Abstract
Group B Streptococcus (GBS) is a common commensal bacterium in adults, but is also the leading cause of invasive bacterial infections in neonates in developed countries. The β-hemolysin/cytolysin (β-h/c), which is always associated with the production of an orange-to-red pigment, is a major virulence factor that is also used for GBS diagnosis. A collection of 1,776 independent clinical GBS strains isolated in France between 2006 and 2013 was evaluated on specific medium for β-h/c activity and pigment production. The genomic sequences of nonhemolytic and nonpigmented (NH/NP) strains were analyzed to identify the molecular basis of this phenotype. Gene deletions or complementations were carried out to confirm the genotype-phenotype association. Sixty-three GBS strains (3.5%) were NH/NP, and 47 of these (74.6%) originated from invasive infections, including bacteremia and meningitis, in neonates or adults. The mutations are localized predominantly in the cyl operon, encoding the β-h/c pigment biosynthetic pathway and, in the abx1 gene, encoding a CovSR regulator partner. In conclusion, although usually associated with GBS virulence, β-h/c pigment production is not absolutely required to cause human invasive infections. Caution should therefore be taken in the use of hemolysis and pigmentation as criteria for GBS diagnosis in routine clinical laboratory settings.
INTRODUCTION
Streptococcus agalactiae, or group B Streptococcus (GBS), is a Gram-positive bacterium that is usually an asymptomatic member of the human intestinal and vaginal microbiota. However, GBS is also the leading cause of neonatal sepsis and meningitis in developed countries (1–3) and an emerging pathogen in adults, mostly in the elderly (4, 5). Neonatal infections occur predominantly during delivery by inhalation or ingestion of contaminated secretions of the mother's vagina. Rapid invasive infections may follow, with a mortality rate of up to 10% during the first week of life (early-onset disease [EOD], age of 0 to 6 days) or during the first 3 months of life (late-onset disease [LOD], age of 7 to 89 days). Systematic GBS screening prior to delivery and intrapartum antibiotic prophylaxis have efficiently reduced EOD but not LOD (6), highlighting the need to further improve the diagnosis and treatments.
GBS is an extracellular pathogen and one of its characteristics is the secretion of a potent β-hemolysin/cytolysin (β-h/c) (7–9). This β-h/c has a central role in balancing the pro- and anti-inflammatory responses of the infected host and is necessary to breach the epithelial and endothelial barriers and the phagolysosome membrane (10–15). Remarkably, the GBS β-h/c is unique among Gram-positive pathogens and its production is linked to that of a characteristic orange-to-red pigment. This dual and specific β-h/c and pigment phenotype is routinely used in clinical settings to identify GBS isolates (16, 17).
It was initially proposed that the GBS protein CylE was, in fact, β-h/c and that the pigment was a distinct carotenoid-like molecule (18, 19). However, it was recently established that the β-h/c and the pigment correspond to a unique molecule called granadaene (9, 11), a 676-Da ornithine rhamno-polyene (11, 20). Granadaene may be entirely synthesized by the 12 genes of the cyl operon (11, 19, 21). It involves the initial synthesis of the β-h/c core lipid by AcpC, CylD, CylI, and CylG (i.e., the Cyl homologues of fatty acid synthesis enzymes) followed by addition of one ornithine and one rhamnose residue by CylE and CylJ, respectively (11). Finally, export of the β-h/c toxin requires the ABC-type transporter encoded by cylA and cylB (22).
Transcription of the cyl operon is controlled by CovSR (Control of virulence Sensor and Regulator), a two-component system and the major regulator of GBS virulence (23, 24). The transcriptional regulator CovR binds directly to the cyl promoter, thereby inhibiting operon transcription (23, 25). CovR activity is itself regulated by phosphorylation by its cognate histidine kinase CovS. In addition, two other proteins control CovR: the serine/threonine kinase Stk1, which also phosphorylates CovR, but not at the same site as that modified by CovS (25, 26), and the transmembrane protein Abx1, which interacts with CovS to control the balance between its kinase and phosphatase activities (27). This regulatory network has been proposed to be the central mediator of the switch between the commensal and the virulent GBS lifestyles (26, 27).
Independent studies clearly demonstrated that β-h/c pigment production is associated with virulence (9–15). However, it is currently unclear if β-h/c pigment production correlates strictly with GBS virulence. To date, it has been estimated that 85 to 98% of strains isolated from humans produce the β-h/c pigment (16, 28, 29), and it is assumed that nonhemolytic and nonpigmented (NH/NP) strains rarely cause infection (30, 31). Four NH/NP GBS clinical isolates have been characterized in which the cyl operon was inactivated by the insertion element IS1381 (21, 30). In this study, we report the screening of a large collection of human GBS isolates for the presence or absence of the β-h/c pigment. Among 1,776 isolates, 63 (3.5%) were NH/NP. The causative mutations of these NH/NP GBS were identified by whole-genome sequencing or by PCR sequencing of genes known to be involved in β-h/c pigment production. Several as yet uncharacterized genes of the cyl operon were also inactivated to accurately characterize and compare the phenotypes of the mutants generated.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
GBS clinical strains from noninvasive and invasive infections (positive blood culture, cerebrospinal fluid, or other normally sterile sites) were obtained from the French National Reference Center for Streptococci (www.cnr-strep.fr; Paris, France), and their origins are summarized in Table S1 in the supplemental material. GBS cultures were grown at 37°C in Todd-Hewitt (TH) agar or broth (Difco Laboratories) under static conditions or anaerobically on Granada agar (bioMérieux, France) (16) or Columbia horse blood (5%) agar plates (bioMérieux) When appropriate, erythromycin (10 μg/ml) or kanamycin (1,000 μg/ml) was added to the medium. Escherichia coli XL1-Blue (Stratagene) and TOP10 (Invitrogen) were cultured in Luria-Bertani (LB) broth at 37°C with shaking. When appropriate, erythromycin (150 μg/ml) or ticarcillin (100 μg/ml) was added to the medium.
Confirmation of NH/NP GBS strains.
