ABSTRACT
Natural influenza A virus infections elicit both virus-specific antibody and CD4+ and CD8+ T cell responses. Influenza A virus-specific CD8+ cytotoxic T lymphocytes (CTLs) contribute to clearance of influenza virus infections. Viral CTL epitopes can display variation, allowing influenza A viruses to evade recognition by epitope-specific CTLs. Due to functional constraints, some epitopes, like the immunodominant HLA-A*0201-restricted matrix protein 1 (M158–66) epitope, are highly conserved between influenza A viruses regardless of their subtype or host species of origin. We hypothesized that human influenza A viruses evade recognition of this epitope by impairing antigen processing and presentation by extraepitopic amino acid substitutions. Activation of specific T cells was used as an indication of antigen presentation. Here, we show that the M158–66 epitope in the M1 protein derived from human influenza A virus was poorly recognized compared to the M1 protein derived from avian influenza A virus. Furthermore, we demonstrate that naturally occurring variations at extraepitopic amino acid residues affect CD8+ T cell recognition of the M158–66 epitope. These data indicate that human influenza A viruses can impair recognition by M158–66-specific CTLs while retaining the conserved amino acid sequence of the epitope, which may represent a yet-unknown immune evasion strategy for influenza A viruses. This difference in recognition may have implications for the viral replication kinetics in HLA-A*0201 individuals and spread of influenza A viruses in the human population. The findings may aid the rational design of universal influenza vaccines that aim at the induction of cross-reactive virus-specific CTL responses.
IMPORTANCE Influenza viruses are an important cause of acute respiratory tract infections. Natural influenza A virus infections elicit both humoral and cellular immunity. CD8+ cytotoxic T lymphocytes (CTLs) are directed predominantly against conserved internal proteins and confer cross-protection, even against influenza A viruses of various subtypes. In some CTL epitopes, mutations occur that allow influenza A viruses to evade recognition by CTLs. However, the immunodominant HLA-A*0201-restricted M158–66 epitope does not tolerate mutations without loss of viral fitness. Here, we describe naturally occurring variations in amino acid residues outside the M158–66 epitope that influence the recognition of the epitope. These results provide novel insights into the epidemiology of influenza A viruses and their pathogenicity and may aid rational design of vaccines that aim at the induction of CTL responses.
INTRODUCTION
Influenza viruses are among the leading causes of acute respiratory tract infections worldwide (1). Classification of influenza A viruses (IAVs) is based on their surface glycoproteins hemagglutinin (HA) and neuraminidase (NA). At present, 18 HA subtypes (H1 to H18) and 11 NA subtypes (N1 to N11) have been identified (2, 3). IAVs of the H3N2 and H1N1 subtype together with influenza B viruses cause yearly epidemics in the human population (1). Other IAV subtypes circulate in animal reservoirs, like aquatic birds and pigs (4), but can occasionally cross the species barrier into the human population (5). Genetic reassortment between animal and human IAVs has resulted in the emergence of pandemic strains in the last century (6–9).
Natural influenza virus infections elicit both humoral and cellular immune responses. Virus-neutralizing antibodies are mainly directed against the highly variable globular head of the HA protein and prevent reinfection with the same virus (10). However, most antibodies have limited cross-reactivity against influenza viruses of another subtype (11, 12) and may afford little protection against the development of severe disease caused by infection with antigenically distinct viruses, including those of novel subtypes.
Influenza virus-specific CD8+ T cells (cytotoxic T lymphocytes [CTLs]), on the other hand, are directed predominantly against more conserved internal proteins (13, 14) and recognize their epitopes as major histocompatibility complex (MHC) class I/peptide complexes (15). The recognition of conserved proteins results in a high degree of cross-reactivity with antigenically distinct IAVs (13, 14, 16, 17). Although CTLs do not afford sterilizing immunity, they contribute substantially to viral clearance and reduce the severity of infections with influenza viruses, including those with antigenically distinct HA or NA (18–20). However, the high mutation rate of influenza viruses and the selective pressure exerted by virus-specific CTLs drive the accumulation of amino acid substitutions that are associated with evasion from recognition by CTLs specific for some epitopes. Indeed, significantly more nonsynonymous mutations are observed in CTL epitopes than in the rest of the viral nucleoprotein (NP) (21, 22). Amino acid substitutions in T cell receptor (TCR) contact residues have been identified that result in loss of recognition by epitope-specific CTLs (13, 23), as has been described for the human leukocyte antigen (HLA)-B*3501-restricted NP418–426 epitope (24). In addition, mutations at anchor residues of CTL epitopes have been identified (13, 23), which resulted in complete loss of the CTL epitope, as has been described for the HLA-B*2705-restricted NP383–391 epitope (25, 26). Both types of CTL escape mutations were observed during natural evolution of seasonal IAVs (H3N2) (23, 25). Similar CTL evasion strategies have been described for viruses that cause chronic infections, like human immunodeficiency virus (HIV) (27), hepatitis C virus (HCV) (28), Epstein-Barr virus (EBV) (29), and lymphocytic choriomeningitis virus (LCMV) (30).
In contrast, some IAV CTL epitopes are highly conserved even between different subtypes of IAV, like the HLA-A*0201/HLA-C*0801-restricted M158–66 (GILGFVFTL) epitope (31, 32). Matrix protein 1 (M1) of seasonal A/H3N2 viruses originates from the 1918 pandemic A/H1N1 virus (Fig. 1) (6–8, 33, 34). Most likely, the selective pressure against the M158–66 epitope is high, considering the immunodominant nature of the epitope (35) and the high prevalence of the HLA-A*0201 allele in the Caucasian population (>40%) (36). However, mutations at TCR contact or anchor residues were not tolerated in this epitope without loss of viral fitness (21, 37), which coincides with the presence of a highly conserved nuclear export signal overlapping the M158–66 epitope (38).
FIG 1.
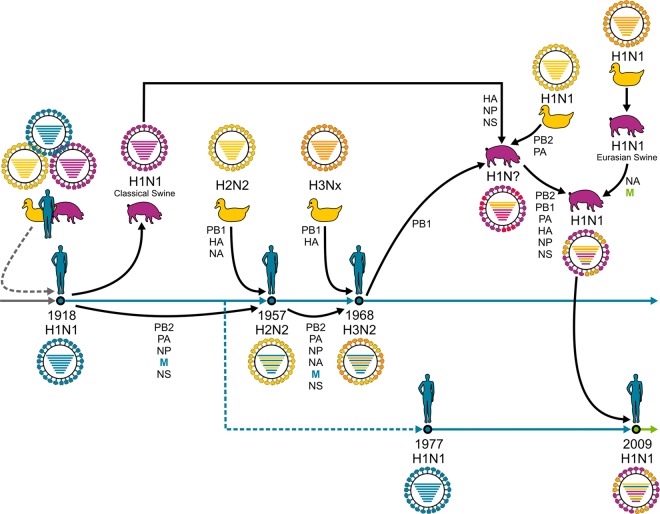
Reassortment events of pandemic influenza A viruses. The 1918 A/H1N1 virus possibly originated from multiple reassortment events between avian, swine, and human viruses. This A/H1N1 virus continued to circulate, causing seasonal epidemics, until 1957, when a novel A/H2N2 virus emerged after a reassortment event with an avian A/H2N2 virus. This virus circulated until 1968, when it reassorted with an avian A/H3Nx virus, and caused seasonal epidemics ever since. A/H1N1 was reintroduced in the human population in 1977 and cocirculated with A/H3N2 viruses until 2009, when it was replaced by H1N1pdm2009, which originated after multiple reassortment events between avian, swine, and human viruses. Although it is unknown whether the M gene segment originated from viruses that were newly introduced into humans or had circulated in humans prior to 1918, viruses with this gene segment continued to circulate in the human population in A/H2N2 and A/H3N2 viruses until today (blue arrow). Also, the A/H1N1 virus that was reintroduced into the human population in 1977 contained the M gene segment of 1918 origin, but this virus was replaced by a virus with a swine derived M gene segment during the A/H1N1 pandemic outbreak of 2009 (green arrow).
We hypothesized that IAVs may have adopted other escape mechanisms for highly conserved CTL epitopes, like the M158–66 epitope, based on the observation that avian IAVs of the H5N1 and H7N9 subtype are better recognized by polyclonal IAV-specific CTLs than human seasonal IAVs (16, 17). It is well known that amino acid substitutions flanking an epitope can affect antigen presentation by changing the cleavage motifs used by the proteasome, alter trimming of the N- and C-terminal sequence by cytosolic or endoplasmic reticulum (ER)-resident proteases or impair the translocation via TAP (transporter associated with antigen presentation) (15, 39, 40). So far, an effect of extraepitopic mutations on T cell recognition has been demonstrated only for CTLs directed to viruses that cause chronic infections, including for HIV (27), HCV (41), and EBV (42, 43).
