Abstract
Wild nonhuman primates are immediate sources and long-term reservoirs of human pathogens. However, ethical and technical challenges have hampered the identification of novel blood-borne pathogens in these animals. We recently examined RNA viruses in plasma from wild African monkeys and discovered several novel, highly divergent viruses belonging to the family Arteriviridae. Close relatives of these viruses, including simian hemorrhagic fever virus, have caused sporadic outbreaks of viral hemorrhagic fever in captive macaque monkeys since the 1960s. However, arterivirus infection in wild nonhuman primates had not been described prior to 2011. The arteriviruses recently identified in wild monkeys have high sequence and host species diversity, maintain high viremia, and are prevalent in affected populations. Taken together, these features suggest that the simian arteriviruses may be “preemergent” zoonotic pathogens. If not, this would imply that biological characteristics of RNA viruses thought to facilitate zoonotic transmission may not, by themselves, be sufficient for such transmission to occur.
INTRODUCTION
Novel human pathogens are emerging from wildlife at increasing rates (1, 2). Recent efforts to understand this phenomenon have focused on identifying microorganisms with zoonotic potential in their “preemergent” state, i.e., within their “natural” host(s) (1, 3). Even with advances in pathogen discovery technology, a complete global assessment of animal pathogens is not currently feasible (3). However, not all pathogens possess equal zoonotic potential. In particular, multihost pathogens and RNA viruses are significantly more likely to cause zoonoses than pathogens of other classes (4). Additionally, nonhuman primate hosts in Africa are historically important sources of zoonotic pathogens (5–8).
Recently, we used unbiased deep sequencing to identify and characterize RNA viruses in the plasma of African nonhuman primates. Our survey included 10 or more primates from populations representing eight cercopithecoid (i.e., Old World monkey) species. These samples were collected from multiple distinct geographic regions, spanning over 3,000 km. In nearly half of these populations, we discovered novel highly divergent viruses belonging to the family Arteriviridae (9–12) (Fig. 1). Along with coronaviruses, roniviruses, and mesoniviruses, arteriviruses belong to the order Nidovirales and infect a variety of mammals, including pigs, horses, mice, possums, and Old World monkeys (13). Notably, the arteriviruses are the only family of RNA viruses that infect mammals for which human infection has never been documented (14).
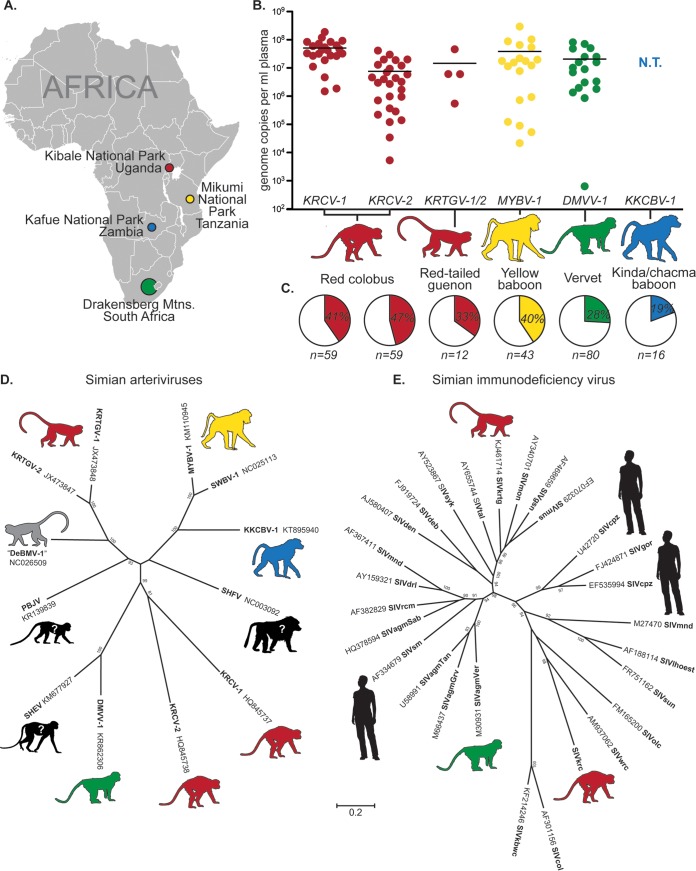
FIG 1.