The NH/NP phenotypes of GBS strains were confirmed by spotting serial dilutions of an overnight culture on blood and Granada agar plates, as described previously (27). The NH/NP strains were further confirmed as GBS by colony morphology, Lancefield grouping with type B antisera (Oxoid), and matrix-assisted laser desorption ionization−time of flight (MALDI-TOF) mass spectrometry (Bruker) according to the manufacturer's recommendations. Molecular capsular typing was performed by multiplex PCR as described previously (32). Multilocus sequence typing (MLST) was performed in silico from whole-genome sequences or by PCR amplification and sequencing as described previously (33); the sequence type (ST) assignment was determined using the reference GBS MLST website (http://pubmlst.org/sagalactiae/).
Genome sequencing.
Genomic DNAs were purified (DNeasy blood and tissue kit [Qiagen] or MasterPure Gram-positive DNA purification kit [Epicentre Biotechnologies]) from 10 ml of overnight cultures in TH agar. Whole-genome sequencing was performed at the Sequencing Core Facilities of Institut Pasteur (Paris, France) and at Institut Cochin (Paris, France) using, respectively, Illumina HiSeq and Life Technology Ion Proton systems. Libraries were prepared using an Illumina TrueSeq DNA LT sample prep kit or an Ion Plus fragment kit according to the manufacturers' instructions. We obtained genomic paired-end reads of 2 × 95 nucleotides (HiSeq) or 150-nucleotide reads (Ion Proton). After trimming and quality assessments, short reads were assembled with Geneious software (Biomatters Ltd. [34]) using the NEM316 genome (NCBI NC_004368.1 [35]) as a reference. Contigs encompassing the cyl, abx1, covS, covR, and stk1 locus were extracted and screened for mutations resulting in frameshifts (microdeletions or microinsertions), nonsense (SNPs leading to premature stop codons), or protein length modifications (insertion elements and deletions). Relevant mutations were confirmed by PCR with high-fidelity Taq polymerase (Phusion; Thermo Scientific) and sequencing.
DNA manipulations and mutant constructions.
The bacterial strains, plasmids, and primers used in this study are listed in Supplemental Tables S2, S3, and S4, respectively, in the supplemental material. GBS mutants were constructed as described previously (27, 36). Briefly, in-frame deletions of cylX, cylD, cylG, cylAB, and cylI were constructed in the reference strain NEM316 (35). Deletion of the IS1381 element was constructed in the clinical isolate CCH1084. The expected ΔcylX, ΔcylD, ΔcylG, ΔcylAB, ΔcylI, and ΔIS1381 mutants were confirmed by sequencing the relevant loci after PCR amplification.
With use of specific primers (see Table S4 in the supplemental material), the IS1381 element and flanking chromosomal sequences were amplified by PCR from CCH1084 DNA and cloned into pG1. Integration of the IS1381 element into the NEM316 chromosome at the same location as in the parental CCH1084 strain was done by homologous recombination as described previously (27, 36).
The pTCVΩPcyl+_abx1 vector that constitutively expresses a wild-type (WT) abx1 allele (27) was used to complement the NH/NP strains producing a truncated Abx1 protein. The complementing vector was transferred by conjugation using the E. coli HB101/pRK24 donor strain to clinical NH/NP GBS strains as described previously (37), and transconjugants were selected with nalidixic acid (50 μg/ml) for the donor and kanamycin (1000 μg/ml) for the recipient.
RESULTS
Prevalence of NH/NP GBS strains among clinical isolates.
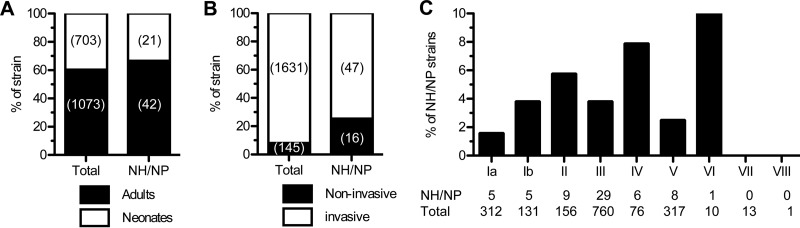
A total of 1,776 nonredundant clinical GBS strains isolated in France between 2006 and 2013, from various geographical areas were included in this study. All strains isolated from adults and neonates were screened for β-h/c activity and pigment production. Sixty-three (3.5%) strains, whose prevalence was slightly higher in adults (42/1,073 = 3.9%) than in neonates (21/703 = 3%) were NH/NP (Fig. 1A). Notably, NH/NP strains were about four times more frequently associated with noninvasive diseases or colonization (16/145 = 11%) than with invasive infections (47/1,631 = 2.9%) (Fig. 1B). The NH/NP strains were isolated from all types of clinical manifestations caused by GBS (Table 1 and Table S1 in the supplemental material). Most strains from adult invasive infections were from bacteremia (22/31 = 71%). Sixteen strains from neonates had elicited invasive infection syndromes (n = 5 EOD and 11 LOD) and, among these, 4 (n = 1 EOD and 3 LOD) were responsible for meningitis (Table 1 and Table S1 in the supplemental material).
FIG 1.
Distribution of NH/NP GBS clinical isolates. Percentages and numbers of clinical GBS strains according to the age of the patient (A), the strain invasiveness (B), and the capsular serotype (C) for the whole collection and for NH/NP strains.
TABLE 1.
