, which is the average of measurements of the different scrapie (n = 10) or BSE (n = 8) Q/Q samples; the fraction of the 171R-PrP product (the ARR-PrP level) could be deduced from the formula (ratioQ/Q − ratiox)/ratioQ/Q. For the SAF84/Sha31 combination, the same formulas were applied but the L42 values were replaced with those for Sha31.
The validity of the approach was confirmed by mixing, in loading buffer, samples from a VRQ/VRQ and an ARR/ARR sheep, both infected with BSE, at volume ratios of 9/1, 8.5/1.5, 8/2, 7.5/2.5 7/3, 6/4, 5/5, 4/6, 3/7, 2/8, and 1/9 (for both antibody combinations). To exclude the possibility that the outcomes were influenced by the concentration of the PrPres signal, a further check was performed by calculating the PrPres signal per sample in nanograms of PrP as observed from detection of L42 and Sha31 and using as a reference the recombinant PrP signal, 15 ng of which was run in a lane of each gel.
RESULTS
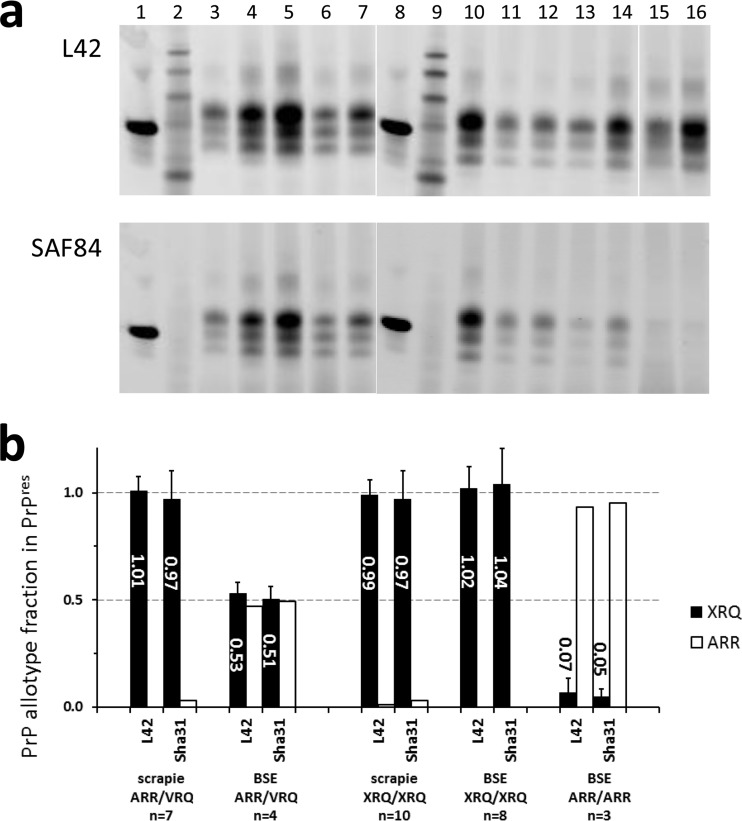
PrPres samples from sheep homozygous for the 171Q codon allele (genotypes VRQ/VRQ and ARQ/ARQ) exhibited full reactivity with the antibodies L42 and SAF84 in both BSE- and scrapie-infected animals (Fig. 1a, respectively, lanes 3 to 5 and lanes 10 and 11). As expected, the PrPres from ARR/ARR BSE-infected sheep reacted with antibody L42 but not at all with SAF84 (Fig. 1a, lanes 15 and 16). Scrapie-infected ARR/ARR sheep were not available since these animals remained TSE negative throughout their experimental lifetime, which is indicative of the high scrapie resistance contributed by the 171R codon (>2,000 days) (F. Houston and N. Hunter, unpublished data). The analyses from the heterozygous ARR/VRQ sheep with scrapie and BSE yielded contrasting results in that the staining with SAF84 relative to that with L42 on scrapie-infected sheep samples produced results very similar to each other while that with SAF84 on the BSE samples was reduced. Similar results were observed when the SAF84/Sha31 antibody duplex combination (Fig. 1b) was used. A further calculation of the fraction of VRQ-PrP in the PrPres samples from the heterozygous animals using the SAF84/L42 combination yielded in scrapie-infected ARR/VRQ sheep a VRQ-PrP fraction, Fr(171Q-PrP), of 1.01 ± 0.07 (average ± standard deviation; n = 7) (Fig. 1b). This result compared fairly well with previous estimations using two-dimensional (2D) gel electrophoresis on isolated PrPres fragments and two different Western blotting techniques (an enzymatically enhanced chemiluminescence immunodetection method and a triplex WB-based fluorescence immunolabeling method) (33). This result further implied that the ARR-PrP fraction varied between different ARR/VRQ sheep-derived samples from 0 to only 0.1. In contrast, for BSE-infected ARR/VRQ sheep, the VRQ-PrP fraction was 0.53 ± 0.05 (n = 4), indicating that PrPres of the BSE-infected ARR/VRQ animals contained nearly equal amounts of both VRQ-PrP and ARR-PrP allotype products. Similar values were obtained when samples were tested with the SAF84/Sha31 combination (Fig. 1b).
FIG 1.
