Abstract
Cell-associated HIV unspliced RNA is an important marker of the viral reservoir. HIV gag RNA-specific assays are frequently used to monitor reservoir activation. Because HIV preferentially integrates into actively transcribed genes, some of the transcripts detected by these assays may not represent genuine HIV RNA but rather chimeric host-HIV readthrough transcripts. Here, we demonstrate that in HIV-infected patients on suppressive combination antiretroviral therapy, such host-derived transcripts do not significantly contribute to the HIV gag RNA level.
TEXT
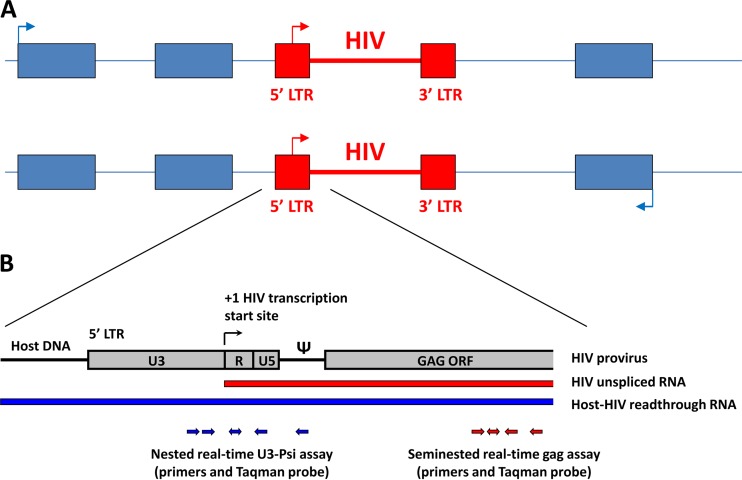
Cell-associated (CA) HIV unspliced RNA is an important marker of the viral reservoir and the response to combination antiretroviral therapy (cART) (1). Recently, there has been considerable interest in the utilization of CA HIV RNA as a surrogate marker of virus activation by latency-reversing agents (LRA) (2), and CA HIV RNA has been used as a main output measure in several clinical trials aimed at reduction of the HIV reservoir (3–6). Primers specific for the HIV gag region are frequently used in PCR-based assays that quantify unspliced RNA (7, 8). However, because HIV integrates preferentially within actively transcribed host genes (Fig. 1A) (9), it has been suggested that some of the transcripts detected by the gag-specific assays may not represent genuine HIV RNA but rather chimeric host-HIV readthrough transcripts that are transcribed from host promoters (10). In this case, an effect of LRA measured by induction of gag RNA transcription could represent activation of a host gene instead of HIV latency reversal. Therefore, to properly interpret the results of the gag assays, it is necessary to determine the relative contribution of such readthrough transcripts to the total HIV gag RNA signal in cART-treated patients.
FIG 1.
(A) HIV proviruses integrate in intronic regions of transcriptionally active host genes, in the same (upper panel; sense) or opposite (lower panel; antisense) orientation with regard to local host gene transcription. (B) A closeup of the 5′ HIV region, with a schematic representation of real-time PCR assays for detection of readthrough and gag RNA. ORF, open reading frame; Ψ, HIV packaging signal (Psi).
We developed a sensitive nested real-time PCR assay that amplifies the 5′ long terminal repeat (LTR)-encoded U3 packaging signal region (U3-Psi) of HIV-1. As the forward primers are located 5′ of the HIV LTR transcription start site, this assay specifically detects host-HIV readthrough transcripts but not genuine HIV-1 unspliced RNA (Fig. 1B). The assay has a linear range of 5 orders of magnitude and sensitivity of 4 copies per reaction (Fig. 2). For this study, we used peripheral blood mononuclear cells (PBMC) of 48 cART-treated patients visiting the HIV outpatient clinic of the Academic Medical Center of the University of Amsterdam from 2011 to 2013 and participating in the Co-morBidity in Relation to AIDS (COBRA) cohort and whose plasma viremia had been undetectable (<40 copies/ml) for a median of 7 years prior to the time of sampling. The median CD4+ T-cell count was 675.5 cells/μl. Total DNA and total RNA were isolated from the patient PBMC by using the Boom isolation method (11), and CA HIV DNA and RNA were separately quantified using both the U3-Psi assay and a seminested real-time PCR assay specific for the HIV gag region (Fig. 1B) (7, 8). Cellular RNA was treated with DNase (DNA-free kit; Ambion) to remove DNA that could interfere with the quantitation and then was reverse transcribed using random primers and SuperScript III reverse transcriptase (all from Invitrogen). As HIV integrates in a random orientation with regard to the host genes, we used random primers to allow detection of readthrough RNA transcribed in both directions, i.e., from upstream and downstream host promoters (Fig. 1A). Same-volume aliquots of the same DNA or cDNA preparations were used as input for U3-Psi and gag assays. HIV DNA and RNA amounts were normalized to the cellular inputs, as described previously (12).