Features of simian arterivirus infections among African monkeys. (A) Map of Africa depicting the geographic locations where wild monkeys harboring simian arteriviruses have been sampled. Colors correspond to the respective host species and virus throughout the figure. (Map from Lokal_Profil [https://commons.wikimedia.org/wiki/File:BlankMap-Africa.svg].) (B) Plasma viral loads, as measured by quantitative reverse transcription-PCR, showing simian arterivirus viremia in infected monkeys. N.T., not tested. (C) Prevalence of simian arterivirus-positive monkeys in affected populations, as determined by quantitative reverse transcription-PCR, reverse transcription-PCR, and/or unbiased deep sequencing. (D and E) Phylogeny of known simian arteriviruses (D) with a simian immunodeficiency virus (SIV) phylogeny shown on the same scale for comparison (E). Maximum likelihood trees were generated using MEGA6.06 (1,000 bootstrap replicates, GTR+I+γ model) from codon-based alignments (via MAFFT) of 12 simian arterivirus ORF1b sequences or 27 simian immunodeficiency virus gag sequences. Bootstrap values of less than 70 are not shown. DeBMV-1, De Brazza's monkey virus 1; DMVV-1, Drakensberg Mountain vervet virus 1; KKCBV-1, Kafue kinda-chacma baboon virus 1; KRCV-1/2, Kibale red colobus virus 1/2; KRTGV-1/2, Kibale red-tailed guenon virus 1/2; MYBV-1, Mikumi yellow baboon virus 1; PBJV, Pebjah virus; SHEV, simian hemorrhagic encephalitis virus; SHFV, simian hemorrhagic fever virus; SWBV-1, Southwest baboon virus 1.
The arteriviruses that we discovered in wild African monkeys—here referred to as simian arteriviruses—are monophyletic, and all possess genomic features that indicate common ancestry with simian hemorrhagic fever virus (SHFV). These viruses all contain a duplication of 3 to 4 open reading frames (ORFs) in the 3′-proximal half of the genome that is not observed in other arteriviruses (13). These additional ORFs putatively express additional structural proteins that are thought to be required for replication, but their precise function is not understood.
A history of cross-species transmission.
SHFV was identified in 1964 following an outbreak of simian hemorrhagic fever (SHF) which affected Asian macaques of several species (Macaca fascicularis, Macaca mulatta, and Macaca arctoides) in a quarantine facility at the National Institutes of Health (NIH; Bethesda, MD) (15). Clinically, SHF is characterized by fever, facial edema, cyanosis, anorexia, adipsia, vomiting, dehydration, and signs of hemorrhagic disease (e.g., melena, petechiae, subcutaneous hematoma, retrobulbar hemorrhage, and epistaxis) (16). Internal hemorrhages affecting the lungs, liver, kidneys, and gastrointestinal tract are found on pathological examination (17). Laboratory tests from monkeys with SHF reveal hematologic and urinary changes characteristic of viral hemorrhagic fever (VHF) in humans, including lymphopenia with a left shift followed by neutrophilic leukocytosis, thrombocytopenia, prolonged coagulation time, and proteinuria indicative of renal failure (18). The case-fatality rate of SHFV infection in the 1964 NIH outbreak was initially reported to be 100%; however, this rate may be an overestimate as subclinical infection in a small number of individual macaques has since been documented (16). Additional outbreaks of SHF (affecting macaques of the species M. fascicularis, Macaca nemestrina, M. mulatta, and Macaca radiata) occurred throughout the 1960s to 1990s (19–23), with transmission among macaques occurring via direct contact and indirect contact and possibly via aerosol routes (24).
Although the source of virus in these outbreaks was never definitively identified, serological studies of captive African monkeys implicated primates of several species—namely, patas monkeys (Erythrocebus patas), grivets (Chlorocebus aethiops), and Guinea baboons (Papio papio)—as subclinical carriers and likely sources (24). However, captive patas monkeys inoculated experimentally with SHFV developed significant signs of disease (25), demonstrating that simian arterivirus-induced pathology is not specific to macaques. Moreover, although it was originally presumed that all SHF outbreaks were caused by a single agent (i.e., SHFV), we recently showed that at least three highly divergent simian arteriviruses were responsible for past outbreaks of SHF, suggesting that the simian arteriviruses as a group possess features that facilitate transmission among primates of different species (26).