PrP allotype fraction estimates in PrPres from brain of PrP scrapie- and BSE-infected sheep with different PRNP genotypes. (a) Western blot of scrapie and BSE PrPres samples of infected sheep with heterozygous and homozygous genotypes as tested with the L42/SAF84 antibody combination. Lanes: 1 and 8, recombinant ovine PrP; 2 and 9, molecular mass standards; 3 to 5, VRQ/VRQ sheep with scrapie; 6 and 7 ARR/VRQ sheep with scrapie; 10 to 12, VRQ/VRQ sheep with BSE; 13 and 14, ARR/VRQ with BSE; 15 and 16 ARR/ARR sheep with BSE. Blotting procedures followed the triplex WB method as described previously (38, 39). Tissue equivalents per each brain sample applied were 0.5 mg per lane. (b) VRQ- or ARQ-PrP and ARR-PrP allotype fractions per genotype group of sheep with scrapie or BSE. Genotypes are given for PrP amino acid residue positions 136, 154, and 171; XRQ means combined data from either three (scrapie ARQ/ARQ, VRQ/VRQ, and ARQ/VRQ) or two (BSE ARQ/ARQ and VRQ/VRQ) genotypes, respectively. The results shown of the two antibody combinations, SAF84/L42 and SAF84/Sha31, appeared very similar. Black bars, VRQ- and/or ARQ-PrP fraction; open bars, ARR-PrP fraction. The numbers within the bars reflect the average XRQ-PrP fraction values, and vertical lines indicate the standard deviation of the XRQ fraction. n, number of individual samples.
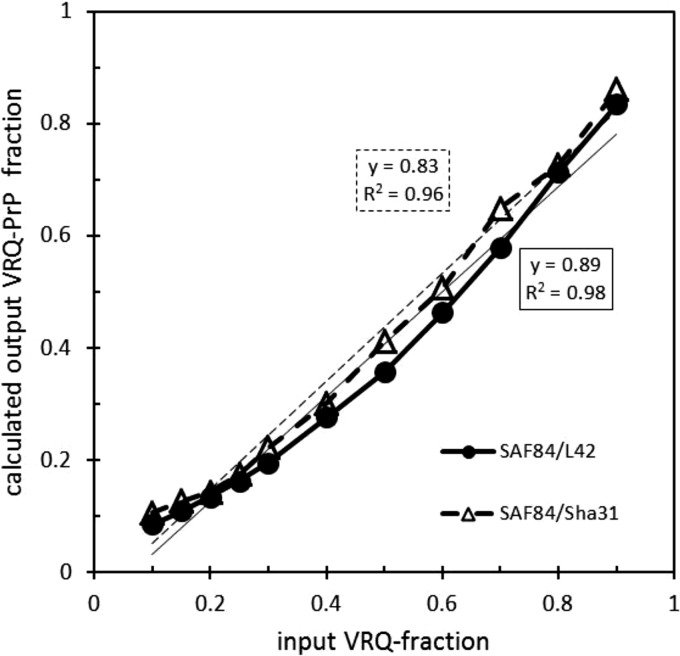
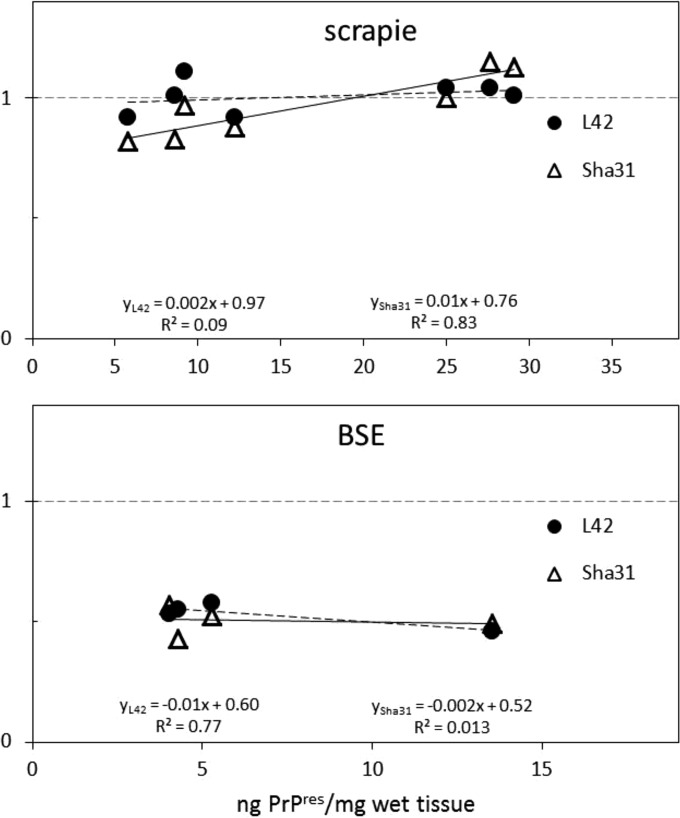
The validity of this approach was confirmed by mixing a VRQ/VRQ sample with an ARR/ARR BSE sample in loading buffer in different proportions from 9/1 to 1/9. The output-versus-input curves for the VRQ-PrP fraction of PrPres were concave but approached linearity rather well when either the SAF84/L42 or the SAF84/Sha31 antibody combination was used (Fig. 2). The final data shown in Fig. 1b represent adjusted values based on these concave curves. Finally, an effect on the outcomes of the PrPres concentration in the tissue digest was estimated. The regression curves obtained for scrapie and BSE samples were approaching a horizontal line, pointing to negligible effects from the PrPres concentration on the Fr(171Q-PrP) values (Fig. 3). For all individual and overall sample data, the outcomes with the SAF84/L42 and SAF84/Sha31 antibody combinations were very comparable. Also, the current scrapie data confirm our previous results from ARR/VRQ scrapie-infected sheep, determined in different ways, and prove the quantitative value of the current immunochemical Western blotting methodology used (33).