Origins of GBS NH/NP isolates according to their capsular serotype
| Origin | Results for capsular serotypea: |
|||||||
|---|---|---|---|---|---|---|---|---|
| Ia | Ib | II | III | IV | V | VI | Total | |
| From neonates | ||||||||
| EOD meningitis | 1 (1) | 1 (1) | ||||||
| EOD bacteremia | 1 (0) | 2 (1) | 1 (0) | 4 (1) | ||||
| LOD meningitis | 3 (3) | 3 (3) | ||||||
| LOD bacteremia | 1 (1) | 6 (4) | 1 (1) | 8 (6) | ||||
| Colonization | 3 (0) | 1 (0) | 1 (0) | 5 (0) | ||||
| Total neonates | 4 (1) | 1 (0) | 0 | 13 (9) | 2 (1) | 0 | 1 (0) | 21 (11) |
| From adults | ||||||||
| Meningitis | 1 (1) | 1 (1) | ||||||
| Bacteremia | 3 (1) | 6 (4) | 5 (2) | 2 (1) | 6 (2) | 22 (10) | ||
| Endocarditis | 1 (1) | 1 (1) | 2 (2) | |||||
| Bone and joint infection | 1 (1) | 2 (0) | 1 (0) | 1 (1) | 5 (2) | |||
| Others | 1 (1) | 1 (1) | 7 (5) | 2 (1) | 11 (8) | |||
| Total adults | 1 (1) | 4 (2) | 9 (5) | 16 (9) | 4 (2) | 8 (4) | 0 | 42 (23) |
| Total NH/NP | 5 (2) | 5 (2) | 9 (5) | 29 (18) | 6 (3) | 8 (4) | 1 (0) | 63 (34) |
Data are the numbers of strains for a given capsular serotype and, in parentheses, the numbers of strains selected for sequencing.
To determine if NH/NP strains comprise a specific subpopulation of GBS, molecular capsule (capsular serotype [CPS]) typing was performed on the whole collection, and the ratio of NH/NP isolates per CPS type was determined (Fig. 1C). For the 6 clinically representative CPS types (Ia, Ib, II, III, IV, and V), all containing at least 76 isolates, the percentage of NH/NP strains ranged between 1.6% (CPS type Ia, 5/312) and 7.9% (CPS type IV, 6/76) (Fig. 1C). Most strains were CPS type III, the major CPS type associated with neonatal invasive infection (1–3), among which 3.8% (29/760) were NH/NP. The remaining 24 strains were CPS type VI (n = 10), VII (n = 13), and VIII (n = 1) and the single NH/NP was CPS type VI. Finally, molecular typing by MLST on a subset of 34 strains representative of the 63 NH/NP clinical isolates revealed several sequence types (ST), including 12 strains belonging to the hypervirulent ST17 lineage responsible for the majority of neonatal meningitis cases (Table 2 and Table S1 in the supplemental material). These analyses show that NH/NP strains are distributed among the major CPS and sequence types responsible for infection.
TABLE 2.
Loss-of-function mutations in the NH/NP sequenced GBS strains
| Isolate | CSPa | STb | Mutated gene | Type of mutation | Size or characteristic | DNA positionc | Protein positiond |
|---|---|---|---|---|---|---|---|
| CCH1334 | III | 17 | cylX-D | Deletion | X1 → D849 | ||
| CCH1084 | III | 1 | cylX-D | Insertion | IS1381 | X306 → D1 | |
| CCH1318 | III | 17 | cylG | Substitution | Stop codon | 636/723 | 213/241 |
| CCH1338 | III | 366 | cylG | Deletion | 4 bp | 300/723 | 103/241 |
| CCH1347 | III | 17 | acpC | Substitution | Stop codon | 42/306 | 15/102 |
| CCH1340 | IV | 459 | cylZ | Deletion | 1 bp | 92/477 | 55/159 |
| CCH1321 | III | 106 | cylA | Insertion | IS1381 | 471/930 | |
| CCH1323 | III | 27 | cylA | Insertion | IS1381 | 471/930 | |
| CCH1337 | III | 27 | cylA | Insertion | IS1381 | 466/930 | |
| CCH1326 | III | 17 | cylE | Deletion | 5 bp | 241/2004 | 81/668 |
| CCH1339 | Ia | 136 | cylE | Insertion | ISSa4 | 1582/2004 | |
| CCH1341 | III | 17 | cylE | Substitution | Stop codon | 1335/2004 | 446/668 |
| CCH1342 | III | 27 | cylE | Insertion | 1 bp | 897/2004 | 304/668 |
| CCH1352 | III | 17 | cylE | Deletion | 1 bp | 1514/2004 | 523/668 |
| CCH1345 | II | 10 | cylF | Substitution | Stop codon | 595/954 | 199/318 |
| CCH1333 | III | 17 | cylF-I | Deletion | 692 pb | F749 → I492 | |
| CCH1319 | III | 17 | cylI | Deletion | 1 bp | 1121/2196 | 386/732 |
| CCH1320 | III | 17 | cylI | Insertion | 17 bp | 1817/2196 | 619/732 |
| CCH1322 | Ia | 23 | cylI | Deletion | 1 bp | 883/2196 | 306/732 |
| CCH1327 | II | 28 | cylI | Insertion | ICESp1108 | 1863/2196 | |
| CCH1329 | II | 12 | cylI | Insertion | 1 bp | 434/2196 | 158/732 |
| CCH1343 | II | 22 | cylI | Substitution | Stop codon | 936/2196 | 313/732 |
| CCH1344 | Ib | 8 | cylI | Insertion | IS1381 | 1844/2196 | |
| CCH1349 | Ib | 1 | cylI | Insertion | IS1381 | 1842/2196 | |
| abx1 | Deletion | 1 bp | 176/921 | 73/307 | |||
| CCH1346 | IV | 1 | cylI | Insertion | 12 bp | 1172/2196 | |
| abx1 | Substitution | Stop codon | 504/921 | 169/307 | |||
| CCH1324 | V | 1 | cylI | Deletion | 83 bp | 1893/2196 | |
| abx1 | Deletion | 1 bp | 176/921 | 73/307 | |||
| CCH1325 | III | 1 | abx1 | Deletion | 1 bp | 176/921 | 73/307 |
| CCH1330 | III | 17 | abx1 | Insertion | 1 bp | 101/921 | 53/307 |
| CCH1331 | V | 1 | abx1 | Deletion | 1 bp | 176/921 | 73/307 |
| CCH1350 | V | 1 | abx1 | Deletion | 1 bp | 176/921 | 73/307 |
| CCH1328 | V | 1 | abx1 | Deletion | 1 bp | 176/921 | 73/307 |
| CCH1351 | II | 2 | NDe | ||||
| CCH1348 | IV | 459 | ND | ||||
| CCH1336 | III | 17 | ND |
CSP, capsular serotype.
ST, sequence type by MLST.
Nucleotidic position of the mutation/wild-type gene length.
Amino acid position of the truncation/wild-type protein length.
ND, not determined.