Here, we investigated whether differences in extraepitopic amino acid residues between avian and human IAVs could be responsible for differential recognition of the M158–66 epitope. It was demonstrated that naturally occurring amino acid differences at positions in the region flanking the otherwise fully conserved M158–66 epitope affect recognition by M158–66-specific CD8+ T cells. The reduced recognition of human IAVs by M158–66-specific CD8+ T cells by extraepitopic amino acid substitutions indicates the existence of an immune evasion strategy additional to variation in CTL epitopes and may help the virus to perpetuate in the human population in the presence of pre-existing virus-specific CD8+ T cell immunity. Furthermore, these results are of interest for the development of vaccines that aim at the induction of virus-specific CTL responses.
MATERIALS AND METHODS
Cells.
An HLA-A*0101/A*0201/B*0801/B*2705 B lymphoblastoid cell line (BLCL) was prepared as described previously (44). BLCs were cultured in RPMI 1640 medium (Lonza, Basal, Switzerland) supplemented with 100 μg/ml penicillin, 100 U/ml streptomycin, 2 mM l-glutamine (P/S/G) (Lonza), and 10% fetal bovine serum (FBS) (Sigma-Aldrich, Zwijndrecht, The Netherlands) (R10F medium).
The previously described A549-HLA-A*0201+ human lung carcinoma cell line (45) was cultured in Ham's F-12 medium (Gibco Life Technologies, Bleiswijk, The Netherlands) containing P/S/G and 10% fetal calf serum (FCS) (Hycone) (Sigma) (H10F) and in the presence of 1 μg/ml puromycin (Invivogen, Toulouse, France). HLA-A*02 expression was confirmed by staining with anti-HLA-A*02-FITC (BD Biosciences, Breda, The Netherlands) and using a FACSCanto II flow cytometer and FACSDiva software (Becton Dickinson B.V., Breda, The Netherlands), prior to each experiment.
Peptides.
Synthetic immunograde peptides (>85% purity) of the HLA-A*0201-restricted M158–66 (GILGFVFTL) and the HLA-B*2705 NP383–391 (SRYWAIRTR) epitopes were purchased (Eurogentec, Seraing, Belgium). Peptides were dissolved in dimethyl sulfoxide (5 mg/ml), diluted to 100 μM in RPMI 1640 medium, and stored at −20°C until further use.
Plasmids.
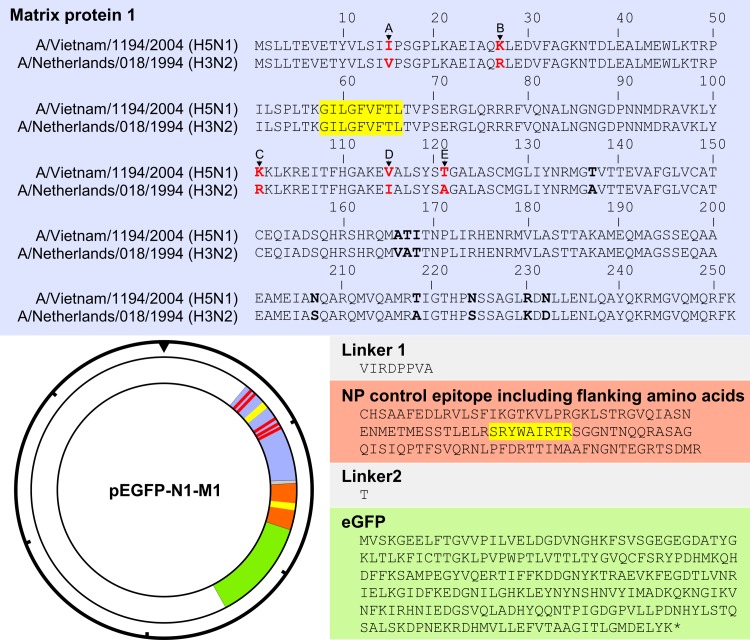
The open reading frames (ORF) of the M1 protein of influenza viruses A/Netherlands/018/1994 (H3N2) and A/Vietnam/1194/2004 (H5N1) without their stop codons were cloned in frame with the ORF of enhanced green fluorescent protein (eGFP) into the pEGFP-N1 plasmid (Becton Dickinson) as described previously (46). Next, the ORF of the NP383–391 epitope (SRYWAIRTR), including 50 N- and C-terminal amino acids (all derived from A/Puerto Rico/8/1934), was cloned in frame between the ORFs of the M1 and eGFP protein (Fig. 2). Briefly, the NP insert (nucleotide [nt] positions 997 to 1323) was created by PCR amplification of A/Puerto Rico/8/1934-derived NP cDNA using a forward and reverse primer that encompassed 20 nt of the vector and 20 nt of the desired NP insert. These primers were used for PCR, as follows: the mixture was 10 pmol of each primer, 5 μl of PFU Ultra II buffer, 1 μl of PFU Ultra II enzyme (Agilent Technologies, Amstelveen, The Netherlands), deoxynucleoside triphosphates (dNTPs; 10 mM each) (Roche, Woerden, The Netherlands), and 100 ng A/Puerto Rico/8/1934 NP gene segment cDNA in a final volume of 50 μl, which was subsequently incubated at 95°C for 3 min (min), followed by 40 cycles of 95°C for 1 min, 1 min at 45°C, and 72°C for 2 min. PCR products were loaded on a 1% agarose gel, and DNA was isolated using the Minelute gel extraction kit from Qiagen according to the manufacturer's instructions (Qiagen, Venlo, The Netherlands). This purified PCR product served as a “megaprimer.” The second PCR was performed as described above, except that 100, 300, and 500 ng of the megaprimer were combined with 50 and 100 ng of vector DNA and 3 μl Quik solution (Agilent). The PCR product was digested for 1 h at 37°C with 20 U of DpnI (New England BioLabs, Ipswich, MA, USA). The DpnI-digested PCR product was transformed using Z-competent XL-10 Gold cells (Zymo Research, Irvine, CA, USA). Plasmid DNA was purified using a Genopure plasmid maxikit (Roche). Reciprocal exchange of the extraepitopic amino acids in the M1 protein at positions 15, 27, 101, 115, and 121 was introduced using the QuikChange Multi site-directed mutagenesis kit according to the manufacturer's instructions (Agilent Technologies) (Fig. 2). These plasmids were used in the fluorescent antigen-transfected target cell-CTL (FATT-CTL) assay.
FIG 2.
Amino acid sequences of viral M1-NP-eGFP fusion proteins and expression plasmid map. The amino acid sequence of the chimeric M1-NP-eGFP fusion construct is shown with the avian IAV A/Vietnam/1194/2004 (H5N1) and human IAV A/Netherlands/018/1994 (H3N2) M1 sequences in blue, NP in orange, and eGFP in green. Linker sequences are shown in gray. The locations of the M158–66 (GILGFVFTL) and NP383–391 (SRYWAIRTR) epitopes are highlighted in yellow. Amino acid differences studied here are in red type, and additional amino acid differences are in bold. The insert was cloned into the pEGFP-N1 vector as indicated. The hash marks around the perimeter of the plasmid map indicate 1,000-nucleotide increments.
A bidirectional reverse genetic system based on influenza virus A/Netherlands/178/1995 (H3N2; M), A/Vietnam/1194/2005 (H5N1; M) and A/Puerto Rico/8/1934 (H1N1; PB2, PB1, PA, HA, NP, NA, and NS) was used for the generation of recombinant influenza viruses as described previously (21, 47, 48).
Sequences of all recombinant plasmids were confirmed by sequence analysis using a BigDye Terminator v3.1 cycle sequencing kit and a 3130xl genetic analyzer (Applied Biosystems, Bleiswijk, The Netherlands) prior to use.
Viruses.
293T cells were transfected with the recombinant bidirectional plasmids (M derived from A/Netherlands/178/1995 or A/Vietnam/1194/2005; other gene segments derived from A/Puerto Rico/8/1934) as described previously (47). Culture supernatants were harvested after 48 h and used for a subsequent inoculation of Madin-Darby canine kidney (MDCK) cells (26). After 3 days, culture supernatants were harvested and passaged twice in MDCK cells. Culture supernatants were clarified by low-speed centrifugation and subsequently purified by ultracentrifugation through a sucrose gradient. Sequence analysis was used to confirm the sequence of the M gene segments as described above, and their infectious-virus titers were determined as described previously (49).
It is important to note that the M gene segment of the recombinant virus was derived from an alternative A/H3N2 virus (A/Netherlands/178/1995) which differed from the A/Netherlands/018/1994 virus at amino acid positions 227 and 239 (A227T and A239T). However, we argue that due to the large C-terminal distance (>160 amino acids) from the M158–66 epitope, these amino acid differences are unlikely to interfere with the processing of this epitope.
T cell clones.
CD8+ T cell clones directed against the HLA-A*0201-restricted M158–66 (GILGFVFTL) epitope and the HLA-B*2705-restricted NP383–391 (SRYWAIRTR) epitope were generated as described previously (25, 50).
FATT-CTL assay.