FIG 2.
Probing the VRQ-PrP allotype level between input and calculated output levels in PrPres samples in dose-response mixing experiments. See Materials and Methods for the design of the experiment. For both duplex antibody combinations, similar concave curves were obtained. These hollow curves were used for calculation of the final data shown in Fig. 1b. Thus, a sample with an output value of 20, 40, 60, or 80% VRQ-PrP allotype yielded, in case of the SAF84/L42 combination, respectively, 30, 55, 72, and 87% and, for the SAF84/Sha31 combination, 29, 51, 67 and 86% VRQ-PrP. The inset presents the values of the calculated regression lines derived from the data points.
FIG 3.
Relation between PrPres concentration and VRQ-PrP level of ARR/VRQ sheep brain. For individual samples from ARR/VRQ sheep, the PrP concentration in the samples was calculated using recombinant PrP as a standard in both blots probed with the SAF84/L42 (filled circles) and SAF84/Sha31 (open triangles) antibody combinations (see Materials and Methods). The VRQ-PrP levels in all individual samples were around 1 in the scrapie samples and 0.5 in the BSE samples. The linear regression formulae for the data from the two antibody combinations point to nearly horizontal curves, indicative of the absence of a concentration effect on the Fr(171Q-VRQ) values in the triplex WB methodology used. Vertical axis, Fr(171-VRQ) fraction.
DISCUSSION
The analyses of the PrP allotype composition of prion material in heterozygous ARR/VRQ sheep yielded for BSE-infected sheep a VRQ-PrP fraction approaching 0.5. This contrasted with the fraction determined in scrapie-infected sheep, where the VRQ-PrP fraction approximated 1, thus representing nearly all of the PrPres mass. Since in the ARR/VRQ scrapie PrPres only one allotype is found while both alleles, because of diploidy, can and do express PrP (34, 48), it is surprising that the ARR-PrP fraction in the PrPres material of the scrapie cases is nearly zero. This is in contrast to the ∼50% ARR-PrP fraction in ARR/VRQ BSE PrPres mass. This wide difference in VRQ-PrP and ARR-PrP contents in the prion material of these sheep with scrapie and BSE infection is unique for three reasons. First, two different acquired (infectious) conditions of prion disease were studied in these animals. Second, individual animals carrying two non-wild-type PrP alleles with very contrasting TSE-type susceptibilities were investigated; on the one hand, the VRQ-PrP makes them highly susceptible to scrapie, on the other hand the ARR-PrP makes them resistant to both BSE and scrapie. Third, the study was performed on tissues obtained from infected animals; thus, the prions studied are products of in vivo conditions. These data from heterozygous animals carrying two different TSEs, scrapie and BSE, confirm in vitro conversion data that a certain PrP polymorphism of the host can be less prone to conversion to PrPSc than another (15, 26). Or, as an alternative to the species barrier concept, on infection with scrapie, only ARR-PrP forms a polymorphism barrier, whereas with primary infection with BSE both ARR- and VRQ-PrP contribute to this barrier. Importantly, these new data also strongly support the concept that the type (or strain) of the infecting agent itself has an influence on this conversion event.
The role a certain prion type plays in susceptibility and resistance of the sheep host is strikingly reflected in in vivo situations, as exemplified with three different TSE types. With BSE infection, ARR/ARR and VRQ/VRQ sheep have long incubation times to clinical disease following intracerebral challenge at, respectively, >1,400 days and >1,000 days, compared to that in the wild-type ARQ/ARQ sheep (around 600 days) (N. Hunter and F. Houston, personal communication). With classical scrapie infection with the agent derived from VRQ-rich sheep flocks, ARR/ARR sheep are nearly fully resistant to challenge, whereas VRQ/VRQ sheep with scrapie have very short incubation times (180 to 720 days), and the wild-type (ARQ/ARQ) sheep have intermediate incubation times (14, 27, 36, 37, 40, 49–51). Interestingly with atypical/Nor98 scrapie, a prion disease that is nonspreading and may be of spontaneous origin, VRQ/VRQ animals appear highly insensitive based on genotype frequency, while ARR/ARR sheep can be affected but are less frequently so than ARQ/ARQ sheep with this scrapie type (Table 2) (52). Though the susceptibilities to prion diseases may also be influenced by route of infection, prevailing PrP polymorphism of the flock, extent of involvement of the lympho-reticular system, and other pathogenic aspects, the above mutual differences in susceptibilities are relatively consistent. A breed effect between the Cheviot and Texel sheep used in this study cannot be excluded as another factor for the potential difference in allotype ratios between BSE- and scrapie-infected ARR/VRQ animals, but susceptibilities to TSE within a breed (in casu Romanovs) are expected to be largely independent of polygenic effects, and this view may also apply to between-breed effects (14, 53). Therefore, the allotype PrP composition in prion material found in our results reflects the effect of the type of TSE or prion agent rather than variation in the host.