FIG 2.
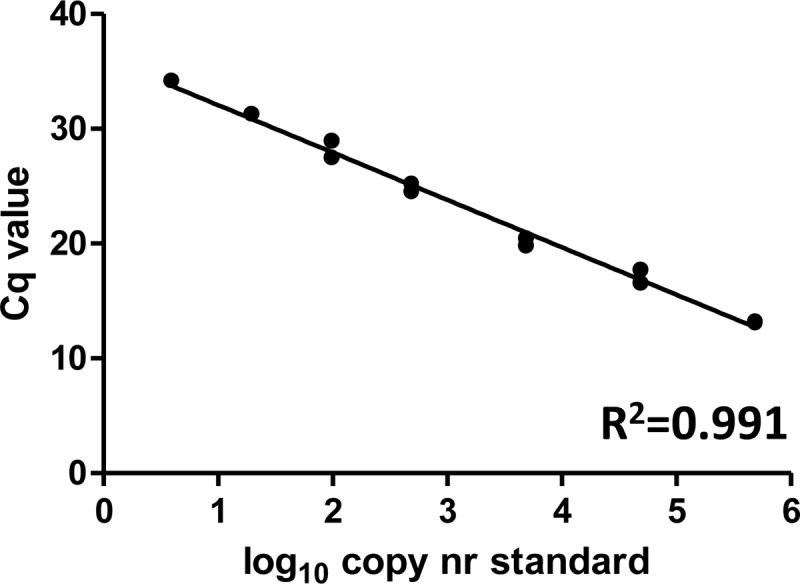
Quantitation of the serially diluted plasmid, pLAIΔRT, which is a molecular clone of HIV-1 that harbors a deletion of the reverse transcriptase gene (7), by using the U3-Psi nested real-time PCR assay. Preamplification (15 cycles) was performed with the forward primer (5′-AGTGGCGAGCCCTCAGATG-3′) and reverse primer (5′-CAGCAAGCCGAGTCCT-3′) in a volume of 25 μl. Two microliters of this PCR was used as input for a nested real-time PCR performed with the forward primer (5′-CAGATGCTGCATATAAGCAGCTG-3′) and reverse primer (5′-CACAACAGACGGGCACACAC-3′) (10) and probe (5′–6-carboxyfluorescein–GAGCTCTCTGGCTAACTAGGGAACCC–6-carboxytetramethyl rhodamine–3′) in a total volume of 50 μl. Cq, quantitation cycle.
The study was approved by the ethics commission of the Academic Medical Center and was conducted in accordance with the ethical principles of the Declaration of Helsinki. All patients provided written informed consent.
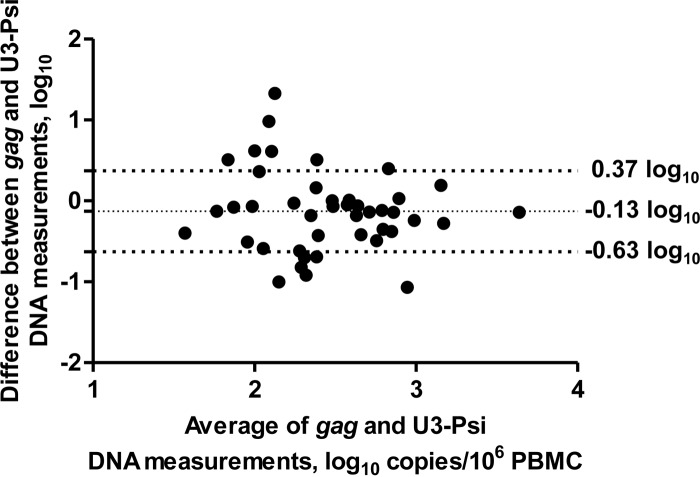
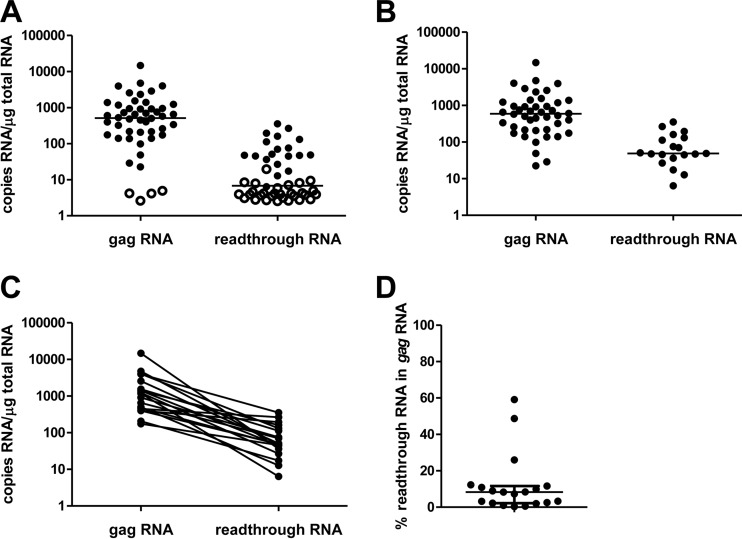
As expected, both the U3-Psi and gag assays detected HIV DNA in >90% of the patients (44/48 and 46/48, respectively) with no significant quantitative bias between the assays (0.13 ± 0.50 log10; P > 0.05 for comparison of the difference to 0) (Fig. 3), and a highly significant correlation between the two measurements was observed (P = 0.001), demonstrating the functionality of the U3-Psi assay. However, a major difference in detectability of HIV RNA was observed. HIV gag RNA was detected in 44/48 of these patients (92%) with a median copy number of 590 (interquartile range, 217 to 1,194) copies/μg total RNA. However, the detectability of readthrough RNA was only 40% (19/48 patients) (Fig. 4A). In the 19 patients where the readthrough RNA was detected, its median copy number was 49 (interquartile range, 41 to 122) copies/μg total RNA (P = 0.0001 for the paired comparison with HIV gag RNA) (Fig. 4B and C). This represented only 8.3% (2.4% to 11.2%) of the HIV gag RNA (Fig. 4D). Notably, this is a large overestimation, and the real readthrough/gag RNA ratio is much lower, as patients with undetectable readthrough RNA (60% of all patients) were excluded from this calculation. No significant correlation was observed between HIV gag RNA and the readthrough RNA (P = 0.64).
FIG 3.
Bland-Altman plot of the gag and U3-Psi HIV DNA measurements. Horizontal lines indicate the average difference between the measurements and average difference ± standard deviations.