Association of the NH/NP phenotype with mutations in the cyl operon and abx1 gene.
To elucidate the molecular basis of the naturally occurring NH/NP phenotype, genome sequences of the 34 representative selected isolates were determined and compared to the genomes of GBS reference strains (Table 2 and Fig. 2A). Until now, NH/NP GBS mutants were constructed by introducing mutations within the cyl operon (11, 19, 27, 36, 38) or in genes coding its cognate transcriptional regulators, CovSR, Stk1, or Abx1 (27, 38). We therefore focused our analysis on this group of genes (Fig. 2B).
FIG 2.
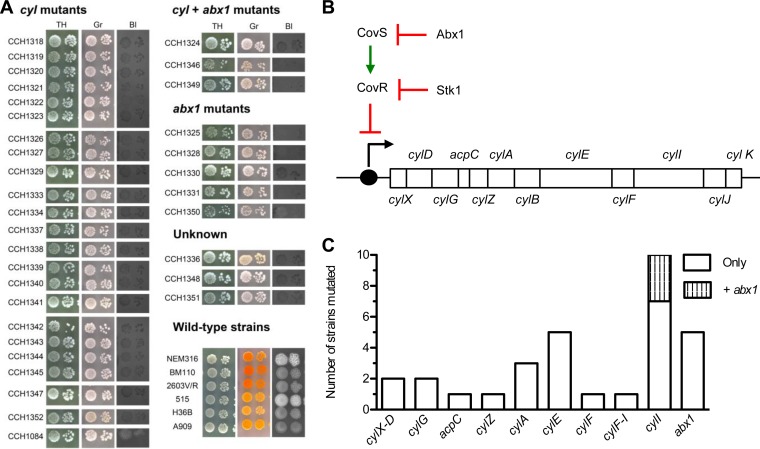
Phenotypes and genotypes of NH/NP clinical isolates. (A) Hemolytic and pigmentation phenotypes of the 34 GBS clinical isolates selected for sequence analysis, organized according to the identified mutation, and of GBS wild-type strains NEM316, BM110, 2603V/R, 515, H36B and A909. TH, Todd-Hewitt agar; Gr, Granada agar; Bl, Columbia agar supplemented with 5% horse blood. (B) Structure and regulation of the cyl operon. Red crossed line, transcriptional repression; green straight arrow, activation; black bent arrow, initiation of transcription. (C) Distribution of identified mutations in the cyl operon and in the cyl regulator gene abx1 among 31 GBS strains; note that no mutation was associated to the NH/NP phenotype among 3 sequenced strains.
Sequence comparison revealed that 26 (76.5%) NH/NP strains had at least one mutation in the cyl operon, potentially leading to synthesis of truncated proteins (Table 2 and Fig. 2C). These mutations were diverse and include the following: insertion of an insertion element (n = 8), deletions or insertions of various lengths (n = 13) including 1-bp frameshift insertions or deletions (n = 6), and single nucleotide polymorphisms yielding premature stop codons (n = 5) (Table 2). Mutations were detected in 9 out of the 12 genes of the cyl operon (Table 2 and Fig. 2C). The most frequently mutated genes were cylA, cylE, and cylI, which were all inactivated in at least 3 strains. Interestingly, all characterized mutations were different, except two IS1381 insertions that occurred twice at the same location in cylA (strains CCH1321 and CCH1323) and cylI (strains CCH1344 and CCH1349) genes (Table 2).
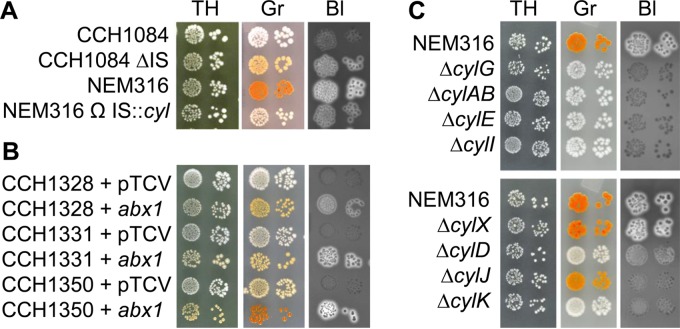
The cyl operon of 8 strains was inactivated by insertion sequences IS1381 (n = 6), ISSa4 (n = 1), and ICESp1108 (n = 1). Insertional inactivation of cylA by IS1381 was previously described in four clinical isolates (21, 30). Consistently, we identified insertions of IS1381 within cylA (3 strains) and additionally in cylI (2 strains). Interestingly, in strain CCH1084 (Table 2), the IS1381 insertion occurred between cylX and cylD, although neither gene was disrupted. To confirm that this insertion was responsible for the NH/NP phenotype, we demonstrated that restoration of the WT cyl sequence in CCH1084 restored pigmentation and hemolysis, whereas introduction of the same IS1381 insertion in NEM316 yielded a very weakly β-h/c mutant strain (Fig. 3A). In strain CCH1339, cylE was inactivated by ISSa4, whereas in strain CCH1327, cylI was disrupted by ICESp1108. These results uncover numerous different types of events occurring at the cyl locus that are responsible for the NH/NP phenotype in GBS clinical strains.
FIG 3.
Hemolytic and pigmentation phenotypes in GBS mutants. (A) Effects of deletion of the IS1381 element from the CCH1084 strain and of the insertion of the same element at the cognate locus in the NEM316 strain. (B) Effects of the genetic complementation of clinical strains mutated only in abx1 with an empty vector (+ pTCV) and the same vector containing a wild-type copy of abx1 (+ abx1). (C) Pigmentation and hemolysis of the NEM316 reference strain and corresponding in-frame deletion mutants ΔcylG, ΔcylAB, ΔcylE, ΔcylI, ΔcylX, ΔcylD, ΔcylJ, and ΔcylK. Bacterial growth, hemolysis, and pigmentation were observed on Todd-Hewitt agar (TH), Granada agar (Gr), and blood agar (Bl), respectively.