TABLE 2.
Susceptibility dependence on TSE/prion type and host PrP polymorphism
| Disease type | PrP allotype susceptibilitya |
||
|---|---|---|---|
| Most | Medium | Least | |
| BSE | Wild type | V136 | R171 |
| Classical scrapie | V136 | Wild type | R171 |
| Atypical/Nor98 scrapie | Wild type | R171 | V136 |
Susceptibility is presented in a qualitative way for the single amino acid allotype. Wild-type represents the A136R154Q171 allele. Data about BSE are from experimental infections, classical scrapie data are from natural and experimental infections, and atypical/Nor98 scrapie data are from active monitoring in a number of European countries.
With respect to animal species other than sheep, some results have been obtained with TSE infections in heterozygous TSE-infected bank voles. One polymorphism has been described which, if present in 109M/I animals, leads to 20 to 30% differences in incubation times for the heterozygous animals compared to that in the wild-type carriers after intracerebral infection with sheep or goat scrapie but to equal incubation times after infection with mouse scrapie strain 139A (23, 54). In these models deposition of both wild-type and non-wild type PrP allotypes was observed in significant amounts, pointing to equal allotype levels in the prions. This equal deposition of both PrP allotypes in heterozygous bank voles might indicate that incubation times alone are not sufficiently indicative of a great difference in convertibility of PrPC to PrPSc, and this therefore may lead to 100% attack rates. Thus, the situation in these bank vole experiments is different from that in ARR/VRQ sheep where two non-wild-type PrP allotypes have been studied, each of which has a proven influence on susceptibility and PrPC-to-PrPSc convertibility.
In contrast to infectious conditions, in inherited human TSEs, the patients carry a PrP gene-linked predisposition to develop disease by a mutation in the coding region of the PRNP gene. The patients are nearly always heterozygous (55, 56). Depending on the polymorphism, the non-wild-type variant is frequently the dominant PrP variant present in the PK-resistant or detergent-insoluble PrPSc material, but in some instances both wild-type and non-wild-type PrPs are present in significant amounts (55, 57–63). The PrP allotype prevalence in the deposited prion PrP material is supposed to depend on the position and nature of the amino acid in the PrP sequence. In these spontaneous prion diseases, PrPC can be considered to be the main host factor determining the PrP allotype ratio of the prion material. However, the role of non-PrP host factors should also be taken into consideration (64). Under infectious conditions, such as those studied in animals, the agent itself can have an equally important role as that of host PrP and non-PrP host factors. Probably, binding of PrPSc to PrPC (at least for sheep PrP) does not discriminate between different polymorphic PrP variants, while the PrPC-to-PrPSc conversion efficiency clearly is related to PrP-linked genotype-dependent susceptibilities, as was shown for sheep prions (12, 15, 27, 36, 65).
The example of possibly different allotype compositions in prion material between two TSE types, scrapie and BSE, as exemplified in the ARR/VRQ sheep of this study is a novel finding for in vivo situations and confirms the in vitro studies that show that different TSE types have different PrP polymorphism variant preferences in the PrPC-to-PrPSc conversion (13, 14, 36). It also shows that, in disease, the prion type can determine the ability of certain host PrP allotype sequence variants to be converted from PrPC to PrPSc. The critical issue of how the conversion process works and of whether other factors than the PrP amino acid sequence of the host can influence it is still uncertain. The species source from which the infection is derived is one determinant (36), as in our case the BSE material used to infect the sheep is of bovine origin. Bovine PrP differs from sheep PrP in having an extra octarepeat in the PrP N terminus and six further amino acid codon differences (sheep PrP codons 98, 100, 146, 158, 189, and 208) (48, 66). Further structural differences in the folding of the prions of BSE and different scrapie types might well have a role in susceptibility of the host, as has been hypothesized in sheep challenge experiments with BSE, CH1641 scrapie, and SSBP1 scrapie (13). Whether a non-PrP factor in the agent could play a role remains to be investigated. However, considering the major role of PrPSc structure in TSEs, our data suggest that further studies on PrP allotype heterozygosity in agent and host are needed in order to understand the factors determining the fate of prion diseases.
ACKNOWLEDGMENTS
This article is in memory of our colleague Alan Rigter, who died in April 2014 at the stage in his life when he was going to apply his education as a molecular biologist as a full-time Ph.D.