Characteristics of natural simian arterivirus infection.
Despite the widely held suspicion that African monkeys were the natural reservoir for SHFV, it was not until 2011 that the first SHFV-like viruses were discovered in a wild animal: a red colobus monkey (Procolobus rufomitratus tephrosceles) in Kibale National Park, Uganda (the viruses were referred to as KRCV-1 and KRCV-2, to indicate Kibale red colobus viruses 1 and 2, respectively) (11). We have since discovered simian arteriviruses in red-tailed guenons (Cercopithecus ascanius schmidti) from Kibale (Kibale red-tailed guenon viruses 1 and 2 [KRTGV-1 and -2], respectively) (12), yellow baboons (Papio cynocephalus) from Mikumi National Park in Tanzania (Mikumi yellow baboon virus 1 [MYBV-1]) (10), hybrid kinda × grayfooted-chacma baboons (Papio kindae × Papio ursinus griseipes) from Kafue National Park in Zambia (Kafue kinda-chacma baboon virus [KKCBV-1]), and vervets (Chlorocebus pygerythrus) from the Drakensberg Mountains in South Africa (Drakensberg Mountain vervet virus 1 [DMVV-1]) (Fig. 1A). These viruses were detected at high titers in the blood of infected individuals (≈1 × 107 genome copies/ml [Fig. 1B]) and were prevalent in affected populations (≈40% of monkeys tested [Fig. 1C]). Sequence analysis revealed that simian arteriviruses from each species share only ≈50% nucleotide identity with one another or SHFV (Fig. 1D). These discoveries also demonstrate that all major clades of cercopithecoid monkeys, including both recognized cercopithecid subfamilies, naturally harbor simian arteriviruses.
Although simian arteriviruses have been identified to date in monkeys from only four locations, the distribution of these sampling sites suggests that simian arterivirus infections occur in monkeys across sub-Saharan Africa. Alternatively, the lack of detection of simian arterivirus infection in black-and-white colobus monkeys (Colobus guereza) (n = 10) or olive baboons (Papio anubis) (n = 23) from Kibale, as well as sooty mangabeys (Cercocebus atys) (n = 12) from the Moa and Mabole rivers in Sierra Leone, implies that the occurrence of simian arterivirus infection in African monkey populations may be variable. Regardless, given the sequence, geographic, and host species diversity of these viruses in our limited data set—and that the natural host(s) of the viruses responsible for past outbreaks of SHF in macaques is still not known—many more simian arteriviruses are likely to be discovered.
Simian arterivirus biology and zoonotic potential.
The zoonotic potential of a virus cannot be inferred from its biological properties alone. However, certain biological features are thought to potentiate zoonotic transmission of RNA viruses. These features include, but are not limited to, high genetic diversity, the ability to overcome host restriction factors, high virus production within infected animals, infection of primates, high prevalence of infection within naturally infected animal populations, and intense interactions between infected animals and humans (1, 27, 28). Although not all viruses known to be transmitted from animals to humans possess all of these features (7), various combinations of these characteristics are observed in RNA viruses that have emerged (or are emerging) to threaten human health.
Theoretically, high genetic diversity may facilitate cross-species transmission of a virus by providing a variety of phenotypically unique viral variants that may interact with host factors in different ways (27–29). Classically, phylogenetic analyses have been used to quantify the diversity of a virus (or group of viruses) at the family, genus, species, strain, or isolate level. At each of these levels, the simian arteriviruses display an impressive degree of diversity, even in comparison to simian immunodeficiency virus (SIV), a virus known for its high genetic diversity also found in African primates (Fig. 1E). Indeed, a motivating force behind newly proposed taxonomic revisions of the Arteriviridae has been the recent recognition of diversity within the simian arterivirus clade (30). Recent advances in sequencing technology have also allowed for detailed analysis of viral diversity within infected animals. The effect of this “intrahost” viral diversity on cross-species transmission is not yet well understood and warrants further investigation. Intuitively, one might expect that high viral diversity in the source host would increase the likelihood that one or more viral variants will be capable of replicating in the recipient host. In wild monkeys studied to date, simian arteriviruses display high levels of intrahost diversity (9). While it remains to be seen whether this diversity includes variants capable of establishing a successful infection in humans, it may increase the likelihood that such a simian arterivirus variant exists.