The analysis of abx1, stk1, covS, and covR sequences revealed mutations in abx1 only (Fig. 2 and Table 2). These mutations occurred in 8 strains (23.5%), 4 being of CPS type V (ST-1), which had the same microdeletion, suggesting a mutation hot spot. Three out of the 8 strains had an additional mutation in the cyl operon. A plasmid harboring a wild-type copy of abx1 was introduced in 3 strains in which abx1 was mutated (CCH1328, CCH1331, and CCH1350). β-h/c pigment production was restored in all three strains (Fig. 3B), confirming that abx1 mutations were responsible for the NH/NP phenotype.
Overall, of the 34 NH/NP strains analyzed, mutations were present in the cyl operon (23/34, 67.6%), in abx1 (5/34, 14.7%), or in both loci (3/34, 8.8%) (Fig. 2 and Table 2). No mutation that might account for the NH/NP phenotype was found in the 3 (8.8%) remaining strains. This suggests that at least one uncharacterized gene(s) controls β-h/c pigment production in GBS.
Phenotypic characterization of isogenic cyl mutants.
To complete the analysis of the cyl operon in the same genetic background, we chose the reference NEM316 strain (35) to generate in-frame deletions of the cylX, cylD, cylG, and cylI genes, which encode proteins putatively required for synthesis of the lipid core backbone of the β-h/c pigment (9, 11). We similarly deleted the cylAB genes encoding the two subunits of an ABC-like transporter supposedly involved in β-h/c pigment export (22). These mutant strains, as well as previously constructed cylE, cylJ, and cylK NEM316 deletion derivatives for β-h/c pigment maturation (11, 36), enabled us to perform a thorough phenotype-genotype correlation study (Fig. 3C).
Comparison of these isogenic cyl mutants revealed various hemolytic and pigmentation phenotypes (Fig. 3C). While the cylX mutant was indistinguishable from the parental strain, the cylE, cylG, cylAB, and cylI mutants were all nonhemolytic and nonpigmented (Fig. 3C). The cylD and cylK mutants were faintly pigmented and displayed reduced hemolytic activity. Interestingly, the cylJ mutant exhibited slightly reduced pigmentation, contrasting with very weak hemolysis (Fig. 3C). Thus, the critical cyl genes for β-h/c pigment production in GBS are cylE, cylG, cylAB, and cylI and to a lesser extent cylD and cylK. Mutations in all these genes except cylK were found in the sequenced NH/NP GBS clinical isolates (Table 2).
DISCUSSION
The β-hemolytic/cytotoxic activity and the orange-to-red pigmentation are the hallmarks of group B Streptococcus. These two phenotypic characteristics have been used historically for GBS diagnosis, and the current view is that they are essential for virulence (8, 9). We tested this assumption by analyzing a large collection of documented GBS human clinical isolates. Sixty-three of the 1,776 isolates were NH/NP, a proportion in agreement with those in previous smaller scale studies (16, 28, 29). Although it is currently thought that NH/NP strains rarely cause infection in humans (30, 31), we observed that NH/NP isolates were found in the most prevalent GBS capsular serotypes and sequence types, including the neonatal hypervirulent ST17 clone (39–41). Remarkably, 47 of the 63 NH/NP strains originated from invasive diseases, including bacteremia and meningitis. These results suggest that β-h/c pigment production is not critical for GBS virulence although numerous in vitro and in vivo studies using cellular or animal models indicated that greater β-h/c pigment production is associated with increased virulence (10–15, 18).
The pathogenicity of GBS is multifactorial and combines the expression of several factors to result in an overall virulence phenotype that may vary for each isolate (42). In addition, host factors, at the genetic or microbiota level, might contribute to virulence of NH/NP strains that do not express one of the most potent GBS virulence-associated factors. The necessity, but dispensability, of β-h/c pigment production for GBS virulence is also illustrated by the greater proportion of NH/NP strains isolated from noninvasive clinical cases (11%) than from invasive disease cases (2.9%). This may suggest that β-h/c pigment production is important for eliciting invasion, but dispensable for the establishment of colonization, in agreement with previous studies on GBS urinary tract infections in animal models (43, 44).
At the genetic level, we identified independent mutations responsible for the NH/NP phenotype, in agreement with the fact that NH/NP strains are independent and not associated with a specific GBS subpopulation. Three-quarters (26/34) of the NH/NP strains harbored a mutation in the cyl operon, confirming its central function for the synthesis, maturation, and export of the β-h/c pigment (9, 11, 22, 36). Eight of those strains carry an IS element in the cyl operon, among which six correspond to an IS1381 element previously identified in four clinical NH/NP isolates (21, 30). The different locations and nature of the insertion and the different sequence types of the two pairs of strains with an insertion at the same position (in cylA genes for CCH1321 and CCH1323 and in cylI genes for CCH1344 and CCH1349) suggest that these mutations were acquired independently and that the IS1381 insertion is a fairly common event. The remaining 20 strains have independent mutations in at least 1 of the 12 cyl operon genes. While cylE inactivation has been shown to systematically yield NH/NP mutants in several unrelated isolates, the phenotype resulting from the inactivation of the other cyl genes had not been systematically studied (11, 18, 19, 21, 36). Seven of the 12 genes of the cyl operon are thought to be involved in the synthesis of the lipid backbone of the carotenoid-like molecule granadaene (cylX, cylD, cylG, acpC, cylZ, cylI, and cylK) (11, 18, 19, 21, 36). While inactivation of cylG and cylI results in a complete loss of hemolysis and pigmentation, as previously observed with a cylE mutant (11, 19, 27, 36), the cylD and cylK mutants displayed a faint pigmentation and reduced hemolytic activity. CylX, which is involved in the synthesis of malonyl-CoA, is dispensable for hemolysis and pigmentation. These results suggest that CylG and CylI are essential for synthesis of the β-h/c pigment lipid backbone, whereas functional redundancy might exist between CylX, CylD, or CylK and enzymes from other metabolic pathways, in particular the type II fatty acid biosynthetic pathway. The lipid backbone is conjugated to ornithine by CylE and glycosylated by addition of a rhamnose by CylJ. Surprisingly, while cylE inactivation yields NH/NP mutants, a cylJ mutant exhibits a slightly reduced pigmentation but very weak hemolysis (Fig. 3C). Based on the proposed synthesis pathway (9, 11), this might suggest that the presence of ornithine is essential for both pigmentation and hemolytic activity, whereas the absence of rhamnose mostly affects hemolysis. Further chemical analysis would greatly improve our understanding of the structure-function of this rhamnolipid toxin (9, 11, 20), which is unrelated to the known Gram-positive pore-forming toxins (45–48). Lastly, we confirmed that the ABC transporter encoded by cylAB is critical for hemolysis and pigmentation (22).