The fluorescent antigen-transfected target cell-CTL (FATT-CTL) assay was used for the detection of lytic activity of the M158–66- and NP383–391-specific CD8+ T cell clones as described previously (46). Briefly, the cell line Nucleofector kit V (Lonza), program T16, was used to transfect 5 × 106 BLCs with 8 μg plasmid DNA and incubated in R10F for 4 h at 37°C. The number of viable eGFP-positive cells was determined after TOPRO-3 iodide (Invitrogen, Breda, The Netherlands) staining using the FACSCanto II flow cytometer and FACSDiva software. Quadruple samples consisting of 1,500 viable eGFP-positive target cells were cocultured for another 3.5 h with 20,000, 40,000, or 80,000 M158–66- or NP383–391-specific CD8+ T cells, and the number of viable eGFP-positive cells was determined as described above. (For the gating strategy performed with FlowJo software [FlowJo, Ashland, OR, USA], see Fig. 3A.) The percent epitope-specific lysis was then calculated using the following formula: 100 × [(number of viable eGFP-positive cells in the sample without effector − number of viable eGFP-positive cells in the sample with effector)/number of viable eGFP-positive cells in the sample without effector].
FIG 3.
Lytic activity of M158–66- and NP383–391-specific CD8+ T cells against target cells transfected with various M1-NP-eGFP encoding plasmids. (A) Gating strategy used to assess the number of viable eGFP+ target cells. The first dot plot demonstrates a gate for the transfected target cells, the second gate demonstrates the viable cells, and the third gate demonstrates the eGFP+ cells. (B) Percent lytic activity exerted by the M158–66-specific CD8+ T cell clone. (C) Percent lytic activity exerted by the NP383–391-specific CD8+ T cell clone. Target cells were transfected with chimeric M1-NP-eGFP fusion plasmids that encode the M1 protein of the avian A/H5N1 virus (WT avian), the M1 protein of the human A/H3N2 virus (WT human), the M1 protein of avian A/H5N1 virus with extraepitopic amino acid residues of the human A/H3N2 virus (A→H ABCDE) and the M1 protein of human A/H3N2 virus with extraepitopic amino acid residues of the avian A/H5N1 virus (H→ A ABCDE). Data points represent means and error bars indicate standard deviations (SD) for quadruplicates (n = 4). **, all groups were statistically significantly different from each other after correction for multiple hypothesis testing using a false discovery rate (FDR) of 0.01; *, only the value for the A→H ABCDE group was significantly lower.
Kinetics of CD8+ T cell activation in the FATT-CTL assay.
BLCs were transfected and counted as described above. Quadruple samples consisting of 3,000 viable eGFP-positive target cells were cocultured for another 7 h with 10,000 or 20,000 M158–66- or NP383–391-specific CD8+ T cells in the presence of Golgistop [4 μl/6 ml] (BD Biosciences) and 0.5 μl/100 μl CD107a-V450 (BD Biosciences) and subsequently stained in phosphate-buffered saline (PBS) supplemented with 2% FBS and Golgistop (P2FG) with CD3 conjugated with peridinin-chlorophyll proteins-cyanine5.5 (PerCP-Cy5.5), allophycocyanin (APC)-conjugated CD8 (eBioscience, Vienna, Austria), CD137 conjugated with phycoerythrin-cyanine7 (PE-Cy7) (BioLegend, London, United Kingdom), CD69-APC-H7 (BD Biosciences), and Live/Dead fixable aqua dead-cell stain (L/D) (Invitrogen). Next, cells were fixed using Cytofix (BD Biosciences) and stored in PBS supplemented with 0.5% bovine serum albumin (BSA) and 2 mM EDTA (Sigma) at 4°C until they were analyzed using a FACSCanto II flow cytometer and FACSDiva software. (For the gating strategy using FlowJo software, see Fig. 4A.) BLCs pulsed with or without 100 μM GILGFVFTL or SRYWAIRTR peptide were used as a positive control (data not shown).
FIG 4.
Activation of M158–66- and NP383–391-specific CD8+ T cells after stimulation with target cells transfected with various M1-NP-eGFP-encoding plasmids. (A) Dot plots on top from left to right gate the transfected target cells, the viable cells, the CD3− CD8− cells, and finally the eGFP+ cells. This gating was used to assess the lytic activity of the CD8+ T cell clone in a FATT-CTL-dependent manner. Lower dot plots gate the lymphocytes, single cells, viable cells, and CD3+ CD8+ cells followed by gating for the upregulation of activation markers CD137, CD69, and CD107a after stimulation with target cells transfected with the M1-NP-eGFP plasmids (colored histograms) or the eGFP only plasmid (gray histogram). (B) Percent lytic activity exerted by the M158–66-specific CD8+ T cells (top) and NP383–391-specific CD8+ T cells (bottom). Upregulation of the activation markers CD137, CD69, and CD107a on M158–66-specific CD8+ T cells (C) or the NP383–391-specific CD8+ T cells (D) after stimulation with target cells transfected with M1-NP-eGFP plasmids that encode the M1 protein of the avian A/H5N1 virus (WT avian), the M1 protein of the human A/H3N2 virus (WT human), the avian A/H5N1 M1 protein with extraepitopic amino acid residues of the human A/H3N2 virus (A→H ABCDE), the human A/H3N2 M1 protein with extraepitopic amino acid residues of the avian A/H5N1 virus (H→A ABCDE) or eGFP only (mock). Data points represent means and error bars indicate SD for quadruplicates (n = 4). **, differences between groups are statistically significant after correction for multiple hypothesis testing using an FDR of 0.01.
IFN-γ ELISpot assay.
The interferon gamma (IFN-γ) responses of M158–66- and NP383–391-specific CD8+ T cells were determined by enzyme-linked immunosorbent spot (ELISpot) assay, which was performed according to the manufacturer's instructions (Mabtech, Nacka Strand, Sweden). In brief, 3,000 transfected viable eGFP-positive BLCs were incubated with 10,000 M158–66- or NP383–391-specific CD8+ T cells for 7 h, in quadruplicate. The average number of spots was determined using an ELISpot reader and image analysis software (Aelvis, Sanquin Reagents, Amsterdam, The Netherlands).
Virus infection and kinetics CD8+ T cell activation.
The kinetics of M156–66-specific CD8+ T cells activation after stimulation with A549-HLA-A*0201+ cells infected with the avian or human recombinant viruses was studied by assessing expression of activation markers CD137, CD69, and CD107a. Peptide-pulsed A549-HLA-A*0201+ cells were used as a positive control.
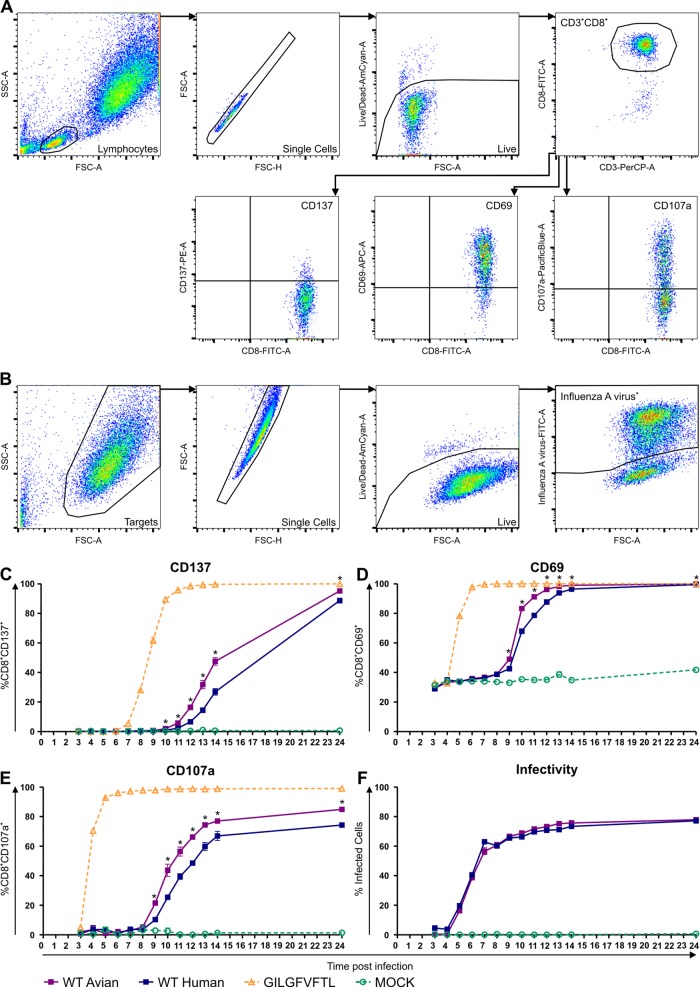
A549-HLA-A*0201+ cells were incubated with or without 100 μM GILGFVFTL in H10F for 1 h at 37°C in an ultralow-attachment plate (Corning, New York, USA). Meanwhile, virus-infected target cells were prepared by inoculating A549-HLA-A*0201+ cells at a multiplicity of infection (MOI) of 3 with the avian or human recombinant virus in ultralow-attachment plates. After 1 h, cells were washed with H10F and cocultured with the M156–66-specific CD8+ T cell clone at an effector-to-target cell (E:T) ratio of 0.2 in the presence of Golgistop [4 μl/6 ml] and 0.5 μl/100 μl CD107a-V450, in triplicate for each time point. Cells were stained each hour from 3 until 14 h postinoculation (p.i.) and at 24 h p.i. with CD8 conjugated with fluorescein isothiocyanate (FITC) (Dako, Glostrup, Denmark), CD137-PE (Miltenyi Biotec, Bergisch Gladbach, Germany), CD3-PerCP, CD69-APC (BD Biosciences), and L/D and subsequently fixed and stored as described above. Virus-infected A549-HLA-A*0201+ cells in the absence of the M156–66-specific CD8+ T cell clone were simultaneously stained with L/D and subsequently fixed and permeabilized with Cytofix and Cytoperm (BD Biosciences), after which the cells were stained for 30 min at 4°C with anti-influenza A virus–FITC (reagent A) (Oxoid, Landsmeer, The Netherlands). Cells were analyzed using a FACSCanto II flow cytometer and FACSDiva software. (For the gating strategy using FlowJo software, see Fig. 6A and B.)