High virus production in naturally infected animal hosts may also facilitate cross-species transmission by increasing the dose of virus transferred to the recipient host. As the dose of virus required to initiate infection is influenced by the route of transmission and several variables intrinsic to the virus and host in question, direct comparisons between unrelated viruses are difficult. However, several zoonotic RNA viruses are known to have high viral loads in their natural hosts, e.g., lentiviruses in primates (31), hantaviruses in rodents (32), and coronaviruses in camelids (33). Although the route(s) by which simian arterivirus infections might be acquired by humans remains unknown, the high titers of virus detected in the blood of infected monkeys (Fig. 1B) suggest that even a small exposure to blood from an infected animal could expose a human to a relatively large quantity of virus. The extent and type of contact between humans and wild nonhuman primates in Africa vary by geographic region, local customs, and the specific primate species in question. However, many groups of humans come into contact with monkeys through hunting, butchering, and consuming bush meat, including primates from populations known to harbor simian arteriviruses (34–36).
The host factors that influence zoonosis are myriad and highly virus specific. Although host phylogenetic relatedness may serve as a rough proxy for susceptibility to cross-species viral infection, some RNA viruses (e.g., filoviruses, rhabdoviruses, and influenza A viruses) have a very broad species tropism while others (e.g., lentiviruses, hepaciviruses, and pegiviruses) appear to be highly host species restricted (37–40). African monkeys are more closely related to macaques than they are to humans (41), but the full extent of simian arterivirus species tropism remains an important open question. Identifying the specific cellular factors utilized by simian arteriviruses should allow for more sophisticated analyses of simian arterivirus infection in its hosts (42). Determining the host factors that influence simian arterivirus pathogenesis and disease severity in various hosts is another potentially valuable, yet unexplored, avenue of research.
Simian arteriviruses appear to cause persistent viremia in African monkeys, suggesting that these viruses may have evolved mechanisms to evade the immune system (9, 10), as has been shown for other, nonsimian arteriviruses (13). While persistence directly influences the prevalence of infection in a given population, preexisting immune evasion mechanisms might also play an important role in establishing infection in a novel host. We speculate that simian arterivirus persistence in natural hosts may be mediated, in part, by genetic plasticity and mutational escape of host immune responses, as has been shown for other persistent viruses such as HIV and hepatitis C virus (9, 43, 44). The ability of simian arteriviruses to cause persistent high-titer viremia and genomic diversity in their natural hosts does not guarantee that these properties would be maintained in zoonotic human infections. However, high-titer replication and genetic plasticity could also facilitate rapid adaptation to a human host, lowering barriers to replication in humans and human-to-human transmission.
A final consideration is the possible effect of simian arterivirus infection on the pathogenesis of other coinfections. For example, Reston virus, a relative of Ebola virus, was discovered during a particularly severe outbreak of SHF (45). Given the high prevalence of infectious diseases in human populations most likely to be exposed to simian arteriviruses, there exists the possibility for synergy among coinfecting pathogens and simian arteriviruses.
Are simian arteriviruses a zoonotic threat?
Our ability to identify viruses in nature and predict their emergence in humans is still in its infancy. Yet, given the features of simian arterivirus biology explored to date, investigation into the zoonotic potential of these viruses seems prudent. Importantly, lack of documented human infection should not be taken as evidence for lack of zoonotic potential. For example, if a human were infected with a simian arterivirus, would that infection cause clinical disease, and if so, would that disease be distinguishable from other, more common infections? Searching for evidence of simian arterivirus infection in people (e.g., animal caretakers and bush meat hunters) or wild great apes (e.g., chimpanzees and gorillas) that interact frequently with infected monkeys could help answer these questions. However, the genetic diversity of known simian arteriviruses (and presumably of those not yet discovered) poses a formidable technical barrier to such an analysis. For this reason, screening of additional primate populations will be essential to resolve the genetic diversity, host range, geographic distribution, and natural history of simian arteriviruses. Technologies such as unbiased deep sequencing will undoubtedly play a major role in this effort. As we gain a greater appreciation for the extent of simian arterivirus diversity, more widely accessible techniques (e.g., PCR and serology) may become useful in screening for simian arterivirus infections in humans.