REFERENCES
- 1.Fast C, Groschup MH. 2013. Classical and atypical scrapie in sheep and goats, p 15–44. In Zou W-Q, Gambetti P (ed), Prions and diseases: animals, humans and the environment, vol 2 Springer, New York, NY. [Google Scholar]
- 2.Oesch B, Westaway D, Walchli M, McKinley MP, Kent SB, Aebersold R, Barry RA, Tempst P, Teplow DB, Hood LE, Prusiner SB, Weissman C. 1985. A cellular gene encodes scrapie PrP 27-30 protein. Cell 40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner SB. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner SB, McKinley MP, Bowman KA, Bolton DC, Bendheim PE, Groth DF, Glenner GG. 1983. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell 35:349–358. doi: 10.1016/0092-8674(83)90168-X. [DOI] [PubMed] [Google Scholar]
- 5.Di Bari MA, Chianini F, Vaccari G, Esposito E, Conte M, Eaton SL, Hamilton S, Finlayson J, Steele PJ, Dagleish MP, Reid HW, Bruce M, Jeffrey M, Agrimi U, Nonno R. 2008. The bank vole (Myodes glareolus) as a sensitive bioassay for sheep scrapie. J Gen Virol 89:2975–2985. doi: 10.1099/vir.0.2008/005520-0. [DOI] [PubMed] [Google Scholar]
- 6.Gambetti P, Kong Q, Zou W, Parchi P, Chen SG. 2003. Sporadic and familial CJD: classification and characterisation. Br Med Bull 66:213–239. doi: 10.1093/bmb/66.1.213. [DOI] [PubMed] [Google Scholar]
- 7.Hunter N, Bossers A. 2006. The PrP genotype as a marker for scrapie susceptibility in sheep, p 640–647. In Ḧornlimann B, Riesner D, Kretzschmar H (ed), Prions in humans and animals. de Gruyter, Berlin, Germany. [Google Scholar]
- 8.Mead S, Whitfield J, Poulter M, Shah P, Uphill J, Campbell T, Al-Dujaily H, Hummerich H, Beck J, Mein CA, Verzilli C, Whittaker J, Alpers MP, Collinge J. 2009. A novel protective prion protein variant that colocalizes with kuru exposure. N Engl J Med 361:2056–2065. doi: 10.1056/NEJMoa0809716. [DOI] [PubMed] [Google Scholar]
- 9.Meade-White KD, Barbian KD, Race B, Favara C, Gardner D, Taubner L, Porcella S, Race R. 2009. Characteristics of 263K scrapie agent in multiple hamster species. Emerg Infect Dis 15:207–215. doi: 10.3201/eid1502.081173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaccari G, Panagiotidis CH, Acin C, Peletto S, Barillet F, Acutis P, Bossers A, Langeveld J, van Keulen L, Sklaviadis T, Badiola JJ, Andreeoletti O, Groschup MH, Agrimi U, Foster J, Goldmann W. 2009. State-of-the-art review of goat TSE in the European Union, with special emphasis on PRNP genetics and epidemiology. Vet Res 40:48. doi: 10.1051/vetres/2009031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams ES, Young S. 1980. Chronic wasting disease of captive mule deer: a spongiform encephalopathy. J Wildl Dis 16:89–98. doi: 10.7589/0090-3558-16.1.89. [DOI] [PubMed] [Google Scholar]
- 12.Bossers A, Belt P, Raymond GJ, Caughey B, de Vries R, Smits MA. 1997. Scrapie susceptibility-linked polymorphisms modulate the in vitro conversion of sheep prion protein to protease-resistant forms. Proc Natl Acad Sci U S A 94:4931–4936. doi: 10.1073/pnas.94.10.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldmann W, Hunter N, Smith G, Foster J, Hope J. 1994. PrP genotype and agent effects in scrapie: change in allelic interaction with different isolates of agent in sheep, a natural host of scrapie. J Gen Virol 75:989–995. doi: 10.1099/0022-1317-75-5-989. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez L, Jeffrey M, Dagleish MP, Goldmann W, Siso S, Eaton SL, Martin S, Finlayson J, Stewart P, Steele P, Pang Y, Hamilton S, Reid HW, Chianini F. 2012. Susceptibility to scrapie and disease phenotype in sheep: cross-PRNP genotype experimental transmissions with natural sources. Vet Res 43:55. doi: 10.1186/1297-9716-43-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raymond GJ, Hope J, Kocisko DA, Priola SA, Raymond LD, Bossers A, Ironside J, Will RG, Chen SG, Petersen RB, Gambetti P, Rubenstein R, Smits MA, Lansbury PT Jr, Caughey B. 1997. Molecular assessment of the potential transmissibilities of BSE and scrapie to humans. Nature 388:285–288. doi: 10.1038/40876. [DOI] [PubMed] [Google Scholar]
- 16.Fraser H, Dickinson AG. 1968. The sequential development of the brain lesion of scrapie in three strains of mice. J Comp Pathol 78:301–311. doi: 10.1016/0021-9975(68)90006-6. [DOI] [PubMed] [Google Scholar]
- 17.Kimberlin RH, Walker C. 1977. Characteristics of a short incubation model of scrapie in the golden hamster. J Gen Virol 34:295–304. doi: 10.1099/0022-1317-34-2-295. [DOI] [PubMed] [Google Scholar]
- 18.Bruce ME, McConnell I, Fraser H, Dickinson AG. 1991. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol 72:595–603. doi: 10.1099/0022-1317-72-3-595. [DOI] [PubMed] [Google Scholar]