FIG 6.
Activation kinetics of M158–66-specific CD8+ T cells after stimulation with cells infected with isogenic influenza A viruses with gene segment 7 of human or avian influenza A viruses. (A) Gating strategy used to assess the upregulation of activation markers on M158–66-specific CD8+ T cells. Dot plots gate the lymphocytes, single cells, viable cells, and CD3+ CD8+ cells, followed by gating for the upregulation of the activation markers CD137, CD69, and CD107a. (B) Gating strategy used to determine infection efficiency of the target cells. Dot plots gate the target cells, the viable cells, and finally the influenza A virus-positive cells. Expression of the activation markers CD137 (C), CD69 (D), and CD107a (E) by M158–66-specific CD8+ T cells after stimulation with A549-HLA-A*0201+ cells infected with recombinant virus A/Puerto Rico/8/1934 with gene segment 7 of avian virus A/Vietnam/1194/2004 (H5N1) (WT avian) or human virus A/Netherlands/178/1995 (H3N2) (WT human) or pulsed with M158–66 peptide (GILGFVFTL) or untreated (mock) is shown. (F) Percent infected A549-HLA-A*0201+ cells at each time point (without T cells). The x axis represent hours postinfection. A549-HLA-A*0201+ cells were infected and peptide pulsed for 1 h and then used to stimulate M158–66-specific CD8+ T cells. Data points represent means and error bars indicate SD for triplicates (n = 3). *, the differences between avian and human derived viruses were statistically significant after correction for multiple hypothesis testing using an FDR of 0.01.
Sequence data.
To assess the frequency of amino acid variations in the M1 protein at positions 15, 27, 101, 115, and 121 in avian (all subtypes available from 2001 to 2015), swine (A/H3N2, 1977 to 2015, and A/H1N1, 1930 to 2015), and human (A/H1N1, 1918 to 1957, A/H2N2, 1957 to 1968, A/H3N2, 1968 to 2015, A/H1N1, 1977 to 2008, and A/H1N1, 2009 to 2015) viruses, all full-length M1 amino acid sequences available in the influenza virus resource database of the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/genomes/FLU) as of 11 April 2015 were downloaded. Due to the large number of avian viruses available, we collapsed all identical sequences prior to analysis. After excluding sequences with large deletions using BioEdit, we analyzed the data set in Ugene 1.16.1 (http://ugene.unipro.ru; Unipro, Novosibirsk, Russia) to assess the frequency of the avian or human amino acids at positions 15, 27, 101, 115, and 121. Viruses were analyzed in Excel to determine whether observed frequencies were the result of cluster formation and whether certain mutations became fixed in time.
Statistical analysis.
Data from the FATT-CTL, ELISpot, and activation assays were analyzed using the independent-sample t test to calculate the respective P value between pairs of groups. These P values were then analyzed using the Benjamini-Hochberg method (a false discovery rate [FDR] of 0.01 was used for all assays) to correct for multiple hypothesis testing (51). Each experiment, with the exception of the IFN-γ ELISpot assay, was performed at least twice.
RESULTS
Differences in lytic activity of M158–66-specific CD8+ T cells against M1 proteins derived from avian and human influenza A viruses.
Viruses were selected based on the previous observation that avian IAV A/Vietnam/1194/2004 (H5N1) was better recognized by IAV-specific CTLs than human IAV A/Netherlands/018/1994 (H3N2) (16), which may be attributable to a yet-unidentified, additional CTL escape mechanism utilized by human A/H3N2 viruses.
The M1 genes of both viruses were cloned in frame with the enhanced green fluorescent protein (eGFP) gene into an expression plasmid as described previously (46). In addition, a region of the NP gene, encoding the HLA-B*2705 NP383–391 (SRYWAIRTR) epitope and 50 N- and C-terminal flanking amino acids, was cloned in frame between the M1 and eGFP genes (Fig. 2). The NP383–391 epitope was included as a control, since CTLs specific for this epitope have functional avidity similar to that of CTLs directed to the M158–66 epitope (35). These plasmids were used in the FATT-CTL assay to monitor the lytic activity by M158–66-specific CD8+ T cells (Fig. 3A), as described previously (46). Lytic activity of the NP383–391-specific CD8+ T cells was used as a control to exclude differences in transfection efficiencies and/or protein expression levels. M158–66-specific CD8+ T cells lysed significantly more target cells expressing the M1 protein derived from avian virus A/Vietnam/1194/2004 (H5N1) than those expressing the M1 protein of human virus A/Netherlands/018/1994 (H3N2) (83% and 45%, respectively, at the highest effector-to-target [E:T] ratio) (Fig. 3B). The lytic activities of NP383–391-specific CD8+ T cells for both M1-NP-eGFP fusion proteins were similar (70% at the highest E:T ratio) (Fig. 3C). These results demonstrate that the M158–66 epitope in the context of an M1 protein derived from a human IAV is less well recognized than its counterpart in the context of an M1 protein derived from an avian IAV. Next, we wished to assess whether differences in amino acids flanking the M158–66 epitope had contributed to the observed difference in lytic activity of epitope-specific CD8+ T cells. Although no amino acid differences were found in close proximity to the M158–66 epitope, we identified five extraepitopic (avian-to-human) amino acid substitutions within a 60-amino-acid distance of the M158–66 epitope, namely, at positions I15V (substitution A), K27R (substitution B), K101R (substitution C), V115I (substitution D), and T121A (substitution E) (Fig. 2). Reciprocal exchange of these extraepitopic amino acid residues in the M1 protein allowed assessment of the effect of these substitutions on recognition by M158–66-specific CD8+ T cells. Exchanging the extraepitopic amino acid residues partially reversed the recognition pattern of the M155–66-specific CD8+ T cells. Introducing the five M1 amino acid residues of the human virus into the M1 protein of the avian virus (A→H ABCDE) significantly reduced the lytic activity of the M158–66-specific CD8+ T cells from 83% to 70% at the highest E:T ratio. Introducing the five M1 amino acid residues of the avian virus into the M1 protein of the human virus (H→A ABCDE) significantly improved the lytic activity of M158–66-specific CD8+ T cells from 45% to 56% (Fig. 3B). The H→A ABCDE exchange did not affect recognition by the NP383–391-specific CD8+ T cells, while the A→H ABCDE exchange affected recognition slightly (Fig. 3C). The five amino acid differences are unlikely to have altered the CTL response by interfering with the splice site, as this would have resulted in a shift of the NP open reading frame (ORF) (52), resulting in comparable patterns of recognition by both CD8+ T cell clones, which was not observed.
Differential activation of M158–66-specific CD8+ T cells by M1 protein derived from avian or human IAV.
Next, we investigated activation of M158–66-specific CD8+ T cells after stimulation with HLA-A*0201/B*2705-positive EBV-transformed B cells expressing the respective chimeric M1-NP-eGFP fusion proteins. Upon stimulation, the expression of the activation markers CD137, CD69, and CD107a by M158–66- and NP383–391-specific CD8+ T cells was determined by flow cytometry (Fig. 4A). Because both the E:T ratio and incubation time had to be adapted for this purpose, we also assessed the lytic activity of the CD8+ T cells in a FATT-CTL assay under these conditions (Fig. 4). Again, only the M158–66-specific CD8+ T cells displayed differential lytic activity against target cells expressing the M1 protein derived from avian or human IAV. Once again, the reciprocal exchange of the extraepitopic amino acid residues partially reversed the lytic activity pattern (Fig. 4B). The lytic activity of the NP383–391-specific CD8+ T cells to the respective M1 proteins was similar for all chimeric M1-NP-eGFP fusion proteins (Fig. 4B).