The recent discovery of multiple highly diverse and prevalent simian arteriviruses in primates of several species across sub-Saharan Africa highlights how little we know about these viruses. Research on these viruses to date has identified the characteristics described here, but further characterization of the molecular biology, evolution, and ecology of these viruses is needed to more fully appreciate the implications of these viruses for human health. The relative rarity with which novel human pathogens emerge and the effectiveness of antiviral host restriction factors might suggest that the simian arteriviruses are unlikely to do so. However, understanding why certain viruses do not emerge in humans despite predisposing biological characteristics may also hold value for refining our understanding of the factors that drive zoonotic transmission of RNA viruses.
ETHICS STATEMENT
All research involving nonhuman primates was conducted according to the relevant national and international guidelines. Briefly, all animals were sedated prior to blood collection and were released back to their social group without incident following sample collection and recovery from anesthesia. All animal research was approved by the appropriate wildlife authorities and institutional animal care and use committees. Collection of samples from nonhuman primates in Uganda was approved by the Uganda Wildlife Authority (permit UWA/TDO/33/02), the Uganda National Council for Science and Technology (permit HS 364), and the University of Wisconsin Animal Care and Use Committee (protocol V01409-0-02-09) prior to initiation of the study, as described previously (11). Sampling of vervet monkeys in South Africa was approved by the Interfaculty Animal Ethics Committee (project no. 13/2010) at the University of the Free State and by the University of Wisconsin—Milwaukee Animal Care and Use Committee (protocol 07-08 #32) as described previously (31). Sampling of yellow baboons in Tanzania was performed in 1985 and 1986 using standard methods for field studies of baboons as described previously and was approved by the appropriate Tanzanian government authorities, Washington University, and Yale University. Sampling of hybrid kinda × grayfooted-chacma baboons in Zambia was performed in compliance with the rules of the Zambian Wildlife Authority and was conducted in compliance with the rules of the animal care and use committees from Baylor College of Medicine (AN-5538), Washington University School of Medicine (protocol 20120269), and New York University (protocol 10-1349) and applicable national laws.
ACKNOWLEDGMENTS
We thank the University of Wisconsin, Department of Pathology and Laboratory Medicine, and the WNPRC for the use of its facilities and services. We thank the Department of Environmental Affairs, South Africa; Department of Tourism, Environmental and Economic Affairs, Free State Province; the Ezemvelo KZN Wildlife in KwaZulu Natal Province; and Department of Economic Development and Environmental Affairs, Eastern Cape. We also thank the Zambian Wildlife Authority for permission to conduct research in Kafue National Park; the Tanzanian National Parks Authority (TANAPA) for permission to work at Mikumi National Park; and the Uganda Wildlife Authority and Uganda National Council for Science and Technology and Makerere University for permission to conduct research in Kibale National Park. We thank IRF-Frederick employee Jiro Wada for artistic rendering of animal silhouettes. We thank Wendy Maury and Laura Bollinger for critical reading and editing of the manuscript. We thank Beatrice Hahn for helpful discussions.
A.L.B. wrote the manuscript. A.L.B., M.L., and S.D.S. performed all assays and data analyses. J.E.P.-C., C.J.J., N.B.F., A.J.J., P.A.M., C.A., J.R., and T.L.G. performed and/or organized collection of primary samples. All authors edited and approved the manuscript.