Mutations relevant to NH/NP isolates were also identified in abx1, whose product interacts with the histidine kinase CovS to control activity of the major regulator CovR and, thus, cyl transcription (27). While Abx1 mutations have been shown to decrease virulence in animals (27), the characterization of clinical NH/NP strains demonstrates that Abx1 is not absolutely required to cause infections in humans. This suggests that one of the other regulators (CovS or Stk1) may still respond to environmental signals to activate virulence gene expression in specific tissues. Characterization of these signals is necessary to decipher the regulatory network controlling host-GBS interactions.
In summary, we report the identification of several independent NH/NP GBS strains that retain the capacity to cause invasive infections in humans, including meningitis in the neonate. These strains lost one of their main virulence-associated characteristics, the specific GBS β-h/c pigment molecule, due to mutations in the cyl operon or in a cyl transcriptional regulator. Thus, while overproduction of the GBS β-h/c pigment is strongly associated with increased GBS virulence, as supported by independent studies, its absence is not synonymous with a loss of virulence, as reported in this study. A direct consequence of this result is the necessity for professionals to be cautious when using pigmentation and hemolysis as the main criteria for GBS diagnosis (17, 29).
Supplementary Material
ACKNOWLEDGMENTS
We thank Alexandra Gruss for critical reading of the manuscript and all of the correspondents of the CNR-Strep for sending GBS strains.
C. Poyart has received reimbursement for attending meetings from bioMérieux, Bio-Rad, Cepheid, Novartis, and Becton Dickinson and has received research funding from Institut Mérieux, bioMérieux, Wyeth, Oxoid, and Siemens. The other authors declare no competing financial interests.
Funding Statement
This work was supported by the Fondation pour la Recherche Médicale (grants DEQ20100318279 and DEQ20130326538), Laboratoire d'Excellence (LABEX) Integrative Biology of Emerging Infectious Diseases (grant ANR-10-LABX-62-IBEID), INSERM, CNRS, Université Paris Descartes, the Institut Pasteur, and Institut de Veille Sanitaire. A. Six was a recipient of a doctoral fellowship from the Ministère de la Recherche et de l'Enseignement Supérieur University Paris Descartes (contract 641 113 10). A. Firon was a recipient of a postdoctoral fellowship from the Fondation pour la Recherche Médicale (grant DEQ20130326538).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02177-15.
REFERENCES
- 1.Edmond KM, Kortsalioudaki C, Scott S, Schrag SJ, Zaidi AK, Cousens S, Heath PT. 2012. Group B streptococcal disease in infants aged younger than 3 months: systematic review and meta-analysis. Lancet 379:547–556. doi: 10.1016/S0140-6736(11)61651-6. [DOI] [PubMed] [Google Scholar]
- 2.Phares CR, Lynfield R, Farley MM, Mohle-Boetani J, Harrison LH, Petit S, Craig AS, Schaffner W, Zansky SM, Gershman K, Stefonek KR, Albanese BA, Zell ER, Schuchat A, Schrag SJ. 2008. Epidemiology of invasive group B streptococcal disease in the United States, 1999−2005. JAMA 299:2056–2065. doi: 10.1001/jama.299.17.2056. [DOI] [PubMed] [Google Scholar]
- 3.Joubrel C, Tazi A, Six A, Dmytruk N, Touak G, Bidet P, Raymond J, Trieu-Cuot P, Fouet A, Kerneis S, Poyart C. 2015. Group B Streptococcus neonatal invasive infections, France 2007−2012. Clin Microbiol Infect 21:910–916. doi: 10.1016/j.cmi.2015.05.039. [DOI] [PubMed] [Google Scholar]
- 4.Skoff TH, Farley MM, Petit S, Craig AS, Schaffner W, Gershman K, Harrison LH, Lynfield R, Mohle-Boetani J, Zansky S, Albanese BA, Stefonek K, Zell ER, Jackson D, Thompson T, Schrag SJ. 2009. Increasing burden of invasive group B streptococcal disease in nonpregnant adults, 1990−2007. Clin Infect Dis 49:85–92. doi: 10.1086/599369. [DOI] [PubMed] [Google Scholar]
- 5.Tazi A, Morand PC, Reglier-Poupet H, Dmytruk N, Billoet A, Antona D, Trieu-Cuot P, Poyart C. 2011. Invasive group B streptococcal infections in adults, France (2007−2010). Clin Microbiol Infect 17:1587–1589. doi: 10.1111/j.1469-0691.2011.03628.x. [DOI] [PubMed] [Google Scholar]
- 6.Van Dyke MK, Phares CR, Lynfield R, Thomas AR, Arnold KE, Craig AS, Mohle-Boetani J, Gershman K, Schaffner W, Petit S, Zansky SM, Morin CA, Spina NL, Wymore K, Harrison LH, Shutt KA, Bareta J, Bulens SN, Zell ER, Schuchat A, Schrag SJ. 2009. Evaluation of universal antenatal screening for group B Streptococcus. N Engl J Med 360:2626–2636. doi: 10.1056/NEJMoa0806820. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell TJ. 2003. The pathogenesis of Streptococcal infections: from tooth decay to meningitis. Nat Rev Microbiol 1:219–230. doi: 10.1038/nrmicro771. [DOI] [PubMed] [Google Scholar]
- 8.Nizet V. 2002. Streptococcal β-hemolysins: genetics and role in disease pathogenesis. Trends Microbiol 10:575–580. doi: 10.1016/S0966-842X(02)02473-3. [DOI] [PubMed] [Google Scholar]