- 19.Le Dur A, Beringue V, Andreoletti O, Reine F, Lai TL, Baron T, Bratberg B, Vilotte JL, Sarradin P, Benestad SL, Laude H. 2005. A newly identified type of scrapie agent can naturally infect sheep with resistant PrP genotypes. Proc Natl Acad Sci U S A 102:16031–16036. doi: 10.1073/pnas.0502296102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nonno R, Di Bari MA, Cardone F, Vaccari G, Fazzi P, Dell'Omo G, Cartoni C, Ingrosso L, Boyle A, Galeno R, Sbriccoli M, Lipp HP, Bruce M, Pocchiari M, Agrimi U. 2006. Efficient transmission and characterization of Creutzfeldt-Jakob disease strains in bank voles. PLoS Pathog 2:e12. doi: 10.1371/journal.ppat.0020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collinge J. 2005. Molecular neurology of prion disease. J Neurol Neurosurg Psychiatry 76:906–919. doi: 10.1136/jnnp.2004.048660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson CJ, Herbst A, Duque-Velasquez C, Vanderloo JP, Bochsler P, Chappell R, McKenzie D. 2011. Prion protein polymorphisms affect chronic wasting disease progression. PLoS One 6:e17450. doi: 10.1371/journal.pone.0017450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cartoni C, Schinina ME, Maras B, Nonno R, Vaccari G, Di Baria MA, Conte M, Liu QG, Lu M, Cardone F, Windl O, Pocchiari M, Agrimi U. 2005. Identification of the pathological prion protein allotypes in scrapie-infected heterozygous bank voles (Clethrionomys glareolus) by high-performance liquid chromatography-mass spectrometry. J Chromatogr A 1081:122–126. doi: 10.1016/j.chroma.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 24.Westaway D, Zuliani V, Cooper CM, Da Costa M, Neuman S, Jenny AL, Detwiler L, Prusiner SB. 1994. Homozygosity for prion protein alleles encoding glutamine-171 renders sheep susceptible to natural scrapie. Genes Dev 8:959–969. doi: 10.1101/gad.8.8.959. [DOI] [PubMed] [Google Scholar]
- 25.Belt PB, Muileman IH, Schreuder BE, Bos-de Ruijter J, Gielkens AL, Smits MA. 1995. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J Gen Virol 76:509–517. doi: 10.1099/0022-1317-76-3-509. [DOI] [PubMed] [Google Scholar]
- 26.Bossers A, de Vries R, Smits MA. 2000. Susceptibility of sheep for scrapie as assessed by in vitro conversion of nine naturally occurring variants of PrP. J Virol 74:1407–1414. doi: 10.1128/JVI.74.3.1407-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bossers A, Schreuder BE, Muileman IH, Belt PB, Smits MA. 1996. PrP genotype contributes to determining survival times of sheep with natural scrapie. J Gen Virol 77:2669–2673. doi: 10.1099/0022-1317-77-10-2669. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda T, Horiuchi M, Ishiguro N, Muramatsu Y, Kai-Uwe GD, Shinagawa M. 1995. Amino acid polymorphisms of PrP with reference to onset of scrapie in Suffolk and Corriedale sheep in Japan. J Gen Virol 76:2577–2581. doi: 10.1099/0022-1317-76-10-2577. [DOI] [PubMed] [Google Scholar]
- 29.Saunders GC, Lantier I, Cawthraw S, Berthon P, Moore SJ, Arnold ME, Windl O, Simmons MM, Andreoletti O, Bellworthy S, Lantier F. 2009. Protective effect of the T112 PrP variant in sheep challenged with bovine spongiform encephalopathy. J Gen Virol 90:2569–2574. doi: 10.1099/vir.0.012724-0. [DOI] [PubMed] [Google Scholar]
- 30.Tan BC, Alejo-Blanco AR, Goldmann W, Stewart P, Gill AC, Graham JF, Manson JC, McCutcheon S. 2010. Codon 141 in ovine PRNP gene modulates incubation time in sheep orally infected with BSE. Prion 4:195. doi: 10.4161/pri.12774. [DOI] [Google Scholar]
- 31.Commission of the European Union. 2003. 2003/100/EC: Commission decision of 13 February 2003 laying down minimum requirements for the establishment of breeding programmes for resistance to transmissible spongiform encephalopathies in sheep. Official Journal of the European Union L41, 46:41–45. [Google Scholar]
- 32.Melchior MB, Windig JJ, Hagenaars TJ, Bossers A, Davidse A, van Zijderveld FG. 2010. Eradication of scrapie with selective breeding: are we nearly there? BMC Vet Res 6:24. doi: 10.1186/1746-6148-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs JG, Bossers A, Rezaei H, van Keulen LJ, McCutcheon S, Sklaviadis T, Lantier I, Berthon P, Lantier F, van Zijderveld FG, Langeveld JP. 2011. Proteinase K-resistant material in ARR/VRQ sheep brain affected with classical scrapie is composed mainly of VRQ prion protein. J Virol 85:12537–12546. doi: 10.1128/JVI.00448-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morel N, Andreoletti O, Grassi J, Clement G. 2007. Absolute and relative quantification of sheep brain prion protein (PrP) allelic variants by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 21:4093–4100. doi: 10.1002/rcm.3317. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Crespo D, Juste RA, Hurtado A. 2005. Selection of ovine housekeeping genes for normalisation by real-time RT-PCR; analysis of PrP gene expression and genetic susceptibility to scrapie. BMC Vet Res 1:3. doi: 10.1186/1746-6148-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Priem J, Langeveld JP, van Keulen LJ, van Zijderveld FG, Andreoletti O, Bossers A. 2014. Enhanced virulence of sheep-passaged bovine spongiform encephalopathy agent is revealed by decreased polymorphism barriers in prion protein conversion studies. J Virol 88:2903–2912. doi: 10.1128/JVI.02446-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houston F, Goldmann W, Chong A, Jeffrey M, Gonzalez L, Foster J, Parnham D, Hunter N. 2003. Prion diseases: BSE in sheep bred for resistance to infection. Nature 423:498. doi: 10.1038/423498a. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs JG, Sauer M, van Keulen LJ, Tang Y, Bossers A, Langeveld JP. 2011. Differentiation of ruminant transmissible spongiform encephalopathy isolate types, including bovine spongiform encephalopathy and CH1641 scrapie. J Gen Virol 92:222–232. doi: 10.1099/vir.0.026153-0. [DOI] [PubMed] [Google Scholar]
- 39.Langeveld JP, Jacobs JG, Erkens JH, Baron T, Andreoletti O, Yokoyama T, van Keulen LJ, van Zijderveld FG, Davidse A, Hope J, Tang Y, Bossers A. 2014. Sheep prions with molecular properties intermediate between classical scrapie, BSE and CH1641-scrapie. Prion 8:296–305. doi: 10.4161/19336896.2014.983396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thuring CM, Erkens JH, Jacobs JG, Bossers A, Van Keulen LJ, Garssen GJ, Van Zijderveld FG, Ryder SJ, Groschup MH, Sweeney T, Langeveld JP. 2004. Discrimination between scrapie and bovine spongiform encephalopathy in sheep by molecular size, immunoreactivity, and glycoprofile of prion protein. J Clin Microbiol 42:972–980. doi: 10.1128/JCM.42.3.972-980.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thuring CM, van Keulen LJ, Langeveld JP, Vromans ME, van Zijderveld FG, Sweeney T. 2005. Immunohistochemical distinction between preclinical bovine spongiform encephalopathy and scrapie infection in sheep. J Comp Pathol 132:59–69. doi: 10.1016/j.jcpa.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 42.Jeffrey M, Gonzalez L, Chong A, Foster J, Goldmann W, Hunter N, Martin S. 2006. Ovine infection with the agents of scrapie (CH1641 isolate) and bovine spongiform encephalopathy: immunochemical similarities can be resolved by immunohistochemistry. J Comp Pathol 134:17–29. doi: 10.1016/j.jcpa.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Feraudet C, Morel N, Simon S, Volland H, Frobert Y, Creminon C, Vilette D, Lehmann S, Grassi J. 2005. Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J Biol Chem 280:11247–11258. doi: 10.1074/jbc.M407006200. [DOI] [PubMed] [Google Scholar]
- 44.Harmeyer S, Pfaff E, Groschup MH. 1998. Synthetic peptide vaccines yield monoclonal antibodies to cellular and pathological prion proteins of ruminants. J Gen Virol 79:937–945. doi: 10.1099/0022-1317-79-4-937. [DOI] [PubMed] [Google Scholar]
- 45.Demart S, Fournier JG, Creminon C, Frobert Y, Lamoury F, Marce D, Lasmezas C, Dormont D, Grassi J, Deslys JP. 1999. New insight into abnormal prion protein using monoclonal antibodies. Biochem Biophys Res Commun 265:652–657. doi: 10.1006/bbrc.1999.1730. [DOI] [PubMed] [Google Scholar]
- 46.Slootstra JW, Puijk WC, Ligtvoet GJ, Langeveld JP, Meloen RH. 1996. Structural aspects of antibody-antigen interaction revealed through small random peptide libraries. Mol Divers 1:87–96. doi: 10.1007/BF01721323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rezaei H, Marc D, Choiset Y, Takahashi M, Hui Bon Hoa G, Haertle T, Grosclaude J, Debey P. 2000. High yield purification and physico-chemical properties of full-length recombinant allelic variants of sheep prion protein linked to scrapie susceptibility. Eur J Biochem 267:2833–2839. doi: 10.1046/j.1432-1033.2000.01347.x. [DOI] [PubMed] [Google Scholar]
- 48.Goldmann W, Hunter N, Foster JD, Salbaum JM, Beyreuther K, Hope J. 1990. Two alleles of a neural protein gene linked to scrapie in sheep. Proc Natl Acad Sci U S A 87:2476–2480. doi: 10.1073/pnas.87.7.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jeffrey M, Martin S, Barr J, Chong A, Fraser JR. 2001. Onset of accumulation of PrPres in murine ME7 scrapie in relation to pathological and PrP immunohistochemical changes. J Comp Pathol 124:20–28. doi: 10.1053/jcpa.2000.0423. [DOI] [PubMed] [Google Scholar]