Funding Statement
This work was funded by the National Institutes of Health (NIH) (R01AI077376-01, R01AI084787, R01RR025781, P01AI088564), the National Science Foundation (NSF) (NSF1029302, NSF1029323, NSF1029451), and the joint NIH-NSF Ecology of Infectious Diseases program and the UK Economic and Social Research Council (TW009237), and by the Wisconsin Partnership Program through the Wisconsin Center for Infectious Diseases. All animal research was approved by the appropriate wildlife authorities and Institutional Animal Care and Use Committees. This publication was made possible in part by grants (P51RR000167, a component of the NIH, to the Wisconsin National Primate Research Center [WNPRC], University of Wisconsin—Madison, and R01OD010980, formerly R01RR016300, to the University of California Los Angeles [UCLA]) from the Office of Research Infrastructure Programs (ORIP). This research was conducted in part at a facility constructed with support from the Research Facilities Improvement Program, grant numbers RR15459-01 and RR020141-01. A.L.B. performed this work with support from the University of Wisconsin's Medical Scientist Training Program (MSTP) (grant T32 GM008692) and a National Research Service Award (NRSA) through the Microbes in Health and Disease (MHD) training program at the University of Wisconsin—Madison (T32 AI55397). We thank the University of Wisconsin, Department of Pathology and Laboratory Medicine, and the WNPRC for funding and the use of its facilities and services. We thank the Department of Environmental Affairs, South Africa; Department of Tourism, Environmental and Economic Affairs, Free State Province; the Ezemvelo KZN Wildlife in KwaZulu Natal Province; and Department of Economic Development and Environmental Affairs, Eastern Cape. This work was also funded in part through Battelle Memorial Institute's prime contract with the U.S. National Institute of Allergy and Infectious Diseases (NIAID) under contract HHSN272200700016I. Subcontractors to Battelle Memorial Institute who performed this work are: J.H.K., an employee of Tunnell Government Services, Inc. This publication's contents are solely the responsibility of the author(s) and do not necessarily represent the official views of ORIP, NIH, U.S. Department of Health and Human Services, or of the institutions and companies affiliated with the authors. The funders of this research had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Morse SS, Mazet JAK, Woolhouse M, Parrish CR, Carroll D, Karesh WB, Zambrana-Torrelio C, Lipkin WI, Daszak P. 2012. Prediction and prevention of the next pandemic zoonosis. Lancet 380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A, Ojeda-Flores R, Arrigo NC, Islam A, Ali Khan S, Hosseini P, Bogich TL, Olival KJ, Sanchez-Leon MD, Karesh WB, Goldstein T, Luby SP, Morse SS, Mazet JA, Daszak P, Lipkin WI. 2013. A strategy to estimate unknown viral diversity in mammals. mBio 4:e00598-13. doi: 10.1128/mBio.00598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleaveland S, Haydon DT, Taylor L. 2007. Overviews of pathogen emergence: which pathogens emerge, when and why? Curr Top Microbiol Immunol 315:85–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies TJ, Pedersen AB. 2008. Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc Biol Sci 275:1695–1701. doi: 10.1098/rspb.2008.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper N, Nunn CL. 2013. Identifying future zoonotic disease threats: where are the gaps in our understanding of primate infectious diseases? Evol Med Public Health 2013:27–36. doi: 10.1093/emph/eot001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvignac-Spencer S, Leendertz SAJ, Gillespie TR, Leendertz FH. 2012. Wild great apes as sentinels and sources of infectious disease. Clin Microbiol Infect 18:521–527. doi: 10.1111/j.1469-0691.2012.03816.x. [DOI] [PubMed] [Google Scholar]
- 8.Hoppe E, Pauly M, Gillespie TR, Akoua-Koffi C, Hohmann G, Fruth B, Karhemere S, Madinda NF, Mugisha L, Muyembe J-J. 2015. Multiple cross-species transmission events of human adenoviruses (HAdV) during hominine evolution. Mol Biol Evol 32:2072–2084. doi: 10.1093/molbev/msv090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailey AL, Lauck M, Weiler A, Sibley SD, Dinis JM, Bergman Z, Nelson CW, Correll M, Gleicher M, Hyeroba D, Tumukunde A, Weny G, Chapman C, Kuhn JH, Hughes AL, Friedrich TC, Goldberg TL, O'Connor DH. 2014. High genetic diversity and adaptive potential of two simian hemorrhagic fever viruses in a wild primate population. PLoS One 9:e90714. doi: 10.1371/journal.pone.0090714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey AL, Lauck M, Sibley SD, Pecotte J, Rice K, Weny G, Tumukunde A, Hyeroba D, Greene J, Correll M, Gleicher M, Friedrich