- 9.Rosa-Fraile M, Dramsi S, Spellerberg B. 2014. Group B streptococcal haemolysin and pigment, a tale of twins. FEMS Microbiol Rev 38:932–946. doi: 10.1111/1574-6976.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whidbey C, Vornhagen J, Gendrin C, Boldenow E, Samson JM, Doering K, Ngo L, Ezekwe EA Jr, Gundlach JH, Elovitz MA, Liggitt D, Duncan JA, Adams Waldorf KM, Rajagopal L. 2015. A streptococcal lipid toxin induces membrane permeabilization and pyroptosis leading to fetal injury. EMBO Mol Med 7:488–505. doi: 10.15252/emmm.201404883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whidbey C, Harrell MI, Burnside K, Ngo L, Becraft AK, Iyer LM, Aravind L, Hitti J, Waldorf KM, Rajagopal L. 2013. A hemolytic pigment of group B Streptococcus allows bacterial penetration of human placenta. J Exp Med 210:1265–1281. doi: 10.1084/jem.20122753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Randis TM, Gelber SE, Hooven TA, Abellar RG, Akabas LH, Lewis EL, Walker LB, Byland LM, Nizet V, Ratner AJ. 2014. Group B Streptococcus β-hemolysin/cytolysin breaches maternal-fetal barriers to cause preterm birth and intrauterine fetal demise in vivo. J Infect Dis 210:265–273. doi: 10.1093/infdis/jiu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta R, Ghosh S, Monks B, DeOliveira RB, Tzeng TC, Kalantari P, Nandy A, Bhattacharjee B, Chan J, Ferreira F, Rathinam V, Sharma S, Lien E, Silverman N, Fitzgerald K, Firon A, Trieu-Cuot P, Henneke P, Golenbock DT. 2014. RNA and beta-hemolysin of group B Streptococcus induce interleukin-1β (IL-1beta) by activating NLRP3 inflammasomes in mouse macrophages. J Biol Chem 289:13701–13705. doi: 10.1074/jbc.C114.548982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bebien M, Hensler ME, Davanture S, Hsu LC, Karin M, Park JM, Alexopoulou L, Liu GY, Nizet V, Lawrence T. 2012. The pore-forming toxin β hemolysin/cytolysin triggers p38 MAPK-dependent IL-10 production in macrophages and inhibits innate immunity. PLoS Pathog 8:e1002812. doi: 10.1371/journal.ppat.1002812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reiss A, Braun JS, Jager K, Freyer D, Laube G, Buhrer C, Felderhoff-Muser U, Stadelmann C, Nizet V, Weber JR. 2011. Bacterial pore-forming cytolysins induce neuronal damage in a rat model of neonatal meningitis. J Infect Dis 203:393–400. doi: 10.1093/infdis/jiq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Rosa M, Perez M, Carazo C, Pareja L, Peis JI, Hernandez F. 1992. New Granada medium for detection and identification of group B streptococci. J Clin Microbiol 30:1019–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joubrel C, Gendron N, Dmytruk N, Touak G, Verlaguet M, Poyart C, Reglier-Poupet H. 2014. Comparative evaluation of 5 different selective media for group B Streptococcus screening in pregnant women. Diagn Microbiol Infect Dis 80:282–284. doi: 10.1016/j.diagmicrobio.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Liu GY, Doran KS, Lawrence T, Turkson N, Puliti M, Tissi L, Nizet V. 2004. Sword and shield: linked group B streptococcal β-hemolysin/cytolysin and carotenoid pigment function to subvert host phagocyte defense. Proc Natl Acad Sci U S A 101:14491–14496. doi: 10.1073/pnas.0406143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pritzlaff CA, Chang JC, Kuo SP, Tamura GS, Rubens CE, Nizet V. 2001. Genetic basis for the beta-haemolytic/cytolytic activity of group B Streptococcus. Mol Microbiol 39:236–247. doi: 10.1046/j.1365-2958.2001.02211.x. [DOI] [PubMed] [Google Scholar]
- 20.Rosa-Fraile M, Rodriguez-Granger J, Haidour-Benamin A, Cuerva JM, Sampedro A. 2006. Granadaene: proposed structure of the group B Streptococcus polyenic pigment. Appl Environ Microbiol 72:6367–6370. doi: 10.1128/AEM.00756-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spellerberg B, Pohl B, Haase G, Martin S, Weber-Heynemann J, Lutticken R. 1999. Identification of genetic determinants for the hemolytic activity of Streptococcus agalactiae by ISS1 transposition. J Bacteriol 181:3212–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottschalk B, Broker G, Kuhn M, Aymanns S, Gleich-Theurer U, Spellerberg B. 2006. Transport of multidrug resistance substrates by the Streptococcus agalactiae hemolysin transporter. J Bacteriol 188:5984–5992. doi: 10.1128/JB.00768-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamy MC, Zouine M, Fert J, Vergassola M, Couve E, Pellegrini E, Glaser P, Kunst F, Msadek T, Trieu-Cuot P, Poyart C. 2004. CovS/CovR of group B Streptococcus: a two-component global regulatory system involved in virulence. Mol Microbiol 54:1250–1268. doi: 10.1111/j.1365-2958.2004.04365.x. [DOI] [PubMed] [Google Scholar]
- 24.Jiang SM, Cieslewicz MJ, Kasper DL, Wessels MR. 2005. Regulation of virulence by a two-component system in group B Streptococcus. J Bacteriol 187:1105–1113. doi: 10.1128/JB.187.3.1105-1113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin WJ, Walthers D, Connelly JE, Burnside K, Jewell KA, Kenney LJ, Rajagopal L. 2009. Threonine phosphorylation prevents promoter DNA binding of the group B Streptococcus response regulator CovR. Mol Microbiol 71:1477–1495. doi: 10.1111/j.1365-2958.2009.06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lembo A, Gurney MA, Burnside K, Banerjee A, de los Reyes M, Connelly JE, Lin WJ, Jewell KA, Vo A, Renken CW, Doran KS, Rajagopal L. 2010. Regulation of CovR expression in group B Streptococcus impacts blood-brain barrier penetration. Mol Microbiol 77:431–443. doi: 10.1111/j.1365-2958.2010.07215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Firon A, Tazi A, Da Cunha V, Brinster S, Sauvage E, Dramsi S, Golenbock DT, Glaser P, Poyart C, Trieu-Cuot P. 2013. The Abi-domain protein Abx1 interacts with the CovS histidine kinase to control virulence gene expression in group B Streptococcus. PLoS Pathog 9:e1003179. doi: 10.1371/journal.ppat.1003179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merritt K, Jacobs NJ. 1978. Characterization and incidence of pigment production by human clinical group