- 50.Langeveld JP, Jacobs JG, Erkens JH, Bossers A, van Zijderveld FG, van Keulen LJ. 2006. Rapid and discriminatory diagnosis of scrapie and BSE in retro-pharyngeal lymph nodes of sheep. BMC Vet Res 2:19. doi: 10.1186/1746-6148-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryder SJ, Dexter GE, Heasman L, Warner R, Moore SJ. 2009. Accumulation and dissemination of prion protein in experimental sheep scrapie in the natural host. BMC Vet Res 5:9. doi: 10.1186/1746-6148-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fediaevsky A, Tongue SC, Noremark M, Calavas D, Ru G, Hopp P. 2008. A descriptive study of the prevalence of atypical and classical scrapie in sheep in 20 European countries. BMC Vet Res 4:19. doi: 10.1186/1746-6148-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diaz C, Vitezica ZG, Rupp R, Andreoletti O, Elsen JM. 2005. Polygenic variation and transmission factors involved in the resistance/susceptibility to scrapie in a Romanov flock. J Gen Virol 86:849–857. [DOI] [PubMed] [Google Scholar]
- 54.Cartoni C, Schinina ME, Maras B, Nonno R, Vaccari G, Di Bari M, Conte M, De Pascalis A, Principe S, Cardone F, Pocchiari M, Agrimi U. 2007. Quantitative profiling of the pathological prion protein allotypes in bank voles by liquid chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 849:302–306. doi: 10.1016/j.jchromb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 55.Principe S, Maras B, Schinina ME, Pocchiari M, Cardone F. 2008. Unraveling the details of prion (con)formation(s): recent advances by mass spectrometry. Curr Opin Drug Discov Devel 11:697–707. [PubMed] [Google Scholar]
- 56.Silvestrini MC, Cardone F, Maras B, Pucci P, Barra D, Brunori M, Pocchiari M. 1997. Identification of the prion protein allotypes which accumulate in the brain of sporadic and familial Creutzfeldt-Jakob disease patients. Nat Med 3:521–525. doi: 10.1038/nm0597-521. [DOI] [PubMed] [Google Scholar]
- 57.Cardone F, Principe S, Schinina ME, Maras B, Capellari S, Parchi P, Notari S, Di Francesco L, Poleggi A, Galeno R, Vinci R, Mellina V, Almonti S, Ladogana A, Pocchiari M. 2014. Mutant PrPCJD prevails over wild-type PrPCJD in the brain of V210I and R208H genetic Creutzfeldt-Jakob disease patients. Biochem Biophys Res Commun 454:289–294. doi: 10.1016/j.bbrc.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 58.Chen SG, Parchi P, Brown P, Capellari S, Zou W, Cochran EJ, Vnencak-Jones CL, Julien J, Vital C, Mikol J, Lugaresi E, Autilio-Gambetti L, Gambetti P. 1997. Allelic origin of the abnormal prion protein isoform in familial prion diseases. Nat Med 3:1009–1015. doi: 10.1038/nm0997-1009. [DOI] [PubMed] [Google Scholar]
- 59.Tagliavini F, Prelli F, Porro M, Rossi G, Giaccone G, Farlow MR, Dlouhy SR, Ghetti B, Bugiani O, Frangione B. 1994. Amyloid fibrils in Gerstmann-Straussler-Scheinker disease (Indiana and Swedish kindreds) express only PrP peptides encoded by the mutant allele. Cell 79:695–703. doi: 10.1016/0092-8674(94)90554-1. [DOI] [PubMed] [Google Scholar]
- 60.Capellari S, Cardone F, Notari S, Schinina ME, Maras B, Sita D, Baruzzi A, Pocchiari M, Parchi P. 2005. Creutzfeldt-Jakob disease associated with the R208H mutation in the prion protein gene. Neurology 64:905–907. doi: 10.1212/01.WNL.0000152837.82388.DE. [DOI] [PubMed] [Google Scholar]
- 61.Kitamoto T, Yamaguchi K, Doh-ura K, Tateishi J. 1991. A prion protein missense variant is integrated in kuru plaque cores in patients with Gerstmann-Straussler syndrome. Neurology 41:306–310. doi: 10.1212/WNL.41.2_Part_1.306. [DOI] [PubMed] [Google Scholar]
- 62.Parchi P, Chen SG, Brown P, Zou W, Capellari S, Budka H, Hainfellner J, Reyes PF, Golden GT, Hauw JJ, Gajdusek DC, Gambetti P. 1998. Different patterns of truncated prion protein fragments correlate with distinct phenotypes in P102L Gerstmann-Straussler-Scheinker disease. Proc Natl Acad Sci U S A 95:8322–8327. doi: 10.1073/pnas.95.14.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monaco S, Fiorini M, Farinazzo A, Ferrari S, Gelati M, Piccardo P, Zanusso G, Ghetti B. 2012. Allelic origin of protease-sensitive and protease-resistant prion protein isoforms in Gerstmann-Straussler-Scheinker disease with the P102L mutation. PLoS One 7:e32382. doi: 10.1371/journal.pone.0032382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crowell J, Hughson A, Caughey B, Bessen RA. 5 August 2015. Host determinants of prion strain diversity independent of prion protein genotype. J Virol doi: 10.1128/JVI.01586-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rigter A, Bossers A. 2005. Sheep scrapie susceptibility-linked polymorphisms do not modulate the initial binding of cellular to disease-associated prion protein prior to conversion. J Gen Virol 86:2627–2634. doi: 10.1099/vir.0.80901-0. [DOI] [PubMed] [Google Scholar]
- 66.Goldmann W, Hunter N, Martin T, Dawson M, Hope J. 1991. Different forms of the bovine PrP gene have five or six copies of a short, G-C-rich element within the protein-coding exon. J Gen Virol 72:201–204. doi: 10.1099/0022-1317-72-1-201. [DOI] [PubMed] [Google Scholar]