Upon stimulation with the M1 protein derived from avian IAV A/Vietnam/1194/2004 (H5N1), a significantly higher percentage of M158–66-specific CD8+ T cells was positive for the activation markers than after stimulation with the M1 protein derived from the human influenza virus A/Netherlands/018/1994 (H3N2) (approximately a 2.5-fold difference) (for CD137, 19% versus 7.6%; for CD69, 2.5% versus 1.1%; and for CD107a, 54% versus 20%, respectively) (Fig. 4C). Such differences were not observed for the NP383–391-specific CD8+ T cells, although CD107a expression was slightly higher after stimulation with M1 protein from the avian virus (1.1-fold), but this difference was far smaller than that observed for the M158–66-specific CD8+ T cells (Fig. 4D). Again, the reciprocal exchange of the extraepitopic amino acid residues partially reversed the pattern of differential activation of the M158–66-specific CD8+ T cells. The introduction of extraepitopic amino acid residues from the human IAV into the M1 protein of the avian IAV reduced activation of the M158–66-specific CD8+ T cells and vice versa (Fig. 4C). The exchange of amino acid residues in the M1 protein resulted in minor differences in activation of the NP383–391-specific CD8+ T cells. Although some of these small differences were statistically significant, they did not correlate with the activation pattern observed for the M158–66-specific CD8+ T cells (Fig. 4D). In addition to assessing the expression of CD107a, a proxy for degranulation and lytic activity, we also assessed IFN-γ production by the CD8+ T cells as an alternative functional property of CD8+ T cell activation by ELISpot assay. Again, stimulation with the M1 protein derived from the avian IAV resulted in a significantly higher number of IFN-γ-producing M158–66-specific CD8+ T cells than stimulation with the M1 protein of human IAV (228 versus 54 IFN-γ+ spots/104 cells) (Fig. 5A). No significant difference was observed for the NP383–391-specific CD8+ T cells (Fig. 5B). Thus, a good correlation was observed between the differential expression of activation markers (including CD107a), lytic activity, and IFN-γ production by M158–66-specific CD8+ T cells, which was dependent on the source of the M1 proteins used for stimulation and their extraepitopic amino acid residues.
FIG 5.
IFN-γ response by M158–66- and NP383–391-specific CD8+ T cells after stimulation with target cells transfected with various M1-NP-eGFP-encoding plasmids. Numbers of IFN-γ positive spots/104 M158–66-specific CD8+ T cells (A) and NP383–391-specific CD8+ T cells (B) after stimulation by target cells transfected with M1-NP-eGFP plasmids that encode the M1 protein of the avian A/H5N1 virus (WT avian) or the M1 protein of the human A/H3N2 virus (WT human) or eGFP only (mock) are shown. Data points represent means and error bars indicate SD of quadruplicates (n = 4). **, the difference between groups was statistically significant after correction for multiple hypothesis testing using an FDR of 0.01.
The M1 protein context determines the kinetics of M158–66-specific CD8+ T cell activation after stimulation with virus-infected cells.
Finally, we wished to assess whether recognition of cells infected with IAVs carrying either of the respective M1 proteins could lead to differential activation of M158–66-specific CD8+ T cells. To this end, isogenic recombinant viruses containing the matrix (M) gene segment of avian virus A/Vietnam/1194/2004 (H5N1) or human virus A/Netherlands/178/1995 (H3N2) were used to infect A549-HLA-A*0201+ target cells. Two hours postinoculation (p.i.), these infected target cells were incubated with the M158–66-specific CD8+ T cells, and the kinetics of CD137, CD69, and CD107a expression was assessed (Fig. 6A).
As shown in Fig. 6, stimulation with virus containing the M gene segment of human influenza virus A/Netherlands/178/1995 (H3N2) resulted in delayed activation of M158–66-specific CD8+ T cells compared to stimulation with virus containing the M gene segment of avian influenza virus A/Vietnam/1194/2004 (H5N1). One of the earliest markers of T cell activation was expression of the degranulation marker CD107a. Upon stimulation with peptide-pulsed A549-HLA*A0201+ cells, M158–66-specific CD8+ T cells degranulated almost immediately. After stimulation with virus-infected cells, CD107a expression was detected as early as 9 h p.i. Similar observations were made for CD137 and CD69, although CD137 expression started at a later time point than CD69 and CD107a. In any case, the proportion of CD8+ T cells that became activated and expressed either activation marker was significantly higher after stimulation with the virus containing the M gene segment of avian A/H5N1 at every time point p.i. (the greatest difference for CD137 was at 14 h p.i. [1.8-fold] and that for CD107a and CD69 was at 10 h p.i. [1.7- and 1.2-fold, respectively]) (Fig. 6C, D, and E). The replication kinetics of both viruses was very similar and resulted in equal numbers of infected target cells, which excluded infection rates as the cause of the differences in the kinetics of CD8+ T cell activation (Fig. 6B and F). These results clearly indicate that extraepitopic amino acid residues of the M158–66 epitope affect the recognition of viruses containing the M gene segment of the seasonal A/H3N2 virus by M158–66-specific CTLs.
Evolution of extraepitopic amino acid residues of the M158–66 epitope.
In order to link immunologic observations with the epidemiology of IAVs that circulated in the human population, we examined the origin of gene segment 7, which encodes the M1 protein, and the evolution of amino acid residues outside the M158–66 epitope. The M1 protein in the human population originates from the 1918 pandemic virus (Fig. 1) (6–8, 33, 34). Of interest, the extraepitopic substitutions described in this paper were present in most human IAVs isolated since 1918 and were maintained over 100 years of viral evolution in the human population (Fig. 7; Table 1). The only exception was the V115I substitution, which was not observed in the only 1918 virus sequence (A/BrevigMission/1/1918) available in the influenza virus resource database (Fig. 7; Table 1). Interestingly, the H1N1 IAVs that caused the pandemic in 2009 (H1N1pdm09) and that replaced the old seasonal H1N1 viruses possess an M1 protein with an avian/swine signature (Fig. 1 and 7; Table 1).
FIG 7.
Frequency of amino acid variations region flanking M158–66. The frequencies of amino acid variations at positions 15, 27, 101, 115, and 121 in the M1 proteins of avian, swine, and human influenza A viruses isolated in the indicated time period are shown. Frequencies were based on the total number of M1 protein sequences present in the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/genomes/FLU) database as of 11 April 2015. Pie charts represent frequencies of avian (purple) and human (blue) amino acid residues based on the variations observed in Fig. 2. The frequencies of other amino acid residues at these positions are in orange. Frequencies of the preferred human IAV amino acid residues are indicated in the pie charts. *, only one 1918 sequence (A/BrevigMission/1/1918) could be obtained from the NCBI database, which was the only virus in this group containing the 115V residue; the other H1N1 viruses in this group were from the 1930s. A more detailed overview of the frequencies of the respective amino acid variations at these positions can be found in Table 1.
TABLE 1.
Frequency of amino acid differences in the region flanking the M158–66 epitopea
“Position” refers to the position in the M1 amino acid sequence; “no.” indicates the absolute number of viruses. Boldface indicates amino acids present in the avian H5N1 (A/Vietnam/1194/04) and human H3N2 (A/Netherlands/018/1994) viruses used in the present study.
b All amino acid sequences present in the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/genomes/FLU) database as of 11 April 2015. Yr, years of isolation that were present in the database.
c The 95K mutation was introduced in 1997 and rapidly fixated in human seasonal A/H3N2 viruses in the following years.
d Only one 1918 sequence (A/BrevigMission/1/1918) could be obtained from the NCBI database, which was the only virus in this group containing the 115V residue; the other H1N1 viruses in this group were from the 1930s.
The extraepitopic amino acid residues of human IAVs were occasionally observed in avian and swine IAVs, although they were mainly present in isolation. The avian variants of these extraepitopic amino acid residues were observed with a higher frequency in both avian and swine IAVs (Fig. 7; Table 1). Avian variants of these extraepitopic amino acid residues were rarely observed in human IAVs (Fig. 7; Table 1). The arginine residue at position 27 (present in human viruses) was the exception, as it was also observed with a high frequency in avian and swine viruses (77.4% and 99.6%, respectively) (Fig. 7; Table 1).
DISCUSSION
In the present study, we demonstrated that extraepitopic amino acid residues affect CD8+ T cell recognition of the highly conserved immunodominant HLA-A*0201-restricted M158–66 IAV epitope. Naturally occurring amino acid variation at positions outside the epitope contributed to the observed differences in epitope recognition of avian and human IAVs. The origin from which the M1 protein was derived, an avian or human IAV, determined the kinetics of CD8+ T cell activation after stimulation with virus-infected cells. Recognition of the M1 protein derived from human IAV delayed and impaired the activation and reduced lytic activity of the M158–66-specific CD8+ T cells compared to recognition of M1 protein derived from an avian IAV. This difference in recognition may have implications for virus clearance in HLA-A*0201 individuals and spread of the IAVs in the human population. The differential recognition of the M158–66 epitope may explain in part previously described differences in recognition of human seasonal A/H3N2 virus and avian A/H5N1 and A/H7N9 viruses (16, 17).
Although, it is not fully clear what the selective pressure is for the preferred use of the amino acid residues under investigation, it is tempting to speculate that evasion of recognition by M158–66-specific CD8+ T cells plays a role. It has been suggested that the immunodominance of the epitope serves as a stealth strategy and that impaired function of M158–66-specific CD8+ T cells explained the virus's ability to evade recognition by these T cells (53). However, this is a matter of debate (54) also because HLA-A*0201-positive individuals display stronger CTL responses after IAV infection (55).