B Streptococci. J Clin Microbiol 8:105–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nickmans S, Verhoye E, Boel A, Van Vaerenbergh K, De Beenhouwer H. 2012. Possible solution to the problem of nonhemolytic group B Streptococcus on Granada medium. J Clin Microbiol 50:1132–1133. doi: 10.1128/JCM.05372-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sigge A, Schmid M, Mauerer S, Spellerberg B. 2008. Heterogeneity of hemolysin expression during neonatal Streptococcus agalactiae sepsis. J Clin Microbiol 46:807–809. doi: 10.1128/JCM.01963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banno H, Kimura K, Tanaka Y, Kitanaka H, Jin W, Wachino J, Yamada K, Shibayama K, Arakawa Y. 2014. Characterization of multidrug-resistant group B streptococci with reduced penicillin susceptibility forming small non-Beta-hemolytic colonies on sheep blood agar plates. J Clin Microbiol 52:2169–2171. doi: 10.1128/JCM.00226-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poyart C, Tazi A, Reglier-Poupet H, Billoet A, Tavares N, Raymond J, Trieu-Cuot P. 2007. Multiplex PCR assay for rapid and accurate capsular typing of group B streptococci. J Clin Microbiol 45:1985–1988. doi: 10.1128/JCM.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones N, Bohnsack JF, Takahashi S, Oliver KA, Chan MS, Kunst F, Glaser P, Rusniok C, Crook DW, Harding RM, Bisharat N, Spratt BG. 2003. Multilocus sequence typing system for group B Streptococcus. J Clin Microbiol 41:2530–2536. doi: 10.1128/JCM.41.6.2530-2536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glaser P, Rusniok C, Buchrieser C, Chevalier F, Frangeul L, Msadek T, Zouine M, Couve E, Lalioui L, Poyart C, Trieu-Cuot P, Kunst F. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol Microbiol 45:1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- 36.Forquin MP, Tazi A, Rosa-Fraile M, Poyart C, Trieu-Cuot P, Dramsi S. 2007. The putative glycosyltransferase-encoding gene cylJ and the group B Streptococcus (GBS)-specific gene cylK modulate hemolysin production and virulence of GBS. Infect Immun 75:2063–2066. doi: 10.1128/IAI.01565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poyart C, Trieu-Cuot P. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in Gram-positive bacteria. FEMS Microbiol Lett 156:193–198. doi: 10.1016/S0378-1097(97)00423-0. [DOI] [PubMed] [Google Scholar]
- 38.Rajagopal L, Vo A, Silvestroni A, Rubens CE. 2006. Regulation of cytotoxin expression by converging eukaryotic-type and two-component signalling mechanisms in Streptococcus agalactiae. Mol Microbiol 62:941–957. doi: 10.1111/j.1365-2958.2006.05431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Musser JM, Mattingly SJ, Quentin R, Goudeau A, Selander RK. 1989. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B Streptococcus) causing invasive neonatal disease. Proc Natl Acad Sci U S A 86:4731–4735. doi: 10.1073/pnas.86.12.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tazi A, Disson O, Bellais S, Bouaboud A, Dmytruk N, Dramsi S, Mistou MY, Khun H, Mechler C, Tardieux I, Trieu-Cuot P, Lecuit M, Poyart C. 2010. The surface protein HvgA mediates group B Streptococcus hypervirulence and meningeal tropism in neonates. J Exp Med 207:2313–2322. doi: 10.1084/jem.20092594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Da Cunha V, Davies MR, Douarre PE, Rosinski-Chupin I, Margarit I, Spinali S, Perkins T, Lechat P, Dmytruk N, Sauvage E, Ma L, Romi B, Tichit M, Lopez-Sanchez MJ, Descorps-Declere S, Souche E, Buchrieser C, Trieu-Cuot P, Moszer I, Clermont D, Maione D, Bouchier C, McMillan DJ, Parkhill J, Telford JL, Dougan G, Walker MJ, Consortium D, Holden MT, Poyart C, Glaser P. 2014. Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat Commun 5:4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landwehr-Kenzel S, Henneke P. 2014. Interaction of Streptococcus agalactiae and cellular innate immunity in colonization and disease. Front Immunol 5:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kulkarni R, Randis TM, Antala S, Wang A, Amaral FE, Ratner AJ. 2013. beta-Hemolysin/cytolysin of group B Streptococcus enhances host inflammation but is dispensable for establishment of urinary tract infection. PLoS One 8:e59091. doi: 10.1371/journal.pone.0059091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kline KA, Schwartz DJ, Lewis WG, Hultgren SJ, Lewis AL. 2011. Immune activation and suppression by group B Streptococcus in a murine model of urinary tract infection. Infect Immun 79:3588–3595. doi: 10.1128/IAI.00122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Los FC, Randis TM, Aroian RV, Ratner AJ. 2013. Role of pore-forming toxins in bacterial infectious diseases. Microbiol Mol Biol Rev 77:173–207. doi: 10.1128/MMBR.00052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosado CJ, Kondos S, Bull TE, Kuiper MJ, Law RH, Buckle AM, Voskoboinik I, Bird PI, Trapani JA, Whisstock JC, Dunstone MA. 2008. The MACPF/CDC family of pore-forming toxins. Cell Microbiol 10:1765–1774. doi: 10.1111/j.1462-5822.2008.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molloy EM, Cotter PD, Hill C, Mitchell DA, Ross RP. 2011. Streptolysin S-like virulence factors: the continuing sagA. Nat Rev Microbiol 9:670–681. doi: 10.1038/nrmicro2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamon MA, Ribet D, Stavru F, Cossart P. 2012. Listeriolysin O: the Swiss army knife of Listeria. Trends Microbiol 20:360–368. doi: 10.1016/j.tim.2012.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.