FIG 4.
Comparison of HIV-1 gag RNA and readthrough RNA in all patients, with undetectable values left-censored at the detection limits of corresponding assays shown by open circles (A) or only in patients with detectable gag or readthrough RNA (B). Also shown are paired comparison in patients with detectable readthrough RNA (C) and the percentage of readthrough RNA in the HIV-1 gag RNA in patients with detectable readthrough RNA (D). Medians are shown in panels A and B, and medians and interquartile ranges are shown in panel D.
Although the existence of host-HIV readthrough transcripts has been demonstrated previously (9, 13), this is the first quantitative comparison of these transcripts with HIV gag RNA in cells from HIV-infected patients. Our results show compellingly that in PBMC of HIV-infected patients on suppressive cART, the contribution of host-derived transcripts to the RNA measured in HIV gag assays is very small. The host-HIV readthrough RNA transcribed in the same direction as HIV (sense) is most probably polyadenylated at the HIV 5′ LTR, whereas HIV has evolved with a number of strategies to suppress polyadenylation of its nascent RNA transcript (14, 15). However, polyadenylation cannot be the only explanation for the scarcity of host-HIV readthrough transcripts that we found, as the readthrough RNA transcribed in the antisense direction is not expected to be polyadenylated at the HIV LTRs. Rather, as introns represent the absolute majority of HIV integration sites within genes (9), the low abundance of host-HIV readthrough transcripts compared to genuine HIV RNA might reflect a combination of the short half-lives of pre-mRNA and intronic RNA in a cell (16) and the relative strength of the HIV LTR promoter.
A limitation of this study is that we only quantified HIV RNA in total PBMC. It is possible that the HIV readthrough/gag RNA ratio is different in resting CD4+ T cells. However, although the HIV transcription level is lower in resting than in activated CD4+ cells (17), host cell transcription is also expected to be lower due to the absence of nuclear forms of key transcription factors (e.g., NF-κB and NFAT) in resting cells (18). In addition, to monitor the efficacy of LRA clinical trials, HIV gag RNA is usually quantified in total CD4+ cells or PBMC (3, 4, 6). Therefore, our report is relevant for the interpretation of the outcome of such trials.