Most likely, extraepitopic amino acid residues affect the processing and presentation of the M158–66 epitope. Differences in translocation by TAP (39, 40, 56) or trimming of peptides by ER-resident proteases like ER amino peptidase 1 or 2 (57) may not have contributed, because TAP typically transports peptides of 8 to 16 amino acids (40) and the amino acid substitutions under investigation are too distant from the epitope to be able to have an effect on these processes. More likely, earlier steps in the antigen processing pathway are involved, like degradation by the proteasome (58). However, because the constitutive proteasome potentially cleaves inside the epitope sequence (59), alternative proteases like the immunoproteasome (60), or nonproteasomal proteases (61), like tripeptidyl peptidase II (TPPII) (62, 63), are more likely candidates.
Based on these studies we hypothesize that the difference in extraepitopic amino acid residues either change the cleavage pattern of the M1 protein and/or define which protease processes the M1 protein, which will eventually determine the extent of M158–66 epitope presentation.
It has been shown that amino acid residues flanking a mouse CTL epitope altered recognition of IAV (64–68). However, these findings were obtained with artificially introduced mutations. In the present study, we show for the first time that naturally occurring variation at positions outside the epitope influences antigen processing resulting in differential CD8+ T cell recognition of human and avian IAVs. Of note, mutations flanking CTL epitopes affecting CD8+ T cell recognition have been observed in viruses causing chronic infections (27, 41, 42). In most cases, these mutations were located in close proximity to the epitope (within 10 amino acids). In contrast, the extraepitopic variation in amino acid residues observed in the present study were more distant from the epitope (over 30 amino acids), and to the best of our knowledge, this has not been observed previously. Since the reciprocal exchange of the five extraepitopic amino acid residues described in this study only partly reverses the CD8+ T cell recognition patterns between the M1 protein of the avian and human IAV, it cannot be excluded that other substitutions more distant from the M158–66 epitope, e.g., at positions 137, 166, 167, 168, 207, 218, 224, 230, and 232 (Fig. 2), also contribute to antigen processing and thus to differential recognition.
Analysis of the extraepitopic residues in all M1 protein amino acid sequences of avian human and swine IAVs available in the influenza virus resource database (http://www.ncbi.nlm.nih.gov/genomes/FLU) revealed that 15V, 27R, 101R, 115I, and 121A were the preferred residues in human IAVs (Fig. 7; Table 1). Residues 15I, 101K, 115V, and 121T were preferred in avian IAVs. However, residue 27K was a minor variant in avian and swine IAVs (Fig. 7; Table 1). It would be of interest to determine the minimal set of amino acid residues that are responsible for the observed differences in recognition. Of note, within the 60-amino-acid distance from the M158–66 epitope, the R95K substitution was rapidly fixed in human A/H3N2 viruses after its initial introduction in 1997 (Table 1). Its rapid fixation suggests that this substitution might also contribute to evasion from recognition by M158–66-specific CD8+ T cells. It has been hypothesized that the preferred avian or human IAV amino acid residues at positions 115 and 121 of the M1 protein reflect viral host adaptation (33). However, as H1N1pdm09 contained an M1 protein with the preferred avian/swine amino acid residue at these positions, this might not be the case (Fig. 1 and 7; Table 1) (9). Evasion of recognition by M158–66-specific CTLs, as demonstrated in the present study, may provide an alternative explanation. It would therefore be of interest to monitor acquisition of these preferred human amino acid residues in the M1 amino acid sequence of H1N1pdm09 IAVs.
The pandemic of 2009 demonstrated that the frequency of pre-existing IAV-specific CD8+ T cells inversely correlated with disease severity (18, 19). Compared to the pandemics of 1918, 1957, and 1968, the pandemic of 2009 was generally considered mild (69, 70). Especially in the elderly, morbidity and mortality were relatively low (70). This was attributed to the presence of antibodies in this age group induced by infection with A/H1N1 viruses that circulated prior to 1957 and that are antigenically related to the H1N1pdm09 virus (71, 72). In contrast to previous pandemic viruses which possess an M1 protein of human signature, the H1N1pdm09 virus possessed an M1 protein of avian/swine signature (Fig. 1 and 7; Table 1). Consequently, our data suggest that in previously infected HLA-A*0201-positive individuals, the 2009 pandemic viruses were better recognized by preexisting M158–66-specific CD8+ T cells, which also contributed to protective immunity.
The continuous pandemic threat posed by avian IAVs of various subtypes and the emergence of drift variants of seasonal IAVs underscore the need for vaccines that could induce broad protective immunity, the so-called universal vaccines. IAV-specific CD8+ T cells are predominantly directed to conserved epitopes, including M158–66, and are considered an important correlate of cross-protective immunity (13, 14, 16–20). Therefore, universal influenza vaccines should aim at the induction of virus-specific CTL responses. Although the extent of exertion of antiviral activity by M158–66-specific CD8+ T cells during subsequent influenza virus infections will depend on the origin of the virus (avian or human), the present study suggests that for the efficient induction of CTL responses, proteins from avian IAVs may be advantageous over those derived from human IAVs. This may also apply to live attenuated influenza vaccines that are known to induce CTL responses (73, 74). These responses may be improved with the use of viral proteins originating from avian IAVs. Alternatively, vaccine approaches that circumvent the antigen-processing pathways are of interest (56, 75).
Collectively, we have demonstrated that the conserved M158–66 epitope is differentially recognized by epitope-specific CD8+ T cells depending on the origin of the M1 protein. Extraepitopic amino acid residues are responsible for the differential recognition, which indicates that differences in antigen processing and presentation are at the basis of these observations. In the context of an M1 protein of human signature the epitope is relatively poorly recognized compared to the M1 protein of avian viruses. It can be speculated that the possession of an M1 protein of human signature offers the virus an advantage by impairing recognition by specific CD8+ T cells. Consequently, these viruses may replicate better in HLA-A*0201-positive individuals. Since HLA-A*0201 has a high prevalence in the human population, this also may impact the spread of the virus in the human population. In addition, our findings may have implications for the development of vaccines that aim at the induction of virus-specific CTL responses.
ACKNOWLEDGMENTS
G.F.R. was financially supported by EU grant FLUNIVAC (602604). R.A.M.F. and M.I.S. received funding from NIAID/NIH (contract HHSN272201400008C).
We thank Miranda de Graaf for excellent technical advice and assistance.
We have no conflicts of interest to declare.
C.E.V.D.S., J.H.C.M.K., M.I.S., A.D.M.E.O., and G.F.R. designed the experiments; C.E.V.D.S., J.H.C.M.K., M.M.G.-M., and N.J.N. performed the experiments; C.E.V.D.S. and D.A.M.C.V.D.V. analyzed the experiments; C.E.V.D.S., R.A.M.F., and G.F.R. wrote the manuscript.
REFERENCES
- 1.WHO. 2015. Influenza (seasonal) fact sheet no. 211. http://www.who.int/mediacentre/factsheets/fs211/en/index.html Accessed 27 June 2015.