TC, Jahrling PB, Kuhn JH, Goldberg TL, Rogers J, O'Connor DH. 2014. Two novel simian arteriviruses in captive and wild baboons (Papio spp.). J Virol 88:13231–13239. doi: 10.1128/JVI.02203-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lauck M, Hyeroba D, Tumukunde A, Weny G, Lank SM, Chapman CA, O'Connor DH, Friedrich TC, Goldberg TL. 2011. Novel, divergent simian hemorrhagic fever viruses in a wild Ugandan red colobus monkey discovered using direct pyrosequencing. PLoS One 6:e19056. doi: 10.1371/journal.pone.0019056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauck M, Sibley SD, Hyeroba D, Tumukunde A, Weny G, Chapman CA, Ting N, Switzer WM, Kuhn JH, Friedrich TC, O'Connor DH, Goldberg TL. 2013. Exceptional simian hemorrhagic fever virus diversity in a wild African primate community. J Virol 87:688–691. doi: 10.1128/JVI.02433-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snijder EJ, Kikkert M, Fang Y. 2013. Arterivirus molecular biology and pathogenesis. J Gen Virol 94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 14.Woolhouse M, Scott F, Hudson Z, Howey R, Chase-Topping M. 2012. Human viruses: discovery and emergence. Phil Trans R Soc Lond Ser B Biol Sci 367:2864–2871. doi: 10.1098/rstb.2011.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tauraso NM, Myers MG, McCarthy K, Tribe GW. 1970. Simian hemorrhagic fever, p 101–109. In Balner H, Beveridge WIB (ed), Infections and immunosuppression in subhuman primates. Munksgaard, Copenhagen, Denmark. [Google Scholar]
- 16.Palmer AE, Allen AM, Tauraso NM, Shelokov A. 1968. Simian hemorrhagic fever. I. Clinical and epizootiologic aspects of an outbreak among quarantined monkeys. Am J Trop Med Hyg 17:404–412. [PubMed] [Google Scholar]
- 17.Allen AM, Palmer AE, Tauraso NM, Shelokov A. 1968. Simian hemorrhagic fever. II. Studies in pathology. Am J Trop Med Hyg 17:413–421. [DOI] [PubMed] [Google Scholar]
- 18.Paessler S, Walker DH. 2013. Pathogenesis of the viral hemorrhagic fevers. Annu Rev Pathol 8:411–440. doi: 10.1146/annurev-pathol-020712-164041. [DOI] [PubMed] [Google Scholar]
- 19.Lapin BA, Pekerman SM, Iakovleva LA, Dzhikidze EK, Shevtsova ZV, Kuksova MI, Dan'ko LV, Krylova RI, Akbroit EI, Agrba VZ. 1967. Hemorrhagic fever in monkeys. Vopr Virusol 12:168–173. (In Russian.) [PubMed] [Google Scholar]
- 20.Espana C. 1971. Review of some outbreaks of viral disease in captive nonhuman primates. Lab Anim Sci 21:1023–1031. [PubMed] [Google Scholar]
- 21.Abildgaard C, Harrison J, Espana C, Spangler W, Gribble D. 1975. Simian hemorrhagic fever: studies of coagulation and pathology. Am J Trop Med Hyg 24:537–544. [DOI] [PubMed] [Google Scholar]
- 22.Renquist D. 1990. Outbreak of simian hemorrhagic fever. J Med Primatol 19:77–79. [PubMed] [Google Scholar]
- 23.Dalgard DW, Hardy RJ, Pearson SL, Pucak GJ, Quander RV, Zack PM, Peters CJ, Jahrling PB. 1992. Combined simian hemorrhagic fever and Ebola virus infection in cynomolgus monkeys. Lab Anim Sci 42:152–157. [PubMed] [Google Scholar]
- 24.London WT. 1977. Epizootiology, transmission and approach to prevention of fatal simian haemorrhagic fever in rhesus monkeys. Nature 268:344–345. doi: 10.1038/268344a0. [DOI] [PubMed] [Google Scholar]
- 25.Gravell M, London WT, Leon ME, Palmer AE, Hamilton RS. 1986. Differences among isolates of simian hemorrhagic fever (SHF) virus. Proc Soc Exp Biol Med 181:112–119. doi: 10.3181/00379727-181-42231. [DOI] [PubMed] [Google Scholar]
- 26.Lauck M, Alkhovsky SV, Bao Y, Bailey AL, Shevtsova ZV, Shchetinin AM, Vishnevskaya TV, Lackemeyer MG, Postnikova E, Mazur S, Wada J, Radoshitzky SR, Friedrich TC, Lapin BA, Deriabin PG, Jahrling PB, Goldberg TL, O'Connor DH, Kuhn JH. 2015. Historical outbreaks of simian hemorrhagic fever in captive macaques were caused by distinct arteriviruses. J Virol 89:8082–8087. doi: 10.1128/JVI.01046-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parrish CR, Holmes EC, Morens DM, Park E-C, Burke DS, Calisher CH, Laughlin CA, Saif LJ, Daszak P. 2008. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev 72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmes EC. 2009. The evolution and emergence of RNA viruses. Oxford series in ecology and evolution. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 29.Apetrei C, Robertson DL, Marx PA. 2004. The history of SIVS and AIDS: epidemiology, phylogeny and biology of isolates from naturally SIV infected non-human primates (NHP) in Africa. Front Biosci 9:225–254. doi: 10.2741/1154. [DOI] [PubMed] [Google Scholar]
- 30.International Committee on the Taxonomy of Viruses. 2015. ICTV files and discussions: animal ssRNA+ viruses. International Committee on the Taxonomy of Viruses. http://talk.ictvonline.org/files/proposals/animal_ssrna_viruses/default.aspx.