In summary, we observed only a minor contribution of host-HIV readthrough transcripts to the level of HIV gag RNA. The vast majority of HIV gag RNA transcripts in cART-treated patients represent genuine HIV unspliced RNA.
ACKNOWLEDGMENTS
We are grateful to the COBRA collaboration for making available the samples from COBRA study participants.
The Co-morBidity in Relation to AIDS (COBRA) collaboration has received funding from the European Union's Seventh Framework Programme for Research, Technological Development and Demonstration under grant agreement 305522. A.O.P. is financially supported by the Dutch Aids Fonds, grants 2011020 and 2012025.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Pasternak AO, Lukashov VV, Berkhout B. 2013. Cell-associated HIV RNA: a dynamic biomarker of viral persistence. Retrovirology 10:41. doi: 10.1186/1742-4690-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruner KM, Hosmane NN, Siliciano RF. 2015. Towards an HIV-1 cure: measuring the latent reservoir. Trends Microbiol 23:192–203. doi: 10.1016/j.tim.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, Smith MZ, Spelman T, McMahon J, Velayudham P, Brown G, Roney J, Watson J, Prince MH, Hoy JF, Chomont N, Fromentin R, Procopio FA, Zeidan J, Palmer S, Odevall L, Johnstone RW, Martin BP, Sinclair E, Deeks SG, Hazuda DJ, Cameron PU, Sekaly RP, Lewin SR. 2014. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 10:e1004473. doi: 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, Winckelmann A, Palmer S, Dinarello C, Buzon M, Lichterfeld M, Lewin SR, Østergaard L, Søgaard OS. 2014. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 1:e13–e21. doi: 10.1016/S2352-3018(14)70014-1. [DOI] [PubMed] [Google Scholar]
- 5.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Søgaard OS, Graversen ME, Leth S, Olesen R, Brinkmann CR, Nissen SK, Kjaer AS, Schleimann MH, Denton PW, Hey-Cunningham WJ, Koelsch KK, Pantaleo G, Krogsgaard K, Sommerfelt M, Fromentin R, Chomont N, Rasmussen TA, Østergaard L, Tolstrup M. 2015. The depsipeptide romidepsin reverses HIV-1 latency in vivo. PLoS Pathog 11:e1005142. doi: 10.1371/journal.ppat.1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasternak AO, Adema KW, Bakker M, Jurriaans S, Berkhout B, Cornelissen M, Lukashov VV. 2008. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J Clin Microbiol 46:2206–2211. doi: 10.1128/JCM.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiselinova M, Pasternak AO, De Spiegelaere W, Vogelaers D, Berkhout B, Vandekerckhove L. 2014. Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PLoS One 9:e85999. doi: 10.1371/journal.pone.0085999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han Y, Lassen K, Monie D, Sedaghat AR, Shimoji S, Liu X, Pierson TC, Margolick JB, Siliciano RF, Siliciano JD. 2004. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol 78:6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. 2014. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med 20:425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. 1990. Rapid and simple method for purification of nucleic acids. J Clin Microbiol 28:495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasternak AO, Jurriaans S, Bakker M, Prins JM, Berkhout B, Lukashov VV. 2009. Cellular levels of HIV unspliced RNA from patients on combination antiretroviral therapy with undetectable plasma viremia predict the therapy outcome. PLoS One 4:e8490. doi: 10.1371/journal.pone.0008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherrill-Mix S, Ocwieja KE, Bushman FD. 2015. Gene activity in primary T cells infected with HIV89.6: intron retention and induction of genomic repeats. Retrovirology 12:79. doi: 10.1186/s12977-015-0205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown PH, Tiley LS, Cullen BR. 1991. Efficient polyadenylation within the human immunodeficiency virus type 1 long terminal repeat requires flanking U3-specific sequences. J Virol 65:3340–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das AT, Klaver B, Berkhout B. 1999. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J Virol 73:81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clement JQ, Qian L, Kaplinsky N, Wilkinson MF. 1999. The stability and fate of a spliced intron from vertebrate cells. RNA 5:206–220. doi: 10.1017/S1355838299981190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser P, Joos B, Niederost B, Weber R, Gunthard HF, Fischer M. 2007. Productive human immunodeficiency virus type 1 infection in peripheral blood predominantly takes place in CD4/CD8 double-negative T lymphocytes. J Virol 81:9693–9706. doi: 10.1128/JVI.00492-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siliciano RF, Greene WC. 2011. HIV latency. Cold Spring Harb Perspect Med 1:a007096. [DOI] [PMC free article] [PubMed] [Google Scholar]