- 2.Palese P, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication, p 1647–1689. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schrauwen EJ, Fouchier RA. 2014. Host adaptation and transmission of influenza A viruses in mammals. Emerg Microbes Infect 3:e9. doi: 10.1038/emi.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith GJ, Bahl J, Vijaykrishna D, Zhang J, Poon LL, Chen H, Webster RG, Peiris JS, Guan Y. 2009. Dating the emergence of pandemic influenza viruses. Proc Natl Acad Sci U S A 106:11709–11712. doi: 10.1073/pnas.0904991106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Worobey M, Han GZ, Rambaut A. 2014. Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc Natl Acad Sci U S A 111:8107–8112. doi: 10.1073/pnas.1324197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. 1978. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 9.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD Jr, Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI, Cox NJ. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiley DC, Wilson IA, Skehel JJ. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 11.Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, Skepner E, Lewis NS, Spronken MI, Russell CA, Eropkin MY, Hurt AC, Barr IG, de Jong JC, Rimmelzwaan GF, Osterhaus AD, Fouchier RA, Smith DJ. 2013. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- 12.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 13.van de Sandt CE, Kreijtz JH, Rimmelzwaan GF. 2012. Evasion of influenza A viruses from innate and adaptive immune responses. Viruses 4:1438–1476. doi: 10.3390/v4091438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinones-Parra S, Loh L, Brown LE, Kedzierska K, Valkenburg SA. 2014. Universal immunity to influenza must outwit immune evasion. Front Microbiol 5:285. doi: 10.3389/fmicb.2014.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neefjes J, Jongsma ML, Paul P, Bakke O. 2011. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev 11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 16.Kreijtz JH, de Mutsert G, van Baalen CA, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2008. Cross-recognition of avian H5N1 influenza virus by human cytotoxic T-lymphocyte populations directed to human influenza A virus. J Virol 82:5161–5166. doi: 10.1128/JVI.02694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van de Sandt CE, Kreijtz JH, de Mutsert G, Geelhoed-Mieras MM, Hillaire ML, Vogelzang-van Trierum SE, Osterhaus AD, Fouchier RA, Rimmelzwaan GF. 2014. Human cytotoxic T lymphocytes directed to seasonal influenza A viruses cross-react with the newly emerging H7N9 virus. J Virol 88:1684–1693. doi: 10.1128/JVI.02843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, Bean T, Barclay W, Deeks JJ, Lalvani A. 2013. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med 19:1305–1312. doi: 10.1038/nm.3350. [DOI] [PubMed] [Google Scholar]
- 19.Hayward AC, Wang L, Goonetilleke N, Fragaszy EB, Bermingham A, Copas A, Dukes O, Millett ER, Nazareth I, Nguyen-Van-Tam JS, Watson JM, Zambon M, Johnson AM, McMichael AJ, Flu Watch G. 2015. Natural T cell-mediated protection against seasonal and pandemic influenza. Results of the Flu Watch Cohort Study. Am J Respir Crit Care Med 191:1422–1431. doi: 10.1164/rccm.201411-1988OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Wan Y, Qiu C, Quinones-Parra S, Zhu Z, Loh L, Tian D, Ren Y, Hu Y, Zhang X, Thomas PG, Inouye M, Doherty PC, Kedzierska K, Xu J. 2015. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8(+) T cells. Nat Commun 6:6833. doi: 10.1038/ncomms7833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berkhoff EG, de Wit E, Geelhoed-Mieras MM, Boon AC, Symons J, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2005. Functional constraints of influenza A virus epitopes limit escape from cytotoxic T lymphocytes. J Virol 79:11239–11246. doi: 10.1128/JVI.79.17.11239-11246.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machkovech HM, Bedford T, Suchard MA, Bloom JD. 2015. Positive selection in CD8+ T-cell epitopes of influenza virus nucleoprotein revealed by a comparative analysis of human and swine viral lineages. J Virol 89:11275–11283. doi: 10.1128/JVI.01571-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berkhoff EG, Geelhoed-Mieras MM, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2007. Assessment of the extent of variation in influenza A virus cytotoxic T-lymphocyte epitopes by using virus-specific CD8+ T-cell clones. J Gen Virol 88:530–535. doi: 10.1099/vir.0.82120-0. [DOI] [PubMed] [Google Scholar]
- 24.Boon AC, de Mutsert G, Graus YM, Fouchier RA, Sintnicolaas K, Osterhaus AD, Rimmelzwaan GF. 2002. Sequence variation in a newly identified HLA-B35-restricted epitope in the influenza A virus nucleoprotein associated with escape from cytotoxic T lymphocytes. J Virol 76:2567–2572. doi: 10.1128/jvi.76.5.2567-2572.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voeten JT, Bestebroer TM, Nieuwkoop NJ, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2000. Antigenic drift in the influenza A virus (H3N2) nucleoprotein and escape from recognition by cytotoxic T lymphocytes. J Virol 74:6800–6807. doi: 10.1128/JVI.74.15.6800-6807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berkhoff EG, Boon AC, Nieuwkoop NJ, Fouchier RA, Sintnicolaas K, Osterhaus AD, Rimmelzwaan GF. 2004. A mutation in the HLA-B*2705-restricted NP383-391 epitope affects the human influenza A virus-specific cytotoxic T-lymphocyte response in vitro. J Virol 78:5216–5222. doi: 10.1128/JVI.78.10.5216-5222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goulder PJ, Watkins DI. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat Rev 4:630–640. doi: 10.1038/nrc1410. [DOI] [PubMed] [Google Scholar]
- 28.Chang KM, Rehermann B, McHutchison JG, Pasquinelli C, Southwood S, Sette A, Chisari FV. 1997. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J Clin Invest 100:2376–2385. doi: 10.1172/JCI119778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khanna R, Burrows SR, Moss DJ. 1995. Immune regulation in Epstein-Barr virus-associated diseases. Microbiol Rev 59:387–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aebischer T, Moskophidis D, Rohrer UH, Zinkernagel RM, Hengartner H. 1991. In vitro selection of lymphocytic choriomeningitis virus escape mutants by cytotoxic T lymphocytes. Proc Natl Acad Sci U S A 88:11047–11051. doi: 10.1073/pnas.88.24.11047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrison J, Elvin J, Latron F, Gotch F, Moots R, Strominger JL, McMichael A. 1992. Identification of the nonamer peptide from influenza A matrix protein and the role of pockets of HLA-A2 in its recognition by cytotoxic T lymphocytes. Eur J Immunol 22:903–907. doi: 10.1002/eji.1830220404. [DOI] [PubMed] [Google Scholar]
- 32.Choo JA, Liu J, Toh X, Grotenbreg GM, Ren EC. 2014. The immunodominant influenza A virus M158-66 cytotoxic T lymphocyte epitope exhibits degenerate class I major histocompatibility complex restriction in humans. J Virol 88:10613–10623. doi: 10.1128/JVI.00855-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furuse Y, Suzuki A, Kamigaki T, Oshitani H. 2009. Evolution of the M gene of the influenza A virus in different host species: large-scale sequence analysis. Virol J 6:67. doi: 10.1186/1743-422X-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437:889–893. doi: 10.1038/nature04230. [DOI] [PubMed] [Google Scholar]
- 35.Boon AC, de Mutsert G, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2006. The hypervariable immunodominant NP418-426 epitope from the influenza A virus nucleoprotein is recognized by cytotoxic T lymphocytes with high functional avidity. J Virol 80:6024–6032. doi: 10.1128/JVI.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidney J, Grey HM, Kubo RT, Sette A. 1996. Practical, biochemical and evolutionary implications of the discovery of HLA class I supermotifs. Immunol Today 17:261–266. doi: 10.1016/0167-5699(96)80542-1. [DOI] [PubMed] [Google Scholar]
- 37.Berkhoff EG, de Wit E, Geelhoed-Mieras MM, Boon AC, Symons J, Fouchier RA, Osterhaus AD, Rimmelzwaan GF. 2006. Fitness costs limit escape from cytotoxic T lymphocytes by influenza A viruses. Vaccine 24:6594–6596. doi: 10.1016/j.vaccine.2006.05.051. [DOI] [PubMed] [Google Scholar]
- 38.Cao S, Liu X, Yu M, Li J, Jia X, Bi Y, Sun L, Gao GF, Liu W. 2012. A nuclear export signal in the matrix protein of Influenza A virus is required for efficient virus replication. J Virol 86:4883–4891. doi: 10.1128/JVI.06586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neisig A, Roelse J, Sijts AJ, Ossendorp F, Feltkamp MC, Kast WM, Melief CJ, Neefjes JJ. 1995. Major differences in transporter associated with antigen presentation (TAP)-dependent translocation of MHC class I-presentable peptides and the effect of flanking sequences. J Immunol 154:1273–1279. [PubMed] [Google Scholar]
- 40.Hofmann M, Nussbaum AK, Emmerich NP, Stoltze L, Schild H. 2001. Mechanisms of MHC class I-restricted antigen presentation. Expert Opin Ther Targets 5:379–393. doi: 10.1517/14728222.5.3.379. [DOI] [PubMed] [Google Scholar]
- 41.Seifert U, Liermann H, Racanelli V, Halenius A, Wiese M, Wedemeyer H, Ruppert T, Rispeter K, Henklein P, Sijts A, Hengel H, Kloetzel PM, Rehermann B. 2004. Hepatitis C virus mutation affects proteasomal epitope processing. J Clin Invest 114:250–259. doi: 10.1172/JCI200420985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frisan T, Zhang QJ, Levitskaya J, Coram M, Kurilla MG, Masucci MG. 1996. Defective presentation of MHC class I-restricted cytotoxic T-cell epitopes in Burkitt's lymphoma cells. Int J Cancer 68:251–258. doi:. [DOI] [PubMed] [Google Scholar]