- 31.Ma D, Jasinska A, Kristoff J, Grobler JP, Turner T, Jung Y, Schmitt C, Raehtz K, Feyertag F, Martinez Sosa N, Wijewardana V, Burke DS, Robertson DL, Tracy R, Pandrea I, Freimer N, Apetrei C, International Vervet Research Consortium. 2013. SIVagm infection in wild African green monkeys from South Africa: epidemiology, natural history, and evolutionary considerations. PLoS Pathog 9:e1003011. doi: 10.1371/journal.ppat.1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korva M, Duh D, Saksida A, Trilar T, Avsic-Zupanc T. 2009. The hantaviral load in tissues of naturally infected rodents. Microbes Infect 11:344–351. doi: 10.1016/j.micinf.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 33.Hemida MG, Chu DK, Poon LL, Perera RA, Alhammadi MA, Ng HY, Siu LY, Guan Y, Alnaeem A, Peiris M. 2014. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg Infect Dis 20:1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hahn BH, Shaw GM, De Cock KM, Sharp PM. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 35.Peeters M, Courgnaud V, Abela B, Auzel P, Pourrut X, Bibollet-Ruche F, Loul S, Liegeois F, Butel C, Koulagna D, Mpoudi-Ngole E, Shaw GM, Hahn BH, Delaporte E. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg Infect Dis 8:451–457. doi: 10.3201/eid0805.010522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paige SB, Frost SD, Gibson MA, Jones JH, Shankar A, Switzer WM, Ting N, Goldberg TL. 2014. Beyond bushmeat: animal contact, injury, and zoonotic disease risk in Western Uganda. Ecohealth 11:534–543. doi: 10.1007/s10393-014-0942-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takada A. 2012. Filovirus tropism: cellular molecules for viral entry. Front Microbiol 3:34. doi: 10.3389/fmicb.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.VandeWoude S, Apetrei C. 2006. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin Microbiol Rev 19:728–762. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shulla A, Randall G. 2012. Hepatitis C virus-host interactions, replication, and viral assembly. Curr Opin Virol 2:725–732. doi: 10.1016/j.coviro.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed). 2013. Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 41.Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MAM, Kessing B, Pontius J, Roelke M, Rumpler Y, Schneider MPC, Silva A, O'Brien SJ, Pecon-Slattery J. 2011. A molecular phylogeny of living primates. PLoS Genet 7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caì Y, Postnikova EN, Bernbaum JG, Yú SQ, Mazur S, Deiuliis NM, Radoshitzky SR, Lackemeyer MG, McCluskey A, Robinson PJ, Haucke V, Wahl-Jensen V, Bailey AL, Lauck M, Friedrich TC, O'Connor DH, Goldberg TL, Jahrling PB, Kuhn JH. 2015. Simian hemorrhagic fever virus cell entry is dependent on CD163 and uses a clathrin-mediated endocytosis-like pathway. J Virol 89:844–856. doi: 10.1128/JVI.02697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246. doi: 10.1093/glycob/cwh106. [DOI] [PubMed] [Google Scholar]
- 44.John M, Gaudieri S. 2014. Influence of HIV and HCV on T cell antigen presentation and challenges in the development of vaccines. Front Microbiol 5:514. doi: 10.3389/fmicb.2014.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jahrling PB, Geisbert TW, Dalgard DW, Johnson ED, Ksiazek TG, Hall WC, Peters CJ. 1990. Preliminary report: isolation of Ebola virus from monkeys imported to USA. Lancet 335:502–505. doi: 10.1016/0140-6736(90)90737-P. [DOI] [PubMed] [Google Scholar]