- 43.Ito Y, Kondo E, Demachi-Okamura A, Akatsuka Y, Tsujimura K, Tanimoto M, Morishima Y, Takahashi T, Kuzushima K. 2006. Three immunoproteasome-associated subunits cooperatively generate a cytotoxic T-lymphocyte epitope of Epstein-Barr virus LMP2A by overcoming specific structures resistant to epitope liberation. J Virol 80:883–890. doi: 10.1128/JVI.80.2.883-890.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rimmelzwaan GF, Nieuwkoop N, Brandenburg A, Sutter G, Beyer WE, Maher D, Bates J, Osterhaus AD. 2000. A randomized, double blind study in young healthy adults comparing cell mediated and humoral immune responses induced by influenza ISCOM vaccines and conventional vaccines. Vaccine 19:1180–1187. doi: 10.1016/S0264-410X(00)00310-8. [DOI] [PubMed] [Google Scholar]
- 45.Rimmelzwaan GF, Boon AC, Geelhoed-Mieras MM, Voeten JT, Fouchier RA, Osterhaus AD. 2004. Human airway epithelial cells present antigen to influenza virus-specific CD8+ CTL inefficiently after incubation with viral protein together with ISCOMATRIX. Vaccine 22:2769–2775. doi: 10.1016/j.vaccine.2004.01.052. [DOI] [PubMed] [Google Scholar]
- 46.van Baalen CA, Kwa D, Verschuren EJ, Reedijk ML, Boon AC, de Mutsert G, Rimmelzwaan GF, Osterhaus AD, Gruters RA. 2005. Fluorescent antigen-transfected target cell cytotoxic T lymphocyte assay for ex vivo detection of antigen-specific cell-mediated cytotoxicity. J Infect Dis 192:1183–1190. doi: 10.1086/444546. [DOI] [PubMed] [Google Scholar]
- 47.de Wit E, Spronken MI, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2004. Efficient generation and growth of influenza virus A/PR/8/34 from eight cDNA fragments. Virus research 103:155–161. doi: 10.1016/j.virusres.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 48.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci U S A 97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rimmelzwaan GF, Baars M, Claas EC, Osterhaus AD. 1998. Comparison of RNA hybridization, hemagglutination assay, titration of infectious virus and immunofluorescence as methods for monitoring influenza virus replication in vitro. J Virol Methods 74:57–66. doi: 10.1016/S0166-0934(98)00071-8. [DOI] [PubMed] [Google Scholar]
- 50.Voeten JT, Rimmelzwaan GF, Nieuwkoop NJ, Lovgren-Bengtsson K, Osterhaus AD. 2000. Introduction of the haemagglutinin transmembrane region in the influenza virus matrix protein facilitates its incorporation into ISCOM and activation of specific CD8(+) cytotoxic T lymphocytes. Vaccine 19:514–522. doi: 10.1016/S0264-410X(00)00179-1. [DOI] [PubMed] [Google Scholar]
- 51.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
- 52.Lamb RA, Lai CJ, Choppin PW. 1981. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc Natl Acad Sci U S A 78:4170–4174. doi: 10.1073/pnas.78.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keskin DB, Reinhold BB, Zhang GL, Ivanov AR, Karger BL, Reinherz EL. 2015. Physical detection of influenza A epitopes identifies a stealth subset on human lung epithelium evading natural CD8 immunity. Proc Natl Acad Sci U S A 112:2151–2156. doi: 10.1073/pnas.1423482112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van de Sandt CE, Rimmelzwaan GF. 2015. Immunodominant responses to the influenza virus M158-66 epitope: stealth or protection? Proc Natl Acad Sci U S A 112:E2417. doi: 10.1073/pnas.1503245112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boon AC, de Mutsert G, Graus YM, Fouchier RA, Sintnicolaas K, Osterhaus AD, Rimmelzwaan GF. 2002. The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype. J Virol 76:582–590. doi: 10.1128/JVI.76.2.582-590.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gueguen M, Biddison WE, Long EO. 1994. T cell recognition of an HLA-A2-restricted epitope derived from a cleaved signal sequence. J Exp Med 180:1989–1994. doi: 10.1084/jem.180.5.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, Greer F, Schomburg L, Fruci D, Niedermann G, van Endert PM. 2005. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol 6:689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 58.Cerundolo V, Benham A, Braud V, Mukherjee S, Gould K, Macino B, Neefjes J, Townsend A. 1997. The proteasome-specific inhibitor lactacystin blocks presentation of cytotoxic T lymphocyte epitopes in human and murine cells. Eur J Immunol 27:336–341. doi: 10.1002/eji.1830270148. [DOI] [PubMed] [Google Scholar]
- 59.Luckey CJ, King GM, Marto JA, Venketeswaran S, Maier BF, Crotzer VL, Colella TA, Shabanowitz J, Hunt DF, Engelhard VH. 1998. Proteasomes can either generate or destroy MHC class I epitopes: evidence for nonproteasomal epitope generation in the cytosol. J Immunol 161:112–121. [PubMed] [Google Scholar]
- 60.Gileadi U, Moins-Teisserenc HT, Correa I, Booth BL Jr, Dunbar PR, Sewell AK, Trowsdale J, Phillips RE, Cerundolo V. 1999. Generation of an immunodominant CTL epitope is affected by proteasome subunit composition and stability of the antigenic protein. J Immunol 163:6045–6052. [PubMed] [Google Scholar]
- 61.Vinitsky A, Anton LC, Snyder HL, Orlowski M, Bennink JR, Yewdell JW. 1997. The generation of MHC class I-associated peptides is only partially inhibited by proteasome inhibitors: involvement of nonproteasomal cytosolic proteases in antigen processing? J Immunol 159:554–564. [PubMed] [Google Scholar]
- 62.Geier E, Pfeifer G, Wilm M, Lucchiari-Hartz M, Baumeister W, Eichmann K, Niedermann G. 1999. A giant protease with potential to substitute for some functions of the proteasome. Science 283:978–981. doi: 10.1126/science.283.5404.978. [DOI] [PubMed] [Google Scholar]
- 63.Seifert U, Maranon C, Shmueli A, Desoutter JF, Wesoloski L, Janek K, Henklein P, Diescher S, Andrieu M, de la Salle H, Weinschenk T, Schild H, Laderach D, Galy A, Haas G, Kloetzel PM, Reiss Y, Hosmalin A. 2003. An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nat Immunol 4:375–379. doi: 10.1038/ni905. [DOI] [PubMed] [Google Scholar]
- 64.Del Val M, Schlicht HJ, Ruppert T, Reddehase MJ, Koszinowski UH. 1991. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell 66:1145–1153. doi: 10.1016/0092-8674(91)90037-Y. [DOI] [PubMed] [Google Scholar]
- 65.Eisenlohr LC, Yewdell JW, Bennink JR. 1992. Flanking sequences influence the presentation of an endogenously synthesized peptide to cytotoxic T lymphocytes. J Exp Med 175:481–487. doi: 10.1084/jem.175.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yellen-Shaw AJ, Wherry EJ, Dubois GC, Eisenlohr LC. 1997. Point mutation flanking a CTL epitope ablates in vitro and in vivo recognition of a full-length viral protein. J Immunol 158:3227–3234. [PubMed] [Google Scholar]
- 67.Mo XY, Cascio P, Lemerise K, Goldberg AL, Rock K. 1999. Distinct proteolytic processes generate the C and N termini of MHC class I-binding peptides. J Immunol 163:5851–5859. [PubMed] [Google Scholar]
- 68.Gileadi U, Gallimore A, Van der Bruggen P, Cerundolo V. 1999. Effect of epitope flanking residues on the presentation of N-terminal cytotoxic T lymphocyte epitopes. Eur J immunology 29:2213–2222. doi:. [DOI] [PubMed] [Google Scholar]
- 69.Mathews JD, Chesson JM, McCaw JM, McVernon J. 2009. Understanding influenza transmission, immunity and pandemic threats. Influenza Other Respir Viruses 3:143–149. doi: 10.1111/j.1750-2659.2009.00089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dawood FS, Iuliano AD, Reed C, Meltzer MI, Shay DK, Cheng PY, Bandaranayake D, Breiman RF, Brooks WA, Buchy P, Feikin DR, Fowler KB, Gordon A, Hien NT, Horby P, Huang QS, Katz MA, Krishnan A, Lal R, Montgomery JM, Molbak K, Pebody R, Presanis AM, Razuri H, Steens A, Tinoco YO, Wallinga J, Yu H, Vong S, Bresee J, Widdowson MA. 2012. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis 12:687–695. doi: 10.1016/S1473-3099(12)70121-4. [DOI] [PubMed] [Google Scholar]
- 71.Ikonen N, Strengell M, Kinnunen L, Osterlund P, Pirhonen J, Broman M, Davidkin I, Ziegler T, Julkunen I. 2010. High frequency of cross-reacting antibodies against 2009 pandemic influenza A(H1N1) virus among the elderly in Finland. Euro Surveill 15:19478. [PubMed] [Google Scholar]
- 72.Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, Brammer TL, Cox NJ, Tumpey TM, Katz JM. 2009. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med 361:1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 73.He XS, Holmes TH, Zhang C, Mahmood K, Kemble GW, Lewis DB, Dekker CL, Greenberg HB, Arvin AM. 2006. Cellular immune responses in children and adults receiving inactivated or live attenuated influenza vaccines. J Virol 80:11756–11766. doi: 10.1128/JVI.01460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hoft DF, Babusis E, Worku S, Spencer CT, Lottenbach K, Truscott SM, Abate G, Sakala IG, Edwards KM, Creech CB, Gerber MA, Bernstein DI, Newman F, Graham I, Anderson EL, Belshe RB. 2011. Live and inactivated influenza vaccines induce similar humoral responses, but only live vaccines induce diverse T-cell responses in young children. J Infect Dis 204:845–853. doi: 10.1093/infdis/jir436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elliott T, Willis A, Cerundolo V, Townsend A. 1995. Processing of major histocompatibility class I-restricted antigens in the endoplasmic reticulum. J Exp Med 181:1481–1491. doi: 10.1084/jem.181.4.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]